Abstract
Background
Chronic venous disease is common and shares some risk factors with venous thromboembolism (VTE). Several genetic loci have been discovered and well-replicated for VTE in European descent populations. We examined associations of a genetic risk score (GRS), comprising known VTE loci, with chronic venous disease.
Methods
The San Diego Population Study (SDPS) is a multi-ethnic cohort that evaluated 2404 men and women aged 29–91 from 1994 – 1998 for chronic venous disease. The current study includes 1447 participants genotyped for 33 variants in 22 established VTE risk loci. Using these variants, unweighted and weighted GRS were constructed. Logistic regression was used to examine associations with venous disease.
Results
In non-Hispanic Whites, African-Americans, Hispanics, and Asians, each standard deviation increment higher of the unweighted 33-SNP GRS was associated with a 1.45-fold (95% CI (1.26, 1.67)), 1.74-fold (1.18, 2.55), a 1.80-fold (1.30, 2.51), and 1.88-fold (1.30, 2.73) greater odds, respectively, for moderate plus severe disease. The difference in c-statistics was significant between a known venous risk factor model and a model adding the 33-SNP GRS for Whites (p=0.008), African-Americans (0.03), and Hispanics (p=0.04), with marginal significance in Asians (p=0.06).
Conclusions
GRS comprising variants primarily from VTE findings in European descent populations were associated with chronic venous disease across all race/ethnic groups, and contributed significantly to prediction, indicating some level of generalizability to other race/ethnic groups. Future work should focus on more in depth examination of racial/ethnic group genetic architecture in relation to chronic venous disease.
Keywords: chronic venous disease, venous thromboembolism, race/ethnicity, genetic risk score, single nucleotide polymorphisms
Introduction
Chronic venous disease is common, with varicose veins affecting up to 25 million people in the US, and 6 million people affected by more advanced venous disease manifesting as trophic changes or ulcers[1]. Venous disease is often overlooked as a chronic condition. However, it can lead to serious complications such as ulcers and edema[1–3], adversely affecting quality of life and mobility[2,4,5]. Prevalence estimates of chronic venous disease can vary quite widely due to differing evaluation methods and definitions[1–3,6–9], with prevalence of varicose veins ranging from 2% to 56% in men and <1% to 73% in various countries around the world[2]. The prevalence estimates for chronic venous insufficiency range from <15 to 17% in men, while the range for women is <1% to 40%[2].
Risk factors for chronic venous disease include age, sex, non-Hispanic White race, smoking, obesity, pregnancy, previous leg injury, time spent sitting or standing, physical inactivity, hypertension, and previous venous thromboembolism (VTE)[7,9–11]. Chronic venous disease is also heritable, as several studies have found family history as a significant risk factor[7,9,12–15]. However, since conditions such as varicose veins are relatively common, familial aggregation studies can easily suffer from misclassification and bias[2].
While several genetic loci have been discovered and replicated for VTE[16–27], primarily in populations of European descent, very little attention has been paid to genes which may contribute to chronic venous disease or whether gene variants related to VTE may also play a role in chronic venous disease. In this regard, chronic venous disease can be a result of post-thrombotic syndrome from a previously unrecognized deep vein thrombosis, and these two conditions share some risk factors[28]. Thus, we examined the association of 33-SNP genetic risk score, comprised of known loci for VTE, with chronic venous disease in a multi-ethnic cohort, the San Diego Population Study (SDPS). We additionally examined the association of a 5-SNP race/ethnic specific genetic risk score with chronic venous disease in this cohort, and sought to determine whether the 33-SNP and 5-SNP genetic risk scores significantly contributed to predictive ability above and beyond known venous risk factors in this multi-ethnic population.
Materials and Methods
Study Participants
The San Diego Population Study (SDPS) is a prospective cohort study of 2404 ethnically diverse men and women aged 29–91 designed to study lower extremity chronic venous disease and peripheral artery disease. Participants residing in San Diego County were chosen randomly within age, race/ethnic and sex strata, resulting in the cohort of approximately 65% women, and 60% that were non-Hispanic White, 13% African-American,15% Hispanic and 12% Asian.
The baseline clinic examination took place in 1994–98 and included evaluation of venous disease. Additional information on demographics, lifestyle factors, family history, personal health history and habits, anthropometrics, comorbidities, lipids and glucose was also collected. Further details of the study design and the clinic examination have been previously published [9,10].
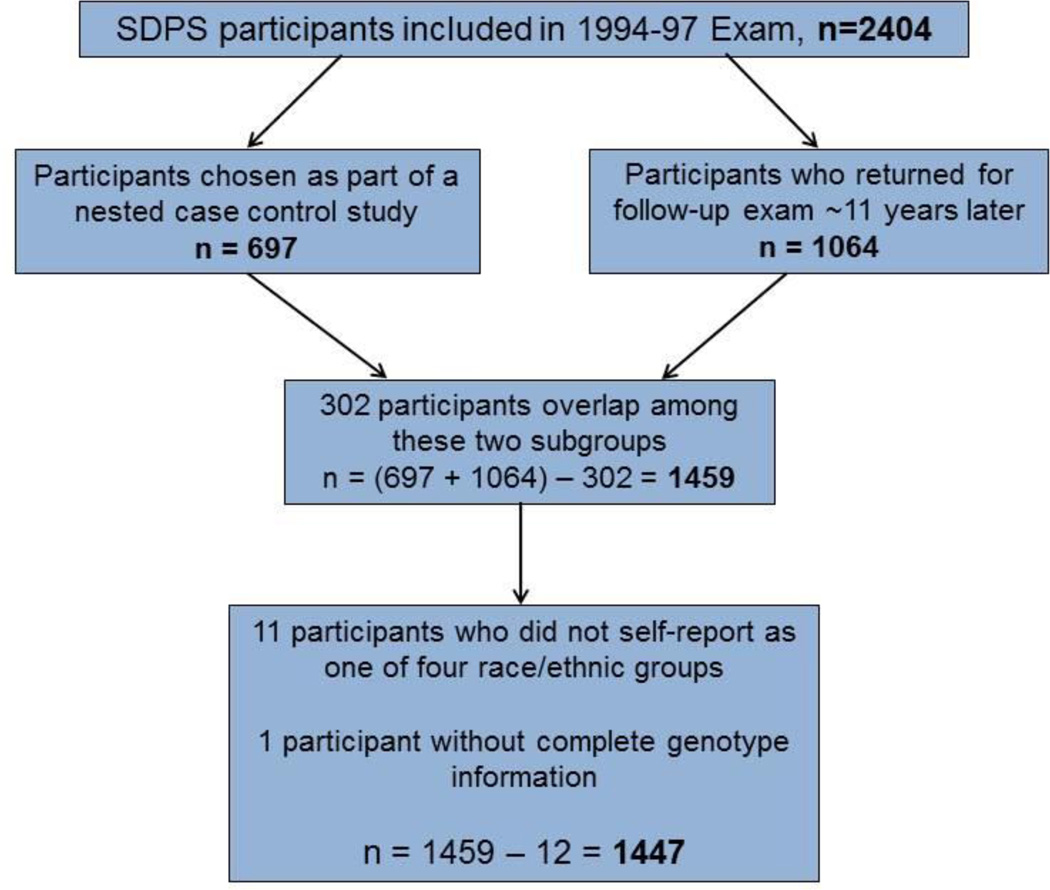
The current study includes a subsample of the original 2404 participants from the baseline exam who had returned for a second follow-up clinic exam approximately 11 years later (n=1064) or who were sampled as part of a nested case control study of peripheral venous disease[28] (n=697) (Figure 1). Participants selected as part of the peripheral venous disease case control study could not have had a previous venous thrombosis[28]. Of these 1761 total participants in the two subsamples, 302 overlapped between the follow-up exam and the nested case control study, leaving 1459 distinct participants with who also had available genotyping for variants previously associated with VTE (Figure 1). Of the 1459 participants, 11 did not self-report as non-Hispanic White, African-American, Hispanic, or Asian and were excluded. One additional participant did not have complete genotyping on all SNPs used in the risk score, leaving a total sample size of 1447 for the current study (Figure 1).
