Abstract
Blastocystis species has been extensively studied in recent few years to establish its pathogenecity. Present study was designed to identify and examine the association of Blastocystis sp. and its subtypes with Irritable Bowel Syndrome (IBS).Blastocystis sp. detected using wet-mount microscopy, trichrome staining, in-vitro culture and Polymerase Chain Reaction (PCR) assay in a cohort of IBS patients (n = 150) and healthy controls (n = 100). Isolates of Blastocystis sp.were subtyped using Sequence Tagged Site and representative samples were sequenced at SSUrRNA locus.A total of sixty five isolates of Blastocystis sp. were identified [IBS (n = 50); Controls (n = 15)] of which 91% belonged to ST3 and 9% belonged to ST1. No other subtypes could be identified. Statistically significant association was observed between Blastocystis sp. and IBS patients; however no particular subtype could be ascertained to any particular clinical type of IBS.The frequency of occurrence of Blastocystis sp. was more in IBS patients as compared to the controls and ST3 being the most prevalent subtype. The genetic polymorphism of SSU-rRNA gene amongst the different Blastocystis sp.isolates found in this study reinforces the fact that these organisms are genetically highly divergent.
Introduction
Blastocystis species,the non-motile Stramenopile, isolated from human gut [1] has four different morphological forms i.e. vacuolar, granular, ameboid and cystic. Among these, vacuolar is the most commonly isolated from human stool specimens. In less than a decade, 17 different subtypes (STs) of Blastocystis sp. have been described on the basis of Sequence Tagged Sites (STS) analysis on SSU-rRNA locus [2]. Subtypes ST1 to ST9 have so far been isolated from humans [1], and ST3 being the commonest subtype. Role of Blastocystis sp. as an etiology of gastrointestinal disorder has been linked to symptoms like chronic diarrhea, skin lesion and functional gastrointestinal disorder i.e. Irritable Bowel syndrome (IBS)which manifests with symptoms of recurrent abdominal pain associated with changes in bowel habit [3,4] Prevalence of Blastocystis as a causal agent of IBS varies with geographical region ranging from 2.6% to 100% [2,5,6].
In Indian subcontinent, clinical studies on IBS so far have been reported in only two studies with a prevalence of 4% to 4.2% [7,8]. Although four symptom-based clinical subgroups of IBS, such as IBS-C for patients with constipation, IBS-D for patients with diarrhea, IBS-M for patients with alternating diarrhea and constipation and IBS-U for patients with unsubtyped IBS have have been characterized, causal association of different subtypes of Blastocystis sp. in IBS patients was difficult to correlate in this part of the world.
The present prospective study was conducted with an aim to find out the causal association of Blastocystis species with IBS and further subtyping these isolates to extrapolate subtype association with that of different clinical types of IBS.
Materials and Methods
Study design
The present study was a hospital based observational study conducted from December, 2012 to October 2014 at All India Institute of Medical Sciences (AIIMS), a tertiary care health centre.
Patients
Study population (cases) comprised of adult patients satisfying Rome III diagnostic criteria [9] for clinical diagnosis of IBS. A thorough clinical examination and investigation was carried out in all suspected IBS patients and sigmoidoscopy was performed in some to rule out Tuberculer colitis and Inflammatory Bowel disease (IBD).The enrolled patients were further examined and assessed for various factors such as stress disorders, particular food habits, menstrual history and any family history as these are known precipitating factors for IBS. Patients’ positive for infectious agents like Salmonella, Shigella, Campylobacter jejuni and gastrointestinal viruses were excluded from the study.Stool samples obtained from all the enrolled individuals were subjected to wet mount microscopy and samples positive for parasites like Giardia intestinalis, Entamoeba dispar/histolytica, Dientamoeba fragilis and Endolimax nana were excluded from the study. It was also ensured that patients who had a history of taking any anti parasitic drugs three weeks prior to sample collection were not included in the study.
A total of one hundred and fifty (n = 150) adult patients with clinical definition of IBS were enrolled. On the basis of clinical symptoms these patients were further categorized into IBS-D, IBS-C, IBS-M and IBS-U.Individuals above 18yrs of age who had no overt clinical signs and symptoms and any other gastrointestinal disorders after thorough clinical investigations were included as controls (n = 100).
