Summary
Introduction.
Recent studies have demonstrated the role of the interleukin 28B (IL28B) polymorphisms in predicting treatment induced and spontaneous clearance from Hepatitis C virus (HCV) infection, suggesting the possibility of tailored therapy in HCV infected patients. Genome-wide association studies have shown that single nucleotide polymorphisms (SNPs) near IL 28B gene on chromosome 19 are strong predictors of sustained virologic response (SVR) to pegylated interferon and ribavirin. This study was aimed at analyzing the co-prevalence of two common and clinically significant SNPs in a cohort of Ligurian patients.
Methods.
Two SNPs (rs12979860, rs8099917) were genotyped in the IL28B locus from 175 DNA samples collected from HCVinfected consecutive patients in a Laboratory of Liguria Region, northern Italy. A real-time polymerase chain reaction in a Corbett Research Termocycler (Rotor Gene 3000A) by fluorescent probes (Fast Set IL 28B©, Arrow Diagnostics) was used for the detection, according to the manufacturer's instructions.
Results.
Carriers of rs12979860CT genotype predominated (87/175, 50%), homozygotes for allele C were 68/175 (39%) and the remaining were homozygotes for IFN-resistant allele T (11%). As for the rs8099917 SNP, genotypes were thus distributed: 96/175 (55%) carried the rs8099917 TT genotype, whereas 70/175 (40%) and 9/175 (5%), were heterozygotes or homozygotes for the G allele. The variants rs12979860CC and rs8099917TT were found in 39% and 54% of overall patients with HCV genotype 1, respectively. The combined assessment of examined SNPs resulted in three most prevalent genotypes (rs12979860CC/rs8099917TT, rs12979860CT/rs8099917TG and rs12979860CT/rs8099917TT) with a frequency of 35%, 31% and 18%, respectively.
Discussion.
Recent findings demonstrated that in carriers of rs12979860CT the determination of additional genotype of rs8099917 SNP could significantly improve the prediction of SVR. In our study cohort carriers of rs12979860CT represented 50% of all patients, who could take advantage with respect to SVR prediction by further determination of the rs8099917 SNP. The simultaneous genotyping of two IL28B SNPs should thus be recommended in HCV infected patients prior to treatment initiation.
Key words: Hepatitis C virus, IL28B, Single nucleotide polymorphism
Introduction
Hepatitis C virus (HCV) infection is a global health problem with 120–180 million carriers worldwide; about 30% of infected individuals develop severe liver disease, as it is the leading cause of hepatocellular carcinoma, cirrhosis and liver transplants in Europe and the USA [1-4].
Combination therapy of long-acting pegylated interferon-α 2a or 2b (PEG-IFN) and oral treatment with ribavirin (RBV) represents the main antiviral tool; however, it is unsuccessful in about 50% of cases [5-8], expensive and associated with significant side effects, such as hematologic abnormalities, flu-like syndrome and adverse neuropsychiatric events, sometimes requiring dose reduction or premature termination of therapy [9].
In order to reduce several side effects and to avoid the heavy medical cost of PEG-IFN/RBV, a clinical tool able to predict an individual response before the treatment would be quite useful [10].
Several studies have demonstrated the role of both viral factors (such as HCV genotype, baseline viremia) and host factors (i.e. age, sex, ethnicity, liver fibrosis, body mass index) in predicting the natural course of hepatitis C and response to therapy.
A number of genome-wide association studies (GWAS) have shown that single nucleotide polymorphisms (SNPs) near the interleukin 28B (IL28B) gene on chromosome 19 coding for IFN-3 (rs12979860, rs8099917, rs12980275, and rs8103142) [11-16] are strong predictors of sustained virologic response (SVR) to PEG-IFN/ RBV as well as of spontaneous viral clearance in HCV infected individuals [17, 18]. Other SNPs of IL28B (rs 8105790, rs 11881222, rs 28416813, rs 4803219 and rs 7248668) have been studied in HCV genotype 1 infected patients [17, 19, 20].
The SNPs rs12979860 and rs8099917 were found to be the most prevalent in different populations and strongly associated with treatment outcome. The first one, identified by Ge et al., has been associated with an approximately 2-fold difference in SVR rates in type 1-infected patients of European, African-American or Hispanic ancestry, with its favorable variant rs12979860CC, representing the best predictor of SVR in these ethnic groups [12, 20, 21-24].
