Abstract
The identification of transcriptional differences has served as an important starting point in understanding the molecular mechanisms behind biological processes and systems. The developmental biology of the inner ear, the biology of hearing and of course the pathology of deafness are all processes that warrant a molecular description if we are to improve human health. To this end, technological innovation has meant that larger scale analysis of gene transcription has been possible for a number of years now, extending our molecular analysis of genes to beyond those that are currently in vogue for a given system. In this review, some of the contributions gene profiling has made to understanding developmental, pathological and physiological processes in the inner ear are highlighted.
Keywords: deafness, hearing, transcription
Introduction
Approximately 20 years ago, the possibility to perform high‐throughput gene expression analysis was invented and, in the intervening period to now, this approach has made a significant and long‐lasting impact on the way the expression and function of genes are studied. Before the advent of this technology, the expression of single or only small groups of genes was studied at the time using techniques such as Northern blot analysis, RNase protection or in situ hybridization. Molecular analysis of the consequences following a loss of gene function in current animal models used for gene inactivation studies, such as the mouse, fish, nematode or fly, were usually confined to a limited set of predicted target genes, and their expression levels were tested one by one using the standard techniques mentioned above. The advent of DNA microarray technology has revolutionized this field, and nowadays allows the rapid and cost‐effective analysis of gene expression levels of the entire transcriptome at the level of the whole organism, or at the tissue‐ and/or cell type‐specific level. As a consequence, many more potential target genes that are regulated by a gene of interest can be rapidly identified, and bioinformatic tools can lead to the prediction of entire molecular pathways and networks that are controlled by this gene under primary study. The results from microarray data are usually deposited in the public domain in databases such as the Gene Expression Omnibus (Barrett et al. 2011) at the National Center of Biotechnology or Arrayexpress (Brazma et al. 2003) run by the European Informatics Institute, and are standardized by the criteria given by the Minimum Information about a Microarray Experiment. Microarray technology is based on the hybridization between a target DNA and a probe. Usually short sequences such as oligonucleotides from known genes are printed on a solid platform that is also termed a gene chip. This chip or microarray is then hybridized against target cDNAs derived from RNAs isolated from the whole organism, tissue or cell type of interest. Due to the labelling of target sequences with fluorescent dyes, following hybridization to the microarray and subsequent washes, the relative intensity of targets identified by the probe can be measured and quantified following normalization of the hybridization signals.
The advance in the technology of sequencing whole genomes termed next‐generation sequencing has led to the development of novel techniques that do not require the prior knowledge of probes and therefore have the potential to detect novel genes with a higher sensitivity. Serial analysis of gene expression (SAGE) generates unique tag sequences from cDNAs that are cleaved and sequenced as concatemeres (Velculescu et al. 1995). This technique has been developed in parallel to DNA microarrays. A more recent development that also applies next‐generation sequencing is RNA‐seq, which is based on the creation and subsequent sequencing of a cDNA library (Wang et al. 2009). Both SAGE and RNA‐seq offer a higher sensitivity to detect changes in gene expression levels compared with DNA microarrays and, moreover, can be used to measure the expression of both known and unknown genes.
The application of gene profiling to the inner ear was initiated more than 10 years ago (Chen & Corey, 2002; Cho et al. 2002), and has created a vast wealth of data on expression levels of genes from the entire organ, and specific tissues or cell types at different stages of development, or following a variety of stimuli, physiological damage or loss of specific genes in mouse mutants. In the present review, the data from these experiments are summarized with the aim to give an overview of the different datasets generated through microarrays (Table 1), and data from the more recently developed technologies that follow on from microarrays such as RNA‐seq in the inner ear and its anatomical substructures. This review mainly focuses on rodent animal models, but in some cases such as regeneration of sensory epithelia, will also refer to key studies performed in chicken and zebrafish models as these species, unlike mammals, do show some regeneration of hair cells and thus serve as excellent models to detect potential candidate genes involved during this process.
Table 1.
Gene profiling studies of the inner ear
| Genotype/tissue/condition | Organism | Microarray/technique | References |
|---|---|---|---|
| Developmental studies | |||
| Pre‐otic placodal ectoderm | Chick | Agilent 4 × 44 k chicken genome array | Paxton et al. (2010) |
| Otic placode | Chick | Affymetrix chicken genome array | Yang et al. (2013) |
| Otic vesicle | Chick | SAGE | Sinkkonen et al. (2011b) |
| Otic vesicle | Mouse | Affymetrix U74Av2 array | Powles et al. (2004) |
| Otic vesicle (ventral and dorsomedial) | Mouse | Affymetrix mouse genome 430 2.0 array | Fujimoto et al. (2010) |
| Developing inner ear E9–E15 | Mouse | Affymetrix mouse genome 430 2.0 array | Sajan et al. (2007) |
| Cochlea at P2 and P12 | Mouse | Affymetrix Mu30K array | Chen & Corey (2002) |
| Cochlear sensory epithelium, modiolus cochlear nucleus, inferior colliculus (8–12 weeks) | Rat | Atlas cDNA rat expression array | Cho et al. (2002) |
| Endolymphatic sac (10 weeks) | Rat | Affymetrix rat genome 230.2 array | Friis et al. (2011) |
| Cochlear sensory epithelium (P3 and adult) | Mouse | Affymetrix mouse genome 430 2.0 array | Smeti et al. (2012) |
| Cochlea (4–5 weeks) | Mouse | Custom‐made inner ear cDNA array | Morris et al. (2005) |
| Cochlea (adult) | Mouse | Custom‐made inner ear cDNA array | Liu et al. (2004) |
| Cochlea and vestibule (adult) | Human | Custom‐made cDNA array | Abe et al. (2003) |
| Modiolus (8 weeks) | Mouse | Affymetrix mouse genome 430 2.0 array | Shah et al. (2009) |
| Inner ear (juvenile) | Mouse | Soares NMIE mouse inner ear cDNA array | Hildebrand et al. (2004) |
| Tonotopic axis | |||
| Cochlea (apical and basal) at P10 | Mouse | Amersham mouse 20K Bioarray chip | Sato et al. (2009) |
| Cochlea (basal, medial, apical) at P0–P8 | Mouse | Affymetrix mouse GeneChip 1.0 ST | Son et al. (2012) |
| Cochlea (basal, medial, apical) at 6 weeks | Mouse | Agilent G3 mouse exon microarrays | Yoshimura et al. (2014) |
| Basilar papilla (proximal, middle, distal) at P0 | Chicken | Affymetrix chicken genome array | Frucht et al. (2011) |
| Basilar papilla (proximal, middle, distal) at E6.5 | Chicken | RNA‐seq | Thiede et al. (2014) |
| Basilar papilla (proximal, middle, distal) at E6.5 | Chicken | RNA‐seq, Affymetrix mouse genome 430 2.0 array | Mann et al. (2014) |
| Aging | |||
| Cochlea at 2 and 8 months | Mouse | Affymetrix mouse expression 430A array | Someya et al. (2007a) |
| Cochlea of young and old CBA mice | Mouse | Affymetrix mouse expression 430A array | D'Souza et al. (2008), Tadros et al. (2008), Christensen et al. (2009), Tadros et al. (2014) |
| Cochlea at 4 and 15 months, caloric restriction | Mouse | Affymetrix mouse genome 430 2.0 array | Someya et al. (2007b) |
| Cochlea at 4, 15 and 45 weeks | Mouse | Illumina mouse WG‐6 expression chip | Marano et al. (2012) |
| Inferior colliculus at 1 and 12–15 months | Mouse | Takara mouse IntelliGene II chip | Osumi et al. (2012) |
| Inferior colliculus of young and old CBA mice | Mouse | Affymetrix mouse expression 430A array | Tadros et al. (2007) |
| Damage | |||
| Noise exposure, cochlea at 8 weeks | Rat | Atlas cDNA rat expression array | Cho et al. (2004) |
| Noise exposure, adult cochlea | Rat | Affymetrix rat expression array 230A | Kirkegaard et al. (2006) |
| Noise exposure, sensory epithelium of CBA mice at 4–8 weeks | Mouse | RNA‐seq | Cai et al. (2014) |
| Noise exposure, cochlea at 8–10 weeks | Rat | Affymetrix rat genome 230.2 array | Han et al. (2012) |
| Noise exposure, cochlea at 7–8 weeks | Mouse | RNA‐seq | Park et al. (2014) |
| Gentamicin exposure, organ of Corti at P5 | Rat | Affymetrix gene chip rat genome U34A array | Nagy et al. (2004) |
| Salicylate exposure, adult cochlea | Mouse | Applied Biosystems mouse genome survey array | Im et al. (2007) |
| Labyrinthectomy of adult vestibular nucleus | Rat | Amersham Codelink UniSet rat bioarray | Horii et al. (2004) |
| Labyrinthectomy of adult vestibular nucleus | Rat | Agilent whole‐rat genome microarray | Park et al. (2012) |
| Pharmacological treatment/cellular signalling | |||
| Vasopressin exposure, adult cochlea | Rat | BiostarR‐40s microarray | Gu et al. (2006) |
| Dexamethasone exposure, cochlea at E15 | Mouse | Filgen mouse 32K array | Maeda et al. (2010) |
| Dexamethasone exposure, cochlea at 8 weeks | Guinea pig | Roche Nimble gene guinea pig expression array | Takumi et al. (2014) |
| Inhibition of NF‐kappa‐B, cochlea at P5 | Rat | Affymetrix rat genome 230.2 array | Nagy et al. (2007) |
| IGF‐1 exposure, cochlear sensory epithelium at P2 | Mouse | Affymetrix mouse genome 430 2.0 array | Hayashi et al. (2014) |
| Regeneration | |||
| Sensory epithelia of basilar papilla and utricle at P10–P21 | Chicken | Custom‐made human cDNA microarrays | Hawkins et al. (2003) |
