Abstract
Background
At present, there are two main types of lung cancer xenograft models: those derived from stable cell lines, and patient‐derived xenograft models established by surgically resected tissues. However, these animal models may not reflect the biological and genetic characteristics of advanced non‐small cell lung cancer (NSCLC). We utilized bronchoscopy‐guided biopsy tissues of NSCLC patients to establish xenograft models and analyzed their histopathologic and genotypic fidelity with parental tumors.
Methods
Tumor tissues of NSCLC patients taken via bronchoscope were subcutaneously implanted into mice with non‐obese diabetic‐severe combined immunodeficiency disease for model establishment and serial passage. The histopathology and genotype of the samples from bronchoscopy‐guided biopsy‐derived xenograft (BDX) models and their parental tumors were detected.
Results
Thirty BDXs out of 114 NSCLC patients (26.32%) were successfully established. Smoking status significantly affected the success rate of NSCLC BDX establishment (P = 0.010). The BDX establishment success rate in squamous cell cancer was higher than in adenocarcinoma, with no significant difference (32.00% vs. 16.21%, P = 0.112). However, the growth rate of passage 1 BDX was slower than that of passages 2 and 3. Almost all NSCLC BDXs maintained similarity to their parental tumor tissues in regard to histologic characteristics, pathological markers, and driver‐gene mutations. Only one BDX model lost the epidermal growth factor receptor mutation contained in tumor parental tissue, as a result of heterogeneity.
Conclusions
NSCLC BDXs maintained high fidelity of histopathology and genotype with their original tumors. NSCLC BDXs that possess the actual status of advanced lung carcinoma should be used in preclinical research.
Keywords: Bronchoscopy, genotype, NSCLC, patient‐derived xenograft model
Introduction
The treatment of non‐small cell lung cancer (NSCLC), especially lung adenocarcinoma (ADC), has entered an era of individualized therapy based on genotyping. Compared with traditional chemotherapy, molecular‐targeted drugs, represented by epidermal growth factor receptor‐tyrosine kinase inhibitors (EGFR‐TKIs), have significantly prolonged survival time and improved quality of life.1, 2, 3, 4, 5, 6, 7 With the identification of more and more driver‐genes related to NSCLC tumorigenesis and development, the use of next‐generation sequencing technology and series of targeted agents have entered preclinical and clinical trials.8, 9, 10, 11 Although a large number of molecular‐targeted anti‐tumor drugs have exhibited good results in preclinical trials, only a small proportion of these are subject to clinical research.12, 13 There are various reasons for this, including the absence of specific target gene and target populations, a lack of accurate and efficient methods to detect target genes, and the difference between traditional animal models and human lung cancer patients.
Currently, there are two main types of animal models used in the preclinical research of lung cancer: xenograft models derived from stable cell lines, and patient‐derived xenograft (PDX) models established by surgically resected tissues. Cell line xenograft models are established with immunodeficient animals that have particular cells injected into them in order to examine these cell lines. This kind of animal model has been widely used for decades in the preclinical evaluation of anti‐tumor drugs. However, research has also proven that the biological characteristics of animal models greatly differ from the original tumors in human beings.14 In addition, without the support of the tumor microenvironment and mesenchyme, the accuracy of these models in research evaluating the efficacy of anti‐tumor drugs is limited.15 The PDX model, derived from the actual tumor tissues of patients, maintains similar histopathologic and molecular genetic characteristics to the original clinical tumors. As a result, PDX models are superior to cell line xenograft models.16, 17, 18, 19
Most NSCLC PDXs are derived from fresh surgical specimens of early stage NSCLC patients.19, 20 The biogenetic features of early and advanced NSCLC, including tumor angiogenesis, invasion, metastasis, drug resistance, and mutation frequency of relevant driver‐genes, are not exactly the same. Researchers have found that the mutation frequency of EGFR mutated patients is closely relevant to tumor node metastasis (TNM) staging, especially to lymphatic metastasis, and the drug‐resistant mutation of T790M occurs most commonly in advanced lung cancer.21, 22 A meta‐analysis containing 27 retrospective studies and 6950 lung cancer patients revealed that the frequency of echinoderm microtubule associated protein like 4‐anaplastic lymphoma kinase (EML4‐ALK) gene fusion in advanced lung cancer is significantly higher than in early stage.23 Data from a study of whole‐genome sequencing in Asian patients demonstrated a large difference in genetic background between early and advanced lung cancer.24 Therefore, xenograft models established from biopsies of advanced NSCLC patients can better reflect the genetic features during tumor progression, and can be better applied to preclinical research.
In this study, biopsy tissues via bronchoscope were used to establish xenograft models for advanced NSCLC, defined as bronchoscopy‐guided biopsy‐derived xenografts (BDXs). The similarities and differences in histopathologic characteristics and driver‐gene mutations between xenograft models and parental tumor tissues are investigated.
Materials and methods
Subjects
Patients diagnosed with “pulmonary mass” by standard radiography who underwent bronchoscopy‐guided biopsy for pathological diagnosis, and were pathologically diagnosed with NSCLC, were enrolled in this study. Patients with benign lesions, such as inflammation, metaplasia, and granuloma were excluded. Patients infected with human immunodeficiency virus and hepatitis B and C were also excluded from this study. Written consent allowing the use of tumor tissues for research was provided by all of the enrolled patients. The institutional ethics and animal care committees of Peking University Cancer Hospital approved this protocol, which included collecting tumor samples via bronchoscopy for BDX establishment.
