Abstract
Elongation factor G (EF-G) is a GTPase that catalyzes tRNA and mRNA translocation during the elongation cycle of protein synthesis. The GTP-bound state of the factor on the ribosome has been studied mainly with non-hydrolyzable analogs of GTP, which led to controversial conclusions about the role of GTP hydrolysis in translocation. Here we describe a mutant of EF-G in which the catalytic His91 is replaced with Ala. The mutant EF-G does not hydrolyze GTP, but binds GTP with unchanged affinity, allowing us to study the function of the authentic GTP-bound form of EF-G in translocation. Utilizing fluorescent reporter groups attached to the tRNAs, mRNA, and the ribosome we compile the velocity map of translocation seen from different perspectives. The data suggest that GTP hydrolysis accelerates translocation up to 30-fold and facilitates conformational rearrangements of both 30S subunit (presumably the backward rotation of the 30S head) and EF-G that lead to the dissociation of the factor. Thus, EF-G combines the energy regime characteristic for motor proteins, accelerating movement by a conformational change induced by GTP hydrolysis, with that of a switch GTPase, which upon Pi release switches the conformations of EF-G and the ribosome to low affinity, allowing the dissociation of the factor.
Keywords: Ribosomes, translational GTPases, protein synthesis, tRNA, molecular motors
Introduction
During the translocation step of protein synthesis the ribosome moves on the mRNA template by one codon (reviews1-5). The two tRNAs, deacylated tRNA and peptidyl-tRNA bound to the P and A sites of the ribosome move to the E and P sites, respectively, followed by rapid spontaneous dissociation of the tRNA from the E site. Translocation can occur spontaneously; however, the rate of the uncatalyzed reaction is very low. In the cell, translocation is accelerated by elongation factor G (EF-G), a large GTP-binding protein that consists of five domains6-9 (Fig. 1A). Domain 1 (or G domain) of EF-G is highly conserved among the translational GTPases and other members of the GTPase superfamily (Fig. 1B). Despite the similarities in the G-domain structures, the nucleotide binding properties of various translational GTPases are quite different. Unlike EF-Tu, EF-G binds GTP and GDP with similar affinity10,11 and exchanges nucleotides spontaneously and rapidly10; in the cell, free EF-G is likely to be in the GTP-bound form due to the high concentration of GTP.10 Upon binding to the ribosome, EF-G rapidly hydrolyzes GTP,12,13 but remains in the GDP-Pi form because of delayed Pi release, which takes place concomitantly with tRNA translocation.12,14,15 Following Pi release, EF-G-GDP dissociates from the ribosome, thereby freeing the A site for the next round of mRNA decoding.
Figure 1. The H91A mutation in EF-G. (A) Structure of EF-G (pdb 2WRI37). The domains of EF-G are numbered 1–5 and color-coded. Residue His91 in domain 1 is in cyan, GTP (orange) was modeled instead of GDP. (B) Sequence conservation within the G domain of translational GTPases. The conserved histidine residue (His91 in EF-G, His84 in EF-Tu) following the G3 motif DXXG (bar) is indicated. (C) Growth curves of E. coli BL21(DE3)pLysS cells transformed with the pET24a plasmid containing the gene coding for either wt EF-G or EF-G(H91A). The induction of expression by IPTG is indicated (arrow).
The role of GTP hydrolysis by EF-G is controversial. All studies that addressed this question so far used non-hydrolyzable GTP analogs to simulate the GTP-bound form of EF-G. Early biochemical studies employing non-hydrolyzable GTP analogs suggested that EF-G was capable of completing a single round of translocation without GTP hydrolysis, whereas the release of EF-G from the ribosome was delayed or inhibited.16,17 These findings led to models in which GTP hydrolysis in EF-G is necessary to switch the factor from the GTP-bound form, in which the factor binds to the ribosome, to the GDP-bound form, which has a low binding affinity to the ribosome, thereby allowing the factor to dissociate after translocation.18 This energy regime is typical for GTPases that switch between the active GTP-bound form capable to interact with an effector and the inactive GDP-bound form and are regulated by proteins which promote nucleotide exchange and GTP hydrolysis.19 These models were challenged by rapid kinetic studies13,20 which showed that (1) GTP is hydrolyzed prior to translocation and (2) translocation is much faster in the presence of GTP than with any of the non-hydrolyzable analogs used. The observed acceleration ranged from 20- to 70-fold, suggesting that the energy of GTP hydrolysis is‑at least in part‑utilized to drive translocation. A similar magnitude of the effect of GTP hydrolysis was observed in single-molecule FRET studies21; however, other biochemical and pre-steady-state studies suggested a much smaller effect, although at different buffer conditions.12,22,23 Furthermore, whereas translocation with EF-G–GTP leads to the dissociation of deacylated tRNA from the E site,24-26 it has been reported that the tRNA remains bound after translocation by EF-G–GDPNP, which would suggest that tRNA dissociation from the E site is promoted by GTP hydrolysis and/or EF-G release.27 Such a stabilization of E-site tRNA has been noted earlier; however, the magnitude of the effect depended on which non-hydrolyzable nucleotide was used, arguing against a fundamental importance of such a mechanism.13
Apart from the fact that non-hydrolyzable GTP analogs are not perfect mimics of GTP, an additional important caveat of using non-hydrolyzable analogs is that these preparations are often not free from traces of GTP; assuming only 1% contamination (which we routinely find in commercial GDPNP preparations), the addition of non-hydrolyzable GTP analogs in millimolar concentration would bring sufficient GTP to drive rapid tRNA translocation, which may constitute one potential source of the discrepancies found in the literature. To overcome this problem, we set out to study the role of GTP hydrolysis in translocation using a GTPase-deficient mutant of EF-G. In EF-Tu from Escherichia coli, the replacement of His84 with Ala resulted in an inhibition of GTP hydrolysis by six orders of magnitude28; the homologous residue in the switch II region of E. coli EF-G is His91, which we exchanged for Ala. We tested the effects of the mutation from different structural perspectives in real time by pre-steady-state kinetic experiments, monitoring the movement of A- and P-site tRNAs as well as of the mRNA, the dissociation of deacylated tRNA from the E site, and the dissociation of the ribosome-nucleotide-EF-G complex.
Results
Construction of the GTPase-deficient EF-G mutant
The conserved histidine at position 91 in the G domain of E. coli EF-G (Fig. 1A and B) was replaced with alanine (Materials and methods). Cells transformed with the pET24a plasmid containing the gene coding for mutated EF-G were impaired in growth after induction with IPTG (Fig. 1C), indicating that the expression of inactive EF-G conferred a dominant lethal phenotype. To avoid the growth inhibition, we designed a construct in which the gene coding for mutant EF-G contained a C-terminal intein domain followed by a chitin-binding domain and a His tag. This construct allowed the expression of EF-G(H91A) without any detrimental effect on cell growth.