Clinical Samples
Three stool samples on three consecutive days were obtained from both cases and controls. Stool samples were first examined by direct microscopy using both saline and iodine wet-mount preparation.Faecal smears were also prepared and were stained with both Trichrome stain [10] as well as modified acid-fast stain [11] In vitro culture was simultaneously done for all stool samples using Modified Jone’s media [12] and were subcultured at interval of 3 and 5 days. The culture media was kept at least for 10 days before reporting the culture as negative for Blastocystis sp.
DNA Extraction and PCR assay
Genomic DNA was extracted from the stool samples of [cases (n = 150) and controls (n = 100)] using Qiagen DNA stool minikit as per manufacturer’s instructions. For molecular detection of Blastocystis sp. Single round Polymerase Chain Reaction (PCR) assay was carried out using SSU-rDNA as the target gene.BhRDr (5’-GAG CTT TTT AAC TGC AAC AAC G-3’) and RD5 (5’-ATC TGG TTG ATC CTG CCA GTA-3’) were used as forward and reverse primer[13]. Subtyping of Blastocystis sp. was carried out based on Sequence Tagged site (STS) of SSU rRNA gene as described by Yoshikawa et al[14].
Sequencing
Representative samples of Blastocystis sp.obtained from both cases as well as from controls were sequenced at SSU rRNA locus using Big Dye Terminator Chemistry in an automated sequencer ABI prism 310. DNA chromatograms were examined using the BioEdit software versions 7.1.3. The forward and reverse sequences were aligned pair-wise using Clustal W software and were manually refined to obtain a better consensus sequence. The sequences were analysed for subtypes as well as alleles using http://www.pubmlst.org/blastocystis database. Reference sequences from Genbank were used for phylogenetic analyses. The phylogenetic tree was constructed using MEGA6 software. Twenty (n = 20) representative isolates of Blastocystis sp. both from cases (n = 14) and controls (n = 6) were deposited in Genbank (Accession no.KP698119 to KP698138) (Table 1).
Table 1. Blastocystis sp. isolates from IBS patients from the present study and their Genbank Accession numbers.
| Isolate | Host | Accession no. | Subtype | Allele |
|---|---|---|---|---|
| BH11 | Human | KP698119 | ST3 | 34 |
| BH28 | Human | KP698120 | ST3 | 34 |
| BH29 | Human | KP698121 | ST3 | 36 |
| BH49 | Human | KP698122 | ST3 | 36 |
| BH13 | Human | KP698123 | ST3 | 36 |
| BH68 | Human | KP698124 | ST3 | 34 |
| BH71 | Human | KP698125 | ST3 | 34 |
| BH65 | Human | KP698126 | ST3 | 36 |
| BH72 | Human | KP698127 | ST3 | 34 |
| BH75 | Human | KP698128 | ST3 | 36 |
| BH88 | Human | KP698129 | ST3 | 36 |
| BH47 | Human | KP698130 | ST3 | 34 |
| BH55 | Human | KP698131 | ST3 | 36 |
| BH66 | Human | KP698132 | ST3 | 36 |
| BH36 | Human | KP698133 | ST1 | 4 |
| BH23 | Human | KP698134 | ST1 | 4 |
| BH51 | Human | KP698135 | ST1 | 4 |
| BH58 | Human | KP698136 | ST1 | 4 |
| BH59 | Human | KP698137 | ST1 | 4 |
| BH61 | Human | KP698138 | ST1 | 4 |
Ethical
The study was ethically approved by institutional Ethical committee of All India Institute of Medical Sciences. Both informed and written consents were obtained from the patients and controls.
Statistical analysis
Statistical analysis was done using STATA 12.2 software and wherever applicable p value, Odds Ratio and 95% confidence interval was calculated.
Results
Cases and Control cohorts
A total of one hundred fifty (n = 150) cases with a clinical diagnosis of IBS including ninety eight (n = 98) males and fifty two (n = 52) females were enrolled. Controls included eighty six (n = 86) males and fourteen (n = 14) females. The mean age of IBS patients and controls was 38 ± 10.5 years and was 33± 8.7 years, respectively. IBS patients were further categorized to different clinical subtypes. A total of 43% (64/150) belonged to IBS-M type whereas 42% (63/150),14% (21/150), and only 1% (2/150) belonged to IBS-D, IBS-C and IBS-U clinical subtypes respectively.
