Abstract
RNA‐binding proteins (RBPs) play important roles for generating various cell types in many developmental processes, including eggs and sperms. Nanos is widely known as an evolutionarily conserved RNA‐binding protein implicated in germ cell development. Mouse NANOS2 interacts directly with the CCR4‐NOT (CNOT) deadenylase complex, resulting in the suppression of specific RNAs. However, the mechanisms involved in target specificity remain elusive. We show that another RBP, Dead end1 (DND1), directly interacts with NANOS2 to load unique RNAs into the CNOT complex. This interaction is mediated by the zinc finger domain of NANOS2, which is essential for its association with target RNAs. In addition, the conditional deletion of DND1 causes the disruption of male germ cell differentiation similar to that observed in Nanos2‐KO mice. Thus, DND1 is an essential partner for NANOS2 that leads to the degradation of specific RNAs. We also present the first evidence that the zinc finger domain of Nanos acts as a protein‐interacting domain for another RBP, providing a novel insight into Nanos‐mediated germ cell development.
Keywords: Dead end, germ cell, Nanos, RNA
Subject Categories: Development & Differentiation, RNA Biology
Introduction
RNA‐binding proteins (RBPs) control all aspects of the “life cycle” of RNA from its transcription to its degradation. RBPs form dynamic ribonucleoprotein particles, often in a cell type‐specific manner, and affect various biological processes, including asymmetric cell division, cell motility, embryonic axis determination, and neuronal plasticity 1, 2. Most importantly, RBPs play vital roles in germ cell development 3.
The differentiation of mammalian germ cells into male and female gametes is a fundamental biological process. In mice, the primordial germ cells (PGCs) are segregated from the somatic cell lineage at an early gastrulation stage 4 and then begin to migrate to the future gonads. Although the migrating PGCs are potent precursors of both oogonia and spermatogonia, they start sexual differentiation depending on the environmental cues after their colonization of the embryonic gonads with somatic cells; germ cells in female gonads (female gonocytes) immediately enter meiosis, whereas the germ cells in male gonads (male gonocytes) undergo cell cycle arrest at the G0/G1 phase 5. To date, it has been reported that at least two somatic factors are required for the masculinization of the PGCs in male embryonic gonads. CYP26B1, a retinoic acid (RA)‐metabolizing enzyme, is expressed in the Sertoli cells and protects male gonocytes from exposure to RA, resulting in the suppression of meiosis 6, 7. In addition, somatically derived fibroblast growth factor 9 induces the expression of the male gonocyte‐specific gene, Nanos2, via activation of the Nodal/Activin pathway 8. Once the expression of NANOS2 protein begins, male‐specific genetic programming is initiated.
Nanos is an evolutionarily conserved RBP implicated in germ cell development 9, 10, 11, 12. Three Nanos homologues, Nanos1‐3, exist in mice, among which Nanos2 is expressed only in male gonocytes and plays a key role during the sexual development of these cells by suppressing meiosis and promoting male‐type differentiation 13. We have previously shown that NANOS2 associates with the CNOT deadenylase complex via direct interaction with CNOT1, a component of the CNOT complex 14. This complex localizes to P‐bodies, the RNA–protein granules for storage of nontranslating RNAs, which likely induce the degradation of specific RNAs 15, 16. However, the mechanism of specific RNA recognition remains unclear since the zinc finger domain of Nanos, which is composed of two highly conserved, consecutive CCHC‐type zinc finger motifs, has been shown to bind RNA in a sequence‐unspecific manner 17, 18. In our current study, we attempted to elucidate the mechanism by which NANOS2 selects target RNAs for degradation and found that DND1 was a partner of NANOS2. We also established a Dnd1‐conditional knockout (Dnd1‐cKO) mouse strain to examine the in vivo function of the NANOS2–DND1 complex in male gonocytes.
Results and Discussion
DND1 directly interacts with NANOS2 via the zinc finger domain
To elucidate the mechanism of NANOS2‐mediated RNA regulation in the sexual differentiation of male gonocytes, we searched for NANOS2‐interacting proteins by immunoprecipitation (IP) from male gonadal extracts, followed by extensive mass‐spectrometric analyses. For this purpose, we took advantage of a transgenic mouse line expressing FLAG‐tagged NANOS2 under the control of the Nanos2 enhancer, which could fully rescue the defects of Nanos2‐knockout (Nanos2‐KO) mice 19. These experiments allowed the identification of several proteins, including CNOT1, Poly (A)‐binding protein1 (PABP1), and DND1 (Fig EV1A–D and Table EV1). We focused on DND1 as a strong candidate.
Figure EV1. Identification of NANOS2‐associated proteins.
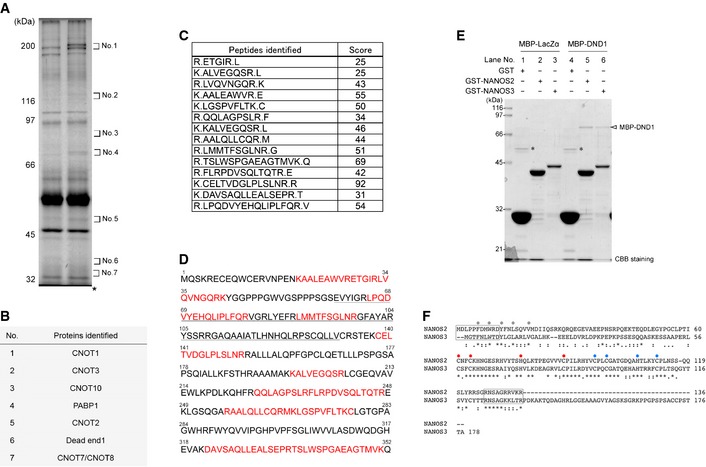
- Silver staining of proteins co‐precipitated with anti‐FLAG antibody from testis extracts of either wild‐type or transgenic embryos expressing FLAG‐tagged NANOS2 at E15.5. Both wild‐type and transgenic proteins in the gel at the location of numbers (No. 1 to 7) were analyzed by mass spectrometry. Note that the gels at No. 3, 6 and 7 were excised and analyzed despite no apparent band in the picture, because slight signals of several bands could be visualized with a backlight source. *Heavy chain of anti‐FLAG antibody.
- Proteins identified exclusively in the transgenic proteins of the gels indicated in (A).
- Scores of DND1 peptides identified by LC–MS/MS and MASCOT.
- Amino acid sequence of DND1. Residues shown in red correspond to peptide sequence obtained by mass spectrometry, and the underlined residues are the RNA recognition motif.
- Results of GST pull‐down assay using E. coli extracts expressing GST, GST‐fused NANOS2, or NANOS3 mixed with MBP‐tagged LacZα or DND1. Precipitates were analyzed by CBB staining. An arrowhead indicates MBP‐DND1 co‐precipitated with GST‐tagged NANOS2 or NANOS3. *Non‐specific band.
