SUMMARY
Fertile offspring from somatic cell nuclear transfer (SCNT) is the goal of most cloning laboratories. For this process to be successful, a number of events must occur correctly. First the donor nucleus must be in a state that is amenable to remodeling and subsequent genomic reprogramming. The nucleus must be introduced into an oocyte cytoplasm that is capable of facilitating the nuclear remodeling. The oocyte must then be adequately stimulated to initiate development. Finally the resulting embryo must be cultured in an environment that is compatible with the development of that particular embryo. Much has been learned about the incredible changes that occur to a nucleus after it is placed in the cytoplasm of an oocyte. While we think that we are gaining an understanding of the reorganization that occurs to proteins in the donor nucleus, the process of cloning is still very inefficient. Below we will introduce the procedures for SCNT, discuss nuclear remodeling and reprogramming, and review techniques that may improve reprogramming. Finally we will briefly touch on other aspects of SCNT that may improve the development of cloned embryos.
INTRODUCTION
The production of offspring by somatic cell nuclear transfer (SCNT) is an inefficient process, but it is required to create genetically modified animals in species that lack successful embryonic stem cell lines. While there are sporadic reports of high efficiency, the overall rate of development to live offspring is on the order of 1–3%. It can be confusing to determine the efficiency of the process because it can be measured in the percentage of clones that cleave, undergo compaction, form a blastocyst, establish a pregnancy, result in a live born animal, or an animal that produces offspring. The confusion can increase because these percentages can be calculated from the number of reconstructed oocytes, fused oocytes, cleaved embryos, or transferred embryos. Throughout this review we will define the endpoints that have been measured and take into consideration the effects that the technique has on producing animals that are fertile. While the review will cover a variety of different species, the review will undoubtedly be domestic animal- and swine-centric due to our background in swine cloning.
There are numerous steps in the process to clone an animal via SCNT. In general, the technique first requires unfertilized oocytes and cells from which to acquire donor nuclei. The unfertilized oocytes can be either in vivo matured or in vitro matured. Better development to the blastocyst stage (Kühholzer et al., 2001; Akagi et al., 2008) and to term can be achieved with in vivo-derived oocytes as compared to in vitro matured oocytes (Wells et al., 1997), and oocytes matured in vitro from sexually mature animals result in better development to the blastocyst stage (Hyun et al., 2003) and to term (Lai et al., 2002) than oocytes from prepubertal animals. In general the percent of development to the blastocyst stage and term are better if the donor cells are from relatively undifferentiated cells as compared to more differentiated cells (Hiiragi and Solter, 2005), and culturing a donor cell line for longer periods of time in vitro generally results in decreased development to blastocyst and to term (Hill et al., 2000b; Roh et al., 2000). While there is evidence that fetal-derived fibroblasts result in better development to the blastocyst stage and to term as compared to fibroblast from newborns—which are better than cells from adolescents, and which are better than cells from adult animals (Hill et al., 2000b; Lee et al., 2003)—this premise has been seriously questioned and some have concluded only that nuclei from cleavage stage blastomeres result in better development to term than any somatic cell (Oback and Wells, 2007).
Explanations for the above suggestions include: (1) It is thought that oocytes from sexually mature animals are more developmentally competent because they have a better supply of the factors that will remodel the nucleus when it is transferred into its cytoplasm, and (2) that the nuclei from less differentiated cells are more plastic and more readily able to remove and/or replace proteins that affect transcription than nuclei from more differentiated cells. With all that said, cells from the same fetus that are cultured in parallel have differing abilities to direct development to the blastocyst stage (Kühholzer et al., 2001), so the above is a generalization. The remainder of this review will focus on what structurally happens to the donor nucleus after transfer to the cytoplasm of the oocyte, and what can be done to improve the remodeling that occurs to the nucleus by the cytoplasm of the oocyte.
BIOLOGY OF SOMATIC CELL NUCLEAR TRANSFER
To understand what is thought to be necessary for development to result from SCNT, one must first understand what regulates gene expression during development. The simple answer to this question is chromatin architecture. Unfortunately chromatin architecture is incredibly complex. A rather long list of specifics can quickly be constructed including: DNA methylation, nuclear lamin composition, histone subunits, and subsequent post-translational modifications such as acetylation, phosphorylation, and methylation, etc. The composition of the nucleus, in terms of these proteins, changes dramatically not only during embryogenesis, but also as nuclei begin to direct specialization of specific tissues. For SCNT to be successful, it is thought that the donor nucleus must be remodeled to resemble the nucleus of a zygote, and the cytoplasm from an oocyte arrested in metaphase can facilitate that remodeling. This change in chromatin architecture, here referred to as nuclear remodeling, is thought to result in a change in the pattern of genes that undergo transcription, here referred to as nuclear reprogramming. This distinction between nuclear remodeling and reprogramming is made because remodeling is a physical repackaging of the DNA while reprogramming is a result of those structural changes.
