Abstract
F+ RNA coliphages (FRNA) are used to source-track fecal contamination and as surrogates for enteric pathogen persistence in the environment. However, the environmental persistence of FRNA is not clearly understood and necessitates the evaluation of the survival of prototype and environmental isolates of FRNA representing all four genogroups in surface waters from the central coast of California. Water temperature played a significant role in persistence–all prototype and environmental strains survived significantly longer at 10°C compared to 25°C. Similarly, the availability of host bacterium was found to be critical in FRNA survival. In the absence of E. coli Famp, all prototypes of FRNA disappeared rapidly with a D-value (days for one log reduction) of <1.2 d from water samples incubated at 25°C; the longest surviving prototype was SP. However, in the presence of the host, the order of persistence at 25°C was QB>MS2>SP>GA and at 10°C it was QB = MS2>GA>SP. Significant differences in survival were observed between prototypes and environmental isolates of FRNA. While most environmental isolates disappeared rapidly at 25°C and in the absence of the host, members of genogroups GIII and GI persisted longer with the host compared to members of GII and GIV. Consequentially, FRNA based source tracking methods can be used to detect phages from recent fecal contamination along with those that persist longer in the environment as a result of cooler temperatures and increased host presence.
Introduction
Fecal contamination of ground and surface waters can cause significant environmental and health hazards. Tracking the sources of contamination is critical in designing pollution prevention and pathogen control measures. Several chemicals and biomarkers were used to track contamination resulting from human or animal sources [1–3]. Of these indicators, F+ RNA coliphages (FRNA) were often used as source identifiers to track fecal contamination in ground and surface waters [4, 5], produce [6], estuarine oysters [3], and as surrogates for enteric viral pathogens [7–9]. Using FRNA, contamination can be sourced to either animals or humans based on the association of four different genogroups with specific hosts. Genogroups I and IV (GI and GIV) are generally associated with animals, while genogroups II and III (GII and GIII) are associated with humans [10, 11]. Thus, human fecal pollution of river and ground waters was predicted based on a high abundance of phages from GII [5, 12, 13]. In a recent study, however, species of GIII predominated in both river and creek waters suspected of human activities nearby [14]. Likewise, contamination of surface waters from nearby farms was linked with the high prevalence of members of GI and GIV [15]. In addition to being source identifiers, some FRNA have shown preferential distribution across geographic distances. Members of GII were found to be prevalent in mainland Japan, whereas phages of GIII were isolated from the southern part of Japan and Southeast Asia (Taiwan, the Philippines, Singapore, and Indonesia) [16, 17].
Although beneficial as fecal indicators, differences in survival characteristics amongst FRNA species make it difficult to discern their proportions in environmental samples. Based on the limited data, members of GI and GII were known to persist longer in the environment [18, 19] and resist disinfection treatments compared to GIII and GIV [20]. In a separate study, phage isolates of GI have shown the highest resistance to environmental stresses and inactivation processes, followed by GII, GIII and GIV [18]. In contrast, the persistence of FRNA of different genogroups varied under different conditions, but temperature and pH were suggested to be major factors responsible for their persistence in river water [21]. At 4°C, phages of GI and GII were detectable even after 110 days, while GIII and GIV were reduced to detection limits after 3 weeks and 10 days, respectively. All of them disappeared rapidly at 20°C [19]. In a separate study, temperature correlated significantly with the decay rate of MS2, a prototype of GI, in ground water [22]. FRNA are known also to persist longer in biofilms compared to wastewater [23]. In addition, environmental FRNA differed in persistence compared to prototypes [24]. Thus, the differences in survival, as influenced by the environmental conditions, seem to alter the prevalence and proportion of FRNA.
In a recent study, we attempted to model source tracking shiga-toxigenic Escherchia coli and E. coli O157:H7 using generic E. coli, coliform bacteria, F-specific DNA coliphages and FRNA [14]. Owing to their scarcity, shiga-toxigenic E. coli could not be correlated with the prevalence of FRNA in river and creek water. In addition, higher prevalence of MS2-like phages (GI) was expected to signify the presence of cattle upstream from sampling sites. Instead, QB-like phages (GIII) were detected to indicate possible human fecal pollution. Since these observations were contrary to previous reports [18, 20], studies were conducted comparing environmental FRNA persistence to prototypes in surface water samples from the produce production region of the central coast of California. Since the availability of a bacterial host is critical for phage survival, persistence tests compared the survival of phages in water samples with or without added host bacteria. In addition, we noticed seasonality in the prevalence of FRNA in surface waters [14]; therefore, persistence measurements also included temperature effects. Survival of prototypes was monitored in waters at two different temperatures representing average summer and winter temperatures for the region. Our results demonstrate the suitability of FRNA to source-track fecal contamination in surface waters.
Materials and Methods
Ethics statement
Water samples were from Gabilan Creek near Salinas, California. Special permits are not required for water collection from this publicly accessible place.
Surface waters
Water was collected from Gabilan Creek at Old Stage Road (GPS co-ordinates: 36.78028084N and -121.5848696W) near the city of Salinas. Surface water from the creek was collected using a bottle attached to a telescopic pole and transported on ice to the laboratory. Water samples were refrigerated for <18 h prior to use in phage persistence studies. The water samples were analyzed for total FRNA and for chemical composition (Table 1).