Establishment of bronchoscopy‐guided biopsy‐derived xenografts (BDXs)
Bronchoscopy‐guided biopsies were sent for pathological diagnosis; all final tumor specimens were collected anonymously. The specimens were placed into ice‐cold transport media (10% fetal bovine serum, Gibco, Grand Island, NY, USA; + 1/100 S/P, Hyclone + RPMI 1640, Hyclone, Logan, UT, USA) using aseptic technique, and were processed at 4°C within four hours. Where available, additional tissues obtained via bronchoscopy were preserved in formalin. In the animal lab, specimens were placed in a 10 cm sterile dish with phosphate buffered saline and rinsed carefully. The tumor tissue was then subcutaneously implanted into female mice with non‐obese diabetic‐severe combined immunodeficiency disease (NOD‐SCID), without disaggregation, to preserve tumor structure. All procedures were conducted in a certified biosafety hood. Mice were numbered according to the order of sample collection, and were observed at least twice a week for six months. Implants showing no growth after six months were deemed unsuccessful, and the corresponding mice were euthanized. When the volume of tumors (which had been verified as growing after engraftment) reached at least 500 mm3, we repeated the implantation into 10–20 naive female NOD‐SCID mice (passage 2) via trocar needle and went down to the third generation (passage 3). During this process, a portion of tumor tissues were frozen in standard cell freezing medium and stored in liquid nitrogen for serial passage, while the others were preserved for detection. Tumor volumes were calculated by measuring two perpendicular diameters using a caliper. Each tumor volume was calculated according to the following formula: D*d*d/2.
Histopathology and immunohistochemistry
Each established xenograft model was bisected, fixed in 10% neutral buffered formalin and embedded in paraffin. Sections 5 um thick were cut from each paraffin‐embedded sample and mounted on glass slides. Routine hematoxylin and eosin staining of xenograft tumor sections was performed under an optical microscope. Tissue sections selected for immunohistochemistry (IHC) were routinely deparaffinized in xylene and rehydrated using a series of graded ethanol solutions, followed by antigen retrieval and background blocking. Primary antibodies against CD56, p63 (Cell Signaling Technology, Danvers, MA, USA), and thyroid transcription factor‐1 (TTF1; Santa Cruz Biotechnology Inc., Dallas, Texas, USA) were used following the manufacturer's instructions. The negative control was prepared following the same steps, except that the primary antibodies were replaced by normal serum from the same species. Two independent pathologists reviewed the tissue sections and compared pathologic similarities between NSCLC BDXs and corresponding patient samples.
Epidermal growth factor receptor and v‐Ki‐ras2 Kirsten rat sarcoma viral oncogene homolog mutation
The tumor tissues or plasma of NSCLC patients and the tumor tissues of BDXs were collected for DNA extraction using a method reported previously.25, 26 The concentration and quality of extracted DNA was determined by NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA), which was then detected for EGFR (deletion in exon 19 or L858R mutation in exon 21) and v‐Ki‐ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations. EGFR mutation was detected by amplification refractory mutation system (ARMS) as reported by Bai et al.26 KRAS mutation was detected by denaturing high‐performance liquid chromatography (DHPLC), as reported by Wang et al.27
Echinoderm microtubule associated protein like 4‐anaplastic lymphoma rearrangement
Tissue sections from BDXs and corresponding patients diagnosed with ADC were subjected to immunohistochemical staining with rabbit monoclonal primary anti‐ALK antibodies (clone D5F3, Cell Signaling Technology) according to product protocol. Tumor samples were considered positive when any percentage of tumor cells exhibited strong intracellular granular cytoplasmic staining.
Positive tissues detected by IHC were also verified using fluorescent in situ hybridization (FISH). The tissue slides of patients and BDXs were processed with the SPEC ALK Dual Color Break Apart probe (Zytovision, Bremen, Germany), which was a mixture of two directly labeled probes hybridizing onto the 2p23 band. This detection was in accordance with manufacturer's instructions. At least 100 representative tumor cells were counted, and the existence of ALK gene rearrangement was concluded if more than 15% of the tumor cells demonstrated a split red and green signal and/or an isolated red signal. Otherwise, the specimen was classified as ALK‐FISH negative. Two independent pathologists who were blinded to all clinical data performed ALK rearrangement‐FISH analyses. When IHC and FISH both illustrated positive results in the same samples, only specimens derived from patients or BDXs were considered to have ALK rearrangement.
Gene amplification of fibroblast growth factor receptor‐1
Fibroblast growth factor receptor‐1 (FGFR1) amplification was conducted by Dual Color Probe (Zytovision) with sections of BDXs and corresponding patients who were diagnosed with lung squamous cell cancer (SCC). The probe contained a mixture of an orange fluorochrome directly labeled CEN 8 probe, specific for the alpha satellite centromeric region of chromosome 8 (D8Z2) and a green fluorochrome directly labeled SPEC FGFR1 probe, specific for the FGFR1 gene at 8p11.23‐p11.22. FISH was performed and analyzed following the manufacturer's instructions.