| Neomycin or laser ablation of hair cells of basilar papilla and utricle at P10–P21 | Chicken | Custom‐made human cDNA microarrays | Hawkins et al. (2007) |
| Streptomycin‐treated utricular sensory epithelia | Chicken | RNA‐seq | Ku et al. (2014) |
| Noise exposure, adult inner ear | Zebrafish | SAGE | Liang et al. (2012) |
| Neomycin exposure, 5 days post‐fertilization, supporting cells | Zebrafish | RNA‐seq | Jiang et al. (2014) |
| Tissue‐ and cell‐specific analysis | |||
| Laser capture of hair cells and supporting cells from adult cristae ampullaris | Rat | Amersham CodeLink rat whole genome bioarrays | Cristobal et al. (2005) |
| Isolated adult hair cells | Zebrafish | Affymetrix zebrafish genome array | McDermott et al. (2007) |
| P0 auditory and vestibular sensory epithelial, neuronal, mesenchymal and vascular endothelial cells | Mouse | Illumina mouse WG6v2 BeadChip arrays | Hertzano et al. (2011) |
| Hair cells, greater and lesser epithelial ridge | Mouse | Illumina MouseRef‐8 version 2.0 chip | Sinkkonen et al. (2011a) |
| Supporting cells, non‐epithelial cells of P3 cochlea, additional comparison with otic vesicle | Hartman et al. (2015) | ||
| Cochlear ganglion neurons from E12 to P15 | Mouse | Affymetrix mouse genome 430 2.0 array | Lu et al. (2011) |
| Inner and outer hair cells, 25–30 days old | Mouse | Affymetrix mouse GeneChip 2.0 ST | Liu et al. (2014) |
| Cochlear hair cells expressing Atoh1 at P0 | Mouse | RNA‐seq | Cai et al. (2015) |
| Hair cells expressing Pou4f3 derived from the utricle and cochlea at E16, P0, P4 and P7 | Mouse | RNA‐seq | Scheffer et al. (2015) |
| Mouse mutant analysis | |||
| Otic placode of Fgf3/Fgf10 double mutants | Mouse | Agilent whole mouse genome microarray | Urness et al. (2010) |
| Otic vesicle of Dlx5 mutant | Mouse | Affymetrix mouse genome 430 2.0 array | Sajan et al. (2011) |
| Periotic mesenchyme of Tbx1 mutant at E12.5 | Mouse | Affymetrix mouse GeneChip 2.0 ST | Monks & Morrow (2012) |
| Inner ear of Pou4f3 mutant at E16.5 | Mouse | Affymetrix mouse U74Av2 array | Hertzano et al. (2004) |
| Cochlear neurons of Gata3 mutant at E13.5 | Mouse | Affymetrix mouse genome 430 2.0 array | Appler et al. (2013) |
| Utricle (gain and loss of Gata3 function) | Chicken | Custom‐made human cDNA microarrays | Alvarado et al. (2009) |
| Inner ear of PTEN mutant at E14.5 | Mouse | Illumina MouseRef‐8 version 2.0 chip | Kim et al. (2014) |
| Cochlea of shaker mutants at 3 weeks and 3 months | Mouse | Affymetrix mouse U74Av2 array | Gong et al. (2006) |
| Cochlea of BK channel subunit mutants at 8 weeks | Mouse | Affymetrix mouse genome 430 2.0 array | Pyott et al. (2007) |
| Cochlea and utricle of Rb1 mutants at P6 and 2 months | Mouse | Affymetrix mouse expression 430 array | Huang et al. (2011) |
| Inner ear of NR2F1/COUP‐TFI mutants at P0 | Mouse | Affymetrix mouse genome 430 2.0 array | Montemayor et al. (2010) |
| Cochlea of IGF‐1 mutants at E18.5 | Mouse | Affymetrix mouse expression 430A array | Sanchez‐Calderon et al. (2010) |
| Stria vascularis of Cx30 mutants at P30 | Mouse | Affymetrix mouse genome 430 2.0 array | Cohen‐Salmon et al. (2007) |
| Stria vascularis of adult Pendrin mutants | Mouse | Affymetrix mouse genome 430 2.0 array | Jabba et al. (2006) |
| Inferior colliculus of Tff3 mutants at 1 year | Mouse | Affymetrix mouse expression 430 array | Lubka‐Pathak et al. (2011) |
Gene expression profiling throughout inner ear development
The earliest phases of inner ear development involve the induction of the otic placode, a localized thickening of the ectoderm flanking the posterior hindbrain (Fig. 1, 8.5 days post‐coitum (dpc; Groves & Fekete, 2012). This placode will next invaginate through the otic cup stage (Fig. 1, 9.0 dpc) and close to form the otic vesicle or otocyst (Fig. 1, 9.5 dpc), which corresponds to the primordium of the future inner ear. Nearly all inner ear tissues derived from here will go on to form the epithelia of the inner ear as well as the sensory nerves that supply the patches of sensory hair cells (Coate & Kelley, 2013). In the chicken embryo, the differential expression between pre‐otic placodal ectoderm (including the adjacent hindbrain and the underlying mesendodermal tissue; Fig. 1) and more anterior positioned non‐specified tissue with otic competence has been examined (Paxton et al. 2010). This led to the identification of 1261 differentially expressed transcripts with a minimum twofold change. Further validation of the changes in gene expression on a selection of these differential transcripts was performed using RNA in situ hybridization, focusing on various transcription factors, including several Hox genes, and members of the fibroblast growth factor (FGF)‐, Wnt‐ and Notch signalling pathways. The differential expression between the otic placode itself and the lateral non‐otic tissue has also been examined by microarrays in chicken embryos (Yang et al. 2013). The genes enriched in the otic ectoderm were further tested for their capacity to be upregulated by FGF signalling, which previously had been shown to be essential for otic induction in the chicken (Schimmang, 2007). Surprisingly, only a minor fraction of the otic placode‐specific genes were shown to be induced by FGF. However, FGF signalling was shown to be required for the expression of otic placode‐specific genes in vivo. These data underline the necessity for FGF signalling during otic induction, but also demonstrate that other signalling pathways must co‐operate during otic placode formation (Yang et al. 2013).
Figure 1.
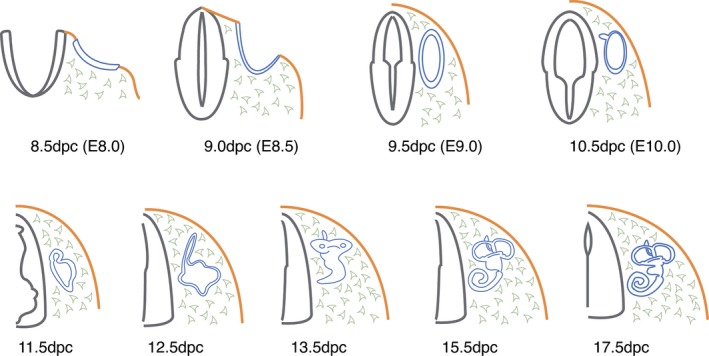
Schematic overview of early inner ear development in the mouse. Transverse sections are taken from the mouse embryo at the level of hindbrain rhombomeres 5/6. Stages of development are shown under each figure as days post‐coitum (dpc), with midnight on Day 0 representing the approximate time of conception. The first emergence of the inner ear is as a thickening of surface ectoderm, the otic placode (blue, 8.5 dpc) from the surrounding surface ectoderm (orange). Neural tube (grey) and mesenchyme (pale grey).
Using SAGE, 4135 genes were identified in the chicken otic vesicle, including signalling proteins and receptors, as well as almost 300 transcriptional regulators (Sinkkonen et al. 2011b). In the mouse, microarray analysis of otic vesicles has been performed to validate otic vesicle‐specific genes identified by cDNA subtractive hybridization against adult liver cDNA (Powles et al. 2004). Using a reporter mouse line expressing enhanced green fluorescent protein in the ventral and dorsomedial region of the mouse otic vesicle and applying fluorescence‐activated cell sorting (FACS), a gene profile of the cells corresponding to these subcompartments has been obtained (Fujimoto et al. 2010).
The most comprehensive gene profiling study covering inner ear development in the mouse from the otic vesicle stage at embryonic day 9 (E9) until differentiation at E15 has been published by Sajan et al. (2007). Samples were taken at half‐day intervals, and included substructures such as the presumptive cochlea, utricle and saccule (Fig. 2 shows the location of these regions in the adult inner ear). Different gene expression signatures for specific timepoints during development and structures could be defined, and pathway analysis identified more than 50 signalling cascades. This dataset is especially useful to define gene clusters that are up‐ or downregulated during development (Hertzano & Elkon, 2012). Before the appearance of this comprehensive analysis of differential gene expression throughout development, previous studies had instead focused only on specific postnatal stages or inner ear substructures. One of the first type of these studies was performed on the cochlea at postnatal day 2 (P2) and P32 (Chen & Corey, 2002; compare the sensory epithelium at P2 in Fig. 3 with that of the mature organ of Corti in Fig. 4). More than 10 000 genes or expressed sequence tags were identified, and the presence of known hair cell genes was confirmed validating the sensitivity of the assay. This publication was accompanied by a web‐based database that has served as a useful tool to evaluate the expression of a given gene in the cochlea. Like most oligonucleotide or gene arrays also used nowadays, these microarrays belonged to the first series of commercially available Gene Chip series from Affymetrix, which at this time comprised approximately 22 000 unique genes. Another commercial array in early use was the Atlas cDNA array from Clontech, containing 588 known rat genes. This array was used to determine the gene profile of the bony portion (modiolus) and sensorineural epithelium of the cochlea (Fig. 4), and the cochlear nucleus and inferior colliculus of the brainstem (Cho et al. 2002; Fig. 5). Insulin‐like growth factor‐binding proteins and matrix metalloproteinases were found to be expressed at higher levels in the cochlea compared with central nervous system (CNS) regions. The endolymphatic sac of the inner ear (Fig. 2) is required for fluid homeostasis and immune defense (Lo et al. 1997). Comparing the expression profile between the endolymphatic sac and the surrounding dura, 463 specific genes could be identified and were annotated to 29 functional clusters (Friis et al. 2011). A recent study in the mouse compared the expression profile of early postnatal cochlear sensory epithelia at P3 and adult age (Figs 3 and 4) with the aim to identify genes that are involved in regeneration or repair (Smeti et al. 2012). These properties are present at postnatal stages but completely lost in adult mammals. This screen identified novel differentially expressed genes so far not associated with the cochlea, such as high‐mobility group AT‐hook 2 (Hmga2) and Notch‐regulated ankyrin repeat protein (Nrap) at P3, and prolactin and the androgen receptor at adult stage. These genes may serve as potential markers for the regenerative capacity of the cochlea during development.
