Abstract
The emergence of multidrug-resistant bacterial strains has heightened the need for new antimicrobial agents based on novel chemical scaffolds that are able to circumvent current modes of resistance. We recently developed a whole-animal drug-screening methodology in pursuit of this goal and now report the discovery of 3-(phenylsulfonyl)-2-pyrazinecarbonitrile (PSPC) as a novel antibacterial effective against resistant nosocomial pathogens. The minimum inhibitory concentrations (MIC) of PSPC against Staphylococcus aureus and Enterococcus faecium were 4 μg/ml and 8 μg/ml, respectively, whereas the MICs were higher against the Gram-negative bacteria Klebsiella pneumoniae (64 μg/ml), Acinetobacter baumannii (32 μg/ml), Pseudomonas aeruginosa (>64 μg/ml), and Enterobacter spp. (>64 μg/ml). However, co-treatment of PSPC with the efflux pump inhibitor phenylalanine arginyl β-naphthylamide (PAβN) or with sub-inhibitory concentrations of the lipopeptide antibiotic polymyxin B reduced the MICs of PSPC against the Gram-negative strains by >4-fold. A sulfide analogue of PSPC (PSPC-1S) showed no antibacterial activity, whereas the sulfoxide analogue (PSPC-6S) showed identical activity as PSPC across all strains, confirming structure-dependent activity for PSPC and suggesting a target-based mechanism of action. PSPC displayed dose dependent toxicity to both Caenorhabditis elegans and HEK-293 mammalian cells, culminating with a survival rate of 16% (100 μg/mL) and 8.5% (64 μg/mL), respectively, at the maximum tested concentration. However, PSPC did not result in hemolysis of erythrocytes, even at a concentration of 64 μg/mL. Together these results support PSPC as a new chemotype suitable for further development of new antibiotics against Gram-positive and Gram-negative bacteria.
Keywords: Staphylococcus aureus, MRSA, ESKAPE, C. elegans, high throughput screening, liquid infection assay, efflux pump inhibition, synergism
Graphical abstract
Antibiotic resistance is a looming health crisis as pathogens gain increased resistance to our current arsenal of clinically useful drugs.1 The “golden age” of antibiotic drug discovery in the previous century revolutionized health care and together with vaccination contributed to the steep decline in infectious disease associated mortality, especially in developed countries.2 Since then, further developments in treatment options have mostly relied on incremental improvements of existing structural classes of drugs.3 Meanwhile, rampant overuse of antibiotics in both humans and agriculture has resulted in bacterial populations that are resistant to even the most powerful drugs available, many of which were meant to be used as last resort drugs partly due to their superior activity and their toxic side effects.4 Therefore, it is apparent that new structural classes of drugs and new treatment strategies are urgently needed.
We previously reported the development of a Caenorhabditis elegans-based whole animal infection model for simultaneous high throughput in vivo screening for compounds that are both efficacious against methicillin resistant S. aureus (MRSA) and which exhibit low levels of eukaryotic toxicity.5 In the current study, we used a C. elegans-MRSA assay to screen 21,472 small molecules supplied by Boehringer Ingelheim GmbH (BI) and identified several hits that prolonged the survival of infected worms. In this study we have characterized the antimicrobial properties of the novel compound 3-(phenylsulfonyl)-2-pyrazinecarbonitrile (PSPC). PSPC displays strong antimicrobial activity against Gram-positive bacteria and is also effective against Gram-negative bacteria, especially in combination with an efflux pump inhibitor or lipopeptide antibiotic. In vivo infection studies in C. elegans showed that PSPC is comparable to vancomycin in prolonging survival of MRSA infected worms at a concentration as low as 0.78 μg/mL, though it is also toxic to nematodes at high concentrations. PSPC does not cause hemolysis of erythrocytes but displays cytotoxicity to mammalian cell lines causing almost 69% mortality at a concentration of 4 μg/mL. These findings support the need for further studies to improve the antibacterial potency and diminish the toxicity of PSPC before in vivo testing in mammalian infection models.