In accordance with original research on the definition of high and low levels of FGFR1 amplification types, 100 cells were analyzed in each case. High‐level amplification was defined when: the FGFR1/CEN8 ratio was ≥2.0; or the average number of FGFR1 signals per tumor cell nucleus was ≥6; or the percentage of tumor cells containing FGFR1 signals was ≥15%; or the percentage of large clusters was ≥10%. Low‐level amplification was defined when the number of FGFR1 signals in ≥50% of the tumor cells was ≥5. Two independent pathologists who were blinded to all clinical data performed FGFR1‐FISH analyses.
Other genotype detection
ROS proto‐oncogene 1 (ROS1) gene fusion and c‐MET amplification of ADC BDXs were also detected using IHC with corresponding primary antibodies (Cell Signaling Technology).
Statistical analysis
Statistical analysis was performed to study the relationship between success rates and clinical factors such as gender, smoker status, pathologic type and stage, EGFR and KRAS mutations. Graphpad Prism software (La Jolla, CA, USA) was used for χ2 test or Fisher's exact test, if appropriate. For the growth curve, a Student's t‐test was used to compare differences between groups. Statistical tests were two sided, with P < 0.05 considered significant.
Results
Establishment and passage of non‐small cell lung cancer (NSCLC) BDXs
From April 2012 to February 2014, 114 bronchoscopy‐guided biopsy samples of patients diagnosed with primary NSCLC were subcutaneously implanted into NOD‐SCID mice; 30 of the xenografts survived and could be serially and stably subcultured, with a total tumor‐formation rate of 26.32%. As shown in Table 1, smoking status had a significant effect on the tumor formation rate of NSCLC. Engraftments from smokers (23/69, 33.33%) survived more often than from non‐smokers (4.76%, 1/24, P = 0.010). The success rate of BDXs derived from ADC samples (6/37, 16.21%) was lower than from SCC samples (32.00%, 24/75), but there was no significant difference (P = 0.112, Fig 1a).
Table 1.
The clinical characteristics of bronchoscopy‐guided biopsy tissues and tumor success rate of BDXs in NSCLC
| Patient characteristics | No. of patients | No. of BDXs (%) | No. of no BDXs (%) | P value |
|---|---|---|---|---|
| Total | 114 | 30 (26.32) | 84 (73.68) | |
| Gender | 0.301 | |||
| Male | 90 | 26 (28.89) | 64 (71.11) | |
| Female | 24 | 4 (16.67) | 20 (83.33) | |
| Smoking status | ||||
| Smoker | 69 | 23 (33.33) | 46 (66.67) | 0.010 |
| Non‐smoker | 21 | 1 (4.76) | 20 (5.00) | |
| Histologic type | ||||
| ADC | 37 | 6 (16.21) | 31 (83.78) | |
| SCC | 75 | 24 (32.00) | 51 (68.00) | 0.112 |
| Others | 2 | 0 (0) | 2 (100) | |
| NSCLC pathologic stage | 0.421 | |||
| Stage I | 4 | 0 (0.00) | 4 (100.00) | |
| Stage II | 3 | 2 (66.67) | 1 (33.33) | |
| Stage III/IV | 93 | 26(27.96) | 67 (72.04) | |
| EGFR status | 0.244 | |||
| EGFR MT | 8 | 1 (12.5) | 7 (87.5) | |
| EGFR WT | 55 | 17 (30.91) | 38 (69.09) | |
| KRAS status | 0.518 | |||
| KRAS MT | 2 | 1 (50.00) | 1 (50.00) | |
| KRAS WT | 59 | 17 (28.81) | 42 (71.19) |
Others: large cell lung cancer and unclassified non‐small cell lung cancer (NSCLC). ADC, adenocarcinoma; BDX, biopsy‐derived xenografts; EGFR, epidermal growth factor receptor; KRAS, v‐Ki‐ras2 Kirsten rat sarcoma viral oncogene homolog; MT, mutation; SCC, squamous cell carcinoma; WT, wild‐type.
Figure 1.
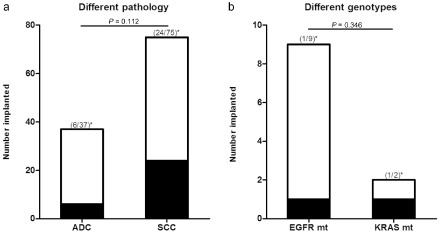
Tumor formation of biopsy‐derived xenografts (BDXs) derived from different pathology or genotypes of non‐small cell lung cancer. (a) The total number of BDXs (successful xenografts) or No‐BDXs (failing xenografts) were divided according to different pathology of parental samples. (b) The total number of BDXs or No‐BDXs were divided according to different genotypes of parental samples. Asterisks denote the corresponding success rates of BDXs. P is the difference between groups.  , BDXs;
, BDXs;  , No‐BDXs. ADC, adenocarcinoma; EGFR mt, epidermal growth factor receptor mutation; KRAS mt, v‐Ki‐ras2 Kirsten rat sarcoma viral oncogene homolog mutation; SCC, squamous cell carcinoma.
