Abstract
Micronutrient deficiencies, including zinc deficiency, are responsible for hundreds of thousands of deaths annually. A key obstacle to allocating scarce treatment resources is the ability to measure population blood micronutrient status inexpensively and quickly enough to identify those who most need treatment. This paper develops a metabolically engineered strain of Escherichia coli to produce different colored pigments (violacein, lycopene, and β-carotene) in response to different extracellular zinc levels, for eventual use in an inexpensive blood zinc diagnostic test. However, obtaining discrete color states in the carotenoid pathway required precise engineering of metabolism to prevent reaction at low zinc concentrations but allow complete reaction at higher concentrations, and all under the constraints of natural regulator limitations. Hence, the metabolic engineering challenge was not to improve titer, but to enable precise control of pathway state. A combination of gene dosage, post-transcriptional, and post-translational regulation was necessary to allow visible color change over physiologically relevant ranges representing a small fraction of the regulator’s dynamic response range, with further tuning possible by modulation of precursor availability. As metabolic engineering expands its applications and develops more complex systems, tight control of system components will likely become increasingly necessary, and the approach presented here can be generalized to other natural sensing systems for precise control of pathway state.
Keywords: Carotenoid pathway, biosensor, pathway regulation, synthetic biology, micronutrients, precision engineering
Introduction
Metabolic engineering has played a key role in advancing a number of critical biotechnological research areas and applications, but there remain numerous areas where control of metabolism has not been leveraged to solve critical problems, such as enabling biosensors for applications like blood micronutrient measurement at a population scale. The biofuels (Atsumi et al., 2008a; Atsumi et al., 2008b; Jin et al., 2005), pharmaceutical (Alonso-Gutierrez et al., 2013; Ro et al., 2006), and specialty and commodity chemicals (Chemler et al., 2010; Raab et al., 2010) applications of metabolic engineering are well-known. Moreover, the techniques of metabolic engineering have been used to engineer plants and foodstocks to help battle problems such as nutrient and micronutrient deficiency, with one of the most prominent examples being the development of Golden Rice (Beyer et al., 2002; Ye et al., 2000). In the battle against micronutrient deficiency, metabolic engineering could also greatly facilitate the development of micronutrient biosensors that would allow targeted interventions with limited treatment resources since engineered crops are not available for most micronutrient deficiencies.
Micronutrients (vitamins and minerals) are a critical component of human health and development, and micronutrient deficiencies are responsible for hundreds of thousands of deaths per year worldwide (Black et al., 2008). One of the most significant challenges in treating micronutrient deficiencies is in the identification of the populations most at risk for deficiencies or most in need of nutritional interventions (which are performed on populations, not individuals, and are prohibitively expensive for continuous application on a wide scale (Bhutta et al., 2013)). While our metabolic engineering-based approach to biosensor design can be broadly applied to many micronutrient deficiencies, we focus here on the essential mineral zinc, since zinc deficiency is estimated to be responsible for 116,000 annual deaths of children younger than five (Black et al., 2013) and contributes to growth stunting and increased incidence of pneumonia, diarrhea, and perhaps malaria (Fischer Walker and Black, 2004). More precise assessment of global zinc deficiency pervasiveness and impacts is lacking, though, due to the high cost and logistical difficulties of measuring zinc status on a regional population scale. In developing countries (which bear the overwhelming burden of nutritional deficiencies) and in the wake of disasters, on-site testing is hindered by the lack of availability of the sophisticated equipment required to measure serum zinc levels (Kelson and Shamberger, 1978), and often by a lack of available electricity. Off-site analyses require continuous cold storage (logistically difficult) during international shipping, can take up to a week for results, and cost tens of dollars each, which is prohibitive for the required epidemiological-scale measurements. This suggests that a low-tech, inexpensive, and easy assay for zinc deficiency could be valuable to help policymakers and non-governmental organizations pinpoint the populations most needing the limited treatment resources.
To address these problems, we engineered a whole-cell biosensor for future application in a point-of-care assay using bacteria to report zinc levels in blood samples. A key innovation in this design is the use of a reporter that requires no equipment for readout. Previous technologies for sensing zinc produced fluorescence or luminescence proportional to zinc concentrations (Date et al., 2010; Gireesh-Babu and Chaudhari, 2012); while such assays give the potential for quantitative results, they require specialized equipment to detect and quantify these readouts that limit application in low-resource areas. Instead, we designed our assay to use pigment production as a readout, removing the need for quantification. Pigment production is performed by heterologous genes expressed in E. coli under the control of zinc-responsive transcription factor/promoter systems. We further sought to make the color outputs as discrete as possible, transitioning sharply from one color to another so as to remove potential ambiguity from the reading.
The pigments used for this biosensor were the red and orange carotenoids lycopene and β-carotene and the purple pigment violacein, each of which have previously been the targets of metabolic engineering efforts whose results and insights have contributed to the engineering efforts described here. There has been extensive work on the engineered overproduction of lycopene and other isoprenoids (Alper et al., 2005; Farmer and Liao, 2001; Yoon et al., 2006), as they have significant industrial value as chemicals, and isoprenoids are a precursor to yet other extremely valuable chemicals, including taxol (Ajikumar et al., 2010). Violacein has also been the subject of significant metabolic engineering efforts (Fang et al., 2015; Lee et al., 2013; Rodrigues et al., 2013), both due to the novelty and complexity of the pathway that produces it, and for its potential value as an antimicrobial or antitumor drug. In these cases, the chief goal of the metabolic engineering efforts to date has been high titer production with the downstream goal of purification. For our application, though, tightly controlled expression rather than high titer was the primary goal of our metabolic engineering efforts.
An effective bacterial-based, pigment-producing diagnostic for our application required (1) sufficient pigment to be visible to the naked eye (2) in a reasonable amount of time (3) with sufficient control over production of all pigments to enable discrete color states. The first two of these criteria directly relate to traditional metabolic engineering efforts, though with a greater emphasis on productivity to meet a minimum (visible) titer rather than maximizing titer. Meeting the third criterion was the most challenging for the final engineering implementation of the sensor, and is the focus of this work. We chose to use two pigments (lycopene and β-carotene) from the linear carotenoid pathway with the expectation that it would facilitate the creation of discrete color states: each molecule of red lycopene could be stoichiometrically converted into orange β-carotene upon induction of the appropriate enzyme by the presence of sufficient zinc, leaving no residual red color in the orange state. This advantage is also a potential challenge, though, if that zinc-inducible promoter has substantial baseline uninduced transcription of that enzyme, which could prevent accumulation of lycopene. However, our application constrains selection of promoters: the promoter must be zinc-inducible, and there are few well-characterized zinc-inducible promoters.
Here, we present a proof-of-principle, metabolically engineered bacterial biosensor with pigment outputs that require no equipment for measurement and interpretation, for potential use in micronutrient (zinc) blood testing. We present the initial results from a straightforward implementation of the desired biosensor, which indicated the need for metabolic engineering in order to achieve the desired discrete color states. We explore multiple avenues to achieve the desired outcome, and we assess which are necessary and sufficient for this specific application. The trends identified in optimizing the construction of the sensor are likely generalizable to similar pigment-based micronutrient sensors or whenever promoter selection is constrained. As metabolic engineering finds more diverse applications, it is likely that techniques to not just optimize production but tightly control it will be increasingly necessary.
Materials and Methods
2.1 Materials
T4 DNA ligase, T5 exonuclease, Taq ligase, Phusion polymerase, Q5 polymerase, and restriction endonucleases were purchased from New England Biolabs (Ipswich, MA, USA). QIAprep Spin Miniprep Kits, QIAquick Gel Extraction Kits, and QIAquick PCR Purification Kits were purchased from QIAGEN (Valencia, CA, USA). Lycopene (98%) was purchased from Cayman Chemical (Ann Arbor, MI, USA). β carotene (97%) was purchased from TCI America (Portland, OR, USA). SC-Ura amino acid supplement was purchased from Sunrise Science (San Diego, CA, USA).
2.2 Cloning and construct assembly
Most construct assembly was carried out via restriction endonuclease digestion of components and subsequent ligation and transformation following the BioBricks idempotent standard assembly. Constructs in pCC1FOS were first assembled as above in BioBrick-compliant vectors. These were then digested with NotI and ligated into pCC1FOS.