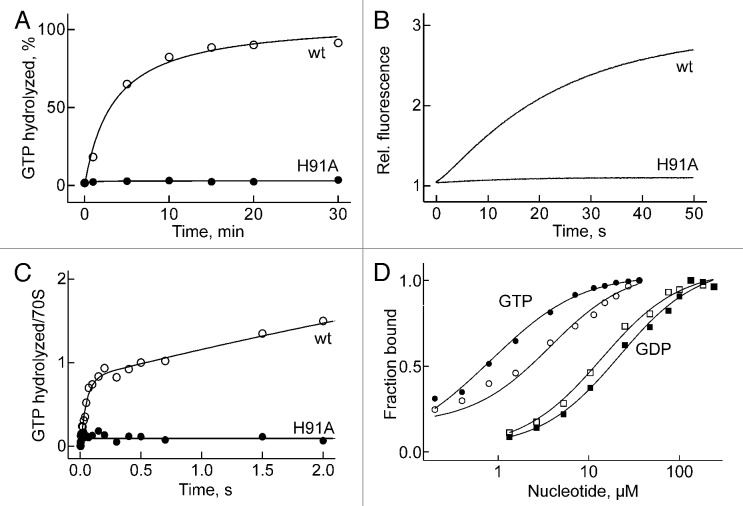
The mutated factor was inactive in GTP hydrolysis on the ribosome under conditions of multiple turnover, i.e., with catalytic amounts of factor relative to ribosomes, as monitored by TLC with [γ-32P]GTP (Fig. 2A) or the release of inorganic phosphate (Pi) monitored by the fluorescence of MDCC-labeled phosphate-binding protein (Fig. 2B). The inhibition of the turnover reaction was caused by the inability of EF-G(H91A) to hydrolyze GTP, rather than its impaired ability to dissociate from the ribosome, as verified by the lack of GTP hydrolysis at single-round GTPase conditions, which reflects the bona fide ability of the factor to cleave GTP. The time course of single-round hydrolysis was measured by quench-flow at an excess of factor over ribosomes, using a fluorescent GTP derivative, mant-GTP. The reaction was monitored by the increasing ratio of mant-GDP to mant-GTP, as determined by HPLC analysis (Fig. 2C). In that assay, the wild-type (wt) factor hydrolyzed close to one equivalent of GTP per ribosome during a rapid burst phase which was followed by a slower turnover reaction, whereas the mutant factor was completely inactive. However, equilibrium titrations with GTP, measuring the fluorescence of tryptophan in EF-G, and with GDP, monitoring fluorescence resonance energy transfer (FRET) from Trp to mant-GDP,10 demonstrated that EF-G(H91A) bound GTP and GDP with wild-type affinity (Fig. 2D), confirming that GTP hydrolysis, rather than GTP binding, was abolished by the H91A mutation.
Figure 2. GTP hydrolysis and GTP binding. (A) Time course of turnover GTP hydrolysis. (B) Turnover GTP hydrolysis monitored by Pi release. (C) Single-round GTP hydrolysis. In the initial burst phase, EF-G hydrolyzed about 0.9 GTP/ribosome. (D) Equilibrium titrations of GTP/GDP binding. EF-G (open symbols) or EF-G(H91A) (closed symbols) (1 µM) was titrated with increasing amounts of GTP, monitoring Trp fluorescence. GDP titrations were performed with mant-GDP, monitoring mant fluorescence excited by FRET from Trp. Kd values were 1–2 µM for GTP and around 20 µM for mant-GDP.
Translocation is slow without GTP hydrolysis
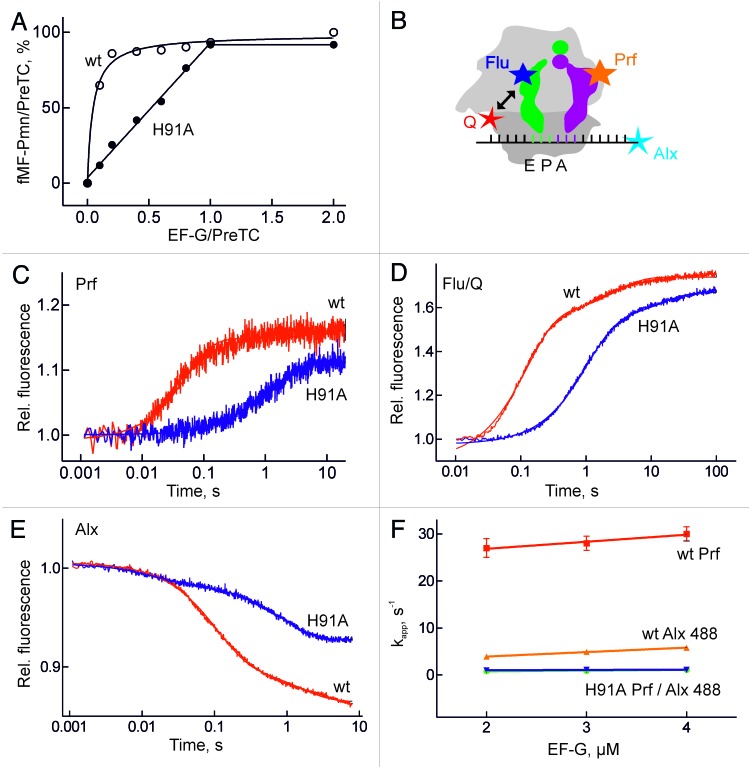
EF-G(H91A) was active in translocation, as shown by the puromycin assay (Materials and methods); by contrast to the wt factor, stoichiometric amounts of mutant EF-G to ribosomes were required to bring about translocation on all ribosomes present (Fig. 3A). This indicated that‑ as expected from previous results obtained with non-hydrolyzable analogs of GTP-EF-G(H91A) catalyzed only a single round of translocation and did not dissociate from the ribosome after one round of translocation.
Figure 3. Translocation. (A) Translocation monitored by the puromycin reaction. Pre-translocation complexes programed with MF-mRNA were incubated with the indicated amount of EF-G or EF-G(H91A) in the presence of GTP and analyzed by the reaction with puromycin (Pmn). The amount of the product fMetPhe-Pmn (fMF-Pmn) is given relative to the initial amount of fMetPhe-tRNA bound to the ribosome. (B) Fluorescence labels in the pre-translocation complex. Prf, proflavin at positions 16/17 in the D loop of fMetPhe-tRNAPhe (magenta); Alx, Alexa488 at position +14 at the 3′ end of MF-mRNA, counting from the A(+1)UG start codon; Flu, fluorescein at 4-thioU(8) in tRNAfMet (green); Q, the non-fluorescent acceptor dye Atto540Q at position 112 of protein S13. (C) Time courses of translocation monitored by Prf fluorescence. (D) Translocation monitored by FRET between Flu in P-site tRNA and Q in protein S13. (E) Translocation monitored by Alx488 at the 3′ end of the mRNA. Stopped-flow traces were evaluated by two-exponential fitting, yielding the values for kapp summarized in Table 1. (F) Saturation of kapp values of translocation with wt and mutant EF-G, monitored by Prf or Alx488.
