Supplemental Digital Content is available in the text
Abstract
Chronic hepatitis B (CHB) remains a global health problem. Therapeutic vaccination has been successfully employed to treat a subpopulation of CHB patients. Personalized treatment can not only improve therapeutic efficacy, but also decrease the cost of medical care. Since microRNAs (miRNAs) are highly conserved and are involved in many cellular processes, exploring their expression profiles in CHB patients in association with responsiveness to therapeutic vaccination may be an approach for personalized treatment. In this study, we examined the kinetic expression profiles of 13 miRNAs in sera and serum-derived hepatitis B surface antigen (HBsAg) particles in 10 CHB patients including 5 responders and 5 nonresponders selected from a large cohort of 136 patients enroled in a phase III clinical trial using antigen-antibody immunogenic complex based therapeutic vaccine (YIC). Eight miRNAs were detected in both sera and HBsAg particles. Among them, the levels of serum miRNAs and serum-derived HBsAg-carried miRNAs (let-7f, miR-22, miR-30a, and miR-122) were significantly lower in the responders group compared to those in the nonresponders group at baseline and throughout the course of treatment. The lower baseline levels of serum miRNAs and HBsAg-carried miRNAs were also associated with hepatitis Be antigen clearance at week 76 and hepatitis Be antigen seroconversion during the study period. In summary, our study suggests that lower baseline levels of serum miRNAs and HBsAg-carried miRNAs (let-7f, miR-22, miR-30a, and miR-122) associated with YIC treatment response and the variation trend of these 4 miRNAs could have a prognostic value for responsiveness to YIC treatment.
INTRODUCTION
Hepatitis B virus (HBV) infection remains a global health problem, with around 300 million chronically infected individuals worldwide who are at risk of developing liver cirrhosis and hepatocellular carcinoma.1 Although several anti-HBV drugs targeting the reverse transcriptase have shown their efficacies in chronic hepatitis B (CHB) patients, rebound of virus replication after withdrawal of drugs and emergence of drug resistance strains are unresolved problems.2 A standard of care for CHB patients includes an immune-modulator interferon-α (IFN-α), which exerts antiviral activity through upregulation of host defense gene expression.3 However, the side effects of IFN-α limit its use in certain population of CHB patients. Therapeutic vaccination designed to improve or modulate host immune responses in CHB patients has been developed as an alternative approach. Several therapeutic vaccines are under preclinical studies and clinical trials.4–7 We have developed an antigen-antibody (hepatitis B surface antigen [HBsAg]-anti-HBs immunoglobulin [HBIG]) immunogenic complex therapeutic vaccine (YIC) for CHB patients who were currently under the second stage of phase III clinical trial.8–10 Analysis of viral genotypes from the first stage of phase III clinical trial showed that patients infected with genotype B of HBV had a higher response rate to YIC than those infected with HBV genotype C. However, the host factors involved in responsiveness to YIC have not been explored.
The microRNAs (miRNAs) are highly conserved short 18 to 25 ribonucleotides noncoding RNAs which regulate many host biological processes, including cellular development, differentiation, apoptosis, proliferation, and metabolism.11 They are also involved in inflammation and cancer.12 To explore the potential host factors that may be associated with responsiveness to YIC, we compared the expression profiles of miRNAs in 10 HBV genotype B–infected CHB patients, including 5 responders and 5 nonresponders, to YIC treatment. In this pilot study, we describe the levels of 13 miRNAs in sera and serum-derived HBsAg particles, collected before treatment (week 0), at the end of treatment (week 52), and 6 months after YIC treatment (week 76). The baseline levels of serum miRNAs and HBsAg-carried miRNAs (let-7f, miR-22, miR-30a, and miR-122) were also correlated with hepatitis Be antigen (HBeAg) clearance and HBeAg seroconversion.
MATERIALS AND METHODS
Ethics Statement
Ethics statements were described previously.9 Briefly, formal approvals from the ethics committees in 21 evaluation centers were completed, and enrolment of patients started at the end of October 2007. A signed written informed consent for participation in this trial was obtained from each patient prior to enrolment.
Clinical and Virological Characteristics of Enroled Patients
One hundred thirty-six patients with CHB were enroled into a phase III clinical trial number ChiCTR-TRC-07000019 assigned by WHO International Clinical Trials Registry Platform, http://www.chictr.org/cn/proj/show.aspx?proj=1369. The inclusion and exclusion standards were described previously.9 In brief, patients were (1) between 18 and 65 years old; (2) HBsAg and HBeAg positive for at least 6 months and anti-HBe negative with HBV viral load >100,000 copies/mL; (3) having alanine aminotransferase (ALT) serum values 2 to 10 times above the normal limit, within 4 weeks before randomization. Patients coinfected with hepatitis A, C, D, E virus or human immunodeficiency virus, patients receiving antiviral, hepatotoxic, or immunosuppressive drug treatment within 6 months preceding the study, patients with liver disease caused by other factors, those suffering from serious medical or psychiatric illness and hepatic cirrhosis or alpha fetal protein >100 ng/mL, patients with abnormal serum creatinine, hemoglobin, thrombocyte count, or serum total bilirubin, and pregnant women were all excluded from participation in the current study.
