 Conformational diversity of 1,4- and 1,5-substituted 1,2,3-triazole amino acids makes them promising building units for novel peptidic foldamers.
Conformational diversity of 1,4- and 1,5-substituted 1,2,3-triazole amino acids makes them promising building units for novel peptidic foldamers.
Abstract
Peptidic foldamers have recently emerged as a novel class of artificial oligomers with properties and structural diversity similar to that of natural peptides, but possessing additional interesting features granting them great potential for applications in fields from nanotechnology to pharmaceuticals. Among these, foldamers containing 1,4- and 1,5-substitued triazole amino acids are easily prepared via the Cu- and Ru-catalyzed click reactions and may offer increased side chain variation, but their structural capabilities have not yet been widely explored. We here describe a systematic analysis of the conformational space of the two most important basic units, the 1,4-substitued (4Tzl) and the 1,5-substitued (5Tzl) 1,2,3-triazole amino acids, using quantum chemical calculations and NMR spectroscopy. Possible conformations of the two triazoles were scanned and their potential minima were located using several theoretical approaches (B3LYP/6-311++G(2d,2p), ωB97X-D/6-311++G(2d,2p), M06-2X/6-311++G(2d,2p) and MP2/6-311++G(2d,2p)) in different solvents. BOC-protected versions of 4Tzl and 5Tzl were also prepared via one step transformations and analyzed by 2D NOESY NMR. Theoretical results show 9 conformers for 5Tzl derivatives with relative energies lying close to each other, which may lead to a great structural diversity. NMR analysis also indicates that conformers preferring turn, helix and zig-zag secondary structures may coexist in solution. In contrast, 4Tzl has a much lower number of conformers, only 4, and these lack strong intraresidual interactions. This is again supported by NMR suggesting the presence of both extended and bent conformers. The structural information provided on these building units could be employed in future design of triazole foldamers.
Introduction
Peptidic compounds with structural properties resembling those of natural amino acids and peptides are interesting because they show great potential for future design of a variety of bioactive molecules.1 Such compounds are nowadays referred to as peptidic foldamers or foldamers 2 due to their ability to fold into versatile secondary structures in oligomeric forms.3 Apart from aliphatic homologues of natural amino acids, such as peptides composed of β-, γ- or δ-amino acids,4 a large subgroup in this area has more exotic backbone structures, including diverse cyclic compounds.5 Peptidic foldamers with cyclic backbones can be built from ACPC,6 ACHC,7 hydrazine,8 oxazolidine,9 cholate,10 pyrrolidone,11 diketopiperazine,12 or dioxolane,13 just to mention a few. Among these, 1,2,3-triazole based peptidomimetics have recently emerged as interesting candidates for several reasons.14 Their relatively easy synthesis using ‘click’ chemistry,15 via either a copper16 (CuAAC) or a ruthenium catalyzed cycloaddition (RuAAC),17 their high in vivo stability, and their conformational flexibility are all very favourable properties in the design of novel bioactive compounds. Furthermore, an important practical advantage is that the backbone of these foldamers is polar enough to retain the same water solubility irrespective of the length of the oligomer.18
There are currently several examples where 1,4- or 1,5-substituted triazoles are either incorporated as monomers into natural peptide sequences,19 or used as oligomers with a completely non-natural peptide composition.20 All these reports demonstrate that this exciting area has promising capacity in terms of providing applications in biotechnology.21 Nevertheless, the number of existing structural examples for foldamers clearly cannot compare to those of the natural proteins. Consequently, estimation of the structural capabilities for these systems has to be made based on the configuration of their monomeric building blocks. It is known that for natural compounds, most of the secondary structures found in nature are constructed from homoconformers, that is from amino acids which have the same structural properties in their backbone.22 For natural peptides and proteins, numerous conformational studies have shown that the abundance of secondary structures in protein databases is closely related to the structural properties and relative energy distribution of the conformers of α-amino acids.23 The same concept holds for non-natural amino acids and peptides as well; the use of molecular modelling techniques, in particular ab initio calculations, in predicting the stability of monomers and thus secondary structures built from them, has proven very effective in the last two decades.24 Recent advances with dispersion energy terms in density functionals have made calculation of energetic properties for larger compounds more accurate.
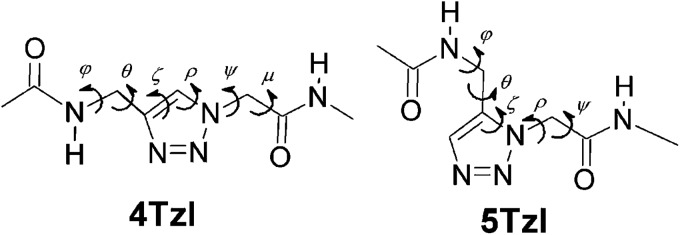
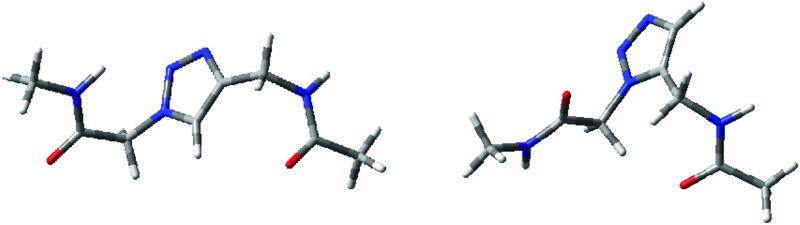
To better estimate the future potential of triazole peptidomimetics as foldamers, we have explored the structural properties and stability of the two simplest monomer units: the 1,4- and 1,5-substituted 1,2,3-triazole peptides, 4Tzl and 5Tzl (Fig. 1).
Fig. 1. Constitution of the 4Tzl and 5Tzl models used for the QM calculations. The nomenclature used for defining torsional angles is highlighted.
Note that when considering these two molecules, the use of 1,5-substituted 1,2,3-triazoles is as yet far less common. Nevertheless we have recently indicated that their conformational properties may be much more diverse than that of 1,4-substituted 1,2,3-triazoles18 and have also shown that by using a microwave-assisted RuAAC reaction, 1,5-substitued 1,2,3-triazoles can be synthesized in excellent yields from an alkyl halide, sodium azide and an alkyne in a sequential one-pot procedure.25
We here investigate the peptidomimetic building units shown in Fig. 1 by employing quantum chemical calculations. To further evaluate these compounds, their synthetically useful BOC-protected versions (BOC-4Tzl and BOC-5Tzl) were prepared and subjected to solution phase characterization by 2D NOESY NMR spectroscopy. Exhaustive systematic analysis of the conformers was achieved by exploring the conformational potential energy hypersurface (PEHS) of 4Tzl and 5Tzl along all their rotatable dihedral angles (Fig. 1). Conformers obtained at lower levels of theory were refined by performing additional calculations at the B3LYP/6-311++G(2d,2p), ωB97X-D/6-311++G(2d,2p), M06-2X/6-311++G(2d,2p) and MP2/6-311++G(2d,2p) levels of theory, and considering effects of solvents with different polarity, i.e. water, dimethylsulfoxide (DMSO) and 1-decanol.
Results and discussion
The synthesis of the investigated compounds, computational calculations, comparison with NMR measurements, as well as a discussion on BOC-4Tzl and on BOC-5Tzl are all addressed separately in this section, organized under their corresponding sub-sections. For methodological details see the Experimental section.