The initial attempted multi-state pigment-producing construct was assembled via Gibson assembly (Gibson et al., 2009). This method was also used to add ssrA tags to eGFP and CrtY and to remove illegal restriction sites for BioBrick standard assembly from coding sequences for eGFP, zur, and tetR. Primers used to generate products for Gibson assembly are presented in supplementary material.
2.3 Cell Culture
For eGFP library expression, freshly transformed DH10B colonies were inoculated in triplicate into 5 mL LB media and were grown at 37°C for 24 hours. 125 µL of each culture was aliquotted into 96-well plates and measured on a Biotek Synergy H4 plate reader for OD600 and fluorescence at 488 nm excitation and 509 nm emission. Samples were also analyzed on a BD Accuri C6 flow cytometer. For zinc carotenoid experiments, each strain was inoculated from freshly transformed colonies in triplicate into LB and minimal media at the following concentrations of supplemented ZnSO4: 0, 50, 500, 1000 µM (LB) and 0, 5, 50, 500 µM (Minimal). LB cultures were incubated at 37°C for 24 hours. To allow zinc titration, a modified minimal media based on M9 but lacking inorganic phosphate was developed containing the following: 2.5 g/L beta-glycerophosphate pentahydrate, 1.64 g/L KCl, 4.5 g/L NaCl, 1 g/L NH4Cl, 3.9 g/L MES, 2 mM MgSO4, 0.1 mM CaCl2, 0.01% thiamine, 0.6% glycerol, and 1.92 g/L SC-Ura amino acid supplement. pH was adjusted to 7.4. The following antibiotics were used as needed for selection: carbenicillin (100 µg/mL), chloramphenicol (34 µg/mL), kanamycin (30 µg/mL). Minimal media cultures were incubated at 37°C for 48 hours. Gradient agar plates were made by allowing supplemented media to gel on inclined petri dishes, forming wedges. The plates were then laid flat and filled in with unsupplemented media. LB gradient plates were made with three sections: supplemented 10 uM TPEN, LB alone, and LB with 250 uM ZnSO4. Minimal gradient plates were made with two sections: plain minimal medium and 250 uM ZnSO4. TPEN was used to simulate zinc deprivation in rich media. Plates were incubated at 37°C for 24 and 48 hours for LB and minimal agar plates, respectively. Color for streaks on LB gradient plates took approximately 24 hours to develop, except for the final fosmid construct and the final pJBEI-supplemented strain with the pSB6A1 construct, which took approximately 14–16 hours to develop color.
2.4 Carotenoid extraction
1 mL of bacterial culture was pelleted at 16,000 rcf for 7 minutes. Cell pellets were resuspended in 50 µl of water from a MilliQ water handling system (EMD Millipore). Carotenoids were extracted with 1 mL of acetone at 55 C for 20 minutes with continuous shaking in a Grant Thermoshaker at 1400 rpm. Cellular debris was pelleted at 16,000 rcf and the supernatant was removed for analysis. All carotenoid extraction was carried out in low light conditions.
2.5 HPLC analysis
All HPLC analysis was conducted on a Shimadzu LC-20AD Liquid Chromatograph using a Shimadzu C18 4.6 mm × 50 mm column with a 5 um particle size and a SPD-20A UV-Vis detector. The instrument was run with a flow rate of 0.6 mL/min and a solvent ratio of 90:10 acetonitrile:THF. Retention times and intensities were compared to analytical standards spiked into control extractions from DH10B cells and calibration curves to convert peak areas to analyte concentrations were constructed.
2.6 Plasmids and regulatory sequences
A list of all vectors used is provided in Table 1, along with their expected copy number. ZntR, Zur, and PznuC were amplified from DH10B genomic DNA. pJBEI-6409 (Alonso-Gutierrez et al., 2013) was obtained from Addgene (Cambridge, MA, USA) and is referred to here as pJBEI-MEV. PzntA sequence was synthesized by Genewiz (South Plainfield, NJ, USA). eGFP was provided by Julie Champion, and pCC1FOS was provided by Brian Hammer. All other parts, including pSB-series plasmids (Shetty et al., 2008), ribosomal binding sites, promoters, and pigment biosynthesis genes were obtained from the Registry of Standard Biological Parts and were sequence-confirmed before use.
Table 1.
Vectors and source plasmids used in this work. A more detailed description of plasmids is included in Supplementary Information S2.
| Vector or plasmid name |
Putative copy number |
Origin of replication |
|---|---|---|
| pJBEI-MEV | Medium | p15A |
| pSB1C3 | High | PUC colE1 |
| pSB1AK3 | High | PUC colE1 |
| pSB3C5 | Medium | p15A Mutated |
| pSB6A1 | Low | pBR322 colE1 |
| pCC1FOS | Approximately single | F |
2.7 Strain naming conventions
Fluorescence library members were named by concatenating the name or Parts Registry part number of the promoter, the last two digits of the Parts Registry part number for the ribosomal binding site, and then potentially three letters indicating whether a strong (LAA) or weak (DAS) ssrA protein degradation tag was added (AANDENYALAA or AANDENYADAS). Carotenoid test parts were named by concatenating the Parts Registry part number of the RBS in front of crtY, an indicator of whether there was a degradation tag on crtY (LAA, DAS, or nothing), and the last three characters of the name of vector the construct was cloned into (1C3, 3C5, 6A1, or FOS). The presence of +J indicates the carotenoid construct was cotransformed with pJBEI-MEV.
Results
3.1 Design of a zinc-responsive, pigment-producing, bacterial biosensor
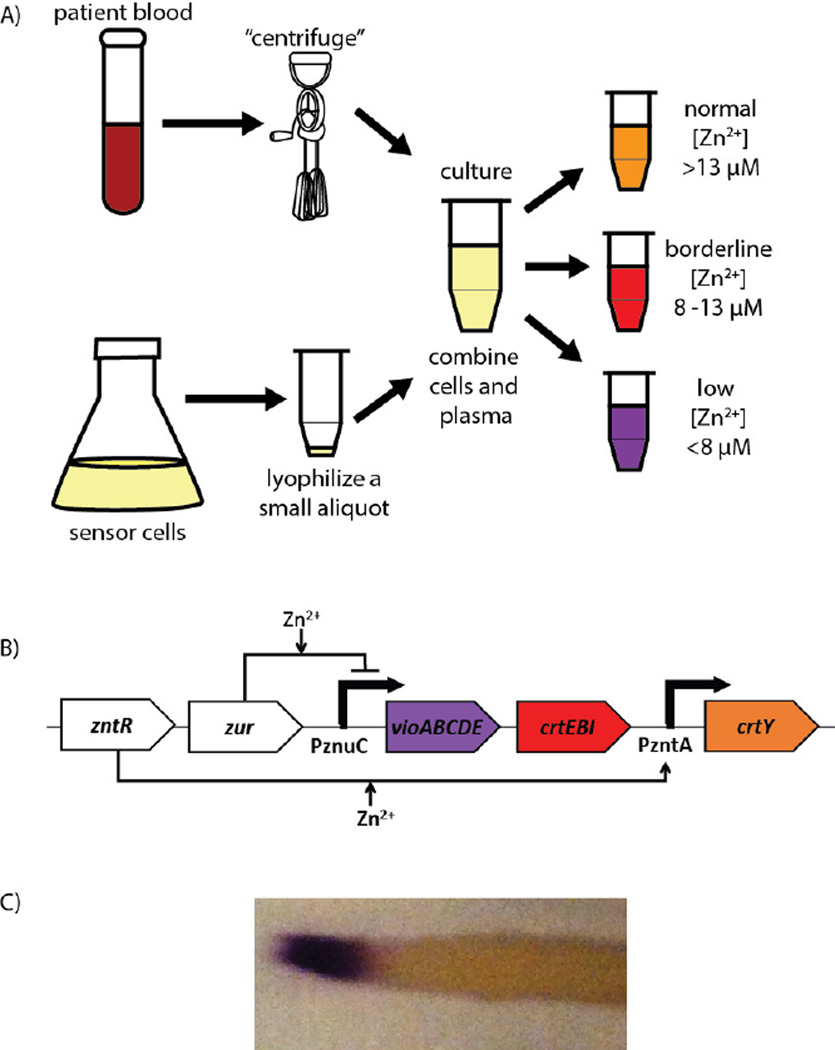
To move towards the goal of population-scale monitoring of blood micronutrient status (specifically zinc), a low-cost, portable, equipment-free approach is necessary. The overall workflow of the final envisioned biosensor is illustrated in Figure 1A: sensor cells would be lyophilized or otherwise preserved for long term storage. A blood sample would be separated in the field via a low-cost, equipment-free approach such as the egg beater centrifuge (Wong et al., 2008). The resulting plasma would then be added to the preserved cells (potentially with additional, zinc-free defined medium), and the cells would produce different pigments in response to the levels of zinc in the physiologically relevant range of 8–15 µM (Hess et al., 2007) corresponding to normal, borderline, and low levels of zinc.