Identification of PSPC as a hit compound that prolongs survival of C. elegans infected with MRSA
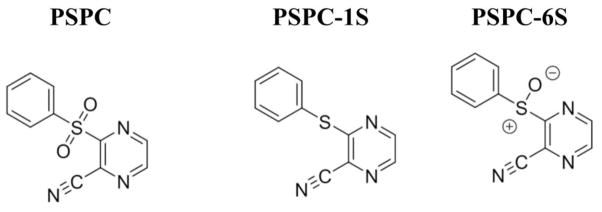
The C. elegans-MRSA liquid infection assay is a well-established high throughput screening platform that allows simultaneous assessment of both the toxicity and antibacterial efficacy of test compounds in 384-well formats.5–7 The end point of the assay is to measure the survival of MRSA-infected worms that have been treated with test compounds. This liquid infection assay was used to identify antibacterial hits from a library of 21,472 pre-selected compounds provided by Boehringer-Ingelheim GmbH (BI). The compounds were pin-transferred to 384-well plates at a final assay concentration of 20 μg/mL. Each sample plate contained 16 wells each of positive control (vancomycin at 10 μg/mL in 1% DMSO) and negative control (1% DMSO). The percent survival of infected worms treated with library compounds was determined at the end of the assay and a Z score was calculated for each well to identify hits. Of the 21,472 compounds tested, 318 (1.4% hit rate) protected worms from MRSA-mediated death. Among the hits, PSPC (Fig. 1) was chosen for follow up studies because the compound is a novel, low molecular weight scaffold that has not previously been reported to have antibacterial activity.
Figure 1.
Chemical structures of 3-(Phenylsulfonyl)-2-pyrazinecarbonitrile (PSPC) and its sulfide (PSPC-1S) and sulfoxide (PSPC-6S) analogues.
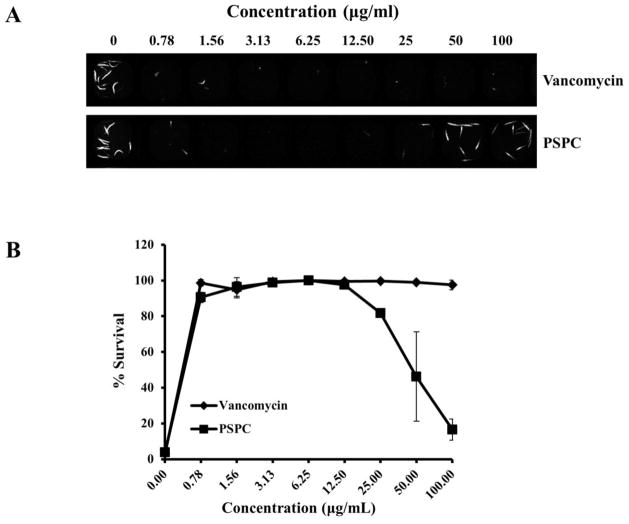
To confirm the activity of PSPC, the compound was resynthesized (see Supporting Information) and the C. elegans-MRSA liquid infection assay was repeated using serial dilutions in the range of 0.78–100 μg/mL. The activity of the resynthesized PSPC was compared to vancomycin over the same concentration range. At the lowest concentration, both PSPC and vancomycin were effective in prolonging survival of more than 90% of infected worms (Fig. 2). However, at the higher concentrations of 50 and 100 μg/mL, worms treated with PSPC showed only 46% and 16% survival, respectively, whereas vancomycin-treated worms showed >97% survival at these concentrations. These findings suggest that although PSPC ameliorates MRSA-associated lethality in worms at low concentration, the compound is toxic to the worms at higher concentrations. The concentration of DMSO in all sample wells did not exceed 1% DMSO, the concentration of DMSO in the negative control, and therefore, the toxicity of PSPC at higher concentrations is not due to DMSO.
Figure 2. PSPC prolongs survival of C. elegans infected with S. aureus (MRSA).
(A) 384-well assay plate was co-inoculated with young adult stage C. elegans glp-4(bn2);sek-1(km4) animals, MRSA strain MW2, and either DMSO (negative control), or 2-fold serial dilutions (0.78–100 μg/mL) of vancomycin (positive control) or PSPC. (A) Sytox Orange-stained and brightfield images of assay wells containing serial dilutions of the drugs. Dead worms take up the vital dye Sytox Orange and fluoresce.
