Abstract
We report the structures of an iridoid synthase, nepetalactol synthase, in the presence of NAD+, NADPH or NAD+/10-oxogeranial. The 10-oxogeranial substrate binds in a transoid-O1-C3 conformation and can be reduced by hydride addition to form S-10-oxo-citronellal, a by-product of the enzyme. Tyr178 Oζ is 2.5 Å from the substrate O1, providing the second proton required for reaction. Nepetalactol product formation requires rotation about C1-C2 to form the cisoid isomer leading to formation of the cis-enolate, together with rotation about C4-C5, enabling cyclization and lactol production. The structure is similar to that of progesterone-5β-reductase with almost identical positioning of NADP, Lys146(147), Tyr178(179) and F342(343) but only Tyr178 and Phe342 appear to be essential for activity. The transoid 10-oxogeranial structure also serves as a model for β-face hydride attack in progesterone 5β-reductases and is of general interest in the context of asymmetric synthesis.
Keywords: Monoterpene, mechanism, catalysis, crystallography, natural products
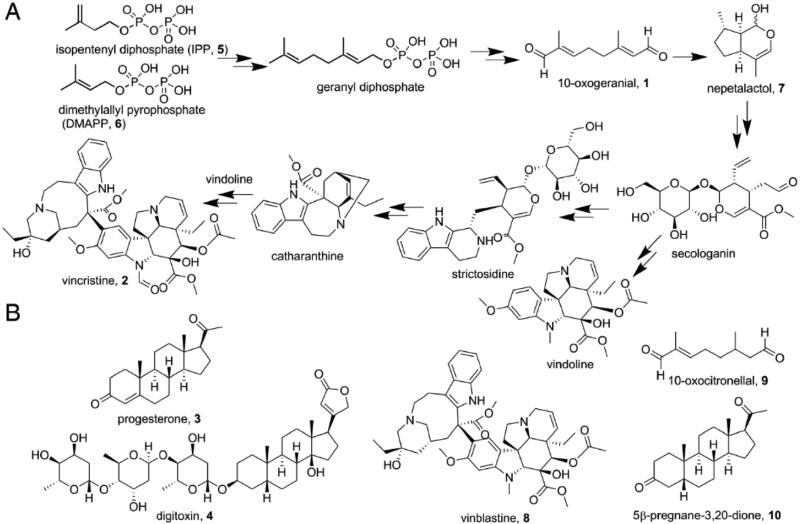
Terpenes and related isoprenoids are the largest class of small-molecule natural products on earth, and the most abundant by mass. [1] Most are made by enzymes that are represented by a relatively small number of folds [2], primarily head-to-tail and head-to-head trans-prenyl transferases, cis-prenyltransferases, and a range of cyclases with one, two or three domains. In most cases, the reactions proceed via formation of carbo-cationic transition states/reactive intermediates. However, in recent work, an alternative route to the formation of terpenes — involving reductive cyclization—has been reported, for iridoid biosynthesis [3]. Iridoids are monoterpenes that are produced from (C10) 10-oxogeranial (1) by an NAD(P)H dependent reduction, followed by a cyclization step that involves either a Diels-Alder cycloaddition or a Michael addition [3a, 4]. There are no X-ray structures of any iridoid synthase (IRIS), with or without substrates, or other ligands. The structures of the iridoid synthases are of interest because their products are converted to important natural products, compounds such as the anti-cancer drug vincristine (2), as well as being of mechanistic interest—not least because their amino-acid sequences have high homology to the plant progesterone (3) 5β-reductases involved in formation of cardiac glycoside drugs such as digitoxin (4). A simplified version of the biosynthesis of vincristine (2) from isopentenyl diphosphate (5) and dimethylallyl diphosphate (6) is shown in Scheme 1A; the structures of other molecules discussed in the Text are shown in Scheme 1B.
Scheme 1.