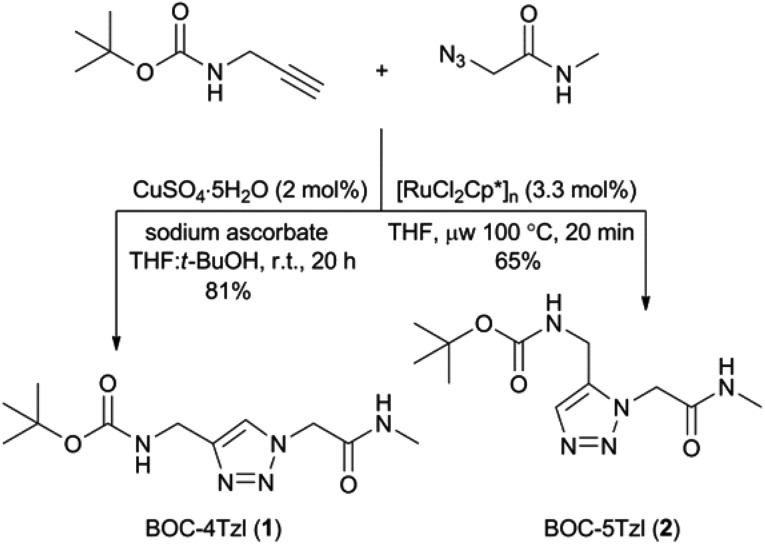
Synthesis of BOC-4Tzl (1) and BOC-5Tzl (2)
To prepare the BOC-protected 1,4- and 1,5-substituted versions of 4Tzl and 5Tzl, the same starting materials could be employed for both reactions while the catalyst was varied to produce the desired regioisomer in each case (Scheme 1). To synthesize BOC-4Tzl (1), commercially available N-BOC-propargylamine and 2-azido-N-methylacetamide were stirred in a 1 : 1 mixture of water and tert-butanol in the presence of catalytic amounts of CuSO4·5H2O and sodium ascorbate, producing the desired 1,4-substituted triazole 1 in 81% yield after 20 h at ambient temperature. Using the same azide and alkyne in conjunction with a ruthenium catalyst ([RuCl2Cp*]n) with THF as the solvent afforded the 1,5-substituted isomer 2 in 65% yield after microwave heating at 100 °C for 20 min. Both compounds are white solids and could be stored at ambient temperature without any significant deterioration.
Scheme 1. Synthesis of BOC-protected 1,2,3-triazole isomers 1 and 2 (BOC = tert-butoxycarbonyl).
Theoretical analysis of 4Tzl conformers
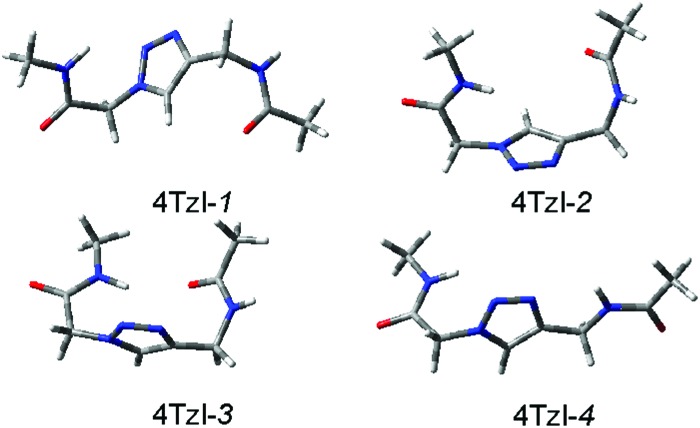
As a result of the systematic scan on the potential energy hypersurface (PEHS) of 4Tzl, the fully relaxed RHF/3-21G level calculations have converged into 12 distinguishable conformers (Table S1, ESI†). For clarity, conformational enantiomers are considered as duplicates and are not discussed. The final optimizations, performed at a higher level of theory or in the presence of solvents, have reduced these into four conformers (Fig. 2 and Tables 1 and 2).
Fig. 2. Conformers of 4Tzl obtained at the ωB97X-D/6-311++G(2d,2p) level of theory.
Table 1. Structural and energetic properties of 4Tzl conformers 1 to 4, located as minima at the B3LYP/6-311++G(2d,2p), M06-2X/6-311++G(2d,2p), ωB97X-D/6-311++G(2d,2p) and RMP2-FC/6-311++G(2d,2p) levels of theory in the gas phase, with solvent effects considered by point energy calculations.
| Conf. | Method | φ | θ | ζ | ρ | ψ | μ | Relative energy
a
|
|||
| Gas | H2O | DMSO | 1-Decanol | ||||||||
| 4Tzl-1 | B3LYP | –78.3 | 79.2 | –178.6 | –177.9 | 107 | 29.1 | 0.0 | 0.0 | 0.0 | 0.0 |
| M06-2X | –72.0 | 77.1 | –177.8 | –176.4 | 98.7 | 28.2 | 0.0 | 0.0 | 0.0 | 0.0 | |
| ωB97X-D | –74.6 | 77.0 | 177.6 | –176.8 | 102.7 | 26.5 | 0.0 | 0.0 | 0.0 | 0.0 | |
| MP2 | –72.4 | 78.4 | –176.1 | –174.2 | 95.7 | 30.8 | 0.0 | 0.0 | 0.0 | 0.0 | |
| 4Tzl-2 | B3LYP | –78.5 | 75.5 | –179.9 | 178.9 | –109.8 | –29.3 | 0.1 | 0.1 | 0.1 | 0.1 |
| M06-2X | –72.9 | 71.4 | –179.7 | 176.7 | –100.5 | –27.8 | 0.1 | 0.0 | 0.0 | 0.0 | |
| ωB97X-D | –74.6 | 76.8 | –179.1 | 176.1 | –100.3 | –29.0 | 0.1 | –0.1 | –0.1 | –0.0 | |
| MP2 | –72.1 | 76.9 | –178 | 174.1 | –96 | –31 | 0.1 | –0.0 | –0.2 | 0.0 | |
| 4Tzl-3 | B3LYP | — | — | — | — | — | — | — | — | — | — |
| M06-2X | –54.2 | 111.0 | –168.6 | 164.6 | –67.1 | 3.3 | –0.1 | 0.0 | 0.0 | 0.0 | |
| ωB97X-D | –55.2 | 109.1 | –168.9 | 165.5 | –67.9 | 4.7 | –0.1 | –0.1 | –0.1 | –0.1 | |
| MP2 | — | — | — | — | — | — | — | — | — | — | |
| 4Tzl-4 | B3LYP | 177.9 | 174.8 | 179.9 | 177.6 | –103.6 | –31.3 | 1.1 | 0.6 | 0.6 | 0.7 |
| M06-2X | 178.5 | –176.0 | 179.6 | 175.8 | –94.5 | –29.0 | 1.6 | 1.0 | 1.0 | 1.1 | |
| ωB97X-D | 177.6 | –178.2 | 179.2 | 176.3 | –97.7 | –29.1 | 1.5 | 0.8 | 0.8 | 0.9 | |
| MP2 | –163 | 141.2 | 177.9 | 173.4 | –92.3 | –32.8 | 2.0 | 1.5 | 1.5 | 1.6 | |
aRelative energies are in kcal mol–1.
Table 2. Energetic properties of 4Tzl conformers located as stable minima optimized in water, DMSO and decanol at the B3LYP/6-311++G(2d,2p) and ωB97X-D/6-311++G(2d,2p) levels of theory.
| Conf. | Method | Relative energy
a
|
||
| H2O | DMSO | 1-Decanol | ||
| 4Tzl-1 | B3LYP | 0.00 | 0.00 | 0.00 |
| ωB97X-D | 0.00 | 0.00 | 0.00 | |
| 4Tzl-2 | B3LYP | 0.01 | 0.02 | 0.05 |
| ωB97X-D | –0.25 | –0.02 | 0.02 | |
| 4Tzl-3 | B3LYP | — | — | — |
| ωB97X-D | 0.30 | 0.50 | 0.37 | |
| 4Tzl-4 | B3LYP | 0.60 | 0.61 | 0.63 |
| ωB97X-D | 0.80 | 1.03 | 1.09 | |
aRelative energies are in kcal mol–1.
Structural properties of the 4Tzl conformers can be described by six dihedral angles (Fig. 1 and Table 1). The two central dihedrals cannot be considered as freely rotating, since these are held fixed in the anti position by the 1,2,3-triazole heterocycle. Considering the four conformers remaining stable at the higher level calculations, the structural properties are rather similar for 4Tzl-1 to 4Tzl-3. For these conformations, the first two dihedrals are in the gauche position, with alternating orientation, i.e. g– and g+. The last two dihedrals are also in a shifted gauche position, although they have the same orientation, i.e. g+g+. The only exception is μ for 4Tzl-3, which is close to zero. The fourth conformer, 4Tzl-4, has a more extended conformation, with the first two dihedrals being anti, and only the last two in the gauche position.