(B) Percent survival of infected worms was calculated from the ratio of fluorescent worm areas to the total area occupied by worms in a well.
In vitro antibacterial activity of PSPC
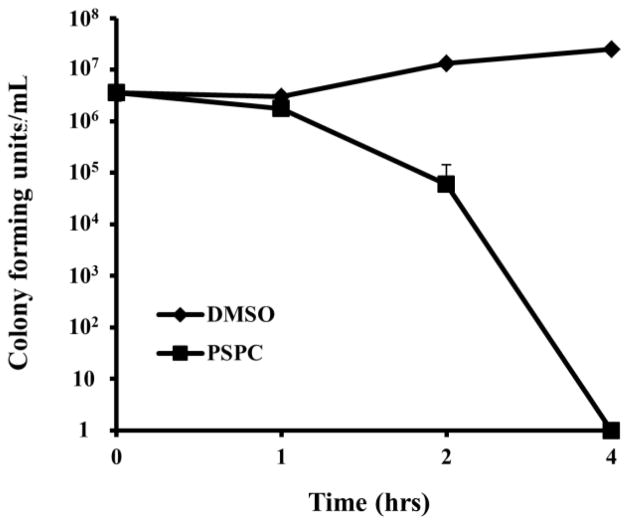
As we have previously described, compounds identified in chemical screens that prevent a pathogen from killing C. elegans, can inhibit the growth of the pathogen, block the virulence of the pathogen, and/or enhance C. elegans immunity.8–10 To help distinguish these possibilities, we determined first whether PSPC could directly affect the growth of the pathogen. We found that PSPC does inhibit the growth of the MRSA strain MW2 in Müller-Hinton broth with an in vitro MIC of 4 μg/ml, compared to an MIC of 2 μg/ml for vancomycin (Table 1). A time-kill assay at 10x MIC (40μg/ml) showed that PSCP is bactericidal, killing essentially 100% of MW2 cells during a 4-hour exposure period (Fig. 3). The minimum bactericidal concentration of PSPC against MW2 is 16 μg/ml (data not shown), 4 times the MIC. These data show that PSPC functions directly to block the growth of MRSA MW2.
Table 1.
MIC of PSPC, PSPC-6S and PSPC-1S against ESKAPE pathogens.
| Bacterial Strains | Vancomycin (μg/mL) | Polymyxin B (μg/mL) | PSPC (μg/mL) | PSPC-1S (μg/mL) | PSPC-6S (μg/mL) |
|---|---|---|---|---|---|
| E. faecium (E007) | 4 | ND | 8 | >64 | 8 |
| S. aureus (MW2) | 2 | ND | 4 | >64 | 4 |
| K. pneumoniae (77326) | ND | 16 | 64 | >64 | 64 |
| A. baumannii (ATCC 17978) | ND | 0.5 | 32 | >64 | 32 |
| P. aeruginosa (PA14) | ND | 2 | >64 | >64 | >64 |
| Enterobacter spp. (KCTC 2625) | ND | 32 | >64 | >64 | >64 |
Figure 3. Time-kill kinetics of PSPC against S. aureus.
The survival of MW2 in a broth culture treated with DMSO or PSPC (10 xMIC, 40 μg/mL).
The ability of PSPC to function as a broad-spectrum antibiotic was determined by measuring its MIC against the ESKAPE pathogens: E. faecium, K. pneumoniae, A. baumannii, P. aeruginosa and Enterobacter spp, in addition to S. aureus. The six ESKAPE pathogens are the main causes of nosocomial infections, which are often difficult to treat due to antimicrobial resistance.11 Similar to S. aureus, the MIC of PSPC against the Gram-positive species E. faecium was 8 μg/mL (Table 1), but PSPC displayed higher MICs against the Gram-negative bacteria K. pneumoniae (64 μg/mL), A. baumannii (32 μg/mL), P. aeruginosa (>64 μg/mL) and Enterobacter spp. (>64 μg/mL).