The C allele distribution presents a geographical pattern with a lower frequency among African individuals than European descents [11].
In other studies a non-coding SNP, rs8099917TT, sited 7.5 kb upstream from the IL28 start codon, has been identified by Tanaka et al. and Suppiah et al. [14, 15]. These GWAS demonstrated that both SNPs (rs12979860, rs8099917) are the most critical for therapy outcome, in particular the favorable variants, homozygosis for the SNPs rs12979860 (CC) and rs8099917 (TT), are significantly associated with SVR in HCV-genotype 1–infected patients treated with Peg-IFN/RBV.
In the present study we describe genotype and allele frequency of IL28B polymorphisms rs12979860 and rs8099917 SNPs in 175 DNA samples collected from HCV-infected consecutive patients in a Laboratory of Liguria Region, northern Italy.
Methods
Genomic DNA was isolated from whole blood samples collected from 175 HCV infected patients. In a subset of these the following clinical data were available: HCV genotype, co-infection with hepatitis B virus (HBV) or human immunodeficiency virus (HIV), biochemical profile and SVR to antiviral therapy, defined as undetectable HCV RNA levels at least 24 weeks after the end of the treatment.
All the DNA samples were genotyped for two sets of IL28B SNPs, rs12979860 and rs8099917, specifying by C or T and T or G allele, using a real-time polymerase chain reaction (PCR) in a Corbett Research Termocycler (Rotor Gene 3000A) by fluorescent probes (Fast Set IL 28B©, Arrow Diagnostics), according to the manufacturer's instructions. The assay discriminated the different genotypes: wild-type homozygote (C/C, T/T), heterozygote (C/T, T/G), replaced homozygote (T/T, C/C), for rs12979860 and rs8099917, respectively. Fluorescence data were measured at the end of every cycle by the following profile: denaturation time of 3 min at 95°C, 35 cycle at 95°C for 15 s and 61°C for 45s. The results were analyzed using Allelic Discrimination Analysis. The samples were tested by duplicate and the IL28B genotypes were assigned by analysis of the reference cycle numbers for each fluorescence curve.
Results
The characteristics of the study cohort are displayed in Table I. All patients in the present study were of Caucasian origin. They had a median age of 49 years (range 22-77 years) and included 121 (69%) men.
Tab. I.
Baseline characteristics of study patients.
| Variable | All patients (n = 175) |
|---|---|
| Age | 49 (22 - 77) |
| Sex, male (%) | 121 (69) |
| HCV genotype (%) (n=139) | |
| 1 | 79 (57) |
| 2 | 8 (6) |
| 3 | 36 (26) |
| 4 | 16 (12) |
| HIV Co-infected (%) (n = 104) | 97 (93) |
| HBV Co-infected (%) (n = 102) | 28 (27) |
| SVR (%) (n = 28) | 13 (46) |
| Platelets (n = 58) (X109 L) | 167x109 (16x109-406x109) |
| AST (IU/L) | 56.5 (16-376) |
| ALT (IU/L) | 78.5 (14-431) |
| WBC (/mmc) | 5300 (2000-12700) |
| Haemoglobin (g/dl) | 14.7 (10.2 - 17.5) |
For categorical data, the number of patients is presented, whereas for continuous data median and range are shown. AST, alanine aminotransferase; AST, aspartate aminotransferase; SVR, sustained virological response; WBC, white blood cells.
HCV genotype 1 was the most frequent (57%), followed by genotypes 3 (26%), 4 (12%) and 2 (6%).
Data on HIV and HBV co-infection were known in 97/104 (93%) and 28/102 (27%) patients, respectively. SVR to antiviral therapy was reached by 13/28 (46%) patients who received PEG-IFN/RBV.
Carriers of rs12979860CT genotype predominated (87/175, 50%), homozygotes for allele C were 68/175 (39%) and the remaining were homozygotes for IFNresistant allele T (11%). As for the rs8099917 SNP, genotypes were thus distributed: 96/175 (55%) carried the rs8099917 TT genotype, whereas 70/175 (40%) and 9/175 (5%), were heterozygotes and homozygotes for the G allele.
