Abstract
The alternative nonhomologous end-joining (alt-NHEJ) machinery facilitates a number of genomic rearrangements, some of which can lead to cellular transformation. This error-prone repair pathway is triggered upon telomere de-protection to promote the formation of deleterious chromosome end-to-end fusions1,2,3. Using next-generation sequencing technology, we found that repair by alt-NHEJ yields non-TTAGGG nucleotide insertions at fusion breakpoints of dysfunctional telomeres. Investigating the enzymatic activity responsible for the random insertions enabled us to identify Polymerase theta (Polθ; encoded by PolQ) as a critical alt-NHEJ factor in mammalian cells. PolQ inhibition suppresses alt-NHEJ at dysfunctional telomeres, and hinders chromosomal translocations at non-telomeric loci. In addition, we found that PolQ loss results in increased rates of homology directed repair (HDR), evident by recombination of dysfunctional telomeres and accumulation of Rad51 at double stranded breaks. Lastly, we show that depletion of PolQ has a synergistic impact on cell survival in the absence of BRCA genes, suggesting that the inhibition of this mutagenic polymerase represents a valid therapeutic avenue for tumors carrying mutations in HDR genes.
Keywords: PolQ, Telomere, Alternative-NHEJ, Homology directed repair, shelterin, BRCA1
Chromosome end-to-end fusions are inhibited by shelterin; a multi-subunit complex anchored to telomeric DNA using two myb-containing proteins – TRF1 and TRF24. Telomere fusions are executed by two independent end-joining pathways. Classical non-homologous end-joining (C-NHEJ), mediated by Lig4 and Ku70/80, is primarily blocked by TRF25. On the other hand, alternative-NHEJ (alt-NHEJ), dependent on Lig36 and PARP17, is repressed in a redundant manner2,3. Alt-NHEJ is fully unleashed following the simultaneous deletion of TRF1 and TRF2, and the creation of shelterin-free telomeres in Ku70/80 deficient cells2. This error-prone end-joining pathway mediates fusion of naturally eroded telomeres1, joining of switch regions during class switch recombination8, and formation of chromosomal translocations in mouse cells9,10.
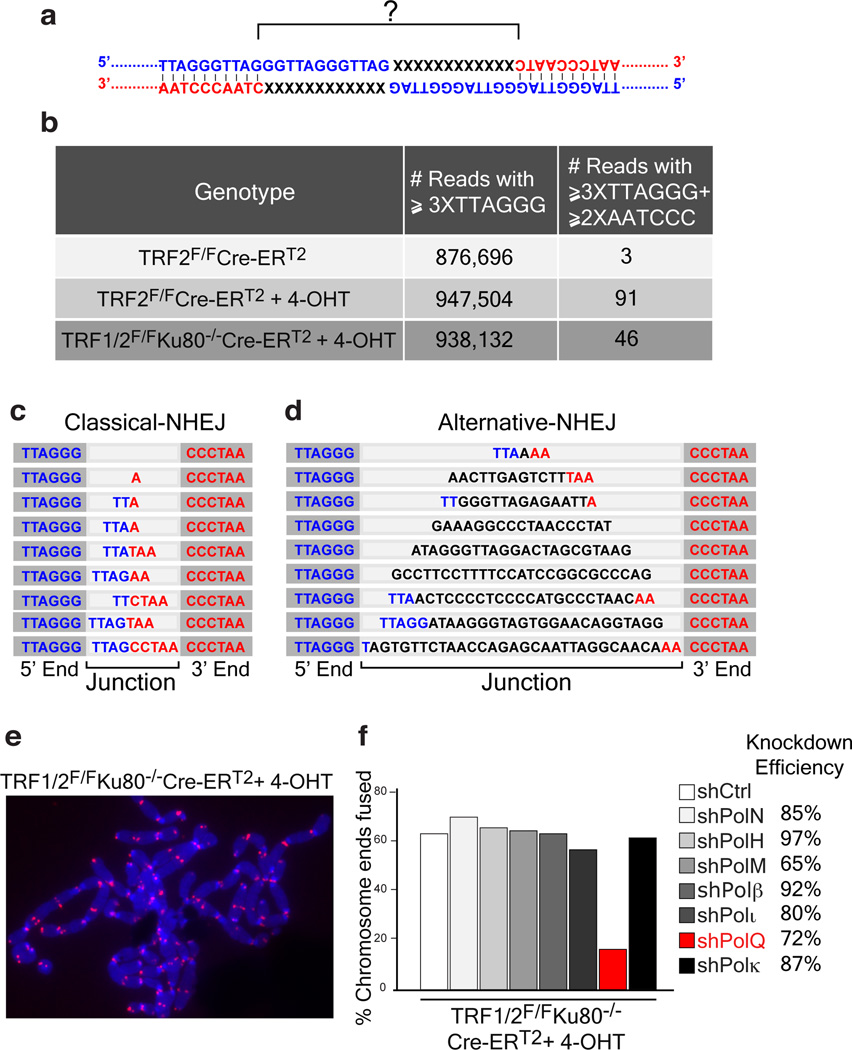
In order to characterize the differences between C-NHEJ and alt-NHEJ at dysfunctional telomeres, we determined whether the sequence of the junction between two fused telomeres differed depending on the type of repair pathway utilized. Telomere fusions by C-NHEJ were triggered by Cre-mediated depletion of TRF2 using previously described mouse embryonic fibroblasts (MEFs) (TRF2F/FCre-ERT2)11(Extended Data Fig.1a). To induce robust fusions by the alt-NHEJ pathway, we depleted the entire shelterin complex by deleting TRF1 and TRF2 from TRF1F/FTRF2F/FKu80−/−Cre-ERT2 MEFs (Extended Data Fig.1a)2. DNA was subjected to next-generation sequencing, and reads corresponding to telomeres were identified based on the presence of at least three consecutive TTAGGG repeats. To detect rare reads containing fusion junctions, we exploited the novel sequence arrangement created by the ligation of the 3’ G-rich strand (TTAGGG-3’) to the 5’ C-rich strand (5’-CCCTAA) (Fig. 1a), and filtered reads that started with at least three G-rich repeats and ended with two or more C-rich repeats. We confirmed that this approach could successfully identify telomere fusions by comparing reads derived from TRF2-proficient and TRF2-deficient cells. Starting with a similar number of telomere-repeat containing reads, we identified >90 fusogenic events in TRF2 knockout MEFs, compared to only three events in wild-type cells (Fig. 1b). Sequence analysis of the junctions highlighted different permutations of TTAGGG/AATCCC sequences. Interestingly, the spectrum of the fusion junctions was different in shelterin-free settings, where frequent non-telomeric nucleotide insertions (9/46 events) were identified at fusion breakpoints (Fig. 1b–d and Supplementary Information).
Figure 1. Random nucleotide insertions at the junction of telomeres fused by alt-NHEJ.
a, Schematic of the junction of a telomere fusion. The 3’ end of the telomeric G-rich strand of a chromosome (Blue) is fused to the 5’ end of the C-rich strand of a different chromosome (Red). b, Illumina sequencing to analyze telomere fusion junctions. Reads ≥3XTTAGGG consecutively were scored as derived from telomeric fragments. Those with ≥3XTTAGGG on the 5’-end and ≥2XCCCTAA at the 3’-end were scored as telomere fusion junctions (see Supplementary Information). c, Examples of telomere fusions generated by C-NHEJ from TRF2 depleted telomeres. Light gray highlights fusion junctions, dark grey marks the flanking telomere repeats. d, Examples of insertions in shelterin-free/Ku80 null MEFs. e, Telomere fusions in metaphase spreads from TRF1F/FTRF2F/FKu80−/−Cre-ERT2 MEFs. Telomeres in red (PNA probe) and chromosomes in blue (DAPI). f, Frequency of telomere fusions following the depletion of candidate polymerases.
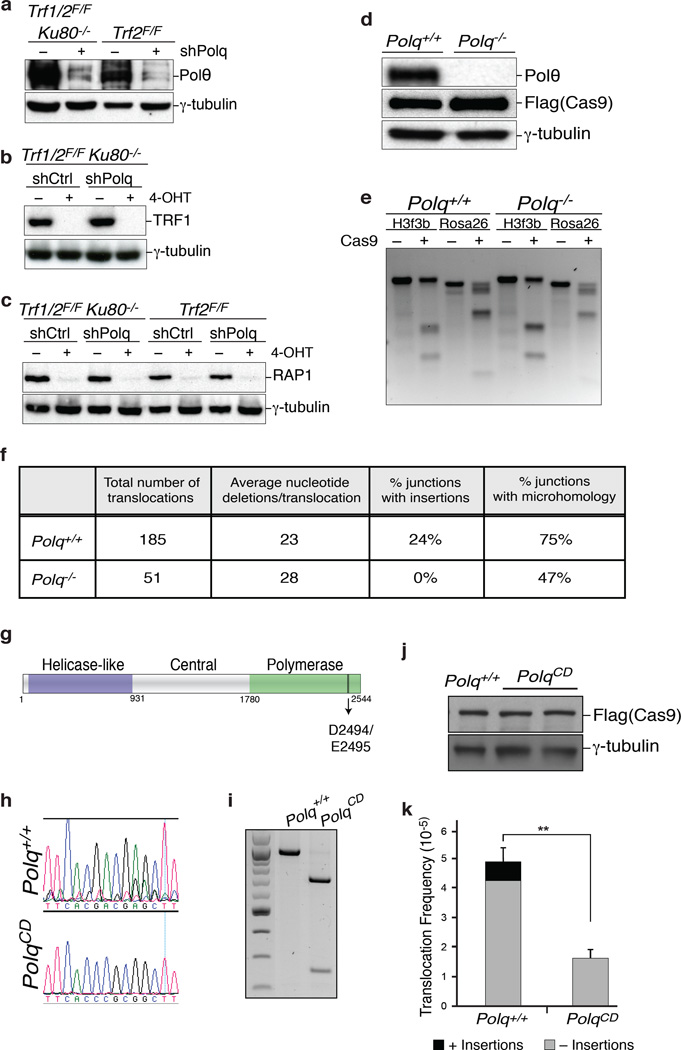
To identify the enzyme that incorporated nucleotides at dysfunctional telomeres, we depleted known low-fidelity DNA polymerases in shelterin-free cells lacking Ku80, and analyzed chromosome-end fusions on metaphase spreads. Strikingly, we observed a reduction in the frequency of telomere fusions in cells with reduced levels of polymerase theta (Polθ, encoded by PolQ) (Fig. 1e–f and Extended Data Fig.1b). The activity of Polθ is specific to alt-NHEJ since its inhibition in TRF2 knockout cells did not impact the frequency of C-NHEJ (Fig. 2a–b and Extended Data Fig.2a–c).