Figure 2.
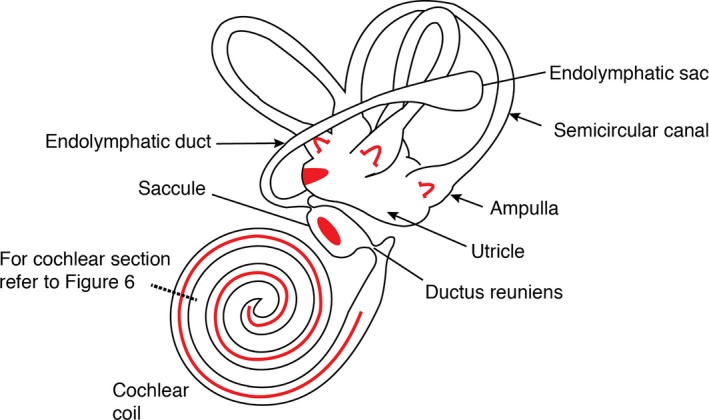
Schematic of the adult inner ear showing the major structural and functional regions. The six patches of sensory hair cells found as cristae in the ampullae of the three semicircular canals (× 3), the maculae in the utricle and saccule (× 2) and the organ of Corti in the cochlea (× 1) are denoted using red. Adapted from fig. 24‐7 from Michael et al. (1995).
Figure 3.
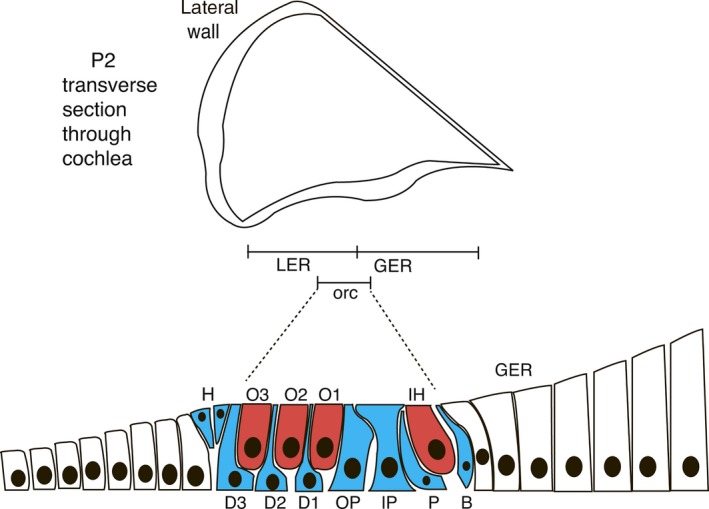
Immature organ of Corti from the P2 mouse inner ear (adapted from Rau et al. 1999) . The middle chamber of the cochlea is expanded to show the organization of supporting and hair cells. A single row of inner hair (IH) cells and three rows of outer hair (O1–3) cells are already evident (red). Supporting cells (blue): B, border cell; D, Deiters’ cells; H, Hensen's cells; IP and OP, inner and outer pillar cells; P, phalangeal cell. Regions of the sensory epithelium: LER and GER, lesser and greater epithelial ridges with the approximate position in the immature organ of Corti (orc) are shown.
Figure 4.
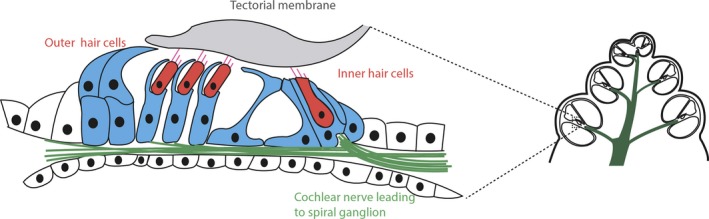
Mature organ of Corti showing the location of inner and outer sensory hair cells complete with the characteristic stereocilia projecting from the apical surface and overlaid by the gelatinous tectorial membrane. The myriad supporting cells found in the mature organ are shown in blue, with the cochlear nerve innervating the hair cells from the spiral ganglion shown in green. The location of the organ of Corti in the cochlea section is shown diagrammatically through dotted lines. Adapted from fig. 24‐14 from Michael et al. (1995).
Figure 5.
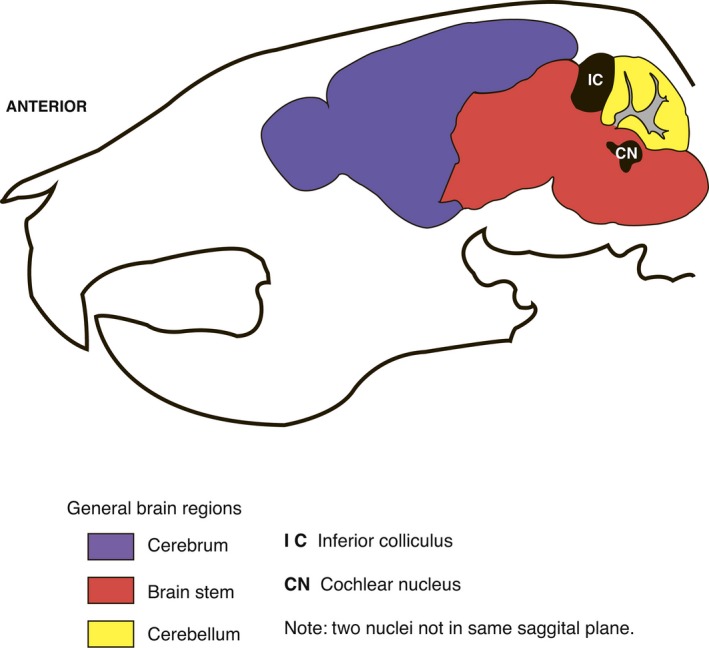
Sagittal schematic section of the adult mouse skull (approximately 8 weeks old) showing the locations of brain regions and nuclei important in auditory processing – the inferior colliculus and the cochlear nucleus. Adapted from information available at p56 sagittal mouse brain atlas; Website: © 2015 Allen Institute for Brain Science. Allen Mouse Brain Atlas [Internet]. Available from: http://mouse.brain-map.org (Lein et al. 2007).
As an alternative to commercial arrays, some researchers have developed custom arrays targeted for different experimental scenarios. For example, non‐redundant cDNA clones were selected from libraries to generate a chip with 2000 cDNAs that are specific for the inner ear (Morris et al. 2005). This array was then used to interrogate the relative expression levels of the selected cDNAs from the organ of Corti, lateral wall (including stria vascularis and spiral ligament) and spiral (auditory) ganglion (Fig. 6). Another example for a custom‐made gene array consisted of the amplification of 100 cDNAs taken from the hereditary hearing loss home page and their hybridization with RNA probes derived from the cochlea or kidney (Liu et al. 2004). The application and the potential usefulness of a custom‐made microarray to identify candidates for genes underlying deafness in humans was first shown by Abe et al. (2003). In this screen, the gene for mu‐crystalin (CRYM) was identified as a highly expressed gene in inner ear tissues. Following a search for mutations of this gene in patients with non‐syndromic deafness, two mutations were identified. In the mouse a screen to identify the nature of neuronal receptors involved in responding to axonal guidance cues was performed using the modiolus of 2‐month‐old animals. This study confirmed the presence of the neuronal transmembrane receptors ephrin, netrin, semaphorin and slit (Shah et al. 2009). For several of the corresponding ligands, essential roles for inner ear innervation or its targeting have recently been confirmed (Defourny et al. 2013; Katayama et al. 2013; Wang et al. 2013). Further screens for genes with transcripts enriched in the human or mouse inner ear have lead to the identification of type IX collagen, which is essential for hearing (Asamura et al. 2005; Van Camp et al. 2006) and DRASIC required for maintenance of hearing (Hildebrand et al. 2004).
Figure 6.
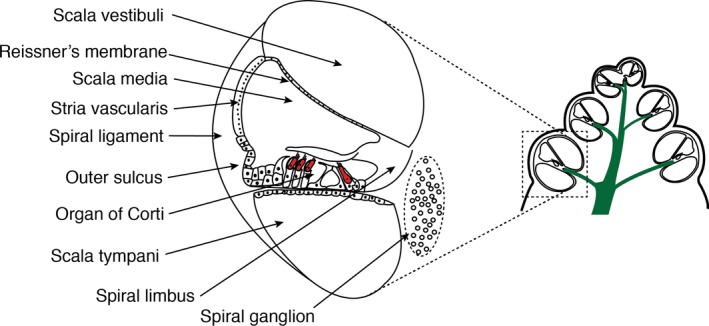
Schematic section through the mammalian cochlea, showing the organization of the three chambers and the position of the organ of Corti hair cells denoted in red. The location of the schematic relative to the cochlea is illustrated by dotted lines. Refer to Fig. 4 for more detailed organization of the organ of Corti. Additional anatomical structures that have been used in gene profiling studies are also highlighted. Adapted from fig. 24‐14 from Michael et al. (1995).
Differential gene expression along the tonotopic axis
The precise discrimination of different frequencies is an important function of the hearing organ in birds and mammals. Individual tones lead to stimulation of mechanosensory hair cells at specific positions along the frequency axis of the cochlea (mammals) or basilar papilla (chicken), and this register of frequency and position along the cochlea is known as the tonotopic axis (Fig. 7). The genes controlling the formation of the tonotopic axis have been investigated by various studies also based on microarrays.