Table 1. Chemical composition of surface waters used.
| Date of water collection | ||||||
|---|---|---|---|---|---|---|
| Chemical componenta | Units | 5-13-15 | 5-26-15 | 6-3-15 | 6-17-15 | 7-20-15 |
| Na | meq/L | 1.43 | 1.48 | 1.48 | 1.48 | 1.52 |
| Ca | meq/L | 6.49 | 5.99 | 5.74 | 6.04 | 6.54 |
| Mg | meq/L | 2.14 | 2.06 | 2.14 | 2.14 | 2.22 |
| HCO3 | meq/L | 5.28 | 4.89 | 4.69 | 5.16 | 5.38 |
| Cl | meq/L | 1.80 | 1.44 | 1.44 | 1.52 | 1.58 |
| Fe | mg/L | 0.04 | 0.04 | <0.04 | <0.04 | <0.04 |
| Mn | mg/L | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| P | mg/L | 0.07 | 0.09 | 0.10 | 0.10 | 0.16 |
| K | mg/L | 1.4 | 1.3 | 1.3 | 1.3 | 1.6 |
| NO3 | mg/L | <2 | <2 | <2 | <2 | 2 |
| SO4 | mg/L | 89 | 87 | 88 | 88 | 93 |
| B | mg/L | 0.05 | 0.06 | 0.07 | 0.06 | 0.07 |
| Dissolved solids | mg/L | 666 | 616 | 601 | 639 | 674 |
| Adjusted SARb | mg/L | 0.96 | 1.00 | 0.99 | 1.00 | 1.01 |
| TSSc | mg/L | 56 | 288 | 92 | 92 | 40 |
| Turbidity | NTU | 0.32 | 0.69 | 0.32 | 0.52 | 0.40 |
| ECd | dS/m | 0.94 | 0.90 | 0.89 | 0.92 | 0.95 |
| pH | 8.1 | 8.1 | 8.1 | 7.9 | 8.2 | |
| Strains assayed | MS2 | QB | SP | GA | All environmental | |
a Analysis performed by A & L Western Agricultural Laboratories, Inc, Modesto, CA
b SAR: sodium adsorption ratio
c TSS: total suspended solids
d EC: electrical conductivity.
Cultures of FRNA
Both prototype and environmental strains of FRNA were evaluated for survival in surface waters. Prototype strains MS2, GA, QB and SP representing genogroups GI, GII, GIII and GIV, respectively, were gifts from Prof. M. Sobsey of the University of North Carolina. Phages previously [14] isolated from surface waters from a produce production region of California (Table 2) representing all four genogroups were included to compare persistence of phages native to these waters to prototypes. Phages were grown in tryptic soy broth (TSB) supplemented with ampicillin/streptomycin (15 μg/mL) and log-phase growth of E. coli Famp (E. coli HS(pFamp)R, ATCC 700891, American Type Culture Collection, Manassas, VA) as described in US EPA method 1601 [25]. Bacterial host cells were initially grown overnight in TSB with ampicillin/streptomycin at 37°C and used as inoculum to prepare fresh batch of log-phase growth. Each FRNA inoculated in 25 mL of log-phase host cell growth was incubated overnight at 37°C to promote phage growth and centrifuged at 10,000 x g for 10 min to remove host cells. The supernatant containing the phage was filtered through 0.2 μm filter and stored at 10°C. The phage preparations were used within 24 h for persistence studies. Concentration of these stocks was 8.7 ± 0.6 log PFU/mL.
Table 2. Environmental strains used.
| Strain | Genogroup | Sourcea | Date of samplingb |
|---|---|---|---|
| GI-1 | GI | ALICAR | 1-2-2013 |
| GII-1 | GII | ALICAR | 1-2-2013 |
| GII-2 | GII | RECVIC | 1-15-2013 |
| GIII-1 | GIII | ALICAR | 1-15-2013 |
| GIII-2 | GIII | ALICAR | 3-20-2013 |
| GIV-1 | GIV | ALICAR | 1-15-2013 |
a ALICAR: Alisal Creek inlet to Carr Lake, Salinas, CA, GPS co-ordinates: 36.67736553N -121.63945327W; RECVIC: Reclamation ditch at Victor street, Salinas, CA; GPS co-ordinates:.68406599N, -121.66718732W [14].
b FRNA were isolated from waters sampled on these dates.
Persistence of prototype strains in surface water
Triplicate 250 mL Erlenmeyer flasks containing 50 mL surface water samples were inoculated with 500 μL portions of prototype FRNA and E. coli Famp host cells. Initial concentrations for FRNA and host cells in each treatment were targeted to be between 6 to 7 log PFU or CFU/mL, respectively. The water was brought to room temperature prior to inoculations and the flasks were kept stationary and in the dark during incubation. Populations of FRNA were monitored for 30 days at two different temperatures and in the presence or absence of the bacterial host. Controls without added FRNA or bacterial host were monitored also to determine the persistence of background populations of FRNA. Survival of each prototype FRNA was monitored at 10°C and 25°C to represent summer and winter temperatures in the Salinas valley region. Average monthly mean temperatures during the past ten years from November to February (winter) was 11.2 ± 0.9°C, March to June (spring) was 14.4 ± 0.6°C and between June to September (summer) was 23.5 ± 2.5°C. The historical data was obtained from the weather station located near KSNS Salinas Municipal Airport, Salinas, CA (http://www.weatherunderground.com/history).
