Abstract
Cell fate decisions require the deployment of distinct transcriptional programmes—how this is controlled and orchestrated is a key question from basic developmental biology to regenerative medicine. In this issue of The EMBO Journal, Pataskar and Jung et al (Pataskar et al, 2015) demonstrate how the transcription factor NeuroD1 acts genome‐wide to elicit a specific neurogenic programme, including differentiation and migration. Much of that activity is due to NeuroD1 acting as a pioneer factor. NeuroD1 is able to bind its targets within repressive chromatin and can induce a more open chromatin state amenable to cell type‐specific regulation.
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Neuroscience; Transcription
How the genome is regulated to give rise to the many distinct cell types of complex organisms is a primary focus of developmental biology today. With the advent of whole genome assays, it has become almost routine to probe cellular differentiation and identity at the genomic scale. This includes the profiling of transcription factors (TFs) to understand which genes they may regulate, but also the profiling of histone and DNA modifications to gain a more general understanding of the regulatory state of the genome.
How, where and when TFs act is as much a function of their own regulation as of the specific cellular environment they must act within, including cofactor availability and chromatin context. Examples of the significance of this “cellular environment” include that binding motif presence is often an extremely poor predictor of actual in vivo binding (e.g. Yang et al, 2006; Wilczynski & Furlong, 2010) and that individual cell types frequently demonstrate stark differences in the genome‐wide distributions for the same sequence‐specific TFs (e.g. Odom et al, 2004; Cao et al, 2010). Several TFs have the capacity to fundamentally shape cellular identity and direct cell fate decisions (Lee et al, 1995; Iwafuchi‐Doi & Zaret, 2014). Understanding the mechanism by which such “pioneer” factors are able to accomplish this feat is a matter of intense investigation. Pataskar and Jung et al (2016) now show how a proneural TF called NeuroD1 navigates and modifies the local chromatin environment of progenitor cells to trigger differentiation and migration programmes.
The bHLH TF NeuroD1 had previously been shown to constitute a potent neuronal differentiation factor (e.g. Lee et al, 1995; Guo et al, 2014). To investigate how NeuroD1 imposes cell fate, the authors establish an embryonic stem cell (ESC) model, where the selective induction of NeuroD1 alone triggers neuronal differentiation, as shown by the loss of pluripotency markers, concomitant gain of neuronal markers and changes in cellular morphology. Up‐regulated genes (URGs) include not only neuronal differentiation factors, but also a significant number of genes implicated in epithelial–mesenchymal transition (EMT) and migration, which is especially striking considering that NeuroD1 is primarily expressed in the subventricular zone (SVZ) of the developing brain, through which differentiating neurons migrate on their path from the progenitor population towards the cortical plate.
Between the ~3,900 genes that change expression in response to NeuroD1 and the ~2,400 binding events detected by ChIP, the authors concentrate on how NeuroD1 interacts with the ~200 genes that NeuroD1 seems to activate directly via binding to cis‐regulatory modules (CRMs), such as promoters and enhancers. Since many of these targets are TFs and chromatin regulators, a large part of the remainder of gene expression changes may be attributable to indirect regulation.
Using Bayesian inference, Pataskar and Jung et al (2016) searched for classifiers among ESC histone modification and TF binding data that distinguish NeuroD1‐bound from non‐bound CRMs near URGs. Compared to unbound URG promoters, bound URG promoters showed significantly higher levels of the chromatin condensation mark H3K27me3 and exhibited lower levels of activity hallmarks such as H3K27ac and chromatin accessibility in ESCs prior to NeuroD1 induction. Upon NeuroD1 induction, the chromatin at target promoters “opens”, marked by a decrease in H3K27me3 and increases in H3K27ac, chromatin accessibility and gene expression. Similarly, target enhancers also gain chromatin hallmarks of activity. Time‐course data at several CRMs reveal the sequence of events: NeuroD1 is bound quickly followed by H3K27 demethylation and acetylation, followed by RNA polymerase II engagement and transcription shortly thereafter. These observations strongly indicate that NeuroD1 is able to find and bind its targets in repressed chromatin, which then allows for chromatin remodelling towards a state more conducive to transcription—the very definition of a pioneer TF (Zaret & Carroll, 2011).