Figure 1.
Figure 1 displays a flow diagram describing inclusions and exclusions from the original study sample size of 2404 participants to the current study sample size of 1447 participants.
All participants provided written, signed informed consent, and the study was approved by the Institutional Review Board at University of California, San Diego.
Single Nucleotide Polymorphism (SNP) selection
Thirty-three SNPs listed in Supplemental Table 1 were chosen from the existing genome-wide association study (GWAS) and candidate gene literature of genetic variants implicated in VTE, i.e. deep vein thrombosis and pulmonary embolism. Studies were largely in participants of European descent, and SNPs were significantly associated with VTE as defined by each study’s criteria.
DNA Extraction and SNP Genotyping
DNA extraction and SNP genotyping was performed at the University of Vermont Laboratory for Clinical Biochemistry Research. DNA was extracted from blood spots on filter paper using a 3mm punch, and eluted DNA was stored at −20C until genotyping. Genotyping was performed using an accurate, flexible, high-throughput genotyping method, the TaqMan 5’ nuclease allelic discrimination assay, on an ABI HT 7900 (Applied Biosystems, Foster City, CA, USA).
Chronic Venous Disease
Chronic venous disease in each leg was assessed by trained vascular technologists using both visual inspection and duplex ultrasound. Participants were categorized into three mutually exclusive disease categories: normal, moderate or severe venous disease. The normal category included participants with no varicose veins or trophic changes (i.e. lipodermatosclerosis, hyperpigmentation, healed ulcer, or active ulcer) upon visual inspection, and no insufficiency or obstruction of the deep or superficial venous systems by duplex ultrasound[9]. The normal category corresponds directly to CEAP classification C0 or C1 with Pn[29]. The moderate disease category included participants with varicose veins or reticular varices in the absence of trophic changes upon visual inspection, or with either insufficiency or obstruction of the superficial system or the perforating veins, but not in the deep system, by duplex ultrasound imaging[9]. The moderate disease category corresponds directly to C2, Ap or An in the CEAP classification[29]. The severe disease category included participants with trophic changes upon visual inspection or insufficiency or obstruction in the deep system by duplex ultrasound imaging[9]. The severe disease category corresponds directly to C4, C5, C6 or Ad in the CEAP classification system[29]. For the current study, we considered venous disease as a combined outcome with moderate plus severe disease, as well as severe only. Further details on the protocol, quality control, and the venous disease classifications for the SDPS have been previously published[9,10].
Covariates
Information on socio-demographics and lifestyle factors, including age, sex, race/ethnicity, smoking, physical activity, and time spent sitting and standing was collected via interviewer-administered questionnaires at the baseline exam. Side-specific information on previous leg injury was obtained via interviewer-administered questionnaire. Prevalent cardiovascular disease, history of previous VTE, and previous hernia surgery were also self-reported at baseline. Participants were asked to self-report flat feet on the study questionnaire. In addition, the standing foot arch height was visually inspected and ranked as flat, small, or normal/high by a trained technologist. Medication use was assessed via medication inventory. Height, weight, and waist circumference were measured using standardized methodology. Body mass index was calculated as the weight in kilograms divided by the height in meters squared. Systolic and diastolic blood pressures were measured in the right arm for each participant after 5 minutes of rest. Prevalent hypertension defined as systolic blood pressure ≥ 140 mmHg, or diastolic ≥ 90 mmHg or use of anti-hypertensive medications. Prevalent diabetes was defined based on self-report or use of anti-diabetic medications, including insulin. Non-fasting total and HDL cholesterol were measured at the Lipid Analytical Laboratory, which is a participant in the ongoing CDC-NHLBI Lipid Standardization Program. The total cholesterol assay was performed on the Abbott VP biochromatic analyzer. The HDL cholesterol procedure utilized heparin-manganese chloride as a precipitant.
Statistical Analysis
Given the sample size constraints and given that the studied SNPs were already known loci for VTE (a related disease) we first used an unbiased approach of constructing a genetic risk score comprising all 33 SNPs. We examined an unweighted score, as well as a weighted score, with each SNP weighted by beta coefficients derived from the literature, primarily from published studies in those of European descent. Next we sought to determine whether a race/ethnic-specific risk score with a reduced number of SNPs would perform better than, or as well as, the 33-SNP score. With this approach, similar to that of deHaan et al[30], the top 5 SNPs associated with moderate plus severe venous disease in each race/ethnic group were selected, regardless of statistical significance threshold. The top 5 SNPs were determined by p-value ranking, i.e. the SNPs with smallest p-values. For the 5-SNP risk score, weights were derived from the beta coefficients from models examining the unadjusted associations of the top 5 SNPs for moderate plus severe venous disease in race/ethnicity stratified models. It should be noted that, given the 5-SNP score is derived from the current study sample, this approach is exploratory. For both the 33-SNP and 5-SNP genetic risk scores, each SNP was assigned a value of 0, 1, or 2 based on the number of alleles conferring risk for VTE in the literature such that the range of possible genetic risk scores for each individual was 0 to 66 for the 33-SNP risk score and 0 to 10 for the 5-SNP risk score. Genetic risk scores only included SNPs not in high LD with each other (r2<0.80) as determined by Haploview V 4.2[31].
Logistic regression was used to examine the associations of the unweighted and weighted genetic risk scores with moderate plus severe, as well as severe venous disease, stratified by race/ethnic group. In order to determine whether the associations of the genetic risk scores with venous disease were linear, we used generalized additive models (GAMs) with a smoother that fits cubic B-splines to the data. Splines indicated no substantial departures from linearity, thus we modeled the genetic risk scores per standard deviation. To determine which venous risk factors should be included in the base model for comparisons with the genetic risk score models, backward stepwise regression was used to create a parsimonious model. Risk factors with p<0.10 in at least one race/ethnic specific model were included for all race/ethnic specific models, resulting in a standard set of covariates across the groups. In order to test whether observed associations were mediated by self-reported history of VTE at the baseline exam, we also adjusted for previous VTE in separate models.
In order to assess whether the genetic risk score contributed significantly to predictive ability and reclassification above and beyond known venous risk factors for moderate plus severe venous disease, we used c-statistics (i.e. area under the curve) and net reclassification improvement (NRI). Given that well-established risk cut points are not available for chronic venous disease, reasonable cut points where chosen for the NRI, including 5, 15, and 25% as well as 5, 10, 15, 20 and 25%.