, No‐BDXs. ADC, adenocarcinoma; EGFR mt, epidermal growth factor receptor mutation; KRAS mt, v‐Ki‐ras2 Kirsten rat sarcoma viral oncogene homolog mutation; SCC, squamous cell carcinoma.
From the perspective of driver‐genes, the success rate of BDXs from bronchoscopy‐guided biopsy tissues with wild type EGFR was 30.91% (17/55), while the success rate in samples carrying mutant EGFR was only 12.50% (1/8). The success rates of BDXs with wild type and mutant KRAS were 50.00% (1/2) and 28.81% (17/59), respectively. However, there was no statistical significance in the difference between wild type and mutant EGFR (P = 0.244), wild type and mutant KRAS (P = 0.518), or mutant EGFR and mutant KRAS (P = 0.346) (Fig 1b).
All NSCLC BDXs were established by subcutaneous implantation, which allowed observation of the survival, size, and growth rates of the xenografts. Figure 2 illustrates the growth curves of xenografts from BDX 17 (ADC) and BDX 33 (SCC) from passage 1 (P1) to passage 3 (P3). The growth rate of the P1 xenograft was slow, because the parental NSCLC samples did not adapt to the microenvironment after implantation. In the second generation, passage 2 (P2) xenografts entered a period of adaption and counter‐adaptation; however, the growth rate was still unstable, leading to a constantly fluctuating tumor volume. In the third generation, P3 xenografts exhibited a stable growth status, with a distinctly faster growth rate than P1 and significant intergroup differences of P = 0.030 and P = 0.009, successively. The successful engraftment of P3 BDXs with a relatively fast growth rate and stable growth status suggested the possibility of successful BDX establishment and stable serial passage.
Figure 2.
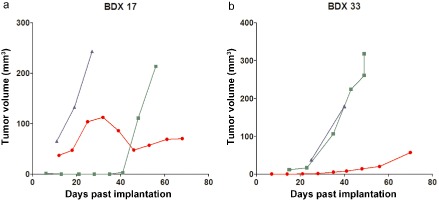
Growth curves show the growth rate of passage 1–3. (a) Biopsy‐derived xenograft (BDX) 17 was derived from adenocarcinoma (ADC), and (b) BDX 33 was derived from squamous cell carcinoma (SCC).  , passage 1;
, passage 1;  , passage 2;
, passage 2;  , passage 3.
, passage 3.
Fidelity of pathomorphology
The histopathologic characteristics and corresponding specific biomarkers of parental tumors were well preserved during the engraftment and passage of BDXs; for example, TTF1 and Napsin A were exclusively expressed in ADC BDXs, while P63 was exclusively expressed in SCC BDXs ( Fig 3a). The pathomorphology of BDXs retained the typical features of parental tissues, for example, glandular ducts could be observed in ADC BDXs, while keratin pearls could be observed in SCC BDXs (Fig 3b).
Figure 3.
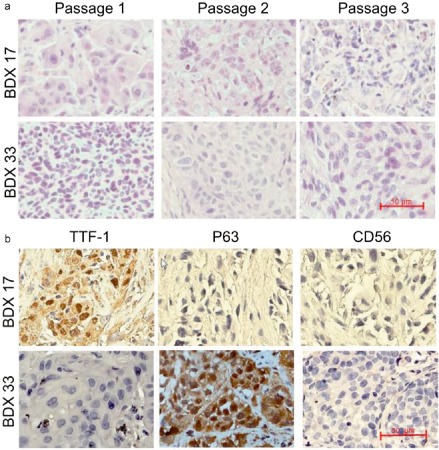
The histopathologic biomarkers and pathomorphology of biopsy‐derived xenografts (BDXs) derived from different pathologic types of non‐small cell lung cancer. BDX 17 was derived from adenocarcinoma and BDX 33 was derived from squamous cell carcinoma. (a) BDX samples were stained for thyroid transcription factor 1, p63, and CD56, respectively. (b) BDX specimens of passages 1–3 were stained for hematoxylin and eosin. The scale bars in (a) and (b) are 50 μm.
Fidelity of known driver‐genes in NSCLC
The difference in mutations of known driver‐genes in NSCLC between BDXs and parental tissues were investigated, including EGFR, KRAS, ALK, c‐MET, ROS1, and FGFR1 mutations. Table 2 summarizes the similarities and differences of the driver‐gene mutation or amplification status between the parental NSCLC tissues that led to successful engraftments and the tissues from corresponding BDXs.
-
EGFR mutations in BDX/parental samples
Sixty‐three out of 114 samples from NSCLC patients underwent EGFR mutation detection. Among these, eight cases carried EGFR mutations, and six of these were ADC patients. Regretfully, establishment of a BDX in these six cases was unsuccessful. The remaining two cases were SCC patients with EGFR mutations; a BDX model was successfully established from one of these samples. EGFR mutation status was detected in the 30 successfully established BDXs. Of 18 BDXs with known patient EGFR mutation status, 17 possessed a consistent EGFR mutation status with their parental tissues. In remaining case, which was SCC BDX (numbered 184), the xenograft possessed a wild‐type EGFR, while its parental tissue harbored an EGFR exon 21 L858R mutation (Fig. 4).
