Abstract
Bruton’s tyrosine kinase (BTK) is a cytoplasmic, non-receptor tyrosine kinase which is expressed in most of the hematopoietic cells and plays an important role in many cellular signaling pathways. B cell malignancies are dependent on BCR signaling, thus making BTK an efficient therapeutic target. Over the last few years, significant efforts have been made in order to develop BTK inhibitors to treat B-cell malignancies, and autoimmunity or allergy/hypersensitivity but limited success has been achieved. Here in this study, 3D QSAR pharmacophore models were generated for Btk based on known IC50 values and experimental energy scores with extensive validations. The five features pharmacophore model, Hypo1, includes one hydrogen bond acceptor lipid, one hydrogen bond donor, and three hydrophobic features, which has the highest correlation coefficient (0.98), cost difference (112.87), and low RMS (1.68). It was further validated by the Fisher’s randomization method and test set. The well validated Hypo1 was used as a 3D query to search novel Btk inhibitors with different chemical scaffold using high throughput virtual screening technique. The screened compounds were further sorted by applying ADMET properties, Lipinski’s rule of five and molecular docking studies to refine the retrieved hits. Furthermore, molecular dynamic simulation was employed to study the stability of docked conformation and to investigate the binding interactions in detail. Several important hydrogen bonds with Btk were revealed, which includes the gatekeeper residues Glu475 and Met 477 at the hinge region. Overall, this study suggests that the proposed hits may be more effective inhibitors for cancer and autoimmune therapy.
Introduction
Bruton’s tyrosine kinase (BTK) is a cytoplasmic, non-receptor tyrosine kinase from a Tec-family kinase, which is expressed in most of the hematopoietic cells and plays an important role in many cellular signaling pathways [1–4]. In the life cycle of B-lineage cells BTK plays a central role in proliferation, development, differentiation, survival and apoptosis [5]. BTK is characterized by five structural domains including N-terminal pleckstrin homology (PH) domain, a proline-rich TEC homology (TH) domain, Src homology 3 (SH3) followed by Src homology 2 (SH2) domain and a C-terminal kinase domain (BTK-KD). The PH domain plays an essential role in the regulation and functioning of the BTK. The PH domain contains the site for binding the transcription factors (BAP-135/TFII-I), inhibitors (PIN 1, 1BTK) [6] and activators (phosphatidylinositol 3,4,5-trisphosphates and G-protein βγ) [7]. The TH domain is stretch of 80 amino acid residues having a conserved region for zinc cofactor binding site and proline-rich segment [8], which serves as a binding site for protein kinase C-beta (PKC-β) [9]. Initially BTK is activated by phosphorylating Tyr551 in the activation loop of C-terminal kinase domain; however further activation occurs in the SH3 domains, were autophosphorylation of Tyr223 occurs [10, 11].
In the lymphoid lineage, Btk is only expressed in B cells and is not found in natural killer or T cells. B cells play a significant role in the pathogenesis of several autoimmune diseases. Clinical studies have shown that depletion of mature B cells can be efficacious in multiple sclerosis, systemic lupus erythematosus (SLE), and rheumatoid arthritis (RA) [12]. Even though Btk is expressed in the myeloid cell lineage, mutations in the Btk gene lead to prominent B cell—specific defects in mice and humans, hence it has been considered as a target for the selective inhibition of B cells [13]. In humans, mutations in the BTK gene is characterized by a B-lymphocyte developmental defect, giving rise to a primary immunodeficiency disease called X-linked agammaglobulinemia (XLA). The individuals suffering from XLA is characterized by lack of circulating B lymphocytes, therefore unable to generate immunoglobulins, and thus cannot stand humoral immune responses. Similarly, mutation in the mouse-Btk gene results in X-linked immunodeficiency (xid), a related but less severe phenotype than XLA [14–18]. B cell expansion and the production of autoantibodies by polyclonal B cell activation is a characteristic of RA [19], thus selective inhibition of Btk may be an attractive therapeutic target for B cell inhibition in RA as well as for B cell lymphoma.
Ibrutinib (PCI-32765), Dasatinib, LFM-A13, CC-292, and ONO-WG-307 are well known Btk inhibitors, with varying specificities [20]. For example, LFM-A13 and Dasatinib not only inhibits Btk with an IC50 value of 2.5 μM and 5 nM, but also binds to other kinases such as PLK3, JAK2 and SRC family members (HCK, SRC, CSK) [21–24]. Also, Ibrutinib (PCI-32765) interferes with B-cell functioning and leads to hypogammaglobulinemia [25]. Though many inhibitors are reported and few are in clinical trials, none are FDA approved and are selective to Btk. Hence, designing potent and specific Btk inhibitors becomes crucial.
Here, we used computer-aided drug design approaches to identify potent and novel inhibitors which can cause inhibition of Btk. A 3D QSAR pharmacophore model was built from the chemical features present in already known inhibitors. The best model, Hypo 1, was validated and used for database screening. The potential compounds were filtered by checking their drug like properties. Binding conformations of the selected hit compounds were predicted by molecular docking studies. Finally, the appropriate binding modes of final hit compounds were revealed by molecular dynamics (MD) simulations and free energy calculation studies.
Materials and Methods
Collection of dataset
To perform pharmacophore modeling calculations a dataset of 85 known inhibitors of Btk with diverse scaffold collected from different literature resources [26–29] and classified into two different data sets: (i) a training set and (ii) a test set. Training set was used to generate the hypothesis while the generated hypothesis was validated by a test set. Among these 85 compounds, 25 were selected as the training set compounds (Fig 1) based on their IC50 values and structural diversity. The remaining 60 compounds were used as a test set for validating the hypothesis. The inhibitory activity value of these compounds was between 0.09 nmol/L to 40570 nmol/L. The data set compounds were classified into active (IC50 < 100 nmol/L, +++), moderately active (100 nmol/L ≤ IC50 < 10000 nmol/L, ++) and inactive (IC50 ≥ 10000 nmol/L, +) based on their IC50 value. ChemSketch was used to sketch the 2D form of all the data set compounds (ACD Inc., Toronto, Canada) and was consequently exported to Discovery Studio v3.5 (DS) for their corresponding 3D structure generation.
Fig 1. 2D Chemical structures of 25 Btk inhibitors in the training set used for hypothesis generation along with their IC50 values.
Generation of pharmacophore model
Before performing the pharmacophore modeling, Feature Mapping protocol was used to identify the chemical features of the training set compounds that are important in inhibition of Btk. The chemical features identified by the feature mapping protocol were used to generate pharmacophore models using the 3D QSAR Pharmacophore Generation protocol available in DS by correlating the experimental activities values of compounds with their chemical structures. BEST algorithm was used to generate low energy conformation of the compounds. Uncertainty value was set to 3 while other parameters had default values. Debnath method was used to identify and evaluate the top ten hypotheses based on the activity values offered by the training set compounds [30]. Debnath method suggests that the model having a high correlation coefficient, the lowest total cost, the lowest RMS deviation, and the total cost close to the fixed cost and far from the null cost is considered as the best quantitative hypothesis [30]. The reliability of hypothesis depends on the difference between the total cost of the generated hypothesis and the null hypothesis.