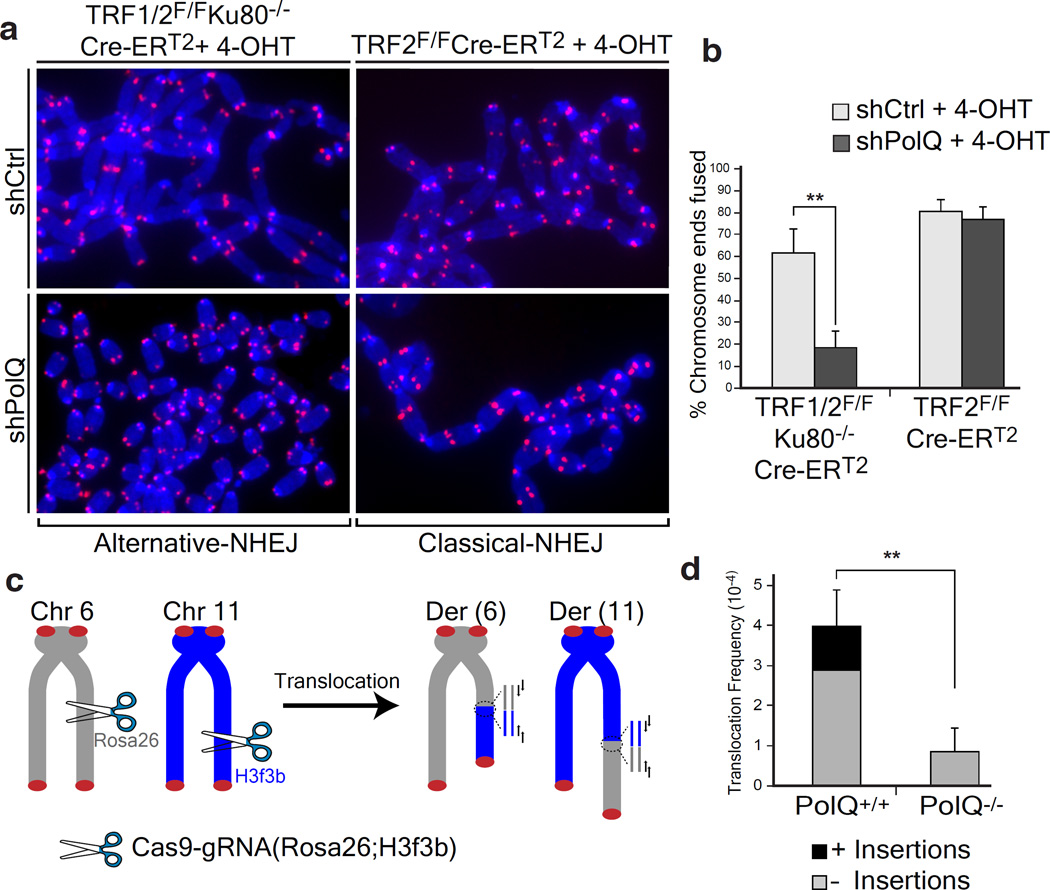
Figure 2. Polθ is required for alt-NHEJ dependent DSB repair in mammalian cells.
a, Metaphases from TRF2 depleted (TRF2F/FCre-ERT2OHT) and shelterin free (TRF1F/FTRF2F/FKu80−/−Cre-ERT2OHT) MEFs infected with the indicated shRNA. b, Quantification of telomere fusions in MEFs with the indicated treatment (Mean ± SD, n=6, **p=0.003; two-tailed student’s t-test). c, Design of the translocation assay in which DSBs are induced by Cas9-gRNA(Rosa26;H3f3b). Joining of DNA ends generates Der(6) and Der(11), detected by nested PCR9. d, Translocation frequency in PolQ+/+ and PolQ−/− cells 60 hours post-Cas9-gRNA(Rosa26;H3f3b) expression. (Mean ± SD, n=3, ** p=0.009; two-tailed student’s t-test).
Polθ is an A-family DNA polymerases that exhibits low-fidelity on templated-DNA12, and also displays a terminal transferase-like activity that catalyzes nucleotide addition in a template-independent manner13. The relevance of these activities in vivo was highlighted in D. melanogaster, where Polθ was shown to stimulate nucleotide insertions during DSB repair by alt-NHEJ14. More recently, Polθ was shown to promote end-joining of replication-associated DSBs in C. elegans15, preventing large deletions around G-rich DNA16. The exact function of Polθ during DSB repair in mammalian cells remains elusive.
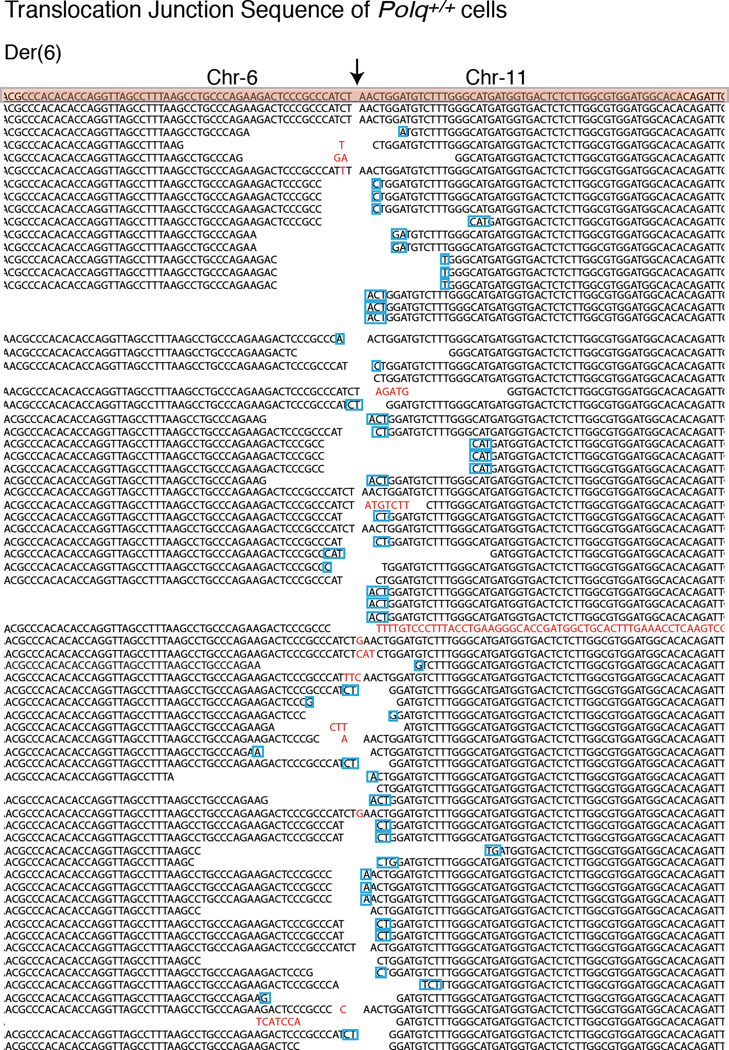
The critical role for Polθ at dysfunctional telomeres prompted us to test whether it is required for DSB repair at non-telomeric loci. To this end, we tested whether depletion of Polθ impacts chromosomal translocations in the context of mouse pluripotent cells, reported to be specifically mediated by alt-NHEJ10 in a Lig3-dependent manner9. To model chromosomal translocations, we induced DSBs in the Rosa26 and the H3f3B loci using CRISPR/Cas9 (Fig. 2c). When introduced into PolQ+/+ and PolQ−/− cells17, the Cas9-gRNA(Rosa26;H3f3b) expression plasmid induced simultaneous cleavage of the both loci with comparable efficiencies (Extended Data Fig.2d–e). Consistent with previous reports, 24% of translocation events in PolQ+/+ cells were scarred by random insertions. Strikingly, the overall frequency of translocations in cells lacking PolQ was significantly reduced (Fig. 2d). Sequence analysis of residual translocations in PolQ−/− cells highlighted the absence of insertions, and a concomitant decrease in micro-homology at junctions (Fig. 2d, Extended Data Fig.2–5). Notably, we observed similar results when assessing translocation frequency in cells expressing a catalytically inactive form of Polθ (Extended Data Fig.2g–k). Altogether, our data indicate that the promiscuous activity of Polθ during DSB repair contributes to the increased mutagenicity of alt-NHEJ. Importantly, our results suggest that mammalian Polθ plays a critical role by stimulating the end-joining reaction.
