Abstract
Certified reference materials (CRMs) are required to guarantee the reliability of analytical measurements. The CRMs available in the field of genetically modified organisms (GMOs) are characterized using real-time polymerase chain reaction (qPCR). This technology has limited application, because of its dependence on a calibrant. The objective of this study was to obtain a method with higher metrological quality, to characterize the CRMs for their contents of T-nos/hmg copy number ratio in maize. A duplex droplet digital PCR (ddPCR) assay was developed and optimized by a central composite design. The developed method achieved an absolute limit of detection (LOD) of 11 cP T-nos, a relative LOD of 0.034%, a limit of quantification (LOQ) of 23 cP (relative LOQ of 0.08%), and a dynamic range of 0.08%–100% T-nos/hmg ratio. The specificity and applicability of the assay were established for the analysis of low T-nos concentrations (0.9%) in several corn varieties. The convenience of DNA digestion to reduce measurement bias in the case of multiple-copy binding was confirmed through an enzymatic restriction assay. Given its overall performance, this method can be used to characterize CRM candidates for their contents of T-nos/hmg ratio.
Cultivation and commercialization of genetically modified organisms (GMOs) are regulated activities around the world. Labeling is a requirement in several countries to identify GMO-derived products when certain thresholds of maximum content are exceeded.26 Reliable analytical quantification methods, as well as certified reference materials (CRMs), are mandatory to verify the fulfillment and correct application of these regulations. The majority of CRMs derived from commercially available GMOs are certified for their content of specific events as a mass fraction, the international measurement unit adopted in the analysis and expression of the results.15,21
In 2004, the European Commission issued a recommendation proposing the expression of GMO quantitative analysis results as a percentage of genetically modified DNA copy number (cp) related to target taxon-specific DNA copy numbers, calculated in terms of haploid genomes.6 For example, this recommendation is based on scientific evidence that have shown the zygosity, parental origin, and ploidy levels of the structural components of corn seed strongly influence the results of GMO quantification, especially when expressed as a mass fraction.22,25,27 However, these biological factors become irrelevant if the employed reference materials (RMs) permit the quantification of GMO content, in terms of the haploid genome copy number.3 Consequently, this measurement unit (copy number ratio) has been used lately in CRM production.15 Nonetheless, CRMs have been characterized using qPCR, which is the most common method for GMO quantification3,15−17 and the characterization of their RMs.
To estimate the concentration of the DNA target sequence, qPCR determines the number of amplification cycles at which the fluorescence exceeds a certain threshold (Cq) and compares it with a calibrant of known concentration. This technique has important drawbacks: it requires a calibrant be available and the possible differences in amplification efficiencies between the calibrant and the material analyzed. In GMO CRM characterization, plasmids have been used as calibrant materials; however, this process is time-consuming and expensive. In addition, this approach does not establish a direct traceability to the International System of Units, which may lead to increases in the uncertainties of the measurement and of certified value assigned to the characterized material. Therefore, the use of methods with higher metrological quality than qPCR is strongly recommended for this purpose.
On the other hand, digital PCR technology is not dependent on the calibrant and presents a less restrictive amplification efficiency, compared to qPCR, allowing for the absolute quantification of DNA molecules from a sample, overcoming several limitations of qPCR. Because of its measurement principle, digital PCR has been suggested as a primary method for certifying GMO RMs for their copy number ratio.5 Although several studies focus on developing methods for nopaline synthase terminator (T-nos) quantification in corn and other matrices have been reported,9,10,14,20,23,28 no method for amplification and simultaneous quantification of this element and the high-mobility group (hmg) reference gene is known, using either real-time PCR or digital PCR. The objective of this work was to develop a reliable analytical method, optimized through appropriate experimental design tools, showing suitable characteristics for its use in the characterization of CRM candidates for their contents of T-nos/hmg copy number ratio. T-nos was chosen because it is a common regulatory element used for GMO screening at analytical laboratories, and it is one of the five sequences proposed for authorized and nonauthorized GMO detection in corn.24
Experimental Section
Experimental Materials
For the development, optimization, and validation of the method, maize seed-powder flour from the DMR 447 series of CRMs from the National Metrology Centre was used (CENAM, México). This series is certified as a mass fraction for the NK603 specific event: DMR 447 IIa (100% mass fraction), DMR 447 Va (10% mass fraction), DMR 447 IVa (5%), and DMR 447 IIIa (1%). The materials employed in the evaluation of the experimental specificity of the method are described in the corresponding paragraph.
DNA Extraction
DNA was extracted from 100 mg of flour of different materials, using a Fast ID Genomic Extraction DNA kit (Genetic ID NA, USA). The extraction protocol followed the instructions indicated by the provider, with some volume modifications (see Annex S1 in the Supporting Information). Genomic DNA concentration was estimated by measuring the absorbance at 260 nm (A260), using a UV-vis spectrophotometer (Jenway, U.K.). It was considered that a value of A260 of 1.00 corresponds to 50 ng/μL of double-stranded DNA. DNA purity was measured by calculating the ratio of absorbance at 260/280 nm. Only samples with a minimum concentration of 45 ng/μL and an absorbance ratio between 1.8 and 2.0 were included in the assays. DNA concentration of samples tested, ranged from 45 ng/μL to 100 ng/μL throughout all assays.