A) Vincristine biosynthesis from isopentenyl diphosphate and dimethylallyl diphosphate. B) Structures of other compounds of interest.
We expressed and purified an iridoid synthase, Cantharanthus roseus (1R, 4aS, 7S, 7aR) nepetalactol (7) synthase, from the corresponding chemically synthesized gene (Genebank accession number K7WDL7.1). C. roseus is the Madagascar periwinkle and produces both vincristine (2) and vinblastine (8), anti-cancer indole terpenes, from iridoids. Attempts to crystallize full length protein were unsuccessful so we attempted crystallization of two N-terminus truncated variants, ΔN13 and ΔN25. ΔN25 crystals were obtained and full protein expression, purification and crystallization details are given in the Supporting Information. The activity of the ΔN25 construct, was (within experimental error) the same as that of the wild type protein, Figure S1. We then co-crystallized this IRIS with either NAD+, NADPH or NAD++10-oxogeranial and obtained their structures. Full data acquisition and refinement details are given in Table S1.
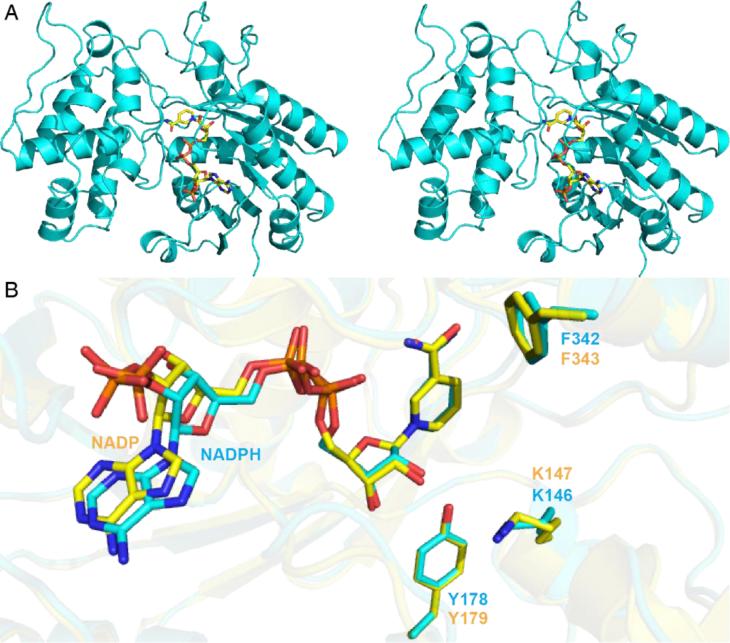
We first obtained the structures of IRIS with either NADPH or NAD+-bound. A stereo view of the NADPH structure is shown in Figure 1A; the NAD+ structure is virtually identical (a 0.21 Å root mean square deviation, rmsd). The protein fold is most similar to that of progesterone 5β-reductase from Digitalis lanata, a plant that produces cardiac glycosides such as 4. There is a 1.0 Å Cα rmsd over 351 residues (using the PDBe Fold Server [5]). An alignment of the active sites in the IRIS protein (+NADPH, PDB ID code 5DBF) and the D. lanata 5β-reductase (+NADP+, PDB ID code 2V6G [6]) is shown in Figure 1B. Both active site structures are very similar and of particular interest, the Tyr179 shown by site-directed mutagenesis to be essential for progesterone 5β-reductase activity [6] occupies the same position in both the IRIS (Tyr178) and progesterone 5β-reductase structures, as do the NADP cofactors and the Lys146 (Lys147 in progesterone 5β-reductase) residues that have been proposed to be involved in progesterone 5β-reductase catalysis. There are, therefore, great similarities between the IRIS and progesterone 5β-reductase structures with the Tyr Oζ being in close proximity to the ribose O2 (~2.8 Å) in both systems. Additional protein-ligand contacts are shown in Figures S2-5. These structures do not, however, give new clues as to the mechanisms of catalysis.
Figure 1.