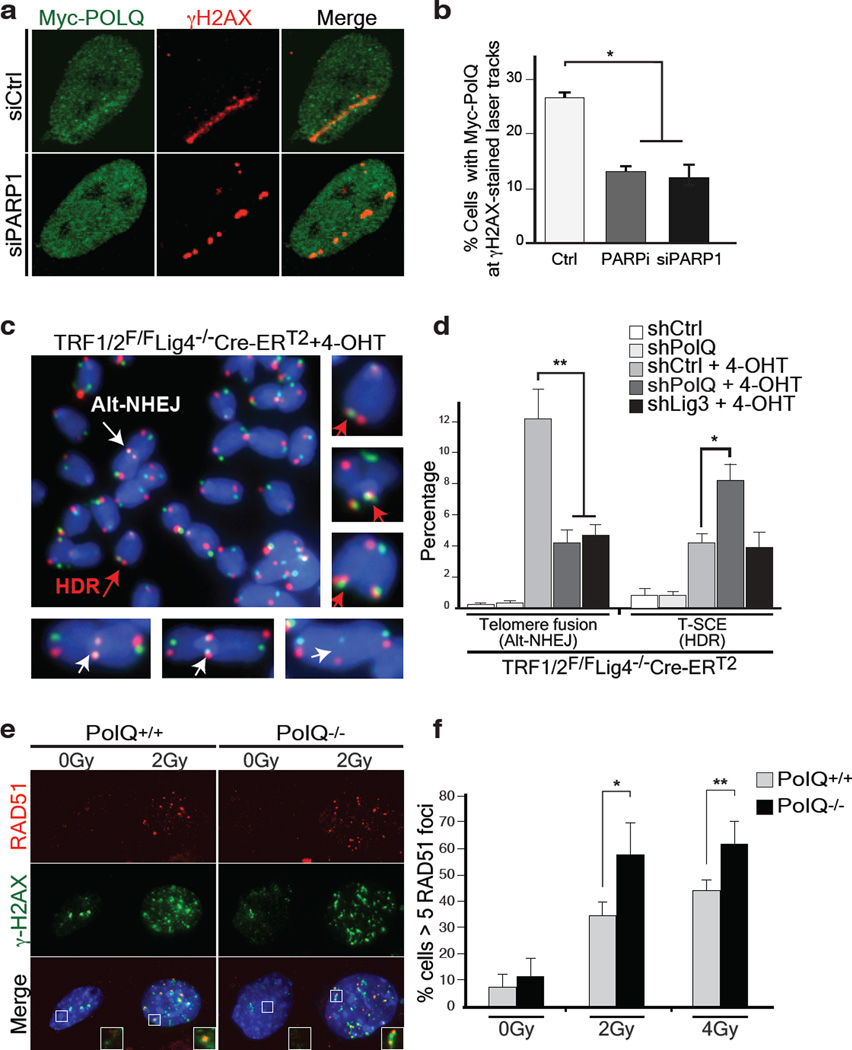
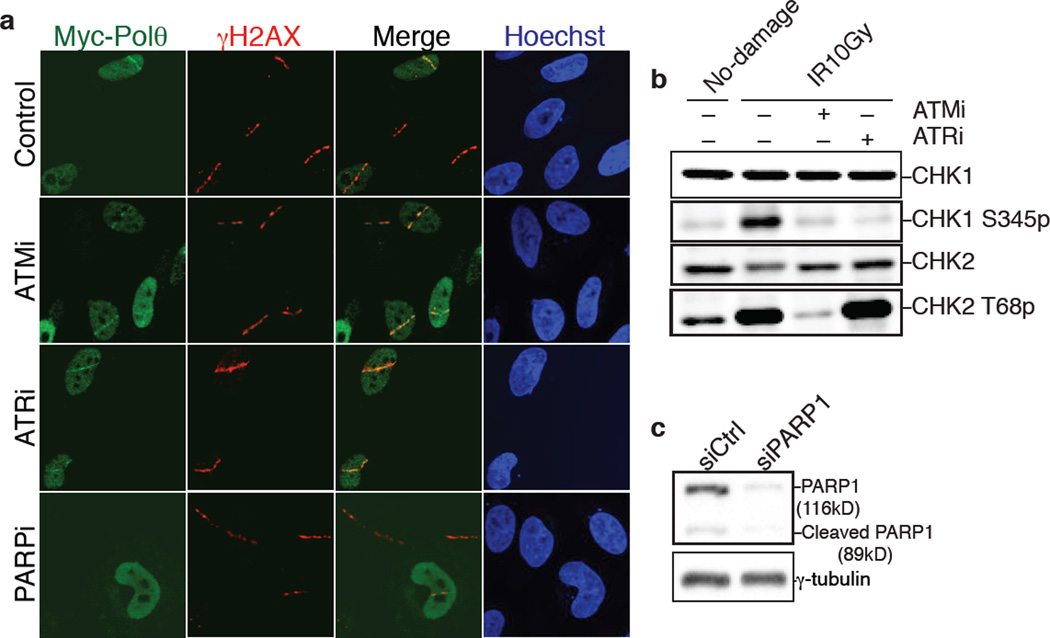
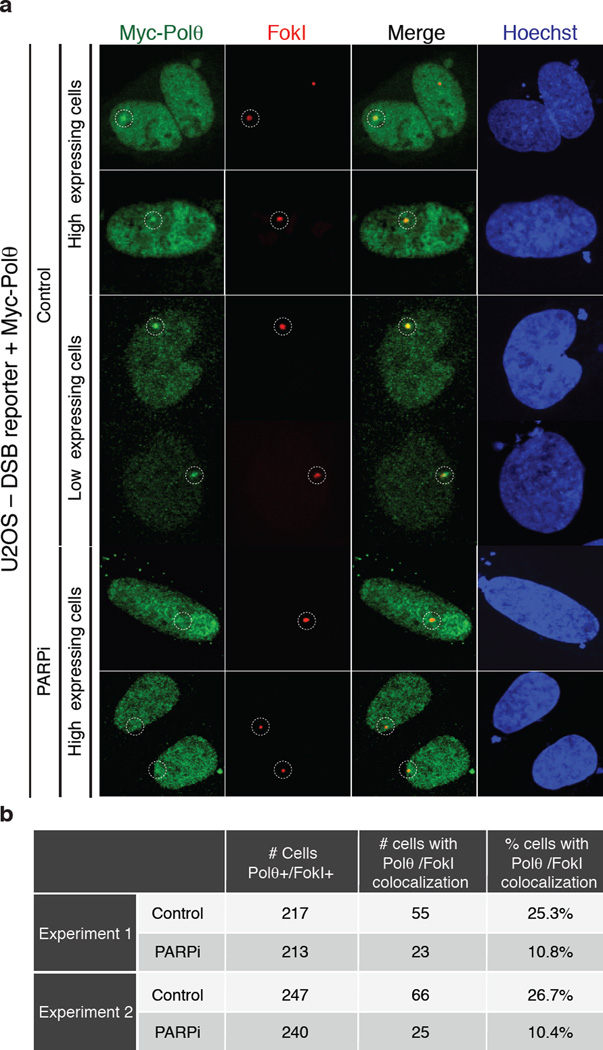
We next investigated the upstream signaling event(s) required for the recruitment of Polθ to DNA damage sites, induced upon micro-irradiation of HeLa cells expressing Myc-tagged PolQ. Accumulation of Polθ at laser-induced DNA breaks, discerned by its co-localization with γ–H2AX, occurred in ~25% of cells that stained positive for Myc (Fig. 3a–b), and was independent of either ATM or ATR signaling (Extended Data Fig.6). Instead, co-localization of Polθ with γ–H2AX was reduced upon depletion of PARP1 with siRNAs, or following the inhibition of PARP1 activity (KU58948) (Fig. 3a–b). In a parallel experiment that employed a recently developed U2OS-DSB reporter cell line18, we were able to ascertain the localization of Myc-PolQ to bona fide DSBs, induced upon Fok1 cleavage of a LacO-tagged genomic locus (Extended Data Fig.7). In conclusion, our data suggest that PARP1, previously known to be required for alt-NHEJ7,19, facilitates the recruitment of Polθ to DSBs.
Figure 3. Polθ is recruited by PARP1 to promote alt-NHEJ at the expense of HDR.
a, Myc-PolQ localization to DNA damage was monitored after laser micro-irradiation of HeLa cells. Cells were fixed and stained for γ–H2AX and Myc, one hour after damage induction. b, Quantification of Polθ accumulation at sites of laser damage (Mean ± s.e.m, n=2). c, To test if Polθ represses recombination at telomeres, we depleted the polymerase in shelterin-free and Lig4 deficient MEFs2, and both repair pathways were monitored using CO-FISH. White arrows indicate alt-NHEJ events, red arrows highlight HDR-mediated T-SCEs. d, Quantification of telomere fusion (alt-NHEJ) and T-SCE (HDR) in cells transduced with PolQ, Lig3, or control shRNA. e, Immunofluorescence for Rad51 and γ-H2AX in the indicated MEFs 3 hours post-irradiation. f, Graph representing quantification of IR-induced RAD51 foci. (Mean ± SD, n=3, *p<0.05, **p<0.01;two-tailed student’s t-test).
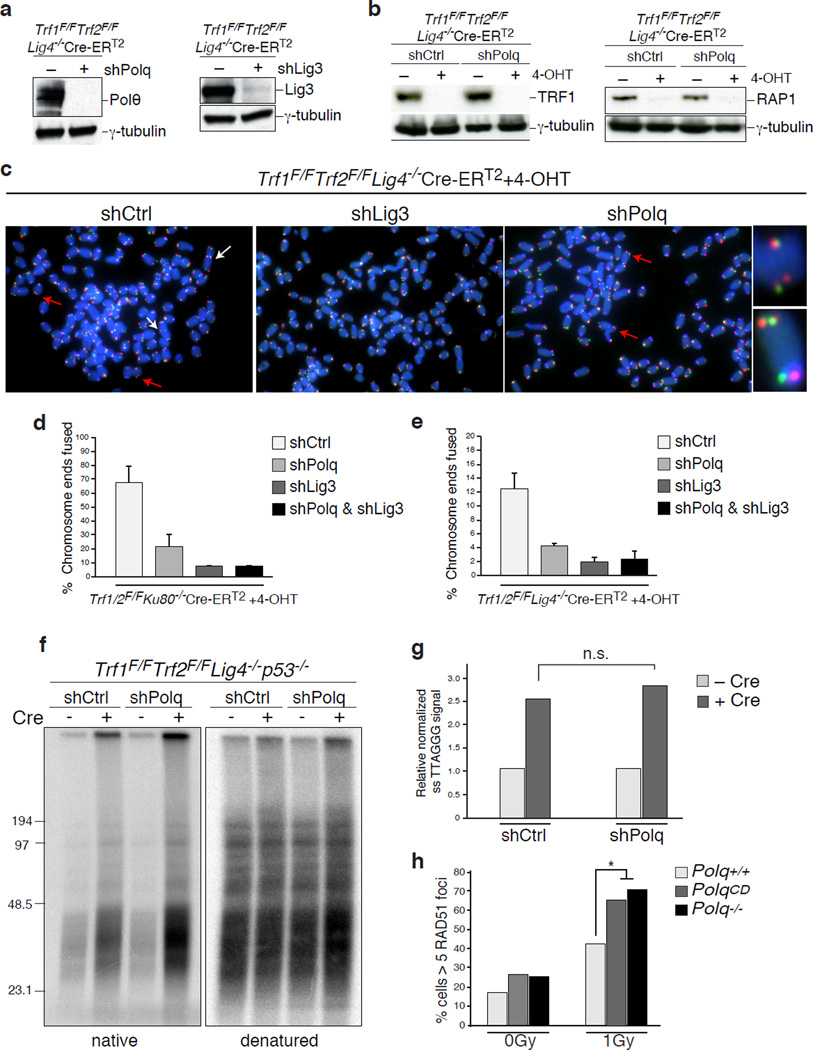
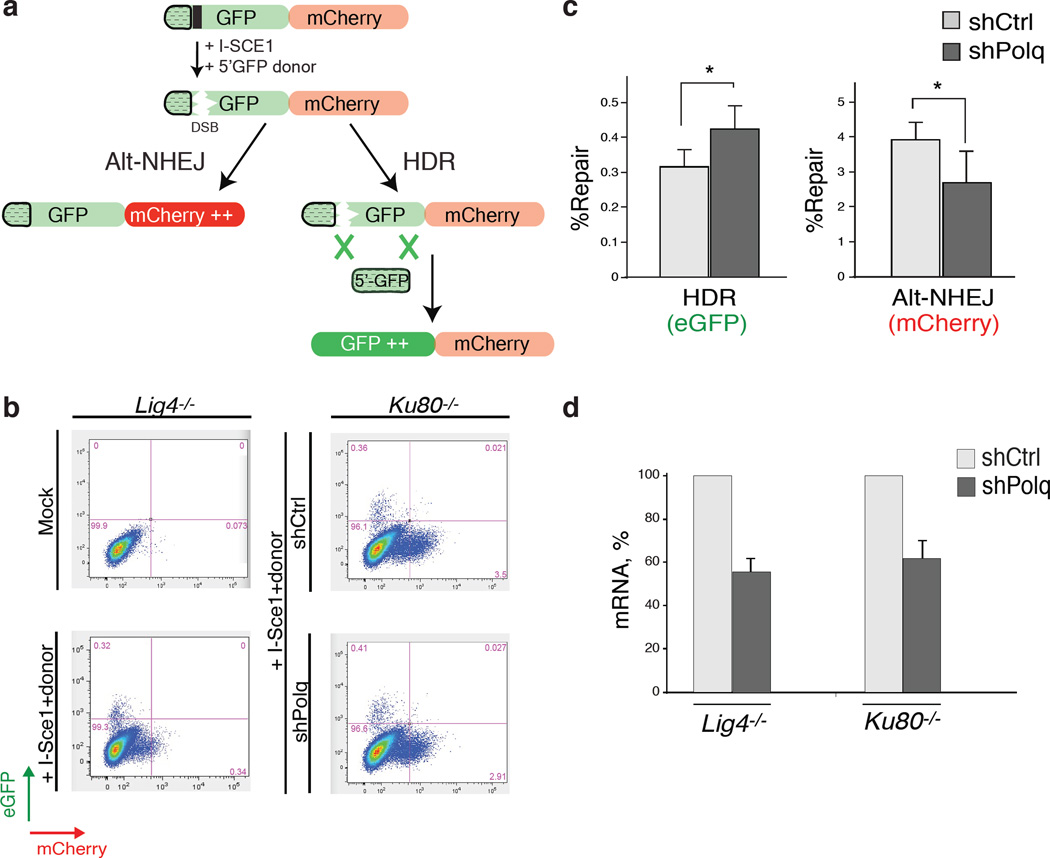
Homology-directed repair (HDR) is prevalent during the S-G2 phases of the cell cycle, which coincides with the peak of alt-NHEJ activity, and these pathways also share the initial resection step mediated by Mre11/CtIP20. To test whether inhibiting alt-NHEJ could potentially result in increased HDR, we depleted shelterin in Lig4 deficient MEFs, a genetic setting that is conducive to the activity of NHEJ as well as HDR2. To investigate the relative contribution of the two repair pathways we used the Chromosome-Orientation FISH (CO-FISH) assay21, and monitored the exchange of telomeres between sister chromatids by HDR (T-SCE: telomere sister chromatid exchange), and at the same time, measured the frequency of chromosome end-end fusion by