Study of Blastocystis sp.in clinical samples
Stool wet-mount microscopy showed the presence of Blastocystis sp. in 32% (48/150) of IBS patients compared to 13% (13/100) in the controls. In contrast, modified Trichrome stained smears were positive for Blastocystis spp in 23.7% (42/150) of IBS patients and 8% (8/100) in controls. In-vitro culture showed a positivity of Blastocystis in 30% (45/150) and 2% (2/100) of the IBS patients and controls respectively. However, PCR assay showed positive amplification for Blastocystis sp. 33.3% (50/150) in case of IBS patients and 15% (15/100) in the control group (Table 2; S1 Table).
Table 2. Comparison of laboratory techniques for detection of Blastocystis sp.
| Wet mount Microscopy | Trichrome staining | In vitro culture | PCR assay | |
|---|---|---|---|---|
| IBS patients(n = 150) | 48 (32%) | 42(28%) | 45(30%) | 50(33.34%) |
| Controls (n = 100) | 13(13%) | 8(8%) | 3(3%) | 15(15%) |
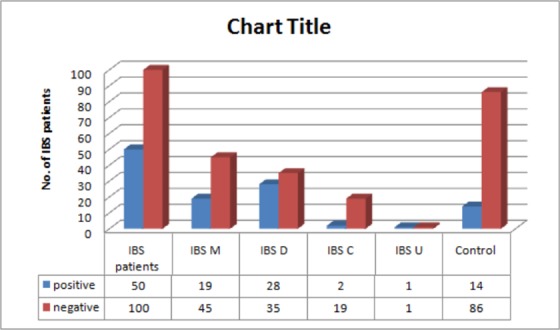
Fig 1. Distribution of Blastocystis sp in clinical types of Irritable Bowel Syndrome.
Subtyping of Blastocystis sp.
All sixty five (n = 65; cases = 50, control = 15) samples positive for Blastocystis sp.by PCR assay were further subtyped. ST 3 was the most common subtype identified in both IBS patients (47/50) as well as in controls (12/15) followed by ST1 (three in both cases and controls) by PCR assay (Table 3; S1 Table). Hence, a total of fifty nine (n = 59) isolates (47 from IBS patients and 12 from controls) belonged to ST3 and only six isolates (3 from IBS patients and 3 from controls) belonged to ST1.
Table 3. Distribution of subtypes in IBS patients and Control group.
| Subtype | IBS patients (n = 50) | Controls (n = 15) |
|---|---|---|
| ST1 | 3 (6%) | 3 (20%) |
| ST3 | 47 (94%) | 12 (80%) |
The frequency of occurrence of ST3 in IBS M and IBS D was 94% (18/19) and 92% (26/28), respectively, followed by ST1 (Table 4; S1 Table). Blastocystis from both IBS C and IBS U belonged to ST3.
Table 4. Distribution of Blastocystis sp subtypes in clinical types of IBS.
| IBS Type | ST1 | ST3 |
|---|---|---|
| IBS M (n = 19) | 1 (6%) | 18 (94%) |
| IBS D (n = 28) | 2 (8%) | 26 (92%) |
| IBS C (n = 2) | - | 2 (100%) |
| IBS U (n = 1) | - | 1 (100%) |
| Total | 3 | 47 |
Sequence analyses
A phylogenetic analysis was done using 21 reference sequences [15] (Table 5). Phylogenetic tree was constructed using neighbor-joining (NJ) and was rooted using Proteromonas lacertae as an outgroup (Fig 2). P. lacertae was chosen as an outgroup since it has shown relationship with Blastocystis sp. in earlier phylogenetic studies [16]. Clusters in the phylogenetic trees were also considered if these had bootstrap support of greater than 50%. Only 20 Blastocystis sp. isolates were randomly chosen and were sequenced at SSU-rRNA locus. Amongst these 14 were subtyped as ST3 and 6 were subtyped as ST1. It was also seen that all ST1 isolates belonged to allele 4, however intra strain variation was observed in ST3. Amongst the 14 ST3 isolates, 8 belonged to allele 36 and 6 belonged to allele 34.