The kinetics of translocation was studied monitoring the signals from fluorophores in different positions in the pre-translocation complex (Fig. 3B and Table 1). Proflavin attached to the D loop of peptidyl-tRNA (fMetPhe-tRNAPhe(Prf)) in the A site reported on tRNA movement from A to P site. The translocation of deacylated tRNA from the P to the E site and its dissociation from the E site was monitored either by FRET from a fluorescein label (Flu), which was attached to 4-thiouracil at position 8 of the tRNA, to a non-fluorescent acceptor attached to position 112 of ribosomal protein S13 (Fig. 3B) or by the fluorescence change of deacylated tRNAfMet(Prf).12,29 The movement of the mRNA on the 30S subunit was monitored by Alexa dyes (Alx488 or Alx405) attached to the 3′ end (position +14) of the mRNA (Fig. 3B). Rates of steps related to tRNA translocation obtained with Bodipy (Bpy) at the N terminus of peptidyl-tRNA29 are also included (Table 1). The step assignment and detailed kinetic analysis for the different reporter groups can be found elsewhere.12-14,29,30
Table 1. Rate constants of elemental steps of translocation (s-1)a.
| Reporter | EF-G | EF-G(H91A) | Reference | ||
|---|---|---|---|---|---|
| Step 1b | Step 2c | Step 1b | Step 2c | ||
| tRNA (A site) | |||||
| Bpy | 28 | –d | 9e | 0.7e | ref. 29 |
| Prf16/17 | 30 | 1.1 | this paper; refs. 13,14,29 | ||
a Rate constants were obtained at saturating EF-G concentration (4 µM) at 37°C. bThe step reports on the concomitant movement of peptidyl-tRNA from the A to the P site, the displacement of deacylated tRNA from the P site, and the translocation of the mRNA. cThe step reports on the final steps of mRNA translocation (Alx488) or the movement of deacylated tRNA through the E site. dThe respective step is not reported by the particular fluorescence label. eWith EF-G(H91A) the translocation on the 50S subunit is biphasic. The amplitude of the step is about 50% of that observed with wt EF-G. fThe difference between these values and those reported previously12 is due to the temperature difference (37°C vs. 25°C). gn.d., not determined. htentative values; the concentration dependence of the respective kapp suggests a linear dependence; however, the amplitude of the step is too small to allow for precise fitting.
Translocation resulted in a major fluorescence change with every label used (Fig. 3). The rate of translocation, as reported by fMetPhe-tRNAPhe(Prf) (Fig. 3C) or mRNA-Alx405,29 with EF-G(H91A) was 1 sec−1, compared with about 30 sec−1 with wt EF-G (Table 1). The 30-fold rate decrease is similar to previously reported values obtained with non-hydrolyzable GTP analogs, consistent with an important contribution of GTP hydrolysis to translocation.13,20 The amplitudes of the fluorescence changes of Prf and Alx405 observed with wt and mutant EF-G were very similar (documented in Fig. 3C for Prf), indicating that despite the different rates the reactions went to completion regardless of whether GTP was hydrolyzed (Fig. 3A). In contrast, the label at the 3′ end of peptidyl-tRNA(Bpy) reported that translocation on the 50S subunit was biphasic with EF-G(H91A), with a fast step that did not require GTP hydrolysis (9 sec−1) and a second, slow step that was dependent on 30S translocation (1 sec−1) (Table 1; data from ref.29). The apparent rate of deacylated tRNA release from the E site in the presence of wt EF-G was about 10 sec−1, based on both tRNAfMet(Prf) fluorescence and S13–tRNAfMet(Flu) FRET assays (Fig. 3D, Table 1). Notably, the rate of A to P site translocation on ribosomes carrying labels in S13 and tRNAfMet(Flu) monitored by the reporter in mRNA–Alx405 was the same as on unmodified ribosomes (30 sec−1; traces not shown), indicating that the labels per se did not affect the kinetics of translocation. The 10 sec−1 rate represents a global rate of tRNA displacement from the P to the E site followed by the dissociation of the E-site tRNA from the ribosome, because at our concentrations and buffer conditions (low Mg2+ concentrations and no polyamines) essentially all deacylated tRNA dissociated from the ribosome following translocation to the E site. Deconvoluting the rates of the individual reactions yielded an elemental rate constant of E-site clearance of about 15 sec−1. With EF-G(H91A) the amplitude of the FRET change was the same as with wt EF-G (Fig. 3D), suggesting that tRNA release from the E site was not blocked when GTP was not hydrolyzed. The release rate (1 sec−1) was limited by the preceding tRNA translocation step, which was slow with the mutant factor. Essentially the same kinetics of P to E-site translocation was observed with tRNAfMet(Prf) (Table 1).
Unexpectedly, translocation monitored by the Alx488 label at the 3′ end of the mRNA reported a step that did not follow the pattern of tRNA-mRNA translocation observed with the other labels (Fig. 3E,F). With wt EF-G, the rate of the dominant step reported by Alx488 was significantly slower (6 sec−1) than the rates of translocation on either 30S or 50S subunits reported by the other labels (30 sec−1) (Fig. 3F). With EF-G(H91A), the reaction was as slow as translocation (1 sec−1) (Table 1); however, the fluorescence change did not reach the full amplitude (Fig. 3E). This may suggest that Alx488 at the 3′ end of the mRNA reports on a step in translocation which is physically distinct from mRNA-tRNA displacement itself and is affected by GTP hydrolysis. Notably, the reduced amplitude was not due to EF-G remaining bound after translocation, because the full fluorescence amplitude due to mRNA-Alx488 translocation was observed in the presence of fusidic acid (FA) (data not shown), which freezes EF-G on the ribosome in a post-translocation state.31
The complex of ribosomes with EF-G(H91A)-GTP is very stable
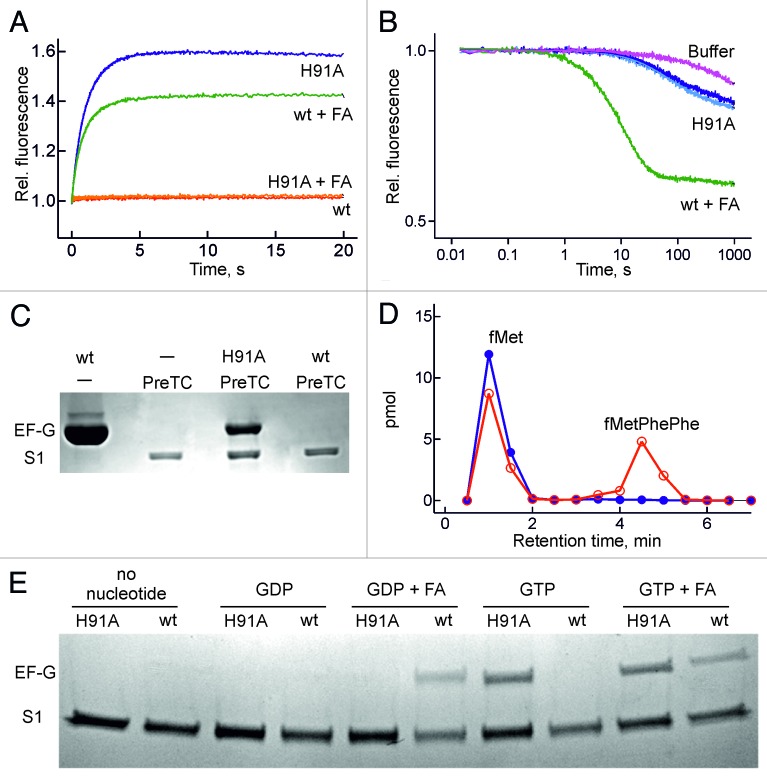
With wt EF-G, the initially formed ribosome-EF-G-GTP complex, which is unstable and dissociates rapidly (koff ≈100 sec−1),20 rearranges to form the GTPase-activated complex, which subsequently hydrolyzes GTP.13 The resulting ribosome-EF-G-GDP-Pi complex does not accumulate, as monitored by the fluorescence of mant-GTP that was excited via FRET from a tryptophan residue in EF-G (Fig. 4A), likely because it is consumed in the following steps that lead to factor dissociation. By contrast, when GTP hydrolysis is blocked, as in EF-G(H91A)-GTP, the increase of mant-GTP fluorescence reports the formation of a ribosome–EF-G-mant-GTP complex at a rate of around 1 sec−1 (Fig. 4A), whereas with mant-GDP the stable complex was not formed, even though EF-G(H91A) could bind mant-GDP (Fig. 2D). When the dissociation of wt EF-G from the ribosome was blocked by FA the high-fluorescence intermediate was stabilized, although the amplitude of fluorescence change was smaller than with EF-G(H91A) (Fig. 4A). The smaller amplitude may either be due to the fact that FA is not very effective as an inhibitor,31,32 allowing partial dissociation of EF-G–mant-GDP from the ribosome, or indicate that the complexes with EF-G(H91A)–GTP and EF-G–GDP–FA are structurally different. To examine that possibility, we tested the stability of mant-GTP/mant-GDP binding to EF-G on the ribosome by competition with excess unlabeled GTP or GDP; the observed rates represent the dissociation rate constant of mant-GTP/mant-GDP from EF-G bound to the ribosome. The complex was almost 10 times more stable with EF-G(H91A) than with EF-G-FA (Fig. 4B), which suggests that the ribosome-EF-G complex formed with GTP is structurally different from that formed after GTP hydrolysis and Pi release in the presence of FA.