- Amino acid sequence alignment of NANOS2 and NANOS3. Grey circles indicate conserved amino acids in NIM, while red and blue circles indicate conserved CCHC residues in the former and latter zinc finger motif. Amino acids enclosed with a square in N‐ and C‐terminus are highly similar sequences deleted in Fig 1D.
DND1 is an RBP initially identified in zebrafish and conserved among vertebrates 20. Mouse Dnd1 is identified as a causable gene for the Ter mutation 21, by which most of the PGCs disappear at the migration stage, resulting in complete male sterility 22. However, it remains unclear whether DND1 plays a role in germ cell development after the migration stage. Therefore, we first generated a specific antibody to examine the expression profile. As expected from a functional study of Ter mutants showing a decreased germ cell number in the early embryonic stage, DND1 was highly expressed in the migrating PGCs (Fig EV2A–L). However, its expression became restricted to male gonocytes after colonization of the embryonic gonads, while such signals became undetectable in female gonocytes (Fig EV2M–R). Western blotting analyses confirmed the expression profile; DND1 expression gradually increased in the male gonads and was absent in the female gonads by embryonic day (E) 14.5 (Fig EV2S), raising the possibility that, together with NANOS2, DND1 is involved in the sexual differentiation of male gonocytes.
Figure EV2. Expression profile of DND1.
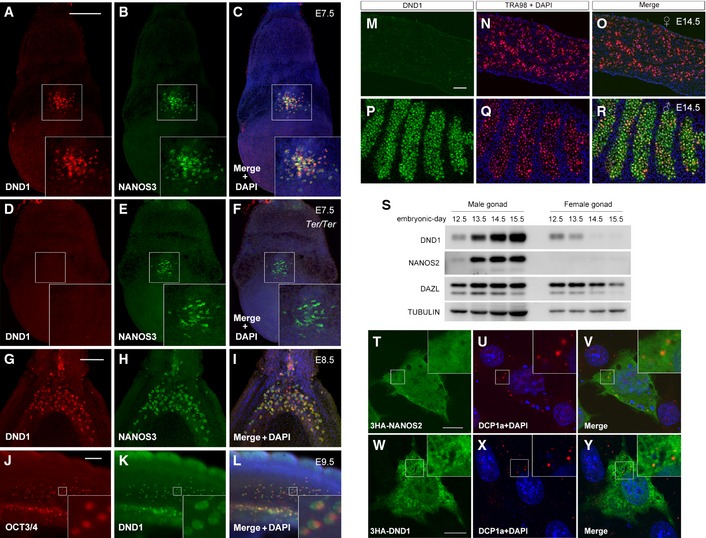
-
A–LWild‐type whole embryos at E7.5 (A–C), E8.5 (G–I) and E9.5 (J–L), and a Ter/Ter embryo at E7.5 (D–F) were stained with antibodies against DND1 (A, D, G, K), NANOS3 (B, E, H), or OCT3/4 (J). Note that the immunostaining signal of DND1 was not detected in a Ter/Ter embryo, indicating the specificity of the antibody against DND1. Insets in (A–F) and (J–L) show an enlarged version of each picture to better depict PGCs.
-
M–RSections of ovary (M–O) and testes (P–R) from embryos at E14.5 were stained with antibodies against DND1 (green) (M, P) and TRA98 (red) (N, Q).
-
SWestern blotting analyses of proteins in 1/3 of a male gonad and female gonad per lane at E12.5, E13.5, E14.5, and E15.5 using the antibodies indicated. Note that the amount of loading control protein TUBULIN in male gonads is larger than that in female gonads because of the larger size of male gonads compared to female gonads.
-
T–YNIH3T3 cells were transfected with HA‐tagged Nanos2 (T–V) or Dnd1 (W–Y) and then stained with antibodies against NANOS2 (green) (T) or DND1 (green) (W) and DCP1a (red) (U, X). Note that both HA‐tagged NANOS2 and DND1 do not show clear localization to P‐bodies.
To confirm the interaction between NANOS2 and DND1, we conducted IP assays from E15.5 male gonadal extracts using the transgenic mouse line expressing FLAG‐tagged NANOS2, followed by Western blotting analyses (Fig 1A). This assay revealed that CNOT proteins co‐precipitated with FLAG‐tagged NANOS2 independently of RNA, as reported previously 19. Factors localizing to P‐bodies, DEAD box polypeptide 6 (DDX6) and Argonaute 2 (AGO2) 23, 24, also co‐precipitated with FLAG‐tagged NANOS2 but were removed by RNase treatment, indicating that the interaction is RNA‐dependent. On the contrary, DND1 co‐precipitated efficiently with NANOS2 even when the extracts were treated with RNase, demonstrating that NANOS2 interacts with DND1 in vivo, independent of the presence of RNA.
Figure 1. DND1 interacts directly with NANOS2 via its zinc finger motif.
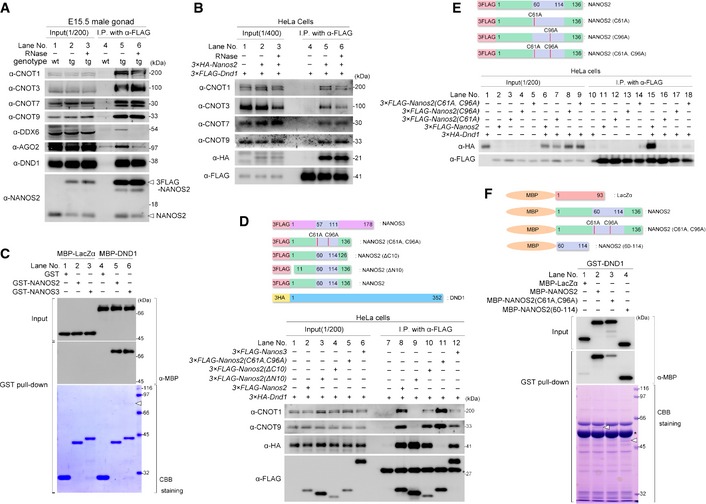
-
A, BWestern blotting analysis of proteins co‐precipitated with anti‐FLAG antibody from testis extracts of E15.5 wild‐type embryos and transgenic embryos expressing FLAG‐tagged NANOS2 with or without RNase (A) or from extracts of HeLa cells transfected with FLAG‐tagged DND1 and with or without HA‐tagged NANOS2 (B). Precipitates were analyzed with the indicated antibodies.
-
CGST pull‐down assay using E. coli extracts expressing GST, GST‐fused NANOS2, or NANOS3 mixed with MBP‐tagged LacZα or DND1. Precipitates were analyzed by Coomassie Brilliant Blue (CBB) staining or Western blotting with an anti‐MBP antibody. An arrowhead indicates MBP‐DND1 co‐precipitated with GST‐tagged NANOS2 or NANOS3. See also Fig EV1E.