-
KRAS mutations in BDXs/parental samples
Sixty‐one out of 114 parental samples (tissues or plasma samples) underwent KRAS mutation detection. Thirty ADC cases possessed no KRAS mutation, two (6.67%) of 30 SCC cases carried KRAS mutations with one of these leading to the successful establishment of a BDX model (BDX 54), while the large cell lung cancer case possessed no KRAS mutation. The KRAS mutation status of the 30 successfully established BDXs was detected. The BDX samples from SCC patients with mutant KRAS possessed KRAS mutations, and the samples from BDXs all possessed a consistent KRAS mutation status with their parental tissues during model establishment and passage.
-
EML4‐ALK rearrangement in BDXs/parental samples
Samples from six ADC parental tissues and their corresponding BDXs underwent EML4‐ALK rearrangement detection. According to FISH and IHC (clone D5F3, Cell Signaling Technology), only BDX37 and its parental tissue possessed EML4‐ALK rearrangement (Fig. 5). This case was relatively unique, with a pathologic diagnosis of high‐grade mucoepidermoid carcinoma with sarcomatoid carcinoma.
-
ROS1 fusion and c‐MET amplification in BDXs/parental samples
Samples from six ADC parental tissues, as mentioned above, and their corresponding BDXs underwent ROS1 fusion and c‐MET amplification detection. No positive sample was detected.
-
FGFR1 amplification in BDXs/parental samples
Samples from 22 SCC BDXs and the corresponding parental tissues underwent FGFR1 amplification detection. Five (22.73%) BDXs harbored FGFR1 amplification, including three high‐level amplifications (BDX 27, BDX 84, BDX 133) and two low‐level amplifications (BDX 68, BDX 74). As illustrated in Figure 6, the samples from BDXs all possessed a consistent FGFR1 status with their parental tissues.
Table 2.
Driver‐gene mutation status in successfully established BDXs and their corresponding parental tumors
| Patient information | BDX information | ||||||
|---|---|---|---|---|---|---|---|
| ID | Age (years) | Gender† | Smoking status‡ | Stage | Pathology | Genotype status | Genotype status |
| 27 | 59 | 1 | 1 | IIIA | SCC | FGFR1 AMP | FGFR1 AMP |
| 37 | 55 | 2 | 0 | IV | ADC | EML4‐ALK | EML4‐ALK |
| 54 | 63 | 1 | 1 | IV | SCC | KRAS G34T | KRAS G34T |
| 68 | 76 | 1 | 1 | IIIB | SCC | FGFR1 AMP | FGFR1 AMP |
| 74 | 53 | 1 | 1 | IV | SCC | FGFR1 AMP | FGFR1 AMP |
| 84 | 67 | 1 | 1 | IV | SCC | FGFR1AMP | FGFR1 AMP |
| 133 | 62 | 1 | 1 | IIIA | SCC | FGFR1AMP | FGFR1 AMP |
| 184 | 64 | 1 | 1 | IIB | SCC | EGFR L858R | EGFR WT |
†1: male, 2: female. ‡0: non‐/mild smokers, 1: former/current smokers. ADC, adenocarcinoma; AMP, amplification; BDX, biopsy‐derived xenograft; EGFR, epidermal growth factor receptor; EML4‐ALK, echinoderm microtubule associated protein like 4‐anaplastic lymphoma kinase; FGFR1, fibroblast growth factor receptor‐1; KRAS, v‐Ki‐ras2 Kirsten rat sarcoma viral oncogene; SCC, squamous cell carcinoma; WT, wild‐type.
Figure 4.
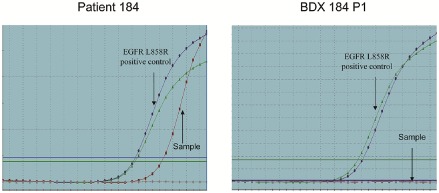
Epidermal growth factor receptor (EGFR) mutation status of samples. (a) The EGFR mutation status of bronchoscopy‐guided biopsy from squamous cell carcinoma patient (No.184). (b) EGFR mutation status of P1 biopsy‐derived xenograft 184.
Figure 5.
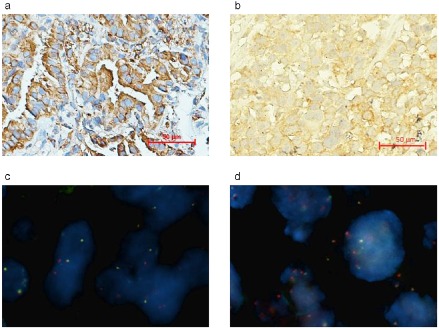
Detection of anaplastic lymphoma kinase rearrangement (ALK) in samples of biopsy‐derived xenograft (BDX) 37 and its parental tumor. (a) ALK (D5F3) detection of tumor biopsies from patient No.37 using immunohistochemistry (IHC). (b) The ALK (D5F3) detection of P3 BDX 37 using IHC. (c) Echinoderm microtubule associated protein like 4‐anaplastic lymphoma kinase (EML4‐ ALK) rearrangement detection of tumor biopsies from patient No.37 using fluorescent in situ hybridization (FISH). (d) The EML4‐ALK rearrangement detection of P3 BDX 37 using FISH.
Figure 6.