Primers and Probes
The selected primers and probes (Table S1 in the Supporting Information) were described by Reiting et al.20 and ISO 2157013 and were synthesized by Applied Biosystems (USA). The quencher used in the probes was modified to comply with ddPCR usage requirements.
Droplet Digital PCR
The analysis of samples (Annex S2 in the Supporting Information) was based on the Droplet Generator and Droplet Reader QX200 manuals (ddPCR, Bio-Rad, USA). All the primers, probes, and reaction mixes were prepared gravimetrically. For this process, MIQE guides were consulted.1,12 Positive and negative droplets were differentiated by applying a manual fluorescence amplitude threshold (Figure S1 in the Supporting Information). Copy numbers reported by the droplet digital reader, were corrected to copy numbers/μL of DNA sample, considering the dilution factor of the ddPCR reaction mix.
Optimization
A central composite design (Table 1) for 3 factors and 20 trials was used. The factors were T-nos primer/probe concentration (T-nos), hmg primer/probe concentration (hmg), and annealing temperature (Ta). Two experiments were run; one with the material DMR 447 IIa and the other with the material DMR 447 Va. A homogeneous mix of DNA extracts was prepared for each material. From preliminary explanatory results previously obtained at the laboratory (data not shown), a high value (900/350 nM and 80/140 nM for T-nos and hmg respectively) and a low value (600/250 nM and 40/80 nM for T-nos and hmg, respectively) were established for the T-nos and hmg factors. For factor Ta, temperatures of 61 and 59 °C were taken as high and low values, respectively. A design matrix with 20 independent trials per experiment was constructed (see Table 1, as well as Tables S2 and S3 in the Supporting Information). The response variables were the T-nos/hmg copy number ratio (T-nos/hmg); the hmg copy number (hmg cp); and droplet separation for each analyte, measured as the fluorescence difference between positive and negative droplets (T-nos ΔF and hmg ΔF). The average of three replicates per treatment and a nontemplate control (NTC) were registered for each dependent variable.
Table 1. Central Composite Design To Optimize the T-nos/hmg ddPCR Duplex Assay.
| factor | axial (−1.68) | low (−1) | central (0) | high (1) | axial (1.68) |
|---|---|---|---|---|---|
| T-nos primer/probe concentration, T-nos (nM) | 498/216 | 600/250 | 750/300 | 900/350 | 1002/384 |
| hmg primer/probe concentration, hmg (nM) | 25/60 | 40/80 | 60/110 | 80/140 | 95/160 |
| annealing temperature, Ta (°C) | 58 | 59 | 60 | 61 | 62 |
A quadratic linear regression model was estimated for each dependent variable; then, optimal experimental conditions and/or acceptable value ranges for each response variable were found, using a multiresponse optimal analysis (by contour plots and the desirability function). DNA concentrations estimated by spectrophotometry, maize genome weight, and T-nos expected concentrations in the analyzed samples, were used as reference to define acceptable values for T-nos/hmg and hmg cp variables. The optimal experimental conditions of the method (primer and probe concentrations and annealing temperature) were defined as those measuring the expected T-nos/hmg copy number ratio and hmg copy number, with the highest droplet separation. The goal of the study was to find target values for T-nos/hmg and hmg cp, and maximum values for T-nos ΔF and hmg ΔF within the established ranges. All collected data were analyzed using the statistical package Minitab v. 16.
Performance Assessment of the Optimized Method
Specificity
The theoretical specificity of T-nos and hmg primer sequences used in this study was evaluated in silico, using the local alignment tool (BLAST) from NCBI. To evaluate the experimental specificity of the hmg detection method, DNA was extracted and analyzed from two replicates of popping corn, blue corn, red corn, white corn, yellow corn, soybean, wheat, and rice. Seeds of popping corn, blue corn, red corn, and rice samples were acquired from the local markets, while seeds of white corn were provided by the International Maize and Wheat Improvement Center. The seeds were ground, sieved, and dried at a particle size of 125–425 μm and humidity lower than 2%. For the yellow corn, CENAM DMR 436 Ia (negative control material for MON 810 event) and DMR 453 Ia (negative control material for MON 88017 event) RMs were used. In the case of soybean and wheat, DMR 495 IIa, DMR 495 IIIa (soybean positive control material for MON-04032-6), DMR 496 IIa, and DMR 496 IIIa (wheat positive control material for DREB1A) RMs were used.
To evaluate the specificity of the T-nos detection method, two replicates of RMs lacking the sequence of interest—either for not being included in the transformation event structure (DMR 436 Vb, MON810) or because the analyzed batch matched the nontransgenic material of the series (DMR 451 Ia MON 863, DMR 453 Ia MON 88017 and DMR 482 negative control material to p35S and T-nos)—were evaluated. Materials corresponding to events regulated by T-nos (DMR 447 Va (NK603), DMR 451 Va (MON863), DMR 452 IIa (MON89034), and DMR 453 Va (MON 88017)) were also analyzed.