Pharmacophore validation
The best hypothesis selected from the top ten hypotheses was subjected to validation by Fisher’s randomization and the external test set method. The statistical significance of the model was computed by employing Fischer’s randomization method [31]. This method is used to verify that the selected hypothesis is not generated by chance and also to checks if there is a strong correlation between the biological activities and the chemical structures. The confidence level was set to 95% and nineteen random spreadsheets were generated [32]. This was done by randomizing the activity of these compounds by using the same features and parameters used to generate the original pharmacophore hypothesis. During this process if any of the random pharmacophore hypotheses showed better statistical values than Hypo1, then the Hypo1 was generated by random correlation [33]. Test set was used to determine whether Hypo1 can predict and classify the compounds correctly in their activity scale of molecules other than the training set compounds. External test set contained 60 chemically diverse compounds with wide range of inhibitory activity values when compared to the training-set compounds. The selected pharmacophore hypothesis (Hypo1) was used to predict the activity values of test-set compounds. The predicted and experimental activity values were plotted to observe the range of correlation between them.
Virtual screening and drug-likeness prediction
Virtual screening of chemical databases is done to identify new scaffolds that can trigger or inhibit the activity of a particular target. The benefit of virtual screening is that the hit compounds can be gained easily for biological testing as compared to de novo design methods [34]. In this work, Hypo1 was used as a 3D structural query in virtual screening to retrieve a novel lead compound for Btk inhibition from four different chemical databases: Chembridge, NCI, Asinex, and Maybridge. The screened compounds that mapped all the features of Hypo1 were selected as hit compounds. Estimated activity values, geometric fit values ADME properties and Lipinski’s rule of five were used as a filter for further refinement of mapped compounds. During ADME investigation the compounds were checked for low blood—brain barrier (BBB), optimal solubility, good absorption, non-inhibition to CYP2D6 and non-hepatotoxicity; if the molecule had values of 3, 3, and 0 for BBB, solubility, and absorption, respectively, it was considered that the molecule had good solubility, absorption, and BBB [35]. Lipinski’s rule of five [36] estimates the absorption and intestinal permeability of a compound. Lipinski’s rule states that, the compounds that are well absorbed have a logP value less than 5, less than 5 hydrogen-bond donors, less than 10 hydrogen-bond acceptors, molecular weight of less than 500, and fewer than ten rotatable bonds. The compounds having better estimated activity values and filtered by drug-like properties was conceded further for molecular docking.
Molecular docking
In the drug designing process, molecular docking is used as a filtering method, as it is used to find the most appropriate conformation and interactions of each hit compound at the active site of protein. Docking studies were performed using GOLD program version 5.2.2. For molecular docking calculation a high resolution (1.80 Å) crystal structure (PDB code 3OCS) of Btk bound with an inhibitor was selected as protein molecule [37]. The water molecules and hetero atoms were removed from protein. CHARMm force field was used to add hydrogen atoms to the protein molecule. The binding site was identified based on the volume occupied by the co-crystallized ligand in the protein. The hit compounds along with training set compounds were docked into the active site of protein. ND1H protonation state was kept for all the histidine tautomers as observed in the crystal structure. To predict the binding affinity of the ligand to the target protein, Gold fitness score function was used as the default scoring function while rescoring was done using Chemscore. Based on the scoring functions (high Goldscore and low Chemscore), molecular interactions, and the formation of hydrogen bonds between the ligand and the active site residues of protein the best docked poses were selected.
Molecular dynamics simulations
The molecular dynamic (MD) simulations of Btk in complex with the final hit compounds obtained from docking studies and the most active compound from the training set were performed using GROMACS 4.5.7 package with CHARMm27 force field [38]. Topology files for ligands were generated by SwissParam [39]. The system were solvated in a dodecahedron box containing TIP3P water model to form an aqueous environment and neutralized with Na+ counter ions. 10000 minimization steps were carried out with steepest descent algorithm to remove possible bad contacts from initial structures until tolerance of 2000 kJ/mol. The energy minimized system was then subjected to equilibration in three different steps. A constant temperature controlled by V-rescale thermostat [40] was applied for 100 ps at 300k in the first phase of equilibration. Later, 100 ps NPT ensemble was applied at 1 bar of pressure followed by 20 ns of production run under the same ensembles. During this process, Parrinello-Rahman barostat was used to maintain the pressure of the system [41]. In the equilibration process the solvent molecules with counter ions were allowed to move while protein backbone was restrained. SETTLE and LINCS algorithm were used to constrain the geometry of water molecules and bond involving hydrogen atoms respectively [42, 43]. Periodic boundary conditions were applied to avoid edge effects. Particle Mesh Ewald (PME) algorithm were applied to calculate the long range electrostatic interactions [44]. A cut off distance of 9Å and 10Å was set for Coulombic and van der Waals interactions. Each simulation was run for 20 ns and the coordinate data was stored at every picosecond (ps). All the analysis of MD simulations was carried out by VMD [45] and DS software.
Binding free energy calculations
The binding free energy calculations were performed using Molecular Mechanics/Poisson-Boltzmann Surface Area (MM/PBSA) method as described previously [46, 47]. For calculating binding free energy 40 snapshots of protein-ligand complex were selected evenly from 0 to 20 ns of MD trajectories as per earlier studies [46, 48]. Different energy parameters have been calculated using MM/PBSA method by using the same snapshots [46, 48– 49]. The binding interaction between protein and ligand was calculated in three terms such as solvation contribution (ΔEsol), van der Waals contribution (ΔEvdw) and the electrostatic contribution (ΔEele).
Results and Discussion
Pharmacophore modeling
HypoGen algorithm was used to build the quantitative hypotheses by correlating the estimated and the experimental activity values of the Btk inhibitors. The hypothesis was generated by using the training set of 25 chemically diverse compounds (Fig 1) with activity values ranging from 0.09 nmol/L to 40570 nmol/L by selecting hydrogen bond acceptor lipid (HBAL), hydrogen bond donor (HBD), hydrophobic (HYP), hydrogen bond acceptor (HBA), and ring aromatic (RA) features as suggested by Feature Mapping protocol. A total of 10 hypotheses were generated each having five chemical features. Hypo1 is the representative hypothesis, showing a good geometric spatial arrangement consisting five chemical features namely 1 HBAL, 1 HBD, and 3 HYP (Fig 2). Hypo1 fulfilled all the statistical parameters such as the configuration cost of 12.21; total cost (125.42) which was close to the fixed cost (116.43) and away from the null cost (238.29) indicates that Hypo1 was the best hypothesis. There was a high correlation coefficient of 0.981 along with large cost difference of 112.87 and lowest RMS value of 0.68 (Table 1). By considering all the above parameters it was revealed that the statistical values of Hypo1 was best as compared to the other hypothetical structures. As a result, Hypo1, was selected as the best hypothesis for further analysis (Fig 2). The 3D spatial relationship and distance constraint of Hypo1 is depicted in Fig 2.
Fig 2. Chemical features of the best pharmacophore 'Hypo 1' with its distance constraints.