NUCLEAR REMODELING
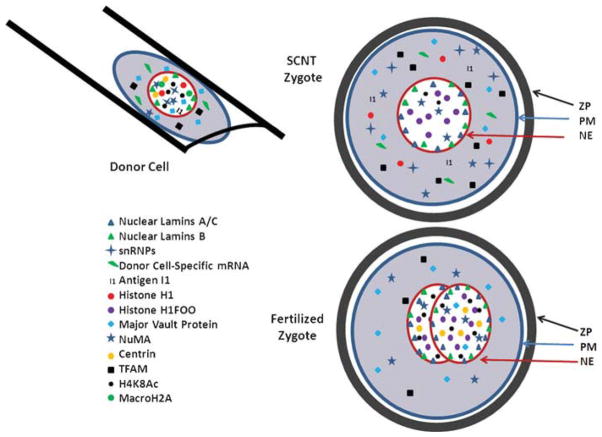
To understand the structural changes that occur to a nucleus during SCNT, the basic structure of the donor nucleus as well as the structure of the pronuclei must be described. The structure and function of the pronuclei in a zygote are rather unique in that they are organized in such a way that in the environment of the zygote cytoplasm there is very little, if any, transcription occurring. The first few cleavage divisions are directed by factors stored in the cytoplasm of the oocyte. At a species-specific cell stage, the embryo begins producing significant amounts of RNA. The point at which significant amounts of transcription occur is called embryonic genome activation (EGA). EGA occurs during the two-cell stage in the mouse (Latham, 1999), four-cell stage in the human (Braude et al., 1988), rat (Zernicka-Goetz, 1994), and pig (Jarrell et al., 1991), and during the 8- to 16-cell stage in the sheep and cow (Telford et al., 1990). This is in contrast to other well-studied species such as Xenopus where the EGA occurs at the 12th cleavage division (~4,000 cells) around the time of the mid-blastula transition (Newport and Kirschner, 1982a,b). At this point the nuclei in the developing embryo begin truly controlling the development of the embryo. As the mammalian embryo goes through the first few cleavage divisions and initiates the EGA, the proteins associated with the nucleus change (Prather and Schatten, 1992). A few examples include the nuclear lamins, SnRNPs (Fig. 1), and the types of histones and their associated acetylation and methylation (reviewed by Zhao et al., 2010b), etc. During formation of the first two distinct cell types (inner cell mass: ICM; trophectoderm: TE) not only do a different set of genes undergo transcription, but a different set of proteins are associated with the nuclei (e.g., CDX2 and POU5F1, Reik, 2007). As development proceeds and various tissues form and begin to specialize, each tissue type has its specific nuclear structure and repertoire of genes that are turned-on. An analogy to the differentiation process is that of dominos falling over, with each domino that falls representing a difference in transcription. Fertilization might start the dominos (no transcription), with EGA knocking over another (the first set of genes that are turned-on during development). That domino might hit two dominos; one representing the ICM and the other the TE, each with their own specific genes that are expressed in that tissue. This would continue on through development with more and more dominos triggering more and more pathways until all the tissues in the body are represented. This analogy extends to fetal-derived fibroblast cells cultured in vitro that likely have a different repertoire of genes that are turned-on than is found anywhere in the body. When a nucleus from a fetal-derived fibroblast cell is transferred to the cytoplasm of the oocyte and the structure of the nucleus is changed to be like that of a zygote, then all the dominos from that pathway are reset to their upright position. The transferred nucleus is then poised to recapitulate the same pattern of development observed in a normally fertilized embryo. Thus, structural remodeling of the chromatin should result in subsequent reprogramming of the developmental pattern of gene expression, such that the pattern recapitulates that observed in a normally fertilized embryo.
Figure 1.