Figure 1.
An initial synthetic biology-based zinc biosensor for application to equipment-free blood tests. A) A schematic of the workflow envisioned for the eventual final implementation of the diagnostic biosensor. Cells from general culture (bottom) are lyophilized for long-term storage. A blood sample (top) is centrifuged using an egg beater (Wong et al., 2008) to separate plasma, which is then added to the stored cells. These cells then produce pigment (right) based on the level of zinc in the plasma sample. Physiological concentrations of zinc are included for reference. B) Schematic of the circuit design intended for implementation of the initial biosensor. Transcriptional regulators ZntR and Zur are expressed on the same plasmid as the construct with violacein synthesis driven by the zinc-repressible PznuC promoter, lycopene synthesis driven by a weak promoter, and the gene catalyzing the transformation of lycopene to β-carotene driven by the zinc-inducible PzntA promoter. The bolded zinc annotation for PzntA indicates that activation by zinc happens at higher concentrations than for pZnuC. C) Streak from a zinc gradient plate demonstrating direct transition from purple to orange without an intervening lycopene-dominated, red state. (Image contrast was adjusted to reduce the background from the agar plate; the original image is included as Supplementary Figure 1.)
To implement the desired functionality, we designed a circuit using heterologous pigment production pathways driven by zinc-responsive transcription factor/promoter pairs present in E. coli (Figure 1B). The first heterologous pigment production pathway was the purple violacein biosynthesis pathway, which uses the vioABCDE operon (originally derived from Chromobacterium violaceum) to produce violacein from endogenous tryptophan. The second pigment production pathway was carotenoid biosynthesis, which uses the crtE, crtB, and crtI genes to produce red lycopene starting from endogenous FPP, and uses crtY to produce orange β-carotene from lycopene; these genes were originally derived from Pantoea ananatis. Expression of these pigment production pathways was driven by two endogenous E. coli zinc-responsive promoter/transcription factor pairs, PznuC/Zur and PzntA/ZntR, that control expression of zinc importer and exporter genes, respectively. (Zur is a zinc-activated transcriptional repressor and ZntR is a zinc-activated transcriptional activator.) Previous work (Yamamoto and Ishihama, 2005) suggested that Zur is able to fully repress expression from PznuC at approximately 10 µM zinc, while induction of transcription from PzntA should begin at least by100 µM zinc (preliminary results suggested induction by 10 µM) with full induction not until approximately 1.1 mM (Brocklehurst et al., 1999).
These pieces were then combined together such that the circuit would provide discrete color states of purple, red, and orange depending on the zinc concentration of the sample. PznuC was used to drive expression of vioABCDE and PzntA was used to drive expression of crtY; zntR and zur were expressed from the same plasmid using their genomic promoters, while crtEBI was expressed from a mutated pλ-based, extremely weak promoter (avoiding toxic expression; details in Supplementary Table 1). At low zinc concentrations (below 10 µM), transcription from PzntA would be low and transcription from PznuC would be induced, resulting in violacein biosynthesis (which is so dark as to visibly overwhelm minor lycopene production). At borderline levels of zinc (approximately 10 µM), Zur would become activated and repress expression from PznuC. This would repress expression of the violacein biosynthesis operon, but expression of crtY to produce β-carotene would not be sufficiently induced, resulting in the production of only the red pigment lycopene. At normal levels of zinc (over 10 µM), violacein production would still be repressed, but ZntR would then be activated and crtY expression would increase, facilitating the transformation of lycopene to β-carotene and yielding only orange pigment.
3.2 Upon implementation, the designed circuit only exhibited two distinct color states
We implemented the circuit design and expressed it on a high copy number plasmid in E. coli. The entire construct was sequence confirmed. The behavior of the implemented construct is shown in Figure 1C, as a streak on a zinc gradient plate. There is a clear, fairly sharp transition between two color states, purple (low zinc) and orange (high zinc), but no observable red (intermediate zinc) state. This behavior provides a number of key insights. First, the fact that crtEBI is very weakly and constitutively expressed is not a problem, as the dark color produced by violacein overwhelms any potential background signal of carotenoid production. Next, the repression of violacein production at higher zinc concentrations is sufficient to allow for the carotenoid production to be visible without any interference from leakiness of the PznuC promoter. Finally, the basal uninduced expression of crtY from PzntA at low and intermediate zinc levels is too great to allow precise control over the pathway, as lycopene is converted to β-carotene even at zinc levels where crtY should not be strongly induced.
3.3 Absence of a lycopene-only state is due to overlap in expression regimes
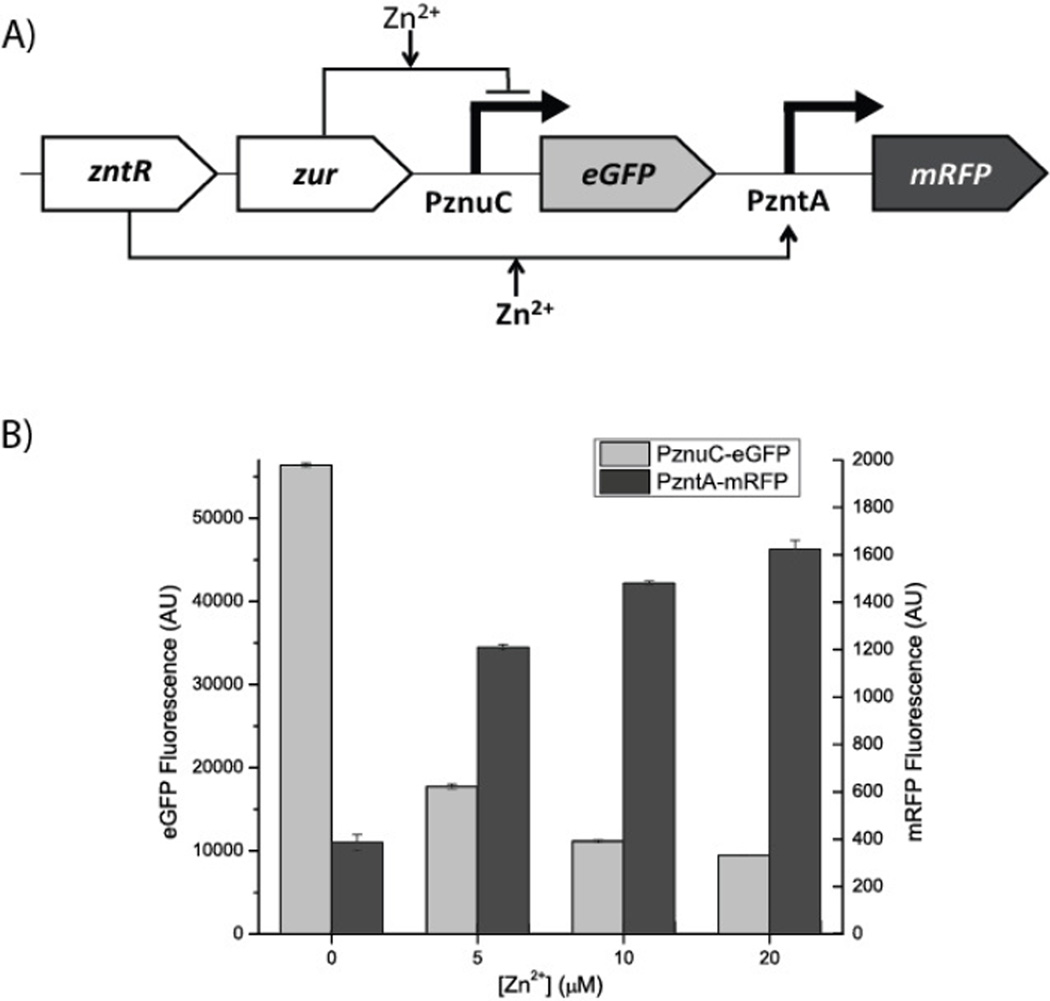
To investigate the cause of the absence of a lycopene-only state, we implemented a fluorescence-based construct to assess the induction and repression of PzntA and PznuC, respectively. Figure 2a shows the structure of this construct: RFP expression is driven by the zinc-inducible PzntA promoter, while GFP expression is driven by the zinc-repressible PznuC promoter, with the transcription factors for each promoter (ZntR and Zur, respectively) constitutively expressed from the same high-copy plasmid. Figure 2b shows the quantitative results of GFP and RFP expression from the promoters. It is thus clear that while PzntA and PznuC can be significantly induced and repressed, respectively, there is some overlap in their expression regimes, whether due to their response curves to zinc or due to baseline leaky expression. It is worth noting that most of the repression from PznuC occurs over the low concentrations of zinc, yet there is clearly (Figure 1c) a regime where violacein is not visible. Since violacein is much easier to detect visually than the carotenoids, this suggests that even though there is measurable fluorescence (and thus measurable leakiness) for this test construct, the production of violacein corresponding to that level of fluorescence would not be visually detectable. Combined with knowledge that the maximum repression of PznuC has been shown to happen at the bottom end of the PzntA induction range, this supports the idea that the reason why no lycopene-only state is detectable is that crtY expression is too great at low zinc levels, rather than vioABCDE expression being too great at higher zinc levels. Supplementary Figure 2, showing the expression of mRFP from PznuC and PzntA on parallel regulated constructs, suggests that visibly there is likely to be a significant region of non-overlap between the repression of PznuC and the induction of PzntA.