Table 5. GenBank reference sequences used in the construction of phylogenetic tree.
| S. No. | Host | Accession No. |
|---|---|---|
| 1. | Human | AB070989 |
| 2. | Human | AB070987 |
| 3. | Human | AB070997 |
| 4. | Human | AB070988 |
| 5. | Human | AB070990 |
| 6. | Human | AB070991 |
| 7. | Human | AF408427 |
| 8. | Monkey | AB107967 |
| 9. | Monkey | AB070997 |
| 10. | Monkey | AB107969 |
| 11. | Pig | AB107963 |
| 12. | Pig | AB070999 |
| 13. | Pig | AB107964 |
| 14. | Pig | AB070998 |
| 15. | Rat | AB071000 |
| 16. | Rat | AY135408 |
| 17. | Partridge | AB107972 |
| 18. | Quail | AB070995 |
| 19. | Duck | Y135412 |
| 20. | Cattle | AB107965 |
| 21. | Guinea pig | U51152 |
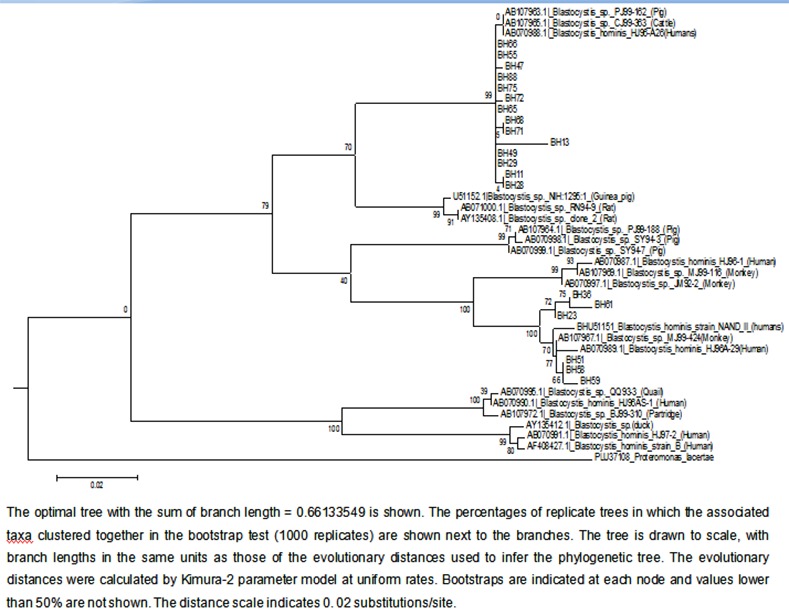
Fig 2. Phylogenetic tree of Blastocystis sp based on SSUrRNA gene sequence.
Discussion
Blastocystis sp.have been established as a probable causal agent in patients suffering from IBS [5], however, the exact pathogenic mechanisms have not been clearly delineated, except some protease enzymes of Blastocystis sp. that cause mucosal disruption and dysbiosis [17]. Most of the studies related to association of Blastocystis sp.in IBS cohorts have been conducted in Middle-East, Southeast Asia and South-America.
In the present study amongst IBS patients, Blastocystis sp.could be detected at a frequency of 32% (48/150), 33.34% (50/150), 28% (42/150) and 30% (45/150) using wet-mount, PCR assay, modified trichrome stain and in-vitro culture respectively. In this study, sensitivity of microscopy was more than in-vitro culture, which is in contrast to studies reported by Tan and Dogruman[1,18]. Relatively higher sensitivity of microscopy in the present study can be attributed to examination of three consecutive stool samples on three consecutive days. Concerning the importance of detection of the Blastocystis sp.and PCR being an objective method has been advocated as the method of choice for the detection [19].
Blastocystis sp.could be detected at a frequency of 33.34% (50/150) and 15% in cases and controls respectively and this difference was statistically significant [p = 0.0016; OR = 2.833; 95% CI = 1.48 to 5.40]. Studies from Turkey [12] Pakistan [20] and Italy [21]have also reported significant association of Blastocystis sp. in IBS patients compared to healthy controls. Nevertheless, contrasting results from Thailand [22] and Mexican studies [23] have shown no difference in the frequency of Blastocystis sp isolation between IBS patients and the controls
Blastocystis sp.from IBS patients in the present study suggests the possible causal role for IBS. Among fifty IBS patients positive for Blastocystis sp, 56% (28/50) belonged to IBS-D followed by 38% (19/50) IBS-M and 4% (2/50) IBS-C type.Upon statistical analysis a significant association between Blastocystis sp. and IBS-D could be made (p = 0.015; OR = 2.36 with 95% CI of 1.16 to 4.72) but not with other clinical subtypes of IBS.Using conventional PCR assay, Yakoob et al. [20], had reported a higher prevalence of Blastocystis sp. in an IBS-D clinical subtypes in Pakistan (44% versus 21% in controls, p<0.001). On the contrary, study by Nourrisson et al.,[24] from France, did not show any The disparity in prevalence of Blastocystis sp.in each clinical subtype of IBS may possibly be explained by the number of IBS patients enrolled in the respective studies.