Nuclear remodeling in donor cells, zygotes, and SCNT zygotes (not drawn to scale as the donor cell and nucleus would be smaller). The donor cell is drawn in a pipette. Different figures and colors are used to depict the location and concentration of a variety of proteins that have been localized in donor nuclei, zygotes and SCNT zygotes (ZP, zona pellucida; PM, plasma membrane; NE, nuclear envelope). Data are based upon the following manuscripts: using a blastomere as a nuclear donor cell (Prather et al., 1989, 1990, 1991, 2000; Kubiak et al., 1991; Prather and Rickords, 1992; Parry and Prather, 1995) or fetal fibroblast as a nuclear donor cell (Park et al., 2001b; Moreira et al., 2003; Gao et al., 2004; Mananadhar et al., 2004; Novak et al., 2004; Teranishi et al., 2004; Sutovsky et al., 2005; Liu et al., 2006; Antelman et al., 2008; Chang et al., 2010; Zhao et al., 2010a). A more complete description of differences in histone acetylation and DNA methylation in donor cells, zygotes and SCNT embryos has recently been published (Zhao et al., 2010b).
Structural remodeling can be measured in a number of different ways including gross changes in the structure of the nucleus after transfer to oocyte cytoplasm and subsequent activation, include swelling of the nucleus to a size similar to a pronucleus (Prather et al., 1990) and reformation of the nucleoli to a compact state, again similar to the nucleoli in pronuclei (Ouhibi et al., 1996; Prather, 1999; Martin et al., 2006). The pioneering studies of Gurdon in Xenopus showed that many proteins located in the nucleus of the donor cell are released into the cytoplasm of the oocyte, and other proteins present in the cytoplasm of the oocyte are acquired by the transferred nucleus (reviewed by Prather et al., 2000). More recently in mammals a battery of antibodies has detected the movement (possibly unmasking/masking) of numerous proteins between the cytoplasm and the nucleus that appear to perform as described in Xenopus back in the 1960s, that is, the release of donor cell-specific proteins into the cytoplasm, and the acquisition of oocyte cytoplasmic proteins by the transferred nucleus (e.g., see Fig. 1).
One well-characterized exchange of proteins involves histone H1. Histone H1 links the histone subunits together to form the nucleosome. There is an oocyte-specific variant of histone H1 called H1FOO. When somatic nuclei are transferred to oocyte cytoplasm, the somatic histone H1 is rapidly replaced with the oocyte (Gao et al., 2004). Interestingly this exchange can occur in the presence of a nuclear envelope and appears to be an active process. In contrast to H1FOO, another histone variant is MacroH2A. MacroH2A is absent from the nuclei of zygotes and the first few cleavage divisions, but present in somatic cells. When a somatic cell is transferred to oocyte cytoplasm, MacroH2A is removed from the chromatin and degraded. It is then re-synthesized and assembled into the chromatin structure when the embryo reaches the morula stage, coinciding with the normal appearance of MacroH2A in fertilized embryos (Chang et al., 2010).
In addition to the exchange of proteins between the donor nucleus and oocyte cytoplasm there are modifications to DNA and post-translationally to proteins that occur after SCNT. Two prominent examples include methylation of DNA and modifications of histones (methylation, acetylation, etc.). While these topics have been recently extensively reviewed (Fuks, 2005; Reik, 2007; Yang et al., 2007; Zhao et al., 2010b) a brief description is in order. First a disclaimer is warranted: while the description of the changes in histones and DNA methylation that occur during normal embryo development is relatively well characterized in the mouse there are numerous species-specific differences that have been described. So the reader should beware when learning about the developmental pattern described in one species as it does not always translate to other species. Because these patterns are not precisely replicated in different species, it brings into question the developmental significance of the particular modification. Nevertheless, global DNA demethylation occurs during preimplantation development in the mouse and is facilitated by active demethylation of the paternal genome and passive demethylation during cleavage of the maternal genome (Mayer et al., 2000; Santos et al., 2002). De novo DNA methylation begins around the time of ICM and TE differentiation, followed by lineage-specific methylation as the cells become more and more differentiated. Similarly, bovine, rat, and pig zygotes have a demethylated paternal genome, suggesting active demethylation. In porcine embryos, de novo methylation can be detected as early as the blastocyst stage (Bonk et al., 2008). In contrast, sheep embryos undergo limited demethylation after fertilization, or remethylation from the zygote to the blastocyst stage (Young and Beaujean, 2004).