Figure 7.
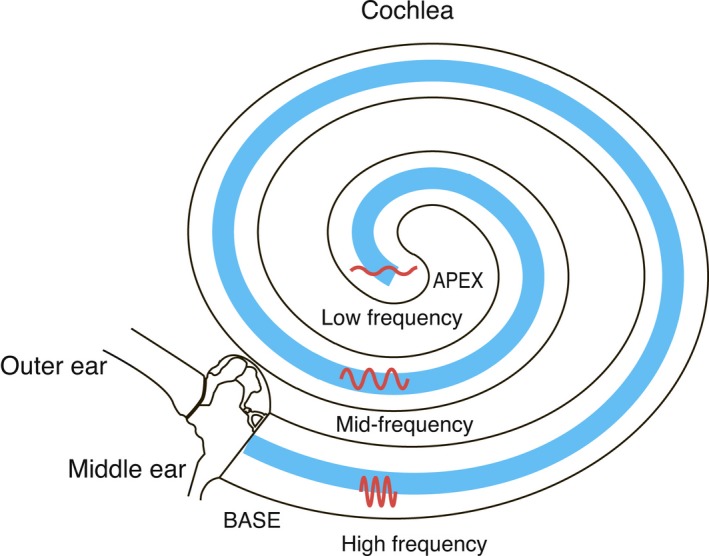
Schematic diagram of the cochlea illustrating the tonotopic axis along the cochlear coil. Hair cells (found within the organ of Corti; blue) respond to different frequencies of sound waves dependent on their position along the cochlear coil as illustrated. High‐frequency waves are detected at the base of the cochlea whilst low frequencies are detected at the apex of the cochlea, with a gradient of frequencies detected between.
In the mouse, the gene expression profile of the apical and basal portions of the cochlea, containing the organ of Corti and spiral ganglion neurons, was firstly examined by Sato et al. (2009). In the apical portion, 64 genes were found to be more than fivefold more abundantly expressed than in the basal portion; 77 genes showed the reciprocal pattern. In the basal cochlea, beta2 and gamma2 subunits of γ‐aminobutyric acid (GABA)A receptors showed high levels of expression. In a more recent study, the basal, middle and apical part of the cochlea, comprising the organ of Corti, spiral limbus and lateral wall (Fig. 6), were analysed by microarrays at P0 and P8 (Son et al. 2012). Tenascin C and nephroblastoma overexpressed gene (Nov) showed increasing expression levels towards the apex in the basilar membrane. Likewise, Follistatin, an antagonist of transforming growth factor (TGF)‐beta/bone morphogenetic protein (BMP) signalling, which is expressed in the lesser epithelial ridge (Fig. 3), increased its expression levels towards the apex. Finally, a microarray analysis of the cochlea, including the lateral wall of the cochlear duct, the organ of Corti and the spiral ganglion neurons of 6‐week‐old mice (Fig. 6) identified a gradient for Pou4f3, Slc17a8, Tmc1 and Crym with a higher expression level in the apex (Yoshimura et al. 2014). Interestingly, mutations of these genes cause autosomal dominant deafness in humans. It would be interesting to understand whether their role in deafness is separable from any role in frequency discrimination.
In the chicken, an initial analysis for gene expression gradients along the tonotopic axis was performed in the isolated auditory epithelium (basilar papilla) at birth (Frucht et al. 2011). Ion channels such as KCNMA1 (Slo) were found to be differentially expressed and potentially associated with the activities of protein kinase A and C. Very recently, analysis of the chicken embryo transcriptomes from the proximal, middle and distal regions of the chicken basilar papilla (Goodyear & Richardson, 1997) at E6.5 have been defined by RNA‐seq (Mann et al. 2014; Thiede et al. 2014). Retinoic acid‐synthesizing enzymes and retinoid receptors were found to be expressed at a high level in the proximal portion along the longitudinal gradient of the basilar papilla (Thiede et al. 2014). Additional experiments showed that retinoic acid was necessary and sufficient to induce hair cells corresponding to this region in the chicken embryo. In a similar study, using both microarrays and RNA‐seq, BMP7 was shown to be expressed in a gradient with its highest level in the distal portion (Mann et al. 2014). Interestingly, blocking of BMP signalling caused a loss of the tonotopic organization due to a change in the morphology of hair cells, which resulted in a sensory epithelium with uniform frequency characteristics.
Age‐related changes in gene expression
Sensory deficits associated with aging show a growing prevalence worldwide, thanks to the increased life expectancy of humans as a result of medical advances, better understanding of risk factors and lifestyle choices. Age‐related hearing loss, termed presbycusis, is one of the most frequent sensory deficits related with aging in humans. An early study compared cochlear gene expression between mice aged 2 months vs. 8 months when severe age‐related hearing loss in the DBA/2J mouse occurs, a strain that carries a mutation in the cadherin gene Cdh23 present in stereocilia of hair cells that is associated with the early‐onset deafness phenotype (Someya et al. 2007a). A gene array analysis of the cochlea showed that age‐related hearing loss in these mice was associated with profound downregulation of genes regulating the mitochondrial respiratory chain complexes in the cochlea. The expression levels of different GABAA receptor units have also been shown to be differentially modulated in CBA mice, another mouse model for human presbycusis (D'Souza et al. 2008), because it loses its hearing progressively over its lifespan. Further studies in this mouse model revealed the influence of apoptosis‐related genes and, more recently, the influence of antioxidant systems during presbycusis (Tadros et al. 2008, 2014). Interestingly, studies in wild‐type mice revealed that caloric restriction prevented presbycusis presumably by suppressing apoptosis (Someya et al. 2007b). Age‐related changes in gene expression leading to strong upregulation of prolactin and growth hormone have also been reported (Marano et al. 2012). Finally, studies in the auditory CNS during aging revealed changes in glutamate‐related gene expression (Tadros et al. 2007; Osumi et al. 2012).
Genes modulated upon damage to the inner ear
Damage to the sensorineural function of the inner ear may occur via a variety of insults, including direct lesions, excessive noise or during medical interventions that are based on drugs that show unwanted side‐effects on the survival or function of sensory hair cells or spiral ganglion neurons of the inner ear (Figs 4 and 6). Several studies have addressed the role of noise exposure via gene profiling in rodents. Studies in the cochlea of rats identified the upregulation of immediate‐early genes such as transcription factors, cytokines or genes related to oxidative stress a few hours after noise exposure (Cho et al. 2004; Kirkegaard et al. 2006). Twenty‐four hours after the trauma, members of antioxidant and inflammatory pathways were upregulated (Kirkegaard et al. 2006). Very recently a molecular profile of genes related to the immune system has been obtained by RNA‐seq in mice (Cai et al. 2014). Transcripts from genes belonging to the Toll‐like receptor signalling pathway were localized to supporting cells (Fig. 4) and their expression levels were found to be changed upon noise damage. Genes involved in the immune system or induced as a response to stress or stimuli were also altered in a study where gene expression was monitored in rats 1 h after cessation of noise exposure (Han et al. 2012). Finally, the role of cysteinyl leukotriene signalling, which plays an important role during inflammation, has recently been studied in a mouse model of noise damage (Park et al. 2014). Expression of the cysteinyl leukotriene type 1 receptor (CysLTR1) was found to be increased in the organ of Corti 3 days after noise exposure. Treatment with a leukotriene receptor antagonist (LTRA) after noise exposure reduced the hair cell damage and threshold shifts were observed. By using RNA‐seq, matrix metalloproteinase‐3 (MMP‐3) was also found to be upregulated upon noise induction and this upregulation was significantly inhibited by LTRA application.
Gentamicin is an aminoglycoside used as an antibiotic against bacterial infections, but unfortunately a side‐effect of high dosing levels is the induction of hair cell death in the cochlea. Explant cultures from rats treated with gentamicin and analysed by microarrays revealed the downregulation of genes associated with cellular stress mediated by reactive oxygen species and N‐methyl‐d‐aspartate receptors (Nagy et al. 2004). The frequently used drug salicylate may also cause temporary hearing loss or tinnitus when administered. Microarray analysis of mice 3 h after injection with salicylate revealed the upregulation of 87 genes in the cochlea (Im et al. 2007).
Unilateral vestibular deafferentiation in the bilateral vestibular nuclear complex (VNC; Khan & Chang, 2013) is followed by vestibular compensation of the contralateral VNC. To monitor the asymmetric gene expression between both VNCs, microarrays were applied and more than 200 genes were found to be differentially regulated in rats (Horii et al. 2004). In a similar study, differential expression of zinc finger protein 307, zinc metallopeptidase, P34, calcitonin receptor, insulin‐like growth factor‐binding protein 5, GATA‐binding protein 3 and CD151 was observed (Park et al. 2012).
Modulation of gene expression during pharmacological interventions or cellular signalling
Vasopressin or anti‐diuretic hormone (ADH) is a hormone secreted by the neurohypophysis, which regulates water levels in the body. Hormone levels are elevated in patients suffering from Meniere's disease that is caused by an excess of fluid (endolymph) in the inner ear (Takeda et al. 1995). Using microarrays, the effects on gene expression following injection of vasopressin in rats were examined. This led to the identification of 25 genes whose expression was deregulated (Gu et al. 2006). The change in expression of aquaporins (water channels) was suggested to be responsible for the increased production and reduced absorption of endolymph. Changes in aquaporin gene expression levels have also been associated with the development of presbycusis (Christensen et al. 2009).
Due to its anti‐inflammatory effects, glucocorticoids such as dexamethasone are also used in treating cases of acute sensorineural hearing loss. The effect of dexamethasone on gene expression has been monitored in cultured cochleae of mice, and the expression of genes with a potential cytoprotective effect, operating by preventing inflammation, cellular stress or oxidative damage was shown to be significantly changed (Maeda et al. 2010). The effects of dexamethasone have also been evaluated in the context of a dexamethasone‐eluting electrode as a part of a cochlear implant in guinea pigs (Takumi et al. 2014). The gene profile within the cochlea was compared with that of a normal electrode and a non‐surgically treated control. The group of animals carrying the dexamethasone‐eluting electrode showed a tendency to downregulate genes associated with the immune response caused by the implant.
