Abstract
Integrator is a multi-subunit complex stably associated with the C-terminal domain (CTD) of RNA polymerase II (RNAPII) 1. Integrator is endowed with a core catalytic RNA endonuclease activity, which is required for the 3′-end processing of non-polyadenylated RNAPII-dependent uridylate-rich small nuclear RNA genes (UsnRNAs) 1. Here, we examined the requirement of Integrator in the biogenesis of transcripts derived from distal regulatory elements (enhancers) involved in tissue- and temporal-specific regulation of gene expression 2–5. Integrator is recruited to enhancers and super-enhancers in a stimulus-dependent manner. Functional depletion of Integrator subunits diminishes the signal-dependent induction of eRNAs and abrogates the stimulus-induced enhancer-promoter chromatin looping. Global nuclear run-on and RNAPII profiling reveals a role for Integrator in 3′-end cleavage of eRNAs primary transcripts leading to transcriptional termination. In the absence of Integrator, eRNAs remain bound to RNAPII and their primary transcripts accumulates. Importantly, the induction of eRNAs and gene expression responsiveness requires the catalytic activity of Integrator complex. We propose a role for Integrator in biogenesis of eRNAs and enhancer function in metazoans.
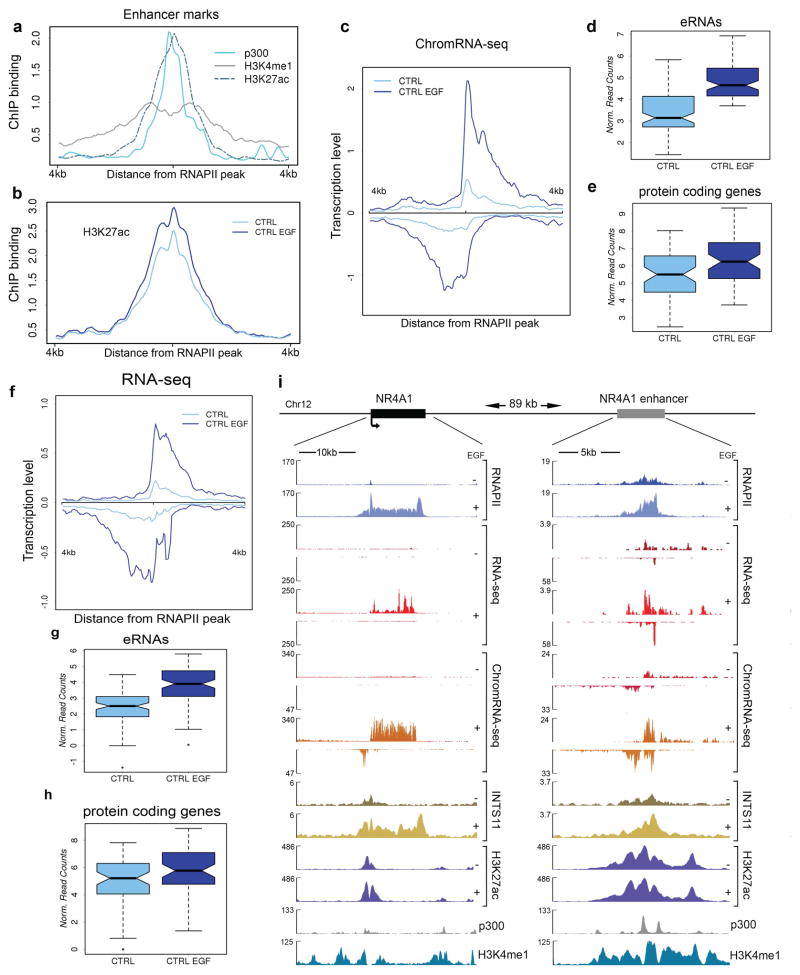
To assess the role for Integrator in the biogenesis of eRNAs, we examined the signal-dependent recruitment of Integrator complex to enhancer sites. HeLa cells were starved of serum for 48 hours following which they were stimulated with epidermal growth factor (EGF) to induce immediate early genes (IEGs). We identified 2029 enhancers based on their occupancy by RNAPII, CBP/p300 and containing acetylated histone H3 lysine 27 (H3K27ac) chromatin modification (see Methods). We found that while assessing steady-state levels of eRNAs provided a measure of EGF-induced eRNAs, we obtained a better read out of eRNAs following sequencing of the chromatin-enriched RNA fractions (ChromRNA-seq) 6. We focused on 91 enhancers that displayed EGF-induced eRNAs in the proximity of EGF-responsive genes following 20 minutes of induction (Extended Data Fig. 1, Table 1 and see Methods). Interestingly, the chromatin surrounding these enhancers displayed H3K27ac in starved cells and following EGF stimulation there was a small increase in H3K27ac levels (Extended Data Fig. 1b). In order to assess the polyadenylation state of eRNAs, total RNA was enriched for polyadenylated and non-polyadenylated fractions and was subjected to high throughput sequencing. Similar to prior reports, EGF-induced enhancers displayed bi-directional eRNAs that were predominantly not polyadenylated (Extended Data Fig. 2)5,7.
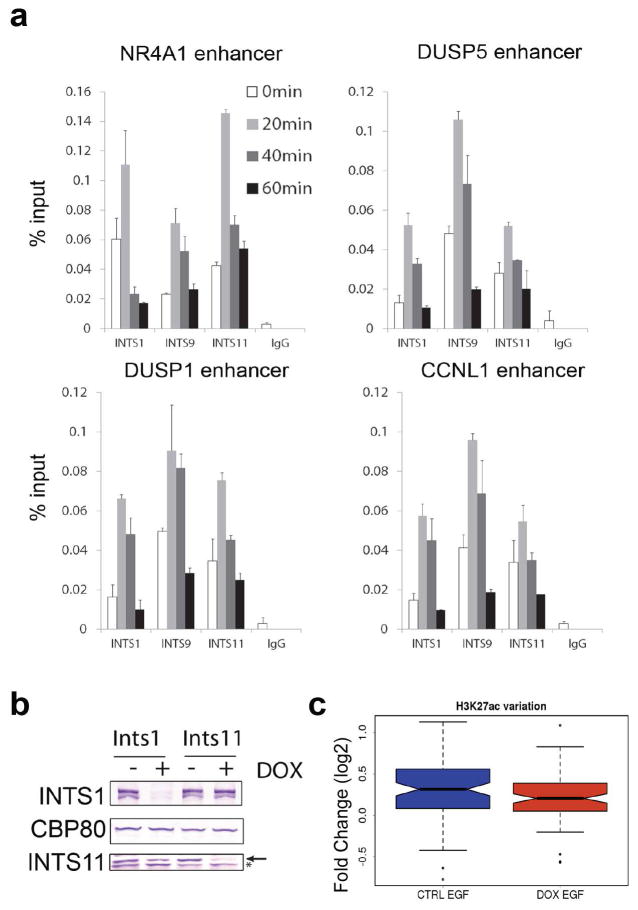
We next analyzed Integrator occupancy at these enhancers by using antibodies against the INTS11 subunit of the Integrator complex prior to and following EGF stimulation. While these enhancers were occupied by a detectable amount of Integrator prior to EGF induction, addition of EGF resulted in a further recruitment of Integrator complex (Fig. 1a–c). RNAPII displayed a similar pattern of stimulus-dependent chromatin residence (Fig. 1d and e). The stimulus-dependent recruitment of Integrator at enhancers was further confirmed using two additional antibodies against INTS1 and INTS9 subunits of the Integrator complex (Extended Data Fig. 3a). These results demonstrated the stimulus-dependent recruitment of the Integrator complex at EGF-responsive enhancers.