The clinical profile of CHB patients was as previously described.9 Briefly, HBsAg, HBeAg, and anti-HBe levels were assayed by Abbott EIA AxSYM kit (Abbott, Abbott Park, IL). HBV deoxyribonucleic acid (DNA) levels were measured by fluorescent Phosphate Buffered Saline (PCR) assay with detection limit of 500 copies/mL (PiJi, Shenzhen Co, China). Levels of HBsAg in serum were quantified by Architect HBsAg QT assay (Abbott, Abbott Park, IL).
From this large cohort of patients, we selected for the current study. These patients have undergone the complete course of treatment with YIC for 54 weeks (60 μg of hepatitis B surface antigen complexed with high titer of human HBIG was intramuscularly injected every 4 weeks with 12 injections, while the interval between the 7th and 8th injection was 8 weeks) and followed up for additional 24 weeks. HBeAg seroconversion at week 76 was used as the criterion for responsiveness to YIC treatment.13,14 Ten selected patients were divided into 2 groups based on the HBeAg seroconversion, the responders group (Figure 1, A-E) and the nonresponders group (Figure 1, F-J). The dynamic changes of serum HBV DNA, HBsAg, HBeAg, anti-HBe, and ALT (liver functions test) of these 10 selected patients during and after treatment are also shown in Figure 1 and Supplementary Figure 1.
FIGURE 1.
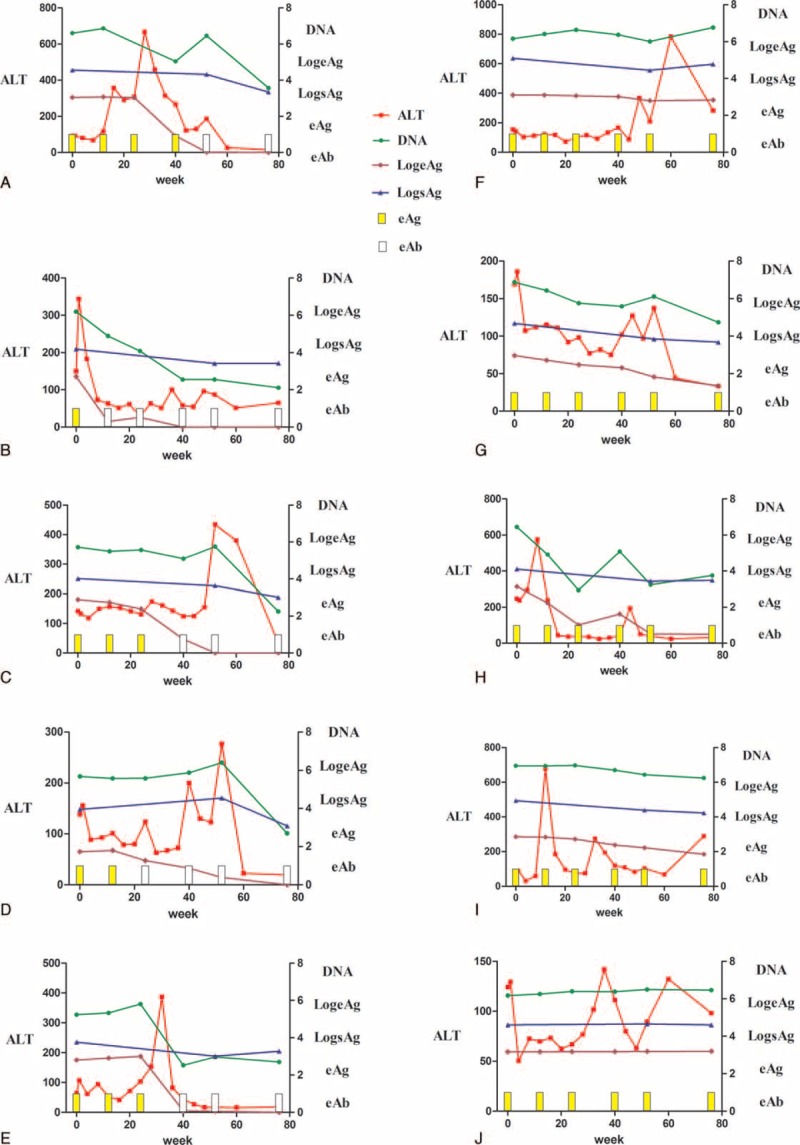
Comparison of clinical and virological data from individual CHB patients receiving YIC treatment over the study period. The levels of ALT, HBV DNA, HBsAg, HBeAg, and HBeAb were compared between responders and nonresponders groups. Data of 5 patients in the responders group are listed on the left (A–E, patient IDs: 2, 29, 85, 87, 307, respectively), and data of 5 patients in the nonresponders group are listed on the right (F–J, patient IDs: 19, 24, 44, 58, 88, respectively). The levels of HBeAg, DNA, and HBsAg were log10 transformed. The yellow bars represent HBeAg positivity, while the white bars represent HBeAg negativity. ALT = alanine aminotransferase, CHB = chronic hepatitis B, HBeAb = Hepatitis Be antibody, HBeAg = hepatitis Be antigen, HBsAg = hepatitis B surface antigen, HBV DNA = hepatitis B virus deoxyribonucleic acid, YIC = antigen-antibody immunogenic complex based therapeutic vaccine.
Preprocessing of Serum Samples
Thirty serum samples collected at weeks 0, 52, and 76 from these 10 patients were used to quantify total miRNA and HBsAg-encapsidated miRNAs. Sera were first centrifuged at 2000 × g for 20 minutes at 4°C to remove dead cells and supernatants were again centrifuged at 10,000 × g for 30 minutes followed by centrifugation at 14,000 × g for 10 minutes at 4°C to completely remove cell debris.