NF‐kappaB complexes are involved in the immediate response of cells to insulting stimuli, and of interest is the demonstration that NF‐kappaB signalling has been shown to be required for the survival of postnatal auditory hair cells in vitro (Nagy et al. 2005). Transcriptional changes downstream of this pathway were explored by using microarrays, and the regulatory subunit of phosphatidylinositol 3‐kinase (PI3K), a critical intracellular signalling effector molecule, was found to be upregulated (Nagy et al. 2007). Inhibition of the PI3K pathway in cochleae treated with a NF‐kappaB inhibitor lead to reduced caspase‐3 activation (a pro‐apoptotic gene), indicating a link between these pathways during hair cell survival. Inhibition of the PI3K and MEK/ERK pathways has also been shown to be involved in the upregulation of growth‐associated protein 43 (Gap43) and netrin 1 (Ntn1). Both these genes were detected in a microarray screen based on the protective effects of insulin‐like growth factor 1 (IGF‐1) against aminoglycosides in the postnatal murine cochlea (Hayashi et al. 2014). However, it is worth bearing in mind that other growth factors may also lead to activation of MEK/ERK (MAPK) pathways, such as FGF signalling (Goetz & Mohammadi, 2013).
Accessing hair cell regeneration through gene profiling
The utility of the avian system as a model to identify potential deafness loci in mammals including man was first demonstrated by interrogating custom‐made cross‐species microarrays containing conserved human cDNA sequences, using probes generated from sensory epithelia of avian utricles and basilar papillae (Hawkins et al. 2003). Using this approach, differentially expressed transcription factors that mapped to so far unknown deafness loci could be identified where the genetic lesion has yet to be defined. They therefore represent good candidates for further investigation for the underlying cause of these deafness disorders. The same approach was used for gene profiling of the regenerating sensory epithelia of the avian utricle and basilar papilla following treatment with neomycin or laser ablation of hair cells (Hawkins et al. 2007). A great variety of genes was found to be differentially regulated during regeneration, such as components of many conserved signalling pathways, apoptosis, the cell cycle and transcription factors. Recently the chick utricle was analysed using RNA‐seq to identify the transcriptome of hair cell regeneration following treatment with aminoglycosides (Ku et al. 2014). Almost 500 new putative hair cell‐specific genes with more than 200 transcription factors differentially expressed during regeneration were identified.
Next to the chick, the zebrafish has emerged as a model to study hair cell regeneration. In the adult zebrafish, using next‐generation sequencing, a network of genes involved in hair cell regeneration following noise damage has been defined (Liang et al. 2012). The stat3/socs3 signalling pathway was identified as a key regulator during this process. Hair cells are also present in the lateral line organ of fish, a sensory organ for the detection of movement, which also serves as a valuable model to study hair cell regeneration (Lush & Piotrowski, 2014). Supporting cells of the lateral line that will give rise to hair cells were recently purified by FACS, and a RNA‐seq analysis was performed (Jiang et al. 2014). Interestingly, and in stark contrast to mammals, the Notch signalling pathway was downregulated after injury (Mizutari et al. 2013). The identification of these key differences between the responses of non‐mammalian vs. mammalian species may reveal important insights in potential therapies for hair cell regeneration, and highlight the transcriptional differences between species that are able to regenerate hair cells and those that cannot.
Gene expression profiling in specific inner ear tissues and cell types
A prerequisite to fully understand the molecular function of the inner ear is to acquire a thorough understanding of the transcriptional profiles at the tissue‐ or cell‐specific level. This task is especially complicated in the inner ear due to: (i) the paucity of tissue; and (ii) the great variety of cell types found in the inner ear. In order to achieve a more specific transcriptional analysis of subcompartments or cell types of the inner ear, laser capture microdissection was first used in the cristae ampullaris of adult rats (Cristobal et al. 2005). Using this technique, differences between the expression of more than 400 genes in supporting cells compared with hair cells could be defined. Next, in the zebrafish by hybridizing mRNAs from isolated hair cells vs. liver to oligonucleotide microarrays, more than 1000 genes were identified comprising the hair cell transcriptome of this species (McDermott et al. 2007). More recently in the same species, FACS was applied to study regenerating supporting cells of the lateral line by RNA‐seq (Jiang et al. 2014; see above). However, it must always be considered that any mechanical disaggregation or collection of tissue can lead to errors of interpretation where the process of collection leads to identification of transcripts due to the techniques employed rather than any particular discrete cellular population. In the mouse, FACS has also been used to separate different cell types such as hair cells, supporting cells, neurons, mesenchyme, other epithelial cells and blood vessels. Sorting of cells was achieved by using specific cell surface markers (Hertzano et al. 2011; Sinkkonen et al. 2011a). In one of these studies, a computational promoter analysis of genes preferentially expressed in inner ear sensory epithelia revealed an enrichment for the binding site of the zinc finger transcription factor Zeb1 (Hertzano et al. 2011). Interestingly, mouse mutants for Zeb1 have been shown to have malformations of the inner ear arising during development (Kurima et al. 2011), and many of the genes containing Zeb1‐binding sites were shown to be dysregulated in the mutants (Hertzano et al. 2011). Another study also used FACS to purify hair cells and different non‐sensory populations from the neonatal mouse cochlea (Sinkkonen et al. 2011a). Purified cochlear supporting cells and cells of the lesser epithelial ridge (Fig. 3) were shown to proliferate and differentiate illustrating their regenerative potential unlike the nearby hair cells. With the aim to identify genes specific for the otic sensory lineage, the microarray data from this study were compared with gene profiles from otic vesicles and non‐otic tissue, and the gene encoding F‐box ubiquitin ligase F‐Box 2 (Fbx2) was shown to have the highest specificity (Hartman et al. 2015). The developmental profile of gene expression from initiation of neurite extension until the onset of hearing in spiral ganglia neurons purified by FACS has also been determined (Lu et al. 2011). This led to the identification of unique transcription and axon guidance factors. Recently, hair cells characterized by the expression of the transcription factor Atoh1 or Pou4f3, which are required for their formation or maintenance, respectively, have been purified via FACS and analysed by RNA‐seq (Cai et al. 2015; Scheffer et al. 2015). Various Atoh1 target genes and genes showing specific expression in vestibular hair cells, cochlear hair cells, before or after maturation of mechanosensitivity could be defined and this will permit gene regulatory networks for hair cell development to be constructed. Finally, the targetome from individual inner and outer hair cells has been described, and both common and differentially expressed genes between these cell types were identified in their apical, basolateral and synaptic membranes (Liu et al. 2014). Clearly much transcriptional information on individual cells and tissues in the inner ear has been obtained, and the next goal is to make some functional sense from this.
Gene expression profiling in mutant mice
One of the most frequent applications of gene profiling has so far been the comparative analysis of mouse mutants vs. wild‐type animals in order to identify potential target genes whose expression is altered as a consequence of the gene mutation. Such studies allow the underlying molecular mechanisms of the different processes involved in inner ear development, hair cell regeneration or the pathogenesis of deafness to be elucidated.
As mentioned earlier in this review, several studies have used gene profiling to study the molecular mechanisms underlying otic placode induction. As a model to discover genes relevant for otic induction, the double mutants for Fgf3 and Fgf10 that fail to initiate inner ear development were used (Urness et al. 2010). Using the otic region, including the prospective placodal ectoderm, neural ectoderm and underlying mesendoderm as a probe (Fig. 1), the downregulation of several placode‐specific genes could be validated by RNA in situ hybridization. Loss of Fgf3 and Fgf10 was also associated with downregulation of wnt8 expressed in the hindbrain neighbouring the otic placode, and was suggested to provide a link between FGF‐induced formation of the otic placode and its restriction to the posterior extent of the hindbrain at rhombomeres 4 or 5–6.
At the otic vesicle stage, the homeobox transcription factor Dlx5 is expressed in the dorsal compartment, and its loss in mouse mutants leads to defects in the vestibular portion of the mature inner ear (Acampora et al. 1999; Depew et al. 1999). To identify target genes for Dlx5, the transcriptional profile of gene expression for otic vesicles deficient for this gene was carried out (Sajan et al. 2011). Several genes that overlapped with the Dlx5 expression domain were identified and their binding to the Dlx5 promoter was confirmed by chromatin immunoprecipitation (ChIP). The T‐box transcription factor Tbx1 is expressed in the otic vesicle and surrounding mesenchyme, and is required for inner ear morphogenesis (Vitelli et al. 2003). In a similar approach to that above, to identify target genes for Tbx1 in the mesenchyme of Tbx1 mutants, the periotic area was isolated and examined by microarrays (Monks & Morrow, 2012). This led to the identification of several retinoic acid responsive genes.
The first mouse mutants analysed by microarrays in the inner ear were mice lacking the transcription factor Pou4f3, a gene required for hair cell survival (Hertzano et al. 2004). From this analysis the growth factor independent factor 1 (Gfi1), which encodes a transcriptional repressor, was identified and confirmed as a target for Pou4f3. The zinc finger transcription factor Gata3 is expressed in spiral ganglion neurons (Fig. 6), and loss of its expression in mouse mutants leads to defects in the normal innervation patterns of these neurons (Appler et al. 2013). Expression profiling of spiral ganglia isolated from these mutants revealed a change in the expression profile to encompass genes more associated with a later neuronal phenotype. Downstream targets for Gata3 have also been identified using custom‐made cross‐species microarrays containing conserved human cDNA sequences in the avian utricle where Gata3 is expressed in a specific zone containing morphologically distinct hair cells (Alvarado et al. 2009). Additional gene profiles were obtained after treatment with Gata3 RNAi or Gata3 overexpression, and led to the identification of two direct targets of Gata3 that were validated in vitro using ChiP.