FRNA were enumerated from ten-fold serial dilutions of 100 μL sub-samples of treated and untreated waters at various intervals during the incubation. Double agar layer (DAL) method [25] was used to enumerate FRNA from dilutions prepared in sterile 0.01M phosphate-buffered saline (pH 7.2). Briefly, 100 μL of each of serial dilution of waters and log-phase (4 h growth) E. coli Famp host cells were added to 5 mL of 0.7% tryptic soy agar (TSA) with ampicillin/streptomycin at 45°C, mixed thoroughly and added on to the top of 1.5% TSA plates with ampicillin/streptomycin. The soft agar was allowed to set and the inverted plates were incubated for 18 h at 37°C for plaque formation. The plaques were counted and D-values (days for one log reduction) were calculated based on the log PFU decline from day zero to the last day on which measurable phage growth was observed. D-values were calculated for each replicate separately, and the significant differences in persistence of prototypes of FRNA at two different temperatures and in the presence or absence of E. coli host were determined using three way analysis of variance (SigmaPlot 13, Systat Software, Inc., San Jose, CA).
Persistence of environmental strains in surface water
Survival of native FRNA (Table 2) strains, previously isolated from waters from the same geographical region, was monitored in freshly collected surface water incubated at 25°C. The influence of added E. coli Famp host was also compared during the stationary incubations for 30 d in the dark. The treatments were in triplicate and populations of environmental FRNA were monitored at various intervals from 100 μL serial dilutions in 0.01M phosphate-buffered saline as described above. Data on D-values of environmental and prototype FRNA in the presence or absence of E. coli Famp host was analyzed to determine the differences in persistence of FRNA using two-way analysis of variance (SigmaPlot 13).
Results
Water chemistry
Since freshly collected waters were used for determining the persistence of each prototype strain of FRNA, waters were analyzed to find out if differences in chemical/physical composition alter phage survival. The chemical composition was similar in waters collected on all occasions, except for an elevated level of suspended solids along with increased turbidity for water collected on May 26, 2015 (Table 1). Waters were alkaline. Survival of each prototype phage was monitored using water collected on separate days and all environmental strains were assayed using water collected on one day. FRNA were not detected in these waters prior to inoculations.
Persistence of prototype strains of FRNA
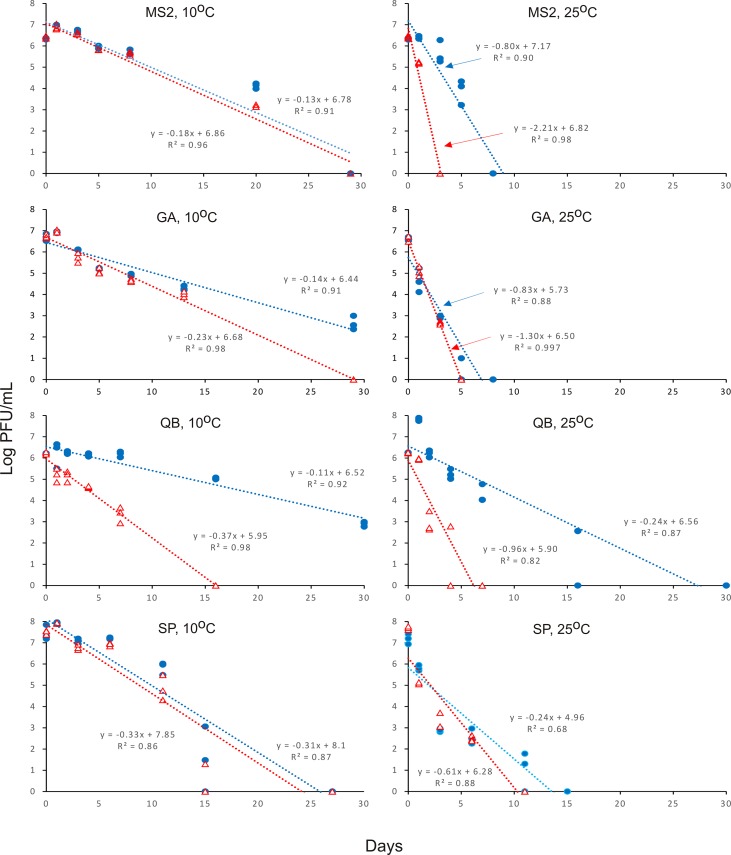
The survival of prototype strains of FRNA was monitored in freshly collected surface water to determine the differences in persistence at temperatures representing the summer and winter conditions in the central coast of California. An initial concentration of 6.4 ± 0.2 PFU/mL was measured from waters treated with MS2, GA or QB. The concentration of SP was somewhat higher at 7.5 ± 0.3 PFU/mL. These high concentrations for FRNA were chosen to obtain several experimental points to aid in calculating D-values. The treatments supplemented with the host received 6.4 ± 0.2 CFU/mL of log phase growth of E. coli Famp.
An increase in phage growth of 0.4 to 0.7 log PFU/mL was observed within one day of incubation at 10°C in treatments of MS2, GA and SP supplemented with the host (Fig 1). A substantially higher increase in growth of 1.5 log PFU/mL on day one was observed with QB plus host at 25°C. Host cells disappeared rapidly with D-values of 5.9 d and 4.0 d at 10°C and 25°C, respectively, from FRNA-free waters.