Structure of C. roseus IRIS+NADPH and a comparison with D. lanata progesterone 5β-reductase. A) stereo view of IRIS in complex with NADPH (PDB ID: 5DBF). B) Superimposition of IRIS+NADPH structure (cyan) and progesterone 5β-reductase (PDB ID: 2V6G[6]) in complex with NADP+ (yellow).
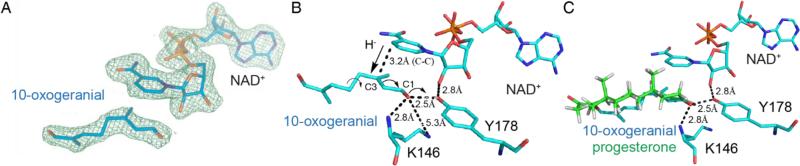
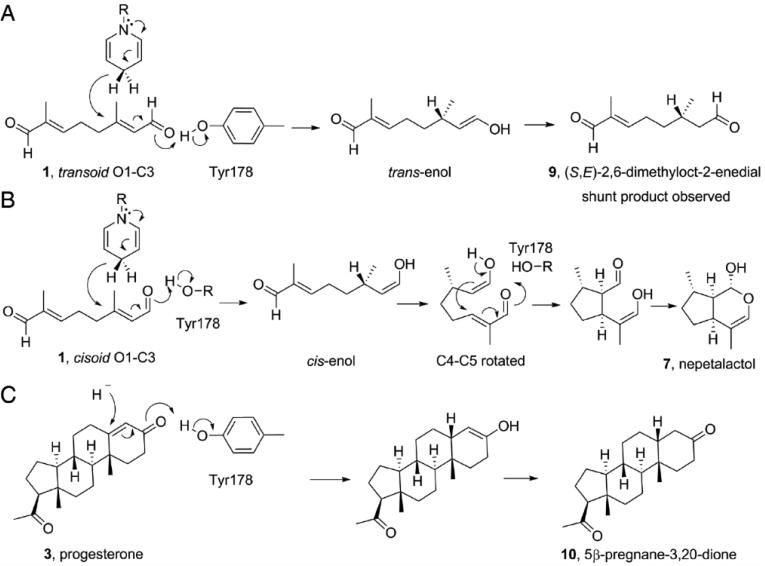
We thus soaked 10-oxogeranial into crystals of IRIS co-crystallized with NAD+ (to prevent 10-oxogeranial substrate reduction) and obtained an IRIS+NAD++10-oxogeranial complex structure (PDB ID code 5DBI). Attempts to obtain structures with nepetalactol or progesterone (+/− NADPH) were not successful. The bound NAD+ and 10-oxogeranial electron-density map is shown in Figure 2A, and the active site region together with distances of interest is shown in Figure 2B. This structure is essential for a detailed analysis of the IRIS (and progesterone reductase) mechanism of action. As can be seen in Figure 2 (and the ligand interaction plots in SI Figures S4, S5), the NAD+ (NADH, when reduced) nicotinamide, 10-oxogeranial and Tyr178 OζH groups are perfectly aligned for a 1,4 hydride addition. The pro-4(S) proton from NADH attacks the C2,3 double bond from “above” (in Figure 2B) to generate the transenol, the Tyr178 OζH providing the enolate hydrogen. This reaction is shown schematically in Figure 3A. The Tyr178-Oζ is only 2.5 Å from the 10-oxogeranial O1 in the crystal structure, suggesting that there could be a hydrogen-bond present, even prior to reduction. On hydride attack, the tyrosinate formed can be stabilized by interacting with the ribose O2H (2.8 Å distant in the X-ray structure, at least prior to reduction). The enol that forms on hydride-attack has a 3(S) center, as found in nepetalactol. However, the product of this reaction is actually S-10-oxocitronellal (9), a known [4] shunt reaction product observed experimentally, not nepetalactol, as shown in the Figure 3A. In order for nepetalactol to be formed, there needs to be a 180° rotation about C1-C2 to form the cisoid isomer of 1, which on reduction forms the cis-enol, Figure 3B. Rotation about C4-C5 is also required (as indicated in Figure 2B and the SI video), then cyclization and lactol formation can proceed, as illustrated in Figure 3B.