end-joining (Fig. 3c). Following depletion of shelterin from TRF1F/FTRF2F/FLig4−/−Cre-ERT2 MEFs, ~10% of the telomeres were processed by alt-NHEJ, while ~5% of chromosome ends showed T-SCEs2 (Fig. 3c–d and Extended Data Fig.8a–c). As expected, we observed a substantial reduction in the frequency of alt-NHEJ at shelterin-free telomeres in Lig4−/− cells that lack Polθ or Lig3 (Fig. 3d and Extended Data Fig.8a–e). Remarkably, PolQ depleted cells exhibited a concomitant increase in T-SCE, which was not evident in cells lacking Lig3 (Fig. 3d), thereby highlighting a unique role for Polθ in counteracting HDR. To gain insight into this novel Polθ function, we show that the promiscuous polymerase is not required for end-resection of DSBs (Extended Data Fig.8f–g). Instead, its activity counteracts the accumulation of Rad51 foci (Fig. 3e–f and Extended Data Fig.8h). To corroborate these findings, we employed the traffic light reporter (TLR) system, designed to generate a flow-cytometric readout for HDR and end-joining at a site-specific DNA break induced by I-Sce122. We observed that upon knocking down PolQ in Lig4−/− cells, resolution of the I-Sce1 induced DNA break by HDR is increased, in conjunction with a significant reduction in the frequency of alt-NHEJ (Extended Data Fig.9).
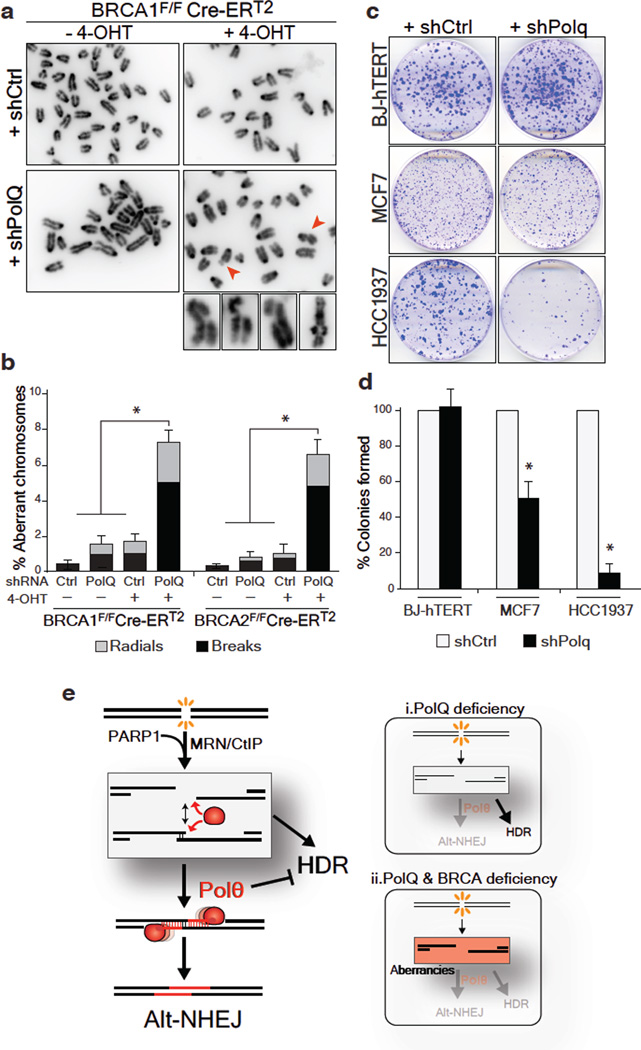
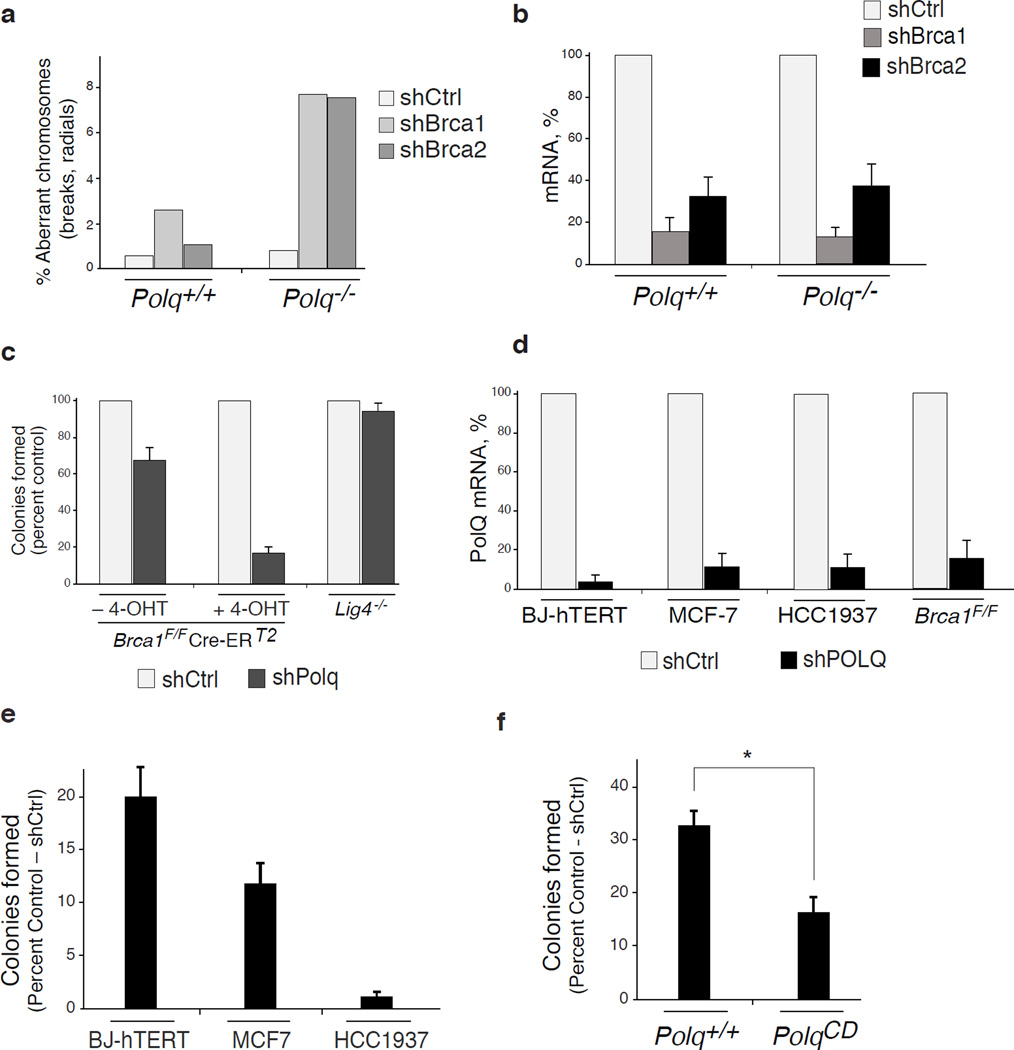
Alt-NHEJ is often considered as a back-up choice for DSB repair, operating at the expense of genomic stability. Circumstantial evidence suggests that this pathway could be enhanced when HDR is impaired23,24. We therefore postulated that this error-prone mode of repair plays an essential role in cells with compromised HDR activity. We tested this hypothesis by inhibiting PolQ in cells lacking the breast cancer susceptibility genes – BRCA1 and BRCA2. Chromosome analysis revealed a 4-fold increase in chromosomal aberrancies following PolQ depletion in MEFs lacking either BRCA1 or BRCA2. Such aberrancies included chromatid and chromosome breaks, in addition to radial chromosome structures characteristic of Lig4-mediated processing of chromatid breaks via the C-NHEJ pathway (Fig. 4a–b and Extended Data Fig.10a–b). Ultimately, the increased genomic instability in cells co-depleted for BRCA and PolQ compromised cellular survival. We observed that BRCA1 mutated human cells (Fig. 4c–d), and mouse cells lacking BRCA1 (Extended Data Fig.10c–f) displayed significantly reduced colony forming capabilities upon PolQ impairment. Although we cannot exclude that Polθ performs additional activities required for the survival of BRCA deficient cells25, our data suggest that Polθ-mediated alt-NHEJ promotes survival of cells with a compromised HDR pathway. One can envision that in the absence of a safer means to repair breaks alt-NHEJ prevents genomic havoc by resolving unrepaired lesions.
