Abstract
In yeast Saccharomyces cerevisiae, 25S rRNA makes up the major mass and shape of the 60S ribosomal subunit. During translation initiation, the 60S subunit joins the 40S initiation complex, producing the 80S initiation complex. During elongation, the 60S subunit binds the CCA-ends of aminoacyl- and peptidyl-tRNAs at the A-loop and P-loop, respectively, transferring the peptide onto the α-amino group of the aminoacyl-tRNA. To study the role of 25S rRNA in translation in vivo, we randomly mutated 25S rRNA and isolated and characterized seven point mutations that affected yeast cell growth and polysome profiles. Four of these mutations, G651A, A1435U, A1446G and A1587G, change a base involved in base triples crucial for structural integrity. Three other mutations change bases near the ribosomal surface: C2879U and U2408C alter the A-loop and P-loop, respectively, and G1735A maps near a Eukarya-specific bridge to the 40S subunit. By polysome profiling in mmslΔ mutants defective in nonfunctional 25S rRNA decay, we show that some of these mutations are defective in both the initiation and elongation phases of translation. Of the mutants characterized, C2879U displays the strongest defect in translation initiation. The ribosome transit-time assay directly shows that this mutation is also defective in peptide elongation/termination. Thus, our genetic analysis not only identifies bases critical for structural integrity of the 60S subunit, but also suggests a role for bases near the peptidyl transferase center in translation initiation.
Keywords: subunit joining, translation initiation, ribosome, 60S subunit, 25S rRNA, yeast
During the initiation phase of protein synthesis, the ribosome forms an 80S initiation complex comprising the initiator methionyl-tRNA base-paired to the mRNA start codon (AUG) in the peptidyl-tRNA binding site (P-site). During the elongation phase, the methionyl group or nascent peptide attached to its tRNA in the P-site is transferred to the α-amino group of the incoming aminoacyl-tRNA bound in the A-site (peptidyl transferase reaction). When the ribosome encounters a stop codon (UAA, UGA or UAG), peptidyl-tRNA is hydrolyzed, releasing the polypeptide (the termination phase).1-4
A ribosome is made up of a large 60S (LSU) and small 40S (SSU) subunit, the former containing the peptidyl transferase center (PTC), the catalytic site of peptide bond formation (the peptidyl transferase reaction). This reaction occurs spontaneously through entropic effects – i.e., by correct positioning of the peptidyl- and aminoacyl-moieties attached to P- and A-tRNAs, respectively. This positioning is administrated by base-pairing between the conserved rRNA guanine residues in the LSU PTC and the conserved cytosine residues of the 3`-terminal CCA sequence of both the P- and A-tRNAs.1 Additional functional sites on the LSU include the factor-binding center and the interface to the SSU. The factor-binding center binds distinct GTP-bound translation factors during all three phases of translation, initiation, elongation and termination. The SSU interface is crucial for subunit joining during the initiation phase and for maintaining the ribosome’s integrity during elongation.
We have been studying the 25S rRNA component of the LSU by yeast genetics. Three of the four rRNA components, 18S rRNA, 5.8S rRNA and 25S rRNA are transcribed by RNA polymerase I from repetitive genomic rDNA (RDN) as a single transcript, 35S rRNA, which is then processed to maturate final products. Together with 5S rRNA transcribed by RNA polymerase III from another part of the RDN, the four rRNAs are incorporated into the 80S ribosome.5 Taking advantage of the yeast strain deleted for all the genomic RDN repeats but carrying a high-copy RDN plasmid as the sole source of rDNA,6 we randomly mutated this RDN locus. We isolated dozens of point mutations (rdn) that affected yeast cell growth and polysome profiles, and reported previously the characterization of 10 rdn mutations altering 18S rRNA in the 40S subunit.7 Here we have characterized seven point mutations altering 25S rRNA and studied their effects on the initiation and elongation phases of translation in vivo.
Results and Discussion
Isolation and initial characterization of 25S rRNA rdn mutants
In the yeast Saccharomyces cerevisiae, all of the rRNA components are encoded by ~100 tandem copies of rDNA repeats (RDN). To study the effect of point mutations in 25S rRNA, we utilized yeast strains deleted for all of the genomic RDN repeats (rdnΔΔ::HIS3) but carrying a high-copy (hc) number plasmid containing a single RDN.7 KAY488 (NOY890) is one such strain that carries a hc RDN URA3 plasmid.6,7 To generate yeast strains expressing mutant rdn as the sole source for generating 25S rRNA, we randomly mutated the RDN in a LEU2 hc plasmid and performed a plasmid shuffle to introduce the mutated rdn plasmid into KAY488, as described in Materials and Methods. Mutant cells were then screened for the slow growth phenotype (Slg-) and those identified were subjected to polysome profile analysis as described previously.7 Table 1 lists seven 25S rRNA rdn strains thus prepared.
Table 1.S. cerevisiae strains carrying 25S rdn mutations.
| rdn allele | 25S rRNA base change | E. coli nucleotide | Mutated domain | Plasmid introduced | GCN2+ strainsa | mms1Δ strainsb |
|---|---|---|---|---|---|---|
| WT | Wild-type | pNOY373 | NOY908, KAY761 | KAY762 | ||
| 40 | C2879U | C2510 | V (PTC, A-loop) | pNOY373–40 | KAY766, KAY968c | KAY763 |
| 47 | G1735A | C1507 | III | pNOY373–47 | KAY507, KAY779, KAY971c |
KAY782 |
| 49 | A1446G | A1265 | II | pNOY373–49 | KAY499, KAY758, KAY964c | KAY753 |
| 56 | A1435U | A1254 | II | pNOY373–56 | KAY504, KAY777 | KAY772 |
| 67 | G1377A, U2408C | G1197, C2066 | II, V (PTC, P-loop) | pNOY373–67 | KAY503, KAY877, KAY878 | KAY879, KAY880 |
| 67b | U2408C | C2066 | V (PTC, P-loop) | pNOY373–67b | KAY908, KAY1016 | N. C.d |
| 69 | G651A | G577 | II | pNOY373–69 | KAY545, KAY759, KAY963c | KAY754 |
| 77 | A1587G | A1378 | III | pNOY373–77 | KAY882, KAY883 | KAY885, KAY886 |
a Derivatives of NOY908 (MATa ade2–1 his3–11 leu2–3,112 ura3–1 trp1–1 can1–100 rdnΔΔ::HIS3 pNOY373 [2μ RDN LEU2]) carrying indicated mutations. bDerivatives of KAY762 (MATa ade2–1 his3–11 leu2–3,112 ura3–1 trp1–1 can1–100 rdnΔΔ::HIS3 mms1Δ::kanMX pNOY373 [2μ RDN LEU2]) carrying indicated mutations. cpNOY373 derivatives used to generate these strains were prepared by re-introducing each mutation by site-directed mutagenesis (see Supplementary materials). dN. C., not constructed for this study.