'Hypo 1' consists of one hydrogen bond acceptor lipid (HBAL: Green), one hydrogen bond donor (HBD: Magenta), three hydrophobic (HYP: Cyan) features.
Table 1. Statistical data of ten pharmacophore hypotheses generated by HypoGen.
| Hypo No. | Total Cost | Cost Differencea | RMSDb | Correlation (R2) | Max Fit | Featuresc |
|---|---|---|---|---|---|---|
| Hypo 1 | 125.42 | 112.87 | 0.68 | 0.981 | 11.68 | 1 HBAL, 1 HBD, 3 HYP |
| Hypo 2 | 122.71 | 115.58 | 0.84 | 0.970 | 10.97 | 1 HBA, 1 RA, 3 HYP |
| Hypo 3 | 122.07 | 116.22 | 0.87 | 0.968 | 11.19 | 1 HBAL, 3 HYP, 1 RA |
| Hypo 4 | 121.84 | 116.45 | 0.88 | 0.967 | 10.78 | 1 HBD, 1 HBA, 3 HYP |
| Hypo 5 | 119.49 | 118.8 | 0.95 | 0.962 | 11.97 | 1 HBAL, 1 HBD, 3 HYP |
| Hypo 6 | 118.64 | 119.65 | 1.02 | 0.956 | 10.83 | 1 HBAL, 3 HYP, 1 RA |
| Hypo 7 | 118.45 | 119.84 | 1.03 | 0.955 | 10.47 | 1 HBA, 3 HYP, 1 RA |
| Hypo 8 | 117.74 | 120.55 | 1.05 | 0.953 | 10.86 | 1 HBAL, 3HYP, 1 RA |
| Hypo 9 | 117.38 | 120.91 | 1.06 | 0.953 | 11.25 | 1 HBAL, 1 HBD, 3 HYP |
| Hypo 10 | 117.33 | 120.96 | 0.99 | 0.959 | 10.44 | 2 HBA, 1 HYP, 1 RA |
a Cost difference, difference between the null cost and the total cost. The null cost of ten scored hypotheses is 238.29, the fixed cost value is 106.43. All costs are represented in bit units.
b RMSD: deviation of the log (estimated activities) from the log (experimental activities) normalized by the log (Uncertainties).
c HBAL, hydrogen bond acceptor lipid; HBD, hydrogen bond donor; HYP hydrophobic; HBA, hydrogen bond acceptor; and RA, ring aromatic.
To elucidate the predictive accuracy of Hypo1 the training set compounds were classified into active (IC50 < 100 nmol/L, +++), moderately active (100 nmol/L ≤ IC50 < 10000 nmol/L, ++) and inactive (IC50 ≥ 10000 nmol/L, +) based on their IC50 value. Regression analysis was used to estimate the activity of each compound. Hypo1 estimated the inhibitory activity value in the same order of magnitude for all the training set compounds (Table 2) except two moderately active and two inactive compounds which were overestimated as active and moderately active compounds, respectively.
Table 2. Experimental and estimated activity of training set compounds based on Hypo 1.
| Compound No. | Fit Value | Exp IC50 nmol/L | Pred IC50 nmol/L | Errora | Exp Scaleb | Pred Scaleb |
|---|---|---|---|---|---|---|
| 1 | 11.16 | 0.09 | 0.22 | +2.4 | +++ | +++ |
| 2 | 11.33 | 0.12 | 0.15 | +1.3 | +++ | +++ |
| 3 | 10.57 | 0.87 | 0.87 | -1 | +++ | +++ |
| 4 | 10.07 | 1 | 2.8 | +2.8 | +++ | +++ |
| 5 | 10.33 | 1.4 | 1.5 | +1.1 | +++ | +++ |
| 6 | 9.76 | 1.9 | 5.5 | +2.9 | +++ | +++ |
| 7 | 10.39 | 2 | 1.3 | -1.5 | +++ | +++ |
| 8 | 10.01 | 3.8 | 3.2 | -1.2 | +++ | +++ |
| 9 | 9.78 | 7.7 | 5.3 | -1.5 | +++ | +++ |
| 10 | 9.43 | 8 | 12 | +1.5 | +++ | +++ |
| 11 | 9.45 | 17 | 12 | -1.5 | +++ | +++ |
| 12 | 8.79 | 33 | 52 | +1.6 | +++ | +++ |
| 13 | 8.63 | 100 | 76 | -1.3 | +++ | +++ |
| 14 | 8.93 | 140 | 38 | -3.7 | ++ | +++ |
| 15 | 8.40 | 142 | 130 | -1.1 | ++ | ++ |
| 16 | 7.91 | 320 | 390 | +1.2 | ++ | ++ |
| 17 | 6.98 | 670 | 3400 | +5 | ++ | ++ |
| 18 | 7.97 | 1000 | 350 | -2.9 | ++ | ++ |
| 19 | 7.27 | 3400 | 1700 | -1.9 | ++ | ++ |
| 20 | 7.48 | 4100 | 1100 | -3.9 | ++ | ++ |
| 21 | 6.92 | 5800 | 3800 | -1.5 | ++ | ++ |
| 22 | 6.33 | 9200 | 15000 | +1.7 | ++ | +++ |
| 23 | 6.54 | 14000 | 9200 | -1.6 | + | ++ |
| 24 | 6.57 | 15000 | 8600 | -1.8 | + | ++ |
| 25 | 5.40 | 41000 | 130000 | +3.2 | + | + |
a Error, ratio of the predicted activity (Pred IC50) to the experimental activity (Exp IC50) or its negative inverse if the ratio is <1.
b Activity scale: IC50 < 100 nmol/L = +++ (active), 100 nmol/L ≤ IC50 < 10000 nmol/L = ++ (moderate active), IC50 ≥ 10000 nmol/L = + (inactive).
Hypo 1 aligned with the most active (IC50 = 0.09 nmol/L) compound and the least active (IC50 = 40570 nmol/L) compound in the training set as depicted in Fig 3. Clearly, all the hypothetical features were perfectly mapped by the most active compound (Fig 3A), whereas the least active compound (Fig 3B) failed to fit on one HBA and one HYP feature. This reveals the difference in activities among the most active and the least active compounds. This analysis suggests that Hypo1 was able to differentiate the compounds based on the activity values with high accuracy (Table 2). Hypo1 was further validated using the test-set and Fischer randomization method.
Fig 3.
The best pharmacophore model Hypo1 aligned to training set compounds: A) most active compound 1 (IC50 0.09 nmol/L) and B) least activity compound 20 (IC50 40570 nmol/L). The most active compound mapped to all four features in Hypo 1, whereas the least active compound mapped only two features.
Pharmacophore validation
Test set validation
A good pharmacophore should have the ability to predict and classify the compounds according to their activities scale. Hypo1 was validated by external validation (test set) process which consist of 60 structurally diverse compounds other than the training set compounds (S1 Table) and were classified into active (IC50 < 100 nmol/L, +++), moderately active (100 nmol/L ≤ IC50 < 10000 nmol/L, ++) and inactive (IC50 ≥ 10000 nmol/L, +) respectively. One moderately active compound was underestimated as being inactive, and two inactive molecules were overestimated as moderately active compounds. The remaining compounds were classified correctly, indicating that Hypo1 was able to predict the activities of compounds in their own activity scales as depicted in Table 3. The linear regression between the Hypo1-predicted activities and experimental inhibitory activities of the test-set compounds showed a correlation coefficient (r) value of 0.96 (Fig 4). This result shows the predictive capacity of Hypo1 to discriminate between the active and moderately active compounds.