Figure 4. Formation of a stable ribosome-EF-G(H91A) -GTP complex. (A) Time courses monitored by mant-nucleotide fluorescence. Red trace, wt EF-G + mant-GTP; green trace, wt EF-G with mant-GDP and FA (0.2 mM); blue trace, EF-G(H91A) + mant-GTP; orange trace, EF-G(H91A) + mant-GDP with FA. (B) Stability of ribosome-EF-G-mant-GTP complexes. The complex with wt EF-G + FA was chased with excess GDP (green trace), the complex with EF-G(H91A) with excess GDP (dark blue trace) or GTP (light blue trace). (C) EF-G binding to pre-translocation complex (PreTC). EF-G and ribosomal protein S1 were visualized by Coomassie staining. PreTC, pre-translocation complex. (D) Peptide formation. Initiation complexes programed with MFF-mRNA and containing fMet-tRNAfMet in the P site were pre-incubated with wt EF-G (blue) or EF-G(H91A) (red) in the presence of GTP and mixed with EF-Tu–GTP-Phe-tRNAPhe. fMetPhePhe tripeptide was separated from unreacted fMet by HPLC (Materials and methods). (E) eF-G binding to vacant ribosomes. conditions as in (C) but with 5 µM mant-nucleotides and 0.2 mM FA for comparison with Fig. 4A and 4B.
The high stability of ribosome-EF-G(H91A)-GTP complex could also be demonstrated by pull-down experiments. The complex remained intact during centrifugation through a sucrose cushion, in contrast to the complex of ribosomes with wt EF-G, which was not observed (Fig. 4C). Retention of EF-G(H91A) to the ribosome required GTP, suggesting that it is specifically the GTP-bound conformation of the factor that remained stably bound (Fig. 4D). In contrast, the stable binding of wt EF-G to the ribosome required addition of FA and was observed with both GDP and GTP (Fig. 4D); the letter is hydrolyzed to GDP within the time of experiment.
In keeping with these results, in the complex of EF-G(H91A)–GTP with ribosomes the mutant factor was bound strongly enough to prevent the binding of the ternary complex EF-Tu-GTP-Phe-tRNAPhe. This is evident from the lack of fMetPhe and fMetPhePhe di- and tripeptide formation on ribosomes programed with MFF-mRNA when EF-G(H91A) was preincubated with the ribosome prior to the addition of the EF-Tu complex (Fig. 4D). Thus, following (slow) translocation, EF-G in the GTP-bound form is stalled on the ribosome in a conformation that differs from that induced by FA.
Discussion
EF-G hydrolyzes GTP very rapidly, much faster than the displacement of mRNA and tRNA takes place,12,13 which makes it difficult to deconvolute the potential effects of GTP hydrolysis on the elemental steps of translocation. The use of a GTPase-deficient mutant of EF-G allows the dissection of the EF-G cycle into GTPase-dependent and independent steps, using the authentic GTP-bound form of the factor, thereby providing an insight into the energy regime of translocation. The advantage of the H91A mutation is that it does not change the affinity of GTP or GDP binding, indicating that the mutation does not affect the structure and dynamics of the nucleotide binding pocket. Due to the lack of GTP hydrolysis, the conformational switch from the GTP- to the GDP-bound form of EF-G is blocked, and the GTP-bound conformation of the factor is favored; therefore, GTP can be used rather than GTP analogs which may not mimic GTP perfectly.
By analogy to EF-Tu, where the mutation of the catalytic histidine reduces the rate of GTP hydrolysis by five orders of magnitude,28 the H91A mutation in EF-G inhibits GTP hydrolysis, in this case virtually completely, as there is no measurable GTPase activity of the factor, even when it is bound to the ribosome. The complete loss of the GTPase activity of EF-G(H91A) suggests that the detailed mechanism of GTP hydrolysis, and the contributions of different groups to catalysis, may differ in EF-G and EF-Tu, despite the high sequence conservation in their GTP-binding domains. Consistent with this notion, the intrinsic GTPase activities of EF-Tu and EF-G also differ, ranging from the low, but measurable activity of EF-Tu (10−5 s−1) to practically no activity of EF-G. Biochemical evidence suggests that without GTP hydrolysis EF-G brings about one round of translocation, but is not capable of turnover; therefore, to promote translocation on a given amount of pre-translocation complexes, stoichiometric amounts of EF-G(H91A) are required, rather than the catalytic amounts of wt EF-G that suffice to promote translocation on excess ribosomes in a turnover reaction.
The use of fluorophores at different positions in the pre-translocation complex revealed not only movements of the tRNAs and the mRNA from A to P and P to E sites, but also reported on tRNA dissociation from the ribosome and on rearrangements within the 30S subunit which have not been observed before. Thus, the present work provides a comprehensive velocity map of translocation with and without GTP hydrolysis. Several labels robustly report translocation of mRNA and tRNAs from A to P and P to E sites, including 30S translocation monitored by mRNA-Alx405, 50S translocation of tRNA by Bpy, and the movement of the tRNA elbow regions by Prf labels in peptidyl-tRNA and deacylated tRNA. For translocation catalyzed by wt EF-G with GTP, all these labels reported a rate of about 30 sec−1 (Table 1). The Prf label in tRNAfMet additionally reported on the movement of deacylated tRNA out of the E site,12,29 taking place at a rate of 10–13 sec−1. A similar rate was observed when FRET between deacylated tRNA and the ribosome, labeled at protein S13, was monitored. This lower rate can be explained by the step-wise nature of the dissociation of the P-site tRNA, which has to move from the P to the E site (likely with the same rate as the A to P site movement of peptidyl-tRNA) before it dissociates from the E site into solution; this requires additional time and thus lowers the overall rate of the process. In translocation promoted by EF-G(H91A)-GTP all steps proceed at a uniform rate of 1 sec−1, suggesting that the lack of GTP hydrolysis impairs a step that precedes, or takes place concomitantly with, tRNA movement and is rate-limiting for the following steps. The only exception is the Bpy label on the 3′ end of peptidyl-tRNA on the 50S subunit which apparently proceeds in two steps, reaching an intermediate position independently of GTP hydrolysis and then moving to the post state at the rate of slow 30S translocation.29 In contrast to previous results reported for translocation with EF-G and GDPNP,27 in translocation promoted by EF-G(H91A)–GTP the deacylated tRNA did not accumulate in the E site, suggesting that GTP hydrolysis is not required for E-site clearance and indicating that, at least for this step, GDPNP is not a perfect mimic of GTP. Thus, part of the energy of GTP hydrolysis is used to drive a conformational change of the ribosome that controls translocation; the following tRNA release itself is spontaneous.