-
D, EWestern blotting analyses of proteins co‐precipitated with anti‐FLAG antibody from extracts of HeLa cells transfected with FLAG‐tagged NANOS2, NANOS2 (ΔN10), NANOS2 (ΔC10), NANOS2 (C61A, C96A) or NANOS3, and HA‐tagged DND1 (D) or with FLAG‐tagged NANOS2, NANOS2 (C61A), NANOS2 (C96A) or NANOS2 (C61A, C96A), and HA‐tagged DND1 (E). Precipitates were analyzed with the indicated antibodies.
-
FGST pull‐down assay using E. coli extracts expressing GST‐fused DND1 mixed with MBP‐tagged LacZα, NANOS2, NANOS2 (C61A, C96A) or NANOS2 (60–114). Arrowheads indicate MBP‐tagged NANOS2 (lane 2) or NANOS2 (60–114) (lane 4).
Subsequently, we asked whether DND1 associated directly with NANOS2, since the association of DND1 with NANOS2 might be mediated by CNOT proteins. However, IP assays in HeLa cells revealed that the CNOT proteins co‐precipitated with FLAG‐tagged DND1 only when HA‐tagged NANOS2 was co‐expressed, regardless of whether RNase was added (Fig 1B). Furthermore, we found that purified recombinant maltose‐binding protein (MBP)‐tagged DND1 was co‐precipitated with bacterially expressed glutathione‐S‐transferase (GST)‐tagged NANOS2 or NANOS3 in a pull‐down assay using antibodies against MBP (Fig 1C, lanes 5 and 6) or Coomassie Brilliant Blue (CBB) staining (Fig EV1E, lanes 5 and 6), indicating that DND1 interacts directly with both NANOS2 and NANOS3, independently of the CNOT complex. This result also implied that a common amino acid sequence between NANOS2 and NANOS3 might be involved in the interaction with DND1. The N‐terminus of NANOS2 had already been reported to be structurally essential for the direct interaction with the CNOT1, which has been recently named as NOT1‐interacting motif (NIM) 25 (Fig EV1F). We then focused on the conserved zinc finger motifs. IP assays in HeLa cells revealed that substitution of the first cysteine residues in the two CCHC motifs of NANOS2 (C61 and C96) with alanine, yielding NANOS2 (C61A, C96A), drastically reduced and almost abolished the interaction with DND1, while the interactions with the CNOT proteins were intact (Fig 1D, lane 11). In contrast, the deletion of the N‐ or C‐terminal residues had a limited, or almost no effect, on the interaction with DND1, whereas NANOS2 (ΔN10) could not associate with the CNOT complex as previously reported (Fig 1D, lanes 9 and 10) 14, 25. In addition, single mutations in either CCHC motif (C61A or C96A) also drastically reduced this interaction (Fig 1E, lanes 16 and 17), indicating that both of the CCHC‐type zinc finger motifs are structurally essential for the interaction of NANOS2 with DND1. We next tested whether or not these two zinc finger motifs alone were sufficient for effective interaction of NANOS2 and DND1. To this end, MBP‐tagged zinc finger motifs (NANOS2 (60–114)) were subjected to a pull‐down assay with GST‐tagged DND1. Western blotting analyses revealed that the interaction between NANOS2 (C61A, C96A) and DND1 was much weaker than that of wild‐type NANOS2, and rather similar to that of negative control (Fig 1F, lanes 1–3), which could reflect the interaction observed in HeLa cells (Fig 1D, lane 11). On the other hand, NANOS2 (60–114) showed the same degree of co‐precipitation as wild‐type NANOS2 (Fig 1F, lanes 2 and 4). These data indicate that the zinc finger domain of NANOS2 is essential and sufficient for the direct interaction with DND1; thus, the two conserved, consecutive CCHC‐type zinc finger motifs of NANOS2 constitute a protein‐interacting domain for DND1.
Interactions between NANOS2 and PUMILIO1/PUMILIO2 are not detected in male gonocytes
Because we found that DND1 interacted with all NANOS family member (NANOS1–3) in HeLa cells (Fig 2A), DND1 is presumed to be a major partner of mouse NANOS. On the other hand, it is well known that Nanos works together with Pumilio in many species 26, 27, 28; the interaction between human NANOS1 and PUMILIO1 was reported in cultured cells via co‐transfection and IP assay 29. However, the mouse homologues PUMILIO1 and PUMILIO2 were not detected in our mass‐spec analyses of NANOS2‐IP (Fig EV1A and B). To ask more directly the relationship between mouse NANOS and PUMILIO, we transfected HeLa cells with HA‐tagged NANOS (NANOS1, NANOS2, or NANOS3) and Flag‐tagged PUMILIO (PUMILIO1 or PUMILIO2). IP assays revealed that mouse NANOS1 seemed to interact with both PUMILIO1 and PUMILIO2 (Fig 2B, lanes 13 and 14) as was the case in human NANOS1 and PUMILIO1, whereas such interactions were not detected or very weak for NANOS2 and NANOS3 (Fig 2B, lanes 15–18). However, we found obvious co‐precipitation of NANOS2 and NANOS3 with either PUMILIO1 or PUMILIO2 when MYC‐tagged DND1 was co‐expressed (Fig 2B, lanes 21–24), raising the possibility that NANOS2 interacts with PUMILIO1/PUMILIO2 via DND1 in vivo. Therefore, we conducted IP assay using gonadal extracts from E15.5 embryos expressing Flag‐tagged NANOS2, but we found no interaction between NANOS2 and PUMILIO1 or PUMILIO2 (Fig 2C and D), consistent with our mass‐spec analyses. These results suggest that NANOS2 does not interact with either PUMILIO1 or PUMILIO2 in male gonocytes even in the presence of DND1, although the reason is currently unknown.
Figure 2. Relationship between NANOS and PUMILIO .
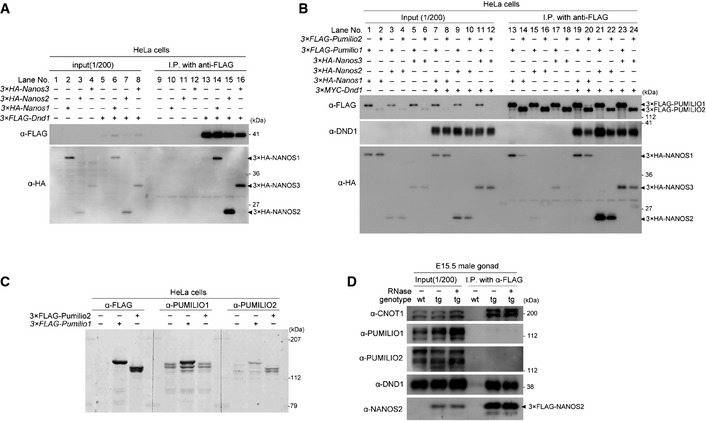
-
A, BWestern blotting analyses of proteins co‐precipitated with anti‐FLAG antibody from extracts of HeLa cells transfected with HA‐tagged NANOS1, NANOS2, or NANOS3, and FLAG‐tagged DND1 (A) or with FLAG‐tagged PUMILIO1 or PUMILIO2, and HA‐tagged NANOS1, NANOS2, or NANOS3 with or without MYC‐tagged DND1 (B).