Mouse mutants lacking the lipid phosphatase PTEN also show defects in spiral ganglion neurons, such as irregular migration of nerve fibres and apoptosis (Kim et al. 2013). A microarray analysis of the inner ear at E14.5 confirmed deregulation of networks of genes involved in these processes (Kim et al. 2014).
Many studies have addressed the consequences of the loss of genes expressed in hair cells using mouse mutants. An early study was performed in shaker2 mutant mice that carry a mutation in the myosin XV gene (Gong et al. 2006). This mutation results in defects of hair cell stereocilia (Fig. 4), and causes deafness in both humans and mice (Libby & Steel, 2000). Gene profiling at 3 weeks and 3 months revealed only very few genes whose expression was affected by the shaker2 genotype, but a large number of genes whose expression was changed dependent on the age of the different groups studied, including genes involved in bone mineralization and components of the extracellular matrix or collagen. The molecular consequences of interfering with the function of large‐conductance voltage‐ and calcium‐activated potassium (BK) channel alpha present in inner hair cells has been addressed in Slo mouse mutants that lack this channel (Pyott et al. 2007). Slo mouse mutants showed normal cochlear function, and in addition revealed no compensatory changes of other ion channels or transporters following the analysis of microarrays, but interestingly these mice showed protection against noise‐induced hearing loss. Mouse mutants lacking the retinoblastoma gene (Rb1), a gene required for proper cell cycle exit, show a loss of cochlear hair cells whereas the survival of hair cells in the utricle is not affected (Sage et al. 2006). A comparative analysis using microarrays revealed that the Rb1 mutant cochlea showed enrichment of transcripts from pathways involved in Wnt and Notch signalling whereas, in the utricle, transcripts associated with cellular proliferation and survival were enriched (Huang et al. 2011). The orphan nuclear receptor Nr2f1 (COUP‐TF1) is expressed during hair cell differentiation (Tang et al. 2005), and mouse mutants lacking this gene show an increased number of hair cells in the cochlea (Tang et al. 2006). Gene expression profiles of mutant mice and additional bioinformatic analysis identified and validated the fatty acid‐binding protein 7 (Fabp7) as a direct target for COUP‐TF1 (Montemayor et al. 2010).
Another set of gene profiling studies has addressed transcripts that are expressed in non‐sensory areas of the cochlea. One of these genes is IGF‐1, which is expressed in the stria vascularis, the spiral limbus, the outer sulcus and Reissner's membrane (Fig. 6; Sanchez‐Calderon et al. 2010). Loss of IGF‐1 results in deafness in humans and mice (Varela‐Nieto et al. 2013). Gene profiling of the cochlea of mutant IGF‐1 mice revealed upregulation of the transcription factors FoxM1, Six6 and Mash1. The stria vascularis is responsible for the generation of the endocochlear potential by maintaining the mechanoelectrical transduction current in auditory hair cells (Zdebik et al. 2009). Loss of the gap junction protein connexin 30 (Cx30) disrupts the function of the stria vascularis and results in deafness in mutant mice (Cohen‐Salmon et al. 2007). Microarray analysis of Cx30 mutant mice revealed an increase in homocysteine, a factor previously associated with dysfunction of the endothelium (Eberhardt et al. 2000), which acts as a barrier of the capillaries that supply the stria vacularis. The function of the stria vascularis is also affected in Pendred syndrome, which is caused by a mutation of the anion exchanger pendrin (SLC26a4; Reardon et al. 2000). The stria vascularis of a mouse mutant for this gene is hyperpigmented and its marginal cells are disorganized (Royaux et al. 2003). Microarray analysis revealed an increase in the expression of macrophage markers, suggesting their involvement in the degeneration of the stria vascularis perhaps as a secondary event to the initial dysfunction (Jabba et al. 2006).
Finally, gene profiling has also been performed in parts of the central auditory system such as the inferior colliculus (Fig. 5). Mice lacking the trefoil factor 3 (Tff3) peptide were shown to suffer from age‐related hearing loss, and microarray analysis showed deregulated expression of the transcriptional regulator securin (Pttg1) and the protease inhibitor serpina3n (Lubka‐Pathak et al. 2011).
These examples demonstrate the powerful technologies that we already have at hand to identify molecular changes during normal or aberrant processes in any species. This has lead to advances in the molecular details underlying these processes, or at least the nature of the candidate genes and pathways involved. These do require verification in vivo. However, as well as the technically complex nature of the technology that will bear fruit in such investigations, it should be noted that each particular experiment needs to be carefully established in the first instance to provide informative data. For example, inappropriate choice of the genetic background in mouse microarray analyses can confound later analysis due to significant variation, as the goal of most studies will be to identify differential transcripts due to the processes under investigation rather than between different genetic backgrounds of animals. However, notwithstanding such a note of care, gene expression profiling has already revealed many of the molecular details that will allow us to further explore the processes of inner ear development, hair cell regeneration, hearing and deafness.
Concluding remarks
The number of studies using gene profiling based on microarrays or next‐generation sequencing has increased dramatically over the recent years (Smith & Rajadinakaran, 2013). These techniques are nowadays commonly used to efficiently analyse the complexity of gene expression levels under different conditions at the tissue‐ or cell type‐specific level. Microarrays have particularly been shown to be cost‐effective, and provide an easy means to rapidly acquire an overview of the changes of gene expression levels. The subsequent usage of bioinformatics will also allow rapid identification of candidate genes or potential pathways up to single‐cell resolution. A good example for the inner ear is in the recent transcriptional profile reconstruction of the mouse otocyst (Durruthy‐Durruthy et al. 2014). In this case, following quantitative RT‐PCR measurements of established otic markers and applying multivariate clusters, principal component and network analysis to the data, a three‐dimensional model of the otocyst that showed the mapping of each individual cell was accomplished.
The present review has shown the wealth of information on gene expression data generated for the inner ear. Next to gene expression studies based on mRNA levels, changes at the protein level also need to be an important focus as mRNA levels do not necessarily reflect protein expression levels. Further changes may also occur at the post‐translational level by, for example, protein phosphorylation. Large‐scale proteomic analyses are conducted using two‐dimensional difference gel electrophoresis (2D‐DIGE), antibody microarrays or liquid chromatography coupled with mass spectrometry (Smith & Rajadinakaran, 2013; Darville & Sokolowski, 2014). Applying these techniques, several interesting studies have already been conducted, such as defining the proteome of the hair cell's ribbon synapse (Uthaiah & Hudspeth, 2010) and stereocilia (Shin et al. 2013), or of the cochlea following ototoxic or noise damage (Jamesdaniel et al. 2008, 2011). In conclusion, the future of inner ear research is likely to see a combination of gene profiling based on microarrays or RNA‐seq together with proteomics; following this with bioinformatic analyses will give a much more complete picture of the complex functions carried out by the inner ear.
Acknowledgements
This work was supported by the Spanish MinEco (BFU2010‐15477, BFU2013‐40944), Fundació La Marató de TV3, TerCel (RD06/0010/0000) and Red de Terapia Célular de la Junta de Castilla y León.