All analyses were performed in SAS V 9.3 (SAS Institute, Cary, NC).
Results
Participant Characteristics
Overall mean ± SD age of the 1447 participants was 60 ± 11 years, with 64% being female, and 62% White, 11% African-American, 14% Hispanic, and 12% Asian. Approximately 38% of participants in this sample had either moderate or severe venous disease (Table 1). Of the 312 (22%) participants with severe venous disease, 65% were non-Hispanic white, 10% were African-American, 12% Hispanic, and 12% Asian.
Table 1.
Participant Characteristics by Venous Disease*
| No Venous Disease n=893 |
Venous Disease n=554 |
|
|---|---|---|
| Age, years | 59 ± 11 | 61 ± 11 |
| Female Gender, n(%) | 555 (62%) | 367 (67%) |
| Ethnicity, n(%) | ||
| White | 542 (61%) | 356 (64%) |
| Hispanic | 128 (14%) | 79 (14%) |
| African-American | 106 (12%) | 59 (11%) |
| Asian | 117 (13%) | 60 (11%) |
| Ever Smoker, n(%) | 436 (49%) | 260 (47%) |
| Physical Activity, n(%)† | ||
| Less Active | 117 (13%) | 86 (16%) |
| Same Active | 197 (22%) | 123 (23%) |
| More Active | 567 (64%) | 330 (61%) |
| Weight, kg | 73 ± 16 | 76 ± 17 |
| Waist Circumference, cm | 88 ± 14 | 91 ± 15 |
| Body Mass Index, kg/m2 | 26.3 ± 4.9 | 27.5 ± 5.2 |
| Prevalent Hypertension, n(%)‡ | 394 (44%) | 275 (50%) |
| Systolic BP, mmHg | 130 ± 19 | 132 ± 20 |
| Diastolic BP, mmHg | 77 ± 11 | 76 ± 11 |
| Total cholesterol, mg/dL | 209 ± 39 | 212 ± 39 |
| HDL cholesterol, mg/dL | 55 ± 17 | 55 ± 17 |
| Prevalent Diabetes, n(%)‡ | 48 (5%) | 33 (6%) |
| Prevalent Cardiovascular Disease, n(%) | 22 (2%) | 22 (4%) |
| Previous Venous Thromboembolism, n(%) | 8 (1%) | 34 (6%) |
| Previous Hernia Surgery, n(%) | 82 (9%) | 65 (12%) |
| Flat Feet, n(%) | 119 (13%) | 93 (17%) |
| Previous Leg Injury, n(%) | 191 (21%) | 136 (25%) |
| Time spent walking or moving/day§ | 4.9 ± 2.6 | 5.0 ± 2.6 |
| Time spent sitting/day§ | 7.4 ± 2.8 | 7.0 ± 2.7 |
Moderate + severe venous disease, mean ± SD or n(%) presented
Compared to other persons your age would you describe your level of physical activity as less active, more active, or similarly active?
Hypertension defined as systolic blood pressure ≥ 140 mmHg, or diastolic ≥ 90 mmHg or use of anti-hypertensive meds; diabetes defined as use of anti-diabetic medications or casual glucose > 200 mg/dL
Currently, in a 24 hour period
Additional detail on the baseline characteristics for participants with and without moderate or severe venous disease and p-values for univariate differences is provided in Table 1. Age, weight, waist circumference, BMI, systolic and diastolic blood pressure, and time spent sitting per day appeared to differ between those with and without venous disease. A larger percentage of participants with venous disease had hypertension and previous VTE compared with participants without venous disease.
SNP Characteristics
Most SNPs were common (MAF≥5%), but many varied in allele frequency across race/ethnic groups (Supplemental Table 1). Two SNPs, rs6025 (Factor V Leiden) and rs1799963 (F2), were low frequency (1%≤MAF<5%) in non-Hispanic Whites and Hispanics, and monomorphic in African-Americans and Asians. All SNPs were in Hardy Weinberg Equilibrium within all race-ethnic groups (all p>0.01).
Single SNP Associations with Venous Disease
Single SNP associations for the top 5 SNPs with moderate plus severe venous disease by race/ethnic group are shown in Table 2. These SNPs were included in the race-specific 5-SNP genetic risk scores for venous disease. Several loci and SNPs overlapped in their associations with venous disease among the race/ethnic groups, primarily those in PROC, F11, and STXBP5. No single SNP association met a Bonferroni significance criterion (0.05/33=0.0015) in any race/ethnic group; however, the top few SNPs in each race/ethnic group did meet a nominal significance criterion (p<0.05).
Table 2.
Unadjusted Associations for Top 5 Single SNPs Contributing to Race/Ethnicity-specific Genetic Risk Scores
| Gene | Coded/ Non-coded Allele |
Moderate + Severe Venous Disease OR (95% CI) |
p-value | |
|---|---|---|---|---|
| White, n = 898 | ||||
| rs1799810 | PROC | A/T | 1.28 (1.06, 1.54) | 0.01 |
| rs2192824 | TFPI | C/T | 1.23 (1.01,1.50) | 0.04 |
| rs1039084 | STXBP5 | A/G | 1.20 (0.99.1.46) | 0.06 |
| rs670659 | RGS7 | C/T | 1.19 (0.97, 1.45) | 0.09 |
| rs1613662 | GP6 | A/G | 1.26 (0.96,1.64) | 0.09 |
| African-Americans, n = 165 | ||||
| rs3756008 | KLKB1 | A/G | 2.89 (1.09.7.67) | 0.03 |
| rs2036914 | F11 | T/C | 1.62 (0.98, 2.66) | 0.06 |
| rs1800788 | FGB | T/C | 1.83 (0.84, 4.01) | 0.13 |
| rs1799810 | PROC | A/T | 1.39 (0.87, 2.22) | 0.16 |
| rs2069915 | PROC | A/G | 1.38 (0.84, 2.27) | 0.20 |
| Hispanic, n = 207 | ||||
| rs5985 | F13A1 | A/C | 2.11 (1.27, 3.51) | 0.004 |
| rs1039084 | STXBP5 | A/G | 1.52 (1.03, 2.26) | 0.04 |
| rs2289252 | F11 | C/T | 1.48 (0.96, 2.28) | 0.07 |
| rs8176720 | ABO | C/T | 1.32 (0.89, 1.99) | 0.17 |
| rs7853989 | ABO | G/C | 1.70 (0.79, 3.69) | 0.18 |
| Asians, n = 177 | ||||
| rs2227564 | PLAU | C/T | 1.62 (1.02, 2.55) | 0.04 |
| rs1799810 | PROC | A/T | 1.84 (0.98, 3.47) | 0.06 |
| rs1039084 | STXBP5 | A/G | 1.65 (0.97, 2.82) | 0.06 |
| rs253061 | F2R | A/C | 1.48 (0.88, 2.48) | 0.14 |
| rs2069915 | PROC | A/G | 1.38 (0.87, 2.21) | 0.17 |
Association of Race/Ethnic Specific Genetic Risk Scores with Venous Disease
For the unweighted 33-SNP genetic risk score, the mean ± SD was 27.9 ± 4.6 for non-Hispanic whites, 32.1 ± 3.6 for African-Americans, 31.7 ± 4.7 for Hispanics and 35.1 ± 4.3 for Asians. The mean ± SD for the weighted 33-SNP race/ethnic specific genetic risk score was 7.9 ± 1.6 for non-Hispanic whites, 9.1 ± 1.3 for African-Americans, 9.5 ± 1.7 for Hispanics and 8.9 ± 1.3 for Asians. For the unweighted 5-SNP genetic risk score, the mean ± SD was 6.4 ± 1.4 for non-Hispanic whites, 2.8 ± 1.6 for African-Americans, 5.2 ± 1.3 for Hispanics and 7.1 ± 1.6 for Asians. The mean ± SD for the weighted 5-SNP race/ethnic specific genetic risk score was 0.8 ± 0.2 for non-Hispanic whites, 0.5 ± 0.3 for African-Americans, 1.7 ± 0.4 for Hispanics and 1.4 ± 0.4 for Asians.