Detection and Quantification Limit
The goal of the proposed method was to quantify the T-nos concentration (in terms of copy number ratio) in maize flour. The copy number of the reference gene (hmg) in the samples analyzed was expected to be at least 18 000 for a DNA extract of ∼50 ng/μL; therefore, the detection and quantification limits were performed only for T-nos, both in copy number and copy number ratio in relation to hmg. The absolute detection limit (LODabs) and the relative detection limit (LODrel) were determined following the AFNOR (LOD6) guidelines.1 Four serial dilutions were prepared during 3 days, based on DMR 447 IIIa RM, using non-GM maize as a diluent. These dilutions were analyzed in six replicates, together with DMR 453 IIIa RM, covering a T-nos concentration range from 0.710% (corresponding to ∼90 copies) to 0.007% (∼2 copies), measured in maize samples with DNA concentrations of 65–100 ng/μL. Given the evaluation conditions, this range could also be considered the asymmetric detection limit (LODasy), because the setup was performed with hmg background levels of 25 000–37 000 cP, corresponding to an approximate ratio of 1:2000 to 1:3500.
Dynamic Range (Linearity, Precision, and Trueness)
The dynamic range of the method was evaluated from the concentration defined as the limit of quantification (LOQ) (0.08% T-nos/hmg ratio) to the highest available concentration of T-nos, using DNA extracts of 45–90 ng/μL. A five-point calibration curve was generated and analyzed in three replicates obtained over 3 days, based on DNA extracted from DMR 447 IIa, DMR Va, DMR IVa, and DMR IIIa materials and a dilution of the DMR 447 IIIa, corresponding to an approximate final concentration of T-nos/hmg copy number ratio from 100% (16 500 cP) to 0.08% (23 cP). Nowadays, there is no reference material certified for its T-nos content; therefore, in order to evaluate the accuracy of the method over the analyzed concentration range, the following analytical approach was used: the event-specific NK603 was measured previously for DMR 447 IIa (100% mass fraction) by digital PCR (data not shown), showing a content of 50% NK603/hmg copy number ratio. Considering this result and the presence of two linked T-nos copies in the genetic construct of the experimental material (Figure 1), it was expected to measure a 100% T-nos/hmg copy number ratio. Accordingly, the certified mass fraction (event-specific) was selected as the theoretical value for T-nos/hmg copy number ratio. Values obtained from the three calibration curves analyzed were plotted against the theoretical concentration, and the correlation coefficient was determined. The dynamic range of the method was defined as the concentration range where the response was linear (correlation coefficient of R2 ≥ 0.98), precise (relative standard deviation of ≤25%) and accurate (±25%).4,8
Figure 1.
Genetic construct of the NK603 transformation event. [Legend: Ctp 2, chloroplast transit peptide; cp4-epsps, 5-enolpyruvylshikimate-3-phosphate synthase, derived from the cp4 strain of Agrobacterium tumefociens; T-nos, nopaline synthase terminator; CaMV, cauliflower mosaic virus; enzyme restriction sites are indicated above the genetic construct. (This figure was constructed based on the information available on the Biosafety Cleaning-House webpage: http://bch.cbd.int/database/record.shtml?documentid=14776).
Applicability Assessment
Since the development, optimization, and validation of the method was performed using yellow corn, the applicability of the method to quantify T-nos/hmg was assessed in four additional corn varieties. To this end, the DNA extracted from non-GM popping corn, blue corn, red corn, and white corn was mixed with an aliquot of DMR 447 IIa DNA to obtain a final concentration close to 0.9% T-nos with hmg background levels of 28 000–37 000 cP. The analyzed variety represented at least 97.5% of the final mix. Each mix was analyzed in three replicates; the precision and accuracy criteria were applied to establish the method applicability for every variety.
Comparison of Duplex and Single Plex Assay
To amplify T-nos and hmg using the single plex method, two reaction mixes were prepared. They were subsequently used to analyze three replicates of the following materials: DMR 447 IIa, DMR 452 IIa, DMR 447 Va, DMR 447 IIIa, and a dilution of the latter to a concentration near the LOQ value (0.08%). The copy number estimated for each analyte, together with the T-nos/hmg copy number ratio, calculated based on these data, was compared with the results of the dynamic range curves generated using the duplex method, through the bias assessment of each parameter.
Restriction Enzyme Digestion
The material (DMR 447) used for optimization matched the NK603 transformation event, whose genetic construct is formed by two expression cassettes, each regulated by one T-nos copy (Figure 1). To selectively separate the linked T-nos copies and to investigate the effects of fragmentation on the quantification of the T-nos cp and T-nos/hmg copy number ratio, an enzymatic digestion assay was performed. Three endonucleases (BamHI, EcoRI, and XhoI; Thermo Scientific, USA) were selected through an in silico analysis; according to the literature and databases, they could have restriction sites outside the sequence of the analytes of interest and between the two copies of T-nos, inside the NK603 structure, except XhoI, which was used as a negative control. To select the endonucleases, the online tool Restriction Mapper v.3 was used, together with the known sequences of T-nos (GenBank: AX342369.1; Section 8 from European Patent No. EP1167531),100hmg (GenBank: AJ131373.1), hsp70 (GenBank: X03714.1), Ctp2-cp4-epsps (GenBank: KJ787649.1) and P35S (GenBank: AJ007626.1); the theoretical restriction sites on the genetic construct are shown in Figure 1. Prior to the preparation of the amplification reaction and droplet generation mix, a DMR 447 Va DNA aliquot was digested, first separately with each enzyme and next with all enzymes simultaneously, according to the manufacturer’s suggestion (see Annex S3 in the Supporting Information). Based on the results of this assay, BamHI was chosen to digest DNA aliquots from DMR 447 IIa and DMR 447 IIIa materials. The fragmentation and analysis of DMR 452 IIa (MON 89034, 100% mass fraction), which corresponded to a transformation event with a single copy of T-nos, were used as controls. The copy numbers reported by the droplet digital reader were corrected considering the dilution factor of the digestion and/or the dilution factor of the reaction mix, for digested and undigested samples, respectively. Each extract was analyzed in three replicates; a comparison between these results and the ones generated by the analysis of the undigested DNA was performed by applying a Student’s t-test, based on mean differences analysis. The completion of the digestion was verified using electrophoresis in a 1% agarose gel.