References
- Abe S, Katagiri T, Saito‐Hisaminato A, et al. (2003) Identification of CRYM as a candidate responsible for nonsyndromic deafness, through cDNA microarray analysis of human cochlear and vestibular tissues. Am J Hum Genet 72, 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acampora D, Merlo GR, Paleari L, et al. (1999) Craniofacial, vestibular and bone defects in mice lacking the Distal‐less‐related gene Dlx5. Development 126, 3795–3809. [DOI] [PubMed] [Google Scholar]
- Alvarado DM, Veile R, Speck J, et al. (2009) Downstream targets of GATA3 in the vestibular sensory organs of the inner ear. Dev Dyn 238, 3093–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appler JM, Lu CC, Druckenbrod NR, et al. (2013) Gata3 is a critical regulator of cochlear wiring. J Neurosci 33, 3679–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamura K, Abe S, Imamura Y, et al. (2005) Type IX collagen is crucial for normal hearing. Neuroscience 132, 493–500. [DOI] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, et al. (2011) NCBI GEO: archive for functional genomics data sets – 10 years on. Nucleic Acids Res 39, D1005–D1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Parkinson H, Sarkans U, et al. (2003) ArrayExpress – a public repository for microarray gene expression data at the EBI. Nucleic Acids Res 31, 68–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Vethanayagam RR, Yang S, et al. (2014) Molecular profile of cochlear immunity in the resident cells of the organ of Corti. J Neuroinflammation 11, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T, Jen HI, Kang H, et al. (2015) Characterization of the transcriptome of nascent hair cells and identification of direct targets of the atoh1 transcription factor. J Neurosci 35, 5870–5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, Corey DP (2002) An inner ear gene expression database. J Assoc Res Otolaryngol 3, 140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Gong TW, Stover T, et al. (2002) Gene expression profiles of the rat cochlea, cochlear nucleus, and inferior colliculus. J Assoc Res Otolaryngol 3, 54–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Gong TW, Kanicki A, et al. (2004) Noise overstimulation induces immediate early genes in the rat cochlea. Brain Res Mol Brain Res 130, 134–148. [DOI] [PubMed] [Google Scholar]
- Christensen N, D'Souza M, Zhu X, et al. (2009) Age‐related hearing loss: aquaporin 4 gene expression changes in the mouse cochlea and auditory midbrain. Brain Res 1253, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coate TM, Kelley MW (2013) Making connections in the inner ear: recent insights into the development of spiral ganglion neurons and their connectivity with sensory hair cells. Semin Cell Dev Biol 24, 460–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen‐Salmon M, Regnault B, Cayet N, et al. (2007) Connexin30 deficiency causes instrastrial fluid‐blood barrier disruption within the cochlear stria vascularis. Proc Natl Acad Sci USA 104, 6229–6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristobal R, Wackym PA, Cioffi JA, et al. (2005) Assessment of differential gene expression in vestibular epithelial cell types using microarray analysis. Brain Res Mol Brain Res 133, 19–36. [DOI] [PubMed] [Google Scholar]
- Darville LN, Sokolowski BH (2014) Bottom‐up and shotgun proteomics to identify a comprehensive cochlear proteome. J Vis Exp 2014 Mar 7;(85). doi: 10.3791/51186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defourny J, Poirrier AL, Lallemend F, et al. (2013) Ephrin‐A5/EphA4 signalling controls specific afferent targeting to cochlear hair cells. Nat Commun 4, 1438. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Liu JK, Long JE, et al. (1999) Dlx5 regulates regional development of the branchial arches and sensory capsules. Development 126, 3831–3846. [DOI] [PubMed] [Google Scholar]
- D'Souza M, Zhu X, Frisina RD (2008) Novel approach to select genes from RMA normalized microarray data using functional hearing tests in aging mice. J Neurosci Methods 171, 279–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durruthy‐Durruthy R, Gottlieb A, Hartman BH, et al. (2014) Reconstruction of the mouse otocyst and early neuroblast lineage at single‐cell resolution. Cell 157, 964–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt RT, Forgione MA, Cap A, et al. (2000) Endothelial dysfunction in a murine model of mild hyperhomocyst(e)inemia. J Clin Invest 106, 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis M, Martin‐Bertelsen T, Friis‐Hansen L, et al. (2011) Gene expression of the endolymphatic sac. Acta Otolaryngol 131, 1257–1263. [DOI] [PubMed] [Google Scholar]
- Frucht CS, Uduman M, Kleinstein SH, et al. (2011) Gene expression gradients along the tonotopic axis of the chicken auditory epithelium. J Assoc Res Otolaryngol 12, 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto C, Ozeki H, Uchijima Y, et al. (2010) Establishment of mice expressing EGFP in the placode‐derived inner ear sensory cell lineage and FACS‐array analysis focused on the regional specificity of the otocyst. J Comp Neurol 518, 4702–4722. [DOI] [PubMed] [Google Scholar]
- Goetz R, Mohammadi M (2013) Exploring mechanisms of FGF signalling through the lens of structural biology. Nat Rev Mol Cell Biol 14, 166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong TW, Karolyi IJ, Macdonald J, et al. (2006) Age‐related changes in cochlear gene expression in normal and shaker 2 mice. J Assoc Res Otolaryngol 7, 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyear R, Richardson G (1997) Pattern formation in the basilar papilla: evidence for cell rearrangement. J Neurosci 17, 6289–6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves AK, Fekete DM (2012) Shaping sound in space: the regulation of inner ear patterning. Development 139, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu FM, Han HL, Zhang LS (2006) Effects of vasopressin on gene expression in rat inner ear. Hear Res 222, 70–78. [DOI] [PubMed] [Google Scholar]
- Han Y, Hong L, Zhong C, et al. (2012) Identification of new altered genes in rat cochleae with noise‐induced hearing loss. Gene 499, 318–322. [DOI] [PubMed] [Google Scholar]
- Hartman BH, Durruthy‐Durruthy R, Laske RD, et al. (2015) Identification and characterization of mouse otic sensory lineage genes. Front Cell Neurosci 9, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Bashiardes S, Helms CA, et al. (2003) Gene expression differences in quiescent versus regenerating hair cells of avian sensory epithelia: implications for human hearing and balance disorders. Hum Mol Genet 12, 1261–1272. [DOI] [PubMed] [Google Scholar]
- Hawkins RD, Bashiardes S, Powder KE, et al. (2007) Large scale gene expression profiles of regenerating inner ear sensory epithelia. PLoS One 2, e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Yamamoto N, Nakagawa T, et al. (2014) Insulin‐like growth factor 1 induces the transcription of Gap43 and Ntn1 during hair cell protection in the neonatal murine cochlea. Neurosci Lett 560, 7–11. [DOI] [PubMed] [Google Scholar]
- Hertzano R, Elkon R (2012) High throughput gene expression analysis of the inner ear. Hear Res 288, 77–88. [DOI] [PubMed] [Google Scholar]
- Hertzano R, Montcouquiol M, Rashi‐Elkeles S, et al. (2004) Transcription profiling of inner ears from Pou4f3(ddl/ddl) identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum Mol Genet 13, 2143–2153. [DOI] [PubMed] [Google Scholar]
- Hertzano R, Elkon R, Kurima K, et al. (2011) Cell type‐specific transcriptome analysis reveals a major role for Zeb1 and miR‐200b in mouse inner ear morphogenesis. PLoS Genet 7, e1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand MS, de Silva MG, Klockars T, et al. (2004) Characterisation of DRASIC in the mouse inner ear. Hear Res 190, 149–160. [DOI] [PubMed] [Google Scholar]
- Horii A, Masumura C, Smith PF, et al. (2004) Microarray analysis of gene expression in the rat vestibular nucleus complex following unilateral vestibular deafferentation. J Neurochem 91, 975–982. [DOI] [PubMed] [Google Scholar]
- Huang M, Sage C, Tang Y, et al. (2011) Overlapping and distinct pRb pathways in the mammalian auditory and vestibular organs. Cell Cycle 10, 337–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im GJ, Jung HH, Chae SW, et al. (2007) Differential gene expression profiles in salicylate ototoxicity of the mouse. Acta Otolaryngol 127, 459–469. [DOI] [PubMed] [Google Scholar]
- Jabba SV, Oelke A, Singh R, et al. (2006) Macrophage invasion contributes to degeneration of stria vascularis in Pendred syndrome mouse model. BMC Med 4, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamesdaniel S, Ding D, Kermany MH, et al. (2008) Proteomic analysis of the balance between survival and cell death responses in cisplatin‐mediated ototoxicity. J Proteome Res 7, 3516–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamesdaniel S, Hu B, Kermany MH, et al. (2011) Noise induced changes in the expression of p38/MAPK signaling proteins in the sensory epithelium of the inner ear. J Proteomics 75, 410–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Romero‐Carvajal A, Haug JS, et al. (2014) Gene‐expression analysis of hair cell regeneration in the zebrafish lateral line. Proc Natl Acad Sci USA 111, E1383–E1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama K, Imai F, Suto F, et al. (2013) Deletion of Sema3a or plexinA1/plexinA3 causes defects in sensory afferent projections of statoacoustic ganglion neurons. PLoS One 8, e72512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Chang R (2013) Anatomy of the vestibular system: a review. NeuroRehabilitation 32, 437–443. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Woo HM, Ryu J, et al. (2013) Conditional deletion of pten leads to defects in nerve innervation and neuronal survival in inner ear development. PLoS One 8, e55609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Ryu J, Woo HM, et al. (2014) Patterns of gene expression associated with Pten deficiency in the developing inner ear. PLoS One 9, e97544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard M, Murai N, Risling M, et al. (2006) Differential gene expression in the rat cochlea after exposure to impulse noise. Neuroscience 142, 425–435. [DOI] [PubMed] [Google Scholar]
- Ku YC, Renaud NA, Veile RA, et al. (2014) The transcriptome of utricle hair cell regeneration in the avian inner ear. J Neurosci 34, 3523–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurima K, Hertzano R, Gavrilova O, et al. (2011) A noncoding point mutation of Zeb1 causes multiple developmental malformations and obesity in Twirler mice. PLoS Genet 7, e1002307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, et al. (2007) Genome‐wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. [DOI] [PubMed] [Google Scholar]
- Liang J, Wang D, Renaud G, et al. (2012) The stat3/socs3a pathway is a key regulator of hair cell regeneration in zebrafish. [corrected]. J Neurosci 32, 10 662–10 673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby RT, Steel KP (2000) The roles of unconventional myosins in hearing and deafness. Essays Biochem 35, 159–174. [DOI] [PubMed] [Google Scholar]
- Liu X, Mohamed JA, Ruan R (2004) Analysis of differential gene expression in the cochlea and kidney of mouse by cDNA microarrays. Hear Res 197, 35–43. [DOI] [PubMed] [Google Scholar]
- Liu H, Pecka JL, Zhang Q, et al. (2014) Characterization of transcriptomes of cochlear inner and outer hair cells. J Neurosci 34, 11 085–11 095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WW, Daniels DL, Chakeres DW, et al. (1997) The endolymphatic duct and sac. Am J Neuroradiol 18, 881–887. [PMC free article] [PubMed] [Google Scholar]