Purification of HBsAg Particles
HBsAg particles were immunoprecipitated from 100 μL of patients’ sera. To do this, first, 2 μL of anti-HBs antibody (Kehua biotech, Shanghai, China) and 50 μL of Protein A/G PLUS-Agarose were conjugated in a volume of 1 mL Phosphate Buffered Saline (PBS) for 24 hours at 4°C followed by centrifugation at 100 × g for 3 minutes at 4°C and the precipitate was washed in PBS (0.1% Tween20) 3 times, 15 minutes each time. One hundred microliter of preprocessed sera was added to the anti-HBsAg antibody conjugated beads and incubated for 24 hours at 4°C. After centrifugation at 100 × g for 3 minutes at 4°C, HBsAg particles were purified and further washed in PBS (0.1% Tween20) 3 times. The purity of HBsAg particles, as determined by Western immunoblotting using anti-HBsAg antibodies, is shown in Supplementary Figure 3.
MiRNA Extraction, Reverse Transcription, and Quantitative PCR
RNAs from sera and serum-derived HBsAg particles were all extracted with TRIzol LS reagent (Invitrogen) according to the recommended protocol. Briefly, miRNAs were reverse transcribed using specific Bulge-Loop reverse transcription (RT) primers (RiboBio, Shenzhen, China) and Multiscribe reverse transcriptase (Applied Biosystems, San Diego, CA) in RT reaction (16°C, 30 minutes, 42°C, 1 hour, 85°C, 5 minutes, 4°C∞). The RT products were used for real-time PCR using Thunderbird SYBR qPCR Mix (Toyobo, Japan) with predesigned primers (RiboBio, Shenzhen, China) according to the recommended protocols. Briefly, a 10-μL PCR reaction included 2 μL of 5-fold-diluted RT product, 5 μL of SYBR Green PCR Master Mix, 0.8-μL ddH2O, 0.2-μL ROX Reference Dye, 1-μL forward primer, and 1-μL reverse primer. The reactions were performed in 384-well plates at 95°C for 10 minutes, followed by 40 cycles at 95°C for 10 s and 60°C for 20 s. All cDNA samples were quantified in duplicate and mean Ct was calculated from the duplicate PCRs. Real-time PCR was performed on a ViiA7 real-time PCR system (Applied Biosystems). Melting curve analysis was performed following 40 cycles of amplification in each run. The relative quantity of each gene was calculated using the equation 2-△Ct.15
Statistical Analysis
Statistical analysis was performed by GraphPad Prism software. The relative quantification of miRNAs was calculated by the equation 2-△Ct. The concentration of miRNAs, HBsAg, HBeAg, and HBV DNA was logarithmically converted to allow the assessment of geometric mean titers. Data are presented as means ± standard deviation (SD) of the responders group or nonresponders group. For dimensional outcomes, 1-way analysis of variance (ANOVA) was employed. Kaplan–Meier analysis was applied to determine the correlation between baseline miRNAs levels and HBeAg seroconversion along with time. P < 0.05 was considered statistically significant.
RESULTS
Clinical and Virological Data Over the Study Period
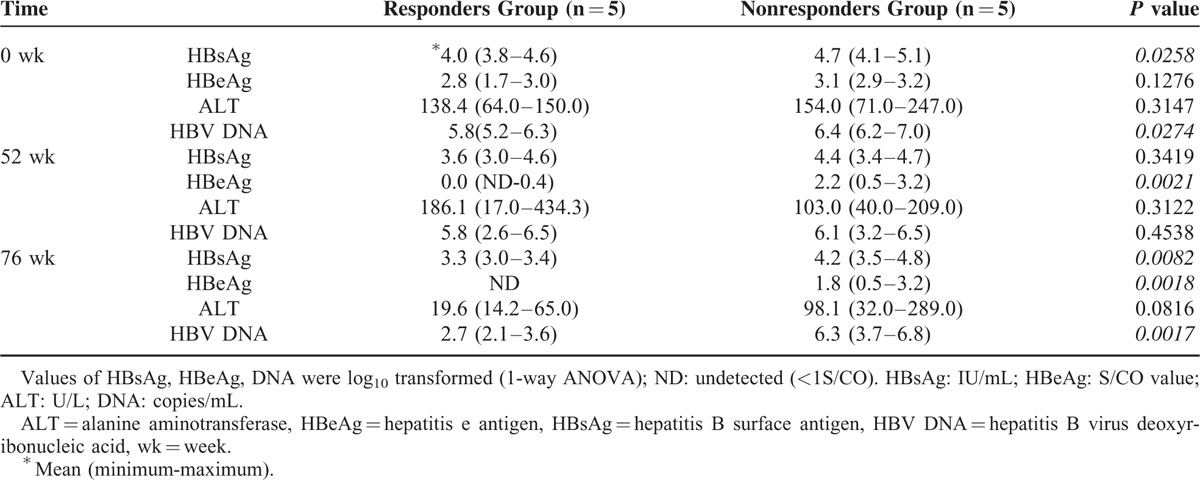
At baseline, the levels of HBsAg and HBV DNA were slightly lower in the responders group compared to those in the nonresponders group (P = 0.0258, P = 0.0274, respectively, Supplementary Figure 1A, D and Table 1), while the levels of HBeAg and ALT were similar in these 2 groups (P = 0.1276, P = 0.3147, respectively, Supplementary Figure 1B, C and Table 1). Before YIC treatment, all patients had abnormal liver function (ALT >40 IU/L, Table 1). YIC monotherapy for 52 weeks led to a significant decline in the levels of HBsAg, HBV DNA, and especially in HBeAg (P = 0.0021, Supplementary Figure 1A, B, D and Table 1) in the responders group compared to those in the nonresponders group. It is worth noting that in the nonresponders group, the levels of HBeAg declined while responsive patients manifested HBeAg clearance (Supplementary Figure 1B). At week 76, the HBV DNA levels further declined in the responders group while no significant changes were observed in the nonresponders group (P = 0.0017, Supplementary Figure 1D and Table 1). Interestingly, although liver inflammation was observed in the responders group as evidenced by abnormal ALT levels at week 52, their liver functions became normalized at week 76 (P = 0.05, weeks 76 vs 52, Supplementary Figure 1C and Table 1). On the other hand, the ALTs of nonresponsive patients did not show these trends (Supplementary Figure 1C).