Fig 1. Survival of prototype strains of FRNA in surface waters.
FRNA populations of four different genogroups were monitored at two temperatures and in the presence (filled blue circles) or absence of (open red triangles) the host E. coli Famp. Data on survival of prototypes in surface water samples is shown in S1 File.
In general, phages disappeared linearly with time and survived longer at 10°C compared to 25°C (Fig 1). In the absence of the host, they all disappeared in less than 10 d from waters at 25°C. MS2 disappeared more rapidly in <3 d. With the host, both GA and QB survived longer at 10°C and nearly 3 log PFU/mL remained after 30 days. QB survived longer with the host at both temperatures.
D-values were calculated based on phage levels on day zero and the initial growth spurts on day one were ignored. Three-way analysis of variance revealed significant differences (P < 0.001) in persistence between prototype strains, in the presence or absence of the host and at different temperatures of incubation (Table 3). In general, FRNA persisted significantly longer in the presence of E. coli Famp compared to no host and at 10°C compared to 25°C. In the presence of the host, the order of persistence at 25°C was QB>MS2>SP>GA; without the host, prototype strains disappeared rapidly. SP survived significantly longer in the absence of host compared to other prototypes. However, the survival spectrum changed at 10°C and MS2 persisted longer both in the presence or absence of the host and the order of persistence without the host was MS2>GA>SP>QB, whereas the order of persistence with the host was MS2 = QB >GA>SP.
Table 3. Influence of temperature of incubation and presence of the host E. coli Famp on the persistence of prototype strains of FRNA in surface water.
| D-value, da | |||||
|---|---|---|---|---|---|
| Prototype strain | Genogroup | 10°C___________ | 25°C____________ | ||
| No host | Host | No host | Host | ||
| MS2 | GI | 6.21 c | 9.05 a | 0.83 hi | 2.11 f |
| GA | GII | 4.94 d | 7.20 b | 0.77 hi | 0.81 hi |
| QB | GIII | 2.46 f | 9.09 a | 0.62 i | 3.95 e |
| SP | GIV | 3.94 e | 4.05 e | 1.15 gh | 1.46 g |
a Three-way analysis of variance statistics: SEM 0.415; P<0.001 for strains, temperature of incubation and presence or absence of the host; P = 0.049 for interactions between these factors; F-values, Fstrains = 15.4; Ftemperature = 450; Fhost = 102. Numbers followed by the same letters are not significantly different from each other. Statistical analysis of D-values is shown in S3 File.
Persistence of environmental strains
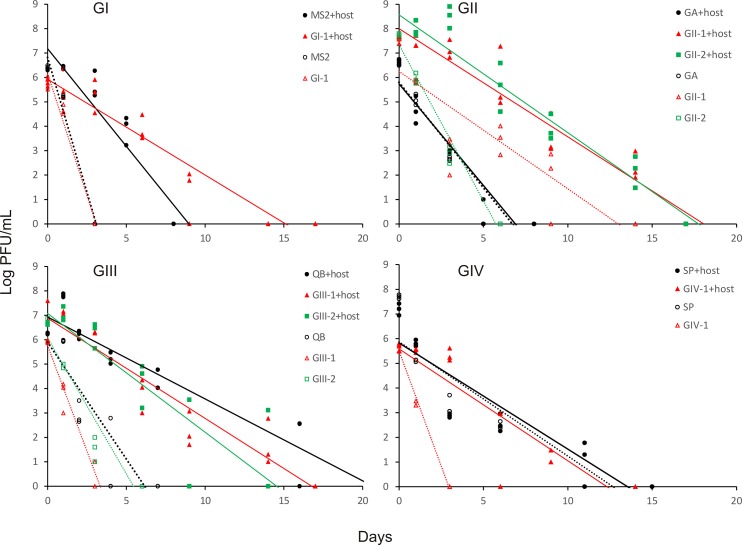
The survival of environmental strains of FRNA isolated from surface waters at different locations and time periods (Table 2) was compared with that of prototype strains incubated at 25°C. Similar to prototypes, an initial surge in growth was observed in the presence of the host with environmental strains of GII and GIII. Absence of the host generally resulted in rapid disappearance of all phages in <10 d (Fig 2) and the environmental strain GI-1 behaved similar to MS2 by rapidly disappearing in <3 d. With the host, environmental strains of GII and GIII survived similarly or longer than their respective prototypes; these were detected even after 14 d.
Fig 2. Persistence of environmental and prototype strains of four different genogroups of FRNA in surface waters.
The survival of phages was monitored at 25°C and in the presence (filled symbols) or absence (open symbols) of the host E. coli Famp. Phage persistence for prototype strains are shown in black. Regression equations are shown in S1 Table. Data on survival of environmental strains in surface water samples is shown in S2 File.
Significant differences (P< 0.001) in D-values were observed between environmental and prototype strains in the presence or absence of the host. Environmental strains of GI, GII and GIV persisted longer compared to prototypes in treatments amended with the host (Table 4). However, environmental strains GIII-1 and GIII-2 disappeared somewhat faster compared to QB. With the exception of GII-1, absence of the host resulted in rapid disappearance of all environmental strains from surface water samples. In the presence of the host, however, significant differences in persistence of strains were found and the order of persistence was GIII-1 = GIII-2 = GI-1>GII-1 = GII-2>GIV-1.