Results and Discussion
Optimization
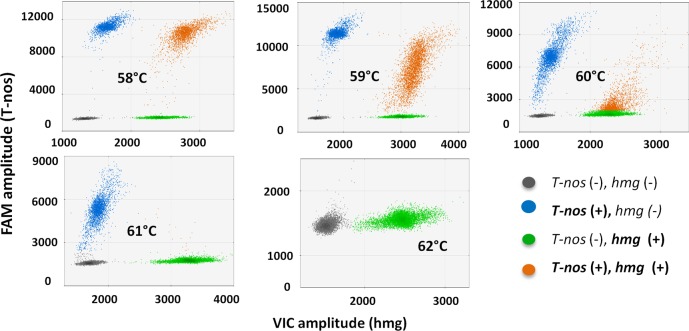
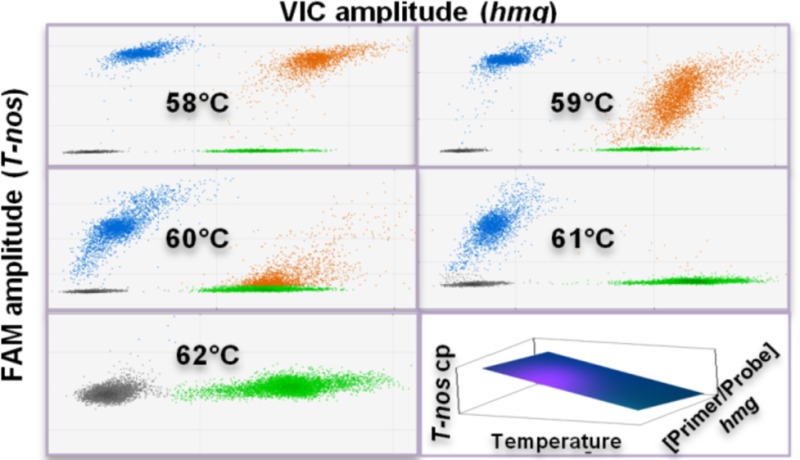
The linear regression analysis for 100% and 10% (DMR 447 IIa and DMR 447 Va) showed significative models for three of the four response variables (T-nos/hmg, T-nos ΔF, and hmg ΔF) (see Tables S4 and S5 in the Supporting Information). In both cases, the most influencing factor for T-nos/hmg and T-nos ΔF was the annealing temperature (Ta). T-nos/hmg and T-nos ΔF increased when the temperature approached 58 °C, whereas, at 60 °C, T-nos/hmg started to decrease, until reaching 0 during the 62 °C thermocycle (see Table S6 in the Supporting Information). The hmg droplet resolution (hmg ΔF) was primarily affected by the hmg primer/probe concentration, although the annealing temperature also showed an inverse effect on this variable. In this regard, two-dimensional (2D) plots from QuantaSoft (see Figure 2) showed that, the T-nos/hmg positive droplet group (orange) was the most sensitive to temperature; above 60 °C, the resolution began to decrease. In this group, where T-nos and hmg coexisted in a single droplet, they might be competing for the amplification reagents. Apparently, increasing the annealing temperature above 58 °C was detrimental to T-nos, leading to lower amplification efficiency, lower fluorescence intensity and resolution, and, eventually, lower estimated copy number.
Figure 2.
Two-dimensional (2D) plots generated during the optimization of the T-nos/hmg duplex ddPCR assay. Analysis on OMR 447 Ha (NK603 100% mass fraction). Representative plots for each of the five evaluated temperature levels (58–62 °C) are shown. Each point represents a droplet with a given fluorescence level; droplet colors indicate which target was amplified: T-nos (blue), hmg (green), none of the two (gray) or both (orange). The x-axis shows the fluorescence amplitude corresponding to the VIC fluorophore (hmg), and the y-axis represents the fluorescence amplitude corresponding to the FAM fluorophore (T-nos).
The regression analysis for DMR 447 IIa showed that the hmg copy number remained constant over the entire experimental region, showing that the reference gene (hmg) can be accurately measured from 58 °C to 62 °C, and the measured value is not affected for the T-nos primer/probe concentration. On the other hand, the regression analysis for DMR 447 Va showed that only hmg primer/probe concentration had a significant influence (p = 0.066) on the hmg cp variable. However, the independent results of the three measurements from the runs corresponding to different levels of hmg (−1.68, −1, 0, 1, 1.68) show variation in hmg cp at low levels (−1.68 and −1) (see Table S7 in the Supporting Information); this could be attributed to low droplet resolution, which could lead to misclassification of hmg-positive droplets in some cases. In this sense, hmg primer/probe concentration could be considered to be an influencing variable on hmg cp, although not in the sense suggested by its regression coefficient; the predicted increase on hmg cp when hmg primers/probe are diluted may have been due to an artifact resulting from erroneous droplet discrimination.