TABLE 1.
Basic Characteristics of Enroled Patients
The Levels of miRNAs Detected in Sera and Serum-derived HBsAg Particles at Baseline
We previously reported the profile of miRNAs in 94 CHB patients following an IFN therapy and found that levels of 11 miRNAs (let-7a, miR-30a, miR-1290, miR-106b, miR-1224–5p, miR-939, miR-1281, let-7f, miR-198, miR-22–3p, and miR-638) correlated with responsiveness to IFN treatment.16 We therefore hypothesized that some of these miRNAs associated with prediction of IFN treatment outcome may also serve as predictors of responsiveness to YIC therapy since the immune-modulatory function of YIC is similar to that of type I interferons(s).17 Moreover, we also previously demonstrated that exosome released from IFN-α–treated hepatic nonparenchymal cells contained increased amounts of miR-638, miR-4284, and miR-1260a and these exosomal miRNAs could suppress the replication of HBV.18 Hence, we included these miRNAs in our study in addition to miR-122, which is a liver-specific and the most abundant miRNA in the liver.19
With the exception of miRNA let-7a, 13 of the 14 candidate miRNAs passed the quality control (Supplementary Table 2). These 13 miRNAs were present in serum samples while only 8 were encapsidated in HBsAg particles. MiRNAs, namely let-7f, miR-22, miR-30a, miR-106b, miR-122, miR-1224, miR-1281, and miR-1260a, were present in both types of samples (Supplementary Table 2). Based on the Ct values of quantitative RT-PCR assay, the most abundant miRNA in both sera and HBsAg particles was miR-122 (Supplementary Table 2).
Different Levels of Serum miRNAs in the Responsive and Nonresponsive CHB Patients
To examine if selected candidate miRNAs are involved in responsiveness to YIC, we compared the levels of serum miRNAs at baseline between these 2 groups. Levels of 5 serum miRNAs (let-7f, miR-22, miR-30a, miR-106b, and miR-122) were significantly lower in the responders group compared with those in the nonresponders group at baseline (week 0) (P = 0.0116, 0.0273, 0.0054, 0.0158, and 0.0091, respectively, Supplementary Table 3). Furthermore, with the exception of miR-106b (Figure 2D), the levels of 4 other miRNAs manifested a decline trend in the responders group compared to those in the nonresponders group over the study period (Figure 2A–C, E). Despite a slight increase in the levels of the serum miRNAs observed at the end of treatment (52 weeks) in the responders group, the overall levels of these miRNAs (except miR-106b) remained lower in the responders group compared to those in the nonresponders group at the end of follow-up (76 weeks) (Figure 2A–C, E).
FIGURE 2.
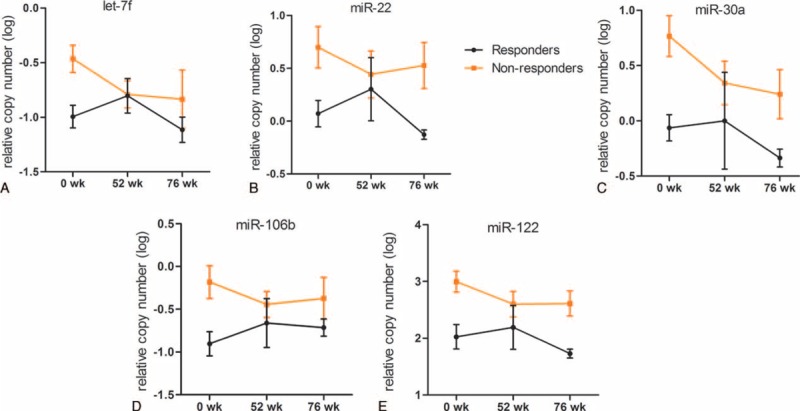
Differences in levels of miRNAs in sera of the responsive and nonresponsive patients at different time points. Kinetic changes of serum let-7f (A); miR-22 (B); miR-30a (C); miR-106b (D); miR-122 (E) in the responders and nonresponders groups during the study period. The relative levels of miRNAs were log10 transformed. Data were presented as means ± SD. miRNA = microRNA, SD = standard deviation.