Table 4. Persistence of prototype and environmental strains of FRNA as influenced by the presence of the host E. coli Famp in surface water incubated at 25°C.
| D-value, da | |||
|---|---|---|---|
| Strain | Genogroup | No host | Host |
| MS2 | GI | 0.83 ghij | 2.11 d |
| GI-1 | GI | 0.87 ghi | 3.25 b |
| GA | GII | 0.77 ij | 0.81 hij |
| GII-1 | GII | 1.60 e | 2.72 c |
| GII-2 | GII | 0.61 ijk | 2.59 c |
| QB | GIII | 0.62 ijk | 3.95 a |
| GIII-1 | GIII | 0.52 ijk | 3.19 b |
| GIII-2 | GIII | 0.58 ijk | 2.88 bc |
| SP | GIV | 1.15 fg | 1.46 ef |
| GIV-1 | GIV | 0.43 k | 2.05 d |
a Numbers in bold face are D-values for prototype strains. Numbers followed by the same letters are not significantly different from each other. Two way analysis of variance statistics: SEM = 0.330; D-values are significantly different (P<0.001) between strains, and presence or absence of E. coli host; There is statistically significant interaction between strains and host levels; F-values: Fstrains = 3.963, Fhost = 133 and Finteraction = 4.96. Statistical analysis of D-values is shown in S3 File.
Discussion
Source tracking of foodborne pathogens is critical for development of pathogen control measures at the source. Water is an obvious source of transport of pathogens from animal raising operations, if they are close to waterways. Indeed, numerous E. coli O157:H7 and other shiga-toxigenic E. coli were isolated from surface waters from the produce production region of the central coast of California [26–28]. Since the levels of these pathogens are transient, they are often undetectable during labor-intensive and expensive trace-back studies in the environment. Detection of FRNA as fecal source indicators holds promise, since the contamination of waters can presumably be sourced to animals or humans. However, the differences in persistence of environmental phages make it difficult to distinguish the proportion of FRNA that can be sourced to animals or humans. In addition, seasonal and environmental factors play a significant role in the environmental persistence of FRNA.
Persistence at 25°C
Summers in Salinas valley don’t appear conducive for the growth of FRNA in surface waters. Both prototype and environmental strains disappeared rapidly with D-values <1.6 d and their infectivity was lost in <10 d. Such rapid inactivation was also observed with prototypes in ground, river and sea waters tested at similar temperatures [11, 21, 22, 29, 30]. To aid the comparison of these published results, D-values were calculated from the reported decay rates. However, others have reported decay rates that give a D-value of >15 d (calculated) for MS2 in lake, mineral and sea waters; and the persistence for prototypes and environmental strains were in the order of GI>GII>GIII>GIV [18–20].The current study differs in that the longest surviving phage is an environmental strain GII-1 (GA-like) with a D-value of 1.6 d and the longest surviving prototype is SP of GIV with a D-value of 1.2 (Table 4). We speculate that the absence of host bacterium resulted in rapid disappearance of phages. Since the inactivation of all phages was rapid, the infectivity assays can detect only recent fecal contamination during the summers in California. Such rapid inactivation, as the waters warm up during the spring, could be a reason for the sparse detection of FRNA after April [14]. Similarly, higher water temperatures were linked with rapid inactivation of FRNA [19, 31], resulting in their infrequent detection from surface waters [32].
Persistence at 10°C
Cooler temperatures were responsible for high prevalence of FRNA in waters during the months of January to March in Salinas valley [14] and in Massachusetts bay [33]. Thus, prototypes were evaluated for survival at 10°C and were observed to survive significantly longer compared to 25°C (Table 3). In the absence of the host, significant differences in survival between prototypes were observed at 10°C. MS2 and GA were more persistent compared to SP and QB. The order of persistence is comparable to results obtained by others at higher temperatures [18–20]. However, in a separate study at 15°C in river water, no such differences in persistence between genogroups were found [21]. Thus, fluctuations in temperatures influence the persistence and distribution of coliphages in surface waters. Consequently, elevated levels of phages might not always be from recent fecal inputs but could be more indicative of their environmental persistence.
Persistence in the presence of host bacterium
‘Bacteriophages (phages) are the most abundant replicating entities on the planet and thrive wherever their bacterial hosts exist’ [34]. Indeed, E. coli strains sensitive to infection by FRNA were commonly found in human and animal feces [35], and a significant correlation was observed between the prevalence of FRNA and E. coli in tropical surface waters [36]. Accordingly, this study compared the survival of prototype and environmental strains of FRNA in the presence of the host E. coli Famp.
FRNA survived significantly longer (P<0.001) in the presence of the host and the persistence spectrum of the phage types differed as compared to no host. Prototypes QB followed by MS2 persisted longer compared to the other prototypes incubated at either 10°C or 25°C (Table 3). Similarly, QB-like (GIII-1 and GIII-2) and MS2-like (GI-1) phages persisted with the host compared to other environmental phages. To the best of our knowledge, there are no other reports that evaluated the significance of the presence of the host on FRNA survival in the environment, although both phage and host are released together in the feces of animals and humans. Determining host prevalence along with FRNA may aid in improved prediction of fecal sources, since the presence of E. coli was positively correlated with FRNA [36] while its absence resulted in the rapid disappearance of coliphages from surface waters.