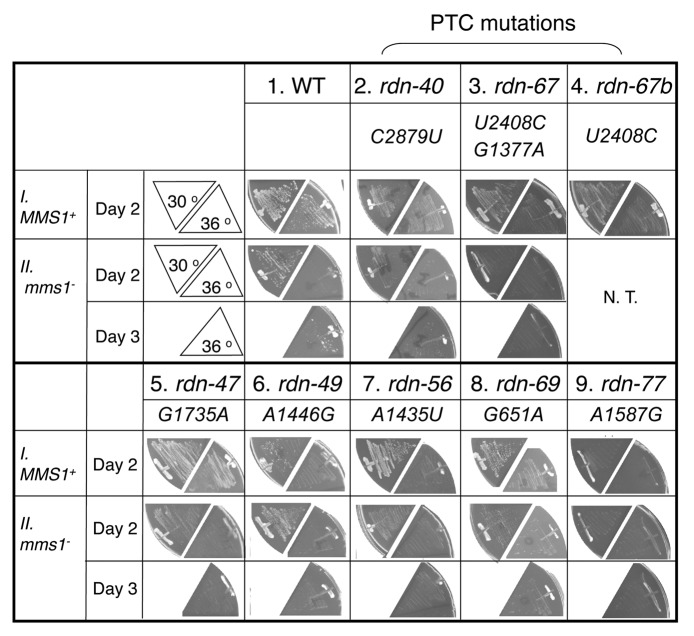
As shown in Figure 1, row I, the seven rdn strains grew slowly compared with the wild-type strain. rdn-40, rdn-67, rdn-47 and rdn-77 grew slowly both at 30° and 36°C, with rdn-77 displaying the severest growth defects (columns 2, 3, 5, and 9). rdn-49, rdn-56 and rdn-69 displayed typical temperature sensitive (Ts-) growth (columns 6–8).
Figure 1.Conditional growth phenotypes of 25S rdn mutant yeasts. Indicated strains (Table 1) are streaked out onto YPD plates and incubated for 2 or 3 d.
In order to better interpret the phenotypes of the mutant strains, we first sequenced their rdn DNAs as described in Materials and Methods. Six of the seven rdn mutations changed a single nucleotide in the region encoding the 25S rRNA, whereas the seventh (rdn-67) contained two base changes, as identified in Table 1. To test if the more conserved U2408C mutation is responsible for the phenotype caused by rdn-67, we created a new mutated rdn (called rdn-67b) that contains only the U2408C mutation, and found that it showed the same Slg- phenotype as rdn-67 (Fig. 1, row I, columns 3 and 4). We also confirmed that the identified 25S rdn mutations are different from naturally occurring base changes (polymorphisms) found in 25S rRNA-coding regions of different strains or isolates of S. cerevisiae or its closely related species (Table S2).
To verify that the mutant yeast express ribosomes with the corresponding 25S rRNA mutations, rRNA was isolated from the mutant ribosomes and used to generate cDNA by RT-PCR (Fig. S1), and then the cDNA products were sequenced. As shown in Figure S2, all of the cDNAs generated from the mutant 25S rRNA except rdn-77 contained the mutated nucleotide, confirming that the majority of the ribosomes found in these rdn mutants contains the expected mutation. In the case of the rdn-77 mutant, we observed both the mutant (G) and wild-type nucleotide (A) at position 1587 of the cDNA generated from its ribosomes (Fig. S2A, panel 7). This suggests a relatively high frequency of reversion, as previously found for some other strong conditionally-lethal 18S rRNA mutations.8 Since we observed a reproducible Slg- phenotype when the strain is maintained on a solid medium, we believe that the reversion may occur more often when it is grown in liquid medium. Accordingly, the rdn-77 mutant was dropped from further biochemical studies.
Five of the seven mutants contained additional mutations outside of the rRNA-coding regions (Table S1). All of these changes are the deletion or insertion of one or two bases in consecutive A- or T-stretches, common polymorphisms found in different strains of S. cerevisiae or its closely related species (Table S4 of ref. 7; see also Table S3-S4). Nevertheless, we re-introduced four of the identified 25S rRNA mutations, G651A, A1446G, G1735A, and C2879U, found in rdn-69, rdn-49, rdn-47 and rdn-40, respectively, into the parental rDNA plasmid (pNOY373), generated yeast carrying these site-specific mutations (Table 1, also see Materials and Methods), and confirmed that the Ts- and Slg- phenotypes are identical to those displayed by the original mutants (data not shown).
In conclusion, the seven point mutations altering the highly conserved 25S rRNA residues, C2879U, G1735A, A1446G, A1435U, U2408C, G651A and A1587G are responsible for the Slg- or Ts- phenotypes, as observed in the rdn-40, rdn-47, rdn-49, rdn-56, rdn-67(b), rdn-69 and rdn-77 mutants, respectively. Thus, in the present study, we will use the rRNA base change (C2879U etc) as the primary designation of the mutants, with the rdn allele designation in parenthesis.
Location of 25S rdn mutation sites within the three-dimensional structure of the 60S subunit
We wished to determine if the altered residues in the rdn mutants are conserved in S. cerevisiae 25S rRNA, as expected since they generate a Slg- phenotype. As shown in Figure 2, the locations of the mutations in the 25S rRNA sequence were identified, and six of the seven rdn mutations changed a residue at a conservation level 0 or 1, the highest or second highest degree of conservation, respectively, according to the 7-scale small subunit (SSU) RNA variability map (http://bioinformatics.psb.ugent.be/webtools/rRNA/).9,10 However, rdn-67 altered two bases, one highly conserved, called U2408C (rdn-67b), and the other much less conserved, called G1377A (rdn-67a) (Fig. 2). As described above, it is the conserved mutation in rdn-67 that is responsible for the Slg- phenotype.