Unfortunately when the DNA methylation pattern of cloned embryos is compared to that of normal embryos, the pattern is not always replicated (Kang et al., 2001; Mann et al., 2003; Yamazaki et al., 2006). In cloned one-cell murine embryos there is reduced methylation relativeto zygotes, but additional demethylation that occurs in fertilized embryos does not occur. Similarly in cattle SCNT embryos, the passive demethylation process appears to be defective (Bourc’his et al., 2001). The pattern of DNA methylation in these early cloned embryos was more similar to those of differentiated cells (Bourc’his et al., 2001; Dean et al., 2001; Bonk et al., 2007). This aberrant DNA methylation continues beyond the blastocyst stage (Sawai et al., 2010). Numerous specific examples can be listed including some imprinted genes such as SNRPN (Suzuki et al., 2009), H19, and IGF2 (Wei et al., 2010). While this issue has already been reviewed (Zhao et al., 2010b) it is probably worth reiterating that if the methylation pattern of the imprinted genes in the donor cell is not set then it appears to be very difficult for SCNT embryos to reestablish these absent imprints. A prime example is when the parental genomic imprint is erased during development of the germ cells and after it is maternally or paternally imprinted, then the clones from these donor cells do not develop (Lee et al., 2002). In the germ cells where the parental imprints have been erased and in the mature germ cell where only the maternal or paternal imprint is present the SCNT embryo is not capable of reestablishing those imprints. Thus, beginning with the correct maternal and paternal imprint is necessary for development of the SCNT embryo.
This incomplete remodeling of the methylation pattern likely contributes to the low efficiency of development of cloned embryos. It is encouraging to note that there are some patterns of DNA methylation that permit discrimination between in vitro- and SCNT-derived embryos (Bonk et al., 2008; Niemann et al., 2010) and these may be useful for identifying high-quality blastocysts and targeting treatments to bring the methylation pattern more in line with in vivo produced embryos.
Changes in the structure of the chromatin caused by an exchange of proteins, changes in DNA methylation, and histone post-translational modifications result in different patterns of gene expression. Ideally, the change in the pattern of gene expression (see below) results in a zygote pattern followed by a recapitulation of the normal developmental pattern, that is, reprogramming.
NUCLEAR REPROGRAMMING
Nuclear remodeling (as described above) is thought to result in an architectural change of the chromatin resulting in a reprogramming of the pattern of gene expression in the cloned embryos. Such a modification of chromatin structure is thought to result in a recapitulation of the expression of many genes. Some of the first such genes studied in Xenopus included the 5Sooc gene (Wakefield and Gurdon, 1983) and muscle-specific actin (Gurdon et al., 1984) and these serve to illustrate the remarkable fidelity of the reprogramming. The 5Sooc gene is transcribed for only a short period of time during the late blastula stage. If a donor nucleus from an embryo beyond the blastula stage is transferred to an oocyte, the 5Sooc gene remains off until the late blastula stage when it is briefly transcribed and shut off. Similarly, muscle-specific actin is produced only in the developing myotome cells. If a nucleus is taken from a muscle cell and transferred to an oocyte then muscle-specific actin is shut off. When the resulting embryo reaches the stage at which the myotome cells are differentiating, then the muscle-specific actin turns on again, but only in the developing myotome cells of the embryo.
While there have been many studies that evaluated gene expression in cloned mammalian embryos and tissues, each set of donor cells appears to be different, as some can readily be reprogrammed and others not (Daniels et al., 2000; Winger et al., 2000; Wrenzycki et al., 2001; Boiani et al., 2002; Bortvin et al., 2003; Li et al., 2005; Pfister-Genskow et al., 2005; Smith et al., 2005; Inoue et al., 2006; Somers et al., 2006; Jouneau et al., 2006; Jiang et al., 2007a; Aston et al., 2010). While the majority of genes appear to be correctly reprogrammed, there is a different subset of genes in many different donor cells that are not reprogrammed and are expressed at inappropriate times. Thus, a consistent pattern of aberrant gene expression has not been identified. This suggests that after SCNT there are numerous flaws in the genomic architecture. For example, there is mounting evidence of more variation in DNA methylation within clones than between non-clonal controls (de Montera et al., 2010) and this also corresponds to phenotypic variation (Lee et al., 2004).
Recently it has been argued that therapeutic cloning, in spite of this aberrant gene expression, could move forward because the developmental defects are most likely to show up in the placenta (Yang et al., 2007). We would argue that if remodeling is not sufficient during the first cell cycle, then all descendants would be affected. Just because the placenta has a stricter requirement for maintaining the fidelity of gene expression does not mean that the cells contributing to the ICM are normal, and thus they might be suspect for therapeutic uses.