-
CCharacterization of antibodies against PUMILIO1 and PUMILIO2. Western blotting analyses of Flag‐tagged PUMILIO1 and PUMILIO2 in HeLa cells transfected with FLAG‐tagged PUMILIO1 or PUMILIO2 using antibodies against FLAG, PUMILIO1 or PUMILIO2. Note that the antibodies against PUMILIO1 specifically recognized FLAG‐tagged PUMILIO1, while the anti‐PUMILIO2 antibody could detect both Flag‐tagged PUMILIO1 and PUMILIO2.
-
DFLAG‐tagged NANOS2 was immunoprecipitated with an anti‐FLAG antibody from E15.5 testicular extracts from transgenic mice expressing FLAG‐tagged NANOS2. Precipitates were analyzed by Western blotting with the indicated antibodies. Note that co‐precipitation of both PUMILIO1 and PUMILIO2 were not detected even though both CNOT1 and DND1 were clearly co‐precipitated.
DND1 co‐localizes with NANOS2 in P‐bodies
As NANOS2 localizes to P‐bodies 19, we expected to find co‐localization of DND1 in P‐bodies. Immunostaining analyses of male gonadal sections at E15.5 revealed that DND1 was distributed in both the nucleus and cytoplasm (Fig 3A). Furthermore, cytoplasmic DND1 seemed to form discrete foci, which co‐localized with DCP1a, a well‐known marker for P‐bodies (Fig 3B and C) 23. To confirm this, we conducted immunostaining using squash preparations of male gonocytes at E16.5 and found clear localization of DND1 to the P‐bodies (Fig 3D–F). Therefore, DND1 and NANOS2 co‐localized in the cytoplasm, particularly in the P‐bodies (Fig 3G–I), as expected. We next asked whether such an interaction is required for the localization to P‐bodies, using a co‐transfection method in cultured cells (NIH3T3 cells). Interestingly, DND1 and NANOS2 localized to P‐bodies only when both proteins were concurrently expressed (Fig 3J–M), whereas expression of each protein alone was not sufficient for clear localization to P‐bodies (Fig EV2T–Y). To further determine whether the direct interaction between DND1 and NANOS2 contributes to their P‐body localization, we conducted a biomolecular fluorescence complementation assay using VENUS‐C‐fused DND1 and VENUS‐N‐fused wild‐type NANOS2 or mutant‐type NANOS2 (C61A, C96A). In the case of wild‐type NANOS2, the fluorescence of VENUS protein was observed throughout the cytoplasm with some granular structures in which both NANOS2 and DND1 co‐localized (Fig 3N–Q). Additionally, these granular VENUS signals were merged with granules of DDX6, a P‐body marker (Fig 3R–U), indicating localization of the NANOS2–DND1 complex to P‐bodies. In contrast, no VENUS signal was detected when NANOS2 (C61A, C96A) was used (Fig 3V–Y), although the assembly of P‐bodies itself was observed in these cells (Fig 3Z–γ). These data demonstrate that the zinc finger domain of NANOS2 is required for the interaction with DND1 and that this interaction is essential for localization of the complex to the P‐bodies.
Figure 3. DND1 co‐localizes with NANOS2 in P‐bodies.
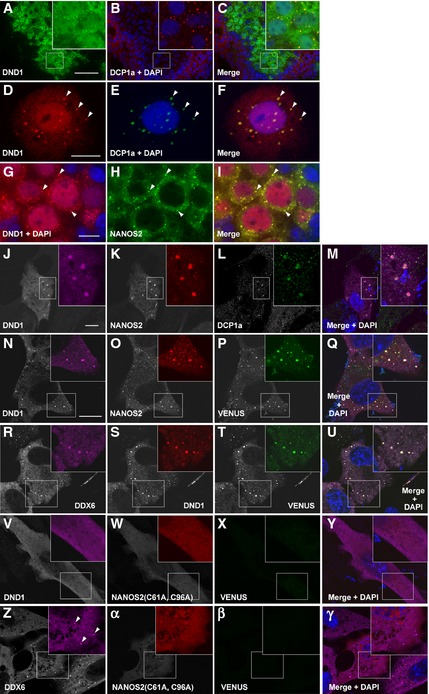
-
A–CSections of male gonads from E15.5 embryos were immunostained with antibodies against DND1 (green) and DCP1a (red).
-
D–FSquash preparation of a male gonocyte from E16.5 embryo immunostained with antibodies against DND1 (red) and DCP1a (green). Arrowheads indicate co‐localization of DND1 and DCP1a.
-
G–ISections of male gonads from E15.5 embryos were immunostained with antibodies against DND1 (red) and NANOS2 (green). Arrowheads indicate co‐localization of DND1 and NANOS2.
-
J–MNIH3T3 cells transfected with HA‐tagged Dnd1 and FLAG‐tagged Nanos2 were then immunostained with antibodies against DND1 (J) (magenta), NANOS2 (K) (red), and DCP1a (L) (green).
-
N–γBiomolecular fluorescence complementation assay. NIH3T3 cells transfected with VENUS‐C‐fused Dnd1 and VENUS‐N‐fused Nanos2 (N–U) or Nanos2 (C61A, C96A) (V–γ) were immunostained with antibodies against DND1 (N, S, V), NANOS2 (O, W, α) or DDX6 (R, Z). Then, the signals of VENUS fusion protein were visualized (P, T, X, β). Arrowheads in (Z) indicate P‐bodies.