Table 3. Evaluation of estimated and experimental activity (IC50) values of test set compounds using Hypo 1.
| Compound number | Fit Value | Experimental IC50 (nmol/L) | Predicted IC50 (nmol/L) | Errora | Experimental Scaleb | Predicted Scaleb |
|---|---|---|---|---|---|---|
| 1 | 11.31 | 0.15 | 0.43 | +0.36 | +++ | +++ |
| 2 | 11.08 | 0.26 | 0.24 | -1.10 | +++ | +++ |
| 3 | 11.01 | 0.30 | 0.51 | +0.60 | +++ | +++ |
| 4 | 10.99 | 0.32 | 0.56 | +0.57 | +++ | +++ |
| 5 | 10.92 | 0.37 | 0.57 | +0.66 | +++ | +++ |
| 6 | 10.88 | 0.41 | 0.95 | +0.43 | +++ | +++ |
| 7 | 10.85 | 0.44 | 0.83 | +0.53 | +++ | +++ |
| 8 | 10.78 | 0.52 | 0.36 | -1.45 | +++ | +++ |
| 9 | 10.40 | 1.27 | 2.2 | +0.58 | +++ | +++ |
| 10 | 10.39 | 1.29 | 3 | +0.43 | +++ | +++ |
| 11 | 10.39 | 1.29 | 2.8 | +0.46 | +++ | +++ |
| 12 | 10.26 | 1.74 | 3.6 | +0.48 | +++ | +++ |
| 13 | 10.26 | 1.74 | 2.1 | +0.83 | +++ | +++ |
| 14 | 10.18 | 2.10 | 2.6 | +0.80 | +++ | +++ |
| 15 | 10.16 | 2.17 | 3.4 | +0.64 | +++ | +++ |
| 16 | 10.12 | 2.41 | 5 | +0.48 | +++ | +++ |
| 17 | 10.10 | 2.51 | 3.3 | +0.76 | +++ | +++ |
| 18 | 10.07 | 2.68 | 3.4 | +0.78 | +++ | +++ |
| 19 | 10.06 | 2.78 | 4 | +0.69 | +++ | +++ |
| 20 | 10.00 | 3.17 | 6.2 | +0.51 | +++ | +++ |
| 21 | 9.97 | 3.43 | 4.7 | +0.73 | +++ | +++ |
| 22 | 9.94 | 3.65 | 4.9 | +0.74 | +++ | +++ |
| 23 | 9.86 | 4.37 | 6 | +0.72 | +++ | +++ |
| 24 | 9.86 | 4.41 | 8.7 | +0.50 | +++ | +++ |
| 25 | 9.83 | 4.70 | 5.2 | +0.90 | +++ | +++ |
| 26 | 9.77 | 5.38 | 9.8 | +0.54 | +++ | +++ |
| 27 | 9.69 | 6.47 | 4.0 | -1.61 | +++ | +++ |
| 28 | 9.68 | 6.67 | 8.1 | +0.82 | +++ | +++ |
| 29 | 9.58 | 8.41 | 13.0 | +0.64 | +++ | +++ |
| 30 | 9.57 | 8.60 | 19.0 | +0.45 | +++ | +++ |
| 31 | 9.55 | 8.90 | 6.1 | -1.45 | +++ | +++ |
| 32 | 9.48 | 10.52 | 6.12 | -1.72 | +++ | +++ |
| 33 | 9.43 | 11.78 | 10.1 | -1.16 | +++ | +++ |
| 34 | 9.42 | 12.06 | 8.0 | -1.50 | +++ | +++ |
| 35 | 9.42 | 12.06 | 9.0 | -1.34 | +++ | +++ |
| 36 | 9.33 | 14.80 | 17.1 | +0.86 | +++ | +++ |
| 37 | 9.32 | 15.21 | 17.1 | +2.14 | +++ | +++ |
| 38 | 9.30 | 15.76 | 22.9 | +0.68 | +++ | +++ |
| 39 | 9.23 | 18.75 | 11.04 | -1.69 | +++ | +++ |
| 40 | 9.15 | 22.64 | 43.0 | +0.52 | +++ | +++ |
| 41 | 8.99 | 32.22 | 14.28 | -2.25 | +++ | +++ |
| 42 | 8.93 | 37.14 | 16.6 | -2.23 | +++ | +++ |
| 43 | 8.91 | 38.61 | 67.0 | +0.57 | +++ | +++ |
| 44 | 8.91 | 38.71 | 16.6 | -2.41 | +++ | +++ |
| 45 | 8.79 | 50.90 | 39.35 | -1.29 | +++ | +++ |
| 46 | 8.73 | 58.77 | 47.0 | -1.25 | +++ | +++ |
| 47 | 8.26 | 174.33 | 129.9 | -1.34 | ++ | ++ |
| 48 | 8.25 | 178.33 | 333.06 | +0.53 | ++ | ++ |
| 49 | 8.24 | 181.87 | 280.83 | +0.64 | ++ | ++ |
| 50 | 8.17 | 215.87 | 267.74 | +0.80 | ++ | ++ |
| 51 | 7.75 | 570.61 | 3700 | +0.15 | ++ | ++ |
| 52 | 7.71 | 620.37 | 518 | -1.19 | ++ | ++ |
| 53 | 7.71 | 620.87 | 287 | -2.16 | ++ | ++ |
| 54 | 7.60 | 798.80 | 3142 | +0.25 | ++ | ++ |
| 55 | 7.37 | 1359.21 | 1255 | -1.08 | ++ | ++ |
| 56 | 6.84 | 4587.11 | 1270 | -3.61 | ++ | ++ |
| 57 | 6.67 | 6787.21 | 3330 | -2.03 | ++ | ++ |
| 58 | 6.61 | 7821.13 | 12700 | +0.61 | ++ | + |
| 59 | 6.37 | 13667.60 | 1050 | -13.01 | + | ++ |
| 60 | 6.03 | 29655.90 | 2687 | -11.03 | + | ++ |
a Error, ratio of the predicted activity to the experimental activity or its negative inverse if the ratio is <1.
b Activity scale: IC50 < 100 nmol/L = +++ (active), 100 nmol/L ≤ IC50 < 10000 nmol/L = ++ (moderate active), IC50 ≥ 10000 nmol/L = + (inactive).
Fig 4. Correlation plot between Hypo1 predicted Btk inhibitory activities and experimental activities of 60 test set compounds and 25 training set compound.