In models adjusting for known venous disease risk factors, each SD increment higher of the unweighted 33-SNP genetic risk score was significantly associated with a 1.45-fold greater odds of moderate plus severe venous disease in non-Hispanic whites (p<0.001) and a 1.74-fold greater odds in African-Americans (p=0.005), a 1.80-fold greater odds in Hispanics (p<0.001), and a 1.88-fold greater odds in Asians (p=0.001) (Table 3). Results were similarly significant for the association of the weighted 33-SNP risk score with moderate plus severe venous disease (Table 3). The unweighted and weighted race/ethnic specific 5-SNP genetic risk scores were also significantly associated with moderate plus severe venous disease for non-Hispanic whites, Hispanics, and Asians (Table 3). However, the associations of the unweighted and weighted 5-SNP risk score for African-Americans were only marginally significant (p=0.06, p=0.13, respectively). The strength of the association was greater for 5-SNP vs. the 33-SNP scores for Hispanics, but similar between the 33-SNP and 5-SNP genetics risk scores in non-Hispanic whites and Asians. However, the 5-SNP risk score is exploratory in nature and results should be viewed with caution.
Table 3.
Association of Unweighted and Weighted Genetic Risk Scores with Moderate + Severe Venous Disease*
| Unweighted 33-SNP GRS† OR (95% CI) |
p | Weighted 33-SNP GRS† OR (95% CI) |
p | Unweighted 5-SNP GRS† OR (95% CI) |
p | Weighted 5-SNP GRS† OR (95% CI) |
p | |
|---|---|---|---|---|---|---|---|---|
| Whites | 1.45 (1.26, 1.67) | <0.001 | 1.41 (1.23, 1.63) | <0.001 | 1.41 (1.22, 1.64) | <0.001 | 1.41 (1.22, 1.62) | <0.001 |
| African-Americans | 1.74 (1.18, 2.55) | 0.005 | 1.55 (1.06, 2.26) | 0.025 | 1.37 (0.96, 1.95) | 0.08 | 1.31 (0.92, 1.87) | 0.13 |
| Hispanics | 1.80 (1.30, 2.51) | <0.001 | 1.63 (1.18, 2.27) | 0.003 | 2.15 (1.52, 3.04) | <0.001 | 1.75 (1.28, 2.40) | <0.001 |
| Asians | 1.88 (1.30, 2.73) | 0.001 | 1.56 (1.09, 2.22) | 0.01 | 1.88 (1.27, 2.79) | 0.002 | 1.61 (1.10, 2.34) | 0.01 |
In race/ethnicity-stratified models adjusted for age, sex, weight, waist, ever smoking, time sitting, time walking, regular movement, hypertension, flat feet, previous leg injury, previous hernia surgery
Per standard deviation increment of the GRS; the 33 SNP is based on all genotyped SNPs, while the 5-SNP GRS is based on the top 5 associated SNPs within each race/ethnic group
Additional adjustment for previous VTE as a potential mediator did not change the results, nor did removing the n=42 participants with previous VTE from the analysis. When adjusting for previous VTE, the unweighted 33-SNP genetic risk score was associated with a 1.45-fold greater odds of moderate plus severe venous disease (95% CI (1.25, 1.68); p<0.001) in non-Hispanic Whites, 1.69-fold greater odds ((1.14, 2.49); p=0.009) in African-Americans, 1.81-fold greater odds ((1.29, 2.53); p<0.001) in Hispanics, and 1.97-fold greater odds ((1.34, 2.91); p<0.001) in Asians. The unweighted 5-SNP genetic risk score was associated with a 1.40-fold greater odds of moderate plus severe venous disease (95% CI (1.21, 1.63); p<0.001) in non-Hispanic Whites, 1.39-fold greater odds ((0.97, 1.98); p=0.07) in African-Americans, 2.16-fold greater odds ((1.53, 3.05); p<0.001) in Hispanics, and 1.90-fold greater odds ((1.28, 2.81); p=0.002) in Asians. Results were similar when adjusting for previous VTE for the weighted 33-SNP and 5-SNP risk scores (data not shown).
The unweighted and weighted 33-SNP and 5-SNP genetic risk scores were also significantly associated with increased odds of severe venous disease, with the magnitude of the associations for the 33-SNP scores slightly larger for severe disease across race/ethnic groups as compared to moderate plus severe disease (Supplemental Table 2). The associations of the 5-SNP scores were less strong as compared to the 33-SNP risk scores, although in general still statistically significant for severe venous disease (Supplemental Table 2). It should be noted that the 5-SNP risk score is exploratory in nature and results should be viewed with caution.