Figure 4. PolQ inhibition in BRCA-mutant cells leads to increased chromosomal aberrancies and reduced cellular survival.
a, Analysis of genomic instability in metaphase spreads from BRCA1F/FCre-ERT2 MEFs treated with shRNA against PolQ or vector control. b, Quantification of breaks (chromatid and chromosome) and radials in BRCA1F/FCre-ERT2 and BRCA2F/FCre-ERT2 MEFs with the indicated treatment. c, Clonogenic survival following PolQ depletion. Crystal violet staining of BJ-hTERT, MCF7, and HCC1937 cells treated with shRNA against PolQ or vector control. d, Quantitative analyses of colony formation assay. Colonies in each control shRNA cell line were set to 100%. Colonies in shPolQ expressing cells normalized to shCtrl (Mean ± SD, n=3’ *p<0.05’ **p<0.01;two-tailed student’s t-test). e, Schematic depicting our model for the function of Polθ during DSB repair (see Supplementary Information).
In summary, we provide direct evidence linking Polθ to alt-NHEJ repair in mammalian cells (Fig. 4e). We also show that while Polθ hinders error-free repair by HDR (Fig. 4e-i), its activity is essential for the survival of HDR-deficient cells (Fig. 4e-ii). The question remains as to how this promiscuous polymerase orchestrates DSB repair. Following DSB formation in S/G2 phase of the cell cycle, resection of DSBs by Mre11/CtIP20 potentially exposes micro-homology which allows spontaneous annealing of broken DNA ends (Fig. 4e). The binding of RPA antagonizes this annealing step26 to promote HDR-mediated repair27. An opposing activity is likely to be exerted by Polθ. Placing our finding in the context of recent biochemical experiments and genetic studies in model organisms13–16, we ultimately propose a model whereby Polθ exploits both its template-independent and template-dependent activities to stabilize the annealed intermediate structure and channel repair towards the alt-NHEJ pathway (Fig. 4e). Our findings that Polθ is critical for alt-NHEJ support this model, which provides a potential explanation as to how this polymerase counteracts HDR.
Finally, it is intriguing that while PolQ expression in normal human tissues is generally repressed28, it is up-regulated in a wide range of human cancers and associates with poor clinical outcome in breast tumors29,30. Our findings that cells with compromised HDR activity depend on this mutagenic polymerase for survival establish a rationale for the development of Polθ-targeted approaches for cancer treatment.
METHODS
Cell culture procedures
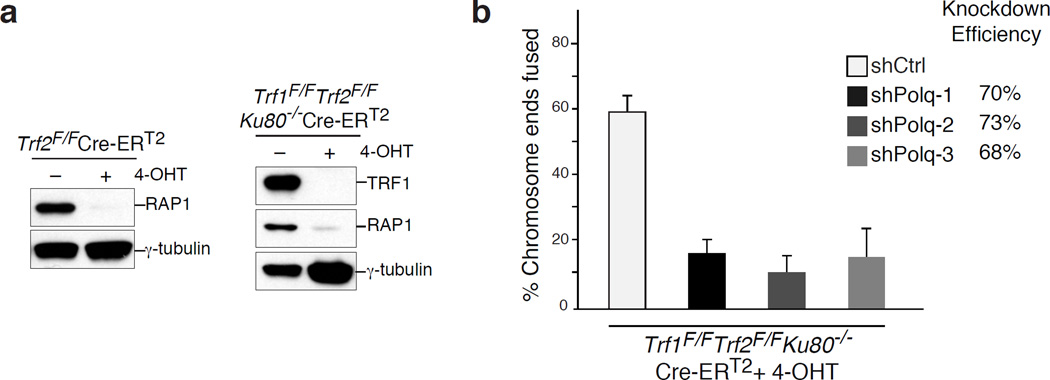
TRF2F/FCre-ERT2, TRF1F/FTRF2F/FKu80−/−Cre-ERT2, TRF1F/FTRF2F/FLig4−/−Cre-ERT2 mouse embryonic fibroblast (MEF) lines were previously described2,11. PolQ+/+ and PolQ−/− MEFs were a generous gift from N. Shima17. BRCA1F/FCre-ERT2, BRCA2F/FCre-ERT2 MEFS and U2OS-DSB reporter cells12 were a gift from R. Greenberg. TRF1F/FTRF2F/FLig4−/−Cre-ERT2 MEFs were derived from mice that were deficient for p53. The remaining MEF lines were immortalized with pBabeSV40LargeT. MEFs were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10–15% fetal bovine serum (FBS) (Gibco), 2 mM L-glutamine (Sigma), 100 U/ml penicillin (Sigma), 0.1 µg/ml streptomycin (Sigma), 0.1 mM non-essential amino acids (Invitrogen), and 1 mM sodium pyruvate (Sigma). Expression of Cre recombinase was induced by treating MEFs carrying Cre-ERT2 allele with 0.5 µM 4-OH tamoxifen (4-OHT; Sigma H7904) for 12 hours. t=0 time-point was set at the time of treatment with 4-OHT. BJ-hTERT and MCF7 cells were grown in DMEM supplemented with 10% FBS. HCC1937 cells were grown in RPMI medium (Gibco) containing 15%FBS. U2OS-DSB reporter cells were grown in DMEM supplemented with 10% BCS. Human HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 µg/ml streptomycin and 2 mM L-glutamine. Mouse embryonic stem cells were grown in DMEM supplemented with 15% fetal bovine serum (ES qualified FBS) (Gibco), 2 mM L-glutamine (Sigma), 100 U/ml penicillin (Sigma), 0.1 µg/ml streptomycin (Sigma), 0.1 mM non-essential amino acids (Invitrogen), leukemia inhibitory factor (LIF) and 2-Betamercaptoethanol (Gibco 21985). For inhibitor experiments, PARPi (KU58948, Axon medchem), ATMi (KU-55933, Tocris), and ATRi (VE-821, Selleckchem) were all used at a final concentration of 10 µM, and were applied to culture medium 2–4 hours prior to irradiation. For ionizing radiation (IR) treatment, cells were exposed to 1–10 Gy IR by a Faxitron X-ray system (120kV, 5mA, does rate 5Gy/min) and recovered for 4 hours before immunofluorescence analysis.
Lentiviral delivery of shRNA
shRNA treatments were carried out prior to 4-OHT treatment. shRNAs (see table for a list of sequences) were introduced by 2 lentiviral infections at 12hr intervals using supernatant from transfected 293T cells. Parallel infection with pLK0.1 was used as a negative control. Cells were selected with puromycin for three days.
Traffic Light Reporter (TLR) assay
Lentiviral constructs coding for TLR (#31482) and I-Sce1 with donor e-GFP (#31476) were purchased from Addgene22. To avoid the confounding impact of classical-NHEJ on the repair of I-Sce1 induced DNA breaks, we stably integrated the TLR construct into Ku80−/− and Lig4−/− MEFs. The plasmid was transduced by 2 lentiviral infections at 12 hr intervals using supernatant from transfected 293T cells. Cells with integrated TLR were selected with puromycin for 5 days. Cells were then co-transduced with concentrated I-Sce1 and shPolQ lentiviral particles. Cells were collected 72 hours later without further antibiotic selection and analyzed on a BD LSRII. eGFP fluorescence which reflects HDR repair was measured using a 488-nm laser for excitation and a 530/30 filter for detection. mCherry fluorescence, indicative of alt-NHEJ was measured by using a 561-nm laser for excitation and a 610/20 filter for detection. Data were analyzed using FloJo software.
Detection of telomeric fusions
To enrich for telomeric DNA, genomic DNA was digested with 2 frequent cutters (AluI and MboI) and fragments greater than 10Kb were isolated. The resulting DNA was used to generate a library using the NEBNext Ultra library prep Kit and the NEBNext Multiplex Oligos for Illumina (NEB) following the manufacturer instructions. The resulting library was run on an Illumina HiSeq platform generating 100 bp indexed pair-end reads.
Transient transfection of cells and laser micro-irradiation
Full-length human PolQ was cloned into pLPC-Myc vectors. HeLa cells were plated on glass-bottomed dishes (Willco Wells). Myc-PolQ constructs were transfected into HeLa and U2OS-DSB reporter cells cell with HilyMax (Dojindo) according to the manufacture’s instruction. 24 hours-post transfection, cells were pre-sensitized with 10 µM 5-bromo-2’ deoxyuridine (BrdU) in normal DMEM medium for 20 hours. After indicated treatments, cells were damaged by laser micro-irradiation as previous described31. After laser micro-irradiation, cells were incubated for 1 hour, then fixed and analyzed by immunofluorescence and microscopic imaging as described below. For PARP siRNA experiments, cells were transfected with siCtrl (Non-Targeting pool, Thermo Scientific) or siPARP1 (GGGCAAGCACAGUGUCAAAUU, Sigma), for 24 hours prior to PolQ transfections and subsequent treatments as described above.