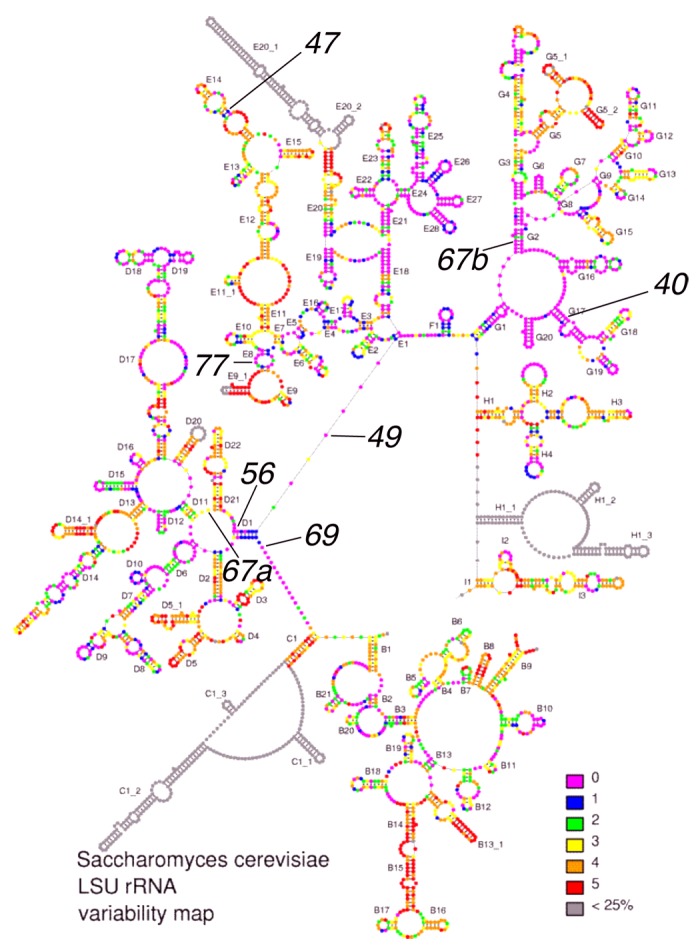
Figure 2.Locations of 25S rdn mutations (in black) within the 25S rRNA variability map (http://bioinformatics.psb.ugent.be/webtools/rRNA/). Lines indicate base changes found in the identified mutants. 40, C2879U; 47, G1735A; 49, A1446G; 56, A1435U; 67a, G1377A; 67b, U2408C; 69, G651A; 77, A1587G.
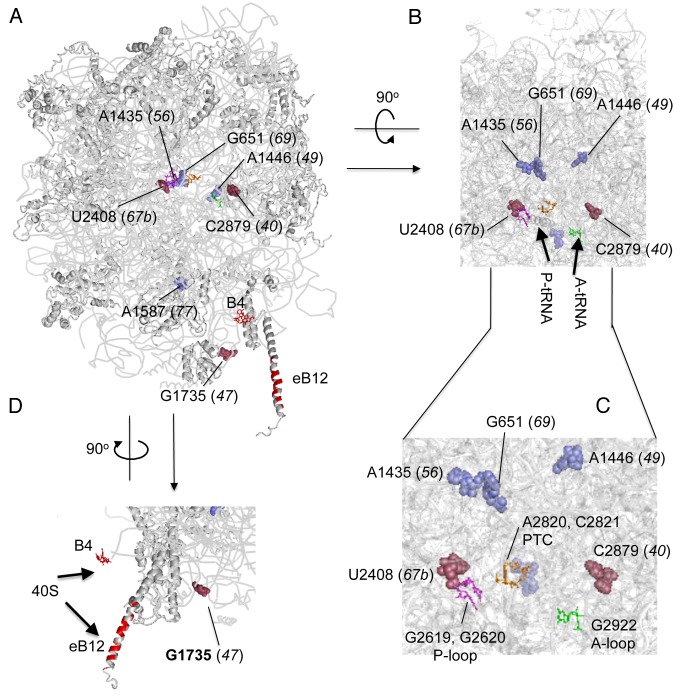
We determined the location of the mutated residues within the three dimensional structure of the yeast LSU, based on its X-ray crystal structures (Fig. 3).11,12 Table 1, column 3, lists the homologous nucleotide in the E. coli 50S subunit structure, while Table 1, column 4, lists the 25S rRNA domain altered by each mutation. As shown in Figure 3, the sites of three Ts- mutations G651A (rdn-69), A1435U (rdn-56), and A1446G (rdn-49) are located in close proximity to each other within an internal core of Domain II right beneath the PTC. The strongest Slg- mutation A1587G (rdn-77) is also located deep inside the domain III structure. Interestingly, C2879U (rdn-40) and U2408C (rdn-67b), appear to change bases near the A-loop (G2922 in green sticks) and P-loop (G2619 and G2620 in magenta sticks) of the PTC, respectively. G1735A (rdn-47) is located at the edge of the interface side of the LSU.
Figure 3. Location of point mutations mapping in yeast 25S rRNA. (A) Yeast 60S subunit (PDB 3O58) with RNA and protein backbones painted in gray and viewed from the 40S interface-side. 25S rRNA residues altered by rdn mutations (rdn allele numbers in parentheses) are highlighted as raspberry or slate spheres. Nucleotides painted raspberry are located near the surface, whereas those painted slate are located deep inside the ribosome structure. Functional rRNA bases relevant to this study are shown as colored sticks; A2820 and C2821 (A2486 and C2487 in H. marismortui; A2451 and C2452 in E. coli) in yellow, G2619 and G2620 in magenta, G2922 in green and A846 and A847 in red. Amino acids of Rpl19e involved in eB12 are painted red. (B) The structure shown in (A) was visualized from the top with the same scale. RNA and protein backbones are 80% transparent, such that the five rRNA mutation sites (indicated) and bases potentially responsible for PTC activities are arranged in the same plane. Arrows indicate the location of 3` ends of tRNAs in the P site and A site. (C) A part of panel B is enlarged to label bases potentially involved in the peptidyl transferase reaction. (D) The structure shown in (A) was visualized from the right with the same scale, in order to show the distance between G1735 and nearby bridges to the 40S subunit (arrows). PYMOL was used to draw this figure.