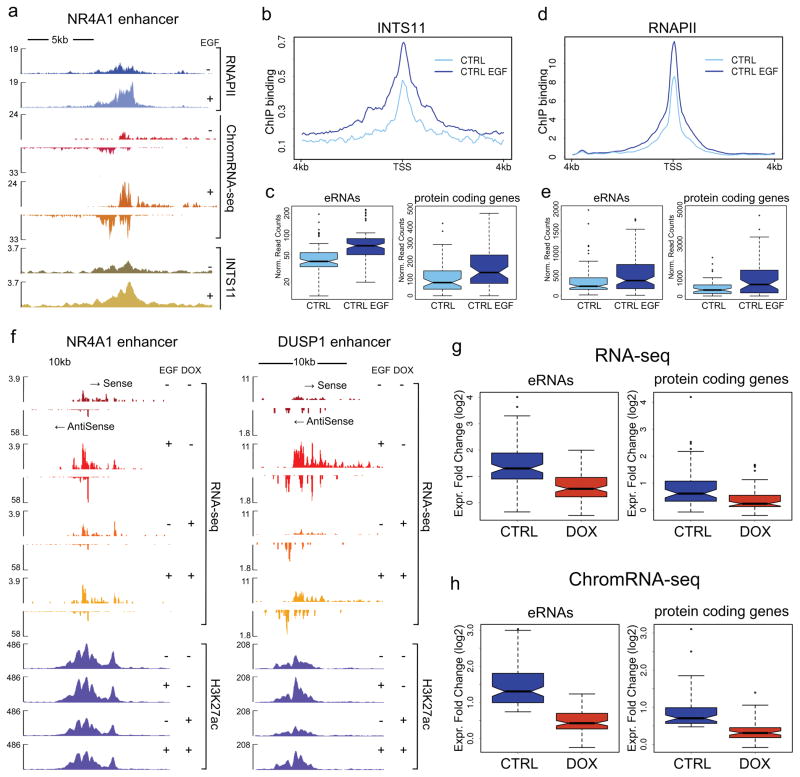
Figure 1. Integrator mediates induction of eRNAs.
a, EGF induction of an enhancer in the vicinity of the NR4A1 gene (see Extended Data Fig. 1i). RNAPII and INTS11 are recruited to the enhancer after 20 min of stimulation and eRNAs are transcribed bi-directionally from the locus (as revealed by deep sequencing of chromatin-associated RNA, ChromRNA-seq). b, Average profile of Integrator recruitment to 91 EGF-responsive enhancers. c, Increased Integrator occupancy at enhancers and their corresponding protein coding genes (mean density was calculated as follows: 6kb surrounding the peak of RNAPII for eRNAs; from −0.5kb to + 2.5kb for coding genes; p<0.001). d, Average profile of RNAPII upon EGF treatment at enhancers. e, Increased RNAPII occupancy following EGF stimulation at enhancers and their corresponding protein coding genes (p<0.005). f, Inducible knockdown of INTS11 (DOX) dramatically reduces steady state levels of eRNAs (as measured by total RNA-seq). Data were obtained using a tet-inducible shRNA system, stably transduced in HeLa cells. Acetylation of H3K27 is also shown. g–h, Average expression levels of 91 eRNAs and their neighboring (<500kb) 57 protein coding genes indicate a significant impairment of activation. Box plots represent the expression fold change (log2) before and after EGF treatment in normal conditions (CTRL) and upon depletion of Integrator (DOX) (t-test, p<0.0005 for all panels). Fold change of RPKM values was calculated from RNA-seq (f) and ChromRNA-seq (g) data.
To examine the functional importance of Integrator at enhancers and its role in the biogenesis of eRNAs, we developed HeLa clones expressing doxycycline (DOX) inducible shRNAs against INTS11 and INTS1 subunits of the Integrator complex (Extended Data Fig. 3b). Within the time course of these experiments the mature levels of snRNAs were not perturbed (data not shown). Twenty minutes of EGF stimulation resulted in the induction of bi-directional eRNAs similar to previous reports (Fig. 1a and f, Extended Data Fig. 1c–h) 5,8–11. Depletion of INTS11 diminished the eRNA induction following EGF stimulation (Fig. 1f; as shown at two enhancer loci; enhancers were named following their proximity to an EGF-responsive gene). The fold induction of eRNAs at all EGF-induced enhancers decreased significantly (Fig. 1g and h). We also observed a significant decrease in the transcriptional induction of EGF-responsive protein coding genes in the proximity of these EGF-induced enhancers (Fig. 1g and h). Interestingly, there was a subtle increase (statistically not significant) in H3K27 acetylation at enhances following EGF stimulation, which was reduced after Integrator depletion (Fig. 1f and Extended Data Fig. 3c).
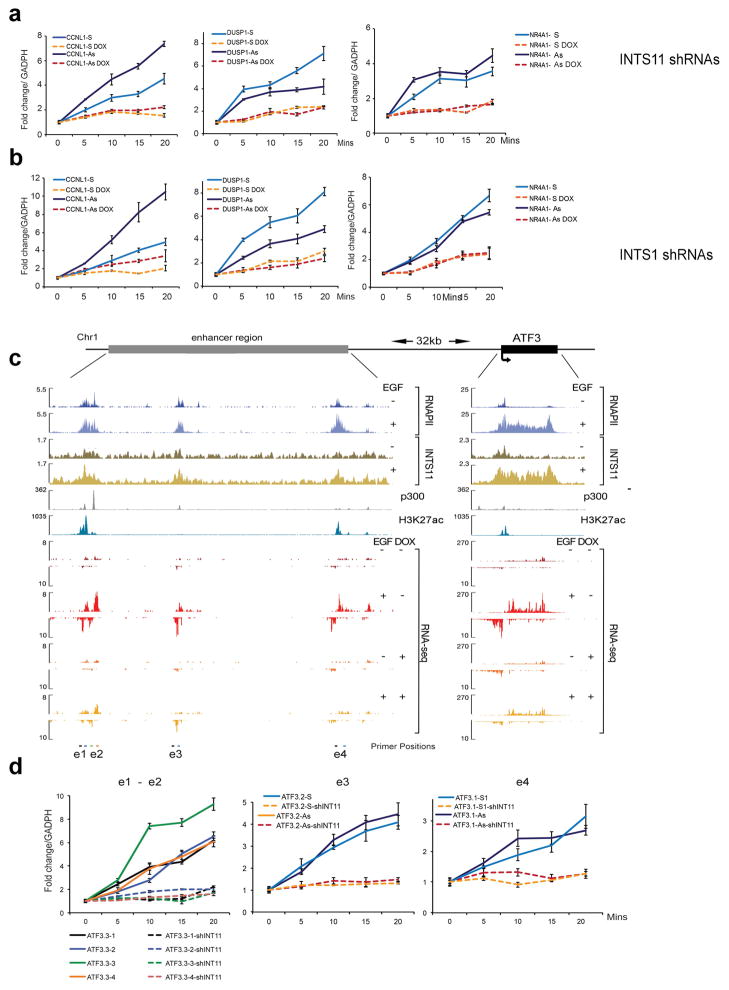
To gain further insight into quantitative changes in eRNAs following depletion of Integrator, we depleted INTS11 or INTS1 and performed a time-course analysis of eRNA induction using specific primer sets for each strand. Depletion of either Integrator subunits diminished the EGF-induced increase in eRNA levels from both strands of the enhancers (Extended Data Fig. 4a and b). Analysis of regulatory landscape in the proximity of the EGF-responsive gene ATF3 (Activating Transcription Factor 3) revealed the presence of clusters of acetylated H3K27 and p300 binding sites similar to that described for super-enhancers 12–14 (Extended Data Fig. 4c). This region also displayed occupancy by RNAPII at multiple sites and we could detect additional recruitment of RNAPII and Integrator to these sites following EGF stimulation (Extended Data Fig. 4c). Analysis of eRNA synthesis using strand-specific RNA-seq and real-time PCR (during a time-course experiment) demonstrated a requirement for Integrator in the induction of eRNAs at the super enhancer sites following EGF stimulation (Extended Data Fig. 4d). Collectively, these results highlight a requirement for Integrator in stimulus-dependent induction of eRNAs from individual enhancers and enhancer clusters.
An important component of enhancer function is the formation of stimulus-dependent chromatin looping allowing enhancer and promoter communication 15–17. We measured chromatin looping between NR4A1 and DUSP1 enhancers and their respective promoters using chromosome conformation capture (3C) following stimulation with EGF (Fig. 2a). We observed a robust association between the enhancer and the promoter regions of NR4A1 and DUSP1 following EGF stimulation (Fig. 2b). Remarkably, depletion of Integrator abrogated the EGF-induced chromatin looping without any effect on non-stimulus induced chromosomal interactions (Fig. 2b and c, Extended Data Fig. 5a and b). These results demonstrate that Integrator regulates enhancer function as reflected by the physical association between enhancers and their respective promoters.