Host presence also caused growth of FRNA. This is predictable as the waters treated with the host received 6.4 log CFU/mL of E. coli Famp. This resulted in replication of phages for 1 to 3 d (Figs 1 and 2). Further phage growth was likely hampered as the host populations decreased with D-values of 4 to 6 d. It was reported that QB failed to replicate when host cells were fewer than 4 log CFU/mL [37]. However, using QB as a model, it was also reported that replication ceases below 25°C [38]. In contrast, this study observed growth with 3 out of 4 prototypes in the presence of the host within a day of incubation at 10°C. Availability of host bacterium significantly influences the survival, growth and prevalence of FRNA in surface waters.
Persistence of environmental strains
Significant differences in survival were observed between prototype and environmental strains of FRNA. A majority of the environmental strains persisted longer than the prototypes when the host was present and strain differences in survival were observed between genogroups. Similar strain differences in persistence were also reported for other environmental isolates [19, 24]. While a majority of the environmental strains disappeared rapidly in the absence of added host, members of GIII and GI followed by GII persisted longer with the host. This is consistent with our earlier, preferential isolation of members of GIII followed by GII and GI from surface waters from the same geographical region [14]. Therefore, survival of environmental and prototype strains of FRNA varies within and between genogroups and is influenced by prevalence of hosts native to the environment.
Persistence and water chemistry
Differences in the chemical constituents of waters did not influence the persistence of prototype strains. The absence of effect is evident as the differences in D-values, at 25°C without the host, between MS2, GA and QB were not significantly different from each other and so as the differences between MS2, GA and SP (Table 3). This might have been predictable as salt concentrations of up to 42 g/L in artificial sea water failed to alter the survival of FRNA compared to mineral water [18]. In addition, it was reported that suspended solids have a protective effect on the survival of viruses [39], while removal of solids results in rapid inactivation [21]. However, in the present study, relatively high amount of solids had no effect, as QB was inactivated at 25°C as rapidly as the other phages incubated in waters with less solids (Tables 1 and 3). Furthermore, FRNA were reported to persist longer as pH decreases from 8.4 to 5.5 [21], and the higher pH (8.1) of surface waters in this study might have aided the rapid disappearance of all FRNA.
Conclusions
Significant differences exist in the persistence of environmental and prototype strains of FRNA. Persistence fluctuated with seasonal temperatures and the availability of bacterial host. Therefore, source tracking attempts would likely detect phages from recent fecal inputs compared to those that persist under fluctuating environmental conditions. Future studies that expand to different geographic locations and include infectivity assays with natural hosts may aid in determining an accurate assessment of prevalence of FRNA in surface waters. Since other fecal indicators (E. coli, enterococi, Bacteroides thetaiotaomicron and others) fluctuate similarly as FRNA [36], multiple indicators may be chosen for improved source tracking of fecal inputs to develop strategies to limit the hazards of fecal contamination of ground and surface water.
Supporting Information
Excel 2007 file.
(XLSX)
Excel 2007 file.
(XLSX)
SigmaPlot 13 notebook.
(JNB)
(DOCX)
Acknowledgments
We thank Anita Liang and Diana Carychao for their assistance with water collections from Salinas valley, and Amarnath Ravva and Michael Cooley for their critical review.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The work was funded by the U.S. Department of Agricuture (USDA), Agricultural Research Service CRIS project 2030-42000-046.
References
- 1.Blanch AR, Belanche-Munoz L, Bonjoch X, Ebdon J, Gantzer C, Lucena F, et al. Integrated analysis of established and novel microbial and chemical methods for microbial source tracking. Appl Environ Microbiol. 2006;72(9):5915–26. 10.1128/AEM.02453-05 ; PMCID: PMC1563622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gourmelon M, Caprais MP, Mieszkin S, Marti R, Wery N, Jarde E, et al. Development of microbial and chemical MST tools to identify the origin of the faecal pollution in bathing and shellfish harvesting waters in France. Water Res. 2010;44(16):4812–24. Epub 2010/08/17. 10.1016/j.watres.2010.07.061 . [DOI] [PubMed] [Google Scholar]
- 3.Mieszkin S, Caprais MP, Le Mennec C, Le Goff M, Edge TA, Gourmelon M. Identification of the origin of faecal contamination in estuarine oysters using Bacteroidales and F-specific RNA bacteriophage markers. J Appl Microbiol. 2013;115(3):897–907. Epub 2013/05/25. 10.1111/jam.12260 . [DOI] [PubMed] [Google Scholar]
- 4.Wilkes G, Brassard J, Edge TA, Gannon V, Gottschall N, Jokinen CC, et al. Long-term monitoring of waterborne pathogens and microbial source tracking markers in paired agricultural watersheds under controlled and conventional tile drainage management. Appl Environ Microbiol. 2014;80(12):3708–20. 10.1128/AEM.00254-14 ; PMCID: PMC4054145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haramoto E, Otagiri M, Morita H, Kitajima M. Genogroup distribution of F-specific coliphages in wastewater and river water in the Kofu basin in Japan. Lett Appl Microbiol. 2012;54(4):367–73. Epub 2012/02/14. 10.1111/j.1472-765X.2012.03221.x . [DOI] [PubMed] [Google Scholar]