Figure 2.
Fluorescence reporter construct shows overlap in PznuC and PzntA expression. A) Schematic of the reporter construct used to simultaneously measure expression from PznuC and PzntA promoters. The transcription factors regulating expression from each promoter were included on the plasmid. The bolded zinc annotation for PzntA indicates that activation by zinc happens at higher concentrations than for PznuC. B) Expression from each promoter in arbitrary units specific to each fluorescent protein (RFP is right axis, GFP is left axis). There is significant induction of expression from PzntA even at low levels of added zinc and significant repression of expression from PznuC, suggesting that the baseline expression of crtY may be the cause of a lycopene-only state being difficult to capture. Error bars represent standard error of the mean.
3.4 A library of fluorescence constructs enables characterization of posttranscriptional control
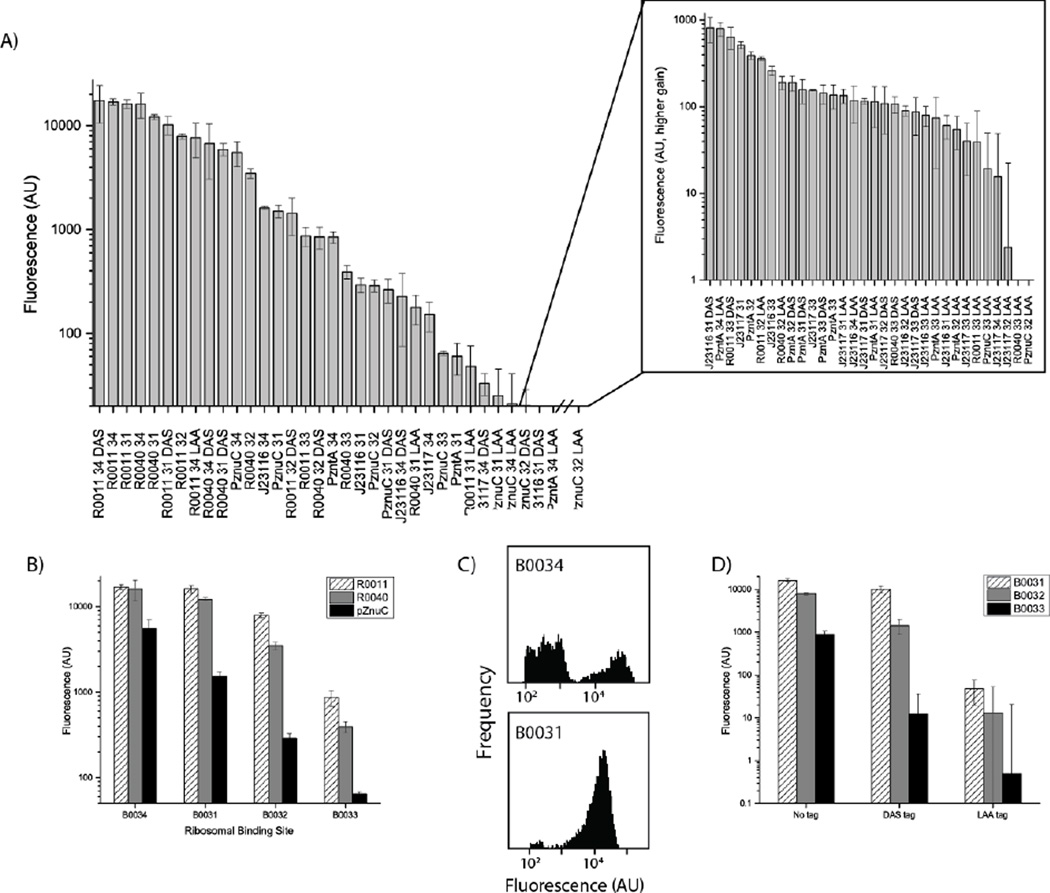
In order to gain sufficient control over the lycopene to β-carotene reaction step to generate a three-color-state sensor, we then sought to adjust post-transcriptional regulatory elements. Specifically, modification of ribosomal binding sites and tagging of proteins for degradation were identified as the simplest approaches to decrease the level of CrtY at low levels of zinc. To do this most efficiently, we characterized the impact of multiple regulatory elements within our specific host strain using an eGFP reporter. We also sought to characterize the strength of multiple potentially constitutive promoters (which could drive expression of the zinc-responsive transcription factors) and the strength of our zinc-responsive promoters (without coexpressed regulators). This characterization was intended to facilitate more rational and focused selection of regulatory elements for the final sensor construct rather than performing combinatorial syntheses that would be challenging based on product toxicities; the results of the characterization are presented in Figure 3.
Figure 3.
A library of regulatory elements provides five orders of magnitude of control over expression. A) Fluorescence measurements of the library of regulatory element combinations. Inset illustrates fluorescence levels at a higher gain setting for library members with insufficient fluorescence to be measured accurately at the same gain setting as in the main plot. Construct names are the promoter name (R0011 & R0040 known strong promoters, J23116 &J23117 known weak promoters, and the PnzuC and PzntA promoters), followed by an indicator of the ribosomal binding site part number (31–34), and then the degradation signal added to the protein sequence (no signal, weak DAS signal, or strong LAA signal). Error bars represent the standard error of the mean. B) Changing ribosomal binding sites for strong or weak promoters provides orders of magnitude reduction in expression. Error bars represent the standard error of the mean. C) Flow cytometry plots showing increasingly mixed populations for higher-strength (B0034) ribosomal binding sites (top) as opposed to weaker ribosomal binding sites (bottom), indicating that the bulk fluorescence measured for B0034 RBS under the strong promoters used here is an underestimate of the actual per-cell fluorescence for B0034 under unaltered promoters. Sequencing of plasmid DNA from cultures with bimodal populations yielded mixed traces, indicating mutation of the plasmids to reduce fluorescence production. D) Degradation tags can also yield orders of magnitude reduction of protein levels; the strong LAA tag typically provides two or more orders of magnitude of decrease in fluorescence, while the weaker DAS tag is more variable in its reduction of fluorescence. Error bars represent standard error of the mean; only positive error is indicated due to the logarithmic scale of the plot because the lowest values are near the detection limits of the instrument and would include zero in the opposite direction.