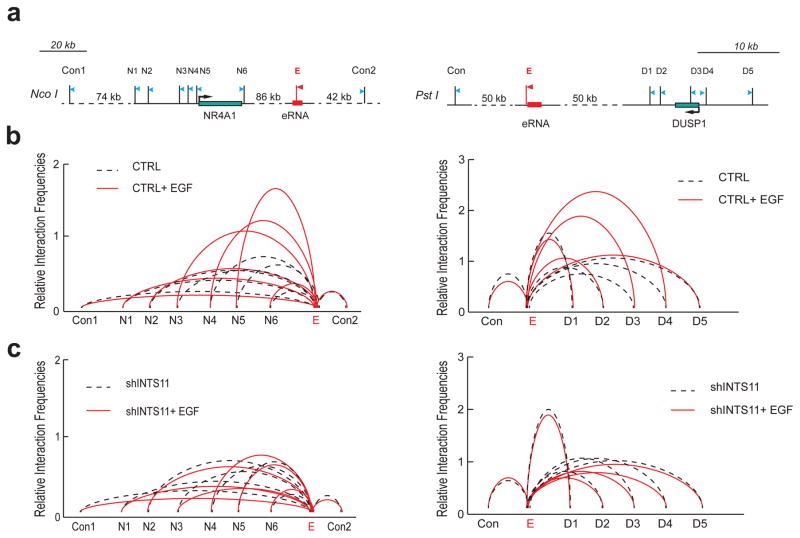
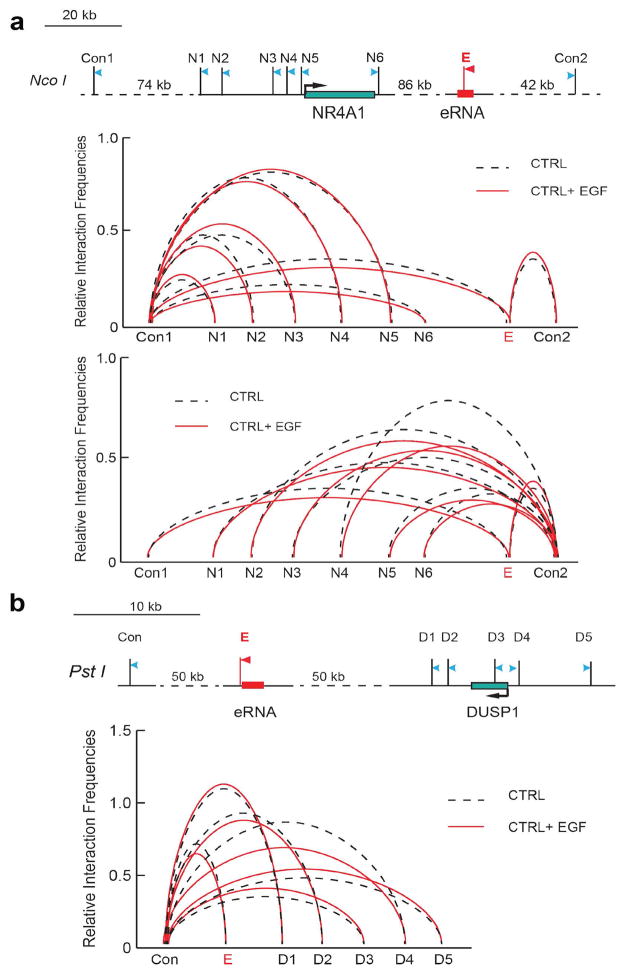
Figure 2. Integrator is required for enhancer-promoter interaction.
a, Diagrams of NR4A1 (left) and DUSP1 (right) genomic regions with their respective enhancers (shown in red). The arrows depict the position of primers for detection of chromatin looping and the stick bars indicate enzyme digestion sites (named N1-6 and D1-5). E refers to the anchor primer at the enhancer sites, control sites are also indicated. b, Looping events between the promoter region of NR4A1 and its enhancer were detected at N3, N4 and N5 sites after EGF induction (left). A similar interaction was also captured between sites D3 and D4 of DUSP1 promoter and its downstream enhancer after EGF induction (right). c, Knockdown of Integrator abolished chromosomal looping events at both NR4A1 and DUSP1 sites. The interaction frequency between the anchoring points and the distal fragments were determined by Real-time PCR and normalized to BAC templates. All sites were assayed in three independent experiments (p<0.01, two-sided t-test). Control anchors are displayed in Extended Data Fig. 5.
To gain an insight into the mechanism by which Integrator regulates enhancer function and eRNA biogenesis, we depleted Integrator and performed RNAPII profiling and global nuclear run-on followed by high throughput sequencing (GRO-Seq) after EGF induction. Interestingly, Integrator depletion resulted in the increase and spreading of GRO-Seq reads throughout the body of eRNA transcripts at both enhancers and super enhancers which was mirrored by a concomitant increase and spreading of RNAPII localization (Fig. 3a and b). Indeed, the average profile of depth normalized reads of 91 EGF-induced enhancers showed a significant accumulation of GRO-seq and RNAPII ChIP-seq reads (Extended Data Fig. 6a and b). Measurement of RNAPII traveling ratio revealed that in contrast to EGF-responsive protein coding genes, which experience a block in productive elongation following Integrator depletion 18, there is increased RNAPII occupancy in the body of eRNA transcripts (Extended Data Fig. 6c and d). The accumulation of RNAPII at eRNA loci following Integrator depletion occurred despite the decreased recruitment of super elongation complex (SEC) to enhancers (Extended Data Fig. 7a and b).
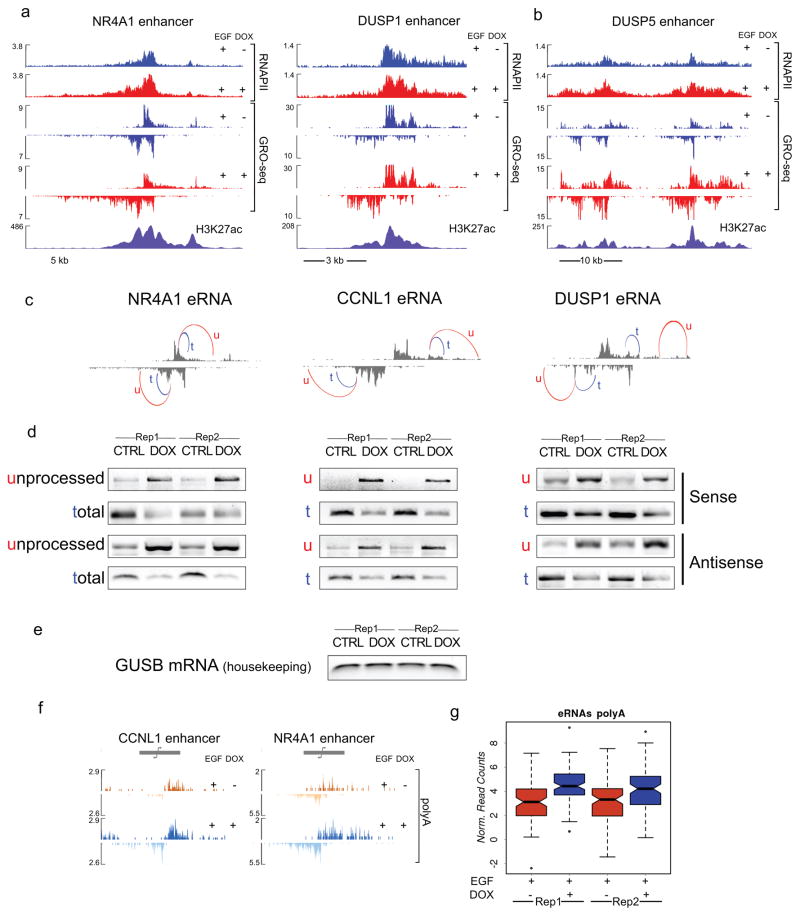
Figure 3. Integrator plays a role in termination of eRNAs.