Figure 1. Local chromatin remodelling by pioneers.
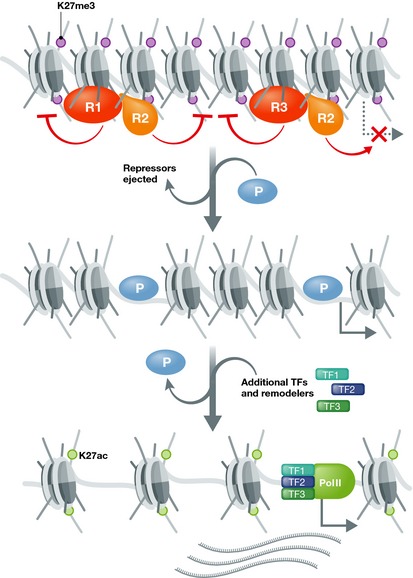
A genomic region is condensed, and genes within are repressed. Repressor proteins (R1‐3) such as Mbd3, Utf1 and Tbx3 maintain the repressive chromatin state and reinforce transcriptional silence. Once available, pioneer factor complexes (P) like NeuroD1 + partners find and bind their target sites, including sites within repressive chromatin. This then recruits enzymatic activities to modulate repression locally. For example, repressor proteins like Tbx3 and Mbd3 are ejected and histone trimethylation of H3K27 is exchanged for acetylation. Chromatin locally decondenses in the process and “opens” for targeting by settler TFs (TF1‐3) to regulate gene expression either in conjunction with, or independent of the pioneer complex. Note that each nucleosome contains two H3K27 positions and is modified on a multitude of histone tail residues; for simplicity, only trimethylation or acetylation on H3K27 is shown. Absence of modifications in the middle panel is meant to indicate uncertainty regarding the exchange kinetics. Complex context‐dependent regulation is achieved by, for example, (i) availability of pioneer binding partners / cofactors, (ii) aspects of the chromatin environment, including recruiting factors already present, (iii) the mode of pioneer interaction with the chromatin (e.g. displacement of proteins, delivery of HAT activities, etc.) and (iv) the specific availability of TFs capable of translating the regulatory information encoded in open chromatin into local gene activity.
Interesting is the temporal requirement of pioneer factors: Is transient activity sufficient to switch chromatin state (and differentiation programmes) long term, or are they required to maintain the differentiation state? NeuroD1 expression is limited largely to the entry into and migration through the SVZ and is absent from differentiated neurons. The authors argue that a pulse of NeuroD1 induction likely suffices to reorganize chromatin longer term: target sites retain marks of chromatin activity state days after ectopic NeuroD1 expression is no longer detectable. However, to what degree this may be attributable to autoregulation of endogenous NeuroD1 remains to be resolved.
NeuroD1's effects on gene activity and chromatin state hold up markedly well in vivo. Not only are individual URGs identified in vitro found to be co‐expressed with NeuroD1 in the SVZ, but the actual binding of NeuroD1 to target CRMs was confirmed in the embryonic cortex. In an elegant assay of NeuroD1 overexpression in the developing brain, Pataskar and Jung et al (2016) confirm that NeuroD1 increases H3K27 acetylation at most target CRMs, coupled to an increase in target gene expression. Moreover, even the EMT stimulation identified in cultured cells is appreciable in the brain: within 48 h after NeuroD1 is induced, cells have left the ventricular zone and lost expression of the neural progenitor marker Pax6.