EFFECT OF NUCLEAR REPROGRAMMING ON PLACENTAL DEVELOPMENT
Abnormal placental development appears to be a phenotype common to many different species, as has been reported in mice (Tanaka et al., 2001; Suemizu et al., 2003), cattle (Hill et al., 2000a; Heyman et al., 2002; Constant et al., 2006), sheep (De Sousa et al., 2001), and pigs (Chae et al., 2009). Most placental defects found in cloned bovine concepti are associated with the placentome. One study tracked SCNT fetuses from Days 30 to 90 of development and found a decrease in both the vascular development and the number of cotyledons. Some placentas had an irregular chorionic epithelium and a decrease in allantoic vascularization as well. The authors concluded that the early loss of cloned bovine fetuses was due to placental defects resulting in smaller fetuses (Chavatte-Palmer et al., 2006). However, bovine fetuses that do survive to term are often larger than normal (Wilson et al., 1995). Another group identified that abnormal placental development in cloned bovine embryos was correlated with aberrant expression of the MHC that may result in rejection by the maternal system (Ramsoondar et al., 1999; Chavatte-Palmer et al., 2007).
The impact of SCNT has also been studied in mouse placentas. One study that analyzed placentas at 10.5 days post-coitis (dpc) identified poor development of the spongiotrophoblast layer in cloned placentas (Wakisaka-Saito et al., 2006). However, cloned mouse pups at term often have a larger placenta than those from normal matings. This overgrowth of the placenta (placentomegaly) occurs as a result of hyperplasia of the spongiotrophoblast layer after days 12.5 dpc (Tanaka et al., 2001). Interestingly, this large placental defect is not transferred to subsequent progeny (Shimozawa et al., 2002). Other specific problems in SCNT-derived placentas include a non-cell autonomous epiblast defect which can be rescued by injection of SCNT blastocysts with normal embryonic stem cells or ICM cells (Jouneau et al., 2006). Defective growth regulation of the extraembryonic ectoderm was also observed in Day 7 dpc SCNT embryos. Aggregation of SCNT embryos with tetraploid embryos(which will only contribute to the trophoblastic cell lineage) corrected the growth abnormalities of the extraembryonic ectoderm observed during gastrulation (Jouneau et al., 2006).
In SCNT-derived pig embryos, the extraembryonic membranes are smaller than control placentas (Chae et al., 2009). In addition to being smaller, there is quite a bit of variation in physical appearance among SCNT placentas (unpublished results, Fig. 2G,H). In one study, global transcript profiling between cloned and in vivo Day 26 extraembryonic membranes was analyzed by using a 13 K oligonucleotide array; 7 elevated and 27 suppressed transcripts were identified in the cloned samples relative to normal in vivo samples (Chae et al., 2009). The aberrantly regulated pathways in this study included RNA splicing-related genes, RNA processing -related genes, and RNA metabolism-related genes suggesting the cloning process was affecting transcriptional regulation in the resulting placentas.
Figure 2.
A comparison of placental development in pigs created by SCNT. A,B: Histological sections comparing the fetal maternal interface at Day 35 of gestation between normal in vivo (IVV) (A) and SCNT (B). Sections are stained with hematoxylin and eosin and show similar structural morphology between the two. C,D: Immunohistochemistry with primary antibodies specific to porcine pregnancy-associated glycoprotein-2 (pPAG2). The IVV section (C) shows a clear delineation between the trophoblast (TE) and luminal epithelium (LE) at the microvillar junction (MVJ) with PAG2 protein specific to the placental side. The SCNT section (D) shows a premature accumulation of PAG2 at the microvillar junction and even transversing into the maternal LE. This observation has only been observed in SCNT sections. E: Both PAG2 message, measured by real-time PCR relative to a reference cDNA and housekeeping gene (YWHAG, described previously Whitworth et al., 2005), and its associated acid peptidase activity (Telugu and Green, 2008) were decreased in placentas derived from SCNT when compared to IVV (P <0.05). F: Miniature piglet created by treating reconstructed SCNT zygotes with the HDACi, Scriptaid which has been shown to improve cloning efficiency. G,H: Fetuses and their associated extraembryonic membranes derived from SCNT (G) compared to their in vivo counterparts (H) collected at Day 30 of gestation. Notice all 4 IVV extraembryonic membranes have an established vasculature and the placentas are much larger (H) than from SCNT. All microscopic images were obtained with a 40× objective.