Figure 2.
The active-site region of the C. roseus iridoid synthase from the IRIS+NAD++10-oxogeranial structure (PDB^^ID: 5DBI). A) Electron-density map of bound NAD+ and substrate 10-oxogeranial (2 Fo-Fc map contoured at 1 sigma). B) Interaction of active-site residues K146 and Y178 with NAD+ and 10-oxogeranial substrates. The C4-C5 rotation to enable cyclization is indicated. Dashed lines indicate distances of interest. C) Progesterone modeled into the IRIS active site.
Figure 3.
Proposed catalytic mechanism of iridoid synthase and progesterone reductase based on the IRIS+ NAD++10-oxogeranial structure (PDB ID: 5DBI). A) transoid 1 gets reduced to S-10-oxocitronellal (9). B) Rotation about C1-C2 enables cis-enol formation, and with C4-C5 rotation, nepetalactol formation. C) Progesterone reacts as in (A).
These results are also of interest in the context of the mechanism of action of the homologous progesterone 5β-reductase in plants such as D. lanata. The reduction of progesterone to 5β-pregnane-3,20-dione (10) by progesterone 5β-reductase is expected to be similar to that proposed for reduction of the transoid form of 1, Figure 3A, with hydride attack on the β-face, at C5, Figure 3C. When compared with the previous structures and mechanistic studies of progesterone 5β-reductase [6], it can be seen that in IRIS the 10-oxogeranial is immediately adjacent to Tyr178, which provides one of the protons required for catalysis, rather than occupying a more distal location, as proposed for progesterone 5β-reductase based on computational modeling. Tyr178 is, however, not the only residue that has been proposed to be involved in progesterone 5β-reductase. Specifically, K147 was suggested to play a role [6], as a proton source. The corresponding residue in IRIS is K146 which, together with Y178, is very well aligned (a 0.171 Å rmsd) between D. lanata progesterone 5β-reductase and the C. roseus IRIS protein. However, the lysine ε-NH3+ group is ~5.3 Å from the Tyr178Oζ, Figure 2B, so it seems unlikely that this Lys plays an essential role in re-protonating Tyr178 after 10-oxogeranial reduction. The backbone N in K146 is, however, only 2.85 Å from the 10-oxogeranial O1, suggesting an H-bond interaction, but this would probably be seen with most amino acids in this position.
Based on the structural data we, it appears that the transoid form simply binds more strongly, and that is why it is seen crystallographically. However, it is reduced relatively slowly (in solution), which is why S-10-oxocitronellal (9) is a minor product. The cisoid form binds more weakly (and we do not see it), but it reacts more rapidly (in solution) which is why, under reducing conditions, nepetalactol is the major product. This is, of course, a post hoc analysis since the X-ray structure is of the transoid form, H-bonded to the (essential) Y178 Cζ-OH.
The similarities between the transoid enone fragment in 10-oxogeranial and the same transoid moiety in progesterone suggested that is should be possible to obtain a good model for progesterone binding simply by aligning the respective enone oxygens and C3 (10-oxogeranial) and C5 (progesterone) carbons. This appears to be the case since as shown in Figure 2C there is a very good overlap of the entire progesterone molecule with 10-oxogeranial, an observation that is expected to facilitate future work on progesterone (and other enone) reductase reactions of interest in asymmetric synthesis.