Out of these conformers, only 4Tzl-3 forms a hydrogen bond with 2.3 Å O···H distance (Fig. 2). For 4Tzl-1 to 4Tzl-3, the energetic properties obtained from the single point energy calculations on the gas phase structure show rather small differences in solvents of different polarities (see Table 1), whereas the more extended conformer 4Tzl-4 is less stable. For 4Tzl-4, all theoretical methods in all three solvents show a higher relative energy, B3LYP: 0.6–0.7 kcal mol–1, ωB97X-D: 0.8–0.9 kcal mol–1, M06-2X: 1.0–1.1 kcal mol–1, MP2: 1.5–1.6 kcal mol–1 (Table 1). The relative difference in stability remains very similar when these four conformer structures are fully optimized in all solvents investigated, with zero-point energy and thermal contributions also considered (Table 2).
The obtained relative energy values indicate that conformers 4Tzl-3 and 4Tzl-4 are somewhat less stable than 4Tzl-1 and 4Tzl-2 in all solvents. However, the highest relative energy is 1.09 kcal mol–1 for 4Tzl-4 which, considering the Boltzmann distribution, may still allow the co-presence of all these conformers in solution. For 4Tzl-1 and 4Tzl-2, the relative energies are nearly identical at the ωB97X-D/6-311++G(2d,2p) level, 0.00 and –0.25 kcal mol–1 for water, 0.00 and –0.02 kcal mol–1 for DMSO and 0.00 and 0.02 kcal mol–1 for decanol, respectively.
Theoretical analysis of 5Tzl conformers
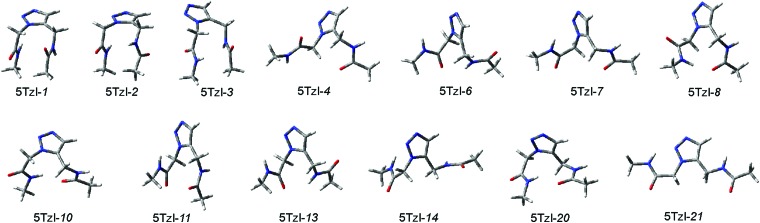
For 5Tzl, the systematic PEHS scan has resulted in 27 distinguishable conformers (Table S2, ESI†). During the higher level optimizations, several conformers vanished and 13 conformers remained (Table 3). Full optimizations in solvents further reduced the number of minima, resulting finally in 9 conformers (Fig. 3 and Table 4). Structural properties of the 5Tzl conformers can be described by five dihedral angles (Fig. 1 and Table 3). The central dihedral angle is fixed in a near zero degree position. The 13 conformers stable at the higher level calculations mostly form “bent” conformers, with the majority of the dihedral angles in the gauche position (Fig. 3 and Table 3). The only extended conformer is 5Tzl-21. Although 5Tzl-3, 5Tzl-4, 5Tzl-8, 5Tzl-10, 5Tzl-11, 5Tzl-14 and 5Tzl-20 have also one or more torsional angles in the anti range, i.e. |120°–180°|, these most probably will not favor extended conformers in oligopeptides. A more detailed overview on potential secondary structures is presented below in the sub-section 5Tzl intraresidual properties. The structures of 5Tzl-6, 5Tzl-10, 5Tzl-11, 5Tzl-14 and 5Tzl-20 are stabilized by a hydrogen bond, with O···H distances of 2.19 Å, 1.91 Å, 1.97 Å, 2.20 Å, and 2.10 Å, respectively (Fig. 3).
Table 3. Structural and energetic properties of 5Tzl conformers obtained at the B3LYP/6-311++G(2d,2p), ωB97X-D/6-311++G(2d,2p) and RMP2-FC/6-311++G(2d,2p) levels of theory in the gas phase, with solvent effects considered by point energy calculations.
| Conf. | Secondary structure a | Method | φ | θ | ζ | ρ | ψ | Relative energy
b
|
|||
| Gas | H2O | DMSO | 1-Decanol | ||||||||
| 5Tzl-1 | B3LYP | –116.3 | 59.6 | –6.8 | –98.4 | –40.3 | 0.00 | 0.00 | 0.00 | 0.00 | |
| M06-2X | –116.4 | 55.9 | –7.9 | –83.8 | –29.2 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| H14 | ωB97X-D | –117.8 | 57.9 | –7.5 | –87.9 | –26.2 | 0.00 | 0.00 | 0.00 | 0.00 | |
| MP2 | –116.5 | 58.9 | –8.5 | –88.2 | –26.7 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| 5Tzl-2 | B3LYP | 103.9 | 75.2 | –5.7 | –83.5 | –73.2 | –0.65 | –0.50 | –0.52 | –0.74 | |
| M06-2X | 68.6 | 52.3 | –9.5 | –74.4 | –29.1 | –0.07 | –0.03 | –0.04 | –0.18 | ||
| C8 | ωB97X-D | 73.4 | 64.0 | –7.2 | –76.6 | –45.4 | –0.17 | 0.45 | 0.41 | 0.12 | |
| MP2 | 68.4 | 63.2 | –8.4 | –77.0 | –40.4 | –0.17 | 0.41 | 0.38 | 0.12 | ||
| 5Tzl-3 | B3LYP | –104.3 | 70.9 | –4.2 | –100.6 | –134.0 | 1.19 | 0.12 | 0.12 | 0.20 | |
| M06-2X | –99.8 | 77.2 | –2.7 | –82.5 | –176.7 | 0.79 | 0.16 | 0.16 | 0.21 | ||
| Turn | ωB97X-D | –102.3 | 73.9 | –3.1 | –85.1 | –174.8 | 1.12 | 0.23 | 0.23 | 0.30 | |
| MP2 | –99.5 | 77.3 | –2.8 | –84.8 | –174.6 | 1.84 | 0.70 | 0.71 | 0.86 | ||
| 5Tzl-4 | B3LYP | –87.6 | 95.9 | 4.6 | 78.0 | 140.3 | 1.00 | –1.15 | –1.13 | –0.83 | |
| M06-2X | –79.5 | 101.4 | 5.8 | 73.9 | 144.5 | 3.52 | 1.58 | 1.60 | 1.82 | ||
| 2-Helix | ωB97X-D | –85.9 | 96.5 | 5.1 | 76.2 | 143.7 | 2.92 | 1.05 | 1.06 | 1.28 | |
| MP2 | –80.0 | 99.5 | 6.8 | 75.0 | 141.7 | 3.56 | 1.60 | 1.62 | 1.85 | ||
| 5Tzl-6 | B3LYP | 103.1 | 42.9 | –7.0 | 66.1 | 76.3 | 1.49 | 0.74 | 0.73 | 0.70 | |
| M06-2X | 83.5 | 49.1 | –4.2 | 57.7 | 59.0 | 2.98 | 2.39 | 2.39 | 2.34 | ||
| H8 | ωB97X-D | 85.0 | 44.4 | –4.5 | 61.7 | 63.7 | 2.42 | 1.95 | 1.94 | 1.85 | |
| MP2 | 79.3 | 48.1 | –4.7 | 57.8 | 58.5 | 2.94 | 1.85 | 1.85 | 1.91 | ||
| 5Tzl-7 | B3LYP | –89.0 | 96.7 | 6.6 | 95.6 | 34.2 | 0.97 | –0.66 | –0.65 | –0.41 | |
| M06-2X | –74.7 | 103.7 | 7.7 | 83.4 | 30.4 | 3.22 | 2.07 | 2.07 | 2.17 | ||
| Spiral | ωB97X-D | –83.0 | 98.5 | 8.3 | 87.3 | 31.7 | 2.68 | 1.49 | 1.49 | 1.61 | |
| H16 | MP2 | –74.0 | 102.0 | 10.0 | 83.1 | 33.5 | 2.68 | 1.71 | 1.71 | 1.79 | |
| 5Tzl-8 | B3LYP | –107.8 | 80.9 | 3.3 | 70.6 | –122.0 | 0.92 | –1.96 | –1.92 | –1.42 | |
| M06-2X | –108.2 | 80.0 | 5.6 | 67.8 | –131.1 | 2.46 | 0.11 | 0.14 | 0.51 | ||
| H10 | ωB97X-D | –109.2 | 78.7 | 4.9 | 70.0 | –120.7 | 1.66 | –0.95 | –0.91 | –0.50 | |
| MP2 | –106.8 | 80.1 | 6.2 | 66.7 | –126.0 | 2.33 | –0.24 | –0.21 | 0.23 | ||