Taken together, manipulating promoters and post-transcriptional regulatory sequences yielded over five orders of magnitude of variation of fluorescence (Figure 3a). Each level alone (transcriptional and post-transcriptional) offered at least three orders of magnitude dynamic range, with low-fluorescence measurements being confounded by limits of detectability even at high plate reader gain settings.
Measurements of promoter strength across multiple sets of post-transcriptional regulatory sequences provided information for later selection of promoters (i.e., for zntR and zur) and provided an estimate of the strength of the PznuC and PzntA promoters. Unsurprisingly, the pλ-based promoters (R0040 and R0011) were the strongest. Known weaker promoters (J23117 and J23116) performed roughly as expected, though the difference between these two promoters was greater than previously reported. We used the expression levels of these promoters to characterize some of the transcriptional behavior of the two inducible promoters. Since these constructs had no supplementary regulator expression (i.e. Zur or ZntR) in order to be consistent with the rest of the library, the expression from PznuC is indicative of essentially unrepressed transcription, while expression from PzntA is indicative of essentially uninduced transcription. We found that baseline expression from PzntA was stronger than at least one known weak constitutive promoter, J23117. This suggests that the underlying reason for the lycopene state being difficult to stabilize may be that the baseline transcription from PzntA is sufficiently high to result in significant production of CrtY.
Changing ribosomal binding sites (RBSs) also enabled many orders of magnitude of reduction for overall gene expression. Figure 3b demonstrates the impact of using four different ribosomal binding sites taken from the Registry of Standard Biological Parts: B0031–B0034. The patterns of RBS strength as measured here were qualitatively consistent with previously measured values, with a few exceptions. First, we found B0031 was slightly stronger than B0032 rather than vice versa; this was consistent across all promoter strengths. Second, we found that expression from B0034 was usually not much stronger than from B0031, whereas previous reports have shown an order of magnitude difference between these two RBS options. Flow cytometry for library members with strong promoters (Figure 3c) showed that the reason for the unexpected low levels of fluorescence from B0034 is a bimodal population of low-expressing and high-expressing cells. This population split was ultimately due to mutation of promoters in library members with the B0034 RBS due to physiological stress from the high levels of protein expression: while measurements of all library members were done from fresh transformants from sequence-verified DNA, plasmid sequencing for B0034 library transformants exhibiting these bimodal populations returned multiple competing signals indicating that the dual operator sites in R0040 and R0011 had combined to heavily mutate the promoter.
Figure 3d shows that LAA and DAS protein degradation tag can significantly curtail the net expression of a gene and that the LAA tag can do so by three orders of magnitude. We tagged the C-terminus of eGFP with one of two ssrA tags, the strong AANDENYALAA tag or the weaker AANDENYADAS tag. These tags induce sspB-mediated binding to the ClpX and ClpA proteases, which increases a protein’s degradation rate (McGinness et al., 2006). The magnitude of the effect of the LAA tag is as much as or greater than the impact of using the lowest-efficiency ribosomal binding site. Of note is the interaction between expression level and the efficiency of repression via tagging for protein degradation. The LAA tag effectively reduces protein levels even for highly-expressed proteins, whereas the DAS tag is most effective for lowly-expressed proteins (giving a reduction of up to 10-fold; for example, B0033 RBS or weak promoters) compared to highly-expressed proteins (with reductions typically closer to 2-fold; for example, B0031 RBS with a strong promoter). This suggests that overall gene dosage may play a role in the effectiveness of post-transcriptional regulation in some cases, though the mechanism by which this would occur is unclear.
3.5 Carotenoid production can be controlled post-transcriptionally
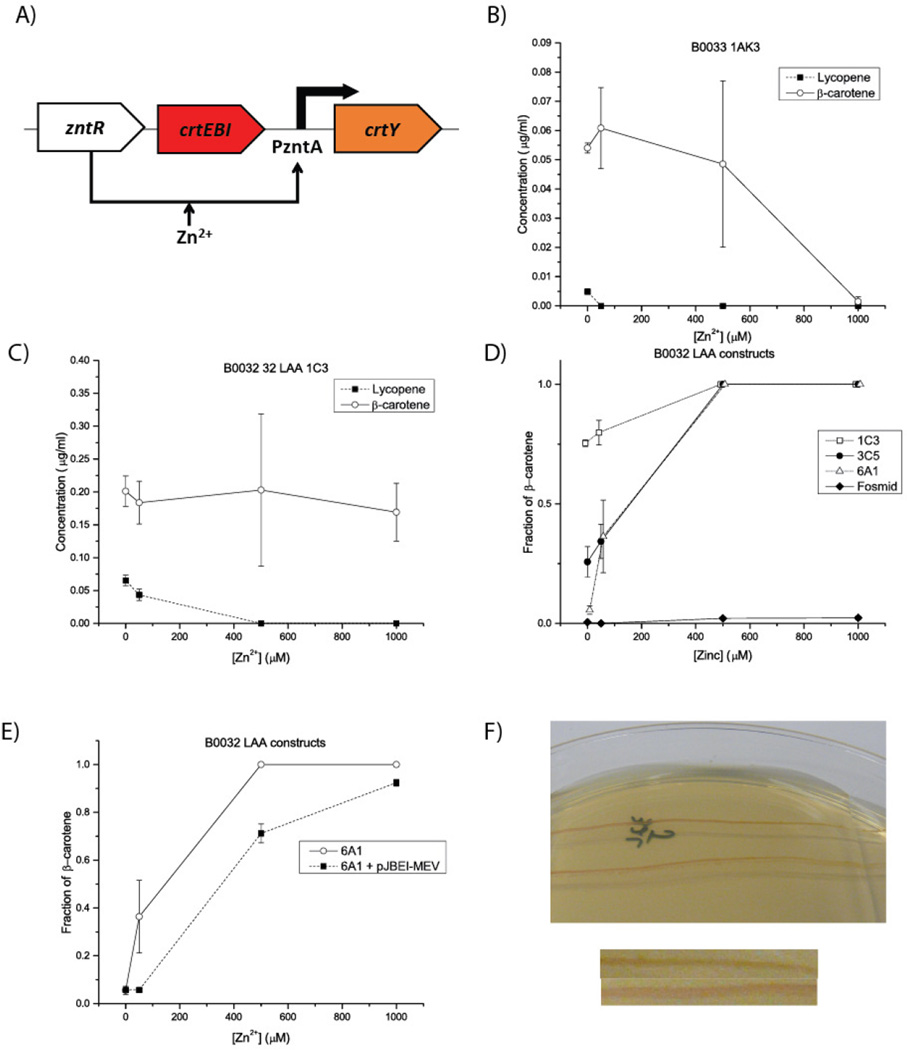
Using this library of regulatory element combinations and the insights gleaned from them, we then assembled thirteen targeted carotenoid-expressing constructs to identify those that could produce two distinct states: one dominated by lycopene and one dominated by β-carotene, which was not observed in our initial attempts. The test constructs consisted of (Figure 4a) crtY being driven by the PzntA promoter and zntR driven by a weak constitutive promoter. crtEBI was cloned in without an explicit promoter (discussed in greater detail in the Discussion and in Supplementary Information S3) so as to yield very low transcription, consistent with previous work where crtEBI had an inducible promoter that was never induced since its expression is detrimental to cell growth (Yoon et al., 2006). Regulatory elements of crtY were varied to control conversion of lycopene to β-carotene.
Figure 4.