A PSPC analogue has antimicrobial activity
Two analogues of PSPC were synthesized (see Supplementary Methods) and tested for antibacterial activity against the ESKAPE pathogens (Table 1). The sulfide analogue PSPC-1S (Fig. 1) was found to be inactive across all species in the panel (MICs > 64 μg/mL). In contrast, the sulfoxide analogue PSPC-6S (Fig. 1) showed identical potency to PSPC against all species. These findings indicate that the activity of PSPC is structure-dependent and requires the polar functionality attached to the sulfur atom, which supports a target-based mechanism.
The efflux pump inhibitor PAβN potentiates the activity of PSPC against Gram-negative bacteria
The peptidomimetic compound phenylalanine arginyl β-naphthylamide (PAβN) is a broad spectrum pump inhibitor that is effective against resistance/nodulation/division (RND) pumps in various Gram-negative bacteria, restoring the antibiotic susceptibility of these strains.12 We examined whether PAβN also possessed the ability to directly inhibit the growth of bacteria, by testing the antimicrobial activity of PAβN against the Gram-negative ESKAPE pathogens in a broth microdilution assay. PAβN did not show any growth inhibitory effects against these strains at the maximum tested concentration of 64 μg/mL (data not shown). Accordingly, we examined whether the activity of PSPC against Gram-negative ESKAPE pathogens could be potentiated in the presence of PAβN at a fixed concentration of 64 μg/mL. In the presence of PAβN, the MIC of PSPC against K. pneumoniae, A. baumannii, P. aeruginosa and Enterobacter spp. decreased by more than 4-fold (Table 2).
Table 2.
PAβN potentiates the antimicrobial activity of PSPC against Gram-negative ESKAPE pathogens.
| Bacterial Strains | MIC (μg/mL) | Fold change | |
|---|---|---|---|
| PSPC | PSPC + PAβN (64 μg/mL) | ||
| K. pneumoniae (ATCC 77326) | 64 | 8 | 8 |
| A. baumannii (ATCC 17978) | 32 | 4 | 8 |
| P. aeruginosa (PA14) | >64 | 16 | >4 |
| Enterobacter spp. (KCTC 2625) | >64 | 8 | >8 |
PSPC acts synergistically with polymyxin B against K. pneumoniae and A. baumannii
We tested the ability of polymyxin B, a lipopeptide antibiotic that is effective against a variety of Gram-negative bacteria, to function synergistically with PSPC against K. pneumoniae and A. baumannii. A microplate checkerboard assay was used to test for growth inhibition of the strains in the presence of PSPC adjusted to final concentrations in the range of 1–64 μg/mL and polymyxin B adjusted to final concentrations of 0.063–64 μg/mL. The minimum concentrations at which the two compounds inhibited bacterial growth, both alone and in combination, were determined in order to calculate the Fractional Inhibitory Concentration (FIC) index.13 The FIC indices of polymyxin B/PSPC against K. pneumoniae and A. baumannii were found to be 0.141 and 0.375, respectively (Table 3). FIC indices <0.5 indicate that polymyxin B exhibits synergistic activity with PSPC against both pathogens.
Table 3.
MIC data for PSPC and polymyxin B alone and in combination against K. pneumoniae and A. baumannii.