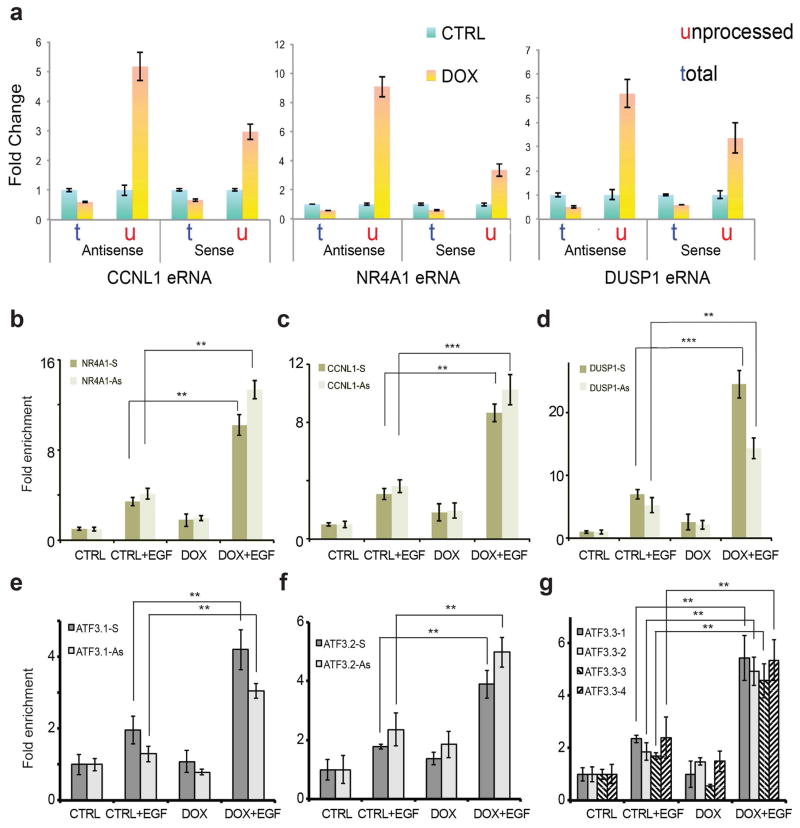
a, RNAPII dynamics was analyzed by ChIP-Seq and GRO-Seq at the enhancer regions adjacent to NR4A1, DUSP1 and b, at the super-enhancer upstream of DUSP5. c, 3′-end cleavage of eRNAs was examined with semi-quantitative PCR. Primer pairs were designed to amplify a portion of the enhancer transcript as detected in the control GRO-seq experiment (t, total) or a longer template further extending into the 3′ of the enhancer region (u, unprocessed). d, PCR analysis was performed in 2 independent replicates, before (CTRL) and after (DOX) depletion of INTS11 at three eRNAs (sense and antisense strand). e, The housekeeping gene GUSB was used as a cDNA loading control. f, Polyadenylation of eRNAs increases after depletion of Integrator at DUSP1 and CCNL1 enhancer loci. The polyadenylated fraction of RNA from whole cell lysates was sequenced after EGF stimulation, before and after depletion of INTS11 (DOX). g, Box plot shows significant increase in polyadenylated RNA reads (p<0.001) across the entire set of EGF responsive enhancers.
The increased RNAPII at eRNA loci suggests a block in 3′-end cleavage of primary eRNA transcripts leading to a defect in termination. To quantitate such a 3′-end cleavage defect, we measured the accumulation of primary levels (or unprocessed levels) of eRNA transcripts following Integrator depletion using semi-quantitative PCR and real-time PCR. We observed a 3 to 10 fold accumulation of unprocessed eRNA transcripts concomitant with the reduction of the processed eRNA levels (Fig. 3c–e and Extended Data Fig. 8a). Previous experiments revealed that the loss of 3′-end cleavage by Integrator led to increased levels of polyadenylated U snRNA transcripts, which are normally not polyadenylated 19. Indeed, analysis of the polyadenylated transcripts revealed a robust increase in polyadenylation of eRNAs in the absence of Integrator (Fig. 3f and g). These results attest to Integrator cleavage of the 3′-end of eRNAs leading to a termination of transcription.
We surmised that such a termination defect might result in the inability of RNAPII to dissociate from the eRNAs leading to accumulation of RNAPII-eRNA complexes and a consequent decrease in mature eRNA levels. We performed ultra violet (UV) cross-linking followed by RNA immunoprecipitation (UV-RIP) using antibodies against RNAPII to examine increased association of eRNAs with RNAPII following depletion of Integrator. Consistent with a role for Integrator in the processing of eRNAs, depletion of Integrator led to a profound increase in eRNA engagement with RNAPII following induction with EGF (Extended Data Fig. 8b–d). We found similar results following analysis of RNAPII interaction with the eRNAs at the ATF3 super enhancer (Extended Data Fig. 8e–g). Taken together, these results implicate the Integrator complex in the termination of eRNAs and highlight Integrator’s role in the release of eRNA transcripts from transcribing RNAPII.
The catalytic subunit of Integrator is composed of the heterodimer of INTS11 and INTS9 enzymes with close homology to CPSF73 and CPSF100, respectively 20. We previously showed that a single point mutation (E203Q) in the catalytic domain of INTS11 leads to impaired processing of small nuclear RNAs 1. To assess the impact of INTS11 enzymatic activity on eRNA biogenesis, we developed wild type and mutant INTS11 (E203Q) that would be refractory to the action of shRNAs against INTS11 and used these constructs to perform rescue experiments. While ectopic expression of wild type INTS11 could substantially rescue the EGF-induced eRNA levels following depletion of INST11, the single point catalytic mutant was without any effect (Figure 4a and Extended Data Fig. 9a). Interestingly, we observed a similar rescue of the transcriptional activation of EGF-induced genes by the wild type INTS11 and not its catalytic mutant (Fig. 4b). These results not only demonstrate the requirement of the INTS11 catalytic activity in regulating the induction of eRNAs but also highlight the defect in eRNA processing as a contributing factor in the loss of transcriptional responsiveness.
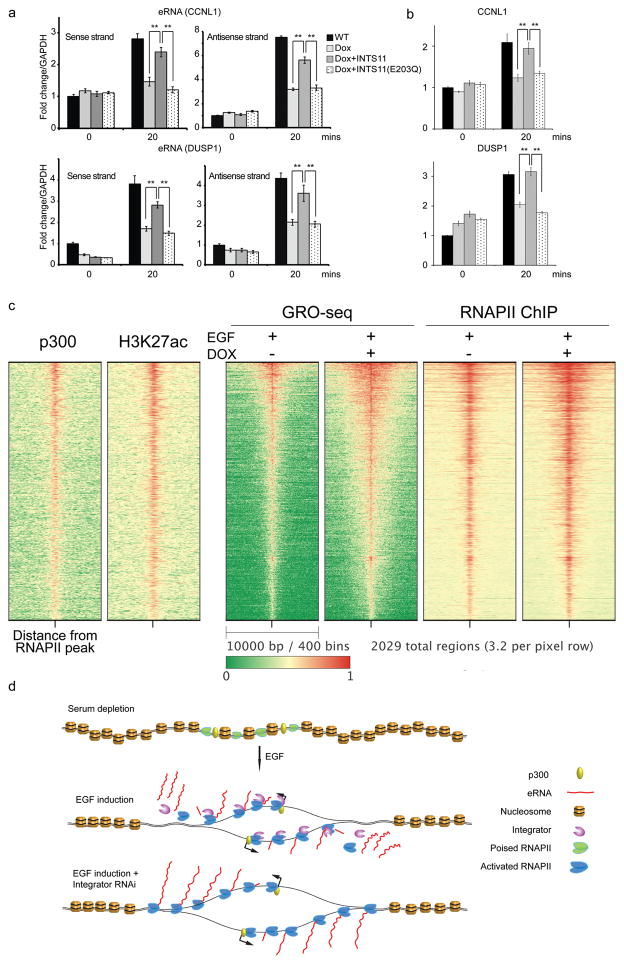
Figure 4. Integrator plays a global role in enhancer regulation.
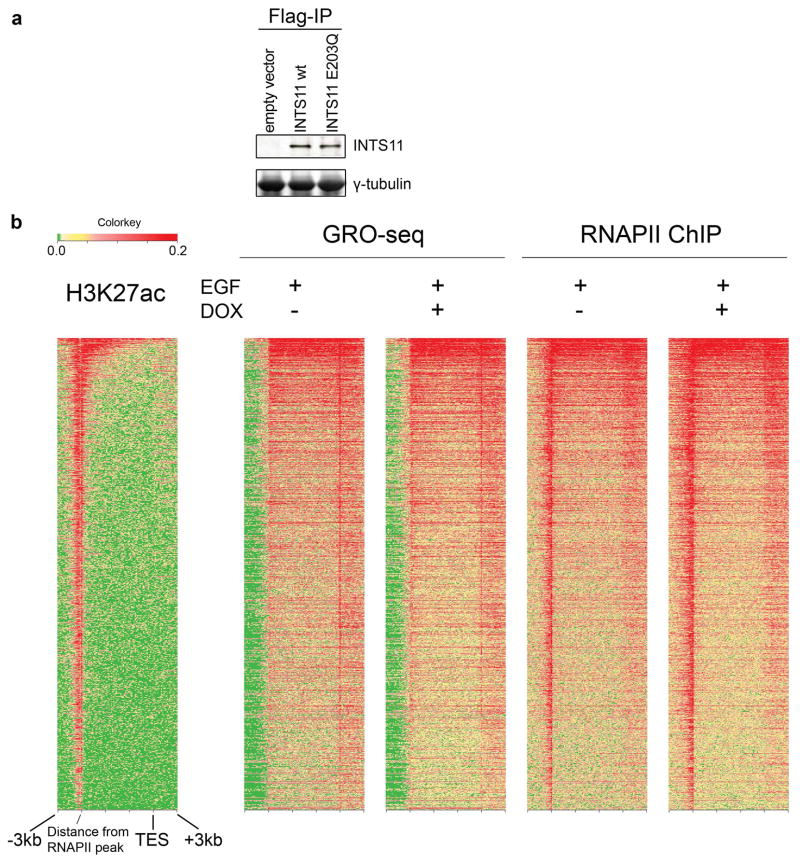
a, Ectopic expression of wild type INTS11, and not its catalytic mutant (E203Q) following Integrator depletion can rescue eRNA induction by EGF. b, A similar rescue was observed for wild type INTS11 on the target protein coding genes. Real time PCR analysis was performed on CCNL1 and DUSP1 eRNAs and their corresponding mRNAs before and after EGF stimulation. Each eRNA was assayed with two sets of primers. Error bars represent ± SEM (n=3 biological independent experiments), **P<0.01 by two-sided t-test. c, The heatmap showcases 2,029 enhancer regions identified using RNAPII extragenic loci enriched in H3K27 acetylation (see Extended Methods). Enhancers were centered at the middle of the RNAPII peak and ranked by transcription activity (GRO-seq). The distribution of p300 and H3K27ac are consistent with a group of active enhancers. Upon Integrator depletion nascent RNA reads and RNAPII profiles spread beyond the normal 3′ end of eRNAs. d, Model for the role of Integrator at eRNAs. Stimulation of serum-starved cells with EGF triggers recruitment of RNAPII and Integrator to enhancer sites and induces bi-directional transcription of non-polyadenylated eRNAs. Upon EGF stimulation Integrator navigates the enhancers along with RNAPII to promote endonucleolytic cleavage of nascent transcripts leading to release of the mature eRNAs. Depletion of Integrator elicits a cleavage defect leading to faulty termination, which results in extended eRNA transcripts and accumulation of RNAPII.