Varying regulatory sequences and vector enables a lycopene-dominated state and allows manipulation of the concentration at which it disappears. All error bars represent the standard error of the mean. All measurements were taken at 0, 50, 500, or 1000 µM zinc, though an offset has been applied in panel D for visualization purposes. A) A simplified version of the construct used to test only carotenoid production for a selection of constructs with varied regulatory sequences and vectors. Ribosomal binding sites and degradation tags are omitted for simplicity. B) Changing only to a very weak ribosomal binding site is insufficient to produce a lycopene-only state, and generally insufficient to produce any lycopene at all. This suggests that multiple levels of control must be used. C) Using a moderate ribosomal binding site and a strong degradation tag (LAA) enables the detectable production of lycopene but still does not enable a lycopene-dominated state. D) Changing the vector carrying the construct from panel C enables the presence of a lycopene-only state. For the lowest-copy plasmid (the fosmid), so little CrtY is accumulated even at full zinc induction that only a lycopene-dominated state can be observed. The low-copy 6A1 vector offers two distinct states. E) Co-transforming with a plasmid containing the mevalonate pathway (pJBEI-MEV) to supplement production of lycopene alters the transition point between the lycopene and β-carotene states. At 50 µM Zn2+, the pJBEI-MEV-supplemented cells have still produced orders of magnitude more lycopene than β-carotene, while the non-supplemented strain is already transitioning to a β-carotene state. Of note is that the lycopene production at no supplemented zinc is three times higher in the supplemented strain, as expected. F) Streak of pJBEI-MEV-supplemented strain from panel E on zinc gradient plates showing a fairly distinct transition between states. The cells change from red to orange going left to right. Below the plates are two parts of the same image cropped next to each other to highlight the color differences.
To manipulate levels of CrtY, we selected variants from the fluorescence characterization presented in Figure 3, as well as additional methods for manipulation of expression levels. We varied the RBS of crtY and added the strong (LAA) degradation tag to crtY. We did not explore further the strong B0034 RBS since preliminary pilot experiments indicated we would need such substantial reduction in expression to allow a lycopene-dominated state that RBS variation would likely be necessary. We also added another level of control over the amount of gene expression via variation of vectors, employing high, medium, low, and essentially-single copy vectors. This would titrate not only the total amount of CrtY in the cell from leaky or mildly-induced expression, but also the expression of crtEBI, which also may cause toxicity and could prevent greater accumulation of carotenoids.
Changing only the RBS of crtY was insufficient to yield a distinct lycopene state. Replacing the original B0034 RBS with the B0033 RBS yielded no visible lycopene in an uninduced state (and essentially no measurable lycopene), but a substantial amount of β-carotene (Figure 4b), despite the B0033 RBS being two to three orders of magnitude weaker than the B0034 RBS used in the original construct. (Again, this supports the earlier assessment that the reason why no lycopene-only state could be detected was due to too much crtY expression at low zinc rather than too much vioABCDE expression at high zinc.) Even combining a lower-strength (B0032) RBS with a strong (LAA) degradation tag, which should yield four or more orders of magnitude less CrtY, was not sufficient to allow lycopene to persist in an uninduced state (Figure 4c).
When the vector was varied, substantial amounts of lycopene were produced (Figure 4d). Medium and low copy number plasmids yielded substantial lycopene at low zinc that is repressed at high zinc, and a fosmid yielded such substantial production of lycopene that CrtY could not be induced enough to consume it (yielding a single state in the pathway dominated only by lycopene instead of by β-carotene). The most promising combinations appear to be the low-copy plasmid with a degradation tag and a moderate RBS, yielding a lycopene-dominated state and a β-carotene dominated state, though with less switch-like behavior than expected. Similar behaviors were observed in minimal medium cultures (Supplementary Figure 3).
Worth noting is that in many constructs, the lycopene and/or β-carotene levels decrease at the 1 mM zinc condition. In previous literature, the full induction level of PzntA was found to be at 1.1 mM zinc, and E. coli has been shown to be capable of normal growth in an excess of 1 mM extracellular zinc. We suspect that the additional metabolic stress of the cells from substantial transcription, translation, and pigment synthesis, combined with the toxicity of carotenoid operon genes and potential interference with zinc homeostasis due to overexpression of native regulators, increases the sensitivity of the engineered cells to zinc. Fortunately, this level of zinc is nowhere near that which would be relevant to the ultimate application of this biosensor in a blood micronutrient diagnostic (Hess et al., 2007).
3.6 Enhancing lycopene production affects sensor response
Another alternative to manipulate the apparent response of PzntA-driven expression is to stimulate the overall production of lycopene in the cells. The root cause of a β-carotene-only state is that there is sufficient CrtY to enzymatically act on all of the lycopene that is present. To this point, we had primarily explored just approaches designed to decrease the levels of CrtY. Instead, we could increase the production of lycopene such that there is insufficient CrtY to act on all of the lycopene in the cell. Lycopene production, or more generally the production of carotenoids and isoprenoids, has long been a target for metabolic engineering studies, with significant advances made in terms of overproducing background strains and identification of multiple independent ways to increase carotenoid titers. One well-known approach is the introduction of a mevalonate pathway to supplement the production of precursors for the carotenoid and isoprenoid pathways (Yoon et al., 2006).
By co-transforming engineered carotenoid constructs with a plasmid containing the mevalonate pathway (pJBEI-MEV), we substantially affected the response of the carotenoid-producing sensor (Figure 4e). A moderate RBS combined with a strong degradation tag and a low-copy plasmid initially yielded a system offering a crossover in production from lycopene to β-carotene. At approximately 50 µM of zinc, the levels of lycopene and β-carotene were of the same order of magnitude. However, by supplementing with pJBEI-MEV, production of lycopene was increased threefold, and at 50 µM lycopene levels remained much higher than β-carotene levels. This effectively changed the “switching point” between lycopene and β-carotene states in the supplemented cells. This behavior was observed both in rich medium and in minimal medium. While in this case it moved the switching point perhaps to a level beyond physiological relevance, application of this approach with other constructs allows for another level of manipulation to engineer the precise response desired.
3.7 Engineered distinct red and orange system states
Figure 4f demonstrates that a transition between red and orange states is now possible after substantial pathway engineering. In order for both states to be possible for the same cells, multiple components needed to be precisely controlled in combination. With a moderate RBS, a strong degradation tag, and the low-copy plasmid, levels of CrtY were limited at low zinc levels (preventing reaction of lycopene) but were adequately inducible to allow a switch to β-carotene at higher zinc levels. Selection of a different plasmid in this configuration would have different results: for example, on the the fosmid, CrtY levels are limited so much that the β-carotene state can never be reached (though for other configurations, for example without LAA tags, the fosmid did allow for accumulation of CrtY and production of β-carotene). Supplementation of lycopene production via the mevalonate pathway enabled further tuning of the zinc levels at which that transition occurred. Taken together, these strategies enabled the ultimate within-pathway state-switching goal of the biosensor.
Discussion
In this work, we present the design and engineering of a biosensor capable of responding to extracellular levels of a micronutrient, zinc, with potential application to blood test diagnostics. Specifically, the biosensor was designed to produce pigment, rather than fluorescence or related readouts, to facilitate equipment-free usage since the color of the readout can be assessed with the naked eye. Our initial efforts yielded only a two-state sensor, despite the design of the construct to have three distinct regimes of pigment production. (A two-color sensor would still be useful, though having a third state provides increased zinc status resolution and thus more information for determining when and where to perform nutritional interventions.) This prompted the need for metabolic engineering of the construct, as baseline levels of the last gene in one pigment production pathway prevented the accumulation of an intermediate intended to represent a distinct readout state in the device.
Our work presents advances in a number of different directions within the purview of developing metabolically engineered pigment-based biosensors. We integrated two different heterologous pigment production pathways, and were able to shut one off completely so as to enable the visibility of the other. Our use of natural zinc-responsive promoters and transcription factors provided one system (Zur/PznuC) that was appropriate for our application, but another system (ZntR/PzntA), with a wide dynamic range and excessively high baseline expression when in an uninduced state, required significant engineering to be usable in the sensor. Engineering efforts were limited to components besides the promoter and regulator themselves, as mutating those would likely have been prone to loss of zinc specificity (Khan et al., 2002) (particularly true given the mechanism of action of ZntR). Without promoter engineering as a viable possibility, we focused on post-transcriptional regulation to harness a very small portion of the dynamic range of the system and to sufficiently suppress expression to allow multiple color states within a single pathway. The system started with no switching capability whatsoever, but we were able to engineer in switching capability as well as demonstrate our ability to change the level at where the switch occurs. This is particularly relevant since our application limits us to the small fraction of ZntR’s dynamic range that is physiologically relevant.