Conditional deletion of DND1 results in similar but not identical phenotypes to those of Nanos2‐KO male gonocytes
Given that the interaction with DND1 is physiologically significant for NANOS2 function, the loss of DND1 in male gonocytes should result in similar phenotypes to those in Nanos2‐KO male gonocytes, including failure of mitotic arrest, entry into meiosis, apoptotic cell death, failure of male‐type differentiation, and upregulation of the PGC characteristics 13. To address this issue, we generated a conditional allele of Dnd1 (Fig EV3A–C) and crossed the Dnd1‐flox mouse with a transgenic mouse expressing CreERT2 under the control of the Oct4ΔPE enhancer (Oct4ΔPE‐CreER T2) 30, followed by tamoxifen injection at E13.5 (Fig EV3D–J). As shown in Western blotting analyses, DND1 expression gradually decreased and almost disappeared at E16.5, whereas the expression level of NANOS2 was unaffected by the deletion of DND1 (Fig 4A). In this condition, the expression of marker proteins for PGC characteristics, NANOS3, NANOG, and OCT3/4 30, 31, 32, was almost unaffected or downregulated in Dnd1‐cKO male gonads regardless of upregulation of these proteins in Nanos2‐KO male gonads at E15.5, indicating that NANOS2 suppresses PGC characteristics in a DND1‐independent manner. On the other hand, the expression of DNMT3L and PIWIL4/MIWI2, marker proteins for male‐type differentiation 33, 34, was strongly compromised. These expressions began at approximately E14.5 and continued in the control male gonads. In the gonads of Dnd1‐cKO mice, however, the expressions were gradually decreased in association with the reduction in DND1 to the same extent as in Nanos2‐KO male gonads at E15.5. These data indicate a failure of male‐type differentiation, as is the case in Nanos2‐KO male gonocytes. To examine germ cell differentiation status more precisely, we used several antibodies to examine mitotic activity, meiotic entry, and apoptosis by immunostaining 13, 35. We found strong signals of phosphorylated histone H3 (pH3) (Fig 4B and C), upregulation of the meiotic markers STRA8 and SYCP3 36, 37 (Fig 4E, F, H and I), and signals of activated caspase‐3 (Fig 4K and L), traits which were all reported to be observed in Nanos2‐KO male gonocytes 13 (Fig 4D, G, J and M). In addition, we found male gonocytes outside the tubules in Dnd1‐cKO male gonads (Fig 4N and O), which is also a phenotype of Nanos2‐KO mice 12 (Fig 4P). Collectively, although there is a difference in the aspect of suppression of PGC characteristics, other phenotypes are similar between these two mutant mice, indicating the physiological significance of DND1 for NANOS2 function.
Figure EV3. Strategy for conditional knockout of Dnd1 .
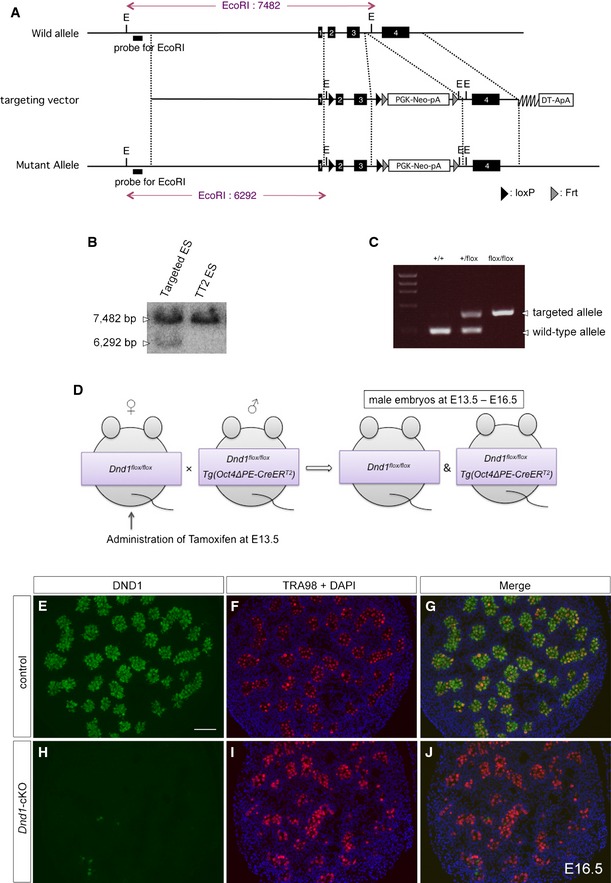
-
ASchematic representation of homologous recombination for the generation of a Dnd1‐flox allele.
-
BGenomic DNAs from a G418‐resistant and wild‐type TT2 embryonic stem (ES) cell clones were digested with EcoRI and analyzed by southern blotting using the probe indicated in (A).
-
CAgarose gel electrophoresis of polymerase chain reaction (PCR) products in the genotyping of Dnd1‐flox allele.
-
DSchematic representation of the mating strategy for conditional knockout of Dnd1.
-
E–JSections of E16.5 testes from Dnd1 flox/flox (control) and Dnd1 flox/flox embryos with the Oct4ΔPE‐CreERT 2 transgene (Dnd1‐cKO), both of which were administered with tamoxifen at E13.5, were immunostained with the antibodies against DND1 (green) (E, H) and TRA98 (red) (F, I). DNA was labeled with DAPI (blue) (F, G, I, J). Scale bar: 50 μm in (E) for (E–J).
Figure 4. Conditional deletion of DND1 causes similar but not identical phenotypes to those of Nanos2‐KO male gonocytes.
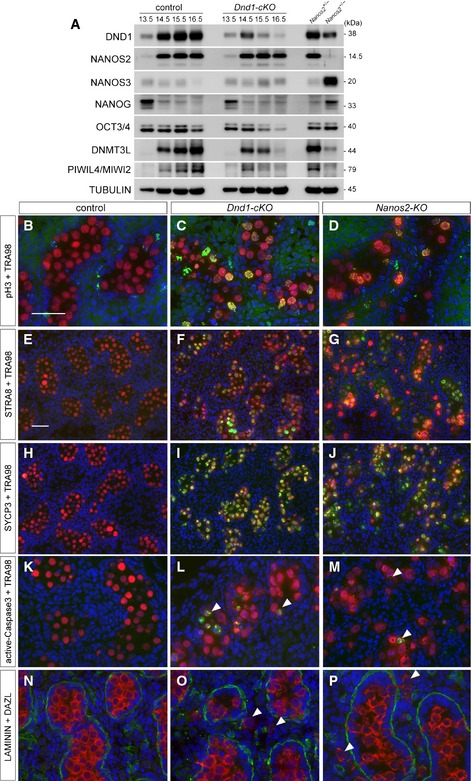
-
AWestern blotting analyses of proteins in testes from E13.5 to E16.5 embryos of Dnd1 flox/flox or Dnd1 flox/flox _Tg(Oct4ΔPE‐Cre ERT 2 ) each administered with tamoxifen at E13.5 and E15.5 embryos of Nanos2 +/− or Nanos2 −/− with the indicated antibodies.