Fischer’s randomization method
To estimate the statistical relevance of Hypo1 Fischer’s test was applied. Here we set a 95% confidence level; as a result 19 random spreadsheets were generated by arbitrarily reassigning the experimental activity values to each compound in the training set, and a hypothesis was created for each spreadsheet (Fig 5). The formula used to calculate the significance of the hypothesis is S = [1–(1+X)/Y]×100, where X denotes total number of hypotheses with total cost that are lower than the original hypothesis, and Y represents the total number of HypoGen runs (initial+random runs). Here, X = 0 and Y = (1+19), hence 95% = {1–[(1+0)/(19+1)]}×100. The generated random spreadsheets showed least total cost value for Hypo1 as compared to other hypothesis, which indicates that Hypo1 is far more superior to all other random hypotheses and was not generated by chance.
Fig 5. Comparison between the total cost of Hypo1 with the total costs of the 19 scrambled runs generated during the Fisher randomization run.
Virtual screening
In the drug discovery process, virtual screening of chemical databases is an alternative method to the high-throughput screening technique. Chemical features of Hypo1 play an important role in mapping and screening out novel compounds from a database. We therefore used Hypo1 as a 3D structural query to screen Asinex, Chembridge, Maybridge, and NCI databases which contains 213262, 50000, 59652 and 238819 compounds, respectively. Among these, Hypo 1 mapped 29249 compounds that had all the chemical features of Hypo1. Further these compounds were filtered down to 3723 by applying a filter of maximum fit value greater than 10. However, even if a molecule passes various filters, it may not be active towards Btk, hence we tested the filtered compounds for their ADME properties and Lipinski’s rule of five. ADME and Lipinski's rule of five plays an important role in sorting the chemical compounds based on drug-like properties. Therefore, ADME and Lipinski's rule of five were used as a filter to sort these molecules. Finally, a total of 23 compounds satisfied the drug-like properties and were subjected to molecular docking to study their critical interactions with the important amino acids present in the active site of Btk.
Molecular docking
The training set compounds along with 23 drug-like hits resulted from pharmacophore modeling were subjected to docking using GOLD program so as to refine the retrieved hit compounds and to eliminate the false positives. To gauge the accuracy of GOLD and to examine the parameters to produce the appropriate binding orientation the co-crystal was docked in the active site of Btk. It resulted in an acceptable RMSD value of 1.04 Å between the predicted structure and the co-crystal (S1 Fig). Therefore, by using the same parameters the candidate compounds were docked. The GOLD fitness scores, molecular interactions with the binding site residues, binding modes, and Chemscore were considered as key components in selecting the best conformation of the docked compounds. GOLD fitness score differentiates molecules based on their interacting ability. GOLD fitness score value greater than that of most active compound was taken as cut-off for the further screening of compounds. Chemscore estimates the total free energy change that occurs upon ligand binding and was used as the rescoring function. The most active compound in the training set has scored a GOLD fitness score of 69.7 and Chemscore of -30.7 (Table 4). Thus, the compounds were selected based on GOLD fitness score greater than 69.7, Chemscore lower than -30.7, and the ligand conformations satisfying the necessary interactions in the active site. Finally, three hit compounds fulfilled the above criteria and also mapped well to the pharmacophoric features of Hypo 1 (Fig 6) were characterized as final hits.
Table 4. Comparison of Gold fitness score, Chemscore and average binding energy of Btk and reference inhibitor/hit1/hit2/hit3 complex.
| Systems | Gold fitness score | ChemScore | Average binding energy (KJ/mol) |
|---|---|---|---|
| BTK + Inhibitor | 69.7 | -30.7 | -84.1 |
| BTK + Hit 1 | 72.4 | -29.3 | -87.9 |
| BTK + Hit 2 | 71.7 | -36.9 | -81.3 |
| BTK + Hit 3 | 70.6 | -36.1 | -92.0 |
Fig 6. Hypo1 mapped onto the hit compounds.
A) Hit 1, (B) Hit 2, (C) Hit 3. The HBAL, HBD and HYP features are displayed in green magenta and cyan, respectively.
Molecular dynamics simulations
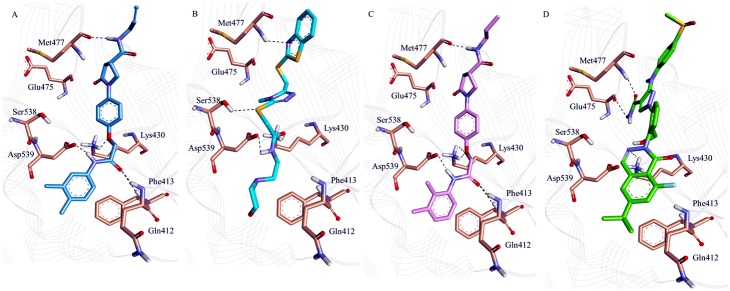
In order to further validate the results and to predict more reliable ligand—receptor interaction MD simulations were performed. The 20 ns MD simulations were done to understand the conformational changes and dynamic behavior with each other by taking the best docked conformation of three hits and a reference compound as the initial structure. All the four systems were subjected to the MD simulation. To explore the dynamic stability of the complexes during simulation the root mean square deviation (RMSD) of protein backbone atoms (Fig 7A) and potential energy (Fig 7B) of the system were calculated. The RMSD values observed for the complexes were in between 0.8 Å to 1.7 Å throughout the simulations which shows that system are well converged. The average RMSD values obtained during simulation for hit1, hit2, hit3, and inhibitor were 1.12 Å, 1.17 Å, 1.05 Å, and 1.45 Å respectively. The potential energy of the system was stable throughout the simulation indicating that no abnormal behavior occurred in the protein. The last 5 ns trajectories were used to analyze the binding mode of the representative structures of four systems. When all the representative structures were superimposed, it was found that the binding pattern of hit compounds was similar to reference compound (Fig 8). The substrate binding pocket of Btk was formed by Gln412, Phe413, Lys430, Glu475, Met 477, Ser538 and Asp539 amino acids. These key residues were also found to interact with the reference inhibitor and hit compounds. In case of Hit compounds, Hit1 formed hydrogen bond interactions with Gln412, Phe413, Lys430, Met477, Asp539 and hydrophobic interaction with Leu408, Gly411, Ala428, Ala478, Gly480, Asp521, Leu528, Leu542, Ser543, Tyr551 (Fig 9A, Table 5). The benzene moiety of hit1 was involved in π -π and σ-π interaction with Phe413 and Val416, respectively. In hit2 binding, hydrogen bonds with Lys430, Met477, Ser538 and Asp539 were observed (Fig 9B, Table 5). Hit2 showed hydrophobic interactions with Leu408, Gln412, Phe413, Val416, Ala428, Met477, Ala478, Gly480, Asn526, Leu528, Ser538, Asp539, Leu542, Ser543, and Tyr551. The benzene moiety of hit2 was involved in π -π interaction with Tyr476 while π-cation interaction with Lys406, and Lys430, respectively. Hit3 formed hydrogen bond interactions with Gln412, Phe413, Lys430, Met477, and Asp539 (Fig 9C, Table 5). Hit3 showed interactions with hydrophobic pocket residues such as Leu408, Gly411, Val416, Ala428, Tyr476, Ala478, Asn479, Gly480, Asn526, Leu528, Leu542, Ser543, Val546, and Tyr551. On the other hand reference compound, inhibitor formed hydrogen bonds with Lys430, Glu475, and Met 477 (Fig 9D, Table 5). Furthermore, Inhibitor was stacked on Lys430 via cation-π interaction. Inhibitor showed hydrophobic interactions with Leu408, Gly411, Gln412, Phe413, Val416, Ala478, Asn479, Gly480, Asn526, Leu528, Leu542, Ser543, Val546, and Tyr551. These results reveals that, the final hit compounds bound to the active site either by forming hydrogen bond interactions, or by σ-π and cation-π interactions and the interacting residues are given in Table 5.