Blastocystis sp.subtype ST 3 was the most common subtype (92%) in the present study. According to the recent study from India [25], ST3 subtype was found in all 27 normal healthy individuals positive for Blastocystis sp.and only in 2 samples showed mixed infection of both ST1 and ST3 In one of the comprehensive study by Yoshikawa et al., [26] ST3 was the dominant subtype in the four different populations from different countries that included Japan, Bangladesh, Pakistan, and Germany, with a frequency ranging from 41.7% to 92.3%. Study from Columbia has also shown marked association of ST1 being commonly isolated from asymptomatic individuals, ST2 from patients presenting with diarrhea and ST3 exclusively in patients with IBS [27] The difference in the relative diversity and prevalence of subtypes of Blastocystis sp. probably reflects epidemiological and demographic differences including climatic conditions, geographical attributes, cultural habits, proximity and exposure to reservoir hosts, and mode of transmission [28] Further, ST3 was relatively common with IBS D (56%, 26/47) followed by IBS M (38%, 18/47) and IBS C (4%, 2/47). Subtype association of Blastocystis sp in IBS patients was not statistically significant [p = 0.7; OR = 0.61; 95% CI = 0.05 to 7.30]. In contrast, a study from neighbouring country Pakistan showed association between ST1 and IBS-D clinical subtype [20].
Phylogenetic studies of Blastocystis sp. based on SSU rRNA [14,16,29,30]) suggested low host specificity of Blastocystis sp. and also the zoonotic potential of the parasite. Molecular phylogenetic study was done to determine the extent to which the parasite can be transmitted among host species and the potential reservoir for infection in humans.
All the Blastocystis sp. SSUrDNA sequences included in the analysis were edited to include only the region under study, and phylogenetic analysis was based on 540 bp barcode region located on SSUrRNA locus, since the complete SSU-rDNA sequence of Blastocystis sp. is not required for assignment of an isolate to its appropriate subtype [13]. Alignment of SSUrRNA sequences of our isolates were compared with those of already reported Blastocystis sp. isolates from human and animal origin and it showed two different clades with a good bootstrap support. Minor differences in the branch order among the subgroups were observed. Isolates subtyped as ST3 (n = 14) were clustered together along with Blastocystis sp. PJ99-162 isolate from pig, Blastocystis sp. CJ99-363 isolate from cattle and Blastocystis sp. isolate from humans. Similary isolates subtyped as ST1 formed a cluster with Blastocystis sp. Strain NAND II from humans, Blastocystis sp. MJ99-424 from monkeys and Blastocystis sp. HJ96A-29 from humans. The phylogenetic tree shows that the same subtypes identified in the present study clustered together, where as the two subtypes clustered as two independent monophyletic groups. Isolate BH13 showed sequence variation at positions 47 to 49 and 52 to 56, and it formed a separate branch in the ST3 cluster of the phylogenetic tree.
Hereby it was clear that Blastocystis sp. subtypes 1 and 3 were shared by isolates from varied hosts and its cross-infectivity among different hosts. The findings clearly show that close association between animals and humans isolates exists, and that the animals can facilitate the transmission of Blastocystis sp. This has been showed by previous studies [16] also that no exclusive ‘human’ clade seems to exist because sequences from human isolates are present in all clades
Conclusion
To best of our knowledge, this is the first molecular study showing correlation of Blastocystis sp. and IBS patients from India. The frequency of occurrence of Blastocystis sp. was highest in IBS-D clinical subtype. The genetic polymorphism of SSU-rRNA gene amongst the different Blastocystis sp.isolates found in this study revealed that these organisms are genetically highly divergent. Among the two identified subtypes, ST3 was predominant. The pathogenic potential of ST3 could not be elucidated in the present study and further studies in this regard will extrapolate various unresolved issues.
Supporting Information
(XLSX)
Acknowledgments
The authors would like to thank Dr. Christen Rune Stensvold, Ph.D, Senior Scientist of Laboratory for Parasitology, Department of Microbiology and Infection Control, Statens Serum Institut, Copenhagen, Denmark for providing the positive control ie. DNA of Blastocystis sp.