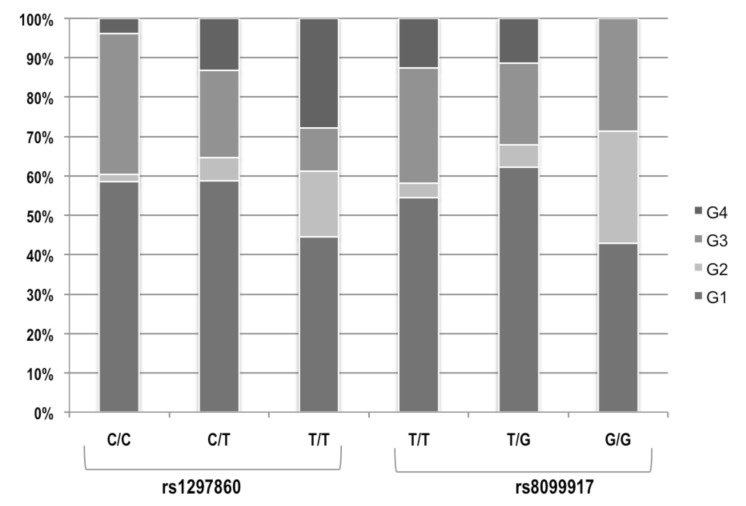
The frequency of HCV genotypes according to IL 28B SNPs (rs1297860 and rs8099917) is depicted in 139 patients (Fig. 1). The preferred variants rs12979860CC and rs8099917TT were found in 39% and 54% of overall patients with HCV genotype 1, respectively. Patients infected with HCV genotype 2 were the minority in both SNPs (6%), whereas those infected with HCV genotype 3 were 36% and 20% carriers of rs1297860CC and non-CC, 29% and 22% of rs8099917TT and non-TT. The frequency of the IFN-resistant allele was 87% for rs1297860 and 37% for rs8099917 among all infected with HCV genotype 4.
Fig. 1.
Frequency of HCV genotypes according to IL 28B SNPs (rs1297860 and rs8099917) in 139 patients.
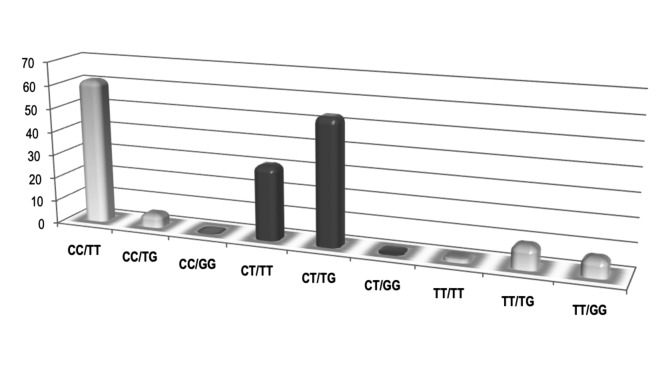
The combined assessment of examined SNPs resulted in three most prevalent genotypes (rs12979860CC/ rs8099917TT, rs12979860CT/rs8099917TG and rs12979860CT/rs8099917TT) with a frequency of 35%, 31% and 18%, respectively (Fig. 2). The remaining genotypes for the combined two loci were less common and, in particular, the variant rs12979860CC/rs8099917GG was not observed.
Fig. 2.
Distribution (%) of combined genotyping of rs1297860 and rs8099917 in 175 HCV infected patients.
Discussion
In the present study we investigated the prevalence of the haplotype and the combined genotyping of rs1297860 and rs8099917 SNPs in a set of samples of Caucasian descendent, by the simultaneous detection of the two IL28B loci. The homozygous variants rs1297860CC and rs8099917TT, the best predictors of SVR, were found in 39% and 55% of our cohort.
Several studies have demonstrated the role of IL28B polymorphisms in predicting treatment induced and spontaneous clearance from HCV infection, suggesting the possibility of tailored therapy and its relevant clinical and pharmacoeconomic implications. For example, carriers of rs1297860CC infected with HCV genotypes 1/4 may be haeded towards treatment with PEG-IFN/ RBV, whereas carriers of the IFN-resistant allele T with HCV genotype 1 and no advance liver fibrosis may defer therapy and wait for new direct-acting antiviral agents [25]. However, further cost-effectiveness analyses of response-guided therapy that include IL28B genotyping are needed for better understanding how current and future genotyping of other polymorphisms may help to evaluate the risk/benefit antiviral treatment profile.