- 6.Allwood PB, Malik YS, Maherchandani S, Vought K, Johnson LA, Braymen C, et al. Occurrence of Escherichia coli, noroviruses, and F-specific coliphages in fresh market-ready produce. J Food Prot. 2004;67(11):2387–90. Epub 2004/11/24. . [DOI] [PubMed] [Google Scholar]
- 7.Rezaeinejad S, Vergara GG, Woo CH, Lim TT, Sobsey MD, Gin KY. Surveillance of enteric viruses and coliphages in a tropical urban catchment. Water Res. 2014;58C:122–31. 10.1016/j.watres.2014.03.051 . [DOI] [PubMed] [Google Scholar]
- 8.Havelaar AH, van Olphen M, Drost YC. F-specific RNA bacteriophages are adequate model organisms for enteric viruses in fresh water. Appl Environ Microbiol. 1993;59(9):2956–62. Epub 1993/09/01. ; PMCID: PMC182392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tung-Thompson G, Libera DA, Koch KL, de Los Reyes FL 3rd, Jaykus LA. Aerosolization of a human norovirus surrogate, bacteriophage MS2, during simulated vomiting. PLoS One. 2015;10(8):e0134277 10.1371/journal.pone.0134277 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Love DC, Vinje J, Khalil SM, Murphy J, Lovelace GL, Sobsey MD. Evaluation of RT-PCR and reverse line blot hybridization for detection and genotyping F+ RNA coliphages from estuarine waters and molluscan shellfish. J Appl Microbiol. 2008;104(4):1203–12. Epub 2007/11/22. 10.1111/j.1365-2672.2007.03646.x . [DOI] [PubMed] [Google Scholar]
- 11.Ogorzaly L, Bertrand I, Paris M, Maul A, Gantzer C. Occurrence, survival, and persistence of human adenoviruses and F-specific RNA phages in raw groundwater. Appl Environ Microbiol. 2010;76(24):8019–25. Epub 2010/10/19. 10.1128/AEM.00917-10 ; PMCID: PMC3008255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogorzaly L, Tissier A, Bertrand I, Maul A, Gantzer C. Relationship between F-specific RNA phage genogroups, faecal pollution indicators and human adenoviruses in river water. Water Res. 2009;43(5):1257–64. Epub 2009/01/06. 10.1016/j.watres.2008.12.011 . [DOI] [PubMed] [Google Scholar]
- 13.Haramoto E, Yamada K, Nishida K. Prevalence of protozoa, viruses, coliphages and indicator bacteria in groundwater and river water in the Kathmandu Valley, Nepal. Trans R Soc Trop Med Hyg. 2011;105(12):711–6. Epub 2011/11/05. 10.1016/j.trstmh.2011.08.004 . [DOI] [PubMed] [Google Scholar]
- 14.Ravva SV, Sarreal CZ, Cooley MB. Male-specific coliphages for source tracking fecal contamination in surface waters and prevalence of Shiga-toxigenic Escherichia coli in a major produce production region of the Central Coast of California. Environ Sci Process Impacts. 2015. 10.1039/c4em00537f . [DOI] [PubMed] [Google Scholar]
- 15.Lee JE, Lee H, Cho YH, Hur HG, Ko G. F+ RNA coliphage-based microbial source tracking in water resources of South Korea. Sci Total Environ. 2011;412–413:127–31. Epub 2011/11/01. 10.1016/j.scitotenv.2011.09.061 . [DOI] [PubMed] [Google Scholar]
- 16.Furuse K, Ando A, Osawa S, Watanabe I. Continuous survey of the distribution of RNA coliphages in Japan. Microbiol Immunol. 1979;23(9):867–75. . [DOI] [PubMed] [Google Scholar]
- 17.Furuse K, Sakurai T, Hirashima A, Katsuki M, Ando A, Watanabe I. Distribution of ribonucleic acid coliphages in south and east Asia. Appl Environ Microbiol. 1978;35(6):995–1002. ; PMCID: PMC242975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaper M, Duran AE, Jofre J. Comparative resistance of phage isolates of four genotypes of f-specific RNA bacteriophages to various inactivation processes. Appl Environ Microbiol. 2002;68(8):3702–7. Epub 2002/07/31. ; PMCID: PMC124048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long SC, Sobsey MD. A comparison of the survival of F+RNA and F+DNA coliphages in lake water microcosms. J Water Health. 2004;2(1):15–22. . [PubMed] [Google Scholar]
- 20.Muniesa M, Payan A, Moce-Llivina L, Blanch AR, Jofre J. Differential persistence of F-specific RNA phage subgroups hinders their use as single tracers for faecal source tracking in surface water. Water Res. 2009;43(6):1559–64. Epub 2009/01/17. 10.1016/j.watres.2008.12.038 . [DOI] [PubMed] [Google Scholar]
- 21.Yang Y, Griffiths MW. Comparative persistence of subgroups of F-specific RNA phages in river water. Appl Environ Microbiol. 2013;79(15):4564–7. Epub 2013/05/21. 10.1128/AEM.00612-13 ; PMCID: PMC3719516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yates MV, Gerba CP, Kelley LM. Virus persistence in groundwater. Appl Environ Microbiol. 1985;49(4):778–81. Epub 1985/04/01. ; PMCID: PMC238444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skraber S, Helmi K, Willame R, Ferreol M, Gantzer C, Hoffmann L, et al. Occurrence and persistence of bacterial and viral faecal indicators in wastewater biofilms. Water Sci Technol. 2007;55(8–9):377–85. . [DOI] [PubMed] [Google Scholar]
- 24.Brion GM, Meschke JS, Sobsey MD. F-specific RNA coliphages: occurrence, types, and survival in natural waters. Water Res. 2002;36(9):2419–25. Epub 2002/07/11. S0043-1354(01)00547-4 [pii]. . [DOI] [PubMed] [Google Scholar]
- 25.Anonymous. 821-R-01-030. Method 1601: Male-specific (F+) and somatic coliphage in water by two-step enrichment procedure Washington, DC.: U.S. Environmental Protection Agency, 2001. http://www.epa.gov/nerlcwww/1601ap01.pdf. Accessed 08/14/2015.