To determine the scope of Integrator function on active enhancers we analyzed the 2,029 transcriptionally active enhancers in HeLa cells. We ranked the enhancers based on their transcriptional activity, which mirrored that of RNAPII occupancy (Fig. 4c). Notably, depletion of Integrator resulted in processing defect at all active enhancers as reflected by the broadening of GRO-Seq and RNAPII ChIP-seq reads commensurate with the transcriptional activity of each enhancer site (Fig. 4c). This was in contrast, to GRO-Seq and RNAPII profiles at transcriptionally active protein coding genes (Extended Data Fig. 9b). These results demonstrate the generality of Integrator in the processing of eRNAs at enhancers (Fig. 4d).
Recent genome-wide studies have revealed the presence of RNAPII at active enhancers coincident with expression of these regulatory elements as long noncoding RNAs 5,21. Importantly, such eRNAs have been shown to play critical roles in transcriptional induction by a variety of signal transduction pathways 7,8,11,16,22. We show that Integrator is the molecular machine that is recruited to enhancers in a signal-dependent manner and is required for the induction of eRNAs. We surmise that the defect in 3′-end processing following Integrator depletion leads to a termination defect reflected in increased levels of primary eRNA transcripts. It is also likely that Integrator affects the stability of the mature transcripts, since its depletion leads to changes in steady-state levels of mature eRNAs.
Similar to other regulatory complexes Integrator is also recruited to the promoters of protein-coding genes including IEGs 18,23,24. Interestingly, recent reports described an association between Integrator and transcriptional pause release factors, negative elongation factor (NELF) and SPT4/SPT5 complexes 18,19,23,24. NELF was also reported to associate with eRNAs in neuronal cells 25. Indeed, we found that Integrator depletion resulted in a defect in transcriptional initiation as well as pause release, which was reflected in the loss of responsiveness of IEGs to EGF stimulation 18. However, depletion of NELF subunits did not affect eRNA induction (Extended Data Fig. 7c and d). Moreover, Integrator depletion did not change NELF occupancy at EGF-induced enhancers (Extended Data Fig. 7e). Taken together, our results point to multiple functions for Integrator at protein coding genes. While Integrator at promoters regulates pause release factors leading to modulation of productive transcriptional elongation, Integrator at enhancers governs eRNA maturation and enhancer-promoter communication.
Methods
Genome-wide data
High-throughput sequencing data analyzed in this study are originally described in 18 and are deposited at the Gene Expression Omnibus with accession number GSE40632. H3K27ac, H3K4me1 and p300 datasets from HeLa-S3 cells are available as part of the ENCODE project 26 and can be retrieved under the following accession numbers: GSM733684, GSM798322, GSM93550. Additional experiments are deposited at GEO (GSE68401) and include RNA-seq data (chromatin-bound RNA, polyadenylated and non polyadenylated fractions of total RNA) as well ChIP-seq experiments (acetylation of H3K27 and occupancy of NELFA). Every genome-wide experiment is performed in two independent biological replicates.
Genome-wide identification of eRNA loci
Peak analysis of RNAPII ChIP-seq data after EGF stimulation was performed using HOMER 4.6 (run in ‘factor’ mode). Next, we used the BEDtools suite to discard any peak overlapping to: i) all exons from Hg19 UCSC Known Genes (with additional 2kb surrounding every exon); ii) RNA Genes (from the Hg18 genome annotation table, plus additional 1kb); iii) tRNA Genes (Hg19, plus additional 1kb). We further selected peaks overlapping (−/+ 400 bp) with H3K27ac peaks from the ENCODE ChIP-seq obtained in HeLa-S3 (GEO GSE31477). The analysis resulted in 2,029 regions that were further examined for their transcriptional response to EGF. Briefly, we centered a 6kb window at the midst of the RNAPII peak and we used HOMER 4.6 to calculate RPKM across the entire eRNA locus using chromRNA-seq data before and after EGF induction. We selected a group of 225 EGF-inducible eRNAs displaying a fold change greater than 2 (CTRL vs EGF) and identified the nearest EGF regulated gene (fold change RPKM>1.6). 91 EGF-induced enhancer RNAs located within 500 kb from the nearest EGF-responsive protein coding genes were selected for further analysis.
ChIP-seq data analysis
ChIP-seq data were obtained using HiSeq 2000 and NextSeq 500. Reads were aligned to the human genome hg19 using bowtie2 27 (end to end alignment, sensitive option). Snapshots of raw ChIP-seq data presented throughout the figures were obtained as follows: BigWiggle files for every ChIP-Seq were generated using samtools, bedtools and RseQC 28, these tracks were then uploaded to the UCSC Genome Browser hg19.
Clustering, heatmaps and average density analysis
ChIP-seq, GRO-seq and RNA-seq data were subjected to read density analysis; seqMINER 1.3.3 29 was used to extract read densities at all enhancer loci with the following parameters: 5′ extension=4kb, 3′ extension=4kb, no read extension, total bin number=180 bins. Mean density profiles were then generated in R 3.0.1 and normalized to sequencing depth. Heatmaps were generated with ChAsE (available at http://chase.cs.univie.ac.at/), using default parameters, a 10kb window and 400 bins and with ngsplot 30.
qChIP
ChIP was performed in HeLa as already described 18. Cells were cross-linked with 1% formaldehyde for 10 minutes at room temperature, harvested and washed twice with 1x PBS. The pellet was resuspended in ChIP lysis buffer (150 mM NaCl, 1% Triton-X 100, 0,7% SDS, 500 μM DTT, 10 mM Tris-HCl, 5mM EDTA) and chromatin was sheared to an average length of 200–400 bp, using a Bioruptor sonication device (20 minutes with 30 seconds intervals). The chromatin lysate was diluted with SDS-free ChIP lysis buffer and aliquoted into single IPs of 2,5x106 cells each. A specific antibody or a total rabbit IgG control was added to the lysate along with Protein A magnetic beads (Invitrogen) and incubated at 4°C overnight. On day 2, beads were washed twice with each of the following buffers: Mixed Micelle Buffer (150 mM NaCl, 1% Triton-X 100, 0,2% SDS, 20 mM Tris-HCl, 5mM EDTA, 65% sucrose), Buffer 500 (500 mM NaCl, % Triton-X 100, 0.1% Na deoxycholate, 25 mM HEPES, 10 mM Tris-HCl, 1mM EDTA), LiCl/detergent wash (250 mM LiCl, 0.5% Na deoxycholate, 0,5% NP-40, 10 mM Tris-HCl, 1mM EDTA) and a final wash was performed with 1x TE. Finally, beads were resuspended in 1x TE containing 1% SDS and incubated at 65°C for 10 minutes to elute immunocomplexes. Elution was repeated twice, and the samples were further incubated overnight at 65°C to reverse cross-linking, along with the untreated input (2,5% of the starting material). After treatment with 0,5 mg/ml proteinase K for 3 hours, DNA was purified with Wizard SV Gel and PCR Clean-up system (Promega). ChIP eluates and input were assayed by real-time quantitative PCR in a 20 μl reaction with the following: 0.4 μM of each primer, 10 μl of iQ SYBR Green Supermix (BioRAD), and 5 μl of template DNA (corresponding to 1/40 of the elution material) using a CFX96 real-time system (BioRAD). Thermal cycling parameters were: 3 minutes at 95°C, followed by 40 cycles of 10 seconds at 95°C, 20 seconds at 63°C followed by 30 seconds at 72°C.