| 5Tzl-10 | H10 | B3LYP | –70.2 | 144.1 | 1.5 | –85.5 | 16.5 | 1.71 | –1.39 | –1.35 | –0.85 |
| M06-2X | –69.1 | 152.5 | –0.9 | –74.5 | 5.4 | 2.13 | –0.11 | –0.08 | 0.21 | ||
| Turn2 | ωB97X-D | –68.9 | 149.1 | 0.8 | –78.1 | –9.8 | 2.02 | –0.47 | –0.44 | –0.10 | |
| Turn3 | MP2 | –66.0 | 150.0 | –0.3 | –78.9 | 9.2 | 2.62 | –0.03 | 0.01 | 0.40 | |
| 5Tzl-11 | B3LYP | –131.7 | 42.5 | –11.6 | 66.6 | 26.3 | 3.88 | 1.93 | 1.96 | 2.38 | |
| M06-2X | –131.0 | 38.9 | –9.8 | 58.1 | 26.1 | 2.53 | 1.84 | 1.85 | 1.98 | ||
| H14 | ωB97X-D | –131.9 | 39.5 | –10.4 | 61.1 | 24.9 | 2.86 | 1.91 | 1.93 | 2.12 | |
| MP2 | –133.1 | 39.8 | –10.6 | 59.8 | 24.7 | 3.21 | 2.20 | 2.21 | 2.42 | ||
| 5Tzl-13 | B3LYP | 84.7 | 94.1 | 6.4 | 94.7 | 37.3 | 2.10 | 0.22 | 0.24 | 0.49 | |
| M06-2X | 82.9 | 49.7 | –4.4 | 57.0 | 58.1 | 2.87 | 1.95 | 1.94 | 1.96 | ||
| ωB97X-D | 85.4 | 45.0 | –4.7 | 61.0 | 61.6 | 2.47 | 1.76 | 1.75 | 1.70 | ||
| MP2 | — | — | — | — | — | — | — | — | — | ||
| 5Tzl-14 | B3LYP | 114.7 | 170.4 | –4.2 | –97.1 | –36.6 | 2.07 | –0.04 | –0.02 | 0.24 | |
| M06-2X | 86.8 | 175.7 | –6.3 | –83.9 | –31.6 | 5.04 | 2.84 | 2.86 | 3.13 | ||
| ωB97X-D | 99.2 | 175.3 | –5.4 | –85.0 | –34.2 | 4.34 | 2.37 | 2.39 | 2.61 | ||
| MP2 | 86.7 | 172.2 | –7.4 | –83.8 | –33.3 | 5.33 | 3.25 | 3.27 | 3.53 | ||
| 5Tzl-20 | B3LYP | 114.6 | 164.7 | 3.6 | 94.0 | 30.80 | 2.25 | –0.27 | –0.25 | 0.11 | |
| M06 – 2X | |||||||||||
| ωB97X-D | — | — | — | — | — | — | — | — | — | ||
| MP2 | — | — | — | — | — | — | — | — | — | ||
| 5Tzl-21 | B3LYP | 180.0 | 180.0 | 0.0 | 180.0 | 0.0 | 6.09 | 4.63 | 4.64 | 4.81 | |
| M06-2X | 180.0 | 180.0 | 0.0 | –180.0 | 0.0 | 9.95 | 9.05 | 9.05 | 9.09 | ||
| ωB97X-D | 180.0 | 180.0 | 0.0 | –180.0 | 0.0 | 9.22 | 8.26 | 8.26 | 8.31 | ||
| MP2 | 180.0 | 180.0 | 0.0 | 180.0 | 0.0 | 10.92 | 10.00 | 10.00 | 10.05 | ||
aPotential secondary structures which could be built for peptidic oligomers using the particular 5Tzl conformer.
bRelative energies are in kcal mol–1.
Fig. 3. The 13 5Tzl conformers as obtained at the ωB97X-D/6-311++G(2d,2p) level of theory.
Table 4. Structural and energetic properties of 5Tzl conformers located as stable minima optimized in various solvents at the B3LYP/6-311++G(2d,2p), ωB97X-D/6-311++G(2d,2p) and RMP2-FC/6-311+G(d,p) levels of theory.
| Conf. | Sec. Struct. a | Method | Relative energy
b
|
Energy level | ||
| H2O | DMSO | Decanol | ||||
| 5Tzl-1 | B3LYP | 0.00 | 0.00 | 0.00 | ||
| H14 | ωB97X-D | 0.00 | 0.00 | 0.00 | Low | |
| MP2 | 0.00 | 0.00 | — | |||
| 5Tzl-2 | B3LYP | –1.65 | –1.64 | –1.60 | ||
| C8 | ωB97X-D | 0.42 | 0.36 | –0.19 | Low | |
| MP2 | 0.94 | 0.95 | — | |||
| 5Tzl-3 | B3LYP | –0.65 | –0.63 | 0.14 | ||
| Turn1 | ωB97X-D | –0.14 | –0.10 | 0.19 | Low | |
| MP2 | 0.23 | 0.22 | — | |||
| 5Tzl-4 | B3LYP | –0.56 | –0.55 | –0.48 | ||
| 2-Helix | ωB97X-D | 1.51 | 1.48 | 1.43 | Moderate | |
| MP2 | 1.27 | 1.27 | — | |||
| 5Tzl-6 | B3LYP | 1.32 | 1.30 | 0.42 | ||
| H8 | ωB97X-D | 2.77 | 2.70 | 2.35 | High | |
| MP2 | 1.88 | 1.85 | — | |||
| 5Tzl-7 | Spiral/ | B3LYP | –0.20 | –0.18 | –0.07 | |
| H16 | ωB97X-D | 1.51 | 1.47 | 1.53 | Moderate | |
| MP2 | 1.62 | 1.60 | — | |||
| 5Tzl-8 | B3LYP | –1.04 | –1.01 | –0.81 | ||
| H10 | ωB97X-D | –0.23 | –0.25 | –0.02 | Low | |
| MP2 | 0.21 | 0.21 | — | |||
| 5Tzl-10 | H10/ | B3LYP | –0.70 | –0.69 | –0.50 | |
| Turn2/ | ωB97X-D | –0.08 | –0.11 | –0.06 | Low | |
| Turn3 | MP2 | 0.29 | 0.29 | — | ||
| 5Tzl-14 | B3LYP | 0.25 | 0.29 | 0.37 | ||
| Spiral | ωB97X-D | 2.24 | 2.28 | 2.49 | High | |
| MP2 | 1.62 | 1.60 | — | |||
aPotential secondary structures which could be built for peptidic oligomers using the particular 5Tzl conformer.
bRelative energies are in kcal mol–1.
For 5Tzl, the energetic distribution of the conformers depends more on the theoretical approach than for 4Tzl, partially due to the significantly higher number of minima, and also because there are discrepancies, e.g. between consideration of dispersive forces in the theoretical methods employed. When solvent effects were only considered by single point energy calculations on the gas phase structures, there are significant changes in the relative stability between the three methods used (Table 3). The calculations with the B3LYP functional indicate that conformers 5Tzl-2, 5Tzl-4, 5Tzl-7, 5Tzl-8, and 5Tzl-10 will be the most stable in the solutions used. These minima are within 1.5 kcal mol–1 in relative energy from the most stable conformer, 5Tzl-8. Conformers 5Tzl-1, 5Tzl-3, 5Tzl-6, 5Tzl-13, 5Tzl-14 and 5Tzl-20 have moderately low relative energies, i.e. lower than 3 kcal mol–1, whereas 5Tzl-11 and 5Tzl-21 have higher energies and are the least stable.
Solvent single point energies with the ωB97X-D functional result in 5Tzl-1, 5Tzl-2, 5Tzl-3, 5Tzl-8, 5Tzl-10 conformers within 1.5 kcal mol–1 counted from the lowest energy structure: 5Tzl-8 (Table 3). Conformers 5Tzl-4, 5Tzl-6, 5Tzl-7, 5Tzl-11, and 5Tzl-13 are moderately low, having smaller than 3 kcal mol–1 relative energy, and 5Tzl-14 and 5Tzl-21 have high relative energy. For the M06-2X functional, the conformers show a very similar energy distribution to that for ωB97X-D, however it results in 5Tzl-10 being the lowest energy conformer. The relative energy distribution of MP2 matches those obtained with the ωB97X-D functional, with only difference being the vanishing conformer 5Tzl-13 at the MP2 level.