We achieved our goals via a directed, rational approach to synthesizing pigment-producing constructs, driven by knowledge gained from initial fluorescence characterization of regulatory elements. We did this due to the difficulty in high-throughput screening for our desired phenotype: screening a library of transformants for a single color at a single zinc concentration does not provide sufficient information, decreasing the potential throughput substantially. Gradient plates as a secondary screen are too qualitative and spatially complex, though a replica plating approach on multiple zinc concentrations could have been a viable strategy. Ultimately, though, our rational approach also provided deeper insight into the requirements for getting the correct level of regulation to allow multiple color states in one pathway, which may not have emerged from a library screen.
To sufficiently control the production of our most downstream enzyme in the carotenoid production pathway, CrtY, multiple levels of regulation were necessary. Combining the use of weaker ribosomal binding sites with protein degradation tags and changing the vector allowed for the establishment of inducible color change and tuning of the zinc concentration at which the color change occurred. While we observed that the lower copy vectors were more effective at allowing a lycopene-dominated state, this phenomenon cannot be solely attributed to copy number since vector-specific differences in readthrough transcription affect the level of transcription of (promoterless) crtEBI and thus the balance of lycopene and crtY. Nonetheless, our data, along with intuition, suggest that copy number likely plays a role in this process; even if not, the fact that selection of vector has such a substantial effect on the metabolic phenotype of the cells is still an important observation.
We used fluorescence measurements on a library of regulatory combinations as the basis for rational exploration of the design space for production of carotenoids. It is possible that the reduction in fluorescence caused by the addition of degradation tags was not just due to increased degradation, but caused also by decreased fluorescence per molecule due to the tag changing the protein conformation. While this was not something controlled for and tempers the interpretations of tag effectiveness from the fluorescence data, the carotenoid production results are consistent with our interpretation of the fluorescence data on the importance of degradation tags for tuning the behavior of the cells.
We also note that while the use of native, rather than orthogonal, sensing (zinc-responsive) elements is not commonplace, it did not prevent successful metabolic engineering of the strain. While this aspect of our approach may have led to more non-linear impacts of the various regulatory changes we implemented due to impacts on host cell behavior or impacts of host cell genomic expression, this disadvantage was outweighed by the many tools available for genetic manipulation in E. coli and the fact that the regulators we used are well-characterized zinc-responsive elements, one of which was already known to respond within a physiologically relevant concentration range.
It is worth noting that these experiments were carried out at a single temperature that is not necessarily the same as the physiological temperature of the organisms used as sources for the heterologous pigment biosynethesis pathways. Since it is known that enzymatic activities of some of the components we have employed are temperature-dependent (i.e., enzymes in the violacein pathway (Rodrigues et al., 2013)), it is possible that some characteristics of our sensor’s behavior may also change as a function of incubation temperature. Exploring the temperature dependence of the switching point of our construct is an important next step, as the results will have design implications for the ultimate implementation of a blood testing device incorporating our sensor.
The desire for tightly controlled but inducible transcription is not unique to our application. For example, there has been recent work developing dynamically controlled metabolic engineering, where the genes for target production remain uninduced during growth phase to avoid toxicity, or where genes crucial for growth are turned off at a specific time to redirect carbon flux towards the production of desired products (Solomon et al., 2012; Soma et al., 2014; Torella et al., 2013; Zalatan et al., 2015). While one way to achieve this would be through small molecule induction after sufficient growth, as might be done for protein expression, there have also been efforts to have automated induction upon sufficient accumulation of cells, for example via response to phosphate limitation or quorum-sensing molecules (Anesiadis et al., 2008; Callura et al., 2012; Kobayashi et al., 2004; Tsao et al., 2010; Williams et al., 2015). These latter cases are the most likely to experience issues similar to the challenges in our biosensor: where the promoter selection is driven by the specific needs of the application and is relatively inflexible, meaning that control of expression must be exerted through other means.
Conclusions
The sensitivity of metabolic pathways to low levels of enzymatic expression – a key aspect of what one could call “precision metabolic engineering” – is not something that has been widely explored in the literature. Nonetheless, it is an extremely important aspect to harnessing metabolism for biosensor applications while minimizing the transcriptional and metabolic burden on cells (and thus using the same pathway to produce multiple different visible outputs). At the outset, we naively implemented the pigment-based sensor without addressing the underlying metabolic pathway engineering problem. It soon became clear that at least some degree of pathway engineering would be critical to make a functional reporter that could express two different carotenoids. While we initially used pathway engineering to allow a lycopene-only intermediate state to persist, we also found that tuning the carotenoid pathway could strongly affect sensor response. Adjusting both upstream and downstream pathway expression controlled the zinc concentration at which pigment production switched, allowing more control over what concentrations would map to ‘intermediate zinc’ and ‘high zinc’ outputs. Overall, the most difficult aspect of the engineering task was compensating for leaky expression of enzymes downstream of the first pigment indicator. This suggests that (when possible) the selection of regulatable promoters with the lowest levels of leakiness may outweigh other important factors (e.g., whether it is inducible or repressible, or the expected response threshold) when designing future pigment-based sensors. However, for natural promoters this will often not be possible, necessitating the precision engineering efforts that we have described. Ultimately, multiple levels of control were necessary to achieve the desired metabolite production given the limitations in inducible regulation and intermediate or product toxicity; we expect that such extensive control will typically be necessary in regulating the response of sensors using metabolite reporters. Perhaps the most interesting lesson for future strain design was that controlling the levels of precursors for the heterologous biosynthetic pathways can affect the switch point of the circuit. This provides a powerful, modular, orthogonal approach to basic pathway engineering of heterologous pigment production pathways, and could be considered an earlier option in strain design if (as in the mevalonate pathway) there are well-established methods to adjust precursor availability to optimize pigment switching around input levels of interest from arbitrary sensors.
The approaches identified and lessons learned could also be generalizable to other “precision” applications where extremely selective production of specific molecules is critical, including the design of multifunctional (potentially portable) microbial cell factories that can produce different products in response to different environmental conditions. A key result of this work is that significant, combinatorial regulation was necessary in order to limit the immediate production of β-carotene in our construct; while it was unsurprising that pigments from two different heterologous pathways could be combined via a switch for a visible output, the level of control necessary to prevent the reaction of all lycopene to β-carotene was unexpected. While the level of control necessary would ultimately be dependent upon enzyme turnover number rate, knowledge that such levels would need to be so precisely regulated for pathway control will likely play a key role in other precision metabolic engineering applications in the future.
Supplementary Material
Highlights.