Differences in HBsAg-Encapsidated miRNAs between the Responders and Nonresponders Groups
As described above, 8 miRNAs were detected in purified HBsAg particles with miR-122 being the most abundant. Our results demonstrated that 4 HBsAg-carried miRNAs, namely let-7f, miR-22, miR-30a, and miR-122, were significantly lower in the responders group compared to those in the nonresponders group at baseline (P = 0.0090, 0.0300, 0.0236, and 0.0075, respectively, Supplementary Table 4 and Figure 3 A-D), which was consistent with the results obtained using sera. In the course of YIC treatment, the levels of HBsAg-carried let-7f, miR-22, miR-30a, and miR-122 showed notable increase in the responders group and exceeded the levels observed in the nonresponders group at week 52 of YIC treatment (Figure 3A–D). This increase, however, ended and the levels of all 4 miRNAs in the responders group decreased and remained lower compared to their levels in the nonresponders group, which exhibited an upward trend at week 76 (Figure 3A–D).
FIGURE 3.
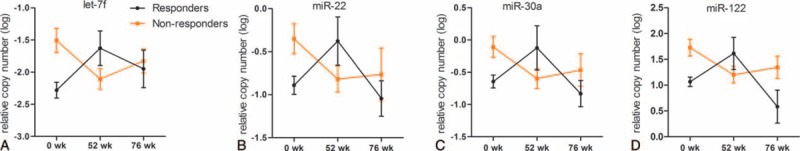
Differences in levels of HBsAg-carried miRNAs between the responders and nonresponders. Kinetic changes of 4 HBsAg-carried miRNAs—let-7f (A); miR-22 (B); miR-30a (C); miR-122 (D)—in the responsive and nonresponsive patients over the study period. The relative levels of miRNAs were log10 transformed. Data were presented as means ± SD. HBsAg = hepatitis B surface antigen, miRNA = microRNA, SD = standard deviation.
Relationships Between Baseline Levels of HBsAg-Carried miRNA and HBeAg Clearance or HBeAg Seroconversion
To further determine the association between HBsAg-carried miRNAs at baseline and HBeAg clearance, patients were separated into 2 groups according to the HBeAg clearance at week 76. Five patients were included in the HBeAg negative group (N = 5) and another 5 in the HBeAg positive group (N = 5). As shown in Figure 4A, the baseline levels of 4 HBsAg-carried miRNAs (let-7f, miR-22, miR-30a, and miR-122) correlated with HBeAg clearance (P = 0.0221–0.0061, Figure 4A). Furthermore, Kaplan–Meier analysis was used to evaluate the correlation between baseline levels of HBsAg-carried miRNAs and HBeAg seroconversion. With the exception of miR-22 (P = 0.3585, Figure 4A), lower baseline miRNAs levels correlated with higher rate of HBeAg seroconversion along with YIC treatment, thus suggesting that baseline levels of let-7f, miR-30a, miR-122 could discriminate HBeAg seroconverted patients from HBeAg nonconverted patients (P = 0.0357, Figure 4B). The relationships between baseline levels of serum miRNAs (let-7f, miR-22, miR-30a, miR-106b, and miR-122) and HBeAg clearance or HBeAg seroconversion were consistent with results obtained from HBsAg-carried miRNAs (Supplementary Figure 2).
FIGURE 4.
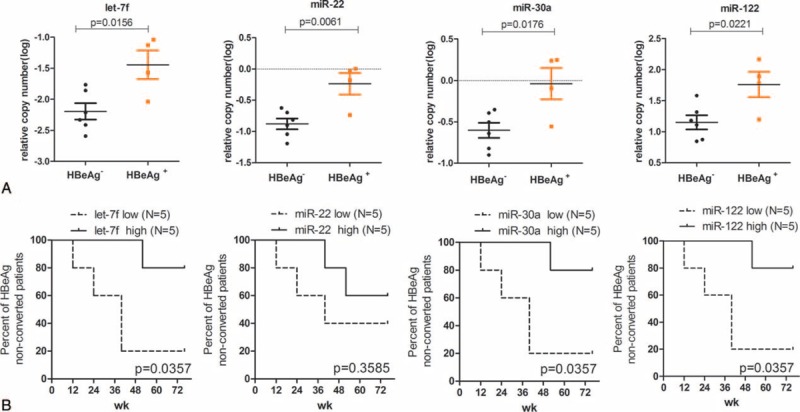
Relationships between baseline levels of HBsAg-carried miRNA and HBeAg clearance or HBeAg seroconversion. Relationships between baseline levels of 4 HBsAg-carried miRNAs and HBeAg clearance (A). Patients were separated into 2 groups based on the HBeAg clearance at week 76. The statistical differences of miRNAs levels between the HBeAg negative (N = 5) and HBeAg positive (N = 5) groups were analyzed by 1-way ANOVA. Kaplan–Meier curves based on differentially expressed 4 HBsAg-carried miRNAs (B). Based on the median value of baseline miRNA levels, patients were divided into 2 groups, expression levels high group (above the median value) and expression levels low group (below the median value), and Kaplan–Meier curves were used to evaluate the relationships of HBeAg seroconversion between the miRNA expression levels high and low groups. Lower baseline miRNA levels inversely correlated with HBeAg seroconversion. ANOVA = analysis of variance, HBeAg = hepatitis Be antigen, HBsAg = hepatitis B surface antigen, miRNA = microRNA.
DISCUSSION
The therapeutic strategy of YIC is to modulate host immune responsiveness through a forced uptake of HBsAg, together with the Fc portion of the complexed HBIG into antigen presenting cells. In this context, HBsAg is recognized by antigen presenting cells, processed and presented to T cells triggering an effective immunity.17 The effects of YIC on induction of cytolytic and noncytolytic responses have been reported.20 Currently, YIC is under the second stage of phase III clinical trial. We observed that CHB patients infected with HBV genotype B responded better to YIC treatment compared with those infected with genotype C, suggesting that there are distinct host factors that determine the responsiveness to YIC therapy. With this in mind, we attempted to identify some of those host factors whose characterization may lead to development of personalized treatments.