As expected, full optimizations in solvents result in smaller energetic differences. At the B3LYP/6-311++G(2d,2p) level of theory, 5Tzl-2 is the most stable conformer and 5Tzl-2, 5Tzl-3, 5Tzl-4, 5Tzl-7, 5Tzl-8, 5Tzl-10 are minima with lower than 1.5 kcal mol–1 energy values relative to 5Tzl-2 (Table 4). However, the ωB97X-D and MP2 calculations again show a significant deviation from the B3LYP results. Here the 5Tzl-1, 5Tzl-2, 5Tzl-3, 5Tzl-8, 5Tzl-10 conformers are the most stable, 5Tzl-4 and 5Tzl-7 being over 1.5 kcal mol–1 in relative energy, and 5Tzl-6, 5Tzl-14 having a ∼3 kcal mol–1 energy difference from the most stable conformer. The most stable conformer is 5Tzl-1 for MP2, and 5Tzl-8 for ωB97X-D calculations (Table 4).
4Tzl intraresidual properties
The small relative energy values obtained for 4Tzl are reasonable if one considers that there are very few possible configurations where this peptidic residue could form intraresidual H-bonds. Only 4Tzl-3 has an H-bond, though the relatively high 2.3 Å distance between the corresponding oxygen and hydrogen indicates that their positioning is not optimal. Furthermore, there are no large differences in relative energies when comparing solvents of very different polarity (Table 2). All these properties suggest that without side chains, 4Tzl is rather flexible, and has small internal preference for a particular conformation. This observation is in close agreement with several experimental examples on conformational properties of 4Tzl, where these are predicted to result in extended configurations, a direct outcome of the extended 1,4-positioning on the triazole heterocycle.20b Although each of the three 4Tzl conformers have four dihedral angles in the gauche position (Table 2), as there are most likely no major barriers for internal conformational transitions, the observed random/extended conformations in experiments may be enforced by the surrounding amino acid residues and the applied side chains. The latter observation is also supported by the fact that the 2D NMR NOESY experiments reveal both H1–H7 and H4–H7 crosspeaks, which suggests that BOC-4Tzl-1, 2, or maybe even 3 have to be present simultaneously in solution (for the list of NOEs, see Fig. S1 in ESI†).
5Tzl intraresidual properties
To compare theoretical results with those obtained by NMR, we have optimized the final 9 5Tzl minima with the same protecting groups as the synthesized compounds, BOC- and N-methyl amide (BOC-5Tzl). When compared to the 5Tzl calculations, there are no significant changes in the structural properties of the BOC-5Tzl conformers. Regarding energetic properties, BOC-5Tzl-1, BOC-5Tzl-2, BOC-5Tzl-3, BOC-5Tzl-8, and BOC-5Tzl-10 have lower relative energies. Conformers 4 and 7 have higher than 1.5 kcal mol–1 values, while BOC-5Tzl-6 and BOC-5Tzl-14 are the highest energy conformers, with values near 2.5 kcal mol–1. The most stable conformer is BOC-5Tzl-1, although 1, 2, 3, 8 and 10 have nearly identical relative energies (Table 5). These conformers obtained for BOC-5Tzl were also used to analyse the NOESY data obtained for the same compound (Table S3 and Fig. S2, ESI†). The number of NOE crosspeaks is relatively large, and they are nearly identical in both DMSO-d6 and D2O, indicating that the same conformer or conformers are present in both solvents. This is in close agreement with the 5Tzl calculations showing nearly identical relative energies for the above two solvents (Table 4).
Table 5. Energetic properties of BOC-5Tzl conformers optimized at the ωB97X-D/6-311++G(2d,2p) level of theory using water as the solvent.
| Conformer | Relative energy (H2O) a |
| BOC-5Tzl-1 | 0.00 |
| BOC-5Tzl-2 | 0.16 |
| BOC-5Tzl-3 | 0.13 |
| BOC-5Tzl-4 | 1.75 |
| BOC-5Tzl-6 | 2.70 |
| BOC-5Tzl-7 | 1.82 |
| BOC-5Tzl-8 | 0.22 |
| BOC-5Tzl-10 | 0.04 |
| BOC-5Tzl-14 | 2.46 |
aRelative energies are in kcal mol–1.
One should highlight the crosspeak H1–H7, which is between protons located on the two end protecting groups. This suggests a bent conformer (Fig. S2 in ESI†). This may apply to several of the located minima; nevertheless, a joint qualitative analysis of available computational and NMR structural data could narrow down the number of probable conformers. Although the presented energy values incorporate only zero point corrections, and thermal corrections to energies and enthalpies, the use of these values to approximate the relative stability of a conformer may be justified in a qualitative analysis. Accordingly, BOC-5Tzl-2 and BOC-5Tzl-10 are most likely to be present in solution. Both conformers have low relative energies (Table 5) and their H–H distances fit to most of the found NOEs (Table S3 in ESI†). Among the other low energy conformers, 1 and 3 would also show a H1–H7 crosspeak, but we conclude that these structures are less probable because both of these would display one proton–proton distance below 3 Å, which, despite the considerable dynamics present in a room temperature solvent, should be seen in the spectra. BOC-5Tzl-8 is also a low energy conformer which could give rise to several NOEs, although not the H1–H7 interaction (Tables 5 and S3 in ESI†). Nevertheless, the H-bond in 8 would constrain the structure to some extent and should give rise to a H2–H7 crosspeak (Fig. 3). BOC-5Tzl-4 and 7, despite having somewhat higher relative energies, have several H–H distances which fit well into the experimental NOEs. At the same time, both have a more extended structure without a stabilizing internal H-bond, and it is likely that in a polar solvent the structure will shift into a conformer where the apolar end groups are less exposed to the solvent. Finally, the two remaining conformers could be present if only the NOEs were considered; nevertheless they have the highest relative energies, making these less likely.
A recent study employing joint theoretical and experimental approaches on trimer–heptamer oligomers composed of achiral 5Tzl demonstrated that several secondary structures may coexist in solution.18 According to calculations on tetramer and heptamer models, H14, H16, H20 helices, T1, T2, T3-type turns, as well as double-stranded constructs all have rather small relative energy differences. Accordingly, the results presented here indicate that several of the 5Tzl conformers would promote a turn in a peptidic oligomer (Fig. 3). Furthermore, our exhaustive search for 5Tzl conformational space suggests a number of other secondary structures which could be built from this residue, especially when they are used as oligomers with the appropriate side chains. Accordingly, the relative energies of the obtained conformers suggest that low energy conformers of 5Tzl would particularly favour turns, Turn1, Turn2, Turn3,18 helices with 10 or 14 atoms in the H-bonded pseudo ring, H10, H14, as well as a zig-zag like conformation with 8-membered H-bond pseudo rings, C8 (Table 4). Furthermore, appropriate side chains would also allow the formation of H16, spiral, or double-helix assemblies, and even H8 or sheet conformations could be achieved. The relationship between the different, theoretically plausible, secondary structures and the conformers which would be most suitable to build them is highlighted (Tables 3 and 4).
Based on the above conformational analysis and the NMR measurements of the BOC protected monomer, we propose that for longer oligomers the most probable secondary structures can be narrowed down to turns, H10, and C8.
Conclusions
We have investigated the conformational properties of two compounds, 1,4-substituted (4Tzl) and 1,5-substituted (5Tzl) 1,2,3-triazole amino acids, which can be considered as basic building units of triazole-based peptidomimetics. We combined exhaustive theoretical conformational analysis with organic synthesis and 2D NMR measurements. BOC-protected 4Tzl and 5Tzl were prepared in good yields from commercially available starting materials in a one-step reaction, using CuSO4 as the catalyst to attain the 1,4-substitution pattern in 4Tzl, and [RuCl2Cp*]n in the case of the 5Tzl isomer. The two isomers were fully characterized and subjected to 2D NOESY NMR analysis to complement the computational studies.
Out of the few (i.e. 4) stable conformers of 4Tzl, in principle none of these has shown a large stabilization relative to the others, with no strong intraresidual H-bonds obtained. The 2D NOESY NMR experiments, in accordance with the calculations, show that extended, BOC-4Tzl-1, and more bent conformers, BOC-4Tzl-2 or BOC-4Tzl-3, may be simultaneously present in solution.