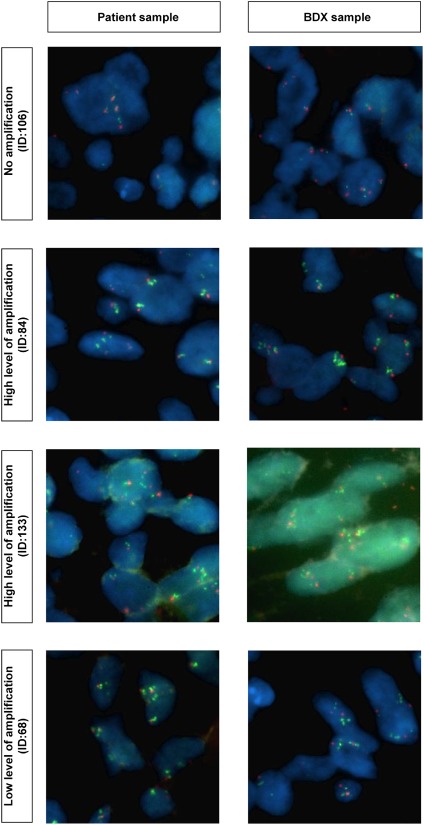
Fibroblast growth factor receptor‐1 amplification status of squamous cell carcinoma biopsy‐derived xenografts (BDXs) and their corresponding parental tumor samples.
Discussion
The advent of individual targeted therapy based on genotyping, such as EGFR‐TKIs and EML4‐ALK inhibitors, has significantly benefited survival duration and quality of life, which is a landmark development in the diagnosis and treatment of advanced lung cancer, particularly lung adenocarcinoma. However, almost every patient that has benefited from targeted therapy will encounter disease progression after a period of remission. In order to explore the molecular mechanisms of tumor relapse, metastasis, and resistance to drugs, and to develop novel targeted drugs, animal models that possess consistent molecular genetics and tumor microenvironment with human advanced lung cancer patients should be established. In this study, xenograft models derived from bronchoscopy‐guided biopsies replicated pathomorphologic and genetic features, such as gene mutations of their parental tumor, and covered the main histologic types of advanced NSCLC.
This type of less‐invasive xenograft model based on biopsy shares many similarities and differences with PDX animal models derived from surgically resected early stage lung cancer tissues, as reported in the literature. The success rate of BDXs in this study was 26.32%, while that of PDXs varies from 25–40% in the literature.16, 17, 28, 29, 30 The success rate in our study was similar for BDXs and PDXs, suggesting that sample size does not influence animal model establishment. We conclude that BDX animal model establishment based on small biopsy tissues is feasible.
Pathologic types of tumor tissues play a critical role in tumor formation, regardless of whether the models are derived from BDX or PDX. Previous research on PDXs reveal a significantly higher success rate of SCC compared with ADC samples.20, 30 However, our study failed to show any statistically significant difference in success rate between SCC and ADC. ADC is mostly peripheral, which means that cases of ADC diagnosed by bronchoscopy‐guided biopsy are limited.
In PDXs, the type of gene mutation is an important factor affecting tumor formation. Previous research on PDXs has revealed that, in immunodeficient mice, it is more difficult to form tumors in surgically resected samples with mutant EGFR than in samples with wild‐type EGFR, while it is easier to form tumors in samples with mutant KRAS than in samples with wild‐type KRAS.31 In this study, tumors could not be formed in six ADC cases with EGFR mutations. In the one SCC case with EGFR mutation in which tumors were successfully formed, the BDX exhibited a loss of EGFR mutation, suggesting that it is, indeed, difficult to form tumors in lung cancer samples carrying EGFR mutations.
Interestingly, data from a relatively special case carrying EML4‐ALK rearrangement and its corresponding BDX model were obtained. The patient was pathologically diagnosed as high‐grade mucoepidermoid carcinoma with sarcomatoid carcinoma. The corresponding BDX model preserved the pathomorphology, as well as the EML4‐ALK rearrangement of the parental tissue. According to statistics, 32, 33 There are controversial opinions on the derivation, classification, and clinical therapeutic strategy of lung sarcomatoid carcinoma.33 In addition, lung sarcomatoid carcinoma with EML4‐ALK rearrangement is even rarer, with only a few cases reported in the literature.34, 35 Therefore, establishment of this BDX model is very valuable in studying rare NSCLC with gene mutation.
Of the SCC BDXs, 22.73% possessed FGFR1 amplification, consistent with parental tissues. FGFR is a transmembrane tyrosine kinase and can combine with EGFR to mediate downstream signals through the RAS/RAF/mitogen activated protein kinase and phosphatidylinositol‐4,5‐bisphosphate 3‐kinase‐protein kinase B pathways. Previous literature has reported that 20% of SCC patients have FGFR1 amplification, and these types of cases exhibit a poorer prognosis and shorter survival time.36, 37 At present, several clinical trials involving targeted drugs against amplified FGFR1 genes, such as AZD4547 and BGJ398, are ongoing.
Although NSCLC BDX models exhibit high fidelity, there is heterogeneity among different passages or between BDXs and parental tumors. For example, in SCC BDX 184, the EGFR mutation existed in the parental tissue but was lost in the corresponding xenograft. This result reveals heterogeneity in tumors and the instability of some subcloning, as a result of adaption to a new environment during engraftment.