Prediction and Reclassification of Venous Disease
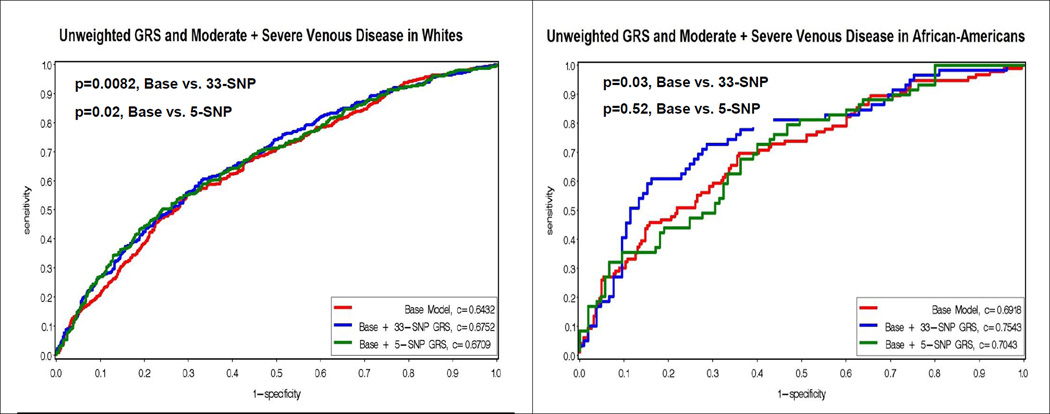
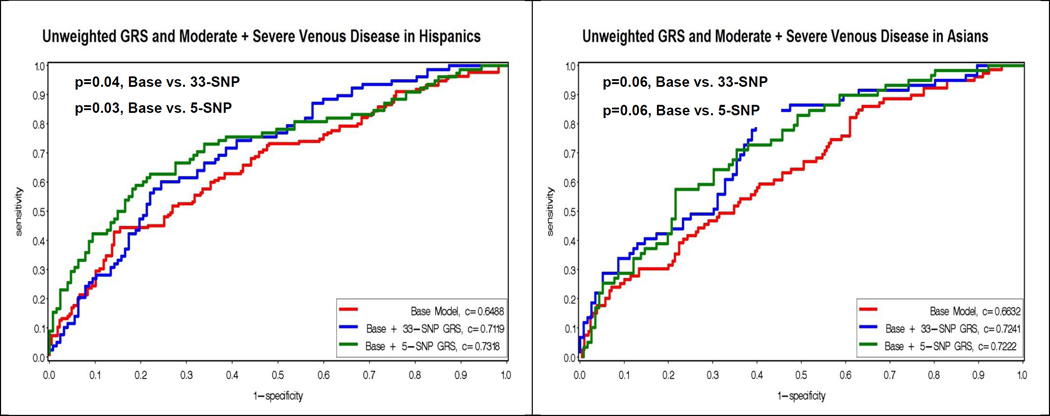
Figure 2 displays the Receiver Operating Characteristic (ROC) curves for a base model with known venous risk factors, and the base model plus the addition of the unweighted 33-SNP and 5-SNP risk scores. The addition of the unweighted 33-SNP genetic risk score resulted in significantly improved c-statistics (i.e. area under the curve) for moderate plus severe venous disease in non-Hispanic whites (0.6432 vs 0.6752, p=0.0082), Hispanics (0.6918 vs 0.7543, p=0.04) and African-Americans (0.6488 vs 0.7119, p=0.03), but was only marginally significant in Asians (0.6632, vs 0.7241, p=0.06). The unweighted race/ethnic specific 5-SNP score added significantly to the area under the curve for non-Hispanic whites (0.6432 vs 0.6709, p=0.02), Hispanics (0.6488 vs 0.7318, p=0.03), and marginally for Asians (0.6632 vs 0.7222, p=0.06), but not African-Americans (0.6918 vs 0.7043, p=0.52).
Figure 2.
Figure 2 displays the Receiver Operating Characteristic (ROC) curves for the unweighted genetic risk scores (GRS) and moderate plus severe venous disease by race/ethnicity. The y-axis corresponds to the sensitivity, and the x-axis corresponds to one minus the specificity. The red curve represents the base model, which contains age, sex, weight, waist, ever smoking, time sitting, time walking, regular movement, hypertension, flat feet, previous leg injury, previous hernia surgery. The blue curve represents the addition of the unweighted 33-SNP GRS to the base model. The green curve represents the addition of the unweighted 5-SNP GRS to the base model. P-values for the comparison of the c-statistics for the curves are presented in the upper left hand corner.
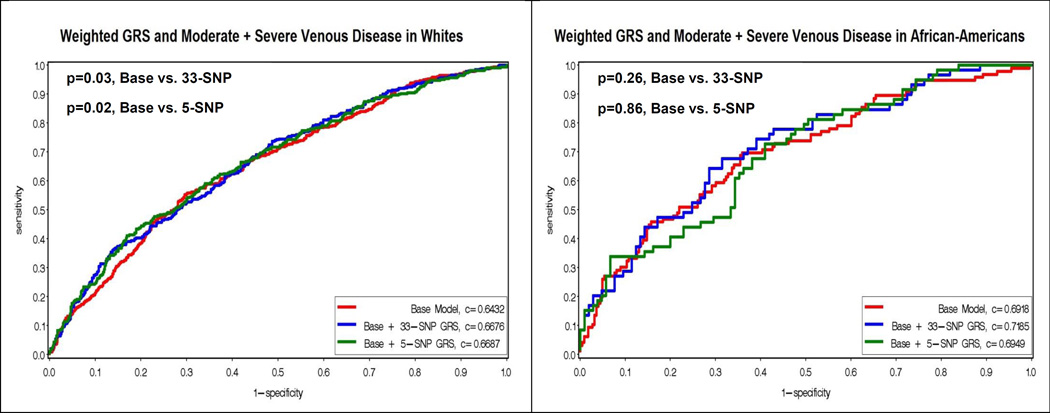
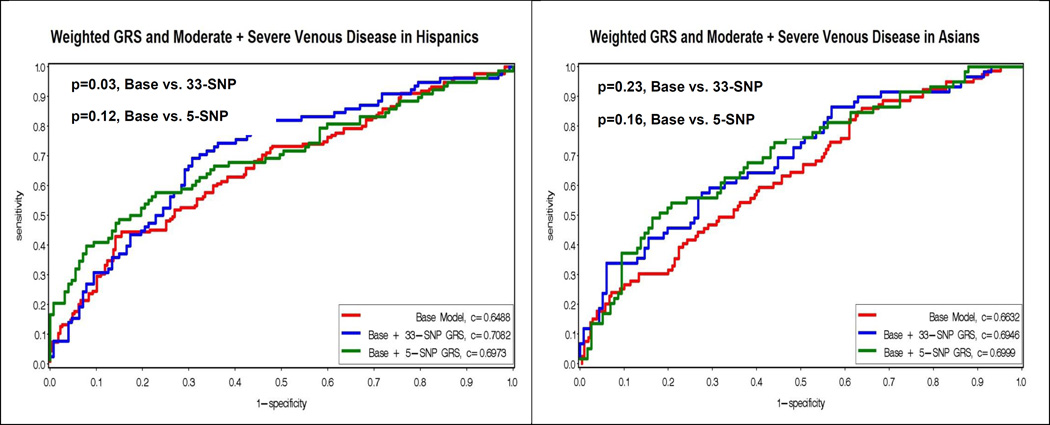
In analyses adding the weighted 33-SNP and 5-SNP risk scores to the base model, the c-statistics were only significantly improved for the non-Hispanic whites (0.6432 vs 0.6676, p=0.03; 0.6432 vs 0.6687, p=0.02, respectively), and for the 33-SNP risk score in Hispanics (0.6488 vs 0.7082, p=0.03) (Figure 3).
Figure 3.
Figure 3 displays the Receiver Operating Characteristic (ROC) curves for the weighted genetic risk scores (GRS) and moderate plus severe venous disease by race/ethnicity. The y-axis corresponds to the sensitivity, and the x-axis corresponds to one minus the specificity. The red curve represents the base model, which contains age, sex, weight, waist, ever smoking, time sitting, time walking, regular movement, hypertension, flat feet, previous leg injury, previous hernia surgery. The blue curve represents the addition of the weighted 33-SNP GRS to the base model. The green curve represents the addition of the weighted 5-SNP GRS to the base model. P-values for the comparison of the c-statistics for the curves are presented in the upper left hand corner.
The net reclassification improvement (NRI) for risk categories of 5, 10, 15, 20, and 25%, as well as 5, 15, and 25% were examined for moderate plus severe venous disease (Supplemental Table 3). Addition of the unweighted 33-SNP and 5-SNP genetic risk scores to models containing known venous risk factors in general resulted in significant reclassification for moderate plus severe venous disease for all race/ethnic groups. The exception was the unweighted 5-SNP score among Hispanics, which was not significant for either the risk categories of 5, 10, 15, 20, and 25% (p=0.40), or for 5, 15, and 25% (p=0.11). The lowest reclassification occurred in non-Hispanic whites and Hispanics, in the range of 5–11% reclassification. More pronounced reclassification was observed for the other race/ethnic groups, in the range of 15–20% net reclassification (Supplemental Table 3). Addition of the weighted 33-SNP and 5-SNP scores resulted in significant reclassification for non-Hispanic whites and African-Americans, but not for Hispanics or Asians (Supplemental Table 3).