| Bacterial Strains | MIC (μg/mL) | FIC | ||
|---|---|---|---|---|
| PSPC | Polymyxin B | PSPC and Polymyxin B combination | ||
| K. pneumoniae (ATCC 77326) | 64 | 16 | 1 and 2 | 0.141 |
| A. baumannii (ATCC 17978) | 32 | 0.5 | 4 and 0.125 | 0.375 |
Cytotoxicity and hemolytic activity of PSPC
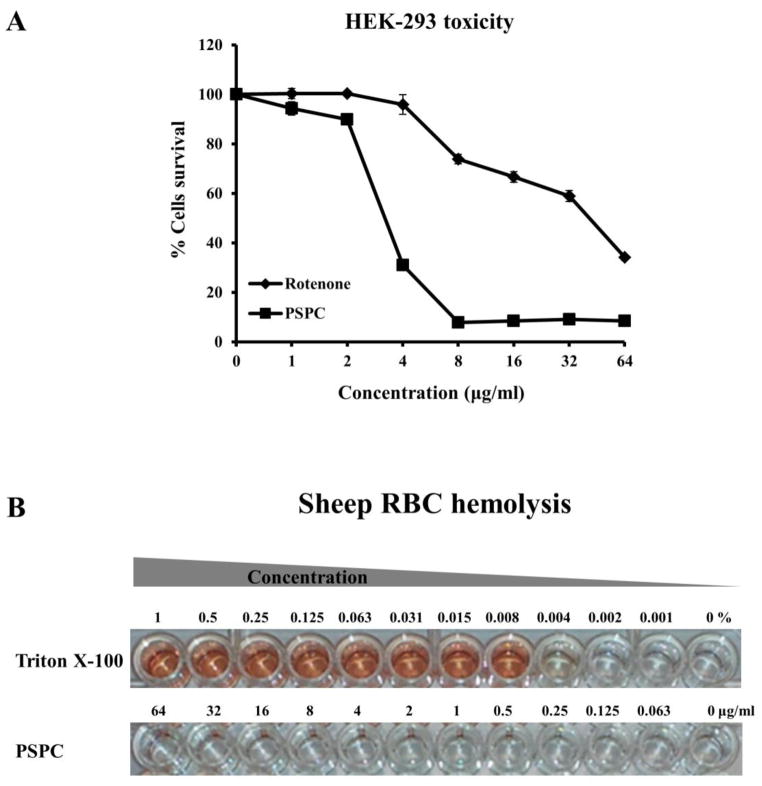
PSPC was found to be toxic to worms at high concentrations (Fig. 2), suggesting the possibility of toxicity to other eukaryotic systems. In vitro cytotoxicity of PSPC was evaluated against HEK-293 cells. The cells were treated with serial dilutions of PSPC over the concentration range 1–64 μg/mL and cell viability determined. The mitochondrial toxin rotenone, which interferes with the electron transport chain in mitochondria, was included as a positive control. At concentrations up to 2 μg/mL of PSPC, almost 90% of treated cells survived (Fig. 4A). Higher concentrations of PSPC caused a dose dependent increase in toxicity to HEK-293 cells, culminating in a survival rate of only 8.5% at 64 μg/mL, which is the maximum concentration tested (Fig. 4A).
Figure 4. Cytotoxicity and hemolytic activity of PSPC.
(A) The viability of HEK-293 cells was measured after treatment with serially diluted concentrations (1–64 μg/ml) of PSPC or the mitochondrial toxin rotenone (positive control). Cell viability was measured spectrophotometrically by detecting degradation of WST-1 dye into formazan by viable cells, which produces measurable color.
(B) Sheep erythrocytes were treated with serial dilutions of Triton X-100 (.001–1%) or PSPC (0.063–64 μg/ml).
We also tested whether PSPC could have lytic activity that would contribute to the toxicity by measuring the hemolysis of sheep erythrocytes. Sheep red blood cells (RBCs) were treated with serial dilutions (0.063–64 μg/ml) of PSPC for 1 hour and cells treated with serial dilutions of Triton X-100 (0.001–1%) were used as a positive control. PSPC did not cause visible RBC hemolysis at the maximum concentration tested (Fig. 4B), which was confirmed by measuring absorbance at 540 nm (data not shown). Triton X-100 caused hemolysis at all concentrations above 0.008%. The findings suggest that PSPC does not cause damage to mammalian cell membranes, but is nonetheless toxic to mammalian cells at high concentrations.