- 26.Benjamin LA, Jay-Russell MT, Atwill ER, Cooley MB, Carychao D, Larsen RE, et al. Risk factors for Escherichia coli O157 on beef cattle ranches located near a major produce production region. Epidemiol Infect. 2014:1–13. Epub 2014/03/22. 10.1017/S0950268814000521 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamin L, Atwill ER, Jay-Russell M, Cooley M, Carychao D, Gorski L, et al. Occurrence of generic Escherichia coli, E. coli O157 and Salmonella spp. in water and sediment from leafy green produce farms and streams on the Central California coast. Int J Food Microbiol. 2013;165(1):65–76. Epub 2013/05/24. 10.1016/j.ijfoodmicro.2013.04.003 . [DOI] [PubMed] [Google Scholar]
- 28.Cooley M, Carychao D, Crawford-Miksza L, Jay MT, Myers C, Rose C, et al. Incidence and tracking of Escherichia coli O157:H7 in a major produce production region in California. PLoS One. 2007;2(11):e1159 10.1371/journal.pone.0001159 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirs M, Smith DC. Multiplex quantitative real-time reverse transcriptase PCR for F+-specific RNA coliphages: a method for use in microbial source tracking. Appl Environ Microbiol. 2007;73(3):808–14. Epub 2006/12/05. 10.1128/AEM.00399-06 ; PMCID: PMC1800770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon C, Toze S. Influence of groundwater characteristics on the survival of enteric viruses. J Appl Microbiol. 2003;95(3):536–44. 10.1046/j.1365-2672.2003.02010.x . [DOI] [PubMed] [Google Scholar]
- 31.Sinton LW, Finlay RK, Lynch PA. Sunlight inactivation of fecal bacteriophages and bacteria in sewage-polluted seawater. Appl Environ Microbiol. 1999;65(8):3605–13. Epub 1999/07/31. ; PMCID: PMC91541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cole D, Long SC, Sobsey MD. Evaluation of F+ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Appl Environ Microbiol. 2003;69(11):6507–14. Epub 2003/11/07. ; PMCID: PMC262259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ballester NA, Fontaine JH, Margolin AB. Occurrence and correlations between coliphages and anthropogenic viruses in the Massachusetts Bay using enrichment and ICC-nPCR. J Water Health. 2005;3(1):59–68. Epub 2005/06/15. . [PubMed] [Google Scholar]
- 34.Mills S, Shanahan F, Stanton C, Hill C, Coffey A, Ross RP. Movers and shakers: influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes. 2013;4(1):4–16. 10.4161/gmic.22371 ; PMCID: PMCPMC3555884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Havelaar AH, Pot-Hogeboom WM, Furuse K, Pot R, Hormann MP. F-specific RNA bacteriophages and sensitive host strains in faeces and wastewater of human and animal origin. J Appl Bacteriol. 1990;69(1):30–7. . [DOI] [PubMed] [Google Scholar]
- 36.Liang L, Goh SG, Vergara GG, Fang HM, Rezaeinejad S, Chang SY, et al. Alternative fecal indicators and their empirical relationships with enteric viruses, Salmonella enterica, and Pseudomonas aeruginosa in surface waters of a tropical urban catchment. Appl Environ Microbiol. 2015;81(3):850–60. 10.1128/AEM.02670-14 ; PMCID: PMCPMC4292481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woody MA, Cliver DO. Replication of coliphage Q beta as affected by host cell number, nutrition, competition from insusceptible cells and non-FRNA coliphages. J Appl Microbiol. 1997;82(4):431–40. Epub 1997/04/01. . [DOI] [PubMed] [Google Scholar]
- 38.Woody MA, Cliver DO. Effects of temperature and host cell growth phase on replication of F-specific RNA coliphage Q beta. Appl Environ Microbiol. 1995;61(4):1520–6. Epub 1995/04/01. ; PMCID: PMC167408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao VC, Seidel KM, Goyal SM, Metcalf TG, Melnick JL. Isolation of enteroviruses from water, suspended solids, and sediments from Galveston Bay: survival of poliovirus and rotavirus adsorbed to sediments. Appl Environ Microbiol. 1984;48(2):404–9. ; PMCID: PMCPMC241526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Excel 2007 file.
(XLSX)
Excel 2007 file.
(XLSX)
SigmaPlot 13 notebook.
(JNB)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.