Next, we sought to see if there were any other residues that might have catalytic activity in both the IRIS and progesterone 5β-reductase reactions, using a SCORECONS analysis [7] of a JPRED3 alignment [8] of C. roseus iridoid synthase. SCORECONS is a computer program that predicts the “essential” nature of a residue for catalysis, based on its occurrence (and physicochemical similarities between different residues) in a set of aligned sequences. A score of 1.000 means really essential, 0, not essential. A list of the top 20 most conserved residues is shown in Table S2. The most conserved residues are Gs, involved in Rossman fold formation. Many of the other conserved residues are hydrophobic, and all conserved residues cluster in the N-terminal domain. Of the polar residues that might be involved in catalysis, R200 and E182 actually form a salt-bridge (2.9 Å Oε2/NH2) located ~10 Å from the substrate C1. D84 forms a salt-bridge with the adenine N6. So, Y178 appears to be most essential for catalysis in both proteins since it is highly conserved and, of course, it is immediately adjacent to the 10-oxogeranial substrate.
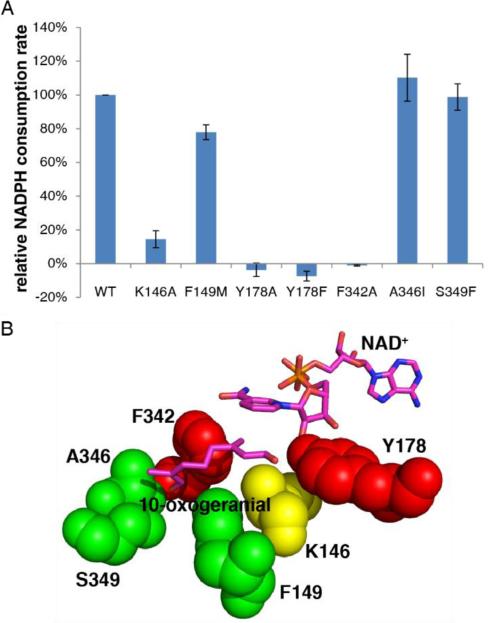
To help test the proposed roles of this and other residues in or near the [putative] catalytic center, we produced 7 IRIS single-point mutants: K146A, F149M, Y178F, Y178A, F342A, A346I, and S349F. Enzyme activity results are shown in Figure 4A and a “heat map” of the results: red=essential; green=non-essential, yellow=desirable) is shown in Figure 4B. These results confirm the essential nature of Y178 for activity. F342 is also essential and this result is of interest since it supports the role of the equivalent Phe residue (in progesterone reductase) for small enol reductase activity [3b], while F149 is not essential-consistent with its non-conserved nature. K146 has reduced activity, but no obvious role for this residue is suggested from the X-ray structure. It might be involved in the second half reaction, but it is clearly not absolutely essential (as also noted in progesterone reductase by Petersen et al. [3e]. A346 and S349, which are close to the substrate, are also not essential for 10-oxogeranial reduction, consistent again with their non-conserved nature. Based on Phe mutagenesis and MD simulations, Petersen et al. propose that several Phes (that are close to the substrate in our structure) play a dual role. In IRIS or IRIS-like reduction reactions involving small 1,4-enones, they help keep ligands inside the protein, while with large enones (e.g. progesterone) they again act as “gatekeepers”, this time impeding access to the active site. Phe>Ala mutations of several PRISEs [3e] favor progesterone reduction, but disfavor small enone reduction. This is consistent with what we see with IRIS in which the F342A mutant is inactive in 10-oxogeranial reduction.
Figure 4.
Mutagenesis results for C. roseus iridoid synthases. A) Activities of wild-type and mutant IRIS proteins for 10-oxogeranial reduction. B) “Heat map” showing the effects of mutations on catalytic activity: green=no effect; yellow=significant effect; red=major effect (no activity, within experimental error).