4Tzl units in foldamers most likely adopt conformations following positioning of their central 1,4-substituted triazole pentacycle and the conformational preference of the surrounding residues in a peptidomimetic oligomer. Based on the joint theoretical analysis and NMR measurements, we conclude that the 1,4-substituted triazoles have promising capabilities in forming sheet-like elongated secondary structures.
In contrast, the 5Tzl shows much higher diversity, with 9 stable conformers found by QM conformational analysis. Surprisingly, the vast majority of 5Tzl conformers have a relative energy lower than 3 kcal mol–1, and they are structurally more diverse than those of 4Tzl. Together with previous NMR results on their corresponding homooligomers,18 this observation suggests that several secondary structures built from these may coexist in solution, a basic prerequisite for larger scale conformational diversity in the case of natural proteins.26 Most likely turns, ten-membered helices (H10) and zig-zag like structures (C8) could be easily manifested for their homooligomers, but with appropriate side chains H8, H14, H16, spiral or even sheet secondary structures may also be achieved.
Four different theoretical methods were tested to see which is the most appropriate to characterize these systems. Based on the close correlation between MP2 and ωB97X-D results, we propose that the latter is a fast and reliable method, also accounting for dispersive effects in triazole foldamers. The M06-2X functional performs nearly equally well, however produces some larger deviations as ωB97X-D in the case of the lower energy conformers. We hope that this analysis of the basic properties of triazole amino acids will help rational computer-aided design of novel oligomers with the desired secondary structures.
Experimental section
Computational methods
All computations were carried out using the Gaussian 09 software package.27 The initial exploration of the conformational space was performed at the RHF/3-21G level of theory, where the dihedral angles were scanned with a resolution of 60°. These have resulted in 1296 and 800 converged constrained conformers for 4Tzl and 5Tzl models, respectively. All the obtained conformers were subjected to fully relaxed optimizations at the RHF/3-21G level of theory, resulting in 12 and 27 conformers for 4Tzl and 5Tzl, respectively. These were subjected to further optimizations using both Becke's three parameter functional with the Lee–Yang–Parr exchange functional (B3LYP),28 Head-Gordon's ωB97X-D functional, which included dispersion correction and long-range electron correlation corrections,29 as well as M06-2X of the Minnesota functionals.30 For all these functionals the 6-311++G(2d,2p) basis set was employed. During these calculations, the number of stable conformers was reduced to 4 (4Tzl) and 13 (5Tzl). The remaining structures obtained from the high level calculations were also subjected to MP2/6-311++G(2d,2p) calculations. Point energy calculations were performed to estimate solvent effects of water, dimethylsulfoxide (DMSO) and 1-decanol. These environments were modelled using the Integral Equation Formalism for Polarizable Continuum Model (IEFPCM).31 To consider the effects of solvent on conformational properties, as well as to confirm the nature of the critical points obtained, all 4 4Tzl and 13 5Tzl conformers were subjected to fully relaxed optimization followed by frequency calculations at the B3LYP/6-311++G(2d,2p), ωB97X-D/6-311++G(2d,2p) and MP2/6-311+G(d,p) levels of theory in the above three solvents using the IEFPCM model. These confirmed that all the obtained critical points are minima. In the case of 5Tzl, these calculations have further reduced the number of stable conformers to 9. To compare theoretical results with the experimental, 5Tzl models containing the same protecting group as the experimental ones, BOC and N-methyl, were also optimized at the ωB97X-D/6-311++G(2d,2p) level with water as the solvent (BOC-5Tzl). During frequency calculations on conformers optimized in solvents, for all three models 4Tzl, 5Tzl and BOC-5Tzl, several low frequency vibrations were found due to internal rotation modes. Such vibrations would not affect significantly the thermal energy and enthalpic contributions, but may cause significant errors when estimating entropic effects at room temperature. Consequently an additional harmonic vibrational analysis, identifying internal rotational modes, was also performed for BOC-5Tzl minima; nevertheless, for several conformers a one-to-one correspondence between vibrational and internal rotation modes could not be achieved. For the compounds studied here, the arising error would be ∼2.6–3.3 kcal mol–1, thus on the same magnitude as the relative energy difference between the lowest and the highest energy conformers. Therefore the presented energies for solvent optimized conformers incorporate only zero-point energy (ZPE) corrections, and thermal corrections to energy and enthalpy.
Precision and accuracy of computations
Although here we focused on the practical applicability of the 4Tzl and 5Tzl residues, as there are no exhaustive studies on which theoretical methods should be the most accurate and computationally least demanding for them, a short discussion is included here on the performance of the applied theoretical methods. Based on previous benchmark studies on peptide conformations,32 alanine and proline dipeptides,33 using among others B3LYP, ωB97X-D, M06-2X, it was expected that ωB97X-D and M06-2X will perform nearly equally well, when compared to reference MP2 calculations.33
Indeed, for 4Tzl and 5Tzl the B3LYP calculations show significant deviations from those of MP2, ωB97X-D and M06-2X methods (Tables 1–4). At the same time, a rather close agreement can be observed between MP2, M06-2X and ωB97X-D, although solvent effects may cause larger deviations. Considering relative energy values in solvents, the latter two is almost equally accurate when compared to the MP2 values. The only difference is that ωB97X-D shows somewhat higher accuracy for the lower energy conformers over M06-2X, which may be important when one aims to estimate structural preferences among the most stable conformers. As the MP2 calculations are rather time consuming, they are not appropriate to handle larger systems such as oligopeptides. The good agreement with ωB97X-D and M06-2X suggests that the latter density functionals are suitable to quickly and accurately assess properties of these foldamers, where we give some preference for the ωB97X-D functional due to the reasons mentioned above. Nevertheless, a systematic analysis on theoretical methods, a task beyond our current focus, is necessary to have solid conclusions on the most suitable theoretical approaches.
General experimental
All reactions were performed under an argon atmosphere. 2-Azido-N-methylacetamide was purchased from Fluorochem (UK). [RuCl2Cp*]n was purchased from Strem Chemicals. Automated flash chromatography was performed using a Biotage Isolera One system. Microwave reactions were carried out in a Biotage Series 60 Initiator (the actual vial temperature was monitored with an IR sensor, using a fixed hold time). Chemical shifts (δ) are given in ppm relative to the solvent residual peak (DMSO-d6: 2.50 ppm for 1H NMR and 39.51 ppm for 13C NMR), or an internal standard (TMS: 0.00 ppm for 1H NMR). 2D NOESY experiments were carried out in a 30–70 mM DMSO-d6 solution at 25 °C, using a NOESY mixing time of 500 ms, and a relaxation delay of 1.5 s, and were acquired with 2048 points in the f2 domain and 256 points in the f1 domain. The data were processed using MestReNova software and baseline correction was applied to both dimensions using Bernstein polynomial fit (3 orders) and a 90 °C sin bell window function was applied in both dimensions.
Synthesis
tert-Butyl ((1-(2-(methylamino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)methyl)carbamate, BOC-4Tzl (1)
2-Azido-N-methylacetamide (115 mg, 1.01 mmol) was dissolved in 2 mL H2O : t-BuOH (1 : 1) in a 4 mL vial equipped with a magnetic stir bar. To the mixture were added N-BOC-propargylamine (188 mg, 1.21 mmol), CuSO4·5H2O (5.0 mg, 0.02 mmol) and sodium ascorbate (12.0 mg, 0.06 mmol). The vial was capped and the orange solution was stirred for 20 h at ambient temperature. Brine (3 mL) was added and the mixture was extracted with EtOAc (5 × 3 mL). The combined organic phases were dried (Na2SO4) and concentrated under vacuum. The crude product was purified by automated flash chromatography on silica gel (eluent 20–80% EtOAc in petroleum ether, followed by 1–30% MeOH in CH2Cl2), affording BOC-4Tzl (1) as a white solid (221 mg, 81%). Anal. Calcd for C11H19N5O3: C, 49.06; H, 7.11; N, 26.01. Found: C, 49.09; H, 7.15; N, 25.91. IR (KBr) 3402, 3332, 3130, 2979, 1669, 1517, 1269, 1176 cm–1; 1H NMR (500 MHz; DMSO-d 6) δ 8.19 (d, J = 4.2 Hz, 1H), 7.82 (s, 1H), 7.32 (t, J = 5.7 Hz, 1H), 5.03 (s, 2H), 4.17 (d, J = 5.9 Hz, 2H), 2.63 (d, J = 4.6 Hz, 3H), 1.39 (s, 9H); 13C NMR (126 MHz; DMSO-d 6) δ 165.8, 155.6, 145.4, 124.0, 77.9, 51.6, 35.6, 28.2, 25.6; m/z (ESI) 270 (M+, 40%), 214 (100).