A more extensive transcriptional profiling study (Whitworth et al., 2010) was conducted comparing Day 30 extraembryonic membranes from not only in vivo samples, but also placentas derived from in vitro fertilization and SCNT from three commonly used activation/fusion methods including standard electrical activation/fusion (Park et al., 2001a), electrical activation followed by a transient treatment with a reversible proteasomal inhibitor, MG132 (Zhou et al., 2003; Sutovsky and Prather, 2004) and fusion in low Ca2+ medium followed by chemical activation with thimerosal/dithiothreitol (DTT) (Machaty et al., 1997, 1999b). All three activation groups have been described in detail previously and have all produced live cloned piglets (Whitworth et al., 2009). This study identified 227 differentially expressed transcripts between the five treatments; however, there were no transcriptional differences identified between the three activation methods. The nuclear transfer groups were pooled and compared to in vivo extraembryonic membranes identifying 34 up- and 19 down-regulated transcripts (>2-fold change, P <0.05) (Whitworth et al., 2010). The significantly up-regulated pathways in SCNT extraembryonic membranes included blood circulation and gas exchange, cell surface receptor-mediated signal transduction, G-protein-mediated signaling, major histocompatibility class 1 (MHCI)-mediated immunity, and ligand-mediated signaling. The significantly down-regulated themes in SCNT extraembryonic membranes also included MHCI-mediated immunity and immunity and defense. Both up- and down-regulated biological processes identified by DAVID included MHCI-mediated immunity. This is interesting because in a normal in vivo pregnancy, pigs lack MHCI antigens on the trophoblast (Ramsoondar et al., 1999); however, 14 (7 up, 7 down) differentially expressed transcripts involved with MHCI-mediated immunity were identified in this study. The presence of MHCI molecules was not examined in the Whitworth et al. (2010) study; however, this observation suggests there are major differences in reproductive tract immunity between normal and cloned pregnancies in the pig, but those specific differences remain to be elucidated.
PAG2 is a trophoblast-specific transcript and a member of the large pregnancy-associated glycoprotein gene family (Szafranska et al., 2001). The function of PAG2 has not been elucidated. Normally PAG2 has very high expression levels in the placenta and also has measurable acid peptidase activity (Telugu and Green, 2008; Telugu et al., 2009). PAG2 message and protein were down-regulated in SCNT placentas compared to in vivo placentas by more than four- and two-fold, respectively (Fig. 2E). SCNT placentas also had decreased acid peptidase activity (Fig. 2E) (Whitworth et al., 2010). Upon further investigation, it was found that PAG2 protein localized prematurely in the microvillar junction and even traverse this junction and was present in the luminal epithelium of the maternal side in the SCNT samples (Fig. 2C,D). In normal Day 35 placentas, PAG2 is only associated with the trophoblast and does not accumulate in the microvillar junction until Day 50 (Wooding et al., 2005; Majewska et al., 2006). Accumulation of PAG2 on the maternal side has only been observed in maternal–fetal interface sections from cloned pigs. Interestingly, hematoxylin and eosin staining of these sections showed that the fetal–maternal interface between IVV and SCNT had indistinguishable structural morphology (Fig. 2A,B) so it is not clear why PAG2 protein would be crossing this junction in cloned placentas. This observation suggests that aberrant gene and protein expression and localization at the fetal–maternal interface, and perhaps not the activation method, are causing defects observed in cloned pig placentas.
X-INACTIVATION AND NUCLEAR TRANSFER
As part of establishing the correct pattern/dosage of gene expression one X chromosome of a female cell must be inactivated in the developing fetus. Female cells have two X chromosomes. As the zygote develops to the blastocyst stage most of the genes on one of the X chromosomes in each cell are inactivated. In the developing ICM cells, X chromosome inactivation occurs independently and either the maternally or paternally derived X chromosome is inactivated. In contrast, the paternal X chromosome is always inactivated in the TE (Dementyeva et al., 2009). For those genes that do not get inactivated, dosage compensation occurs via up-regulation of gene expression to match the autosomal genes (Dementyeva et al., 2009). So the question arises with SCNT as to what happens to the X chromosomes after SCNT. Does the pattern of X chromosome inactivation occur correctly, that is, is the paternal X inactivated in the TE and a random inactivation occurs in the ICM? For many cloned embryos correct reprogramming of X chromosome inactivation is exactly what occurs, that is, both X chromosomes are reactivated after SCNT, and then the paternal X is inactivated in the TE and the maternal or paternal X is inactivated in each cell of the ICM (Eggan et al., 2000), but many X-linked genes are aberrantly expressed at later stages (Senda et al., 2004; Nolen et al., 2005; Jiang et al., 2007b), especially in those cloned neonates that die. The inactivation that occurs is mediated by the X-inactivation center locus located on the X chromosome. At this locus are a number of genes that produce long noncoding RNAs that are thought to coat the to-be-inactivated X chromosome and repress transcription (Okamoto and Heard, 2009). Transcriptional repression progressively increases during development and occurs on a gene-by-gene basis. In mice, complete inactivation does not occur until the end of gestation (Lin et al., 2007; Patrat et al., 2009).