As is well known, the formation and progression of a tumor is a dynamic process, including a consecutive alternation of subcloning motivated by external pressure selection and internal gene mutation. Inside the tumor, subclonal diversification is prominent and different mutations can be found in different parts – as such, most gene mutations only exist in a small fraction of tumor cells.38, 39 The subcutaneous implant method of engraftment suffers from external pressure selection because of a lack of vessels and oxygen. Tumor‐formation of the P1 xenograft is extremely slow, causing the tumor cells to experience consecutive adaptive changes in the new environment. Therefore, the frequency of some gene mutations will be reduced because of a lack of adaptation to the new environment during engraftment. These mutations can even be completely lost during serial passage.
Conclusions
Non‐small cell lung cancer BDX models have a similar tumor‐formation rate of PDXs from surgically resected tumor tissues, and have a high fidelity of pathomorphology and driver gene mutation status with parental tumors. Therefore, BDXs can be used as models for advanced NSCLC research in vivo.
Disclosure
No authors report any conflict of interest.
Acknowledgments
The authors thank BeiGene (Beijing) Co. Ltd., for assistance with technological aspects of xenograft implantation. This work was supported by the National Natural Sciences Foundation Key Program [81330062]; the Education Ministry Innovative Research Team Program [IRT13003]; the Peking University‐Tsinghua University Joint Center for Life Sciences Clinical Investigator; the National High Technology Research and Development Program 863[SS2015AA020403]; the Beijing Technology Project [Z141100000214013]; the Center for Molecular and Translational Medicine (CMTM); and the Special Research Fund for the Doctoral Program of Higher Education.
References
- 1. Paez JG, Jänne PA, Lee JC et al EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 2004; 304: 1497–1500. [DOI] [PubMed] [Google Scholar]
- 2. Lynch TJ, Bell DW, Sordella R et al Activating mutations in the epidermal growth factor receptor underlying responsiveness of non‐small‐cell lung cancer to gefitinib. N Engl J Med 2004; 350: 2129–2139. [DOI] [PubMed] [Google Scholar]
- 3. Pao W, Miller V, Zakowski M et al EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004; 101: 13306–13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mok TS, Wu YL, Thongprasert S et al Gefitinib or carboplatin‐paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009; 361: 947–957. [DOI] [PubMed] [Google Scholar]
- 5. Mitsudomi T, Morita S, Yatabe Y et al Gefitinib versus cisplatin plus docetaxel in patients with non‐small‐cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol 2010; 11: 121–128. [DOI] [PubMed] [Google Scholar]
- 6. Zhou C, Wu YL, Chen G et al Erlotinib versus chemotherapy as first‐line treatment for patients with advanced EGFR mutation‐positive non‐small‐cell lung cancer (OPTIMAL, CTONG‐0802): A multicentre, open‐label, randomised, phase 3 study. Lancet Oncol 2011; 12: 735–742. [DOI] [PubMed] [Google Scholar]
- 7. Rosell R, Carcereny E, Gervais R et al Erlotinib versus standard chemotherapy as first‐line treatment for European patients with advanced EGFR mutation‐positive non‐small‐cell lung cancer (EURTAC): A multicentre, open‐label, randomised phase 3 trial. Lancet Oncol 2012; 13: 239–246. [DOI] [PubMed] [Google Scholar]
- 8. Stacey SN, Sulem P, Jonasdottir A et al A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet 2011; 43: 1098–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xiong D, Li G, Li K et al Exome sequencing identifies MXRA5 as a novel cancer gene frequently mutated in non‐small cell lung carcinoma from Chinese patients. Carcinogenesis 2012; 33: 1797–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu P, Morrison C, Wang L et al Identification of somatic mutations in non‐small cell lung carcinomas using whole‐exome sequencing. Carcinogenesis 2012; 33: 1270–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ignatius Ou SH, Azada M, Hsiang DJ et al Next‐generation sequencing reveals a Novel NSCLC ALK F1174V mutation and confirms ALK G1202R mutation confers high‐level resistance to alectinib (CH5424802/RO5424802) in ALK‐rearranged NSCLC patients who progressed on crizotinib. J Thorac Oncol 2014; 9: 549–553. [DOI] [PubMed] [Google Scholar]
- 12. Gutman S, Kessler LG. The US Food and Drug Administration perspective on cancer biomarker development. Nat Rev Cancer 2006; 6: 565–571. [DOI] [PubMed] [Google Scholar]
- 13. Philip R, Carrington L, Chan M. US FDA perspective on challenges in co‐developing in vitro companion diagnostics and targeted cancer therapeutics. Bioanalysis 2011; 3: 383–389. [DOI] [PubMed] [Google Scholar]
- 14. Daniel VC, Marchionni L, Hierman JS et al A primary xenograft model of small‐cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res 2009; 69: 3364–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Voskoglou‐Nomikos T, Pater JL, Seymour L. Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clin Cancer Res 2003; 9: 4227–4239. [PubMed] [Google Scholar]