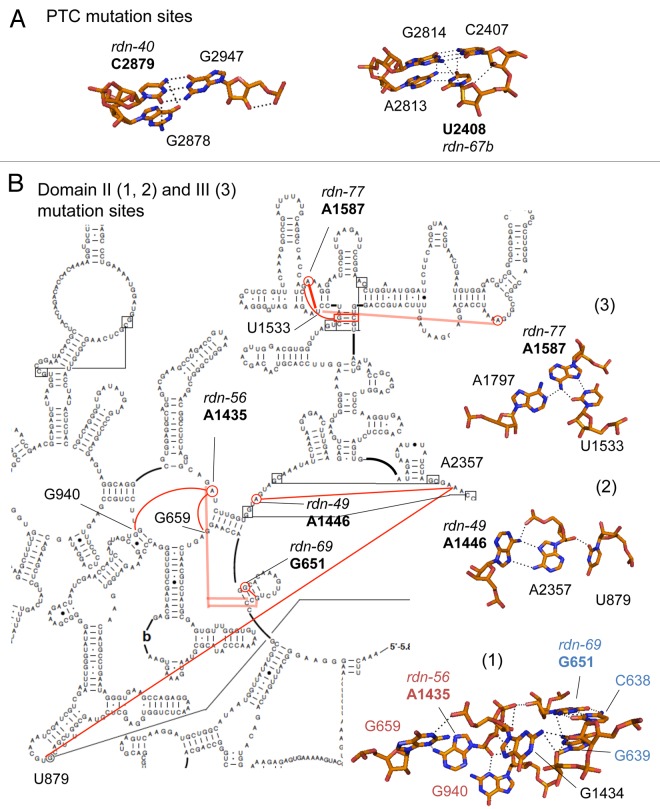
The crystal structure also illustrates the molecular basis of how each mutation affects the ribosome structure. Although C2879U (rdn-40) changes the G:C pair in h90 to a G:U pair, C2879 is hydrogen-bonded to the next base, G2878 (Fig. 4A, left). Therefore, a U at nt. 2879 would not be able to interact with G2878 in a manner that the C at nt. 2879 does, explaining a functional defect by C2879U. Similarly, U2408C (rdn-67b) not only disrupts the A:U base-pair in h74 but it also disrupts hydrogen bonds with G2814 and C2407 in the next base pair (Fig. 4A, right).
Figure 4.Interactions of 25S rRNA nucleotides altered by the rdn mutations with surrounding nucleotides. (A) C2879 and U2408 altered by the PTC rdn mutations are presented in stick models with neighboring residues and dotted lines denoting hydrogen bonds. (B) Left, the rdn mutation sites and neighboring residues are highlighted in red circles within the relevant parts of the secondary structure of yeast 25S rRNA. Red lines indicate the base-base or base-backbone contacts between the indicated residues. Right, panels 1–3 depict stick models of interacting rRNA residues drawn by PYMOL; (1) Two neighboring base triples in Domain II involving A1435 (rdn-56) (labeled orange) and G651 (rdn-69) (labeled blue), (2) the base triple involving A1446 (rdn-49), and (3) the base triple involving A1587 (rdn-77).
G651A (rdn-69) and A1435U (rdn-56) disrupt base-triples located close together (bases labeled in blue and orange, respectively, Figure 4B, panel 1). Similarly, A1446G (rdn-49) disrupts a base-triple formed between A1446 and a long-range, interdomain base-pair U879:A2357 (Fig. 4B, panel 2). All the three base-triples mentioned above are formed by base-backbone contacts. In contrast, the base-triple disrupted by A1587G (rdn-77) is unique, as this involves base-base contacts, which are formed between A1587, U1533 (in h52) and A1797 (Fig. 4B, panel 3). All the base-triples disrupted by the four rdn mutations are conserved in E. coli (bacterial) and H. marismortui (archaeal) 23S rRNA structures. Thus, the phenotypes caused by these mutations provide mutational evidence for the unique base-triples, which are conserved between, bacteria, archaea and eukaryotes.13
G1735A (rdn-47) disrupts a base-pair in h58, which is located at the edge of the interface side of the 60S subunit (Fig. 3A). G1735 is close to the binding site of Rpl19e, which makes a Eukarya-specific bridge eB12 (painted red in Figure 3A and D). The next closest intersubunit bridge is B4 made by A846 and A847 (red sticks in Figure 3A and D). Based on this positioning near an intersubunit bridge, G1735A may affect ribosome subunit joining.
In vivo evidence that the 25S rRNA mutations impair either translation initiation or elongation
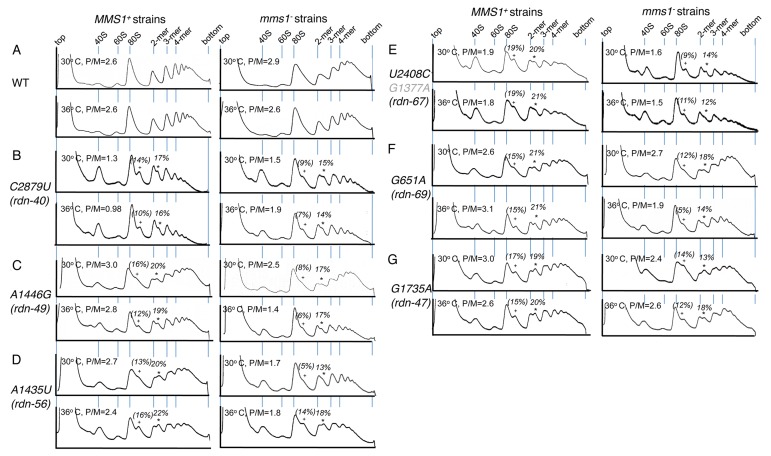
To examine the effect of the rdn mutations on translation in vivo, we analyzed the polysome profiles of the mutant strains by sucrose gradient-velocity sedimentation and calculated the ratio of polysomes to monosomes plus ribosomal subunits (P/M) (see Materials and Methods). As shown in Figure 5B and 5E, left columns, C2879U (rdn-40) and U2408C (rdn-67), altering bases near the PTC, shifted the polysome profile toward smaller polysomes and decreased the P/M ratio at both 30° and 36°C as compared with the wild-type control shown in Figure 5A. The smaller polysomes and decreased P/M ratios suggest that these PTC mutations impair translation initiation. A1435U (rdn-56) did not affect the polysome size or P/M ratio (Fig. 5D,). In contrast, an increase in polysome size and P/M ratio was seen with mutations A1446G (rdn-49), G651A (rdn-69) and G1735A (rdn-47) (Fig. 5C, 5F and 5G, respectively). These increases suggest that the rate of elongation has been diminished in these strains.