One unexpected result concerns the differences in steps reported by the two fluorescence labels in the mRNA, Alx405 and Alx488. Biochemically, the complexes bearing the two mRNAs are very similar, i.e., in both cases the label is attached at position +14 and the extent of complex formation and translocation is the same. However, mRNA-Alx405 reports a step that kinetically coincides with the rate of tRNA movement both on the 50S subunit and at the elbow region (30 sec−1), whereas the major step reported by mRNA-Alx488 is significantly slower (6 sec−1). We hypothesize that on the ribosome the two labels are oriented differently, with Alx405 facing the 30S body and therefore reporting on the mRNA displacement relative to the body of the 50S subunit, and Alx488 following the movements of the 30S head, which are only loosely coupled to the body movements.33,34 If our interpretation is correct, then the rate of backward head movement is the slowest step of EF-G-dependent translocation and may fully or partially determine the rate of EF-G release and EF-G turnover (2–5 sec−1 at our conditions15,35). A similar rate for the backward head movement, 10 sec−1, has been observed by FRET measurements34; however, given the difference in buffer conditions, the comparison should be made with caution. With EF-G(H91A), this step is as slow as all other steps of translocation, 1 sec−1, but occurs only partially, as evident from the 50% smaller amplitude, compared with wt EF-G. This suggests that in the absence of GTP hydrolysis the 30S head may be trapped in a distinct intermediate state between the pre- and post-translocation positions or may reversibly fluctuate between those states. Thus, GTP hydrolysis is not only important for the acceleration of translocation, but may also be essential for the backward rotation of the 30S head.
EF-G(H91A)–GTP forms a very tight complex with the ribosome which may resemble the structure of an intermediate complex with EF-G-GDP-FA revealed by cryo-EM reconstruction.36 In that work, the ribosome complexes contained deacylated tRNA in the P site, and EF-G-GDP was stabilized in the A site by FA. By computational sorting the authors identified two complexes, one with the tRNA in the P/E state, which they denoted as pre-translocation intermediate (TIPRE; we note, however, that the A site of the ribosome contained no tRNA), and the second with the P-site tRNA in intermediate pe/E position, denoted TIPOST. In the latter complex, EF-G with domain 4 reached into the 30S A site, and peptidyl-tRNA could be modeled in an ap/P position without clashing with EF-G.36 In the pe/E position, the tRNA maintained contact with the P site on the 30S head and simultaneously established an interaction with the E site on the 30S platform; domain 4 of EF-G interacted with the 30S head domain, stabilizing the swiveled conformation. While the elbow region and the 3′ end of the tRNA were essentially in their post-translocation position, the translocation of the mRNA with the tRNA anticodons is incomplete with respect to the 30S head. If EF-G(H91A) stabilized the same or a similar state, one would expect that labels at the 3′ end of the mRNA are differently affected by mRNA translocation, depending on their distance to the 30S head and platform, as we hypothesize for mRNA-Alx488 and mRNA-Alx405. This assignment of steps would support the notion that the pe/E state is an authentic translocation intermediate and provide a time line to the cryo-EM results. In contrast to the original assignment of the pe/E state,36 our data indicate that the intermediate referred to as TIPOST is rather a late pre-translocation intermediate. In comparison, TIPRE (which equally well can be described as a post-translocation intermediate, because there is no A-site tRNA), might be closer to the intermediate stabilized by FA, in which the translocation is complete on both 50S and 30S subunit,36-39 consistent with the complex assignment as a post-translocation complex stalled prior to EF-G release.
In agreement with earlier results, our data indicate that GTP hydrolysis is important for the release of EF-G from the ribosome. The ribosome-EF-G(H91A)-GTP complex is extremely stable and blocks the access of other factors. In the GTP-bound form of EF-G, the stability of nucleotide binding in the complex is very high, consistent with the low nucleotide exchange rates observed with GDPNP.10 In comparison, nucleotide exchange in the FA-stalled ribosome-EF-G complex is almost 10 times faster, suggesting that the structure of the nucleotide binding pocket of EF-G is relaxed upon GTP hydrolysis and Pi release, independent of the presence of FA.31
The acceleration of translocation by GTP hydrolysis indicates that in the early phase of translocation EF-G functions as a motor protein that uses the free energy of GTP hydrolysis to drive the movement. Such a function implies that the transition from EF-G-GTP to EF-G-GDP-Pi is accompanied by a structural rearrangement of the factor that is coupled to a conformational change of the pre-translocation complex.14 The energy of GTP hydrolysis is used to accelerate the movement of both tRNAs relative to the 50S subunit and the body of the 30S subunit. The completion of the reaction on the 30S subunit, likely by the backward swivel of the 30S subunit and the dissociation of EF-G, requires another structural change which presumably is driven by the release of Pi.31 The conformational switch is inhibited, and, with that, the dissociation of EF-G from the ribosome, when either the GTPase-inactive EF-G mutant or non-hydrolyzable GTP analogs are used or when the nucleotide binding site retains Pi15 or vanadate mimicking Pi.31 Thus, there are two energy regimes related to different phases of translocation by which EF-G uses GTP hydrolysis. In the first phase EF-G acts as a motor protein which transforms (part of) the energy of GTP hydrolysis into accelerated tRNA–mRNA movement on the ribosome, presumably by driving conformational changes of the ribosome. In the second phase EF-G switches to the GDP-bound conformation, which enables dissociation, and thus acts as a GTPase that couples the energy of GTP hydrolysis to operate conformational switching.
Materials and Methods
Buffers and reagents
Buffer A: 50 mM TRIS-HCl, pH 7.5, 70 mM NH4Cl, 30 mM KCl and 7 mM MgCl2. Chemicals were from Roche Molecular Biochemicals, Sigma Aldrich, or Merck. Radioactive compounds were from Hartmann Analytic.
Ribosomes, mRNAs, tRNAs, and translation factors
Ribosomes from E. coli MRE 600, f[3H]Met-tRNAfMet, f[3H]Met-tRNAfMet(Flu), [14C]Phe-tRNAPhe, EF-Tu, and initiation factors were prepared as described.14,30,40,41 Proflavin-labeled tRNAPhe (yeast) and tRNAfMet (E. coli) were prepared according to published protocols.42,43 The mRNA constructs (IBA) were 30 to 33-nucleotides long and contained one (MF-mRNA) or two (MFF-mRNA) Phe codons following the AUG codon.
Expression and purification of EF-G
The gene coding for EF-G was cloned into pET24a(+) (Novagen) or pTXB1 (NEB). The pET24EF-G plasmid was used for the expression of wt EF-G with an N-terminal His tag. The H91A mutation was introduced into both vectors using the QuikChange protocol. pTXB1EF-G(H91A) was used for expression of EF-G(H91A) with an N-terminal intein cleavage site, a chitin-binding domain, and an additional His tag.