Discussion
In an ethnically diverse sample of 1447 participants from the San Diego Population Study, we found that a 33-SNP genetic risk score and a 5-SNP race/ethnic specific risk score, both generated from known SNPs for VTE, were significantly associated with moderate plus severe chronic venous disease, as well as severe disease only. In general, the genetic risk score exhibited stronger associations in Hispanic and Asian participants as compared to non-Hispanic whites and African-Americans. Among African-American participants, the strength of the association of the 33-SNP score was larger than for the race/ethnic-specific 5-SNP risk score, where associations were only marginally significant. A self-reported history of prior VTE before the baseline exam did not appear to mediate the association between the genetic risk scores and chronic venous disease, despite the fact that the loci chosen were known VTE loci, and further exclusion of participants with previous VTE did not change the results. However, the number of previous VTE cases was quite small, especially in race/ethnicity stratified models, so these results should be interpreted with caution. To our knowledge, this is the first study to examine the role of known loci for VTE in chronic venous disease, as well as to examine this in a multi-ethnic cohort.
While there were statistically significant associations of the unweighted and weighted 33-SNP and 5-SNP with chronic venous disease (except in the case of the 5-SNP score in African-Americans), the significance in terms of improving the area under the curve and reclassification was somewhat inconsistent. It appeared that the unweighted 33-SNP and 5-SNP genetic risk scores contributed in some capacity to improving the area under the curve above and beyond known venous risk factors for all race/ethnic groups, while the weighted risk scores only significantly contributed for non-Hispanic whites. Given the SNP selection and derivation of the weights, which were from studies of European descent populations, this consistency for non-Hispanic whites is expected. Overall, the genetic risk scores, particularly in non-Hispanic whites and African-Americans, did appear to also contribute somewhat to better net classification in each race/ethnic group. The reclassification rate was highest in all race/ethnic groups for the unweighted 33-SNP score.
There could be several reasons for the somewhat inconsistent results across unweighted and weighted risk scores, as well as across race/ethnic groups, in terms of associations, predictive ability and reclassification. While associations of the genetic risk scores with chronic venous disease are statistically significant, this does not necessarily translate to improved predictive ability or reclassification. SNPs were selected from the existing GWAS and candidate gene literature of genetic variants implicated in VTE in primarily European descent populations, thus there could be additional SNPs of greater importance for other race/ethnic groups, particularly for African-Americans, due to differences in linkage disequilibrium structure. Finally, the weights for the risk scores, which were drawn from European descent populations, possibly do not translate quite as well to other race/ethnic groups. The unweighted 33-SNP risk score performs the most consistently for significance of associations, predictive ability and reclassification across all race/ethnic groups, indicating at least some generalizability of findings in European descent populations to other race/ethnic groups.
In 7346 participants from the Multiple Environmental Genetic Assessment (MEGA) study, a genetic risk score comprising 31 known VTE SNPs was successful in increasing the AUC for prediction of VTE, increasing the AUC, or c-statistic, from 0.77 to 0.82[30]. We observed a similar increase in c-statistics in our study. Similar results were found in the MEGA study whether using a 31-SNP or a 5-SNP risk score[30]. This genetic risk score was also predictive of recurrent venous thrombosis[32]. Loci and SNPs in the MEGA study genetic risk score that overlapped with the top SNPs in our genetic risk score for chronic venous disease included SNPs in the ABO, FGG, TFPI, STXBP5, RGS7, GP6, PROC, F11, F13A1 and SERPINC1 loci[30]. Furthermore, similar to our study, this genetic risk score also included SNPs that were not statistically significant per se in the MEGA study[30].
We found that well-established loci for VTE, PROC and F11, had SNPs that were well-represented in the top 5 SNPs for chronic venous disease across race/ethnic groups, and thus contributed to the genetic risk scores. We are not aware of other data linking these to venous insufficiency. PROC SNPs rs1799810 and rs2069915 have been well-replicated for VTE in several studies[16,24,25,33,34], and were top SNPs in non-Hispanic whites, African-Americans, and Asians for chronic venous disease in our study. PROC encodes protein C, a protein well known to contribute to etiology of VT; rs1799810 and rs2069915 are SNPs in the promoter region of the gene, and are implicated in protein C deficiency [33,34]. F11 SNPs rs2036914 and rs2289252 have also been well-replicated for VTE[16,17,21,24,25], or the top most significant SNP in the study was in high LD with either rs2036914 or rs2289252[27]. Rs2036914 fell in the top 5 SNPs for African-Americans, while rs2289252 fell in the top 5 SNPs for Hispanics in our study. Austin et al[17], the only study to our knowledge including African-Americans, found that rs2036914 was significantly associated with VTE. This SNP reached nominal significance in in our study in African-Americans (p=0.06) and was included in the genetic risk score for chronic venous disease. F11 encodes coagulation Factor XI, another well recognized risk factor for VTE.
STXBP5, syntaxin binding protein 5, a more recently discovered locus for VTE, was also represented in the top SNPs for non-Hispanic whites, Hispanics and Asians for chronic venous disease in our study. The SNP rs1039084 has been associated with VTE[20,26,30] but is also an important genetic determinant of levels of von Willebrand factor[35]. However, this SNP did not reach genome-wide significance in a large meta-analysis for VTE[27]. Given these findings, it may be that STXBP5 is related to VTE through mechanisms associated with chronic venous disease as an underlying cause of VTE, and not von Willebrand factor.
Our study has several strengths. To our knowledge, other than reports from the SDPS on Factor V Leiden and prothrombin 20210A, where associations with venous disease were not observed[28], this is the first study to examine the associations of a number of known loci for VTE with chronic venous disease, and in a multi-ethnic population. The SDPS has precise and accurate measurement and classification of chronic venous disease based on both functional and visual assessment. Additionally, we have shown that a set of SNPs in aggregate may be important for a closely related condition or disease, even though each SNP individually may only be of nominal significance in the sample.
Our study also has some limitations. Developing and then implementing a genetic risk score in the same study, such as we did with the race/ethnic specific 5-SNP risk score, can lead to bias and, sometimes an overestimation of associations, thus the 5-SNP risk score should be viewed as exploratory and with caution. We did not validate this 5-SNP genetic risk score in another cohort, but to our knowledge there are no currently available multi-ethnic cohorts, or even cohorts of a single ethnicity, with chronic venous disease measured similarly to the SDPS in addition to having these SNPs already genotyped or imputed from a GWAS. However, since we are using SNPs previously associated with and replicated for VTE, a closely related clinical condition, this seems reasonable and not a major weakness. Furthermore, the 33-SNP risk score, which was implemented using all available SNPs genotyped for VTE, as well as weights from the literature, showed similar strength of associations to the 5-SNP score with chronic venous disease in all race/ethnic groups. Thus, it does not appear there is substantial bias for the 5-SNP genetic risk score.