- 16. Merk J, Rolff J, Becker M, Leschber G, Fichtner I. Patient‐derived xenografts of non‐small‐cell lung cancer: A pre‐clinical model to evaluate adjuvant chemotherapy? Eur J Cardiothorac Surg 2009; 36: 454–459. [DOI] [PubMed] [Google Scholar]
- 17. Bernardo C, Costa C, Amaro T et al Patient‐derived sialyl‐Tn‐positive invasive bladder cancer xenografts in nude mice: An exploratory model study. Anticancer Res 2014; 34: 735–744. [PubMed] [Google Scholar]
- 18. Mattie M, Christensen A, Chang MS et al Molecular characterization of patient‐derived human pancreatic tumor xenograft models for preclinical and translational development of cancer therapeutics. Neoplasia 2013; 15: 1138–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin D, Wyatt AW, Xue H et al High fidelity patient‐derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res 2014; 74: 1272–1283. [DOI] [PubMed] [Google Scholar]
- 20. Zhang XC, Zhang J, Li M et al Establishment of patient‐derived non‐small cell lung cancer xenograft models with genetic aberrations within EGFR, KRAS and FGFR1: Useful tools for preclinical studies of targeted therapies. J Transl Med 2013; 11: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Han X, Zhang Z, Wu D et al Suitability of surgical tumor tissues, biopsy, or cytology samples for epidermal growth factor receptor mutation testing in non‐small cell lung carcinoma based on Chinese population. Transl Oncol 2014; 7: 795–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee Y, Lee GK, Hwang JA, Yun T, Kim HT, Lee JS. Clinical likelihood of sporadic primary EGFR T790M mutation in EGFR‐mutant lung cancer. Clin Lung Cancer 2015; 16: 46–50. [DOI] [PubMed] [Google Scholar]
- 23. Zhao F, Xu M, Lei H et al Clinicopathological characteristics of patients with non‐small‐cell lung cancer who harbor EML4‐ALK fusion gene: A meta‐analysis. PLoS ONE 2015; 10 (2): e0117333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim Y, Hammerman PS, Kim J et al Integrative and comparative genomic analysis of lung squamous cell carcinomas in East Asian patients. J Clin Oncol 2014; 32: 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bai H, Mao L, Wang HS et al Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non‐small‐cell lung cancer. J Clin Oncol 2009; 27: 2653–2659. [DOI] [PubMed] [Google Scholar]
- 26. Bai H, Wang Z, Chen K et al Influence of chemotherapy on EGFR mutation status among patients with non‐small‐cell lung cancer. J Clin Oncol 2012; 30: 3077–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang S, An T, Wang J et al Potential clinical significance of a plasma‐based KRAS mutation analysis in patients with advanced non‐small cell lung cancer. Clin Cancer Res 2010; 16: 1324–1330. [DOI] [PubMed] [Google Scholar]
- 28. Fichtner I, Rolff J, Soong R et al Establishment of patient‐derived non‐small cell lung cancer xenografts as models for the identification of predictive biomarkers. Clin Cancer Res 2008; 14: 6456–6468. [DOI] [PubMed] [Google Scholar]
- 29. Merk J, Rolff J, Dorn C, Leschber G, Fichtner I. Chemoresistance in non‐small‐cell lung cancer: Can multidrug resistance markers predict the response of xenograft lung cancer models to chemotherapy? Eur J Cardiothorac Surg 2011; 40: e29–33. [DOI] [PubMed] [Google Scholar]
- 30. John T, Kohler D, Pintilie M et al The ability to form primary tumor xenografts is predictive of increased risk of disease recurrence in early‐stage non‐small cell lung cancer. Clin Cancer Res 2011; 17: 134–141. [DOI] [PubMed] [Google Scholar]
- 31. Sorensen M, Pijls‐Johannesma M, Felip E. Small‐cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow‐up. Ann Oncol 2010; 21: v120–125. [DOI] [PubMed] [Google Scholar]
- 32. Rossi G, Cavazza A, Sturm N et al Pulmonary carcinomas with pleomorphic, sarcomatoid, or sarcomatous elements: A clinicopathologic and immunohistochemical study of 75 cases. Am J Surg Pathol 2003; 27: 311–324. [DOI] [PubMed] [Google Scholar]
- 33. Travis WD, Travis LB, Devesa SS. Lung cancer. Cancer 1995; 75: 191–202. [DOI] [PubMed] [Google Scholar]
- 34. Ali G, Proietti A, Niccoli C et al EML4‐ALK translocation in both metachronous second primary lung sarcomatoid carcinoma and lung adenocarcinoma: A case report. Lung Cancer 2013; 81: 297–301. [DOI] [PubMed] [Google Scholar]
- 35. Kobayashi Y, Sakao Y, Ito S et al Transformation to sarcomatoid carcinoma in ALK‐rearranged adenocarcinoma, which developed acquired resistance to crizotinib and received subsequent chemotherapies. J Thorac Oncol 2013; 8: e75–78. [DOI] [PubMed] [Google Scholar]
- 36. Weiss J, Sos ML, Seidel D et al Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2010; 2: 62ra93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim HR, Kim DJ, Kang DR et al Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in patients with resected squamous cell lung cancer. J Clin Oncol 2013; 31: 731–737. [DOI] [PubMed] [Google Scholar]
- 38. McFadden DG, Papagiannakopoulos T, Taylor‐Weiner A et al Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell 2014; 156: 1298–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nik‐Zainal S, Van Loo P, Wedge DC et al The life history of 21 breast cancers. Cell 2012; 149: 994–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]