Wt EF-G was overexpressed in BL21(DE3) and EF-G(H91A) in BL21(DE3)pLysS. Cells were grown in LB medium supplemented with kanamycin (30 µg/ml) (wt) or with ampicillin (50 µg/ml) (H91A) at 37°C; expression was induced by the addition of IPTG (1 mM), and cultures were grown further for 4 h. Cells were harvested and pellets were resuspended in Protino buffer (20 mM TRIS-HCl, pH 7.4, 300 mM NaCl) with the addition of Complete Protease Inhibitor (Roche) and a trace of DNaseI. Cells were opened using an Emulsiflex apparatus (Avestin), and the extract was centrifuged for 30 min at 30,000 × g. The supernatant was applied to a Protino gravity-flow column (Macherey-Nagel) for affinity purification using the His tag. The column was washed with Protino buffer and the protein was eluted with Protino buffer containing 250 mM imidazole. The eluted wt protein was concentrated and the buffer was exchanged to 2x buffer A by membrane filtration (Vivaspin 30,000); for storage, one volume of glycerol was added. For EF-G(H91A), the eluate was dialyzed twice against cleavage buffer (20 mM TRIS-HCl, pH 8.5, 500 mM NaCl), 50 mM of sodium 2-mercaptosulfonate (MesNa) were added and the tags were removed by intein cleavage at 4°C for 16 h. The proteins were re-applied to a Protino gravity-flow column and the flow-through was collected. The proteins were concentrated and the buffer was exchanged to 2x buffer A by membrane filtration (Vivaspin 30,000); for storage, one volume of glycerol was added. EF-G concentrations were determined by spectrophotometry at 280 nm using an extinction coefficient of 64,300 M−1cm−1.
Growth curves
BL21(DE3)pLysS cells were transformed with pET24EF-G and pET24EF-G(H91A) and plated on LB agar with 30 µg/ml kanamycin and 1% glucose and grown overnight. Single colonies were picked and overnight cultures were inoculated. Cells were pelleted, resuspended in fresh medium, expression cultures were inoculated to an OD600 of 0.1, and grown at 37°C. After 100 min expression was induced by addition of 1 mM IPTG and the cultures were grown for another 3 h.
Labeling of 30S subunits
The gene coding for ribosomal protein S13 (rpsM) was cloned into a pET24a vector. The single native Cys85 was replaced with Ser and Cys was engineered at position Pro112. Expression, purification and refolding of S13 were performed essentially as described.44 Labeling with Atto540Q was performed under denaturing conditions with 10-fold molar excess of dye for 2 h at 25°C in buffer (6 M urea, 50 mM HEPES. pH 7.1, 300 mM KCl, 10% glycerol). The reaction was stopped by the addition of 6 mM 2-mercaptoethanol, excess dye was removed by cation exchange chromatography (HiTrap SP HP column, GE-Healthcare). The degree of labeling was 100%, as determined spectrophotometrically.
Reconstitution of purified 30S ΔS13 ribosomal subunits (from E.coli strain MG 1655 ΔrpsM::kan, kindly provided by Rachel Green) was performed in buffer (50 mM HEPES, pH 7.5, 400 mM KCl, 20 mM MgCl2, 6 mM 2-mercaptoethanol) with a 1.75-fold molar excess of Atto 540Q-labeled protein S13.45 The mixture was incubated for 60 min at 47°C in the dark. 30S subunits were purified by centrifugation through a 30% sucrose cushion in the same buffer. Pellets were resuspended in buffer A. 30S subunits were labeled to 90–100%, as determined by spectrophotometric analysis.
Ribosome complexes
To prepare pre-translocation complex, ribosomes (0.6–1 µM 70S or labeled 30S together with a 1.5-fold excess of 50S subunits) were incubated with a 3-fold excess of mRNA and a 1.5 to 2-fold excess each of IF1, IF2, IF3, and 1.5-fold excess of f[3H]Met-tRNAfMet or a 3-fold excess of f[3H]Met-tRNAfMet(Flu) in buffer A containing 1 mM GTP for 30 min at 37°C. Ternary complex EF-Tu-[14C]Phe-tRNAPhe-GTP was prepared by incubating EF-Tu (2-fold excess over Phe-tRNAPhe) with GTP (1 mM), phosphoenolpyruvate (3 mM), and pyruvate kinase (0.5 µg/ml) for 15 min at 37°C and then with [14C]Phe-tRNAPhe (2-fold excess over ribosomes) for an additional min. Equal volumes of initiation complex and ternary complex were mixed and incubated for 1 min at 37°C. Pre-translocation complexes used for stopped-flow experiments were purified by centrifugation through a 1.1 M sucrose cushion in buffer A with 20 mM MgCl2. Pellets were dissolved in buffer A with 20 mM MgCl2 and tRNA binding was verified by nitrocellulose filtration.
Turnover GTP hydrolysis
Vacant ribosomes (0.5 µM) were mixed with EF-G (0.5 µM) in buffer A with 1 mM GTP and trace amounts of [γ-32P]GTP at 20°C. Samples were taken and quenched with one volume of 40% formic acid. Samples were analyzed by TLC (Polygram CEL 300, Macherey-Nagel) using 0.5 M potassium phosphate (pH 3.5) running buffer. Radioactivity was detected using a phosphoimager system.
Puromycin reaction
Translocation was tested using the puromycin assay. Pre-translocation complexes (0.1 µM) were incubated with various amount of EF-G or EF-G(H91A) for 3 min at 20°C in buffer A. Samples were then reacted with puromycin (1 mM) for 10 sec before being quenched with 1.5 M sodium acetate saturated with MgSO4. f[3H]Met[14C]Phe-puromycin was extracted with ethyl acetate and quantified by radioactivity counting.
Rapid kinetics methods
Single-round GTP hydrolysis was measured in buffer A at 20°C in a quench-flow device (KinTek Laboratories, Inc.) by rapidly mixing equal volumes of vacant ribosomes (1 µM) and mant-GTP (50 µM) (Jena Biosciences) with EF-G (wt or H91A) (5 µM) and mant-GTP (50 µM). Samples were quenched with 40% formic acid. Following neutralization by KOH, samples were analyzed by HPLC monitoring mant fluorescence. The rates of GTP hydrolysis obtained by this method are identical to those measured with [32P]GTP (Liudmila Filonava, Pohl Milon and Marina V. Rodnina, manuscript in preparation). Pi release was measured in a stopped-flow apparatus (Applied Photophysics).14,46 Vacant ribosomes (0.5 µM) were rapidly mixed with EF-G (0.5 µM) in buffer A, GTP (1 mM), and MDCC-labeled phosphate-binding protein (PBP) (2.5 µM), both pre-incubated with “Pi mop” (0.1 U/ml PNPase, 0.2 µM 7-methylguanosine). MDCC fluorescence was excited at 425 nm and measured after passing a KV450. Rapid kinetics of translocation was measured in buffer A containing 1 mM GTP at 37°C by stopped-flow14,1514,1513,14. Alx488, Prf and Flu fluorescence was excited at 470 nm and detected after passing a KV500 cut-off filter (Schott). Equal volumes of pre-translocation complexes (50–80 nM) were rapidly mixed with EF-G or EF-G(H91A) at saturating concentrations (4 µM) if not stated otherwise. EF-G-ribosome complex formation and dissociation was monitored by the fluorescence of mant-labeled nucleotides. Ribosomes (0.5 µM) were rapidly mixed with 0.5 µM EF-G and mant-GTP or mant-GDP (5 µM) in buffer A at 37°C in a stopped-flow apparatus. Complex formation was monitored by mant fluorescence excited by FRET from tryptophan excited at 290 nm after passing a KV408 filter (Schott). For chase experiments, preformed EF-G-mant-nuclotide-ribosome complexes (0.5 µM) were rapidly mixed with unlabeled GTP or GDP (1 mM) in buffer A at 37°C.