The European Association for the Study of the Liver (EASL) Clinical Practice Guidelines for the management of HCV infection indicate the determination of IL28B variants as an available tool for diagnosis, useful to identify patient's likelihood of response to therapy, but with a low predictive value as other genetic variants may show correlation with disease progression in response to therapy [26]. In the scientific literature there is agreement about the need of integration of IL28B genotyping, which should not be per se considered a determining factor in deciding on a treatment strategy.
In this matter, a significant issue is which IL28B variant choose for diagnosis. A recent review by Soriano et al. summarized the main findings of GWAS conducted to assess the impact of different IL28B polymorphisms [25]: the authors underlined that, even though multiple SNPs around IL28B gene correlated to SVR, such as rs8099917, the current consensus considered rs12979860 as the strongest predictor [27]. The results of a 2013 meta-analysis confirmed that rs12979860CC and rs8099917TT genotypes could be used as independent predictors [28]; however, in consideration of the high frequency of rs12979860 in different populations and its relevant effect on treatment outcome, the determination of this SNP seemed sufficient for predicting response to therapy [29]. Halfon et al suggested that commercial tests for IL28B genotyping should include one simple polymorphism to give a simple message to physicians and, because of its highest predictive value to SVR, rs12979860 determination alone should be sufficient to predict treatment outcome [30]. In contrast with this opinion, Fisher et al. recently demonstrated that in carriers of rs12979860CT the additional genotyping of rs8099917 SNP could significantly improve the prediction of SVR [29]. In our study cohort, carriers of rs12979860CT represented 50% of all patients, who could take advantage with respect to SVR prediction by further determination of the rs8099917 SNP. This finding is line with the frequency observed by Fisher et al.
In conclusion, the simultaneous determination of two IL28B SNPs could be useful in some HCV infected patients to guide therapeutic decisions and improve treatment management and should therefore be included in the panel of pre-treatment evaluations.
ACKNOWLEDGMENTS
We are grateful to Stefano Razeti (Arrow Diagnostics) for his technical support.
References
- 1.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.Hoofnagle J. Course and outcome of hepatitis C. Hepatology. 2002;36:S21–S29. doi: 10.1053/jhep.2002.36227. [DOI] [PubMed] [Google Scholar]
- 3.Micallef JM, Kaldor JM, Dore GJ. Spontaneous viral clearance following acute hepatitis C infection: a systematic review of longitudinal studies. J Viral Hepat. 2006;13:34–41. doi: 10.1111/j.1365-2893.2005.00651.x. [DOI] [PubMed] [Google Scholar]
- 4.Thomas DL, Seeff LB. Natural history of hepatitis C. Clin Liver Dis. 2005;9:383–398. doi: 10.1016/j.cld.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Asselah T, Bieche I, Paradis V, et al. Genetics, genomics, and proteomics: implications for the diagnosis and the treatment of chronic hepatitis C. Semin Liver Dis. 2007;27:13–27. doi: 10.1055/s-2006-960168. [DOI] [PubMed] [Google Scholar]
- 6.Massard J, Ratziu V, Thabut D, et al. Natural history and predictors of disease severity in chronic hepatitis C. J Hepatol. 2006;44:S19–S24. doi: 10.1016/j.jhep.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Thio CL. Host genetic factors and antiviral immune responses to hepatitis C virus. Clin Liver Dis. 2008;12:713–726. doi: 10.1016/j.cld.2008.03.002. xi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yee LJ. Host genetic determinants in hepatitis C virus infection. Genes Immun. 2004;5:237–245. doi: 10.1038/sj.gene.6364090. [DOI] [PubMed] [Google Scholar]
- 9.Fried MW, Shiffman ML, Reddy KR, et al. Peginterferon alfa- 2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 10.Ahlenstiel G, Booth DR, George J. IL28B in hepatitis C virus infection: translating pharmacogenomics into clinical practice. J Gastroenterol. 2010;45:903–910. doi: 10.1007/s00535-010-0287-4. [DOI] [PubMed] [Google Scholar]