Since their discovery in 1993, miRNAs have been shown as critical regulators of gene expression, and their roles have been described widely in physiologic and pathologic processes.21–23 In 2008, detection and characterization of miRNA in sera transformed their use from biological research into clinical practice, and since then, miRNAs have been suggested as biomarkers in a number of diseases.24–29 Although there is a lack of consensus on the expression of miRNA in response to external stimuli and on miRNA-mediated perturbations of signaling networks, a number of population-based clinical studies have been conducted. Among these, miR-122 has been studied in detail in CHB patients.30–32 MiR-122 is the most abundant miRNA in the liver and takes part in the life cycle of HBV.33 Hence, it represented a suitable candidate for our studies. The selection of other candidate miRNAs was based on the results of our previous study, which focused on the involvement of miRNAs in modulation of IFN treatment outcome.16 Moreover, the selection was guided by the fact that the immunomodulatory function of YIC is related to that of the type I interferons(s), critical for innate immune responses and antiviral effects through upregulation of interferon-stimulated genes. In addition, we also reported that exosome released from IFN-α–treated hepatic nonparenchymal cell contained increased amounts of miR-638, miR-4284, and miR-1260a and these 3 exosomal miRNAs could suppress the replication of HBV.18 We therefore also selected these miRNAs for the current study (Supplementary Table 2).
To avoid environmental factors that may influence the host responses, the 10 patients who were among 136 patients involved in the phase III clinical trial were selected from 2 independent hospitals in Shanghai, China. However, because this study was a retrospective assay for miRNAs in sera and HBsAg particles, the number of serum samples available and their volume were very limited. In view of these limitations of the pilot study, we examined the presence of 13 miRNAs in sera and purified HBsAg particles derived from samples of 10 patients at 3 time points. Nonetheless, when the baseline clinical data of selected 5 responders and 5 nonresponders, within HBeAg seroconverted and nonconverted groups, were compared with all other patients infected with HBV genotype B, no statistical significant differences were observed (Supplementary Table 1). Hence, the results of our analysis represented a trend observed in a larger cohort of patients infected with HBV genotype B who responded or did not respond to YIC treatment.
To evaluate the implications of the 13 selected miRNAs in YIC treatment, we analyzed their levels in serum and HBsAg particles derived from CHB patients. Although 13 miRNAs were detected in sera, only 8 miRNAs could be detected in HBsAg particles (let-7f, miR-22, miR-30a, miR-106b, miR-122, miR-1224, miR-1260a, and miR-1281, Supplementary Table 2), suggesting that the miRNAs carried by HBsAg were secreted from hepatocytes, and their expression may be closely associated with biological processes in the liver. In contrast, miRNAs detected in serum could be derived from other sources and may not be directly associated with biological processes in hepatocytes. Our study using a limited number of CHB patients is in agreement with the recent report by Novellino et al32 demonstrating that subviral HBsAg particles are associated with liver-specific miRNAs including miR-27a, miR-30b, miR-122, miR-126, and miR-145 as well as with miRNA manifesting immune regulatory function such as miR-106b and miR-223.
Differences in levels of the 8 miRNAs present in both sera and HBsAg particles were compared between the patients who responded and did not respond to YIC treatment. Our results demonstrated that 5 miRNAs, namely let-7f, miR-22, miR-30a, miR-106b, and miR-122, were present at lower levels in sera of patients in the responders group at baseline (Figure 2). Moreover, the low level of let-7f, miR-22, miR-30a, and miR-122 in serum was mirrored by their association with HBsAg particles purified from samples of patients in the responders group (Figure 3). We also found that HBsAg, HBeAg, and HBV DNA titers were lower in the responders group at baseline and showed a further decline throughout the course of treatment, while the variation trend of HBsAg, HBeAg, and DNA titers in the nonresponders group tended to only modestly level off over the study period (Supplementary Figure 1 A, B, D). However, serum miRNAs (Figure 2) and HBsAg-carried miRNAs (Figure 3) in the responders and nonresponders groups manifested a variation trend that differed from the level of HBsAg, HBeAg, and HBV DNA titers (Supplementary Figure 1). Namely, serum miRNAs and HBsAg-carried miRNAs in the responders group were significantly lower compared to those in the nonresponders group at baseline and increased as YIC treatment for 52 weeks, and then decreased and remained lower compared to the nonresponders group at week 76. Especially, HBsAg-carried miRNAs in the responders group increased and exceeded the miRNAs levels in nonresponders group at the week 52 (Figure 3). But the variation trend of serum miRNAs and HBsAg-carried miRNAs in the nonresponders group was opposite to observed in the responders group. This suggests that levels of miRNAs have no direction correlation with HBsAg, HBeAg, and HBV DNA titers. Our results also demonstrated that the baseline levels of serum miRNAs and miRNAs in HBsAg particles were associated with YIC treatment outcome, such as HBeAg clearance at week 76 (Figure 4A; Supplementary Figure 2A). Besides, with the exception of miR-22 (P = 0.3585); the baseline levels of serum miRNAs (let-7f, miR-30a, miR-122) and miRNAs in HBsAg particles were also correlated with HBeAg seroconversion along with YIC treatment (Figure 4B, P = 0.0357; Supplementary Figure 2B). These results therefore suggest that the baseline levels of serum miRNAs and miRNAs in HBsAg-particles and their variation trends could have a prognostic value for responsiveness to YIC treatment.