Fig 7. The RMSD and potential energy graph for four complex systems.
(A) The RMSD profile for the backbone atoms of Btk protein. (B) The potential energy of the system. These graphs were calculated during 20 ns MD simulations for each complex. Blue, cyan, pink, and green lines represent Hit1, Hit2, Hit3, and Inhibitor respectively.
Fig 8. The binding mode of the three hit compounds and reference inhibitor in the active site of Btk.
All compounds in their representative structures were superimposed (left) and enlarged (right). The Btk protein is shown in gray color solid ribbon while the compounds are depicted by sticks. Blue, cyan, pink, and green sticks represent Hit1, Hit2, Hit3, and Inhibitor respectively.
Fig 9. The binding conformation and hydrogen bonding interactions of the three hit compounds and reference inhibitor in the active site of Btk.
(A) Hit1: blue (B) Hit2: cyan (C) Hit3: pink and (D) Inhibitor: Green. Hydrogen bond interactions between proteins and compounds are shown as black dotted line. Only polar hydrogen atoms are shown for clarity.
Table 5. The molecular interactions between the compounds and Btk protein.
| Compound | Hydrogen bond (<3.0 Å) | Hydrophobic interaction | Cation-π interaction | σ-π interaction |
|---|---|---|---|---|
| Inhibitor | Lys430, Glu475, Met 477 | Leu408, Gly411, Gln412, Phe413, Val416, Ala478, Asn479, Gly480, Asn526, Leu528, Leu542, Ser543, Val546, Tyr551 | Lys430 | |
| Hit 1 | Gln412, Phe413, Lys430, Met 477, Asp539 | Leu408, Gly411, Ala428, Ala478, Gly480, Asp521, Leu528, Leu542, Ser543, Tyr551 | Phe413 | Val416 |
| Hit 2 | Lys430, Met 477, Ser538, Asp539 | Leu408, Gln412, Phe413, Val416, Ala428, Met477, Ala478, Gly480, Asn526, Leu528, Ser538, Asp539, Leu542, Ser543, Tyr551. | Lys406, Lys430 | Tyr476 |
| Hit 3 | Gln412, Phe413, Lys430, Met 477, Asp539 | Leu408, Gly411, Val416, Ala428, Tyr476, Ala478, Asn479, Gly480, Asn526, Leu528, Leu542, Ser543, Val546, Tyr551 |
In order to understand the nature of the binding of drug molecules in the active site, the intermolecular hydrogen bonds between protein and hit compounds were monitored during the simulation period (Fig 10). The average numbers of hydrogen bonds between the Btk protein and hit compounds were 2.6, 1.8, and 1.5 for hit1, hit2, and hit3 respectively. Inhibitor showed almost 1.3 hydrogen bonds throughout the simulation. The reference compound showed relatively less hydrogen bonds than the hit compounds. Further, search by PubChem Structure [50] an online search tool confirmed that these compounds were not tested experimentally for the inhibition of Hck and can be recommended as potential Btk inhibitors. Hence, we suggest that these compounds could be novel as Btk inhibitors (Fig 11).
Fig 10. The number of intermolecular hydrogen bonds between protein and compound during 20 ns MD simulations.
Blue, cyan, pink, and green colors represent Hit1, Hit2, Hit3, and Inhibitor respectively.
Fig 11. 2D structures of the hit compounds.
Analysis of the binding free energy of Btk and reference inhibitor/hit compounds
Calculation of binding energy is a key aspect in understanding the molecular activity of the targeted biomolecules. Estimation of various bonded and non-bonded interactions arbitrating bimolecular association or dissociation offers us supportable information in developing the therapeutic drugs against several biological disorders. MM/PBSA method was used to calculate the binding free energy of each set of protein ligand complex in order to compare the binding affinity between protein with the identified potent lead compounds. The MM/PBSA calculation of Btk-ligand complexes using the reference inhibitor, hit1, hit2 and hit3 as the ligands gave favorable ΔG values in the range of −35 to −137 kJ/mol as depicted in Fig 12. The binding energy showed slight variation in each snapshot as the conformational space was not sampled enough to get converged results. The average binding energy obtained for Btk-ligand complexes were -84.18 kJ/mol (reference inhibitor), -87.96 kJ/mol (hit1), -81.39 (hit2), and -92.09 (hit3) (Table 4). The binding energy obtained from the trajectories produced during the MD simulation, considers the ligand conformation and the fluctuation of the protein in the complex, as a result confirming a proper adjustment of the ligand in the binding site [49, 51]. Btk has charged binding pocket comprising of two Asp, one Glu, two Lys, one Arg, one Gln, and two Asn residues. These amino acid residues form strong ionic interactions with ligands, thus, resulting in strong electrostatic potential in the binding interface of Btk active site. The bound conformation of Btk and ligands shows that ligands get accommodate in the active site of the enzyme through hydrogen bond and hydrophobic interactions.
Fig 12. MM/PBSA estimated binding free energy of Btk and hit 1/hit 2/hit 3/ reference inhibitor complex throughout simulation time.
Color coding; Hit 1: Blue, Hit 2: cyan, Hit 3: pink and Inhibitor: green.
Conclusion
Inhibition of Btk has emerged as a new promising target in the field of B cell malignancies and autoimmunity or allergy/hypersensitivity as it is involved in several signaling pathways. Thus as an attempt, a ligand based pharmacophore modeling was done to find the important chemical features which can inhibit the activity of Btk. The five feature pharmacophore model, Hypo1, was developed consisting of 1 HBAL, 1HBD, 3HYP features. Hypo1 had the highest correlation coefficient (0.98), cost difference (112.87), and low RMS (1.68). It was further validated by the Fisher’s randomization method (95%) and test set (r = 0.96). Hence, the best hypothesis Hypo1 was used as a 3D structural query to screen the chemical databases for retrieving new potent inhibitors of Btk. Fit value, Lipinski’s rule of five, and ADMET properties screening assisted us to discard the non-drug-like compounds. Furthermore, the screened drug-like compounds were identified and were subjected to molecular docking study. Finally, molecular dynamic simulation was employed to study the stability of docked conformation and to investigate the binding interaction in details. Several important hydrogen bonds with Btk were revealed, which includes the gatekeeper residues Glu475 and Met 477 at the hinge region. The analyzed results suggested that the binding mode of hit compounds was similar to the reference compounds. The hit compounds bound to the active site residues by forming hydrogen bond, hydrophobic, σ-π and cation-π interactions. Hence, we propose that the final hit compounds as a possible virtual leads to design novel Btk inhibitors.