tert-Butyl ((1-(2-(methylamino)-2-oxoethyl)-1H-1,2,3-triazol-5-yl)methyl)carbamate, BOC-5Tzl (2)
2-Azido-N-methylacetamide (93 mg, 0.82 mmol, 1.0 equiv.) was dissolved in 2 mL dry THF in a 4 mL vial equipped with a magnetic stir bar under an argon atmosphere. N-BOC-propargylamine (139 mg, 0.90 mmol, 1.1 equiv.) was added, followed by [RuCl2Cp*]n (10.0 mg, 0.033 mmol, 0.04 equiv.). The solution was transferred to a microwave reaction vial using a syringe, equipped with a syringe filter to remove the undissolved catalyst. The reddish reaction mixture was heated for 20 min at 100 °C in a microwave reactor, affording a dark solution. After cooling, the solution was concentrated under vacuum and subsequently purified by automated flash chromatography (eluent 1–30% MeOH in CH2Cl2), affording BOC-5Tzl (4) as a white solid (144 mg, 65%). Anal. Calcd for C11H19N5O3: C, 49.06; H, 7.11; N, 26.01. Found: C, 49.13; H, 7.17; N, 25.71. IR (KBr) 3332, 2984, 1670, 1560, 1520, 1369, 1266, 1163 cm–1; 1H NMR (500 MHz, DMSO-d 6) δ 8.26 (d, J = 4.2 Hz, 1H), 7.51 (s, 1H), 7.36 (s, 1H), 5.09 (s, 2H), 4.22 (d, J = 5.9 Hz, 2H), 2.64 (d, J = 4.6 Hz, 3H), 1.38 (s, 9H); 13C NMR (126 MHz, DMSO-d 6) δ 165.8, 155.5, 136.9, 132.2, 78.4, 49.7, 32.9, 28.1, 25.6; m/z (ESI) 270 (M+, 100%).
Acknowledgments
We would like to thank Prof. Bengt Nordén for inspiring discussions on this project. This work was funded by the King Abdullah University of Science and Technology (KAUST Kuk No1) and by the European Research Council (ERC).
Footnotes
References
- Avan I., Hall C. D., Katritzky A. R. Chem. Soc. Rev. 2014;43:3575. doi: 10.1039/c3cs60384a. [DOI] [PubMed] [Google Scholar]
- (a) Gellman S. H. Acc. Chem. Res. 1998;31:173. [Google Scholar]; (b) Martinek T. A., Fulop F. Chem. Soc. Rev. 2012;41:687. doi: 10.1039/c1cs15097a. [DOI] [PubMed] [Google Scholar]
- Pilsl L. K. A., Reiser O. Amino Acids. 2011;41:709. doi: 10.1007/s00726-011-0894-2. [DOI] [PubMed] [Google Scholar]
- (a) Seebach D., Beck A. K., Bierbaum D. J. Chem. Biodiversity. 2004;1:1111. doi: 10.1002/cbdv.200490087. [DOI] [PubMed] [Google Scholar]; (b) Baldauf C., Gunther R., Hofmann H. J. J. Org. Chem. 2004;69:6214. doi: 10.1021/jo049535r. [DOI] [PubMed] [Google Scholar]
- (a) Horne W. S., Gellman S. H. Acc. Chem. Res. 2008;41:1399. doi: 10.1021/ar800009n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Haldar D. Curr. Org. Synth. 2008;5:61. [Google Scholar]
- (a) Appella D. H., Christianson L. A., Klein D. A., Richards M. R., Powell D. R., Gellman S. H. J. Am. Chem. Soc. 1999;121:7574. [Google Scholar]; (b) Martinek T. A., Mandity I. M., Fulop L., Toth G. K., Vass E., Hollosi M., Forro E., Fulop F. J. Am. Chem. Soc. 2006;128:13539. doi: 10.1021/ja063890c. [DOI] [PubMed] [Google Scholar]
- (a) Schinnerl M., Murray J. K., Langenhan J. M., Gellman S. H. Eur. J. Org. Chem. 2003:721. [Google Scholar]; (b) Mandity I. M., Fulop L., Vass E., Toth G. K., Martinek T. A., Fulop F. Org. Lett. 2010;12:5584. doi: 10.1021/ol102494m. [DOI] [PubMed] [Google Scholar]
- Tsai J. H., Waldman A. S., Nowick J. S. Bioorg. Med. Chem. 1999;7:29. doi: 10.1016/s0968-0896(98)00225-9. [DOI] [PubMed] [Google Scholar]
- (a) Tomasini C., Luppi G., Monari M. J. Am. Chem. Soc. 2006;128:2410. doi: 10.1021/ja056762h. [DOI] [PubMed] [Google Scholar]; (b) Castellucci N., Tomasini C. Eur. J. Org. Chem. 2013:3567. [Google Scholar]
- Zhong Z. Q., Zhao Y. Org. Lett. 2007;9:2891. doi: 10.1021/ol071130g. [DOI] [PubMed] [Google Scholar]
- Porter E. A., Wang X. F., Schmitt M. A., Gellman S. H. Org. Lett. 2002;4:3317. doi: 10.1021/ol0266370. [DOI] [PubMed] [Google Scholar]
- Delatouche R., Durini M., Civera M., Belvisi L., Piarulli U. Tetrahedron Lett. 2010;51:4278. [Google Scholar]
- Kothari A., Qureshi M. K. N., Beck E. M., Smith M. D. Chem. Commun. 2007:2814. doi: 10.1039/b706528k. [DOI] [PubMed] [Google Scholar]
- Holub J. M., Kirshenbaum K. Chem. Soc. Rev. 2010;39:1325. doi: 10.1039/b901977b. [DOI] [PubMed] [Google Scholar]
- Kolb H. C., Finn M. G., Sharpless K. B. Angew. Chem., Int. Ed. 2001;40:2004. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- (a) Tornoe C. W., Christensen C., Meldal M. J. Org. Chem. 2002;67:3057. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]; (b) Rostovtsev V. V., Green L. G., Fokin V. V., Sharpless K. B. Angew. Chem., Int. Ed. 2002;41:2596. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- (a) Zhang L., Chen X. G., Xue P., Sun H. H. Y., Williams I. D., Sharpless K. B., Fokin V. V., Jia G. C. J. Am. Chem. Soc. 2005;127:15998. doi: 10.1021/ja054114s. [DOI] [PubMed] [Google Scholar]; (b) Rasmussen L. K., Boren B. C., Fokin V. V. Org. Lett. 2007;9:5337. doi: 10.1021/ol701912s. [DOI] [PubMed] [Google Scholar]; (c) Boren B. C., Narayan S., Rasmussen L. K., Zhang L., Zhao H. T., Lin Z. Y., Jia G. C., Fokin V. V. J. Am. Chem. Soc. 2008;130:8923. doi: 10.1021/ja0749993. [DOI] [PubMed] [Google Scholar]
- Johansson J. R., Hermansson E., Norden B., Kann N., Beke-Somfai T. Eur. J. Org. Chem. 2014:2703. [Google Scholar]