Immunofluorescence (IF) and confocal microscopy
After the indicated treatments, cells were processed and analyzed for IF as previously described32. Briefly, cells were fixed with 2% (v/v) paraformaldehyde for 15 minutes at room temperature. Cells were washed with PBS, permeabilized with 0.5% (v/v) Triton X for 10 minutes, and blocked with PBS containing 3% BSA. Cells were incubated with the same buffer containing primary antibodies for 1 hour at room temperature followed by secondary antibodies incubations for 1 hour at room temp. Cells were imaged and analyzed with Z-stacked setting using the FV10-ASW3.1 software on a Fluoview 1000 confocal microscope (Olympus). For laser line quantification, > 50 cells were counted for all conditions from two independent experiments. The primary antibodies used for IF were: γH2AX [p Ser139] (Rabbit polyclonal, Novus, NB100-384), c-Myc (Mouse monoclonal, Santa Cruz, sc-40). The secondary antibodies used for IF were: Alexa Fluor 594 (Rabbit) (Invitrogen, A11037) and Alexa Fluor 488 (Mouse) (Invitrogen, A11029). To analyze the recruitment of PolQ to double stranded breaks, U2OS-DSB reporter cells expressing Myc-POLQ were analyzed 4 hours after treatment with shield and tamoxifen. Lastly, to analyze Rad51 foci formation and its co-localization with γH2AX [p Ser139] following IR treatment, cells were treated with 0.2% Triton-X 100 (in PBS) for 5 minutes on ice prior to fixation with paraformaldehyde. The primary antibodies used for Rad51 IF were: γH2AX [p Ser139] (mouse monoclonal, Novus, NB100-384), Rad51 (Rabbit polyclonal, Santa Cruz, sc-8349).
FISH
Cells were harvested at 96 hours following treatment with 4-OHT to analyze the frequency of telomere fusions. Briefly, ~80% confluent MEFs were incubated for 2 hours with 0.2 µg/ml colcemid (Sigma). The cells were harvested by trypsinization, resuspended in 0.075 M KCl at 37°C for 30 minutes, and fixed overnight in methanol/acetic acid (3:1) at 4°C. The cells were dropped onto glass slides and the slides were dried overnight. The next day, the slides were rehydrated with PBS for 15 min then fixed with 4% formaldehyde for 2 min at room temperature. Slides were digested with 1 mg/ml Pepsin (pH 2.2) at 37°C for 10 minutes, washed three times with PBS and fixed again in 4% formaldehyde for 2 min at room temperature. After three PBS washes, the slides were incubated consecutively with 75%, 95%, and 100% ethanol and allowed to air dry for 30 min before applying hybridization solutions (70% formamide, 1 mg/ml blocking reagent (Roche), 10 mM Tris-HCl pH 7.2) containing TAMRA-OO-[TTAGGG]3 PNA probes (Applied Biosystems). Slides were denatured by heating for 3 min at 80°C and hybridized for 2 hours at room temperature. The following day, the slides were washed twice for 15 min each in 70% formamide/10 mM Tris-HCl, followed by three 5 min washes in 0.1 M Tris-HCl, pH 7.0/0.15 M NaCl/0.08% Tween-20. Chromosomal DNA was counterstained with 4,6-diamidino-2-phenylindole (DAPI) during the second PBS wash. Slides were mounted in antifade reagent (ProLong Gold, Invitrogen) and images were captured with a Nikon Eclipse TI microscope. (From http://delangelab.rockefeller.edu/protocols)
CO-FISH
Cells were labeled with BrdU:BrdC (3:1, final concentration: 10 µM) for 14–16 hours. 2 hours prior to harvesting by trypsinization, 0.2 µg/ml colcemid was added to the media. To fix the cells and drop metaphases on a glass slide, the same procedure that was applied for FISH was followed. Slides were treated with 0.5 mg/ml RNase A (in PBS, DNase free) for 10 min at 37°C, incubated with 0.5 µg/ml Hoechst 33258 (Sigma) in 2XSSC for 15 min at room temperature, and exposed to 365-nm UV light (Stratalinker 1800 UV irradiator) for 30 min. The slides were then digested twice with 800 U Exonuclease III (Promega) at room temperature for 10 min each, washed with PBS and dehydrated through an ethanol series of 70%, 95%, 100%. After air-drying, slides were hybridized with Tamra-OO-[TTAGGG]3 PNA probe in hybridization solution (70% formamide, 1 mg/ml blocking reagent (Roche), 10 mM Tris-HCl pH 7.2) for 2 hours at room temperature. The slides were then washed for few seconds with 70% formamide/10 mM Tris-HCl pH 7.2 and incubated with FITC-OO-[CCCTAA]3 PNA probe in hybridization solution for 2 hours. Slides were washed and mounted as described for FISH. (From http://delangelab.rockefeller.edu/protocols)
Western blot analysis
Cells were harvested by trypsinization, lysed in 2X Laemmli buffer (100 mM Tris-HCl pH 6.8, 200 µM DTT, 3% SDS, 20% glycerol, 0.05% bromophenol blue) at 104 cell/µl. The lysate was denatured for 10 min at 95°C, and sheared by forcing it through a 28-gauge insulin needle 10 times. Lysate from 105 cells was loaded on an SDS/PAGE and transferred to a nitrocellulose membrane. The membrane was blocked in 5% milk in TBS with 0.1% Tween-20 and incubated with primary antibody in TBS/5% milk/0.1% Tween-20 for 2 hours at room temperature. The following primary antibodies were utilized: Polθ (ab80906, Abcam); TRF1 (1449, rabbit polyclonal); Rap1 (1252, rabbit polyclonal); Phospho-Chk2 (Thr68) (Rabbit polyclonal, Cell Signaling); Chk2 (rabbit polyclonal, Cell Signaling); Phospho-Chk1 (Ser 345) (mouse monoclonal, Cell Signaling); Chk1 (mouse monoclonal, Santa Cruz); Lig3 (mouse monoclonal, Santa Cruz); Myc (9E10; Calbiochem); γ- tubulin (clone GTU-88, Sigma); PARP1 (Polyclonal, Cell signaling). (From http://delangelab.rockefeller.edu/protocols)
Chromosomal Aberrancies
Cells were harvested and dropped on microscope slides as described for the FISH protocol. After the slides had dried overnight, they were re-hydrated in PBS, stained with 0.25 µg/ml DAPI, dehydrated in 70%, 95%, 100% ethanol series, mounted, and imaged using Nikon Eclipse TI microscope. Aberrancies were scored as a percentage of chromatid breaks, chromosome breaks, and chromosome radial structures compared to total number of chromosomes.
Chromosomal translocation assay
Induced pluripotent stem (iPS) cells were derived from primary PolQ+/+ and PolQ−/− MEFs as according to standard Yamanaka protocol33. To perform the translocation assay, PolQ+/+ and PolQ−/− iPS cells were transfected with 3ug of Cas9-gRNA(Rosa26;H3f3b) using lipofectamine. We constructed Cas9-gRNA(Rosa26;H3f3b) by introducing two guide RNAs (GTTGGCTCGCCGGATACGGG, for H3f3b; ACTCCAGTCTTTCTAGAAGA, for Rosa26) into pX330 (#42230 Addgene). After transfection, 104 cells were seeded per well in a 96-well plate, and lysed 3 days later in 40ul lysis buffer (10mM Tris pH 8.0, 0.45% Nonidet P-40, 0.45% Tween 20). The lysate was incubated with 200ug/ml of Proteinase K for 2 hours at 55°C. Translocation detection was performed according to previously established protocol9, using nested PCR. The primers used for the first PCR reaction: Tr6-11-Fwd:5′-GCGGGAGAAATGGATATGAA-3′; Tr6-11-Rev: 5′- TTGACGCCTTCCTTCTTCTG -3′, and Tr11-6-Fwd: 5′-AACCTTTGAAAAAGCCCACA-3′ and Tr11-6-Rev:5′-GCACGTTTCCGACTTGAGTT-3′, for Der(6) and Der (11) respectively. For the second round of PCR amplification we used the primers: Tr6-11NFwd: 5′-GGCGGATCACAAGCAATAAT-3′; Tr6-11NRev: 5′-CTGCCATTCCAGAGATTGGT-3′ and Tr11-6NFwd:5′-AGCCACAGTGCTCACATCAC-3′ and Tr11-6NRev:5′TCCCAAAGTCGCTCTGAGTT-3′. The number of PCR-positive wells was used to calculate the translocation frequency as described previously described9. Amplified products from positive wells were sequenced to verify translocations and determine the junction sequences.
Surveyor assay
Forty-eight hours after transfection, genomic DNA was extracted with GE Healthcare illustra genomic Prep Mini Spin Kit (28-9042-76). The genomic region encompassing the guide RNA target sites was amplified using Q5 High-Fidelity DNA polymerase (New England BioLabs) with the following primers Rosa26-Fwd: 5′-TAAAACTCGGGTGAGCATGT-3′, Rosa26-Rev: 5′-GGAGTTCTCTGCTGCCTCCTG-3′; H3f3b-Fwd: 5′-GCGGCGGCTTGATTGCTCCAG-3′ and H3f3b-Rev: 5′-AGCAACTTGTCACTCCTGAGCCAC-3′. PCR fragments were gel purified and the surveyor assay was performed using a detection kit (Transgenomic), according to manufacturer’s instructions. 2% agarose gels were used to visualize the bands after surveyor digestion.
Colony Formation Assay
Following lentiviral transduction with shCtrl or shPolQ (sequence listed in table below), cells were selected with Puromycin (BJ: 0.5ug/ml; MCF7 and HCC1937: 1ug/ml; MEFs, 2ug/ml) for 72 hours and plated in 6cm dishes (1000 and 10000 cells per plate). After 10–14 days colonies were fixed with 3% PFA (5 minutes), rinsed with PBS, and stained with crystal violet (Sigma-Aldrich).