Supporting Information
(PNG)
(DOC)
Acknowledgments
Dr. Rohit S. Bavi was financially supported by a postdoctoral fellowship from BK21 PLUS program of Ministry of Education and Human Resources Development, South Korea. This research was supported by the Pioneer Research Center Program through the National Research Foundation (NRF) funded by the Ministry of Science, ICT and Future Planning (NRF-2015M3C1A3023028). This work was also supported by the Next-Generation BioGreen 21 Program (PJ01106202) from Rural Development Administration (RDA) of Republic of Korea. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Dr. Rohit S. Bavi was financially supported by a postdoctoral fellowship from BK21 PLUS program of Ministry of Education and Human Resources Development, South Korea. This research was supported by the Pioneer Research Center Program through the National Research Foundation (NRF) funded by the Ministry of Science, ICT and Future Planning (NRF-2015M3C1A3023028). This work was also supported by the Next-Generation BioGreen 21 Program (PJ01106202) from Rural Development Administration (RDA) of Republic of Korea. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jefferies CA, Doyle S, Brunner C, Dunne A, Brint E, Wietek C, et al. Bruton's tyrosine kinase is a Toll/interleukin-1 receptor domain-binding protein that participates in nuclear factor kappaB activation by Toll-like receptor 4. J Biol Chem. 2003;278(28):26258–64. 10.1074/jbc.M301484200 . [DOI] [PubMed] [Google Scholar]
- 2.Qiu Y, Kung HJ. Signaling network of the Btk family kinases. Oncogene. 2000;19(49):5651–61. 10.1038/sj.onc.1203958 . [DOI] [PubMed] [Google Scholar]
- 3.Uckun FM, Waddick KG, Mahajan S, Jun X, Takata M, Bolen J, et al. BTK as a mediator of radiation-induced apoptosis in DT-40 lymphoma B cells. Science. 1996;273(5278):1096–100. . [DOI] [PubMed] [Google Scholar]
- 4.Kersseboom R, Ta VB, Zijlstra AJ, Middendorp S, Jumaa H, van Loo PF, et al. Bruton's tyrosine kinase and SLP-65 regulate pre-B cell differentiation and the induction of Ig light chain gene rearrangement. J Immunol. 2006;176(8):4543–52. . [DOI] [PubMed] [Google Scholar]
- 5.Maas A, Hendriks RW. Role of Bruton's tyrosine kinase in B cell development. Dev Immunol. 2001;8(3–4):171–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu W, Quinto I, Chen X, Palmieri C, Rabin RL, Schwartz OM, et al. Direct inhibition of Bruton's tyrosine kinase by IBtk, a Btk-binding protein. Nat Immunol. 2001;2(10):939–46. 10.1038/ni1001-939 . [DOI] [PubMed] [Google Scholar]
- 7.Varnai P, Rother KI, Balla T. Phosphatidylinositol 3-kinase-dependent membrane association of the Bruton's tyrosine kinase pleckstrin homology domain visualized in single living cells. J Biol Chem. 1999;274(16):10983–9. . [DOI] [PubMed] [Google Scholar]
- 8.Vihinen M, Nilsson L, Smith CI. Tec homology (TH) adjacent to the PH domain. FEBS Lett. 1994;350(2–3):263–5. . [DOI] [PubMed] [Google Scholar]
- 9.Kang SW, Wahl MI, Chu J, Kitaura J, Kawakami Y, Kato RM, et al. PKCbeta modulates antigen receptor signaling via regulation of Btk membrane localization. EMBO J. 2001;20(20):5692–702. 10.1093/emboj/20.20.5692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nore BF, Mattsson PT, Antonsson P, Backesjo CM, Westlund A, Lennartsson J, et al. Identification of phosphorylation sites within the SH3 domains of Tec family tyrosine kinases. Biochim Biophys Acta. 2003;1645(2):123–32. . [DOI] [PubMed] [Google Scholar]
- 11.Park H, Wahl MI, Afar DE, Turck CW, Rawlings DJ, Tam C, et al. Regulation of Btk function by a major autophosphorylation site within the SH3 domain. Immunity. 1996;4(5):515–25. . [DOI] [PubMed] [Google Scholar]
- 12.Leandro MJ, de la Torre I. Translational Mini-Review Series on B Cell-Directed Therapies: The pathogenic role of B cells in autoantibody-associated autoimmune diseases—lessons from B cell-depletion therapy. Clinical and experimental immunology. 2009;157(2):191–7. 10.1111/j.1365-2249.2009.03978.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jumaa H, Hendriks RW, Reth M. B cell signaling and tumorigenesis. Annual review of immunology. 2005;23:415–45. 10.1146/annurev.immunol.23.021704.115606 . [DOI] [PubMed] [Google Scholar]
- 14.Tsukada S, Saffran DC, Rawlings DJ, Parolini O, Allen RC, Klisak I, et al. Deficient expression of a B cell cytoplasmic tyrosine kinase in human X-linked agammaglobulinemia. Cell. 1993;72(2):279–90. . [DOI] [PubMed] [Google Scholar]
- 15.Vetrie D, Vorechovsky I, Sideras P, Holland J, Davies A, Flinter F, et al. The gene involved in X-linked agammaglobulinaemia is a member of the src family of protein-tyrosine kinases. Nature. 1993;361(6409):226–33. 10.1038/361226a0 . [DOI] [PubMed] [Google Scholar]
- 16.Rawlings DJ, Saffran DC, Tsukada S, Largaespada DA, Grimaldi JC, Cohen L, et al. Mutation of unique region of Bruton's tyrosine kinase in immunodeficient XID mice. Science. 1993;261(5119):358–61. . [DOI] [PubMed] [Google Scholar]
- 17.Thomas JD, Sideras P, Smith CI, Vorechovsky I, Chapman V, Paul WE. Colocalization of X-linked agammaglobulinemia and X-linked immunodeficiency genes. Science. 1993;261(5119):355–8. . [DOI] [PubMed] [Google Scholar]
- 18.Conley ME. B cells in patients with X-linked agammaglobulinemia. J Immunol. 1985;134(5):3070–4. . [PubMed] [Google Scholar]
- 19.Chang BY, Huang MM, Francesco M, Chen J, Sokolove J, Magadala P, et al. The Bruton tyrosine kinase inhibitor PCI-32765 ameliorates autoimmune arthritis by inhibition of multiple effector cells. Arthritis research & therapy. 2011;13(4):R115 10.1186/ar3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aalipour A, Advani RH. Bruton tyrosine kinase inhibitors: a promising novel targeted treatment for B cell lymphomas. British journal of haematology. 2013;163(4):436–43. 10.1111/bjh.12573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahajan S, Ghosh S, Sudbeck EA, Zheng Y, Downs S, Hupke M, et al. Rational design and synthesis of a novel anti-leukemic agent targeting Bruton's tyrosine kinase (BTK), LFM-A13 [alpha-cyano-beta-hydroxy-beta-methyl-N-(2, 5-dibromophenyl)propenamide]. J Biol Chem. 1999;274(14):9587–99. . [DOI] [PubMed] [Google Scholar]
- 22.van den Akker E, van Dijk TB, Schmidt U, Felida L, Beug H, Lowenberg B, et al. The Btk inhibitor LFM-A13 is a potent inhibitor of Jak2 kinase activity. Biological chemistry. 2004;385(5):409–13. 10.1515/BC.2004.045 . [DOI] [PubMed] [Google Scholar]