Data Availability
All relevant data are within the paper and its Supporting Information files. All sequencing files are available from the NCBI database (accession numbers KP698119 to KP698138).
Funding Statement
The work was supported by All India Institute of Medical sciences as a part of MD thesis of Dr. Rojaleen Das.
References
- 1.Tan KS. 2008. New insights on classification, identification, and clinical relevance of Blastocystis sp. Clin Microbiol Rev 21(4): 639–65. 10.1128/CMR.00022-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfellani MA, Stensvold CR, Vidal-Lapiedra A, Onuoha ES, Fagbenro-Beyioku AF, Clark CG. 2013. Variable geographic distribution of Blastocystis subtypes and its potential implications. Acta Trop 126(1):11–8. 10.1016/j.actatropica.2012.12.011 [DOI] [PubMed] [Google Scholar]
- 3.Armentia A, Mendez J, Gomez A, Sanchis E, Fernandez A, de la Fuente R et al. 1993. Urticaria by Blastocystis hominis. Successful treatment with paromomycin. Allergologia et immunopathologia 21(4): 149–51. [PubMed] [Google Scholar]
- 4.Gupta R, Parsi K. 2006. Chronic urticaria due to Blastocystis hominis. Australasian J Dermatol 47(2):117–9. [DOI] [PubMed] [Google Scholar]
- 5.Boorom KF, Smith H, Nimri L, Viscogliosi E, Spanakos G, Parkar U et al. 2008. Oh my aching gut: irritable bowel syndrome, Blastocystis, and asymptomatic infection. Parasite Vectors 1(1): 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Safadi D, Gaayeb L, Meloni D, Cian A, Poirier P, Wawrzyniak I et al. 2014. Children of Senegal River Basin show the highest prevalence of Blastocystis sp. ever observed worldwide. BMC Infect Dis 14:164 10.1186/1471-2334-14-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghoshal UC, Abraham P, Bhatt C, Choudhuri G, Bhatia SJ, Shenoy KT et al. 2008. Epidemiological and clinical profile of irritable bowel syndrome in India: report of the Indian Society of Gastroenterology Task Force. Indian J Gastroenterol 27(1): 22–8. [PubMed] [Google Scholar]
- 8.Makharia GK, Verma AK, Amarchand R, Goswami A, Singh P, Agnihotri A et al. 2011. Prevalence of irritable bowel syndrome: a community based study from northern India. J Neurogastroenterol Motil. 17(1): 82–7. 10.5056/jnm.2011.17.1.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drossman DA. 2006. Functional versus organic: an inappropriate dichotomy for clinical care. Am J Gastroenterol 101(6):1172–5. [DOI] [PubMed] [Google Scholar]
- 10.Wheatley WB. 1951. A rapid staining procedure for intestinal amoebae and flagellates. Am J Clin Path 21: 990–991. [DOI] [PubMed] [Google Scholar]
- 11.Casemore DP, Armstrong M, Sands RL. 1985. Laboratory diagnosis of cryptosporidiosis. J Clin Pathol 38(12): 1337–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dogruman-Al F, Simsek Z, Boorom K, Ekici E, Sahin M, Tuncer C et al. 2010. Comparison of methods for detection of Blastocystis infection in routinely submitted stool samples, and also in IBS/IBD Patients in Ankara, Turkey. PLoS One 5(11): e15484 10.1371/journal.pone.0015484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scicluna SM, Tawari B, Clark CG. 2006. DNA barcoding of blastocystis. Protist 157(1): 77–85. [DOI] [PubMed] [Google Scholar]
- 14.Yoshikawa H, Wu Z, Nagano I, Takahashi Y. 2003. Molecular comparative studies among Blastocystis isolates obtained from humans and animals. J Parasitol 89:585–594. [DOI] [PubMed] [Google Scholar]
- 15.Rivera WL. 2008. Phylogenetic analyses of Blastocystis isolates from human and animal hosts in the Phillipines. Vet Parasitol 156(3–4): 178–82. 10.1016/j.vetpar.2008.06.001 [DOI] [PubMed] [Google Scholar]