CRISPR targeting to mutate PolQ gene in mouse embryonic stem cells
To generate cells carrying a catalytic dead Polθ, two mutations at residues D2494G and E2495S34 were introduced in the endogenous PolQ locus in mouse embryonic stem (ES) cells using CRISPR/Cs9 gene targeting. Two guides RNAs were co-transfected with a Cas9-nickase (pX335-U6-Chimeric_BB-CBh-hSpCas9n(D10A)), and a donor cassette which introduces a SacII restriction site while replacing the two amino acid residues. Clonal cells lines were derived and genotyped to determine successful targeting. Two independent clonally derived lines were used for the analysis of translocation.
shRNA target sequence (pLK0.1 vector)
| POLm: CTCACCTCTCACACACCATAA | |
| POLk: GCCCTTAGAAATGTCTCATAA | |
| POLh: GCTCGATTCTCCAGCTTACAA | |
| POLi: AGTGAAGAAGATACGTTTAAA | |
| POLb: CCAAAGTTGTTACATCGTGTT | |
| POLn: CCTACTCACATGAAGGACATT | |
| Lig3 (mouse): | CCAGACTTCAAACGTCTCAAA |
| Lig3 (human): | CCGGATCATGTTCTCAGAAAT |
| POLQ-1 (mouse): | CGGCGGAGTATGAGAACTATT |
| POLQ-2(mouse): | CCAGGAATCAAAGACGACAAT |
| POLQ-3(mouse): | CCTGGCTGAATGCTGAACTTT |
| hPOLQ (human): | CGGGCCTCTTTAGATATAAAT |
| BRCA1: | GCTCAGTGTATGACTCAGTTT |
Primers for Q-PCR
| BRCA1Fwd: | CTGCCGTCCAAATTCAAGAAGT |
| BRCA1Rev: | CTTGTGCTTCCCTGTAGGCT |
| BRCA2Fwd: | TGTGGTAGATGTTGCTAGTCCGCC |
| BRCA2Rev: | GCTTTTCTCGTTGTAGTACTGCC |
| POLb Fwd: | TGAACCATCATCAACGAATTGGG |
| POLbRev: | CCATGTCTCCACTCGACTCTG |
| POLmFwd: | AGGCTTCCGCGTCCTAGAT |
| POLmRev: | GTGGGGAGAGCATCCATGTT |
| POLkFwd: | AGCTCAAATTACCAGCCAGCA |
| POLkRev: | GGTTGTCCCTCATTTCCACAG |
| POLhFwd: | ATCGAGTGGTTGCTCTTGTAGA |
| POLhRev: | CCAAATGCTCGGGCTTCATAG |
| POLiFwd: | GCAGTCAAGGGCCACCTAC |
| POLiRev: | AGGTCTGTCCTTTAATTCTGGGT |
| POLnFwd: | AGCTGATGGATGCTCTCAAGCAGG |
| POLnRev: | GAGTCAGAGTGCTGTTGCCTACATGG |
| LIG3(mouse)Fwd: | GAAGAAAGCTGCTGTCCAGG |
| LIG3(mouse)Rev: | CAGAGTTGTTGGGTTTTGCTG |
| LIG3(human)Fwd: | GAAGAGCTGGAAGATAATGAGAAGG |
| LIG3(human)Rev: | AGTGGTTGTCAACTTAGCCTGG |
| POLQ(mouse)Fwd: | CAAGGTTTCATTCGGGTCTTGG |
| POLQ (mouse)Rev: | CGAGCAGGAAGATTCACTCCAG |
| POLQ(human) Fwd: | CAGCCCTTATAGTGGAAGAAGC |
| POLQ(human) Rev: | GCACATGGATTCCATTGCACTC |
Extended Data
Extended data figure 1.
(Related to Figure 1). a, Immunoblots for TRF1 and Rap1 after 4-OHT-induced depletion of TRF2 from TRF2F/FCre-ERT2 MEFs and co-depletion of TRF1 and TRF2 from TRF1F/FTRF2F/FKu80−/−Cre-ERT2 cells. Loss of TRF2 is confirmed by the disappearance of Rap1; a TRF2-interacting protein whose stability depends on TRF235,36 b, To validate the impact of PolQ deletion on alt-NHEJ we monitored the frequency of telomere fusions in shelterin-free Ku80 null cells treated with three independent shPolQ vectors. shPolQ-1 was used in Figure 2. Error bars represent s.e.m.
Extended data figure 2.
(Related to Figure 2). a, Immunobloting for Polθ in MEFs with the indicated genotype and treatment. b, Immunoblot for TRF1 in MEFs with the indicated genotype. Cells were analyzed 96 hours following Cre induction. c, Rap1 Immunoblot (similar to B). d, Western blot analysis for Polθ and Flag-Cas9 in lysates prepared from PolQ−/− and PolQ+/+ cells following Cas9 expression. Tubulin serves as a loading control. e, Surveyor nuclease assay for PolQ−/− and PolQ+/+ cells expressing Cas9-gRNA(Rosa26;H3f3b) plasmid. Genomic DNA isolated from cells with the indicated genotype was used as a template to amplify across the cleavage site at either the Rosa26 or the H3f3b locus to assess intra-chromosomal NHEJ. Amplification products were denatured and then re-annealed to form heteroduplexes between unmodified and modified sequences from imprecise NHEJ. The mismatched duplex was selectively cleaved by the Surveyor nuclease at the loops that form at mismatches. f, Signature of translocations in PolQ−/− and PolQ+/+ cells (see Extended Data Fig. 3–5 for complete list of sequences). Table records the total number of translocation events identified following CRISPR-Cas induced-cleavage. On average, the same number of nucleotides was deleted at the fusion junction in PolQ−/− and PolQ+/+ cells. No nucleotide insertions were found in the absence of PolQ. Lastly, the percentage of junctions exhibiting microhomology were significantly reduced in cells lacking PolQ. g, Scheme depicting Polθ domains. CRISPR/Cas9 gene targeting was employed in order to create two base substitutions at D2494G and E2495S, and generate a catalytic dead polymerase34. h, Sequence analysis of targeted cells. i, genotyping PCRs of PolQ+/+ and PolQCD after SacII digestion. j, Immunoblotting to analyze Cas9 expression in PolQ+/+ and two independently derived PolQCD clonal cell lines. k, Frequency of chromosomal translocations (Der-6) in PolQ+/+ and PolQCD cells. Bars represent mean of four independent experiments ± SD (two experiments per clonal cell line). ** represents p=0.006 (two-tailed student’s t test). PCR products were sequenced to confirm translocation and identify possible insertions.
Extended data figure 3.
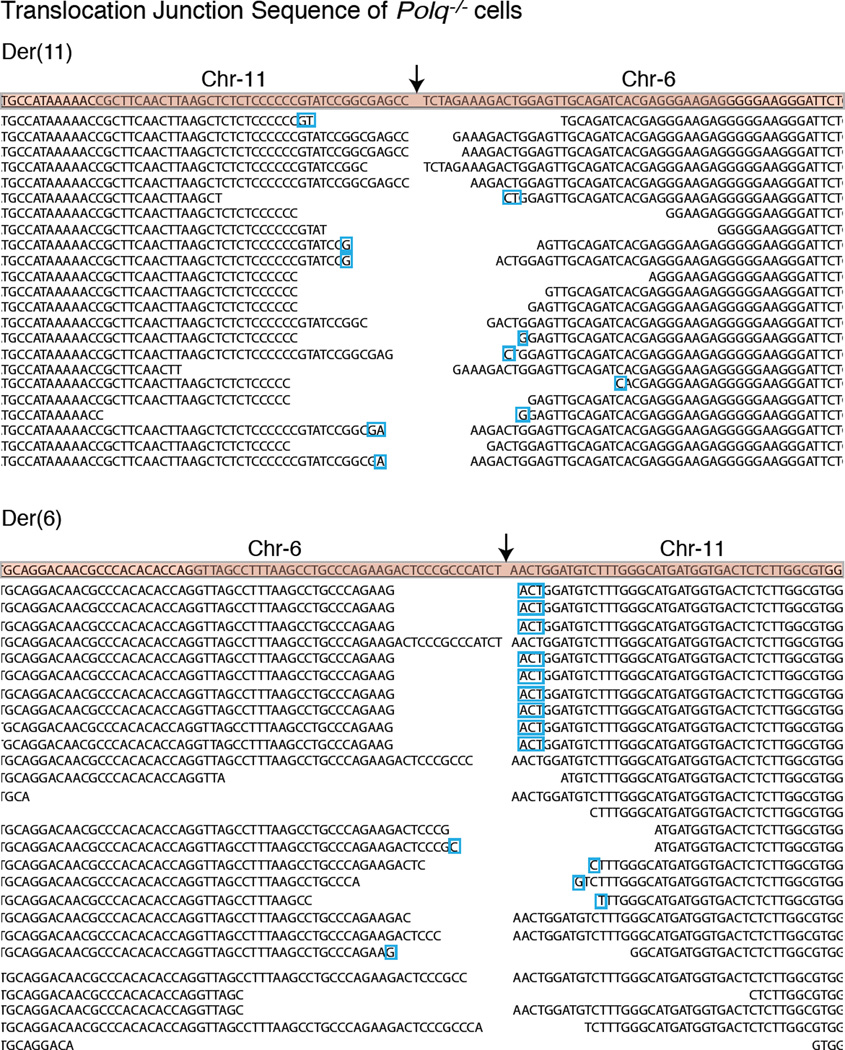
(Related to Figure 2). Sequences of der(6) and der(11) breakpoint junction from PolQ+/+ and PolQ−/− cells. Predicted fusion breakpoint based on CRISPR cutting, is indicated by an arrow. Reference sequence is highlighted at the top. The remaining lines represent individual translocations recovered by PCR and subject to Sanger sequencing. Nucleotide insertions in PolQ+/+ cells are marked in red. In cases where insertions extended beyond the sequence included in the lane, the length of the insertion was noted in parenthesis (red). It is important to note that insertions were completely lacking at the fusions junctions in PolQ−/− cells (Extended Data Fig. 5). Gaps in the sequence represent nucleotide deletions. The average length of the deletions was noted in Extended Data Fig. 2f. Micro-homology is denoted by blue boxes. Micro-homology embedded in DNA extending beyond the enclosed sequence was noted in parentheses (blue).
Extended data figure 4.
(Related to Figure 2). Sequences of der(6) and der(11) breakpoint junction from PolQ+/+ and PolQ−/− cells. Predicted fusion breakpoint based on CRISPR cutting, is indicated by an arrow. Reference sequence is highlighted at the top. The remaining lines represent individual translocations recovered by PCR and subject to Sanger sequencing. Nucleotide insertions in PolQ+/+ cells are marked in red. In cases where insertions extended beyond the sequence included in the lane, the length of the insertion was noted in parenthesis (red). It is important to note that insertions were completely lacking at the fusions junctions in PolQ−/− cells (Extended Data Fig. 5). Gaps in the sequence represent nucleotide deletions. The average length of the deletions was noted in Extended Data Fig. 2f. Micro-homology is denoted by blue boxes. Micro-homology embedded in DNA extending beyond the enclosed sequence was noted in parentheses (blue).
Extended data figure 5.
(Related to Figure 2). Sequences of der(6) and der(11) breakpoint junction from PolQ+/+ and PolQ−/− cells. Predicted fusion breakpoint based on CRISPR cutting, is indicated by an arrow. Reference sequence is highlighted at the top. The remaining lines represent individual translocations recovered by PCR and subject to Sanger sequencing. Nucleotide insertions in PolQ+/+ cells are marked in red. In cases where insertions extended beyond the sequence included in the lane, the length of the insertion was noted in parenthesis (red). It is important to note that insertions were completely lacking at the fusions junctions in PolQ−/− cells (Extended Data Fig. 5). Gaps in the sequence represent nucleotide deletions. The average length of the deletions was noted in Extended Data Fig. 2f. Micro-homology is denoted by blue boxes. Micro-homology embedded in DNA extending beyond the enclosed sequence was noted in parentheses (blue).