- 11.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 13.Rauch A, Kutalik Z, Descombes P, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–1345. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 14.Suppiah V, Moldovan M, Ahlenstiel G, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 16.Honda M, Sakai A, Yamashita T, et al. Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology. 2010;139:499–509. doi: 10.1053/j.gastro.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 17.Balagopal A, Thomas DL, Thio CL. IL28B and the control of hepatitis C virus infection. Gastroenterology. 2010;139:1865–1876. doi: 10.1053/j.gastro.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Jilg N, Shao RX, et al. IL28B inhibits hepatitis C virus replication through the JAK-STAT pathway. J Hepatol. 2011;55:289–298. doi: 10.1016/j.jhep.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tillmann HL, Thompson A, Patel K, et al. IL28B polymorphism is associated with jaundice during acute HCV infection and is a strong predictor for spontaneous clearance in the prospective german anti-D cohort. J Hepatol. 2010;52:S56–S56. [Google Scholar]
- 20.Grebely J, Petoumenos K, Hellard M, et al. Potential role for interleukin-28B genotype in treatment decision-making in recent hepatitis C virus infection. Hepatology. 2010;52:1216–1224. doi: 10.1002/hep.23850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berg T, Weich V, Teuber G, et al. Individualized treatment strategy according to early viral kinetics in hepatitis C virus type 1-infected patients. Hepatology. 2009;50:369–377. doi: 10.1002/hep.22991. [DOI] [PubMed] [Google Scholar]
- 22.Thompson AJ, Muir AJ, Sulkowski MS, et al. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology. 2010;139:120–129. doi: 10.1053/j.gastro.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 23.Stattermayer AF, Stauber R, Hofer H, et al. Impact of IL28B genotype on the early and sustained virologic response in treatment-naïve patients with chronic hepatitis C. Clin Gastroenterol Hepatol. 2011;9:344–350. doi: 10.1016/j.cgh.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 24.Bochud PY, Bibert S, Negro F, et al. IL28B polymorphisms predict reduction of HCV RNA from the first day of therapy in chronic hepatitis C. J Hepatol. 2011;55:980–988. doi: 10.1016/j.jhep.2011.01.050. [DOI] [PubMed] [Google Scholar]
- 25.Soriano V, Poveda E, Vispo E, et al. Pharmacogenetics of hepatitis C. J Antimicrob Chemother. 2012;67:523–529. doi: 10.1093/jac/dkr506. [DOI] [PubMed] [Google Scholar]
- 26. EASL European Association For The Study Of The Liver , author. Clinical Practice Guidelines: Management of chronic hepatitis B virus infection. J Hepatol. 2012;57:167–185. doi: 10.1016/j.jhep.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 27.Afdhal NH, McHutchison JG, Zeuzem S, et al. Pharmacogenetics and Hepatitis C Meeting Participants. Hepatitis C pharmacogenetics: state of the art in 2010. Hepatology. 2011;53:336–345. doi: 10.1002/hep.24052. [DOI] [PubMed] [Google Scholar]
- 28.Luo Y, Jin C, Ling Z. Association study of IL28B: rs12979860 and rs8099917 polymorphisms with SVR in patients infected with chronic HCV genotype 1 to PEGINF/ RBV therapy using systematic meta-analysis. Gene. 2013;513:292–296. doi: 10.1016/j.gene.2012.10.030. [DOI] [PubMed] [Google Scholar]
- 29.Fischer J, Böhm S, Scholz M, et al. Combined effects of different interleukin-28B gene variants on the outcome of dual combination therapy in chronic hepatitis C virus type 1 infection. Hepatology. 2012;55:1700–1710. doi: 10.1002/hep.25582. [DOI] [PubMed] [Google Scholar]
- 30.Fischer J, Böhm S, Scholz M, et al. Combined effects of different interleukin-28B gene variants on the outcome of dual combination therapy in chronic hepatitis C virus type 1 infection. Hepatology. 2012;55:1700–1710. doi: 10.1002/hep.25582. [DOI] [PubMed] [Google Scholar]