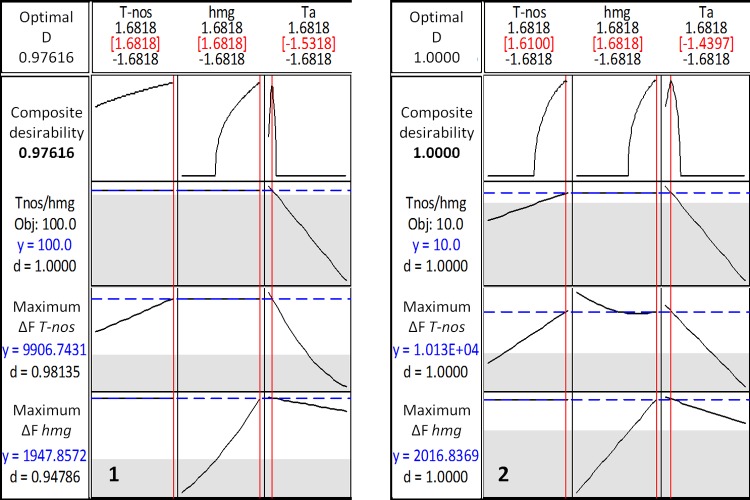
Finally, multiresponse analysis for both materials only included the response variables T-nos/hmg ΔF, T-nos, and hmg ΔF; the hmg cp was excluded from the analysis, since there was no appropriate regression model for this variable. According to the desirability function plots (Figure 3), optimal conditions were found at T-nos = 1.68, hmg = 1.68, and Ta = −1.5; with a composite desirability of >95%. In natural variables, theses values correspond to the 1000:384 nM T-nos primers/probe, the 95:160 nM hmg primers/probe, and an annealing temperature of 58 °C, respectively. This result agrees with the overlaid contour plots (Figure S2 in the Supporting Information), which shows better droplet resolution and the expected T-nos/hmg ratio, at higher levels of the T-nos and hmg factors. From these plots, it can be noticed that, at 58 °C, the T-nos/hmg copy number ratio predicted is relatively constant, even when lowering the T-nos and hmg primer/probe concentration over a defined range, only affecting the droplet resolution. Taking this into account and aiming to save resources, it was decided to set the primer-probe concentrations at the central level (750/300 nM for T-nos and 60:110 nM for hmg), for the performance assessment of the method and the restriction digestion assay. We kept the annealing temperature at 58 °C, since it showed to be the most influencing factor over the response variables. It is noteworthy that the defined optimal conditions were the same for both studied scenarios (100% and 10%), which allowed working with a wide range of concentrations under identical amplification conditions.
Figure 3.
Desirability function plots: (1) DMR 447 Ha, transgenic corn seed (100% mass fraction); (2) DMR 447 Va, transgenic and nontransgenic corn mix (10% mass fraction). [Legend: T-nos, T-nos primer/probe concentration; hmg, hmg primer/probe concentration; Ta, annealing temperature; d, desirability; and D, composite desirability.]
Specificity
Theoretical specificity of primers and probes was evaluated through the NCBI database, using the nucleotide sequences to run BLAST. The results are consistent with an extensive list of records referring to the nopaline synthase terminator, hmg, recombinant vectors, and patents, confirming the complete identity only with target sequences. To assess the experimental specificity, all samples containing at least three positive droplets for the sequence of interest were defined as positive. All the analyzed materials yielded the expected results, in agreement with the information available for each of them (see Table S8 in the Supporting Information). No hmg amplification was observed in materials other than corn, and T-nos amplification was only present in event samples regulated by this element. These results show the method is specific for the detection of T-nos and hmg.
Detection and Quantification Limit
The lowest-concentration dilution at which all replicates were positive (at least three T-nos-positive droplets), regardless of the reading’s accuracy or precision, was set as the detection limit.2 Similarly, the lowest-concentration dilution at which all replicas, in addition to being positive, reached a relative standard deviation of ≤25%, with a percentage accuracy of 75%–125%, was set as the quantification limit.8,18 Under these criteria, the relative limit of detection (LODrel) estimated for T-nos was 0.034%, with an average background level of 32 000 cP of hmg, indicating a fairly low limit, despite the symmetry between the analytes; this corresponded to an absolute limit fo detection (LODabs) of 11 T-nos cp. The LOQ of the method was set at 23 copies. Taking into account the intended purpose of the method, this value was considered as a satisfactory limit, because it allowed for the quantification of T-nos from concentrations as low as 0.08%, with a background level close to 28 000 cP of hmg (assessed condition), or 0.12% with a background level of 18 300 cP of hmg, corresponding to the hmg cp expected in a DNA extract of 50 ng/μL (most common concentration tested in routine assays). In the case of RM production, this limit would allow for the characterization of a material with a T-nos/hmg copy number ratio of ∼0.1%, offering the laboratories the possibility to verify their analysis at this level, as required by the European Union in regulation No. 619/2011.7
Dynamic Range
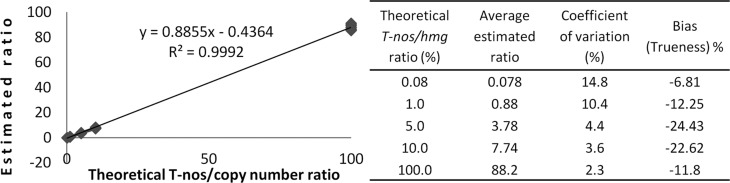
The response (T-nos/hmg copy number ratio) was linear along the entire concentration range evaluated (0.08%–100%), with a correlation coefficient of R2 > 0.99 (see Figure 4). Since the concentration of the highest point of the curve was greater than the rest, this point was removed to verify whether the linear trend was a consequence of a regression damping due to that difference. Even then, R2 was >0.99, indicating the proportionality between the response and the T-nos concentration of the sample. Even though the result variability increased while T-nos concentration decreased, the CV values were far below the criteria (<15%). The method’s accuracy satisfied the criteria established in every evaluated point, although a measurement bias from approximately −6% to −24% was observed. The achieved dynamic range allows for the characterization of RM while covering almost the entire T-nos concentration range that is possible in a GM corn sample, or at least the most probable concentrations.