EF-G-ribosome pull-down assay
Pre-translocation complexes (0.1 µM) and EF-G (wt or H91A) (0.2 µM) were incubated in buffer A with GTP (1 mM) for 5 min at 20°C. Samples were centrifuged through 400 µl sucrose cushions (40% in buffer A with 20 mM MgCl2) at 259,000 g for 2 h. Pellets were resuspended in buffer A and concentrations were measured photometrically at 260 nm. Samples (50 pmol) were analyzed on a pre-casted 4–20% SDS polyacrylamide gradient gel (Serva). Pull-down experiments with vacant ribosomes were performed with mant-GTP/GDP (5 µM) to allow comparisons to Figure 4A and B; otherwise same conditions were used as in Figure 4C.
Tripeptide formation
Initiation complexes (0.5 µM) with f[3H]Met-tRNAfMet in the P site and programed with an mRNA coding for fMetPhePhe were incubated with wt EF-G or EF-G(H91A) (1 µM) for 1 min in buffer A at 37°C before EF-Tu∙[14C]Phe-tRNAPhe∙GTP (2 µM) was added. After 2 min at 37°C samples were quenched with 1/10 volume of 0.5 M KOH and hydrolyzed for 30 min at 37°C. Samples were neutralized with 1/10 volume of acetic acid and peptides were analyzed by HPLC.27
Nucleotide binding
To minimize protein adsorption, cuvettes were treated with a solution of BSA (1 mg/ml) in buffer A for 15 min and rinsed with buffer A. EF-G (1 µM) was titrated with either GTP or mant-GDP in buffer A at 37°C. After each nucleotide addition the sample was equilibrated for 5 min before a reading was taken. Tryptophan fluorescence (GTP) was excited at 290 nm and emission was measured at 350 nm. Mant-GDP fluorescence was excited by FRET from tryptophan as above and measured at 445 nm. Titration curves were evaluated using a quadratic equation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Rachel Green for the kind gift of the strain lacking ribosomal protein S13, Pohl Milon for initiating the cloning work on S13, Liudmila Filonava for providing the non-radioactive GTPase assay, and Anna Bursy, Olaf Geintzer, Sandra Kappler, Christina Kothe, Theresia Uhlendorff, Tanja Wiles, and Michael Zimmermann for expert technical assistance. The work was supported by the Max Planck Society and a grant of the Deutsche Forschungsgemeinschaft (SFB 860, M.V.R.).
References
- 1.Rodnina MV, Wintermeyer W. . The ribosome as a molecular machine: the mechanism of tRNA-mRNA movement in translocation. Biochem Soc Trans 2011; 39:658 - 62; http://dx.doi.org/ 10.1042/BST0390658; PMID: 21428957 [DOI] [PubMed] [Google Scholar]
- 2.Shoji S, Walker SE, Fredrick K. . Ribosomal translocation: one step closer to the molecular mechanism. ACS Chem Biol 2009; 4:93 - 107; http://dx.doi.org/ 10.1021/cb8002946; PMID: 19173642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agirrezabala X, Frank J. . Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q Rev Biophys 2009; 42:159 - 200; http://dx.doi.org/ 10.1017/S0033583509990060; PMID: 20025795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmeing TM, Ramakrishnan V. . What recent ribosome structures have revealed about the mechanism of translation. Nature 2009; 461:1234 - 42; http://dx.doi.org/ 10.1038/nature08403; PMID: 19838167 [DOI] [PubMed] [Google Scholar]
- 5.Noble CG, Song H. . Structural studies of elongation and release factors. Cell Mol Life Sci 2008; 65:1335 - 46; http://dx.doi.org/ 10.1007/s00018-008-7495-6; PMID: 18213444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.AEvarsson A, Brazhnikov E, Garber M, Zheltonosova J, Chirgadze Y, al-Karadaghi S, et al. . Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus.. EMBO J 1994; 13:3669 - 77; PMID: 8070397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.al-Karadaghi S, Aevarsson A, Garber M, Zheltonosova J, Liljas A. . The structure of elongation factor G in complex with GDP: conformational flexibility and nucleotide exchange. Structure 1996; 4:555 - 65; http://dx.doi.org/ 10.1016/S0969-2126(96)00061-5; PMID: 8736554 [DOI] [PubMed] [Google Scholar]
- 8.Laurberg M, Kristensen O, Martemyanov K, Gudkov AT, Nagaev I, Hughes D, et al. . Structure of a mutant EF-G reveals domain III and possibly the fusidic acid binding site. J Mol Biol 2000; 303:593 - 603; http://dx.doi.org/ 10.1006/jmbi.2000.4168; PMID: 11054294 [DOI] [PubMed] [Google Scholar]
- 9.Czworkowski J, Wang J, Steitz TA, Moore PB. . The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution. EMBO J 1994; 13:3661 - 8; PMID: 8070396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilden B, Savelsbergh A, Rodnina MV, Wintermeyer W. . Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc Natl Acad Sci U S A 2006; 103:13670 - 5; http://dx.doi.org/ 10.1073/pnas.0606099103; PMID: 16940356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauryliuk V, Mitkevich VA, Eliseeva NA, Petrushanko IY, Ehrenberg M, Makarov AA. . The pretranslocation ribosome is targeted by GTP-bound EF-G in partially activated form. Proc Natl Acad Sci U S A 2008; 105:15678 - 83; http://dx.doi.org/ 10.1073/pnas.0807912105; PMID: 18836081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan D, Kirillov SV, Cooperman BS. . Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell 2007; 25:519 - 29; http://dx.doi.org/ 10.1016/j.molcel.2007.01.014; PMID: 17317625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. . Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 1997; 385:37 - 41; http://dx.doi.org/ 10.1038/385037a0; PMID: 8985244 [DOI] [PubMed] [Google Scholar]
- 14.Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. . An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell 2003; 11:1517 - 23; http://dx.doi.org/ 10.1016/S1097-2765(03)00230-2; PMID: 12820965 [DOI] [PubMed] [Google Scholar]
- 15.Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. . Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J 2005; 24:4316 - 23; http://dx.doi.org/ 10.1038/sj.emboj.7600884; PMID: 16292341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belitsina NV, Glukhova MA, Spirin AS. . Translocation in ribosomes by attachment-detachment of elongation factor G without GTP cleavage: evidence from a column-bound ribosome system. FEBS Lett 1975; 54:35 - 8; http://dx.doi.org/ 10.1016/0014-5793(75)81062-3; PMID: 1093876 [DOI] [PubMed] [Google Scholar]
- 17.Inoue-Yokosawa N, Ishikawa C, Kaziro Y. . The role of guanosine triphosphate in translocation reaction catalyzed by elongation factor G. J Biol Chem 1974; 249:4321 - 3; PMID: 4605331 [PubMed] [Google Scholar]
- 18.Kaziro Y. . The role of guanosine 5′-triphosphate in polypeptide chain elongation. Biochim Biophys Acta 1978; 505:95 - 127; http://dx.doi.org/ 10.1016/0304-4173(78)90009-5; PMID: 361078 [DOI] [PubMed] [Google Scholar]
- 19.Bourne HR, Sanders DA, McCormick F. . The GTPase superfamily: a conserved switch for diverse cell functions. Nature 1990; 348:125 - 32; http://dx.doi.org/ 10.1038/348125a0; PMID: 2122258 [DOI] [PubMed] [Google Scholar]