Figure 5.Polysome profiles of the isolated 25S rRNA mutants in MMS1+ (left panels) and mms1Δ (right panels) background. A254 profiles of cell extracts prepared from wild-type (panel A) and C2879U (rdn-40), A1446G (rdn-49), A1435U (rdn-56), U2408C (rdn-67), G651A (rdn-69) and G1735A (rdn-47) mutant yeasts (panels B-G) in MMS1+ (left panels) and mms1Δ (right panels) background are shown with the P/M ratio of the strains. KAY761 [MMS1+], KAY766 [C2879U], KAY758 [A1446G], KAY777 [A1435U], KAY877 [U2408C G1377A], KAY759 [G651A], KAY779 [G1735A] or KAY762 [mms1Δ] derivatives in Table 1 were grown in the rich YPD medium at 30° or 36°C for 3 h and processed for density gradient-velocity sedimentation, followed by fractionation with constant A254 monitoring (see Materials and methods). 1.5- and 2.5-mer indexes (in %, italicized) are indicated above each corresponding peaks, labeled + and *, respectively. Lines indicate the locations of free 40S and 60S subunits, monosomes (80S), and polysomes with indicated number of ribosomes. Note that in panel (E) G1377A is labeled in gray, since it is a silent 25S rRNA mutation.
A challenge in studying eukaryotic rRNA functions by mutagenesis is the fact that most rRNA mutations affect the level and/or stability of their ribosomal subunit.7,14,15 Indeed, the polysome profiles shown in Figure 5 show an apparent decrease in the abundance of free 60S subunits for all six rdn strains analyzed, compared with the abundance of free 40S subunits. The possibility that the levels of 60S subunits are diminished was addressed directly by sucrose gradient analysis of mutant rdn cell lysates in the absence of magnesium, resulting in the dissociation of all ribosomes into small and large subunits. As reported in Figure 6A, the 60S subunit abundance relative to 40S subunit abundance (60S/40S ratio) was decreased in all the mutants examined compared with the value from the isogenic wild-type strain (Fig. 6A), as has been observed with other 25S rRNA mutations.11 The lower 60S subunit abundance may be caused by reduced transcription or processing of mutated rRNAs, by affecting the assembly of complete particles, or by the elimination of misfolded or defective particles by surveillance pathways that monitor the function or shape of the ribosome.17 To address this last possibility, we used a mms1Δ strain defective in the Mms1p-dependent non-functional rRNA decay pathway (25S NRD)16,18 to see if the abundance of mutant 60S subunits is altered in this genetic background. As shown in Figure 1, row II, mms1Δ with the wild-type RDN grew slowly compared with the equivalent MMS1+ strain (column 1). Apparently, the Slg- phenotype was stronger for U2408C/G1377A (rdn-67) but not significantly altered for the other rdn strains (Fig. 1, row II, columns 2–9). However, when we measured the 60S/40S ratio in the mms1Δ background, rdn strains showed increased ratios at both 30°C (p = 0.016) and 36°C (p = 0.024) (Fig. S3, n = 5). Although all 60S subunits contain mutant 25S rRNA, apparently only a fraction of such particles are non-functional and thus eliminated by the Mms1p-dependent pathway. Nevertheless, the small enhancement of 60S subunit levels in the mms1Δ background did not significantly affect the polysome profiles and P/M ratios (Fig. 5, right column) for most of the rdn strains. However, in the mms1Δ background, A1436U (rdn-56) showed reduced P/M ratios and shifts to smaller polysomes at both temperatures, and A1446G (rdn-49) behaved similarly at 36°C, suggestive of inhibition of initiation not seen in the wild-type background.
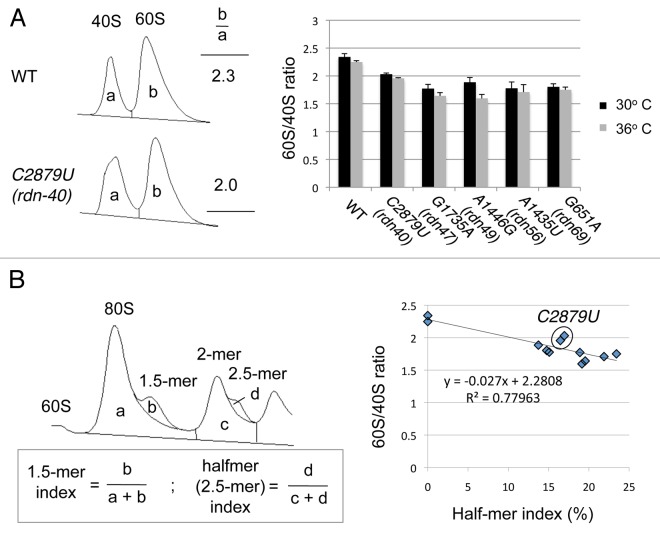
Figure 6.In vivo ribosome assays. (A) 60S/40S abundance ratio. Left, A254 absorbance traces for 40S and 60S peaks isolated from the wild-type (WT) and C2879U (rdn-40) mutant (see Materials and methods). 40S and 60S masses were determined by the areas under the peaks to calculate 60S/40S ratio (b/a). Table to the right shows the 60S/40S ratios determined from the pattern to the left. Right, 60S/40S ratio was shown for the indicated rdn strains grown in YPD for 3 h at 30°C (black) or 36°C (gray). Bars indicate SD (n = 2~3). (B) Halfmer indexes. Left, using the A254 polysome trace for the C2879U (rdn-40) mutant grown at 30°C (Fig. 5B) as an example, the amounts of monosome, disome and halfmers associated with them were determined by the areas under each peak. 1.5-mer index and halfmer (2.5-mer) index were determined by the formula presented in the box below. Right, the relationship between the 60S/40S ratios shown in (A) with the halfmer index, measured for the same strains as used in (A) at 30°C and 36°C. Two points in circle are data from C2879U.
25S rdn mutants produce halfmers
Another feature of the polysome profiles for rdn strains shown in Figure 5 is the clear presence of “halfmers,” polysomes with one or more 80S ribosomes and a single 40S ribosomal subunit. Halfmers are produced when the rate of 60S subunit joining with a 40S initiation complex on a polysome is reduced. Such reduced rates can be caused by a paucity of 60S subunits or by a defect in the joining reaction. In view of the fact that four of the 25S rRNA mutations map in the internal core of the 60S subunit structure, the first model to test is that halfmers are generated by the paucity of mutant 60S subunits. To address this point, we estimated the amounts of halfmers found in disomes (2.5-mers, labeled * in the profiles in Figure 5) as described in Materials and Methods and report the values as the halfmer index (Fig. 6B. and Materials and Methods). The halfmer index for the different rdn strains correlated quite well with the reduction in the 60S/40S subunit ratio in the MMS1+ background (R2 = 0.78, Figure 6B), in support of the notion that halfmers are largely generated by a paucity of 60S subunits.
However, the PTC mutation C2879U (rdn-40) had the smallest effect on the 60S/40S ratio (Fig. 6A), and yet displayed relatively high halfmer indexes (Fig. 5B). In Figure 6B, the two points for C2879U (rdn-40) are outliers of the linear regression (circle). These results suggest that C2879U (rdn-40) directly impairs 60S subunit joining, thereby producing a significant portion of the halfmers observed in the mutant.