So here is a pioneer TF that once activated in pluripotent cells triggers wholesale changes in terms of cellular identity and migration behaviour. It does so in part by recognizing its binding motifs and activating differentiation genes even if they are located in repressive chromatin. This is followed by local remodelling of the chromatin towards a more open state, thus allowing for transcription. How does NeuroD1 achieve this? Part of the underlying mechanism will be interaction with other local TFs. The authors show that NeuroD1 binding correlates with displacement of TFs that were associated with the URG promoters (Tbx3) and enhancers (Mbd3, Tbx3). While the mechanisms of this displacement are not clear, the biological implications are: Mbd3 is a component of the nucleosome remodelling deacetylase complex (NuRD) that would help silence enhancers. Similarly, the sequence‐specific T‐box TF Tbx3 has been described as a potent repressor. Local displacement of factors such as Mbd3 and Tbx3 upon NeuroD1 binding should then allow relief of direct and chromatin‐mediated repression at CRMs and allow for regulation by additional non‐pioneer (aka. “settler”) TFs. Clearly, the hierarchies and mechanisms underlying chromatin remodelling in response to NeuroD1 and other pioneers remain to be elucidated. Since Utf1, Mbd3 and Tbx3 association were found to be predictive for later NeuroD1 binding, one might speculate whether some of these factors may play a role in recruiting NeuroD1 to its targets. Soufi et al recently proposed that some bHLH pioneers (like Myc) bind their targets primarily through cooperative interaction with local factors, while other bHLHs rely on their own binding activity (Soufi et al, 2015). Where NeuroD1 fits on that spectrum is not yet clear.
Among pioneers, the bHLH TFs may be an especially interesting class. Most are able to form homo‐ and heterodimers, which affects their precise binding specificity and their molecular activity. Hence, for bHLH pioneers, the specific dimers formed may well dictate where they modify chromatin state, as well as how they modify it. It is feasible, for example, that the NeuroD1 binding detected in CRMs of up‐regulated genes could reflect dimers distinct from NeuroD1 complexes near repressed genes. Furthermore, NeuroD1 may mediate distinct pioneer activities in other progenitor cell types. NeuroD1 (aka. Beta2) is also an essential component in the differentiation and maintenance of the mature pancreatic β‐cell state (Jia et al, 2015), where it is likely forming complexes distinct from those in neuronal progenitors.
We are only just beginning to understand the complexities of pioneer activity in terms of how such TFs interact with their chromatinized targets, as well as in terms of the molecular activities they deliver to those targets. Once bound, pioneers like NeuroD1 can be thought of as (re‐)partitioners of the regulatable genome: by modulating chromatin state locally in a tissue‐ or condition‐specific manner, they change accessibility for further regulators. A detailed understanding of the hierarchical interactions of “pioneers” and “settlers” with their chromatin environment is an essential prerequisite for predictive modelling of how gene expression programmes are deployed to guide cellular differentiation.
See also: A Pataskar et al (January 2016)
References
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, Gentleman RC, Tapscott SJ (2010) Genome‐wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell 18: 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Zhang L, Wu Z, Chen Y, Wang F, Chen G (2014) In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer's disease model. Cell Stem Cell 14: 188–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi‐Doi M, Zaret KS (2014) Pioneer transcription factors in cell reprogramming. Genes Dev 28: 2679–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Ivanov A, Blasevic D, Muller T, Purfurst B, Sun W, Chen W, Poy MN, Rajewsky N, Birchmeier C (2015) Insm1 cooperates with Neurod1 and Foxa2 to maintain mature pancreatic beta‐cell function. EMBO J 34: 1417–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H (1995) Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix‐loop‐helix protein. Science 268: 836–844 [DOI] [PubMed] [Google Scholar]
- Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, Fraenkel E, Bell GI, Young RA (2004) Control of pancreas and liver gene expression by HNF transcription factors. Science 303: 1378–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pataskar A, Jung J, Smialowski P, Noack F, Calegari F, Straub T, Tiwari VK (2016) NeuroD1 reprograms chromatin and transcription factor landscapes to induce the neuronal program. EMBO J 35: 24–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS (2015) Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161: 555–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynski B, Furlong EE (2010) Dynamic CRM occupancy reflects a temporal map of developmental progression. Mol Syst Biol 6: 383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, Struhl K (2006) Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol Cell 24: 593–602 [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS (2011) Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25: 2227–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]