- Lu CC, Appler JM, Houseman EA, et al. (2011) Developmental profiling of spiral ganglion neurons reveals insights into auditory circuit assembly. J Neurosci 31, 10 903–10 918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubka‐Pathak M, Shah AA, Gallozzi M, et al. (2011) Altered expression of securin (Pttg1) and serpina3n in the auditory system of hearing‐impaired Tff3‐deficient mice. Cell Mol Life Sci 68, 2739–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lush ME, Piotrowski T (2014) Sensory hair cell regeneration in the zebrafish lateral line. Dev Dyn 243, 1187–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y, Fukushima K, Hirai M, et al. (2010) Microarray analysis of the effect of dexamethasone on murine cochlear explants. Acta Otolaryngol 130, 1329–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann ZF, Thiede BR, Chang W, et al. (2014) A gradient of Bmp7 specifies the tonotopic axis in the developing inner ear. Nat Commun 5, 3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marano RJ, Tickner J, Redmond SL (2012) Age related changes in gene expression within the cochlea of C57BL/6J mice. Aging Clin Exp Res 24, 603–611. [DOI] [PubMed] [Google Scholar]
- McDermott BM Jr, Baucom JM, Hudspeth AJ (2007) Analysis and functional evaluation of the hair‐cell transcriptome. Proc Natl Acad Sci USA 104, 11 820–11 825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael R, Romrell L, Kaye G (1995) Histology: A Text and Atlas. Baltimore, MA, USA: Williams and Wilkins. [Google Scholar]
- Mizutari K, Fujioka M, Hosoya M, et al. (2013) Notch inhibition induces cochlear hair cell regeneration and recovery of hearing after acoustic trauma. Neuron 77, 58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks DC, Morrow BE (2012) Identification of putative retinoic acid target genes downstream of mesenchymal Tbx1 during inner ear development. Dev Dyn 241, 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montemayor C, Montemayor OA, Ridgeway A, et al. (2010) Genome‐wide analysis of binding sites and direct target genes of the orphan nuclear receptor NR2F1/COUP‐TFI. PLoS One 5, e8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KA, Snir E, Pompeia C, et al. (2005) Differential expression of genes within the cochlea as defined by a custom mouse inner ear microarray. J Assoc Res Otolaryngol 6, 75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy I, Bodmer M, Brors D, et al. (2004) Early gene expression in the organ of Corti exposed to gentamicin. Hear Res 195, 1–8. [DOI] [PubMed] [Google Scholar]
- Nagy I, Monge A, Albinger‐Hegyi A, et al. (2005) NF‐kappaB is required for survival of immature auditory hair cells in vitro . J Assoc Res Otolaryngol 6, 260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy I, Caelers A, Monge A, et al. (2007) NF‐kappaB‐dependent apoptotic hair cell death in the auditory system. Audiol Neurootol 12, 209–220. [DOI] [PubMed] [Google Scholar]
- Osumi Y, Shibata SB, Kanda S, et al. (2012) Downregulation of N‐methyl‐D‐aspartate receptor zeta1 subunit (GluN1) gene in inferior colliculus with aging. Brain Res 1454, 23–32. [DOI] [PubMed] [Google Scholar]
- Park MK, Lee BD, Lee JD, et al. (2012) Gene profiles during vestibular compensation in rats after unilateral labyrinthectomy. Ann Otol Rhinol Laryngol 121, 761–769. [DOI] [PubMed] [Google Scholar]
- Park JS, Kang SJ, Seo MK, et al. (2014) Role of cysteinyl leukotriene signaling in a mouse model of noise‐induced cochlear injury. Proc Natl Acad Sci USA 111, 9911–9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxton CN, Bleyl SB, Chapman SC, et al. (2010) Identification of differentially expressed genes in early inner ear development. Gene Expr Patterns 10, 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powles N, Babbs C, Ficker M, et al. (2004) Identification and analysis of genes from the mouse otic vesicle and their association with developmental subprocesses through in situ hybridization. Dev Biol 268, 24–38. [DOI] [PubMed] [Google Scholar]
- Pyott SJ, Meredith AL, Fodor AA, et al. (2007) Cochlear function in mice lacking the BK channel alpha, beta1, or beta4 subunits. J Biol Chem 282, 3312–3324. [DOI] [PubMed] [Google Scholar]
- Rau A, Legan PK, Richardson GP (1999) Tectorin mRNA expression is spatially and temporally restricted during mouse inner ear development. J Comp Neurol 405, 271–280. [PubMed] [Google Scholar]
- Reardon W, OMahoney CF, Trembath R, et al. (2000) Enlarged vestibular aqueduct: a radiological marker of pendred syndrome, and mutation of the PDS gene. QJM, 93, 99–104. [DOI] [PubMed] [Google Scholar]
- Royaux IE, Belyantseva IA, Wu T, et al. (2003) Localization and functional studies of pendrin in the mouse inner ear provide insight about the etiology of deafness in pendred syndrome. J Assoc Res Otolaryngol 4, 394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage C, Huang M, Vollrath MA, et al. (2006) Essential role of retinoblastoma protein in mammalian hair cell development and hearing. Proc Natl Acad Sci USA 103, 7345–7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajan SA, Warchol ME, Lovett M (2007) Toward a systems biology of mouse inner ear organogenesis: gene expression pathways, patterns and network analysis. Genetics 177, 631–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajan SA, Rubenstein JL, Warchol ME, et al. (2011) Identification of direct downstream targets of Dlx5 during early inner ear development. Hum Mol Genet 20, 1262–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez‐Calderon H, Rodriguez‐de la Rosa L, Milo M, et al. (2010) RNA microarray analysis in prenatal mouse cochlea reveals novel IGF‐I target genes: implication of MEF2 and FOXM1 transcription factors. PLoS One 5, e8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Doi K, Hibino H, et al. (2009) Analysis of gene expression profiles along the tonotopic map of mouse cochlea by cDNA microarrays. Acta Otolaryngol Suppl 562, 12–17. [DOI] [PubMed] [Google Scholar]
- Scheffer DI, Shen J, Corey DP, et al. (2015) Gene expression by mouse inner ear hair cells during development. J Neurosci 35, 6366–6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmang T (2007) Expression and functions of FGF ligands during early otic development. Int J Dev Biol 51, 473–481. [DOI] [PubMed] [Google Scholar]
- Shah SM, Kang YJ, Christensen BL, et al. (2009) Expression of Wnt receptors in adult spiral ganglion neurons: frizzled 9 localization at growth cones of regenerating neurites. Neuroscience 164, 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JB, Krey JF, Hassan A, et al. (2013) Molecular architecture of the chick vestibular hair bundle. Nat Neurosci 16, 365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkkonen ST, Chai R, Jan TA, et al. (2011a) Intrinsic regenerative potential of murine cochlear supporting cells. Sci Rep 1, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkkonen ST, Starlinger V, Galaiya DJ, et al. (2011b) Serial analysis of gene expression in the chicken otocyst. J Assoc Res Otolaryngol 12, 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeti I, Assou S, Savary E, et al. (2012) Transcriptomic analysis of the developing and adult mouse cochlear sensory epithelia. PLoS One 7, e42987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ME, Rajadinakaran G (2013) The transcriptomics to proteomics of hair cell regeneration: looking for a hair cell in a haystack. Microarrays (Basel), 2(3): 10.3390/microarrays2030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya S, Yamasoba T, Prolla TA, et al. (2007a) Genes encoding mitochondrial respiratory chain components are profoundly down‐regulated with aging in the cochlea of DBA/2J mice. Brain Res 1182, 26–33. [DOI] [PubMed] [Google Scholar]
- Someya S, Yamasoba T, Weindruch R, et al. (2007b) Caloric restriction suppresses apoptotic cell death in the mammalian cochlea and leads to prevention of presbycusis. Neurobiol Aging 28, 1613–1622. [DOI] [PubMed] [Google Scholar]
- Son EJ, Wu L, Yoon H, et al. (2012) Developmental gene expression profiling along the tonotopic axis of the mouse cochlea. PLoS One 7, e40735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros SF, D'Souza M, Zettel ML, et al. (2007) Glutamate‐related gene expression changes with age in the mouse auditory midbrain. Brain Res 1127, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros SF, D'Souza M, Zhu X, et al. (2008) Apoptosis‐related genes change their expression with age and hearing loss in the mouse cochlea. Apoptosis 13, 1303–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros SF, D'Souza M, Zhu X, et al. (2014) Gene expression changes for antioxidants pathways in the mouse cochlea: relations to age‐related hearing deficits. PLoS One 9, e90279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, Kakigi A, Saito H (1995) Antidiuretic hormone (ADH) and endolymphatic hydrops. Acta Otolaryngol Suppl 519, 219–222. [DOI] [PubMed] [Google Scholar]
- Takumi Y, Nishio SY, Mugridge K, et al. (2014) Gene expression pattern after insertion of dexamethasone‐eluting electrode into the Guinea pig cochlea. PLoS One 9, e110238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang LS, Alger HM, Lin F, et al. (2005) Dynamic expression of COUP‐TFI and COUP‐TFII during development and functional maturation of the mouse inner ear. Gene Expr Patterns 5, 587–592. [DOI] [PubMed] [Google Scholar]
- Tang LS, Alger HM, Pereira FA (2006) COUP‐TFI controls Notch regulation of hair cell and support cell differentiation. Development 133, 3683–3693. [DOI] [PubMed] [Google Scholar]
- Thiede BR, Mann ZF, Chang W, et al. (2014) Retinoic acid signalling regulates the development of tonotopically patterned hair cells in the chicken cochlea. Nat Commun 5, 3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urness LD, Paxton CN, Wang X, et al. (2010) FGF signaling regulates otic placode induction and refinement by controlling both ectodermal target genes and hindbrain Wnt8a. Dev Biol 340, 595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uthaiah RC, Hudspeth AJ (2010) Molecular anatomy of the hair cell's ribbon synapse. J Neurosci 30, 12 387–12 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Camp G, Snoeckx RL, Hilgert N, et al. (2006) A new autosomal recessive form of Stickler syndrome is caused by a mutation in the COL9A1 gene. Am J Hum Genet 79, 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela‐Nieto I, Murillo‐Cuesta S, Rodriguez‐de la Rosa L, et al. (2013) IGF‐I deficiency and hearing loss: molecular clues and clinical implications. Pediatr Endocrinol Rev 10, 460–472. [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, et al. (1995) Serial analysis of gene expression. Science 270, 484–487. [DOI] [PubMed] [Google Scholar]
- Vitelli F, Viola A, Morishima M, et al. (2003) TBX1 is required for inner ear morphogenesis. Hum Mol Genet 12, 2041–2048. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M (2009) RNA‐Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SZ, Ibrahim LA, Kim YJ, et al. (2013) Slit/Robo signaling mediates spatial positioning of spiral ganglion neurons during development of cochlear innervation. J Neurosci 33, 12 242–12 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, O'Neill P, Martin K, et al. (2013) Analysis of FGF‐dependent and FGF‐independent pathways in otic placode induction. PLoS One 8, e55011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura H, Takumi Y, Nishio SY, et al. (2014) Deafness gene expression patterns in the mouse cochlea found by microarray analysis. PLoS One 9, e92547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdebik AA, Wangemann P, Jentsch TJ (2009) Potassium ion movement in the inner ear: insights from genetic disease and mouse models. Physiology (Bethesda) 24, 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]