We next assessed the effects of the Mms1p pathway on the generation of halfmers by its influencing 60S subunit levels. To increase sample number, we also measured 1.5-mer indexes (+ in the profiles of Figure 5), as defined in Figure 6B. As is apparent in the polysome profiles shown in Figure 5, right columns, we found that mms1Δ decreased halfmers (1.5-mer and 2.5-mer) at 30° and 36°C (n = 4) for all the mutants tested: the PTC mutations C2879U (rdn-40), p = 0.01 and U2408C (rdn-67), p = 0.003; the Domain II mutations A1446G (rdn-49), p = 0.03, A1435U (rdn-56), p = 0.03 and G651A (rdn-69), p = 0.04; and the subunit interface mutation G1735A (rdn-47), p = 0.02. Given that in the mms1Δ background the level of 60S subunits is increased, the data support the notion that the Mms1p pathway contributes to the generation of halfmers through the degradation of 60S subunits.
In the case of G1735A (rdn-47), the subunit interface mutation, the changes in the amounts of halfmers (Fig. 5G) and 60S/40S ratio (Fig. S3) caused by mms1Δ were the smallest among the mutants tested, suggesting that Mms1p is not primarily involved in the decay of this mutant ribosome. This is in agreement with the idea that Mms1p is more specifically involved in the decay of PTC mutant ribosomes.16
C2879U (rdn-40) altering the PTC directly impairs polypeptide elongation
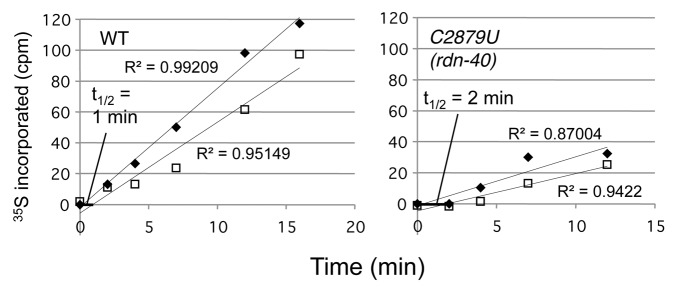
Since C2879U (rdn-40) changes a base near the PTC A-loop and displays a defect in subunit joining during the initiation phase, it was interesting to assess whether it also might inhibit the elongation phase of translation. To directly answer this question, we measured the ribosomal transit time, the average time required following initiation for ribosomes to elongate and release completed polypeptide chains. For this purpose, we labeled yeast proteins with 35S-methionine, withdrew cells at different times, washing and lysing them, fractionated the lysates by ultracentrifugations, and measured the amount of 35S label incorporated into total proteins in the post mitochondrial supernatant (PMS) and into ribosome-free proteins in the post ribosomal supernatant (PRS). As shown in Figure 7, the time course of 35S label incorporation in PMS (filled diamonds) and PRS (open squares) displays good linear regressions (R2 = 0.87~0.99) for both wild-type and C2879U (rdn-40), although they are not in parallel probably due to loss of 35S proteins from PRS.19 Accordingly, the half transit time was defined by the delay in production of ribosome-free 35S-proteins (open squares) compared with total 35S-proteins (filled squares) at y = 0. The half transit time was thereby determined to be 1 min for wild-type and 2 min for C2879U (rdn-40), indicating a defect in translation elongation/termination. Thus, C2879U (rdn-40) impairs both the initiation and elongation/termination phases of translation.
Figure 7. Ribosome transit time assay. Yeast strains NOY908 (WT) and KAY766 (C2879U rdn-40) were grown in a minimal medium at 34°C and allowed to incorporate 35S-methionine. 35S label (in cpm) incorporated into PMS (filled diamonds) and PRS (empty squares) fractions were measured by scintillation counting. Displacement of linear regression for PMS and PRS at y = 0 indicates ribosome half-transit time (thick black line). Shown is a typical result from three independent experiments.
Concluding Remarks
In this paper, our in vivo analyses of 25S rRNA mutations support the notion that a single 25S rRNA mutation can affect both the initiation and elongation phases of translation. C2879U (rdn-40) mapping near the PTC A-loop showed a defect in translation initiation, at least in part resulting from defective subunit joining. While an altered P-loop can be sensed by Met-tRNAiMet during subunit joining, one may wonder how an altered A-loop is detected. A likely protein that can detect it is eIF5B, since cryo-EM studies show that its bacterial counterpart, IF2, can contact the A-loop structure of the 50S subunit.20 We extended this study and recently showed that C2879U (rdn-40) impairs eIF5B-dependent subunjt joining in a reconstituted assay.21 C2879U (rdn-40) also impairs the elongation phase, since the mutation directly increased the ribosome transit time (Fig. 7). It is important to appreciate that a ribosome mutation producing an elongation defect also displays an initiation defect, because, in this way, cells can detect the critical defect before the ribosome enters the elongation phase.
The idea that the rRNA bases in the PTC are directly involved in subunit joining during initiation was first suggested based on the observation of halfmer polysomes in the mutants harboring C2820U and Ψ2922C.15 However, halfmer polysomes can be generated merely due to the reduced 60S subunit levels due to the subunit instability,22 as shown with most 25S rdn mutants in this study (Fig. 6B). This work supports the evidence for PTC involvement in subunit joining by the quantitative analysis of polysome profiles of the PTC mutant, C2879U (rdn-40), in reference to its 60S/40S subunit abundance ratios.
With many ribosomal mutations affecting mRNA translation profiles,24,25 initiation stringency7,8 or even causing cancer26 or diseases called ribosomopathies27 it would be intriguing to determine whether the 25S rRNA mutations analyzed here change specific gene translation or stringency of start codon selection by affecting the 40S initiation complex stalled at start codons.
Materials and Methods
Plasmids and yeast strains
Plasmids and yeast strains carrying rdn mutant 25S rRNA as the sole source are listed in Table 1. Derivatives of pNOY373 (hc LEU2 RDN) carrying Ts- and Slg- 25S rdn mutations were isolated from a mutagenized pool of pNOY373 DNA, as described.7 Details of construction of these and other 25S rdn mutant plasmids and yeast strains carrying them are described in Supplementary text.
Comparative genomics approaches
In order to examine evolutionary conservation of each of the S. cerevisiae rRNA residues, we utilized a large subunit (LSU) RNA variability map.9,10 These maps categorize every residue of LSU rRNA according to the level of conservation throughout eukaryotes, and the map for S. cerevisiae LSU (25S) rRNA was used in Figure 2.