In our study, a 33-SNP genetic risk score and a race-ethnic specific 5-SNP risk score, comprised of known VTE loci, were moderately associated with chronic venous disease, and in the case of the unweighted genetic risk scores, contributed consistently to the predictive ability and net reclassification above and beyond venous risk factors. If confirmed, findings suggest that anticoagulant therapies might have a role in treating or preventing chronic venous disease. Future work should focus on further exploration of the potential overlap of loci for VTE and chronic venous disease in additional, large, genome-wide multi-ethnic studies, as well as examining racial/ethnic group genetic architecture in relation to chronic venous disease through generalization and fine-mapping. Finally, functional studies could better elucidate the role of loci such as STXBP5 in both VTE and chronic venous disease.
Supplementary Material
Highlights.
Genetic loci are well-established risk factors for venous thromboembolism (VTE)
However, these loci were derived primarily from European descent populations
Whether VTE loci contribute to chronic venous disease, a related condition, is unknown
In a multi-ethnic study, a genetic risk score (GRS) associated with venous disease
Addition of this GRS to models increased the prediction for chronic venous disease
Acknowledgements
Funding:
The authors would like to thank the participants of the San Diego Population Study for their contributions, time and effort dedicated to the study. This research was supported by National Institutes of Health–National Heart, Lung, and Blood Institute grant R01 HL53487 to MHC, R01 HL083926 to MC, and National Institutes of Health General Clinical Research Center Program grant M01 RR0827.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eberhardt RT, Raffetto JD. Chronic venous insufficiency. Circulation. 2005 May 10;111(18):2398–2409. doi: 10.1161/01.CIR.0000164199.72440.08. [DOI] [PubMed] [Google Scholar]
- 2.Beebe-Dimmer JL, Pfeifer JR, Engle JS, Schottenfeld D. The epidemiology of chronic venous insufficiency and varicose veins. Ann Epidemiol. 2005 Mar;15(3):175–184. doi: 10.1016/j.annepidem.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 3.Bergan JJ, Schmid-Schonbein GW, Smith PD, Nicolaides AN, Boisseau MR, Eklof B. Chronic venous disease. N Engl J Med. 2006 Aug 3;355(5):488–498. doi: 10.1056/NEJMra055289. [DOI] [PubMed] [Google Scholar]
- 4.Kaplan RM, Criqui MH, Denenberg JO, Bergan J, Fronek A. Quality of life in patients with chronic venous disease: San diego population study. J Vasc Surg. 2003 May;37(5):1047–1053. doi: 10.1067/mva.2003.168. [DOI] [PubMed] [Google Scholar]
- 5.Kahn SR, M'lan CE, Lamping DL, Kurz X, Berard A, Abenhaim LA VEINES Study Group. Relationship between clinical classification of chronic venous disease and patient-reported quality of life: Results from an international cohort study. J Vasc Surg. 2004 Apr;39(4):823–828. doi: 10.1016/j.jvs.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Adhikari A, Criqui MH, Wooll V, Denenberg JO, Fronek A, Langer RD, Klauber M. The epidemiology of chronic venous diseases. Phlebology / Venous Forum of the Royal Society of Medicine. 2000;15(2):3–18. [Google Scholar]
- 7.Fowkes FG, Evans CJ, Lee AJ. Prevalence and risk factors of chronic venous insufficiency. Angiology. 2001 Aug;52(Suppl 1):S5–S15. doi: 10.1177/0003319701052001S02. [DOI] [PubMed] [Google Scholar]
- 8.Evans CJ, Fowkes FG, Ruckley CV, Lee AJ. Prevalence of varicose veins and chronic venous insufficiency in men and women in the general population: Edinburgh vein study. J Epidemiol Community Health. 1999 Mar;53(3):149–153. doi: 10.1136/jech.53.3.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Criqui MH, Denenberg JO, Bergan J, Langer RD, Fronek A. Risk factors for chronic venous disease: The san diego population study. J Vasc Surg. 2007 Aug;46(2):331–337. doi: 10.1016/j.jvs.2007.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Criqui MH, Jamosmos M, Fronek A, Denenberg JO, Langer RD, Bergan J, Golomb BA. Chronic venous disease in an ethnically diverse population: The san diego population study. Am J Epidemiol. 2003 Sep 1;158(5):448–456. doi: 10.1093/aje/kwg166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee AJ, Evans CJ, Allan PL, Ruckley CV, Fowkes FG. Lifestyle factors and the risk of varicose veins: Edinburgh vein study. J Clin Epidemiol. 2003 Feb;56(2):171–179. doi: 10.1016/s0895-4356(02)00518-8. [DOI] [PubMed] [Google Scholar]
- 12.Hirai M, Naiki K, Nakayama R. Prevalence and risk factors of varicose veins in japanese women. Angiology. 1990 Mar;41(3):228–232. doi: 10.1177/000331979004100308. [DOI] [PubMed] [Google Scholar]
- 13.Gourgou S, Dedieu F, Sancho-Garnier H. Lower limb venous insufficiency and tobacco smoking: A case-control study. Am J Epidemiol. 2002 Jun 1;155(11):1007–1015. doi: 10.1093/aje/155.11.1007. [DOI] [PubMed] [Google Scholar]
- 14.Laurikka JO, Sisto T, Tarkka MR, Auvinen O, Hakama M. Risk indicators for varicose veins in forty- to sixty-year-olds in the tampere varicose vein study. World J Surg. 2002 Jun;26(6):648–651. doi: 10.1007/s00268-001-0283-1. [DOI] [PubMed] [Google Scholar]
- 15.Scott TE, LaMorte WW, Gorin DR, Menzoian JO. Risk factors for chronic venous insufficiency: A dual case-control study. J Vasc Surg. 1995 Nov;22(5):622–628. doi: 10.1016/s0741-5214(95)70050-1. [DOI] [PubMed] [Google Scholar]
- 16.Arellano AR, Bezemer ID, Tong CH, Catanese JJ, Devlin JJ, Reitsma PH, Bare LA, Rosendaal FR. Gene variants associated with venous thrombosis: Confirmation in the MEGA study. J Thromb Haemost. 2010 May;8(5):1132–1134. doi: 10.1111/j.1538-7836.2010.03782.x. [DOI] [PubMed] [Google Scholar]
- 17.Austin H, De Staercke C, Lally C, Bezemer ID, Rosendaal FR, Hooper WC. New gene variants associated with venous thrombosis: A replication study in white and black americans. J Thromb Haemost. 2011 Mar;9(3):489–495. doi: 10.1111/j.1538-7836.2011.04185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bezemer ID, Bare LA, Doggen CJ, Arellano AR, Tong C, Rowland CM, Catanese J, Young BA, Reitsma PH, Devlin JJ, Rosendaal FR. Gene variants associated with deep vein thrombosis. JAMA. 2008 Mar 19;299(11):1306–1314. doi: 10.1001/jama.299.11.1306. [DOI] [PubMed] [Google Scholar]