Using a C. elegans – MRSA whole animal high throughput screening assay, we identified a novel antibiotic, PSPC, and its analogue, PSPC-1S, that are effective against the Gram positive bacteria S. aureus and E. faecium at MICs of 4 and 8 μg/ml, respectively. In earlier studies, we described the use of C. elegans infection assays to identify “immunomodulators” in addition to traditional antibiotics that directly inhibit bacterial growth.8, 10, 14 As the name suggests, immune-modulators do not display direct effects on the pathogen, but rather act by modulating the immune system of the host to fight the infection. Although our findings show that PSPC displays direct antibacterial activity against Gram-positive bacteria and to a lesser extent against Gram-negatives, interestingly, the minimum effective concentration (MEC) of PSPC in vivo was ≤ 0.78 μg/mL, which is ≥5-fold lower than its in vitro MIC against S. aureus. This suggests that PSPC might have immunomodulatory properties in addition to its antibacterial effects, similarly to another compound that we have studied, RPW-24, that was identified in a screen for anti-infective compounds against Enterococcus faecalis using a C. elegans liquid infection assay 15. The anti-infective properties of RPW-24 in C. elegans were attributed to the ability of the compound to promote the expression of endogenous immune effector molecules via evolutionarily conserved immune regulators.8 On the other hand, the in vivo MEC of vancomycin was also several fold lower than the MIC, suggesting that the enhanced in vivo activity of PSPC could also be due to bioaccumulation of the molecule in the infected worms. Another possibility for why the MEC in vivo is lower than the MIC in vitro is that the C. elegans immune system (i.e. by modulating the production of antimicrobial peptides) sensitizes certain pathogens to particular classes of antibiotics.
Reasons for the differential activity of PSPC and PSPC-6S against Gram-positive and Gram-negative bacteria could be due to the structural and molecular differences between the two classes of bacteria. Alternatively, the action of efflux pumps in transporting PSPC and PSPC-6S out of Gram-negative bacteria may contribute to the higher MICs. Efflux pumps have been implicated in resistance of P. aeruginosa to tetracycline, chloramphenicol and norfloxacin 16–17, agents that remain effective against Gram-positive bacteria, including S. aureus. Among the several categories of efflux pump systems in Gram-negative bacteria, the RND system is constitutively active and plays a major role in resistance to many antibiotics, including β-lactams, fluoroquinolones, and aminoglycosides.18 It is likely that PAβN potentiated the activity of PSPC against Gram-negative bacteria, through inhibition of efflux pumps. Interestingly, a recent study has shown that PAβN was also capable of permeabilizing bacterial membranes, at a concentration of 50 μg/mL, which can increase uptake of small molecules into bacteria from the surrounding environment.19 Therefore, there could be several ways by which PAβN potentiates the activity of PSPC, including efflux pump inhibition and possibly also by enhancing uptake of PSPC by the bacterial cell. Further studies are therefore needed to confirm the mechanism by which PAβN potentiates the action of PSPC against Gram-negatives.
We also found that polymyxin B synergistically potentiates the activity of PSPC against MRSA. Polymyxin B, which was discovered more than 50 years ago, is effective against a wide range of Gram-negative bacteria, but was rarely used clinically until recently due to its nephrotoxicity and neurotoxicity.20 The rationale for testing polymyxin B in combination with PSPC was that if they functioned synergistically, relatively low concentrations of each drug could be used, reducing their toxicity. The mode of action of polymyxin B involves binding to bacterial outer membranes, causing disruptions that lead to rapid cell death.21 The drug has reentered clinical practice in recent times due to the emergence of multidrug-resistant nosocomial Gram-negative infections and the dwindling antibiotic development pipeline.21 Resistance to polymyxin B is infrequent, although it has been increasing in recent years.22 The primary mechanism of resistance involves modifications to the bacterial outer membrane that reduces membrane affinity for the drug.21