Subcellular Fractionation
Subcellular fractionation was followed as described 6, with minor changes. The cell lysate was re-suspended in cold lysis buffer with 0.15% NP-40, and the sucrose buffer was used to isolate nuclei. 50% glycerol buffer and nuclei lysis buffer contains 1M Urea and 1% NP-40, were performed to isolate nucleoplasmic fraction and chromatin-bound RNA fraction. Chromatin-bound RNA was isolated with Trizol protocol.
RNA isolation for high throughput sequencing
Total RNA or chromatin-bound RNA was extracted using Trizol reagent (Life Technologies). Genomic DNA and ribosomal RNA was removed with Turbo DNA-free kit and RiboMinus Eukaryote Kit (Life Technologies). The polyA and non-polyA fractions were isolated by running RNA samples three times through the Oligo(dT) Dynabeads (Life technologies) to ensure complete separation. The resulting RNA fractions were subjected to strand-specific library preparation using NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs). Sequencing was performed on Nextseq500 (Illumina).
ChIP-seq
ChIP-sequencing was performed as previously described 6. 1x107 cells were crosslinked in 1% formaldehyde for 10 minutes and sonicated with a Bioruptor to obtain chromatin fragments of 200–300 bp. IP was performed overnight with the specific antibodies and Dynabeads Protein A or Protein G beads (Life Technologies). Beads were washed and chromatin fragments were eluted in TE with 1% SDS at 65°C. After de-crosslinking overnight, DNA was extracted using Wizard SV extraction columns (Promega) and Illumina sequencing libraries were prepared using NEBNext ChIP-seq library per reagent set (New England Biolabs) and following manufacturer’s instructions. Libraries were assayed on a BioAnalyzer (High Sensitivity DNA kit) and sequenced on a Nextseq500 (Illumina).
Antibodies
Chromatin Immunoprecipitation was perfomed with polyclonal antibodies against INTS11, INTS9, INTS1 (Bethyl, A301-274A, A300-412A, A300-361A). ChIP-seq of NELFA and H3K27ac were performed with goat polyclonal antibodies (Santa Cruz, sc-23599) and rabbit polyclonal antibodies (Abcam, ab4729), respectively.
Antibodies used for immunoblot analysis were: γ-tubulin (Santa Cruz, mouse monoclonal, sc-17788), CBP80 (Santa Cruz, mouse monoclonal sc-271304), INTS1 (Bethyl, rabbit polyclonal, A300-361A) and a proprietary rabbit polyclonal raised against the C-terminus of INTS11. Flag M2-conjugated beads (Sigma, A2220) were used for immunoprecipitation.
Chromosome Conformation Capture (3C)
3C assay was performed as previous described with minor changes 31. Hela cells were filtered through a 70 μm strainer to obtain single cell preparation. 1×107 cells were then fixed in 1% formaldehyde for 30 min at room temperature for cross-linking. The reaction was quenched with 0.25 M glycine and cells were collected by centrifugation at 240 g for 8 minutes at 4°C. Cell pellet was lysed in 5 ml cold lysis buffer (10 mM Tris-HCl, pH 7.5; 10 mM NaCl; 5 mM MgCl2; 0.1 mM EGTA) with freshly added protease inhibitors (Roche) on ice for 15 minutes. Isolated nuclei were collected by centrifugation at 400 g for 5 min at 4°C then re-suspended in 0.5 ml of 1.2× restriction enzyme buffer (NEB) with 0.3% SDS and incubated for 1 h at 37°C while shaking at 900 rpm. Next, samples were incubated for 1 hr at 37°C after addition of 2% (final concentration) Triton X-100. 400 U of restriction enzyme was added to the nuclei and incubated at 37°C over night. 10 μl of samples were collected before and after the enzyme reaction to evaluate digestion efficiency. The reaction was stopped by addition of 1.6% SDS (final concentration) and incubation at 65°C for 30 minutes while shaking at 900 rpm. The sample was then diluted 10 fold with 1.15× ligation Buffer (NEB) and 1% Triton X-100 and incubated for 1 h at 37°C while shaking at 900 rpm. 400 U of T4 DNA ligase (NEB) were added to the sample and the reaction was carried at 16°C for 4 hrs followed by 30 mins at room temperature. For each sample, 300 μg of Proteinase K were added for protein digestion and de-crosslinking at 65°C overnight. On the next day, RNA was removed by adding 300 μg of RNAse and incubating the sample for 1 h at 37°C. DNA was purified twice by phenol-chloroform extraction and ethanol precipitation. Purified DNA was then analyzed by conventional or quantitative PCR. As control for ligation products the bac-clones were digested with 10 U of restriction enzyme overnight and then incubated with 10 U T4 DNA-ligase at 16°C overnight. The DNA was extracted by phenol-chloroform and precipitated with ethanol. Purified DNA was then analyzed by conventional or quantitative PCR. For Real time PCR, the ΔCt method was applied for analyzing data, using the Bac-clone Ct values as control. Primer sequences for PCR are listed in Supplementary table 2. Bac clone ID: RP-11-294A10, RP-11-1107P14, RP-11-1068G13 (Empire Genomics).
Pol II RNA Immunoprecipitation
RIP was performed as described 31. Hela cells were UV-crosslinked at 254nm (200 mJ/cm-2) in 10 ml ice-cold PBS and collected by scraping. Cells were incubated in lysis solution (0.1% SDS, 0.5% NP40, 0.5% sodium deoxycholate, 400Uml21 RNase Inhibitor (Roche)) and protease inhibitor at 4 °C for 25 min with rotation, followed by DNase treatment (30U of DNase, 15 min at 37 °C). Protein A Dynabeads (Invitrogen) were incubated with 2 ug Pol II antibody (Santa Cruz, N-20) and the cell lysate at 4°C overnight. The purified protein-RNA complex was extracted using TRIzol method for RNA extraction and subjected to RT-qPCR with corresponding primers.
Inducible cell lines
INTS11 and INTS1 knockdown inducible clones were generated from Hela cells using the Tet-pLKO-puro vector. For EGF induction, cells were serum starved in 0.5% FBS for 48 hours and treated with 100 ng/mL EGF (Invitrogen) for the indicated time course.
Transfections
Cells were treated with Dox for 48 hours. 24hrs before EGF induction, INTS11 and INTS11 (E203Q) mutant protein expression plasmids were transfected using Lipofectamine 2000 (Life Technologies, Inc.) according the manufacturer’s instruction. Cells were harvested 0 and 20 minutes after EGF induction.
All the PCR primer sequences are listed in the supplementary table 2.
Extended Data
Extended data Figure 1. Identification eRNAs responsive to EGF.
a, We identified 91 EGF-responsive enhancer regions in HeLa cells. We annotated extragenic RNAPII sites (see Methods) and used the middle of the RNAPII peak as an anchor to display average profiles of p300, H3K27ac and H3K4me1 (data from the ENCODE project). The profiles represent the mean read density of ChIP-seq data. The 91 loci display a typical enhancer signature, with enrichment of p300 and H3K27ac around the TSS and a broader decoration by H3K4me1. b, Profiles of H3K27ac were obtained from ChIP-seq analysis of HeLa cells before and after 20 minutes of EGF induction. Mean read density was normalized to sequencing depth. c, EGF stimulates bi-directional transcription from 91 enhancer regions. We displayed the mean read density obtained from strand-specific sequencing of the chromatin-bound RNA fraction (ChromRNA-seq). Normalized read density (RPKM) was calculated from RNA-seq data for 91 eRNAs. d, and 57 neighboring protein coding genes. e, that responded to EGF stimulation (FC>1.6) and mapped within 500 kb from an EGF-responsive eRNA. f, Average profiles of ChromRNA-seq data at 91 enhancer loci (mean density of reads, normalized to total read number). g–h, Boxplot of 91 eRNAs before and after treatment with EGF shows the average increase of transcription 20 minutes after stimulation (p<0.001), matched by an increase in the neighboring protein coding genes (p<0.02). i, NR4A1 is activated by EGF in HeLa cells: RNAPII and INTS11 are recruited to the NR4A1 locus after 20 min of stimulation, with concomitant accumulation of reads from RNA-seq and ChromRNA-seq. A neighboring eRNA locus also exhibit increased transcription along with RNAPII and INTS11 recruitment. Sequencing tracks are visualized in BigWig format and aligned to the hg19 assembly of the UCSC Genome Browser.