-
B–PSections of testes from Dnd1 flox/flox (B, E, H, K, N), Dnd1 flox/flox _Tg(Oct4ΔPE‐Cre ERT 2 ) (C, F, I, L, O) or Nanos2 −/− (D, G, J, M, P) embryos were prepared at E16.5 and then immunostained with antibodies against pH3 (B–D), STRA8 (E–G), SYCP3 (H–J), activated caspase‐3 (K–M) or LAMININ (N–P) (green). Germ cells were immunostained with TRA98 (B–D) or DAZL (N–P) (red), and DNA was labeled with DAPI (blue). Tamoxifen was administered at E13.5. Arrowheads indicate apoptotic cells (L, M) or cells outside the tubules (O, P). Scale bars: 50 μm in (B) for (B–D) and (K–P), 50 μm in (E) for (E–J). See also Fig EV3.
DND1 is required for loading of the target RNAs into the NANOS2–CNOT complex
To further address the significance of the interaction between DND1 and NANOS2, we next analyzed the localization of NANOS2 in Dnd1‐cKO male gonocytes by immunostaining and found that no cytoplasmic granular structures were observed in the absence of DND1 (Fig 5A–F). Therefore, we checked the status of P‐bodies with an anti‐DCP1a antibody and found that almost all of them had disappeared in Dnd1‐cKO male gonocytes (Fig 5G–L). This dissipation of P‐bodies was confirmed using an anti‐DDX6 antibody (Fig 5M–R), indicating the necessity of DND1 for the assembly of P‐bodies in male gonocytes. This DND1‐dependent mechanism of P‐body assembly may be unique to male gonocytes and different from the mechanism of assembly in cultured cells (Fig 3J–L). Nevertheless, the results suggest that target RNAs may not be loaded into the NANOS2–CNOT deadenylase complex without DND1, since loading of RNAs in the degradation pathway is a prerequisite for P‐body assembly 16, 38. To test this possibility, we generated transgenic mouse lines expressing FLAG‐tagged DND1 (Fig EV4A–L) and examined whether DND1‐associated mRNAs included NANOS2‐associated mRNAs by IP from E15.5 male gonadal extracts of the transgenic embryos. Western blotting analyses revealed co‐precipitation of NANOS2 with FLAG‐tagged DND1 (Fig 5S), and reverse transcription‐quantitative polymerase chain reaction (RT–qPCR) results showed that the known NANOS2‐associated mRNAs, Sycp3, Dazl, Nanog, Stra8, and Taf7l 19, 35 were enriched among the RNAs precipitated with FLAG‐tagged DND1 whereas α‐tubulin mRNA was not (Fig 5T). Finally, to test whether these target RNAs were loaded into NANOS2 in the absence of DND1, we collected Dnd1‐cKO embryonic gonads and conducted IP using antibodies against NANOS2. We used α‐tubulin mRNA as a negative control. Although the amounts of NANOS2 precipitated with the antibodies were almost same between control and Dnd1‐cKO gonadal extracts (Fig 5U), the amounts of the NANOS2‐associated mRNAs, except for Stra8 and Taf7l mRNAs (Sycp3, Dazl and Nanog), were reduced drastically in the absence of DND1 (Fig 5V). These results strongly indicate that DND1 is essential for the association of NANOS2 with certain target mRNAs. Concerning Stra8 and Taf7l mRNAs, however, not all of the target mRNAs appear to interact with NANOS2 in a DND1‐dependent manner. We, therefore, validated what quantity of NANOS2 associated with DND1 in the male gonocytes and vice versa. Immunodepletion analyses using E15.5 male gonadal extracts revealed that the amount of DND1 was slightly affected by NANOS2 depletion (Fig 5W), whereas a large fraction of NANOS2 was eliminated by DND1 depletion (Fig 5X), suggesting that DND1 is a major partner of NANOS2 in male gonocytes. However, a small fraction of NANOS2 seemed to remain in the extracts even after DND1 depletion (Fig 5X), indicating that some NANOS2 is found outside of the DND1–NANOS2 complex. Therefore, certain RNAs, such as Stra8 and Taf7l mRNAs, might associate with a small fraction of NANOS2 that is not associated with DND1; or an alternate RBP regulates the association of these RNAs with the NANOS2–DND1 complex independently of DND1.
Figure 5. DND1 is required for loading of the target RNAs to NANOS2–CNOT complex.
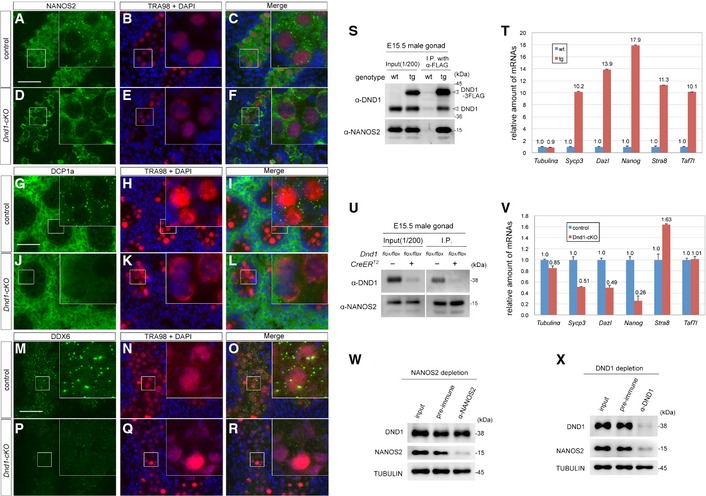
-
A–RSections of testes from Dnd1 flox/flox (A–C, G–I, M–O) and Dnd1 flox/flox _Tg(Oct4ΔPE‐CreERT2) (D–F, J–L, P–R) embryos were prepared at E16.5 and then immunostained with antibodies against NANOS2 (A, D) (green), DCP1a (G, J) (green), DDX6 (M, P) (green) and TRA98 (B, E, H, K, N, Q). DNA was labeled with DAPI (blue). Tamoxifen was administered at E13.5. Scale bars: 50 μm in (A) for (A–F); 50 μm in (G) for (G–L); 50 μm in (M) for (M–R). Insets show an enlarged version of each picture to better depict localization of NANOS2, DCP1a and DDX6.
-
S–VImmunoprecipitation with an anti‐FLAG antibody from E15.5 male gonadal extracts of wild‐type and the transgenic mouse line expressing FLAG‐tagged DND1 (S, T), or with an anti‐NANOS2 antibody from E15.5 male gonadal extracts of Dnd1 flox/flox mice with or without Oct4ΔPE‐CreERT2 (U, V). Precipitates were analyzed by Western blotting (S, U) or by RT–qPCR (T, V). The RT–qPCR data in (T) and (V) are shown as average relative mRNA levels ± SE (n = 3).
-
W, XWestern blotting analyses of proteins in NANOS2‐depleted (W) or DND1‐depleted (X) testis extracts from E15.5 embryos. Proteins were analyzed with the indicated antibodies.
Figure EV4. Characterization of the transgenic mouse line expressing FLAG‐tagged DND1.