TELOMERE LENGTH AND NUCLEAR TRANSFER
One of the questions raised after the first reports of cloning from adult animals regarded the length of telomeres in cloned animals. The telomere is located on the ends of the chromosome arms and is thought to protect the chromosome from degradation. The length of the telomere progressively shortens through fetal development and then from the neonate through adulthood and into old age. So the question becomes what happens when nuclei from adult or aged animals with short telomeres are used as donor cells. Again the answer appears to be species-specific as in pigs and mice the telomeres lengthen during preimplantation development to be similar to normal embryos (Shiels et al., 1999; Wakayama et al., 2000; Jiang et al., 2004). In contrast the telomere length in cloned cattle is reported to be too long (Lanza et al., 2000), too short (Miyashita et al., 2002), or normal (Tian et al., 2000; Betts et al., 2001; Miyashita et al., 2003).
FUSION AND ACTIVATION AFTER NUCLEAR TRANSFER
Oocyte Activation
Implicit in the above description of remodeling and reprogramming is that after the nucleus is transferred to the cytoplasm of the oocyte, initial remodeling of the chromatin must occur. This remodeling is facilitated by proteins that are present in the cytoplasm of the meiotic or mitotic cell. Examples of such oocyte factors that may affect the chromatin structure include nuclear lamins, snRNPs, histone variants, etc. as illustrated in Figure 1. Since the factors that remodel the nucleus associate with the nucleus, the recipient cell must contain condensed chromosomes with those nuclear-associated factors dispersed in the cytoplasm (for example, nuclear lamins, etc). If the cell is in interphase and the factors that affect remodeling are associated with the nucleus, then these factors would be removed during enucleation, and insufficient remodeling would occur (Prather, 2000). Thus, it is thought that the donor nucleus must undergo some degree of dissolution when transferred to the recipient cell cytoplasm so that the nuclear exchange may occur. Nevertheless, after the initial nuclear dissolution, the oocyte must be activated to reduce maturation promotion factor (MPF) activity so that the cell cycle can resume and the developing embryo can proceed to the first interphase. In normal fertilization, intracellular Ca2+ stores are released after the sperm binds to the egg resulting in the release of meiotic arrest and subsequent development (Whitaker and Swann, 1993). Increases in intracellular Ca2+ can be mimicked by applying a high-voltage DC electrical field pulse (Zimmerman and Vienken, 1982) to the oocytes in a fusion chamber in Ca2+ containing medium. After the electrical pulse, pores are created in the plasma membrane allowing for both fusion of the two cells and oocyte activation caused by the influx of Ca2+ (Machaty et al., 1999a).
There are several methods to activate the oocyte after SCNT including electrical activation as discussed above or chemical activation. In chemical activation of pig zygotes, the donor cell is fused to the oocyte in low Ca2+ containing medium to prevent any Ca2+ oscillations. After fusion, the presumptive zygotes are activated by treatment with thimerosal/DTT. Thimerosal, a sulfhydryl-oxidizing compound, will induce Ca2+ transients in metaphase II arrested oocytes (Machaty et al., 1997, 1999b). Subsequent treatment with DTT will then reduce those disulfide bonds and the oocyte will continue to develop as though it were fertilized with sperm. One study compared the efficiency of SCNT using three protocols that all result in the birth of live pigs (Whitworth et al., 2009) including electrical fusion/activation, electrical fusion/activation followed by treatment with a reversible proteasomal inhibitor MG132 (10 μM the for 2 hr after fusion/activation) and electrical fusion in low Ca2+ followed by chemical activation with thimerosal/DTT. The efficiencies of all three methods were then compared to both in vivo and in vitro fertilization (Whitworth et al., 2009). Chemical activation with thimerosal/DTT has a lower fusion rate compared to both of the electrical activation protocols (probably because low Ca2+ results in less adherence between the cells when the electrical pulse is applied); however, there were no differences in mean cell number at the blastocyst stage. The overall pregnancy rate for electrical activation combined with MG132 treatment was 100% (n = 19) at all stages collected (Days 8, 12, 14, and 30) and was significantly higher than electrical activation without MG132 (71.4%, n = 28; P <0.05), but was not significantly higher than chemica activation with thimerosal/DTT (82.6%, n =23; P <0.15). Treatment with MG132 after fusion/activation of reconstructed porcine embryos was the most effective method when comparing the overall pregnancy rates. Interestingly, there were no identifiable differences in gene expression by microarray analysis in Day 30 placentas between the three activation methods analyzed from this study (Whitworth et al., 2010).