In conclusion, we report the identification of PSPC as a hit anti-bacterial from a high throughput screen involving a C. elegans-MRSA liquid infection assay. Antimicrobial susceptibility profiling suggests that PSPC shows moderate activity against the Gram-positive ESKAPE pathogens and exhibits synergism with polymyxin B against the Gram-negative ESKAPE species. The activity of PSPC against Gram-negative bacteria was enhanced by co-treatment with the efflux pump inhibitor PAβN. Although the mechanism of action of PSPC still needs to be resolved, total loss of activity with a simple sulfide analogue of PSPC (i.e. PSPC-1C) supports the hypothesis that the compound may act via a target-based mechanism. Further tests are necessary to evaluate the in vivo activity in mammalian infection models.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health grant P01 AI083214 to EM and FMA, by NIH Grant R01 AI085581 to FMA, and by funding from Boehringer Ingelheim GmbH (Ridgefield, CT) to EM and FMA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pogue JM, Kaye KS, Cohen DA, Marchaim D. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2015;21:302. doi: 10.1016/j.cmi.2014.12.025. [DOI] [PubMed] [Google Scholar]
- 2.Davies J. The Canadian journal of infectious diseases & medical microbiology = Journal canadien des maladies infectieuses et de la microbiologie medicale/AMMI Canada. 2006;17:287. doi: 10.1155/2023/9798013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jabes D. Curr Opin Microbiol. 2011;14:564. doi: 10.1016/j.mib.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 4.McKenna M. Nature. 2013;499:394. doi: 10.1038/499394a. [DOI] [PubMed] [Google Scholar]
- 5.Rajamuthiah R, Fuchs BB, Jayamani E, Kim Y, Larkins-Ford J, Conery A, Ausubel FM, Mylonakis E. PLoS One. 2014;9:e89189. doi: 10.1371/journal.pone.0089189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dolla NK, Chen C, Larkins-Ford J, Rajamuthiah R, Jagadeesan S, Conery AL, Ausubel FM, Mylonakis E, Bremner JB, Lewis K, Kelso MJ. Aust J Chem. 2014;67:1471. doi: 10.1071/CH14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iscla I, Wray R, Blount P, Larkins-Ford J, Conery AL, Ausubel FM, Ramu S, Kavanagh A, Huang JX, Blaskovich MA, Cooper MA, Obregon-Henao A, Orme I, Tjandra ES, Stroeher UH, Brown MH, Macardle C, van Holst N, Ling Tong C, Slattery AD, Gibson CT, Raston CL, Boulos RA. J Antibiot (Tokyo) 2015 doi: 10.1038/ja.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pukkila-Worley R, Feinbaum R, Kirienko NV, Larkins-Ford J, Conery AL, Ausubel FM. PLoS genetics. 2012;8:e1002733. doi: 10.1371/journal.pgen.1002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirienko NV, Kirienko DR, Larkins-Ford J, Wahlby C, Ruvkun G, Ausubel FM. Cell host & microbe. 2013;13:406. doi: 10.1016/j.chom.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong C, Tan MW, Nathan S. Biology open. 2014;3:644. doi: 10.1242/bio.20148334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rice LB. Infect Control Hosp Epidemiol. 2010;31(Suppl 1):S7. doi: 10.1086/655995. [DOI] [PubMed] [Google Scholar]
- 12.Lomovskaya O, Bostian KA. Biochem Pharmacol. 2006;71:910. doi: 10.1016/j.bcp.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Draper LA, Cotter PD, Hill C, Ross RP. BMC microbiology. 2013;13:212. doi: 10.1186/1471-2180-13-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi EJ, Kim HI, Kim JA, Jun SY, Kang SH, Park DJ, Son SJ, Kim Y, Shin OS. Appl Microbiol Biotechnol. 2015;99:4387. doi: 10.1007/s00253-015-6382-y. [DOI] [PubMed] [Google Scholar]
- 15.Moy TI, Conery AL, Larkins-Ford J, Wu G, Mazitschek R, Casadei G, Lewis K, Carpenter AE, Ausubel FM. ACS chemical biology. 2009;4:527. doi: 10.1021/cb900084v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li XZ, Livermore DM, Nikaido H. Antimicrob Agents Chemother. 1994;38:1732. doi: 10.1128/aac.38.8.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li XZ, Ma D, Livermore DM, Nikaido H. Antimicrob Agents Chemother. 1994;38:1742. doi: 10.1128/aac.38.8.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikaido H. Clin Infect Dis. 1998;27(Suppl 1):S32. doi: 10.1086/514920. [DOI] [PubMed] [Google Scholar]
- 19.Lamers RP, Cavallari JF, Burrows LL. PLoS One. 2013;8:e60666. doi: 10.1371/journal.pone.0060666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falagas ME, Kasiakou SK. Critical care. 2006;10:R27. doi: 10.1186/cc3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zavascki AP, Goldani LZ, Li J, Nation RL. J Antimicrob Chemother. 2007;60:1206. doi: 10.1093/jac/dkm357. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE, Liolios L. Antimicrob Agents Chemother. 2006;50:2946. doi: 10.1128/AAC.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.