Extended data figure 6.
(Related to Figure 3). a, Laser micro-irradiation experiment using HeLa cells expressing Myc-PolQ and treated with ATM inhibitor (KU55933). ATR inhibitor (VE-821), or PARP inhibitor (KU58948). b, Western blot analysis for Chk1 and Chk2 phosphorylation. Cells with the indicated treatment were analyzed 2 hours following irradiation. c, Immunoblot for PARP1. HeLa cells were treated with PARP1 siRNA and analyzed 72 hours post-siRNA transfection for efficiency of knockdown.
Extended data figure 7.
(Related to Figure 3). a, Results from immunofluorescence performed at 4 hours after induction (Shield1 ligand, Clontech 631037; 0.5 µM 4-OH tamoxifen) of DSBs by mCherry-LacI-FokI in the U2OS-DSB reporter cells18 transfected with the Myc-PolQ and treated with PARP inhibitor (KU58948). The mCherry signal is used to identify the area of damage and to assess the recruitment of Myc-PolQ to cleaved LacO repeats. b, Table displaying quantification related to a.
Extended data figure 8.
(Related to Figure 3). a, Western blot analysis for Polθ and Lig3 in shelterin-free Lig4 null MEFs. b, Western blot for TRF1 and Rap1 following 4-OHT treatment of shelterin-free Lig4 deficient cells. c, Metaphase spreads from TRF1F/FTRF2F/FLig4−/−Cre-ERT2 MEFs, with the indicated shRNA treatment, 96 hours after Cre expression. CO-FISH assay was performed using a FITC-OO-(CCCTAA)3 PNA probe (green) and a Tamra-OO-(TTAGGG)3 PNA probe (red). DAPI in blue. Examples of alt-NHEJ mediated fusion and T-SCE events (HDR) are indicated by white and red arrows, respectively. Examples of T-SCE events reflective of increased HDR in cells treated with shPolQ are on the right. d–e, Quantification of telomere fusions by alt-NHEJ in MEFs with the indicated genotype and shRNA treatment. Bars represent the mean of two independent experiments ±SD. f, Representative in-gel hybridization to assess 3′ overhang of TRF1F/FTRF2F/FLig4−/−Cre-ERT2 MEFs with the indicate shRNA treatment after Cre deletion. g, Quantification of the gel in f. The ssDNA/total signal ratios of the +Cre samples are expressed relative to the −Cre samples for each shRNA treatment. Mean of two independent experiments. h. Graph representing Rad51 accumulation following IR treatment of PolQCD, Polq+/+ and PolQ−/− embryonic stem cells.
Extended data figure 9.
(Related to Figure 3). a, Polθ represses recombination at DSBs induced by I-Sce1. The Traffic light reporter (TLR) system was used to measure the relative ratio of end-joining (mCherry) and HDR (eGFP) repair of a DSB. A Diagram of the TLR is represented. b, The TLR construct was stably integrated into Lig4−/− and Ku80−/− MEFs to avoid the confounding impact of C-NHEJ, and limit end-joining reactions to the alt-NHEJ pathway. Expression of mCherry and eGFP was assessed by flow cytometry 72 hours after I-Sce1 and 5’eGFP donor transduction in cells with the indicated shRNA construct. Percentages of cells are indicated in the plot. c, Quantification of alt-NHEJ and HDR of TLR containing Ku80−/− MEFs following expression of I-Sce1 and 5’eGFP together with the indicated shRNA construct. Bar graphs represent the mean of three independent experiments ±SD. * represent p=0.03 d, Real-time PCR to monitor the knockdown efficiency of PolQ in Ku80−/− and Lig4−/− MEFs. The FACS analysis reported in e and f was carried it without selecting for cells expressing the shRNA containing plasmid.
Extended data figure 10.
(Related to Figure 4). a, Accumulation of chromosomal aberrancies following BRCA1 and BRCA2 knockdown in PolQ−/− and Polq+/+ MEFs. Quantification of chromosomal aberrancies (chromatid breaks, chromosome breaks, and radials) in MEFs stably transduced with lentiviral vectors expressing the indicated shRNA. b, Real-time PCR to confirm the knockdown of BRCA1 and BRCA2 as in (a). c, Quantitative analysis of colony formation in BRCA1F/FCre-ERT2 and Lig4−/− cells following PolQ depletion. The number of colonies in control shRNA- treated cells was set to 100%. d, Real-time PCR to measure the knockdown efficiency of human PolQ in BJ-hTERT, MCF7, HCC1937, and mouse PolQ in BRCA1F/FCre-ERT2 cells. e, Quantitative analyses of colony formation in BJ-hTERT, MCF7, HCC1937 following Lig3 inhibition. The numbers of colonies in control shRNA treated cells were set to 100%. The knockdown efficiency for Lig3 was ~85%. f. Quantitative analyses of colony formation in PolQCD and Polq+/+ embryonic stem cells following BRCA1 inhibition. The number of colonies in control shRNA-treated cells was set to 100%. The knockdown efficiency for BRCA1 was > 80%.
Supplementary Material
Acknowledgments
We thank Titia de Lange, Roger Greenberg, Jerry Shay, Naoko Shima, Christophe Cazaux, and Richard Wood for providing key reagents for this study. We are grateful to Minhyung Ji, Lillian Walton Masters, Aaron Phillips, Alexandra Pinzaru, Frank Yeung, Peter Tonzi, and Jason Wong for technical assistance. We thank Shaheen Kabir and Francisca Lottersberger for critical reading of the manuscript. This work was supported by a grant from the Breast Cancer Alliance (A.S.), V-foundation (A.S.), Department of Defense Breast Cancer Research Program BC134020 (P.M.G), Pew-Stewart Scholars Award (A.S.), Pew Scholars Award (E.L.D.), Novartis Advanced Discovery Institute (E.L.D.), and a grant from the NIH AG038677 (E.L.D.). The A.S. lab was supported by start-up funds from the Helen L. and Martin S. Kimmel Center for Stem Cell Biology. The K.M.M. laboratory was supported in part by start-up funds from the University of Texas at Austin and from the Cancer Prevention Research Institute of Texas (CPRIT, R116). K.M.M. is a CPRIT scholar.
Footnotes
Author Contributions A.S., E.L.D. and P.M.G. conceived the experimental design. P.M.G. and A.S. performed the experiments and analyzed the data. E.L.D. and N.N. performed telomere-sequencing experiments. F.G. and K.M.M. performed experiments related to PolQ localization at DNA breaks. A.S. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Sequence has been deposited with the BioProject database (ID PRJNA269507).
The authors declare no competing financial interests.
REFERENCES
- 1.Capper R, et al. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 2007;21:2495–2508. doi: 10.1101/gad.439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012;336:593–597. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rai R, et al. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. EMBO J. 2010;29:2598–2610. doi: 10.1038/emboj.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sfeir A. Telomeres at a glance. J. Cell. Sci. 2012;125:4173–4178. doi: 10.1242/jcs.106831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, et al. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 7.Audebert M, Salles B, Weinfeld M, Calsou P. Involvement of polynucleotide kinase in a poly(ADP-ribose) polymerase-1-dependent DNA double-strand breaks rejoining pathway. J. Mol. Biol. 2006;356:257–265. doi: 10.1016/j.jmb.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 8.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 9.Simsek D, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat. Struct. Mol. Biol. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okamoto K, et al. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013;494:502–505. doi: 10.1038/nature11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36:3847–3856. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogg M, Sauer-Eriksson AE, Johansson E. Promiscuous DNA synthesis by human DNA polymerase θ. Nucleic Acids Res. 2012;40:2611–2622. doi: 10.1093/nar/gkr1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan SH, Yu AM, McVey M. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 2010;6:e1001005. doi: 10.1371/journal.pgen.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roerink SF, van Schendel R, Tijsterman M. Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans. Genome Res. 2014;24:954–962. doi: 10.1101/gr.170431.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koole W, et al. A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat Commun. 2014;5:3216. doi: 10.1038/ncomms4216. [DOI] [PubMed] [Google Scholar]
- 17.Shima N, et al. Phenotype-based identification of mouse chromosome instability mutants. Genetics. 2003;163:1031–1040. doi: 10.1093/genetics/163.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Truong LN, et al. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2013;110:7720–7725. doi: 10.1073/pnas.1213431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH. Strand-specific postreplicative processing of mammalian telomeres. Science. 2001;293:2462–2465. doi: 10.1126/science.1062560. [DOI] [PubMed] [Google Scholar]
- 22.Certo MT, et al. Tracking genome engineering outcome at individual DNA breakpoints. Nat. Methods. 2011;8:671–676. doi: 10.1038/nmeth.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bindra RS, Goglia AG, Jasin M, Powell SN. Development of an assay to measure mutagenic non-homologous end-joining repair activity in mammalian cells. Nucleic Acids Res. 2013;41:e115–e115. doi: 10.1093/nar/gkt255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakai W, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez-Vidal A, et al. A role for DNA polymerase θ in the timing of DNA replication. Nat Commun. 2014;5:4285. doi: 10.1038/ncomms5285. [DOI] [PubMed] [Google Scholar]
- 26.Deng SK, Gibb B, de Almeida MJ, Greene EC, Symington LS. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014;21:405–412. doi: 10.1038/nsmb.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H, Lisby M, Symington LS. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol. Cell. 2013;50:589–600. doi: 10.1016/j.molcel.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawamura K, et al. DNA polymerase theta is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int. J. Cancer. 2004;109:9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]
- 29.Higgins GS, et al. Overexpression of POLQ confers a poor prognosis in early breast cancer patients. Oncotarget. 2010;1:175–184. doi: 10.18632/oncotarget.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemée F, et al. DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc. Natl. Acad. Sci. U.S.A. 2010;107:13390–13395. doi: 10.1073/pnas.0910759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 31.Shee C, et al. Engineered proteins detect spontaneous DNA breakage in human and bacterial cells. Elife (Cambridge) 2013;2:e01222–e01222. doi: 10.7554/eLife.01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leung JW, et al. Nucleosome acidic patch promotes RNF168- and RING1B/BMI1-dependent H2AX and H2A ubiquitination and DNA damage signaling. PLoS Genet. 2014;10:e1004178. doi: 10.1371/journal.pgen.1004178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 34.Prasad R, et al. Human DNA polymerase theta possesses 5'-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 2009;37:1868–1877. doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 36.Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science. 2010;327:1657–1661. doi: 10.1126/science.1185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.