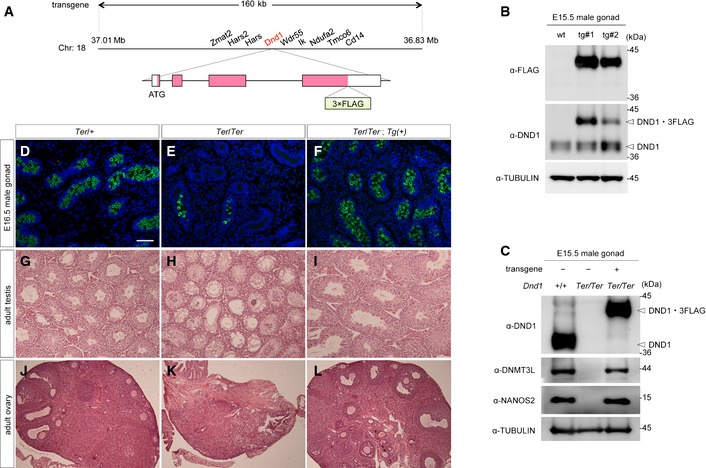
-
ASchematic representation of the bacterial artificial chromosome (BAC) transgene. The DNA sequence encoding 3×FLAG‐tag is inserted at the C‐terminus of Dnd1, resulting in the expression of DND1‐3×FLAG fusion protein under the direct control of the Dnd1 enhancer.
-
BWestern blotting analyses of proteins in E15.5 testes from wild‐type and two independent transgenic embryos (line #1 and #2) with the antibodies indicated. Line #1 was used for further analysis because it produced larger amounts of DND1‐3×FLAG.
-
CWestern blotting analyses of proteins in E15.5 testes from wild‐type, Ter‐homozygous, and from Ter‐homozygous embryos expressing the FLAG‐tagged Dnd1 transgene with the antibodies indicated.
-
D–FSections of testes from E16.5 Ter‐heterozygous, Ter‐homozygous, and from Ter‐homozygous embryos with the FLAG‐tagged Dnd1 transgene were immunostained with anti‐MVH antibodies. DNA was labeled with DAPI (blue). Scale bar: 50 μm in (D) for (D–F).
-
G–LSections of adult testes (G–I) or ovaries (J–L) from Ter‐heterozygous, Ter‐homozygous, and from Ter‐homozygous mice with the FLAG‐tagged Dnd1 transgene were stained with hematoxylin and eosin.
On the other hand, NANOG expression was upregulated in Nanos2‐KO male gonads (Fig 4A) and its mRNA associated with NANOS2 in a DND1‐dependent manner (Fig 5V). However, NANOG expression in Dnd1‐cKO male gonads was almost unaffected (it was similar to that in the wild‐type) (Fig 4A). These data suggest that Nanog is suppressed mainly by an unknown mechanism under the control of NANOS2 but not DND1, and the DND1–NANOS2 complex contributes to only a part of the suppression mechanism for Nanog mRNA. Since DND1 is also implicated in mRNA protection from miRNA‐mediated RNA degradation in other context 39, identification of RNAs associated with the DND1–NANOS2 complex and DND1‐partner proteins except for NANOS2 is required for understanding the Nanos2‐KO and Dnd1‐cKO phenotypes.
Materials and Methods
The detailed experimental procedures are available in the Appendix.
Ethics statement
Experiments were carried out with the permission of the animal experimental committee at the Yokohama National University and the National Institute of Genetics.
Mice
The Nanos2‐KO mouse line, the transgenic mouse line expressing FLAG‐tagged NANOS2 and the Ter‐mutant mouse line were previously described 12, 19, 21.
Immunoprecipitation – mass spectrometry
For the mass‐spectrometric analyses, 100 testes from either wild‐type or transgenic E15.5 embryos expressing FLAG‐tagged NANOS2 were used for the generation of extracts as previously described 19.
Biomolecular fluorescence complementation assay
Both VENUS‐C‐fused Dnd1 and VENUS‐N‐fused Nanos2 were cloned into pcDNA3.1 as described previously 40 and transfected into NIH3T3 cells.
Author contributions
AS and YS developed the concept of this paper. KS carried out the mass‐spectrometric analyses and ZZ conducted the biomolecular fluorescence complementation assay, and MK generated the Dnd1 conditional knockout mouse line. AS and YN carried out all other experiments and data analyses. AS and YS prepared the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Review Process File
Acknowledgements
We thank the following researchers for providing antibodies as a gift: Y. Nishimune (TRA98), S. Chuma and N. Nakatsuji (anti‐SYCP3), H. T. Timmers (anti‐CNOT1), T. Tamura (anti‐CNOT3), A. B. Shyu (anti‐CNOT7/Caf1a), G. J. Hannon (anti‐AGO2), S. Yamanaka (anti‐DNMT3L), and H. Okayama (anti‐CNOT9/Rcd1). We thank Dr. M. Noguchi for providing Ter‐mutant mouse line. We are also grateful to A. Nakamura and S. Hayashi for advice for mass‐spectrometric analysis, and to K. Miyoshi, Y. Nakajima, Y. Morita, T. Ichijo, A. Fukunaga, Y. Odani, T. Naka and N. Nishimaki for their technical assistance. This work was supported in part by the Japan Society for the Promotion of Science KAKENHI Grant Number 21227008, 25112002 and 26251025 to Y. S. and 23112707, 26114505 and 26440120 to A. S.