Figure 4.
Dynamic range for T-nos/hmg by ddPCR. Theoretical T-nos/hmg copy number ratio (x) versus estimated T-nos/hmg copy number ratio (y).
Applicability
The estimate of the T-nos/hmg copy number ratio was satisfactory for precision and accuracy (see Table S9 in the Supporting Information) at the assessed level (0.84%–0.99%) in all the corn varieties analyzed (popping corn, blue corn, red corn, and white corn). It was impossible to determine the method’s applicability to higher concentrations, in these matrices or others, because this type of RM is not yet available. The proposed duplex ddPCR method can be used for T-nos/hmg copy number ratio quantification in yellow corn, at the concentration range from 0.08% to 100%, and DNA extracts of 50–100 ng/μL on average. The method could also be applied to analyze these targets in different corn varieties, at least at low T-nos concentrations (0.9%), complying with labeling limits imposed by the European Union.
Comparison of Duplex and Single-Plex Assay
The estimated bias between duplex and single-plex assay application was <10% (see Table S10 in the Supporting Information). Although a certain bias increase was observed when the T-nos content decreased, the direction of that bias was not maintained and no clear trend was detected. All results, except the evaluated point with high concentration, show that the bias in copy number fraction was produced mainly by the bias in T-nos cp, because the estimated hmg cp was fairly robust, with a maximum bias value of −1.8%. This result is similar to the result reported by Morisset et al.18 In that study, the single-plex version of a MON810/hmg ddPCR assay was compared to its duplex version, obtaining a bias value of −1.8 for hmg cp, and ∼6% for MON810 cP/cp ratio. Although the two versions were compared for a single concentration level (0.7%), the authors concluded that no significant difference could be observed between them. In our study, duplex and single-plex results from five concentration levels, were compared for the T-nos/hmg copy number ratio using the Tukey’s mean test, and no statistically significant difference between the two assays (p = 0.498) was found. On the other hand, no significant increase (or decrease) was observed in the variance of the copy number evaluated by duplex ddPCR. These results show that, in contrast to the single-plex version, the assessment of the T-nos/hmg copy number ratio was not negatively affected by the simultaneous quantification of T-nos and hmg, indicating the reliable application of the duplex version over the entire dynamic range.
Restriction Enzyme Digestion
DNA digestion results showed an increase in the estimated T-nos/hmg copy number ratio in digested DNA, on three out of four enzymatic systems evaluated (Table S11 in the Supporting Information), as expected. The T-nos/hmg ratio estimation was independent of XhoI digestion, proving that, in the tested samples, there are no restriction sites for this enzyme between the two copies of T-nos. The estimated T-nos/hmg copy number ratio became closer to the expected value in the separate digestions with BamHI and EcoRI and in the multiple digestions assay, although a higher increase was obtained using the first system. Sample digestion verification using electrophoresis (Figure S3 in the Supporting Information) revealed that nondigested DNA was also highly fragmented, suggesting the partial unlinking of T-nos copies, as a result of DNA extraction and sample manipulation. Qin et al.19 have reported this phenomenon and suggested the use of DNA preamplification before molecule partitioning through ddPCR, to separate tandem copies. Nonetheless, if the transgenic sequence and the reference gene have different amplification efficiencies, this alternative could introduce measurement bias.11
Since linked T-nos copies are expected to be separated by enzyme digestion, the copy number ratio should approach the certified mass fraction when DMR 447 IIa (100% mass fraction) and DMR 447 IIIa (1% mass fraction) materials are fragmented, with some variation from the expected values being possible, because of he ploidy level and different tissue percentage contributions to the corn seed, as previously reported.22,25,27 Digested DNA from all analyzed DMR 447 batches revealed an increase in the estimated T-nos copy number ratio, which was fairly close to the ratio expected taking into account the aforementioned considerations; the estimated ratio remained unchanged in DMR 452 IIa DNA, suggesting that linked copy separation produced the increase observed in DMR 447 (see Table 2). This increase was heterogeneous between different batches (an increase of 14%, 26%, and 36% for IIa, Va, and IIIa batches, respectively); however, this result may have been altered by the different extents of DNA fragmentation prior to digestion. The DNA enzyme digestion has been shown to reduce the measurement bias, proving it to be a convenient step for quantifying the T-nos/hmg copy number ratio, in materials with linked T-nos copies.