- 16.Noel C, Peyronnet C, Gerbod D, Edgcomb VP, Delgado-Viscogliosi P, Sogin ML et al. 2003. Phylogenetic analysis of Blastocystis isolates from different hosts based on the comparison of small-subunit rRNA gene sequences. Mol Biochem Parasitol 126: 119–123. [DOI] [PubMed] [Google Scholar]
- 17.Poirier P, Wawrzniak I, Vivares CP, Delbac F, El Alaoui H. 2012. New insights into Blastocystis sp.: A potential link with Irritable Bowel Syndrome. Plos Path 8(3): e1002545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dogruman-Al F, Kustimur S, Yoshikawa H, Tuncer C, Simsek Z, Tanyuksel M et al. 2009. Blastocystis subtypes in irritable bowel syndrome and inflammatory bowel disease in Ankara, Turkey. Mem Inst Oswaldo Cruz 104(5):724–7. [DOI] [PubMed] [Google Scholar]
- 19.Stensvold R., Brillowska-Dabrowska A., Nielsen H. V., and Arendrup M. C. (2006). Detection of Blastocystis hominis in unpreserved stool specimens by using polymerase chain reaction. J Parasitol 92, 1081–7. [DOI] [PubMed] [Google Scholar]
- 20.Yakoob J, Jafri W, Beg MA, Abbas Z, Naz S et al. 2010. Irritable bowel syndrome: is it associated with genotypes of Blastocystis hominis. Parasitol Res 106(5): 1033–8. 10.1007/s00436-010-1761-x [DOI] [PubMed] [Google Scholar]
- 21.Giacometti A, Cirioni O, Fiorentini A, Fortuna M, Scalise G. 1991. Irritable bowel syndrome in patients with Blastocystis hominis infection. Eur J Clin Microbiol Infect Dis 18(6): 436–9. [DOI] [PubMed] [Google Scholar]
- 22.Surangsrirat S, Thamrongwittawatpong L, Piyaniran W, Naaglor T, Khoprasert C, Taamasri P et al. 2010. Assessment of the association between Blastocystis infection and irritable bowel syndrome. J Med Assoc Thai 93(6): S119–24. [PubMed] [Google Scholar]
- 23.Ramirez-Miranda ME, Hernandez-Castellanos R, Lopez-Escamilla E, Moncada D, Rodriguez-Magallan A,Pagasa-Melero C et al. (2010) Parasites in Mexican patients with irritable bowel syndrome: a case-control study. Parasit Vectors 3:96 10.1186/1756-3305-3-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nourrisson C, Scanzi J, Pereira B, NkoudMongo C, Wawrzyniak I, Cian A et al. 2014. Blastocystis Is Associated with Decrease of Fecal Microbiota Protective Bacteria: Comparative Analysis between Patients with Irritable Bowel Syndrome and Control Subjects. PLoS One 9(11): e111868 10.1371/journal.pone.0111868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandey PK, Verma P, Marathe N, Shetty S, Bavdekar A, Patole MA et al. 2015. Prevalence and subtype analysis of Blastocystis in healthy Indian Individuals. Infect Genetics Evol 31: 296–299. [DOI] [PubMed] [Google Scholar]
- 26.Yoshikawa H, Dogruman-Al F, Turk S, Kustimur S, Balaban N, Sultan N. 2011. Evaluation of DNA extraction kits for molecular diagnosis of human Blastocystis subtypes from fecal samples. Parasitol Res 109(4): 1045–50. 10.1007/s00436-011-2342-3 [DOI] [PubMed] [Google Scholar]
- 27.Ramírez JD, Sánchez LV, Bautista DC, Corredor AF, Flórez AC, Stensvold CR. 2014. Blastocystis subtypes detected in humans and animals from Colombia. Infect Genetics Evol 22: 223–8. [DOI] [PubMed] [Google Scholar]
- 28.Stensvold CR. 2013. Blastocystis: Genetic diversity and molecular methods for diagnosis and epidemiology. Trop Parasitol 3(1):26-34Abe N. 2004. Molecular and phylogenetic analysis of Blastocystis isolates from various hosts . Vet Parasitol 120: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abe N. 2004. Molecular and phlogenetic analysis of Blastocystis isolates from various hosts. Vet Prasitology 120:235–242. [DOI] [PubMed] [Google Scholar]
- 30.Arisue N, Hashimoto T, Yoshikawa H. 2003. Sequence heterogeneity of the small subunit ribosomal RNA genes among Blastocystisisolates. Parasitol 126:1–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All sequencing files are available from the NCBI database (accession numbers KP698119 to KP698138).