Metabolic engineering of E. coli to enable tight control of pathway intermediates
Application to carotenoid pathway enables intermediates as biosensor outputs
Useful when application requires specific sensor regulator/promoter pairs
Acknowledgments
The authors acknowledge the Bill & Melinda Gates Foundation for funding support (OPP1046289), as well as the National Science Foundation (1254382). MPM was supported by an NIH training grant (T32-EB0064343). The authors thank Julie Champion and Brian Hammer for providing reagents, and Megan Cole for critical review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ajikumar PK, Xiao WH, Tyo KE, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Gutierrez J, Chan R, Batth TS, Adams PD, Keasling JD, Petzold CJ, Lee TS. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metabolic engineering. 2013;19:33–41. doi: 10.1016/j.ymben.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Alper H, Miyaoku K, Stephanopoulos G. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nature biotechnology. 2005;23:612–616. doi: 10.1038/nbt1083. [DOI] [PubMed] [Google Scholar]
- Anesiadis N, Cluett WR, Mahadevan R. Dynamic metabolic engineering for increasing bioprocess productivity. Metabolic engineering. 2008;10:255–266. doi: 10.1016/j.ymben.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Atsumi S, Cann AF, Connor MR, Shen CR, Smith KM, Brynildsen MP, Chou KJ, Hanai T, Liao JC. Metabolic engineering of Escherichia coli for 1-butanol production. Metabolic engineering. 2008a;10:305–311. doi: 10.1016/j.ymben.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008b;451:86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- Beyer P, Al-Babili S, Ye X, Lucca P, Schaub P, Welsch R, Potrykus I. Golden Rice: introducing the beta-carotene biosynthesis pathway into rice endosperm by genetic engineering to defeat vitamin A deficiency. The Journal of nutrition. 2002;132:506S–510S. doi: 10.1093/jn/132.3.506S. [DOI] [PubMed] [Google Scholar]
- Bhutta ZA, Das JK, Rizvi A, Gaffey MF, Walker N, Horton S, Webb P, Lartey A, Black RE Lancet Nutrition Interventions Review, G., Maternal, Child Nutrition Study, G. Evidence-based interventions for improvement of maternal and child nutrition: what can be done and at what cost? Lancet. 2013;382:452–477. doi: 10.1016/S0140-6736(13)60996-4. [DOI] [PubMed] [Google Scholar]
- Black RE, Allen LH, Bhutta ZA, Caulfield LE, de Onis M, Ezzati M, Mathers C, Rivera J. Maternal and child undernutrition: global and regional exposures and health consequences. Lancet. 2008;371:243–260. doi: 10.1016/S0140-6736(07)61690-0. [DOI] [PubMed] [Google Scholar]
- Black RE, Victora CG, Walker SP, Bhutta ZA, Christian P, de Onis M, Ezzati M, Grantham-McGregor S, Katz J, Martorell R, Uauy R. Maternal and child undernutrition and overweight in low-income and middle-income countries. Lancet. 2013;382:427–451. doi: 10.1016/S0140-6736(13)60937-X. [DOI] [PubMed] [Google Scholar]
- Brocklehurst KR, Hobman JL, Lawley B, Blank L, Marshall SJ, Brown NL, Morby AP. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol Microbiol. 1999;31:893–902. doi: 10.1046/j.1365-2958.1999.01229.x. [DOI] [PubMed] [Google Scholar]
- Callura JM, Cantor CR, Collins JJ. Genetic switchboard for synthetic biology applications. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5850–5855. doi: 10.1073/pnas.1203808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemler JA, Fowler ZL, McHugh KP, Koffas MA. Improving NADPH availability for natural product biosynthesis in Escherichia coli by metabolic engineering. Metabolic engineering. 2010;12:96–104. doi: 10.1016/j.ymben.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Date A, Pasini P, Daunert S. Integration of spore-based genetically engineered whole-cell sensing systems into portable centrifugal microfluidic platforms. Anal Bioanal Chem. 2010;398:349–356. doi: 10.1007/s00216-010-3930-2. [DOI] [PubMed] [Google Scholar]
- Fang MY, Zhang C, Yang S, Cui JY, Jiang PX, Lou K, Wachi M, Xing XH. High crude violacein production from glucose by Escherichia coli engineered with interactive control of tryptophan pathway and violacein biosynthetic pathway. Microbial cell factories. 2015;14:8. doi: 10.1186/s12934-015-0192-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer WR, Liao JC. Precursor balancing for metabolic engineering of lycopene production in Escherichia coli. Biotechnol Prog. 2001;17:57–61. doi: 10.1021/bp000137t. [DOI] [PubMed] [Google Scholar]
- Fischer Walker C, Black RE. Zinc and the risk for infectious disease. Annual review of nutrition. 2004;24:255–275. doi: 10.1146/annurev.nutr.23.011702.073054. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gireesh-Babu P, Chaudhari A. Development of a broad-spectrum fluorescent heavy metal bacterial biosensor. Mol Biol Rep. 2012;39:11225–11229. doi: 10.1007/s11033-012-2033-x. [DOI] [PubMed] [Google Scholar]
- Hess SY, Peerson JM, King JC, Brown KH. Use of serum zinc concentration as an indicator of population zinc status. Food Nutr Bull. 2007;28:S403–S429. doi: 10.1177/15648265070283S303. [DOI] [PubMed] [Google Scholar]
- Jin YS, Alper H, Yang YT, Stephanopoulos G. Improvement of xylose uptake and ethanol production in recombinant Saccharomyces cerevisiae through an inverse metabolic engineering approach. Applied and environmental microbiology. 2005;71:8249–8256. doi: 10.1128/AEM.71.12.8249-8256.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelson JR, Shamberger RJ. Methods compared for determining zinc in serum by flame atomic absorption spectroscopy. Clin Chem. 1978;24:240–244. [PubMed] [Google Scholar]
- Khan S, Brocklehurst KR, Jones GW, Morby AP. The functional analysis of directed amino-acid alterations in ZntR from Escherichia coli. Biochemical and biophysical research communications. 2002;299:438–445. doi: 10.1016/s0006-291x(02)02660-8. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ. Programmable cells: interfacing natural and engineered gene networks. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ME, Aswani A, Han AS, Tomlin CJ, Dueber JE. Expression-level optimization of a multi-enzyme pathway in the absence of a high-throughput assay. Nucleic acids research. 2013;41:10668–10678. doi: 10.1093/nar/gkt809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinness KE, Baker TA, Sauer RT. Engineering controllable protein degradation. Molecular cell. 2006;22:701–707. doi: 10.1016/j.molcel.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Raab AM, Gebhardt G, Bolotina N, Weuster-Botz D, Lang C. Metabolic engineering of Saccharomyces cerevisiae for the biotechnological production of succinic acid. Metabolic engineering. 2010;12:518–525. doi: 10.1016/j.ymben.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MC, Withers ST, Shiba Y, Sarpong R, Keasling JD. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- Rodrigues AL, Trachtmann N, Becker J, Lohanatha AF, Blotenberg J, Bolten CJ, Korneli C, de Souza Lima AO, Porto LM, Sprenger GA, Wittmann C. Systems metabolic engineering of Escherichia coli for production of the antitumor drugs violacein and deoxyviolacein. Metabolic engineering. 2013;20:29–41. doi: 10.1016/j.ymben.2013.08.004. [DOI] [PubMed] [Google Scholar]
- Shetty RP, Endy D, Knight TF., Jr Engineering BioBrick vectors from BioBrick parts. J Biol Eng. 2008;2:5. doi: 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon KV, Sanders TM, Prather KL. A dynamic metabolite valve for the control of central carbon metabolism. Metabolic engineering. 2012;14:661–671. doi: 10.1016/j.ymben.2012.08.006. [DOI] [PubMed] [Google Scholar]
- Soma Y, Tsuruno K, Wada M, Yokota A, Hanai T. Metabolic flux redirection from a central metabolic pathway toward a synthetic pathway using a metabolic toggle switch. Metabolic engineering. 2014;23:175–184. doi: 10.1016/j.ymben.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Torella JP, Ford TJ, Kim SN, Chen AM, Way JC, Silver PA. Tailored fatty acid synthesis via dynamic control of fatty acid elongation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11290–11295. doi: 10.1073/pnas.1307129110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao CY, Hooshangi S, Wu HC, Valdes JJ, Bentley WE. Autonomous induction of recombinant proteins by minimally rewiring native quorum sensing regulon of E. coli. Metabolic engineering. 2010;12:291–297. doi: 10.1016/j.ymben.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Williams TC, Averesch NJ, Winter G, Plan MR, Vickers CE, Nielsen LK, Kromer JO. Quorum-sensing linked RNA interference for dynamic metabolic pathway control in Saccharomyces cerevisiae. Metabolic engineering. 2015;29:124–134. doi: 10.1016/j.ymben.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Wong AP, Gupta M, Shevkoplyas SS, Whitesides GM. Egg beater as centrifuge: isolating human blood plasma from whole blood in resource-poor settings. Lab on a chip. 2008;8:2032–2037. doi: 10.1039/b809830c. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Ishihama A. Transcriptional response of Escherichia coli to external zinc. J Bacteriol. 2005;187:6333–6340. doi: 10.1128/JB.187.18.6333-6340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Al-Babili S, Kloti A, Zhang J, Lucca P, Beyer P, Potrykus I. Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
- Yoon SH, Lee YM, Kim JE, Lee SH, Lee JH, Kim JY, Jung KH, Shin YC, Keasling JD, Kim SW. Enhanced lycopene production in Escherichia coli engineered to synthesize isopentenyl diphosphate and dimethylallyl diphosphate from mevalonate. Biotechnology and bioengineering. 2006;94:1025–1032. doi: 10.1002/bit.20912. [DOI] [PubMed] [Google Scholar]
- Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, Lim WA. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.