In order to examine polymorphisms within the S. cerevisiae RDN locus, we undertook systematic BLAST searches with each of the nine segments derived from the 9.15-kb pNOY373 insert sequence as listed across the top of Table S1, as described.7 Naturally occurring base changes (polymorphisms) within the RDN segment upstream of 18S rRNA and the 18S-coding region were reported previously.7 Those in 25S-coding region (Table S2), the region between 18S and 25S-coding regions (Table S3), and the region downstream of 25S-coding region (Table S4) are listed here.
Ribosome analyses
Polysome profiling was performed by sucrose-gradient velocity sedimentation of extracts of yeast treated with cycloheximide, as described.28 The mass ratio of polysome to monosome (P/M ratio) was measured by the ratio of free subunits and monosomes to polysomes from A254 profile of the gradient samples. To measure the degree of halfmer formation in each condition, we focused on disomes and halfmers associated with them (2.5-mer), since the disome forms the best resolved polysomal peak in the polysome profile. Monosome is better resolved, but this is a mixture of vacant ribosomes and mRNA molecules attached to a single ribosome during translation (also see below). We measured the area of the disome peak above the baseline (c) and that of 2.5-mer above the shoulder of the disomal peak (d). The halfmer index was calculated as percentage of c in total disomal peaks (c+d). Since the formation of halfmers should not depend on the size of the polysome, the halfmer index calculated here likely represents the proportion of halfmers formed in total polysomes. In agreement with this assertion, the ratio of each polysomal peak and the halfmer associated with it is roughly identical across each polysome profile (Fig. Five and 6B). To better evaluate formation of halfmers, we also measured 1.5-mer index, based on the areas of monosome and 1.5-mer peaks (Fig. 6B).
To determine the mass ratio of 60S to 40S subunit (60S/40S ratio) independent of polysome profiling, yeast cell extracts were prepared in the absence of free Mg2+ and resolved by sucrose gradient velocity sedimentation followed by monitoring subunit abundance at A254, as described.29
To determine the RNA sequence of the ribosomes, rRNA was isolated by a hot phenol method30 and used to perform reverse-transcriptase (RT)-directed polymerase chain reaction (PCR) after DNase I treatment (DNase I removes any contaminating plasmid DNA). For RT-PCR, we used Access QuickTM RT-PCR system (Promega) and a pair of oligos, as listed in Fig. S1. To design new primers for cDNA synthesis, we avoided m1A(2142) in the area of amplification, as it is known to cause a strong stop in the RT reaction. The resulting cDNA product was used for Sanger dideoxy sequencing.
Ribosome half–transit time measurement
Yeasts were grown in SD medium supplemented with adenine and uracil (2% glucose) at 34°C for 4 h until reached OD600 = 0.8~1.0. The 9.6 A600 units of cells were pelleted and resuspended into 12-ml fresh pre-warmed medium supplemented with 100 μCi 35S methionine. 2-ml sample was taken at 0, 2, 6, 10, 14 min time interval and immediately mixed with 160 μl of cold unlabeled 50 mM methionine. Cells were lysed with acid washed glass beads in lysis buffer (50 mM Tris-HCl [pH.8.0], 1 mM DTT, 200 mM NaCl, 1 mM EDTA, 1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin A) by vortexing for 10 min at 4°C. The lysate was cleared by microcentrifugation at 4°C at 13,200 rpm for 10 min. The half of the lysate was kept as post-mitochondrial supernatant (PMS). The remainder was depleted of ribosome through ultracentrifugation at 90,000 rpm for an hour at 4°C in Beckman OptimaTM MAX-XP to generate post-ribosomal supernatant (PRS). Inc. radioactivity was precipitated by dissolving equal fractions of PMS and PRS in ice-cold 5% TCA for 15 min and applied to glass filters. The filters were washed two times with 5% cold TCA and once with ethanol and dried. Radioactivity from 35S was measured by scintillation counting. The ribosome half transit time was determined as the displacement in time of the PMS (total protein) and PRS (released protein) linear regression trends at y = 0.
Other molecular biology methods
Standard yeast molecular biology methods were used throughout.28
Supplementary Material
Supplementary Material
Supplementary material may be found here: https://www.landesbioscience.com/journals/translation/article/26402/www.landesbioscience.com/journals/
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank Felix Voigts-Hoffmann, Daisuke Takahashi, Rafael Luna and Gota Kawai for discussions and assistance in analyzing ribosome structures, Yuki Kanda for technical support, and Tom Dever for discussions and comments on the manuscript. This work was supported by NIH R01 grant GM64781 and its ARRA supplement (KA), Kansas COBRE-PSF Pilot grant (KA), Innovative Award from KSU Terry Johnson Cancer Center (KA), an internal grant from KSU Division of Biology (KA), and grants from NCRR (5P20RR016475) and NIGMS (8P20GM103418) (SJB).