- 20.Katunin VI, Savelsbergh A, Rodnina MV, Wintermeyer W. . Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry 2002; 41:12806 - 12; http://dx.doi.org/ 10.1021/bi0264871; PMID: 12379123 [DOI] [PubMed] [Google Scholar]
- 21.Munro JB, Wasserman MR, Altman RB, Wang L, Blanchard SC. . Correlated conformational events in EF-G and the ribosome regulate translocation. Nat Struct Mol Biol 2010; 17:1470 - 7; http://dx.doi.org/ 10.1038/nsmb.1925; PMID: 21057527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zavialov AV, Hauryliuk VV, Ehrenberg M. . Guanine-nucleotide exchange on ribosome-bound elongation factor G initiates the translocation of tRNAs. J Biol 2005; 4:9; http://dx.doi.org/ 10.1186/jbiol24; PMID: 15985150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ermolenko DN, Noller HF. . mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol 2011; 18:457 - 62; http://dx.doi.org/ 10.1038/nsmb.2011; PMID: 21399643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertson JM, Wintermeyer W. . Mechanism of ribosomal translocation. tRNA binds transiently to an exit site before leaving the ribosome during translocation. J Mol Biol 1987; 196:525 - 40; http://dx.doi.org/ 10.1016/0022-2836(87)90030-1; PMID: 2824784 [DOI] [PubMed] [Google Scholar]
- 25.Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW, Puglisi JD. . Real-time tRNA transit on single translating ribosomes at codon resolution. Nature 2010; 464:1012 - 7; http://dx.doi.org/ 10.1038/nature08925; PMID: 20393556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C, Stevens B, Kaur J, Smilansky Z, Cooperman BS, Goldman YE. . Allosteric vs. spontaneous exit-site (E-site) tRNA dissociation early in protein synthesis. Proc Natl Acad Sci U S A 2011; 108:16980 - 5; http://dx.doi.org/ 10.1073/pnas.1106999108; PMID: 21969541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spiegel PC, Ermolenko DN, Noller HF. . Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA 2007; 13:1473 - 82; http://dx.doi.org/ 10.1261/rna.601507; PMID: 17630323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daviter T, Wieden HJ, Rodnina MV. . Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J Mol Biol 2003; 332:689 - 99; http://dx.doi.org/ 10.1016/S0022-2836(03)00947-1; PMID: 12963376 [DOI] [PubMed] [Google Scholar]
- 29.Holtkamp W, Cunha CE, Peske F, Konevega AL, Wintermeyer W, Rodnina MV. GTP hydrolysis by EF-G synchronizes tRNA translocation on small and large ribosomal subunits. Submitted for publication 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. . Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol 2004; 343:1183 - 94; http://dx.doi.org/ 10.1016/j.jmb.2004.08.097; PMID: 15491605 [DOI] [PubMed] [Google Scholar]
- 31.Savelsbergh A, Rodnina MV, Wintermeyer W. . Distinct functions of elongation factor G in ribosome recycling and translocation. RNA 2009; 15:772 - 80; http://dx.doi.org/ 10.1261/rna.1592509; PMID: 19324963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seo HS, Abedin S, Kamp D, Wilson DN, Nierhaus KH, Cooperman BS. . EF-G-dependent GTPase on the ribosome. conformational change and fusidic acid inhibition. Biochemistry 2006; 45:2504 - 14; http://dx.doi.org/ 10.1021/bi0516677; PMID: 16489743 [DOI] [PubMed] [Google Scholar]
- 33.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. . Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature 2010; 466:329 - 33; http://dx.doi.org/ 10.1038/nature09206; PMID: 20631791 [DOI] [PubMed] [Google Scholar]
- 34.Guo Z, Noller HF. . Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci U S A 2012; 109:20391 - 4; http://dx.doi.org/ 10.1073/pnas.1218999109; PMID: 23188795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohr D, Wintermeyer W, Rodnina MV. . GTPase activation of elongation factors Tu and G on the ribosome. Biochemistry 2002; 41:12520 - 8; http://dx.doi.org/ 10.1021/bi026301y; PMID: 12369843 [DOI] [PubMed] [Google Scholar]
- 36.Ratje AH, Loerke J, Mikolajka A, Brünner M, Hildebrand PW, Starosta AL, et al. . Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 2010; 468:713 - 6; http://dx.doi.org/ 10.1038/nature09547; PMID: 21124459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. . The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 2009; 326:694 - 9; http://dx.doi.org/ 10.1126/science.1179709; PMID: 19833919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agrawal RK, Penczek P, Grassucci RA, Frank J. . Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc Natl Acad Sci U S A 1998; 95:6134 - 8; http://dx.doi.org/ 10.1073/pnas.95.11.6134; PMID: 9600930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stark H, Rodnina MV, Wieden H-J, van Heel M, Wintermeyer W. . Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell 2000; 100:301 - 9; http://dx.doi.org/ 10.1016/S0092-8674(00)80666-2; PMID: 10676812 [DOI] [PubMed] [Google Scholar]
- 40.Milon P, Konevega AL, Peske F, Fabbretti A, Gualerzi CO, Rodnina MV. . Transient kinetics, fluorescence, and FRET in studies of initiation of translation in bacteria. Methods Enzymol 2007; 430:1 - 30; http://dx.doi.org/ 10.1016/S0076-6879(07)30001-3; PMID: 17913632 [DOI] [PubMed] [Google Scholar]
- 41.Rodnina MV, Wintermeyer W. . GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. Proc Natl Acad Sci U S A 1995; 92:1945 - 9; http://dx.doi.org/ 10.1073/pnas.92.6.1945; PMID: 7892205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wintermeyer W, Schleich HG, Zachau HG. . Incorporation of amines or hydrazines into tRNA replacing wybutine or dihydrouracil. Methods Enzymol 1979; 59:110 - 21; http://dx.doi.org/ 10.1016/0076-6879(79)59073-9; PMID: 440077 [DOI] [PubMed] [Google Scholar]
- 43.Wintermeyer W, Zachau HG. . Fluorescent derivatives of yeast tRNAPhe. Eur J Biochem 1979; 98:465 - 75; http://dx.doi.org/ 10.1111/j.1432-1033.1979.tb13207.x; PMID: 114393 [DOI] [PubMed] [Google Scholar]
- 44.Hickerson R, Majumdar ZK, Baucom A, Clegg RM, Noller HF. . Measurement of internal movements within the 30 S ribosomal subunit using Förster resonance energy transfer. J Mol Biol 2005; 354:459 - 72; http://dx.doi.org/ 10.1016/j.jmb.2005.09.010; PMID: 16243353 [DOI] [PubMed] [Google Scholar]
- 45.Cukras AR, Green R. . Multiple effects of S13 in modulating the strength of intersubunit interactions in the ribosome during translation. J Mol Biol 2005; 349:47 - 59; http://dx.doi.org/ 10.1016/j.jmb.2005.03.075; PMID: 15876367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brune M, Hunter JL, Corrie JE, Webb MR. . Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry 1994; 33:8262 - 71; http://dx.doi.org/ 10.1021/bi00193a013; PMID: 8031761 [DOI] [PubMed] [Google Scholar]