- 19.Bezemer ID, Bare LA, Arellano AR, Reitsma PH, Rosendaal FR. Updated analysis of gene variants associated with deep vein thrombosis. JAMA. 2010 Feb 3;303(5):421–422. doi: 10.1001/jama.2010.57. [DOI] [PubMed] [Google Scholar]
- 20.Germain M, Saut N, Greliche N, Dina C, Lambert JC, Perret C, Cohen W, Oudot-Mellakh T, Antoni G, Alessi MC, Zelenika D, Cambien F, Tiret L, Bertrand M, Dupuy AM, Letenneur L, Lathrop M, Emmerich J, Amouyel P, Tregouet DA, Morange PE. Genetics of venous thrombosis: Insights from a new genome wide association study. PLoS One. 2011;6(9):e25581. doi: 10.1371/journal.pone.0025581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Bezemer ID, Rowland CM, Tong CH, Arellano AR, Catanese JJ, Devlin JJ, Reitsma PH, Bare LA, Rosendaal FR. Genetic variants associated with deep vein thrombosis: The F11 locus. J Thromb Haemost. 2009 Nov;7(11):1802–1808. doi: 10.1111/j.1538-7836.2009.03544.x. [DOI] [PubMed] [Google Scholar]
- 22.Morange PE, Tregouet DA. Lessons from genome-wide association studies in venous thrombosis. J Thromb Haemost. 2011 Jul;9(Suppl 1):258–264. doi: 10.1111/j.1538-7836.2011.04311.x. [DOI] [PubMed] [Google Scholar]
- 23.Reiner AP, Carlson CS, Thyagarajan B, Rieder MJ, Polak JF, Siscovick DS, Nickerson DA, Jacobs DR, Jr, Gross MD. Soluble Pselectin, SELP polymorphisms, and atherosclerotic risk in european-american and african-african young adults: The coronary artery risk development in young adults (CARDIA) study. Arterioscler Thromb Vasc Biol. 2008 Aug;28(8):1549–1555. doi: 10.1161/ATVBAHA.108.169532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reiner AP, Lange LA, Smith NL, Zakai NA, Cushman M, Folsom AR. Common hemostasis and inflammation gene variants and venous thrombosis in older adults from the cardiovascular health study. J Thromb Haemost. 2009 Sep;7(9):1499–1505. doi: 10.1111/j.1538-7836.2009.03522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith NL, Hindorff LA, Heckbert SR, Lemaitre RN, Marciante KD, Rice K, Lumley T, Bis JC, Wiggins KL, Rosendaal FR, Psaty BM. Association of genetic variations with nonfatal venous thrombosis in postmenopausal women. JAMA. 2007 Feb 7;297(5):489–498. doi: 10.1001/jama.297.5.489. [DOI] [PubMed] [Google Scholar]
- 26.Smith NL, Rice KM, Bovill EG, Cushman M, Bis JC, McKnight B, Lumley T, Glazer NL, van Hylckama Vlieg A, Tang W, Dehghan A, Strachan DP, O'Donnell CJ, Rotter JI, Heckbert SR, Psaty BM, Rosendaal FR. Genetic variation associated with plasma von willebrand factor levels and the risk of incident venous thrombosis. Blood. 2011 Jun 2;117(22):6007–6011. doi: 10.1182/blood-2010-10-315473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang W, Teichert M, Chasman DI, Heit JA, Morange PE, Li G, Pankratz N, Leebeek FW, Pare G, de Andrade M, Tzourio C, Psaty BM, Basu S, Ruiter R, Rose L, Armasu SM, Lumley T, Heckbert SR, Uitterlinden AG, Lathrop M, Rice KM, Cushman M, Hofman A, Lambert JC, Glazer NL, Pankow JS, Witteman JC, Amouyel P, Bis JC, Bovill EG, Kong X, Tracy RP, Boerwinkle E, Rotter JI, Tregouet DA, Loth DW, Stricker BH, Ridker PM, Folsom AR, Smith NL. A genome-wide association study for venous thromboembolism: The extended cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium. Genet Epidemiol. 2013 Jul;37(5):512–521. doi: 10.1002/gepi.21731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cushman M, Callas PW, Denenberg JO, Bovill EG, Criqui MH. Risk factors for peripheral venous disease resemble those for venous thrombosis: The san diego population study. J Thromb Haemost. 2010 Aug;8(8):1730–1735. doi: 10.1111/j.1538-7836.2010.03924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eklof B, Rutherford RB, Bergan JJ, Carpentier PH, Gloviczki P, Kistner RL, Meissner MH, Moneta GL, Myers K, Padberg FT, Perrin M, Ruckley CV, Smith PC, Wakefield TW. American Venous Forum International Ad Hoc Committee for Revision of the CEAP Classification. Revision of the CEAP classification for chronic venous disorders: Consensus statement. J Vasc Surg. 2004 Dec;40(6):1248–1252. doi: 10.1016/j.jvs.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 30.de Haan HG, Bezemer ID, Doggen CJ, Le Cessie S, Reitsma PH, Arellano AR, Tong CH, Devlin JJ, Bare LA, Rosendaal FR, Vossen CY. Multiple SNP testing improves risk prediction of first venous thrombosis. Blood. 2012 Jul 19;120(3):656–663. doi: 10.1182/blood-2011-12-397752. [DOI] [PubMed] [Google Scholar]
- 31.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005 Jan 15;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 32.van Hylckama Vlieg A, Flinterman LE, Bare LA, Cannegieter SC, Reitsma PH, Arellano AR, Tong CH, Devlin JJ, Rosendaal FR. Genetic variations associated with recurrent venous thrombosis. Circ Cardiovasc Genet. 2014 Sep 10; doi: 10.1161/CIRCGENETICS.114.000682. [DOI] [PubMed] [Google Scholar]
- 33.Aiach M, Nicaud V, Alhenc-Gelas M, Gandrille S, Arnaud E, Amiral J, Guize L, Fiessinger JN, Emmerich J. Complex association of protein C gene promoter polymorphism with circulating protein C levels and thrombotic risk. Arterioscler Thromb Vasc Biol. 1999 Jun;19(6):1573–1576. doi: 10.1161/01.atv.19.6.1573. [DOI] [PubMed] [Google Scholar]
- 34.Spek CA, Koster T, Rosendaal FR, Bertina RM, Reitsma PH. Genotypic variation in the promoter region of the protein C gene is associated with plasma protein C levels and thrombotic risk. Arterioscler Thromb Vasc Biol. 1995 Feb;15(2):214–218. doi: 10.1161/01.atv.15.2.214. [DOI] [PubMed] [Google Scholar]
- 35.Smith NL, Chen MH, Dehghan A, Strachan DP, Basu S, Soranzo N, Hayward C, Rudan I, Sabater-Lleal M, Bis JC, de Maat MP, Rumley A, Kong X, Yang Q, Williams FM, Vitart V, Campbell H, Malarstig A, Wiggins KL, Van Duijn CM, McArdle WL, Pankow JS, Johnson AD, Silveira A, McKnight B, Uitterlinden AG, Wellcome Trust Case Control Consortium. Aleksic N, Meigs JB, Peters A, Koenig W, Cushman M, Kathiresan S, Rotter JI, Bovill EG, Hofman A, Boerwinkle E, Tofler GH, Peden JF, Psaty BM, Leebeek F, Folsom AR, Larson MG, Spector TD, Wright AF, Wilson JF, Hamsten A, Lumley T, Witteman JC, Tang W, O'Donnell CJ. Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von willebrand factor: The CHARGE (cohorts for heart and aging research in genome epidemiology) consortium. Circulation. 2010 Mar 30;121(12):1382–1392. doi: 10.1161/CIRCULATIONAHA.109.869156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.