EMBO Reports (2016) 17: 37–46
References
- 1. Anderson P, Kedersha N (2009) RNA granules: post‐transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10: 430–436 [DOI] [PubMed] [Google Scholar]
- 2. Martin KC, Ephrussi A (2009) mRNA localization: gene expression in the spatial dimension. Cell 136: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Voronina E, Seydoux G, Sassone‐Corsi P, Nagamori I (2011) RNA granules in germ cells. Cold Spring Harb Perspect Biol 3: a002774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Surani MA, Hayashi K, Hajkova P (2007) Genetic and epigenetic regulators of pluripotency. Cell 128: 747–762 [DOI] [PubMed] [Google Scholar]
- 5. McLaren A (2003) Primordial germ cells in the mouse. Dev Biol 262: 1–15 [DOI] [PubMed] [Google Scholar]
- 6. Bowles J, Knight D, Smith C, Wilhelm D, Richman J, Mamiya S, Yashiro K, Chawengsaksophak K, Wilson MJ, Rossant J et al (2006) Retinoid signaling determines germ cell fate in mice. Science 312: 596–600 [DOI] [PubMed] [Google Scholar]
- 7. Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, Page DC (2006) Retinoic acid regulates sex‐specific timing of meiotic initiation in mice. Proc Natl Acad Sci USA 103: 2474–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu Q, Kanata K, Saba R, Deng CX, Hamada H, Saga Y (2013) Nodal/activin signaling promotes male germ cell fate and suppresses female programming in somatic cells. Development 140: 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kobayashi S, Yamada M, Asaoka M, Kitamura T (1996) Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature 380: 708–711 [DOI] [PubMed] [Google Scholar]
- 10. Koprunner M, Thisse C, Thisse B, Raz E (2001) A zebrafish nanos‐related gene is essential for the development of primordial germ cells. Genes Dev 15: 2877–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Subramaniam K, Seydoux G (1999) nos‐1 and nos‐2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 126: 4861–4871 [DOI] [PubMed] [Google Scholar]
- 12. Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y (2003) Conserved role of nanos proteins in germ cell development. Science 301: 1239–1241 [DOI] [PubMed] [Google Scholar]
- 13. Suzuki A, Saga Y (2008) Nanos2 suppresses meiosis and promotes male germ cell differentiation. Genes Dev 22: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Suzuki A, Saba R, Miyoshi K, Morita Y, Saga Y (2012) Interaction between NANOS2 and the CCR4‐NOT deadenylation complex is essential for male germ cell development in mouse. PLoS One 7: e33558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Decker CJ, Parker R (2012) P‐bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 4: a012286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen CY, Shyu AB (2013) Deadenylation and P‐bodies. Adv Exp Med Biol 768: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Curtis D, Treiber DK, Tao F, Zamore PD, Williamson JR, Lehmann R (1997) A CCHC metal‐binding domain in Nanos is essential for translational regulation. EMBO J 16: 834–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hashimoto H, Hara K, Hishiki A, Kawaguchi S, Shichijo N, Nakamura K, Unzai S, Tamaru Y, Shimizu T, Sato M (2010) Crystal structure of zinc‐finger domain of Nanos and its functional implications. EMBO Rep 11: 848–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Suzuki A, Igarashi K, Aisaki K, Kanno J, Saga Y (2010) NANOS2 interacts with the CCR4‐NOT deadenylation complex and leads to suppression of specific RNAs. Proc Natl Acad Sci USA 107: 3594–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weidinger G, Stebler J, Slanchev K, Dumstrei K, Wise C, Lovell‐Badge R, Thisse C, Thisse B, Raz E (2003) Dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr Biol 13: 1429–1434 [DOI] [PubMed] [Google Scholar]
- 21. Youngren KK, Coveney D, Peng X, Bhattacharya C, Schmidt LS, Nickerson ML, Lamb BT, Deng JM, Behringer RR, Capel B et al (2005) The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature 435: 360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stevens LC (1973) A new inbred subline of mice (129‐terSv) with a high incidence of spontaneous congenital testicular teratomas. J Natl Cancer Inst 50: 235–242 [DOI] [PubMed] [Google Scholar]
- 23. Kedersha N, Anderson P (2007) Mammalian stress granules and processing bodies. Methods Enzymol 431: 61–81 [DOI] [PubMed] [Google Scholar]
- 24. Liu J, Valencia‐Sanchez MA, Hannon GJ, Parker R (2005) MicroRNA‐dependent localization of targeted mRNAs to mammalian P‐bodies. Nat Cell Biol 7: 719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bhandari D, Raisch T, Weichenrieder O, Jonas S, Izaurralde E (2014) Structural basis for the Nanos‐mediated recruitment of the CCR4‐NOT complex and translational repression. Genes Dev 28: 888–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sonoda J, Wharton RP (1999) Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev 13: 2704–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kraemer B, Crittenden S, Gallegos M, Moulder G, Barstead R, Kimble J, Wickens M (1999) NANOS‐3 and FBF proteins physically interact to control the sperm‐oocyte switch in Caenorhabditis elegans. Curr Biol 9: 1009–1018 [DOI] [PubMed] [Google Scholar]
- 28. Nakahata S, Katsu Y, Mita K, Inoue K, Nagahama Y, Yamashita M (2001) Biochemical identification of Xenopus Pumilio as a sequence‐specific cyclin B1 mRNA‐binding protein that physically interacts with a Nanos homolog, Xcat‐2, and a cytoplasmic polyadenylation element‐binding protein. J Biol Chem 276: 20945–20953 [DOI] [PubMed] [Google Scholar]
- 29. Jaruzelska J, Kotecki M, Kusz K, Spik A, Firpo M, Reijo Pera RA (2003) Conservation of a Pumilio‐Nanos complex from Drosophila germ plasm to human germ cells. Dev Genes Evol 213: 120–126 [DOI] [PubMed] [Google Scholar]
- 30. Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, Hubner K, Scholer HR (1996) Germline regulatory element of Oct‐4 specific for the totipotent cycle of embryonal cells. Development 122: 881–894 [DOI] [PubMed] [Google Scholar]
- 31. Suzuki H, Tsuda M, Kiso M, Saga Y (2008) Nanos3 maintains the germ cell lineage in the mouse by suppressing both Bax‐dependent and ‐independent apoptotic pathways. Dev Biol 318: 133–142 [DOI] [PubMed] [Google Scholar]
- 32. Yamaguchi S, Kimura H, Tada M, Nakatsuji N, Tada T (2005) Nanog expression in mouse germ cell development. Gene Expr Patterns 5: 639–646 [DOI] [PubMed] [Google Scholar]
- 33. Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ (2007) MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- 34. Sakai Y, Suetake I, Shinozaki F, Yamashina S, Tajima S (2004) Co‐expression of de novo DNA methyltransferases Dnmt3a2 and Dnmt3L in gonocytes of mouse embryos. Gene Expr Patterns 5: 231–237 [DOI] [PubMed] [Google Scholar]
- 35. Saba R, Kato Y, Saga Y (2014) NANOS2 promotes male germ cell development independent of meiosis suppression. Dev Biol 385: 32–40 [DOI] [PubMed] [Google Scholar]
- 36. Yuan L, Liu JG, Zhao J, Brundell E, Daneholt B, Hoog C (2000) The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol Cell 5: 73–83 [DOI] [PubMed] [Google Scholar]
- 37. Baltus AE, Menke DB, Hu YC, Goodheart ML, Carpenter AE, de Rooij DG, Page DC (2006) In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat Genet 38: 1430–1434 [DOI] [PubMed] [Google Scholar]
- 38. Parker R, Sheth U (2007) P bodies and the control of mRNA translation and degradation. Mol Cell 25: 635–646 [DOI] [PubMed] [Google Scholar]
- 39. Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA et al (2007) RNA‐binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131: 1273–1286 [DOI] [PubMed] [Google Scholar]
- 40. Kerppola TK (2013) Bimolecular fluorescence complementation (BiFC) analysis of protein interactions in live cells. Cold Spring Harb Protoc 2013: 727–731 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Review Process File