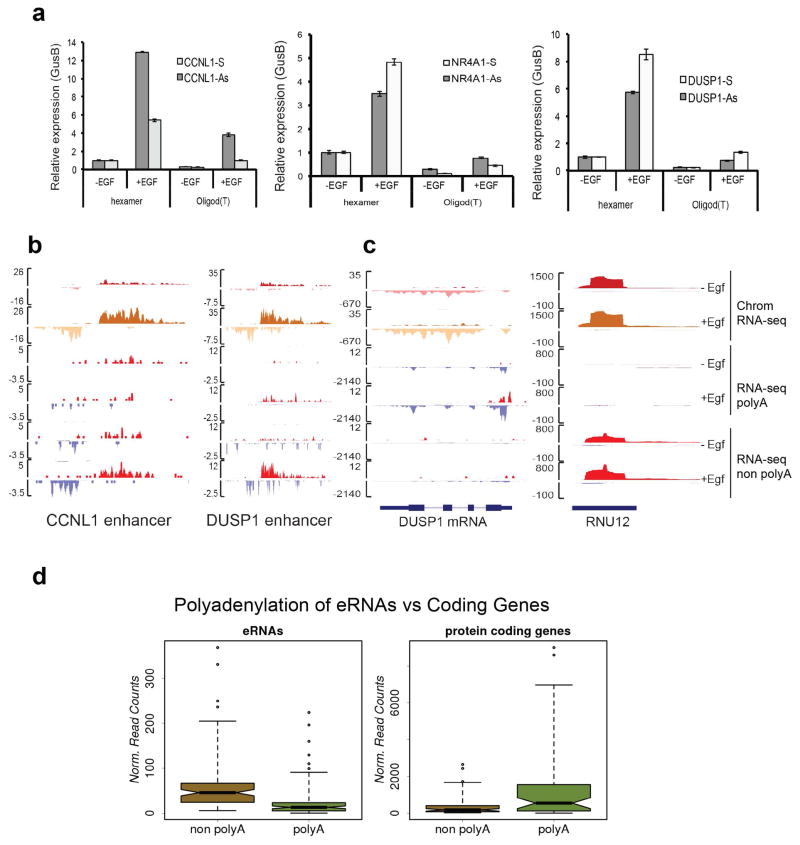
Extended Data Figure 2. EGF-induced eRNAs are predominantly non-polyadenylated.
a, We examined transcription at three enhancers adjacent to EGF-responsive genes CCNL1, NR4A1 and DUSP1. Total RNA samples were collected before and after EGF induction. Reverse transcription was performed with random hexamer primer or oligo d(T) primer. Each eRNA strand was analyzed by Real time PCR with specific primers. Error bars represent ± standard error of the mean (SEM, n=3 biological independent experiments). P<0.01 by two-sided t-test. b–c, RNA-seq was performed on the polyadenylated and non-polyadenylated fraction of total RNA. RNA-seq tracks were visualized in BigWig format and aligned to the hg19 assembly of the UCSC Genome Browser. CCNL1 and DUSP1 enhancers were displayed (b) along with a polyadenylated control (DUSP1 protein coding locus) and a non polyadenylated transcript (snRNA U12) (c). All EGF-induced eRNAs and protein coding genes (RefSeq hg19) were examined for their average RPKM throughout the entire locus. d, We compared polyadenylation levels of 225 eRNAs and 150 protein coding genes (2 fold induction upon EGF, RPKM calculated from ChromRNA-seq data previously described. The boxplot shows predominance of nonpolyadenylated transcripts mapping to eRNA loci, as opposed to transcripts encoding for RefSeq genes.
Extended Data Figure 3. The Integrator Complex is recruited to enhancers upon EGF stimulation.
a, qChIP analysis of Integrator occupancy using INTS11, INTS1 and INST9 antibodies at four eRNA loci. Data were collected during a time course of EGF induction in HeLa cells (0, 20, 40 and 60 minutes). Error bars represent ± standard error of the mean (SEM, n=3 biological independent experiments). P<0.01 by two-sided t-test. b, Depletion of INST1 and INST11 protein levels in tet-inducible HeLa clones. The arrow indicates the INTS11 specific signal, the asterisk shows a non specific band. c, Fold change of H3K27 acetylation (0min/20min EGF) before (CTRL) and after (DOX) depletion of INTS11. Data were calculated from read density of ChIP-seq experiments across EGF-induced enhancers. Depletion of Integrator significantly affects EGF-dependent increase in H3K27ac (p<0.05).
Extended Data Figure 4. Depletion of Integrator impairs activation of eRNAs by EGF.
a–b, Activation of eRNAs near DUSP1, CCNL1 and NR4A1 genes were assayed by qRT-PCR in three independent experiments, using an INTS11 (a) or a INTS1 (b) inducible shRNA clones. Transcription was followed throughout a 20 min time-course experiment. Each eRNA was amplified with two different sets of specific primers to analyze both strands, dashed lines indicate treatment with doxycycline (DOX) to induce shRNAs. Data at every time point are reported as fold change (EGF/non induced). Error bars represent ± SEM (n=3 biological independent experiments), P<0.01 by two-sided t-test. c, Schematic representation of ATF3 and its super enhancer region located 30kb upstream (top). Snapshots of ChIP-seq and RNA-seq tracks show EGF-dependent recruitment of RNAPII and INTS11 at the ATF3 locus and at several upstream enhancers. Depletion of INTS11 nearly abolished transcription of eRNAs and ATF3 mRNA. d, Real time RT-PCR analysis of the ATF3 super enhancer region upon depletion of INTS11. qPCR analysis was performed before and 5, 10, 15, 20 minutes after EGF treatment with strand specific primer sets (indicated below the RNA-seq tracks in panel c). Error bars represent ± SEM (n=3 biological independent experiments), P<0.01 by two-sided t-test.
Extended Data Figure 5. Chromatin conformation capture at Control loci.
a, 3C analysis of NR4A1 promoter and control sites. Con1 site lies 74kb upstream of NR4A1 protein coding gene and Con2 site is located 42kb downstream of enhancer site. There are no looping events between either control sites with the NR4A1 promoter region after EGF induction. b, Similarly, no looping events were detected between the promoter of DUSP1 and a downstream control site (Con). All data were averaged from three independent experiments, p<0.01 by two-sided t-test.
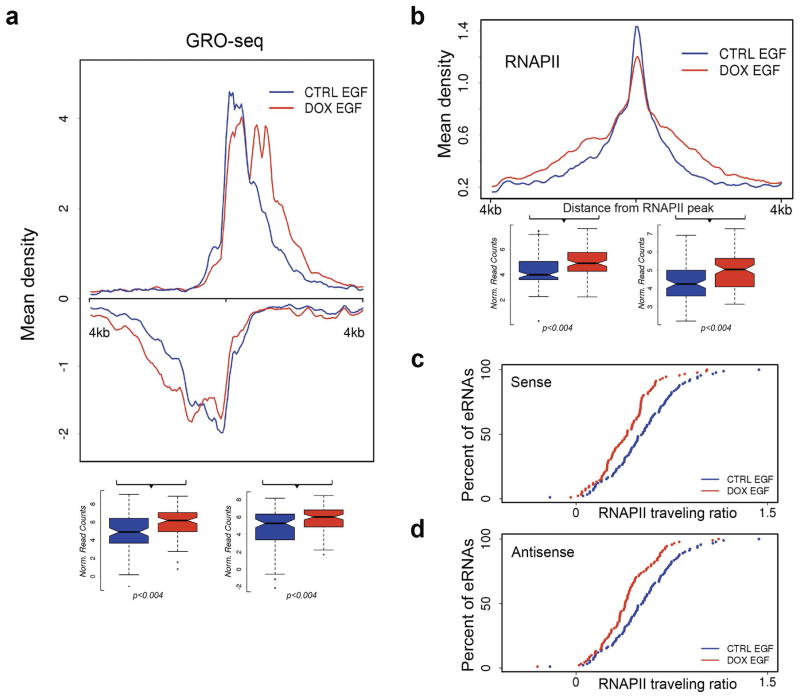
Extended Data Figure 6. Integrator plays a role in eRNA termination.
a, Mean density profiles of GRO-seq data at 91 EGF-induced enhancers. Data are presented as strand-specific mean read density, centered at the middle of the RNAPII peak and normalized to sequencing depth. The underlying box plots were used to quantify the enrichment of GRO-seq reads at the 3′ end of both eRNA transcripts (2kb window, centered 1kb downstream of the RNAPII peak). b, RNAPII profiling at 91 enhancers after INTS11 depletion shows accumulation of ChIP-seq reads towards the 3′ end. Data are presented as mean read density, centered at the middle of the RNAPII peak and normalized to sequencing depth. Box plots represent the enrichment of RNAPII reads of both eRNA transcripts (2kb window, centered 1kb downstream of the RNAPII peak). RNAPII significantly accumulated (p<0.004) after depletion of INTS11. c–d, RNAPII traveling ratio at enhancers was measured as the ratio between RNAPII density close to the transcription start site (the surrounding 300bp) and 3kb downstream. Given the bidirectional nature of transcription at enhancers, traveling ratio was calculated for both sense (c) and antisense (d) transcripts.