- 23.Uckun FM, Dibirdik I, Qazi S, Vassilev A, Ma H, Mao C, et al. Anti-breast cancer activity of LFM-A13, a potent inhibitor of Polo-like kinase (PLK). Bioorganic & medicinal chemistry. 2007;15(2):800–14. . [DOI] [PubMed] [Google Scholar]
- 24.Carter TA, Wodicka LM, Shah NP, Velasco AM, Fabian MA, Treiber DK, et al. Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(31):11011–6. 10.1073/pnas.0504952102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiestner A. Targeting B-Cell receptor signaling for anticancer therapy: the Bruton's tyrosine kinase inhibitor ibrutinib induces impressive responses in B-cell malignancies. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013;31(1):128–30. 10.1200/JCO.2012.44.4281 . [DOI] [PubMed] [Google Scholar]
- 26.Lou Y, Owens TD, Kuglstatter A, Kondru RK, Goldstein DM. Bruton's tyrosine kinase inhibitors: approaches to potent and selective inhibition, preclinical and clinical evaluation for inflammatory diseases and B cell malignancies. Journal of medicinal chemistry. 2012;55(10):4539–50. 10.1021/jm300035p . [DOI] [PubMed] [Google Scholar]
- 27.Kim KH, Maderna A, Schnute ME, Hegen M, Mohan S, Miyashiro J, et al. Imidazo[1,5-a]quinoxalines as irreversible BTK inhibitors for the treatment of rheumatoid arthritis. Bioorganic & medicinal chemistry letters. 2011;21(21):6258–63. . [DOI] [PubMed] [Google Scholar]
- 28.Barf T, Kaptein A. Irreversible protein kinase inhibitors: balancing the benefits and risks. Journal of medicinal chemistry. 2012;55(14):6243–62. 10.1021/jm3003203 . [DOI] [PubMed] [Google Scholar]
- 29.Goldberg DR, Butz T, Cardozo MG, Eckner RJ, Hammach A, Huang J, et al. Optimization of 2-phenylaminoimidazo[4,5-h]isoquinolin-9-ones: orally active inhibitors of lck kinase. Journal of medicinal chemistry. 2003;46(8):1337–49. 10.1021/jm020446l . [DOI] [PubMed] [Google Scholar]
- 30.Debnath AK. Pharmacophore mapping of a series of 2,4-diamino-5-deazapteridine inhibitors of Mycobacterium avium complex dihydrofolate reductase. Journal of medicinal chemistry. 2002;45(1):41–53. . [DOI] [PubMed] [Google Scholar]
- 31.Fischer R., The Principle of Experimentation Illustrated by a Psycho-Physical Expe, Chapter II, 8th ed., Hafner Publishing Co., New York, USA: (1966). [Google Scholar]
- 32.John S, Thangapandian S, Arooj M, Hong JC, Kim KD, Lee KW. Development, evaluation and application of 3D QSAR Pharmacophore model in the discovery of potential human renin inhibitors. BMC bioinformatics. 2011;12 Suppl 14:S4 10.1186/1471-2105-12-S14-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.John S, Thangapandian S, Sakkiah S, Lee KW. Potent BACE-1 inhibitor design using pharmacophore modeling, in silico screening and molecular docking studies. BMC bioinformatics. 2011;12 Suppl 1:S28 10.1186/1471-2105-12-S1-S28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steindl T, Langer T. Influenza virus neuraminidase inhibitors: generation and comparison of structure-based and common feature pharmacophore hypotheses and their application in virtual screening. Journal of chemical information and computer sciences. 2004;44(5):1849–56. 10.1021/ci049844i . [DOI] [PubMed] [Google Scholar]
- 35.Sakkiah S, Thangapandian S, John S, Kwon YJ, Lee KW. 3D QSAR pharmacophore based virtual screening and molecular docking for identification of potential HSP90 inhibitors. European journal of medicinal chemistry. 2010;45(6):2132–40. 10.1016/j.ejmech.2010.01.016 . [DOI] [PubMed] [Google Scholar]
- 36.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced drug delivery reviews. 2001;46(1–3):3–26. . [DOI] [PubMed] [Google Scholar]
- 37.Di Paolo JA, Huang T, Balazs M, Barbosa J, Barck KH, Carano RAD, Darrow J, Davies DR, DeForge LE, Dennis G Jr, Diehl L, Ferrando R. A novel, specific Btk inhibitor antagonizes BCR and Fc[gamma]R signaling and suppresses inflammatory arthritis. [Google Scholar]
- 38.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. Journal of computational chemistry. 2005;26(16):1701–18. 10.1002/jcc.20291 . [DOI] [PubMed] [Google Scholar]
- 39.Zoete V, Cuendet MA, Grosdidier A, Michielin O. SwissParam: a fast force field generation tool for small organic molecules. Journal of computational chemistry. 2011;32(11):2359–68. 10.1002/jcc.21816 . [DOI] [PubMed] [Google Scholar]
- 40.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. The Journal of chemical physics. 2007;126(1):014101 10.1063/1.2408420 . [DOI] [PubMed] [Google Scholar]
- 41.Parrinello M, Rahman A. Polymorphic transitions in single crystals. A new molecular dynamics method. J Appl Phys. 1981;52:7182. [Google Scholar]
- 42.Miyamoto S, Kollman PA. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem. 2004;13(8):952–962. [Google Scholar]
- 43.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 44.Darden T, York D, Pedersen L. Particle mesh Ewald: An N-log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 45.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. Journal of molecular graphics. 1996;14(1):33–8, 27–8. . [DOI] [PubMed] [Google Scholar]
- 46.Hou T, Wang J, Li Y, Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. Journal of chemical information and modeling. 2011;51(1):69–82. 10.1021/ci100275a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spiliotopoulos D, Spitaleri A, Musco G. Exploring PHD fingers and H3K4me0 interactions with molecular dynamics simulations and binding free energy calculations: AIRE-PHD1, a comparative study. PloS one. 2012;7(10):e46902 10.1371/journal.pone.0046902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sonawane KD, Barage SH. Structural analysis of membrane-bound hECE-1 dimer using molecular modeling techniques: insights into conformational changes and Abeta1-42 peptide binding. Amino acids. 2015;47(3):543–59. 10.1007/s00726-014-1887-8 . [DOI] [PubMed] [Google Scholar]
- 49.Vorontsov II, Miyashita O. Crystal molecular dynamics simulations to speed up MM/PB(GB)SA evaluation of binding free energies of di-mannose deoxy analogs with P51G-m4-Cyanovirin-N. Journal of computational chemistry. 2011;32(6):1043–53. 10.1002/jcc.21683 . [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Suzek T, Zhang J, Wang J, He S, Cheng T, et al. PubChem BioAssay: 2014 update. Nucleic acids research. 2014;42(Database issue):D1075–82. 10.1093/nar/gkt978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonnet P, Bryce RA. Molecular dynamics and free energy analysis of neuraminidase-ligand interactions. Protein science: a publication of the Protein Society. 2004;13(4):946–57. 10.1110/ps.03129704 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PNG)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.