Table 2. Effect of DNA Extracts Enzymatic Digestion (BamHI) on T-nos cp, hmg cp, and T-nos/hmg Copy Number Ratio Measured through Duplex ddPCR.
| Undigested
DNA |
Digested
DNA |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| assessed material | mass fraction | DNA (ng/μL) | T-nos cp | hmg cp | estimated copy number ratio | U% (k = 2) | T-nos cp | hmg cp | estimated copy number ratio | U% (k = 2) | undigested/digested ratioa |
| DMR 447 IIa (NK603) | 100% | 45 | 13 966 | 15 830 | 88.22 | 3.1 | 16 190 | 16 054 | 100.84 | 3.2 | –12.5% |
| DMR 447 Va (NK603) | 10% | 55 | 1552 | 20 062 | 7.74 | 3.3 | 1763 | 18 221 | 9.68 | 2.4 | –20.6% |
| DMR 447 IIIa (NK603) | 1% | 55 | 178 | 20 337 | 0.88 | 7.3 | 262 | 22 563 | 1.16 | 9.8 | –26.7% |
| DMR 452 IIa | 100% | 90 | 17 204 | 32 178 | 53.5 | 2.9 | 18 651 | 34 698 | 53.7 | 2.9 | –0.37% |
Difference in percentages between the estimated copy number ratio in undigested DNA, relative to the digested DNA.
Conclusion
This study describes a method for simultaneous quantification of T-nos and hmg and give details of the optimization process using a statistically designed experiment. The proposed assay achieved a limit of detection (LOD) and limit of quantification (LOQ) of 0.034% and 0.08% T-nos/hmg copy number ratio, respectively, with a dynamic range of 0.08%–100%. The measurement principle of the employed technology (ddPCR) and the detection and quantification limits reached in the assay show that this method can be employed to characterize candidates for CRMs for their T-nos/hmg content on maize, in terms of copy number ratio, over a wide T-nos/hmg concentration range. For this purpose, the genetic structure of the transformation event in the analyzed material should be taken into account, because DNA digestion may be needed in the case of linked T-nos copies to avoid underestimation of T-nos copy and copy number ratio. The enzyme selection will be dependent on the transformation event and should be evaluated on a case-by-case basis.
Acknowledgments
The authors thank the National Metrology Center (CENAM) of Mexico, for providing the facilities to conduct this study. The authors also thank the Professional Development System (SIDEPRO) of the same center for fellow support, CNRDOGMs/SENASICA and FONDO CIBIOGEM for providing the necessary reagents for assay implementation, and IBQ Edna Alejandra Matus Cundapí and QFB Héctor Carrillo Yáñez for their support and technical advice during the method development.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.5b03238.
Primers and probes sequences used in the T-nos and hmg amplification (Table S1); central composite design for optimization of a duplex ddPCR assay (Table S2); central composite design for optimization of a duplex ddPCR assay (Table S3); linear regression models built for optimized response variables in a duplex ddPCR method for T-nos/hmg analysis (Table S4); linear regression models built for optimized response variables in a duplex ddPCR method for T-nos/hmg analysis (Table S5); T-nos copy number, T-nos/hmg copy number ratio and relative standard deviation at different annealing temperatures (Table S6); average values and variability for hmg cp at different hmg primer/probe concentrations (Table S7); specificity test of a duplex ddPCR assay (Table S8); applicability assay of the T-nos/hmg ddPCR method, on several corn varieties (Table S9); comparison of quantification using single plex and duplex ddPCR assays (Table S10); and DNA restriction enzyme digestion (Table S11). Figures showing an example of negative and positive results (Figure S1), overlaid contour plots for ddPCR optimization (Figure S2), and DNA enzymatic restriction digestion (Figure S3). Discussions of DNA extraction protocol (Annex S1), enzymatic restriction digestion of genomic DNA (Annex S2), and workflow for digital droplet PCR analysis (Annex S3) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Rep. No. XP V 03e044; Association Francaise de Normalisation (AFNOR), 2008.
- Broeders S.; Huber I.; Grohmann L.; Berben G.; Taverniers I.; Mazzara M.; Roosens N.; Morisset D. Trends Food Sci. Technol. 2014, 37, 115–126. 10.1016/j.tifs.2014.03.008. [DOI] [Google Scholar]
- Chaouachi M.; Bérard A.; Said K. Transgenic Res. 2013, 22, 461–476. 10.1007/s11248-012-9684-1. [DOI] [PubMed] [Google Scholar]
- Rep. No. CAC/GL 74-2010; Codex Committee on Methods of Analysis and Sampling (CCMAS), 2010.
- Corbisier P.; Bhat S.; Partis L.; Xie V. R. D.; Emslie K. R. Anal. Bioanal. Chem. 2010, 396, 2143–2150. 10.1007/s00216-009-3200-3. [DOI] [PubMed] [Google Scholar]
- Off. J. Eur. Union, L: Legis. (Engl. Ed.) 2004, 47 (L 348), 18–26. [Google Scholar]
- Off. J. Eur. Union, L: Legis. (Engl. Ed.) 2011, 40 (L 166), 9–15. [Google Scholar]
- European Network of GMO Laboratories (ENGL), 2008. Available via the Internet at: http://gmo-crl.jrc.ec.europa.eu/doc/Min_Perf_Requirements_Analytical_methods.pdf.