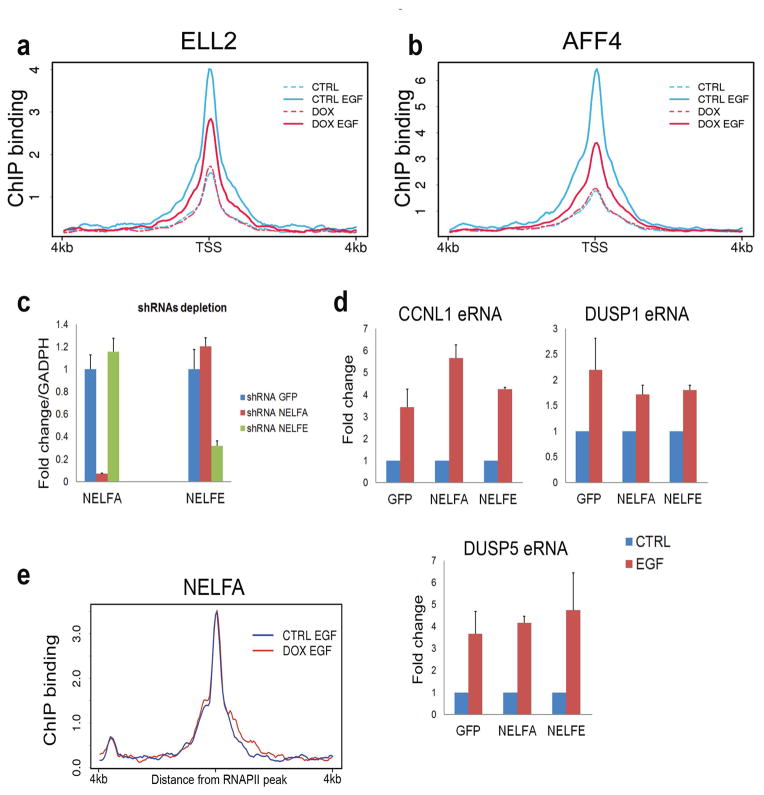
Extended Data Figure 7. Analysis of super elongation complex at enhancers.
a–b, Metagene analysis on 91 eRNA loci shows the effect of EGF stimulation and INTS11 depletion on the recruitment of the ELL2 (a) and AFF4 (b) subunits of the Super Elongation Complex (SEC). SEC was recruited to enhancers upon EGF stimulation. Depletion of Integrator somewhat decreases AFF4 and ELL2 recruitment. Data were visualized as mean read density, normalized to sequencing depth, across 8kb surrounding the center of enhancers. c, To investigate the role of the Negative Elongation Factor (NELF) in induction of eRNAs we infected HeLa cells with lentiviral shRNAs against NELFA, NELFE and a control GFP. Quantitative RT-PCR analysis shows the extent of NELF depletion 72h after infection. Error bars represent ± SEM (n=3 biological independent experiments), P<0.01 by two-sided t-test. d, Depletion of two different NELF subunits does not significantly impact activation of EGF-responsive eRNAs. Data represent fold change of induction (EGF/not induced) after 20 minutes of stimulation and were normalized against GUSB expression. Error bars represent ± SEM (n=3 biological independent experiments), P<0.01 by two-sided t-test. e, ChIP-seq analysis of NELFA before and after depletion of INTS11. Metagene analysis shown mean read density (normalized to sequencing depth) across 91 eRNAs. NELF occupancy at enhancers was not affected by depletion of Integrator.
Extended Data Figure 8. Integrator depletion causes accumulation of unprocessed eRNAs and prevents release of RNAPII.
a, Termination of eRNAs was examined with quantitative RT-PCR. Primer pairs were designed to amplify a portion of the enhancer transcript detected in normal condition (t, total) or a longer template further extending into the 3′ of the enhancer region (u, unprocessed). Q-PCR analysis was performed before (CTRL) and after (DOX) depletion of INTS11 at three eRNAs (sense and antisense strand), after stimulation with EGF. In the absence of INTS11, we observed accumulation of unprocessed eRNA, suggestive of a termination defect. Error bars represent ± SEM (n=3 biological independent experiments), P<0.01 by two-sided t-test. Release of eRNA transcripts from RNA polymerase was investigated by means of RNAPII immunoprecipitation following UV-cross link (UV-RIP). b–d, After RNAPII immuno-precipitation, eRNAs near DUSP1, CCNL1 and NR4A1 genes were assayed by qRT-PCR and showed increased association with RNAPII in the absence of Integrator. Each eRNA was detected by two different sets of specific primers (Sense and Antisense). Error bars represent ± SEM (n=3 biological independent experiments). *P<0.01, **P<0.01, ***P<0.001 by two-sided t-test. e–g, RNAPII UV-RIP analysis was also performed on several eRNAs from the ATF3 super-enhancer. qRT-PCR on the RNA recovered after immunoprecipitation shows increased association between RNAPII and eRNAs in the absence of Integrator. Each eRNA was detected by two different sets of specific primers (Sense and Antisense). Error bars represent ± SEM (n=3 three independent experiments). **P<0.01 by two-sided t-test.
Extended Data Figure 9. Distribution of RNAPII and nascent RNAs across protein coding genes.
a, Expression level of exogenous INTS11 wild type (wt) and its catalytic mutant (E203Q). Nuclear extracts were subjected to Flag immunoprecipitation and probed with a polyclonal antibody raised against the C-terminus of INTS11. b, Heatmap of nascent RNA (GRO-seq) and RNAPII ChIP-seq across the 2,000 most active genes in HeLa cells. Gene loci were analyzed for their entire gene body, with 3 additional kilobases on both ends. H3K27ac data from ENCODE is shown on the left, genes are ranked according to the intensity of RNAPII signal. Depletion of Integrator does not appear to affect termination at protein coding genes.
Supplementary Material
Acknowledgments
We like to thank Jill M. Marinis and Mitchell A. Lazar for technical support for GRO-Seq experiments. Thanks to Deqing Hu in Ali Shilatifard’s laboratory for performing the SEC ChIP-seq experiments. We like to thank the Oncogenomics core facility at Sylvester Comprehensive Cancer Center for performing high-throughput sequencing. Thanks to Shiekhattar laboratory members and Pierre-Jacques Hamard for support and discussions. This work was supported by grant R01 GM078455 and R01 GM105754 (R.S.) from the National Institute of Health.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contribution F.L. and A.G. are co-first authors. R.S., F.L. and A.G. conceived and designed the overall project. F.L., A.G. and A.Z. performed the experiments. R.S., F.L. and A.G. analyzed the data and wrote the paper.
The authors declare no competing financial interests.
References
- 1.Baillat D, et al. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 2.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Santa F, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS biology. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatt DM, et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lam MT, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. nature10006 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sigova AA, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hah N, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. S0092-8674(13)00392-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loven J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. S0092-8674(13)01227-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melo CA, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Molecular cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 17.Mousavi K, et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Molecular cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardini A, et al. Integrator regulates transcriptional initiation and pause release following activation. Molecular cell. 2014;56:128–139. doi: 10.1016/j.molcel.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto J, et al. DSIF and NELF interact with Integrator to specify the correct post-transcriptional fate of snRNA genes. Nature communications. 2014;5:4263. doi: 10.1038/ncomms5263. [DOI] [PubMed] [Google Scholar]
- 20.Albrecht TR, Wagner EJ. snRNA 3′ end formation requires heterodimeric association of integrator subunits. Molecular and cellular biology. 2012;32:1112–1123. doi: 10.1128/MCB.06511-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch F, et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nature structural & molecular biology. 2011;18:956–963. doi: 10.1038/nsmb.2085. [DOI] [PubMed] [Google Scholar]
- 22.Yang L, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stadelmayer B, et al. Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nature communications. 2014;5:5531. doi: 10.1038/ncomms6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skaar JR, et al. The Integrator complex controls the termination of transcription at diverse classes of gene targets. Cell research. 2015;25:288–305. doi: 10.1038/cr.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaukowitch K, et al. Enhancer RNA facilitates NELF release from immediate early genes. Molecular cell. 2014;56:29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. gb-2009-10-3-r25 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang LG, Wang SQ, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 29.Ye T, et al. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 2011;39:e35. doi: 10.1093/nar/gkq1287. gkq1287 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai F, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.