References
- 1.Moore PB, Steitz TA. . The involvement of RNA in ribosome function. Nature 2002; 418:229 - 35; http://dx.doi.org/ 10.1038/418229a; PMID: 12110899 [DOI] [PubMed] [Google Scholar]
- 2.Noller HF. . RNA structure: reading the ribosome. Science 2005; 309:1508 - 14; http://dx.doi.org/ 10.1126/science.1111771; PMID: 16141058 [DOI] [PubMed] [Google Scholar]
- 3.Rodnina MV, Beringer M, Wintermeyer W. . How ribosomes make peptide bonds. Trends Biochem Sci 2007; 32:20 - 6; http://dx.doi.org/ 10.1016/j.tibs.2006.11.007; PMID: 17157507 [DOI] [PubMed] [Google Scholar]
- 4.Asano K, Ito K. in Encyclopedia of Systems Biology eds Werner Dubitzky, Olaf Wolkenhauser, Kwang-Hyun Cho, & Hiroki Yokota) (Springer, 2013). pp. 2259-2263 [Google Scholar]
- 5.Hage AE, Tollervey D. . A surfeit of factors: why is ribosome assembly so much more complicated in eukaryotes than bacteria?. RNA Biol 2004; 1:10 - 5; http://dx.doi.org/ 10.4161/rna.1.1.932; PMID: 17194932 [DOI] [PubMed] [Google Scholar]
- 6.Wai HH, Vu L, Oakes M, Nomura M. . Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo.. Nucleic Acids Res 2000; 28:3524 - 34; http://dx.doi.org/ 10.1093/nar/28.18.3524; PMID: 10982872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nemoto N, Singh CR, Udagawa T, Wang S, Thorson E, Winter Z, Ohira T, Ii M, Valásek L, Brown SJ, et al. . Yeast 18 S rRNA is directly involved in the ribosomal response to stringent AUG selection during translation initiation. J Biol Chem 2010; 285:32200 - 12; http://dx.doi.org/ 10.1074/jbc.M110.146662; PMID: 20699223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong J, Nanda JS, Rahman H, Pruitt MR, Shin BS, Wong CM, Lorsch JR, Hinnebusch AG. . Genetic identification of yeast 18S rRNA residues required for efficient recruitment of initiator tRNA(Met) and AUG selection. Genes Dev 2008; 22:2242 - 55; http://dx.doi.org/ 10.1101/gad.1696608; PMID: 18708582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van de Peer Y, Jansen J, De Rijk P, De Wachter R. . Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res 1997; 25:111 - 6; http://dx.doi.org/ 10.1093/nar/25.1.111; PMID: 9016516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wuyts J, De Rijk P, Van de Peer Y, Winkelmans T, De Wachter R. . The European Large Subunit Ribosomal RNA Database. Nucleic Acids Res 2001; 29:175 - 7; http://dx.doi.org/ 10.1093/nar/29.1.175; PMID: 11125083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. . Crystal structure of the eukaryotic ribosome. Science 2010; 330:1203 - 9; http://dx.doi.org/ 10.1126/science.1194294; PMID: 21109664 [DOI] [PubMed] [Google Scholar]
- 12.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. . The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 2011; 334:1524 - 9; http://dx.doi.org/ 10.1126/science.1212642; PMID: 22096102 [DOI] [PubMed] [Google Scholar]
- 13.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. . The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 2000; 289:905 - 20; http://dx.doi.org/ 10.1126/science.289.5481.905; PMID: 10937989 [DOI] [PubMed] [Google Scholar]
- 14.Velichutina IV, Dresios J, Hong JY, Li C, Mankin A, Synetos D, Liebman SW. . Mutations in helix 27 of the yeast Saccharomyces cerevisiae 18S rRNA affect the function of the decoding center of the ribosome. RNA 2000; 6:1174 - 84; http://dx.doi.org/ 10.1017/S1355838200000637; PMID: 10943896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rakauskaite R, Dinman JD. . rRNA mutants in the yeast peptidyltransferase center reveal allosteric information networks and mechanisms of drug resistance. Nucleic Acids Res 2008; 36:1497 - 507; http://dx.doi.org/ 10.1093/nar/gkm1179; PMID: 18203742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujii K, Kitabatake M, Sakata T, Miyata A, Ohno M. . A role for ubiquitin in the clearance of nonfunctional rRNAs. Genes Dev 2009; 23:963 - 74; http://dx.doi.org/ 10.1101/gad.1775609; PMID: 19390089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graille M, Séraphin B. . Surveillance pathways rescuing eukaryotic ribosomes lost in translation. Nat Rev Mol Cell Biol 2012; 13:727 - 35; http://dx.doi.org/ 10.1038/nrm3457; PMID: 23072885 [DOI] [PubMed] [Google Scholar]
- 18.Fujii K, Kitabatake M, Sakata T, Ohno M. . 40S subunit dissociation and proteasome-dependent RNA degradation in nonfunctional 25S rRNA decay. EMBO J 2012; 31:2579 - 89; http://dx.doi.org/ 10.1038/emboj.2012.85; PMID: 22505030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen PJ, McConkey EH. . Evidence for control of protein synthesis in HeLa cells via the elongation rate. J Cell Physiol 1980; 104:269 - 81; http://dx.doi.org/ 10.1002/jcp.1041040302; PMID: 7419605 [DOI] [PubMed] [Google Scholar]
- 20.Simonetti A, Marzi S, Myasnikov AG, Fabbretti A, Yusupov M, Gualerzi CO, Klaholz BP. . Structure of the 30S translation initiation complex. Nature 2008; 455:416 - 20; http://dx.doi.org/ 10.1038/nature07192; PMID: 18758445 [DOI] [PubMed] [Google Scholar]
- 21.Hiraishi H, et al. . Interaction between 25S rRNA A-loop and eIF5B promotes subunit joining and insures stringent AUG selection. Mol Cell Biol 2013; In press http://dx.doi.org/ 10.1128/MCB.00771-13; PMID: 23836883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foiani M, Cigan AM, Paddon CJ, Harashima S, Hinnebusch AG. . GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae.. Mol Cell Biol 1991; 11:3203 - 16; PMID: 2038326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, et al. . A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 2012; 151:1042 - 54; http://dx.doi.org/ 10.1016/j.cell.2012.10.044; PMID: 23178123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komili S, Farny NG, Roth FP, Silver PA. . Functional specificity among ribosomal proteins regulates gene expression. Cell 2007; 131:557 - 71; http://dx.doi.org/ 10.1016/j.cell.2007.08.037; PMID: 17981122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. . Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 2011; 145:383 - 97; http://dx.doi.org/ 10.1016/j.cell.2011.03.028; PMID: 21529712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N. . Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol 2004; 2:E139; http://dx.doi.org/ 10.1371/journal.pbio.0020139; PMID: 15138505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narla A, Ebert BL. . Ribosomopathies: human disorders of ribosome dysfunction. Blood 2010; 115:3196 - 205; http://dx.doi.org/ 10.1182/blood-2009-10-178129; PMID: 20194897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee B, Udagawa T, Singh CS, Asano K. . Yeast phenotypic assays on translational control. Methods Enzymol 2007; 429:139 - 61; http://dx.doi.org/ 10.1016/S0076-6879(07)29006-8; PMID: 17913622 [DOI] [PubMed] [Google Scholar]
- 29.Decatur WA, Liang XH, Piekna-Przybylska D, Fournier MJ. . Identifying effects of snoRNA-guided modifications on the synthesis and function of the yeast ribosome. Methods Enzymol 2007; 425:283 - 316; http://dx.doi.org/ 10.1016/S0076-6879(07)25013-X; PMID: 17673089 [DOI] [PubMed] [Google Scholar]
- 30.Udagawa T, Nemoto N, Wilkinson CR, Narashimhan J, Jiang L, Watt S, Zook A, Jones N, Wek RC, Bähler J, et al. . Int6/eIF3e promotes general translation and Atf1 abundance to modulate Sty1 MAPK-dependent stress response in fission yeast. J Biol Chem 2008; 283:22063 - 75; http://dx.doi.org/ 10.1074/jbc.M710017200; PMID: 18502752 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.