In summary, the iridoid synthase that produces nepetalactol in C. roseus has a structure that is very similar to that found for progesterone 5β-reductase from D. lanata. This iridoid synthase can produce two different products: reduction of the transoid substrate seen crystallographically results in formation of the side-product S-10-oxocitronellal; reduction of the cisoid form, together with a C4-C5 rotation, enables cyclization and formation of the major product, nepetalactol. The Tyr178 CζOH group provides the proton required for enol formation, the Lys146 HN can H-bond to the substrate O1, and the tyrosinate produced in the first half reaction can be stabilized by H-bonding to the ribose O2H. There are no other acidic groups close-by that appear to be involved in catalysis although F342 is essential for 10-oxogeranial reduction, consistent with the progesterone-5β-reductase mutagenesis results with small enone substrates [3b]. Overall, these results are of broad general interest since they provide new insights into the structures and catalytic mechanisms of iridoid synthases/progesterone 5β–reductases, enzymes that are involved in the biosynthesis of important cancer and heart drugs—the indole terpene alkaloids such as vincristine (2) and cardiac glycosides such as digitoxin (4), in addition to providing structural models for the future development of asymmetric catalysts.
Supplementary Material
Acknowledgements
We thank the National Synchrotron Radiation Research Center of Taiwan for beam time allocation and data collection assistance. This work was supported by National High Technology Research and Development Program of China (2012AA022209), the National Natural Science Foundation of China (31200053, 31400678 and 31300615), the United States Public Health Service (NIH grants CA158191, GM065307), a Harriett A. Harlin Professorship and the University of Illinois Foundation/Oldfield Research Fund.
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- 1.Dictionary of Natural Products on DVD. Chapman and Hall/CRC; 2013. [Google Scholar]
- 2.Oldfield E, Lin FY. Angew. Chem. Int. Ed. Engl. 2012;51:1124–1137. doi: 10.1002/anie.201103110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a Geu-Flores F, Sherden NH, Courdavault V, Burlat V, Glenn WS, Wu C, Nims E, Cui Y, O'Connor SE. Nature. 2012:492, 138. doi: 10.1038/nature11692. [DOI] [PubMed] [Google Scholar]; b Petersen J, Lanig H, Munkert J, Bauer P, Müller-Uri F, Kreis W. J. Biomol. Struct. Dyn. 2015:1–34. doi: 10.1080/07391102.2015.1088797. [DOI] [PubMed] [Google Scholar]; c Miettinen K, Dong L, Navrot N, Schneider T, Burlat V, Pollier J, Woittiez L, van der Krol S, Lugan R, Ilc T, Verpoorte R, Oksman-Caldentey K-M, Martinoia E, Bouwmeester H, Goossens A, Memelink J, Werck-Reichhart D. Nat. Commun. 2014:5. doi: 10.1038/ncomms4606. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Munkert J, Pollier J, Miettinen K, Van Moerkercke A, Payne R, Mueller-Uri F, Burlat V, O'Connor SE, Memelink J, Kreis W, Goossens A. Molecular Plant. 2015;8:136–152. doi: 10.1016/j.molp.2014.11.005. [DOI] [PubMed] [Google Scholar]; e Krithika R, Srivastava PL, Rani B, Kolet SP, Chopade M, Soniya M, Thulasiram HV. Sci. Rep. 2015:5. doi: 10.1038/srep08258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindner S, Geu-Flores F, Braese S, Sherden NH, O'Connor SE. Chem. Biol. 2014;21:1452–1456. doi: 10.1016/j.chembiol.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a Otwinowski WMZ. In: Method. Enzymol. Carter JRMSCW, editor. Vol. 276. Academic Press; New York: 1997. pp. 307–326. [Google Scholar]; b Krissinel E, Henrick K. Acta Crystallogr. D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 6.Thorn A, Egerer-Sieber C, Jaeger CM, Herl V, Mueller-Uri F, Kreis W, Muller YA. J. Biol. Chem. 2008;283:17260–17269. doi: 10.1074/jbc.M706185200. [DOI] [PubMed] [Google Scholar]
- 7.Valdar WSJ. Proteins. 2002;48:227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- 8.Cole C, Barber JD, Barton GJ. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.