- Gaudron T.; Peters C.; Boland E.; Steinmetz A.; Moris G. Eur. Food Res. Technol. 2009, 229, 295–305. 10.1007/s00217-009-1045-9. [DOI] [Google Scholar]
- Huber I.; Block A.; Sebah D.; Debode F.; Morisset D.; Grohmann L.; Berben G.; Stebih D.; Milavec M.; Zel J.; Busch U. J. Agric. Food Chem. 2013, 61, 10293–10301. 10.1021/jf402448y. [DOI] [PubMed] [Google Scholar]
- Hindson B. J.; Ness K. D.; Masquelier D. A.; Belgrader P.; Heredia N. J.; Makarewicz A. J.; Bright I. J.; Lucero M. Y.; Hiddessen A. L.; Legler T. C.; Kitano T. K.; Hodel M. R.; Petersen J. F.; Wyatt P. W.; Steenblock E. R.; Shah P. H.; Bousse L. J.; Troup C. B.; Mellen J. C.; Wittmann D. K.; Erndt N. G.; Cauley T. H.; Koehler R. T.; So A. P.; Dube S.; Rose K. A.; Montesclaros L.; Wang S.; Stumbo D. P.; Hodges S. P.; Romine S.; Milanovich F. P.; White H. E.; Regan J. F.; Karlin-Neumann G. A.; Hindson C. M.; Saxonov S.; Colston B. W. Anal. Chem. 2011, 83, 8604–8610. 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett J. F.; Foy C. A.; Benes V.; Emslie K.; Garson J. A.; Haynes R.; Hellemans J.; Kubista M.; Mueller R. D.; Nolan T.; Pfaffl M. W.; Shipley G. L.; Vandesompele J.; Wittwer C. T.; Bustin S. A. Clin. Chem. 2013, 59, 892. 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- International Standard ISO 21570; International Organization for Standardization: Genéve, Switzerland, 2005. [Google Scholar]
- Lu I. J.; Lin C. H.; Pan T. M. Anal. Bioanal. Chem. 2010, 396, 2055–2064. 10.1007/s00216-009-3214-x. [DOI] [PubMed] [Google Scholar]
- Milavec M.; Dobnik D.; Yang L.; Zhang D.; Gruden K.; Zel J. Anal. Bioanal. Chem. 2014, 406, 6485–6497. 10.1007/s00216-014-8077-0. [DOI] [PubMed] [Google Scholar]
- Miraglia M.; Berdal K. G.; Brera C.; Corbisier P.; Holst-Jensen A.; Kok E. J.; Marvin H. J. P.; Schimmel H.; Rentsch J.; van Rie J. P. P. F.; Zagon J. Food Chem. Toxicol. 2004, 42, 1157–1180. 10.1016/j.fct.2004.02.018. [DOI] [PubMed] [Google Scholar]
- Morisset D.; Demsar T.; Gruden K.; Vojvoda J.; Stebih D.; Zel J. Nat. Biotechnol. 2009, 27, 700–701. 10.1038/nbt0809-700. [DOI] [PubMed] [Google Scholar]
- Morisset D.; Stebih D.; Milavec M.; Gruden K.; Zel J. PLoS One 2013, 8, e62583. 10.1371/journal.pone.0062583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J.; Jones R. C.; Ramakrishnan R. Nucleic Acids Res. 2008, 36, e116. 10.1093/nar/gkn518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiting R.; Broll H.; Waiblinger H. U.; Grohmann L. J. Verbraucherschutz Lebensmittelsicherh. 2007, 2, 116–121. 10.1007/s00003-007-0189-4. [DOI] [Google Scholar]
- Trapmann S.; Emons H. Anal. Bioanal. Chem. 2005, 381, 72–74. 10.1007/s00216-004-2901-x. [DOI] [PubMed] [Google Scholar]
- Trifa Y.; Zhang D. J. Agric. Food Chem. 2004, 52, 1044–1048. 10.1021/jf034574+. [DOI] [PubMed] [Google Scholar]
- Waiblinger H. U.; Ernst B.; Anderson A.; Pietsch K. Eur. Food Res. Technol. 2008, 226, 1221–1228. 10.1007/s00217-007-0748-z. [DOI] [Google Scholar]
- Waiblinger H. U.; Grohmann L.; Mankertz J.; Engelbert D.; Pietsch K. Anal. Bioanal. Chem. 2010, 396, 2065–2072. 10.1007/s00216-009-3173-2. [DOI] [PubMed] [Google Scholar]
- Zel J.; Milavec M.; Morisset D.; Plan D.; Van den Eede G.; Gruden K.. How To Reliably Test for GMOs, 1st Edition; SpringerBriefs in Food, Health and Nutrition; Springer: New York, 2012; 100 pp (ISBN 978-1-4614-1389-9).
- Zhang D.; Guo J. J. Integr. Plant Biol. 2011, 53, 539–551. 10.1111/j.1744-7909.2011.01060.x. [DOI] [PubMed] [Google Scholar]
- Zhang D.; Corlet A.; Fouilloux S. Transgenic Res. 2008, 17, 393–402. 10.1007/s11248-007-9114-y. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Y.; Xu Y. F.; Hussain K.; Liu Y. Z.; Lin F. Middle East J. Sci. Res. 2010, 6, 563–568. [Google Scholar]
- Behr C. F.; Heck G. R.; Hironaka C.; You J.; Corn transformant PV-ZMGT32 (NK603) and compositions and methods for detection thereof, Eur. Patent No. EP1167531, Jan. 2, 2002.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.