MiR-122 accounts for approximately 70% and 52% of all hepatic miRNA in mice and humans, respectively,19 and its expression has been shown to correlate with hepatic necroinflammation in patients with viral hepatitis. Waidmann et al34 observed close correlation between the expression of miR-122 and the levels of ALT, serum HBV DNA, and HBsAg in plasma of HBV-infected carriers and patients. Furthermore, miR-122 levels could be correlated with surrogate markers for viral replication and translation.34 Based on these observations, it was suggested that the levels of miR-122 could be used to differentiate patients with high or low risk for disease progression.34 In this pilot study, we further showed that a low level of miR-122 was associated with favorable response to YIC treatment, suggesting that miR-122 might be used as a marker for responsiveness to therapeutic vaccination. The other miRNAs, such as miR-106b and miR-30a, examined in this study, were also expressed at lower levels in the responders group compared to the nonresponders group. MiR-106b displays multifunctions, mainly involved in oncogenesis and to a less extent associated with angiogenesis, and had been reported to promote cell proliferation and impair TGF-β signaling.35 Overexpression of miR-106b-25 and miR-17–92 clusters has been reported not only during the development of liver cirrhosis but also in the subsequent development of hepatocellular carcinoma.36 Members of miR-17–92 clusters have also been shown to affect the replication of HBVs.37 Exogenous overexpression of miR-30a and miR-30c2 in cultured cells has been shown to play crucial roles in development of hepatobiliary and hepatocellular carcinoma and cholangiocarcinoma.38
In summary, the results of this pilot study identified some candidate miRNAs in serum and HBsAg particles (let-7f, miR-22, miR-30a, and miR-122) whose baseline levels and trends observed in the course of YIC treatment may constitute an important factor in determining the responsiveness to YIC. However, due to the limited number of patients in this study, the levels of identified miRNAs and their implications and association with responsiveness to YIC or other therapeutic vaccinations require verification in more CHB patients in future clinical studies.
Supplementary Material
Footnotes
Abbreviations: CHB = chronic hepatitis B; HBeAg = hepatitis Be antigen; HBIG = anti-HBs immunoglobulin; HBsAg = hepatitis B surface antigen; IFN = interferon; miRNA = microRNA; YIC = antigen-antibody immunogenic complex based therapeutic vaccine.
This work was supported by the National Key Basic Research Program of China (2012CB519000), the “Twelfth Five-Year” National Key Technology Research and Development Programs of China (2012ZX10002007), the German Research Foundation (SFB/Transregio TRR60).
WW, JL, and XZ contributed equally to this work.
The authors have no funding and conflicts of interest to disclose.
REFERENCES
- 1.Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat 2004; 11:97–107. [DOI] [PubMed] [Google Scholar]
- 2.Zoulim F, Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology 2009; 137:1593–1608.e1591-1592. [DOI] [PubMed] [Google Scholar]
- 3.Rijckborst V, Hansen BE, Cakaloglu Y, et al. Early on-treatment prediction of response to peginterferon alfa-2a for HBeAg-negative chronic hepatitis B using HBsAg and HBV DNA levels. Hepatology 2010; 52:454–461. [DOI] [PubMed] [Google Scholar]
- 4.Horiike N, Fazle Akbar SM, Michitaka K, et al. In vivo immunization by vaccine therapy following virus suppression by lamivudine: a novel approach for treating patients with chronic hepatitis B. J Clin VirolV 32 2005; 156–161. [DOI] [PubMed] [Google Scholar]
- 5.Michel ML, Deng Q, Mancini-Bourgine M. Therapeutic vaccines and immune-based therapies for the treatment of chronic hepatitis B: perspectives and challenges. J Hepatol 2011; 54:1286–1296. [DOI] [PubMed] [Google Scholar]
- 6.Vandepapeliere P, Lau GK, Leroux-Roels G, et al. Therapeutic vaccination of chronic hepatitis B patients with virus suppression by antiviral therapy: a randomized, controlled study of co-administration of HBsAg/AS02 candidate vaccine and lamivudine. Vaccine 2007; 25:8585–8597. [DOI] [PubMed] [Google Scholar]
- 7.Yang FQ, Yu YY, Wang GQ, et al. A pilot randomized controlled trial of dual-plasmid HBV DNA vaccine mediated by in vivo electroporation in chronic hepatitis B patients under lamivudine chemotherapy. J Viral Hepat 2012; 19:581–593. [DOI] [PubMed] [Google Scholar]
- 8.Xu DZ, Huang KL, Zhao K, et al. Vaccination with recombinant HBsAg-HBIG complex in healthy adults. Vaccine 2005; 23:2658–2664. [DOI] [PubMed] [Google Scholar]
- 9.Xu DZ, Wang XY, Shen XL, et al. Results of a phase III clinical trial with an HBsAg-HBIG immunogenic complex therapeutic vaccine for chronic hepatitis B patients: experiences and findings. J Hepatol 2013; 59:450–456. [DOI] [PubMed] [Google Scholar]