- (a) Angell Y. L., Burgess K. Chem. Soc. Rev. 2007;36:1674. doi: 10.1039/b701444a. [DOI] [PubMed] [Google Scholar]; (b) Tam A., Arnold U., Soellner M. B., Raines R. T. J. Am. Chem. Soc. 2007;129:12670. doi: 10.1021/ja075865s. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Pokorski J. K., Jenkins L. M. M., Feng H. Q., Durell S. R., Bai Y. W., Appella D. H. Org. Lett. 2007;9:2381. doi: 10.1021/ol070817y. [DOI] [PubMed] [Google Scholar]; (d) Horne W. S., Olsen C. A., Beierle J. M., Montero A., Ghadiri M. R. Angew. Chem., Int. Ed. 2009;48:4718. doi: 10.1002/anie.200805900. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Roux S., Ligeti M., Buisson D. A., Rousseau B., Cintrat J. C. Amino Acids. 2010;38:279. doi: 10.1007/s00726-009-0248-5. [DOI] [PubMed] [Google Scholar]; (f) Tietze D., Tischler M., Voigt S., Imhof D., Ohlenschlager O., Gorlach M., Buntkowsky G. Chem. – Eur. J. 2010;16:7572. doi: 10.1002/chem.200903306. [DOI] [PubMed] [Google Scholar]; (g) Buysse K., Farard J., Nikolaou A., Vanderheyden P., Vauquelin G., Pedersen D. S., Tourwe D., Ballet S. Org. Lett. 2011;13:6468. doi: 10.1021/ol202767k. [DOI] [PubMed] [Google Scholar]; (h) Zhang J. Q., Kemmink J., Rijkers D. T. S., Liskamp R. M. J. Org. Lett. 2011;13:3438. doi: 10.1021/ol201184b. [DOI] [PubMed] [Google Scholar]; (i) Isidro-Llobet A., Murillo T., Bello P., Cilibrizzi A., Hodgkinson J. T., Galloway W., Bender A., Welch M., Spring D. R. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6793. doi: 10.1073/pnas.1015267108. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Castellucci N., Tomasini C. Eur. J. Org. Chem. 2013:3567. [Google Scholar]; (k) Valverde I. E., Bauman A., Kluba C. A., Vomstein S., Walter M. A., Mindt T. L. Angew. Chem., Int. Ed. 2013;52:8957. doi: 10.1002/anie.201303108. [DOI] [PubMed] [Google Scholar]
- (a) Zornik D., Meudtner R. M., El Malah T., Thiele C. M., Hecht S. Chem. – Eur. J. 2011;17:1473. doi: 10.1002/chem.201002491. [DOI] [PubMed] [Google Scholar]; (b) Krause M. R., Goddard R., Kubik S. J. Org. Chem. 2011;76:7084. doi: 10.1021/jo201024r. [DOI] [PubMed] [Google Scholar]; (c) Ke Z. H., Chow H. F., Chan M. C., Liu Z. F., Sze K. H. Org. Lett. 2012;14:394. doi: 10.1021/ol2031685. [DOI] [PubMed] [Google Scholar]; (d) You L. Y., Chen S. G., Zhao X., Liu Y., Lan W. X., Zhang Y., Lu H. J., Cao C. Y., Li Z. T. Angew. Chem., Int. Ed. 2012;51:1657. doi: 10.1002/anie.201106996. [DOI] [PubMed] [Google Scholar]; (e) Lee S., Hua Y. R., Flood A. H. J. Org. Chem. 2014;79:838. doi: 10.1021/jo501595k. [DOI] [PubMed] [Google Scholar]; (f) Wu C. F., Li Z. M., Xu X. N., Zhao Z. X., Zhao X., Wang R. X., Li Z. T. Chem. – Eur. J. 2014;20:1418. doi: 10.1002/chem.201304161. [DOI] [PubMed] [Google Scholar]
- Pedersen D. S., Abell A. Eur. J. Org. Chem. 2011:2399. [Google Scholar]
- (a) Perczel A., Farkas O., Jakli I., Topol I. A., Csizmadia I. G. J. Comput. Chem. 2003;24:1026. doi: 10.1002/jcc.10267. [DOI] [PubMed] [Google Scholar]; (b) Kabsch W., Sander C. Biopolymers. 1983;22:2577. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- (a) Chou P. Y., Fasman G. D. Biochemistry. 1974;13:211. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]; (b) Perczel A., Angyan J. G., Kajtar M., Viviani W., Rivail J. L., Marcoccia J. F., Csizmadia I. G. J. Am. Chem. Soc. 1991;113:6256. [Google Scholar]; (c) Karplus M., McCammon J. A. Nat. Struct. Biol. 2002;9:646. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- (a) Wu Y. D., Wang D. P. J. Am. Chem. Soc. 1998;120:13485. [Google Scholar]; (b) Wu Y. D., Wang D. P. J. Chin. Chem. Soc. 2000;47:129. [Google Scholar]; (c) Baldauf C., Hofmann H. J. Helv. Chim. Acta. 2012;95:2348. [Google Scholar]; (d) Sharma G. V. M., Babu B. S., Ramakrishna K. V. S., Nagendar P., Kunwar A. C., Schramm P., Baldauf C., Hofmann H. J. Chem. – Eur. J. 2009;15:5552. doi: 10.1002/chem.200802078. [DOI] [PubMed] [Google Scholar]; (e) Beke T., Somlai C., Perczel A. J. Comput. Chem. 2006;27:20. doi: 10.1002/jcc.20299. [DOI] [PubMed] [Google Scholar]; (f) Lang A., Fuzery A. K., Beke T., Hudaky P., Perczel A. J. Mol. Struct. (THEOCHEM) 2004;675:163. [Google Scholar]; (g) Shandler S. J., Shapovalov M. V., Dunbrack R. L., DeGrado W. F. J. Am. Chem. Soc. 2010;132:7312. doi: 10.1021/ja906700x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Bisetty K., Corcho F. J., Canto J., Kruger H. G., Perez J. J. J. Pept. Sci. 2006;12:92. doi: 10.1002/psc.692. [DOI] [PubMed] [Google Scholar]
- Johansson J. R., Lincoln P., Norden B., Kann N. J. Org. Chem. 2011;76:2355. doi: 10.1021/jo200134a. [DOI] [PubMed] [Google Scholar]
- (a) Dunbrack R. L., Karplus M. Nat. Struct. Biol. 1994;1:334. doi: 10.1038/nsb0594-334. [DOI] [PubMed] [Google Scholar]; (b) Smith L. J., Mark A. E., Dobson C. M., van Gunsteren W. F. J. Mol. Biol. 1998;280:703. doi: 10.1006/jmbi.1998.1892. [DOI] [PubMed] [Google Scholar]; (c) Csaszar A. G., Perczel A. Prog. Biophys. Mol. Biol. 1999;71:243. doi: 10.1016/s0079-6107(98)00031-5. [DOI] [PubMed] [Google Scholar]
- Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Mennucci B., Petersson G. A., Nakatsuji H., Caricato M., Li X., Hratchian H. P., Izmaylov A. F., Bloino J., Zheng G., Sonnenberg J. L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery Jr. J. A., Peralta J., Oglario F., Bearpark M., Heyd J. J., Brothers E., Kudin K. N., Staroverov V. N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J. C., Iyengar S. S., Tomasi J., Cossi M., Rega N., Millam N. J., Klene M., Knox J. E., Cross J. B., Bakken A. V., Adamo C., Jaramillo J., Gomperts R., Stratmann R. E., Yazyev O., Austin A. J., Cammi R., Pomelli C., Ochterski J. W., Martin R. L., Morokuma K., Zakrzewski V. G., Voth G. A., Salvador P., Dannenberg J. J., Dapprich S., Daniels A. D., Farkas Ö., Foresman J. B., Ortiz J. V., Cioslowski J. and Fox D. J., Gaussian 09 (Revision A.1), Gaussian, Inc., Wallingford CT, 2009.
- (a) Lee C. T., Yang W. T., Parr R. G. Phys. Rev. B: Condens. Matter. 1988;37:785. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]; (b) Becke A. D. J. Chem. Phys. 1993;98:1372. [Google Scholar]
- Chai J. D., Head-Gordon M. Phys. Chem. Chem. Phys. 2008;10:6615. doi: 10.1039/b810189b. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Truhlar D. G. Theor. Chem. Acc. 2008;120:215–241. [Google Scholar]
- Cances E., Mennucci B., Tomasi J. J. Chem. Phys. 1997;107:3032. [Google Scholar]
- (a) Atwood R. E., Urban J. J. J. Phys. Chem. A. 2012;116:1396–1408. doi: 10.1021/jp206152d. [DOI] [PubMed] [Google Scholar]; (b) Mardirossian N., Parkhill J. A., Head-Gordon M. Phys. Chem. Chem. Phys. 2011;13:19325–19337. doi: 10.1039/c1cp21635j. [DOI] [PubMed] [Google Scholar]
- Kang Y. K., Byun B. J. J. Comput. Chem. 2010;31:2915–2923. doi: 10.1002/jcc.21587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.