- 10.Xu DZ, Zhao K, Guo LM, et al. A randomized controlled phase IIb trial of antigen-antibody immunogenic complex therapeutic vaccine in chronic hepatitis B patients. PloS One 2008; 3:e2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 12.Garzon R, Fabbri M, Cimmino A, et al. MicroRNA expression and function in cancer. Trends Mol Med 2006; 12:580–587. [DOI] [PubMed] [Google Scholar]
- 13.Wang XY, Zhang XX, Yao X, et al. Serum HBeAg sero-conversion correlated with decrease of HBsAg and HBV DNA in chronic hepatitis B patients treated with a therapeutic vaccine. Vaccine 2010; 28:8169–8174. [DOI] [PubMed] [Google Scholar]
- 14.Yuen MF, Hui CK, Cheng CC, et al. Long-term follow-up of interferon alfa treatment in Chinese patients with chronic hepatitis B infection: the effect on hepatitis B e antigen seroconversion and the development of cirrhosis-related complications. Hepatology 2001; 34:139–145. [DOI] [PubMed] [Google Scholar]
- 15.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008; 3:1101–1108. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Chen C, Wu M, et al. Plasma microRNA profile as a predictor of early virological response to interferon treatment in chronic hepatitis B patients. Antivir Ther 2012; 17:1243–1253. [DOI] [PubMed] [Google Scholar]
- 17.Wen YM. Antigen-antibody immunogenic complex: promising novel vaccines for microbial persistent infections. Expert Opin Biol Ther 2009; 9:285–291. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Liu K, Liu Y, et al. Exosomes mediate the cell-to-cell transmission of IFN-alpha-induced antiviral activity. Nat Immunol 2013; 14:793–803. [DOI] [PubMed] [Google Scholar]
- 19.Bandiera S, Pfeffer S, Baumert TF, et al. miR-122–a key factor and therapeutic target in liver disease. J Hepatol 2015; 62:448–457. [DOI] [PubMed] [Google Scholar]
- 20.Yao X, Zheng B, Zhou J, et al. Therapeutic effect of hepatitis B surface antigen-antibody complex is associated with cytolytic and non-cytolytic immune responses in hepatitis B patients. Vaccine 2007; 25:1771–1779. [DOI] [PubMed] [Google Scholar]
- 21.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Developmental cell 2006; 11:441–450. [DOI] [PubMed] [Google Scholar]
- 22.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 2008; 9:219–230. [DOI] [PubMed] [Google Scholar]
- 23.Ambros V. The functions of animal microRNAs. Nature 2004; 431:350–355. [DOI] [PubMed] [Google Scholar]
- 24.Gilad S, Meiri E, Yogev Y, et al. Serum microRNAs are promising novel biomarkers. PloS One 2008; 3:e3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wan WN, Zhang YQ, Wang XM, et al. Down-regulated miR-22 as predictive biomarkers for prognosis of epithelial ovarian cancer. Diagn Pathol 2014; 9:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giray BG, Emekdas G, Tezcan S, et al. Profiles of serum microRNAs; miR-125b-5p and miR223-3p serve as novel biomarkers for HBV-positive hepatocellular carcinoma. Mol Biol Rep 2014; 41:4513–4519. [DOI] [PubMed] [Google Scholar]
- 27.Fang C, Zhu DX, Dong HJ, et al. Serum microRNAs are promising novel biomarkers for diffuse large B cell lymphoma. Ann Hematol 2012; 91:553–559. [DOI] [PubMed] [Google Scholar]
- 28.Liu HS, Xiao HS. MicroRNAs as potential biomarkers for gastric cancer. World J Gastroenterol 2014; 20:12007–12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 2009; 58:1375–1381. [DOI] [PubMed] [Google Scholar]
- 30.Tsai WC, Hsu PW, Lai TC, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology 2009; 49:1571–1582. [DOI] [PubMed] [Google Scholar]
- 31.Hao J, Jin W, Li X, et al. Inhibition of alpha interferon (IFN-alpha)-induced microRNA-122 negatively affects the anti-hepatitis B virus efficiency of IFN-alpha. J Virol 2013; 87:137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novellino L, Rossi RL, Bonino F, et al. Circulating hepatitis B surface antigen particles carry hepatocellular microRNAs. PloS One 2012; 7:e31952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S, Qiu L, Yan X, et al. Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G(1) -modulated P53 activity. Hepatology 2012; 55:730–741. [DOI] [PubMed] [Google Scholar]
- 34.Waidmann O, Bihrer V, Pleli T, et al. Serum microRNA-122 levels in different groups of patients with chronic hepatitis B virus infection. J Viral Hepat 2012; 19:e58–65. [DOI] [PubMed] [Google Scholar]
- 35.Yu D, Shin HS, Lee YS, et al. miR-106b modulates cancer stem cell characteristics through TGF-beta/Smad signaling in CD44-positive gastric cancer cells. Lab Invest 2014; 94:1370–1381. [DOI] [PubMed] [Google Scholar]
- 36.Tan W, Li Y, Lim SG, et al. miR-106b-25/miR-17-92 clusters: polycistrons with oncogenic roles in hepatocellular carcinoma. World J Gastroenterol 2014; 20:5962–5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jung YJ, Kim JW, Park SJ, et al. c-Myc-mediated overexpression of miR-17-92 suppresses replication of hepatitis B virus in human hepatoma cells. J Med Virol 2013; 85:969–978. [DOI] [PubMed] [Google Scholar]
- 38.Le Guen CL, Friedman JR, Hand NJ. Novel targets of miR-30, a microRNA required for biliary development. F1000Res 2013; 2:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