In bovine and sheep, SCNT embryos activated and fused simultaneously resulted in poorer development (Campbell et al., 1996) than delayed activation, thus leading to a more complicated activation protocol. Commonly, reconstructed bovine zygotes are fused electrically followed by activation with Ca2+ ionophore or ionomycin to elevate intracellular Ca2+ levels. After activation, zygotes are then treated with compounds such as the broad spectrum protein synthesis inhibitor, cyclyheximide, or protein kinase inhibitor 6-di-methylaminopurine, thus blocking cyclin B from functioning and reducing the level of MPF that is maintaining meiotic arrest (Liu et al., 1998a,b). One study found that delaying activation in reconstructed bovine zygotes for 4 hr after fusion resulted in a much higher blastocyst rate (26% vs. 5%) further suggesting delaying meiotic resumption allows factors within the oocyte to exchange with the metaphase chromosomes and better remodel the donor nuclei (Shen et al., 2008).
NONNUCLEAR REMODELING
In addition to the exchange of proteins between the transferred nucleus and the cytoplasm to restructure the chromatin, there must also be a synchronization of nonnuclear components of the cell such as the machinery required for cell division and mitochondria. For cell division, the chromatin must be configured such that the chromosomes can segregate appropriately during the first mitosis. Components of the spindle include the centrosomes and γ-tubulin. In SCNT embryos, γ-tubulin accumulates at the metaphase spindle poles and is associated with abnormal chromosome segregation at first mitosis (Zhong et al., 2007). The cytoskeleton of the oocyte is important not only for chromosome segregation, but also for movement of organelles such as mitochondria (Katayama et al., 2006). Similarly the nuclear mitotic apparatus and other markers of spindles and chromosome segregation have been observed to be aberrantly distributed in SCNT embryos (Shin et al., 2002; Simerly et al., 2004; Zhong et al., 2005; Liu et al., 2006). While it is not clear how aberrant cellular remodeling directly affects transcription, cellular remodeling is important for chromosome segregation.
LARGE OFFSPRING SYNDROME
When the nucleus is not sufficiently remodeled and reprogrammed, the resulting aberrant gene expression results in abnormal placental (Tanaka et al., 2001; Suemizu et al., 2003) and fetal development (Cassar-Malek et al., 2010). Generally this has been termed large offspring syndrome (LOS) and is manifested by a variety of phenotypes beginning during gestation with hydroallantois, diminished mammary development, and prolonged gestation. At birth, additional phenotypes are noted including large birth weight, contracted tendons, abnormal organ size, loss of motor control, a large tongue, and in the first weeks after delivery the clones can develop respiratory distress (reviewed by Young et al., 1998; Sinclair et al., 2000; Niemann et al., 2002; Constant et al., 2006) and impaired immune response (Carroll et al., 2005) and then develop obesity in adult animals (Tamashiro et al., 2002). Thus, there is a continuum of phenotypes that are consistent within donor cells, that is, a certain set of donor cells generally gives rise to the same phenotypes, again suggesting that a change has occurred to the structure of the nucleus that the cytoplasm of the oocyte is unable to correct or modify. Additionally, there are significant species differences. Bovine clones tend to be overweight at birth, while pigs are underweight (Estrada et al., 2007) with underdeveloped placenta, and mouse concepti have an overgrowth of their placenta (Young et al., 1998; Constant et al., 2006). Presumably, correct remodeling would result in correct reprogramming and correct reprogramming would result in normal concepti and offspring that are capable of reaching puberty and reproducing.
One might assume that all the abnormal phenotypes resulting from cloning would be heritable and passed on to offspring or to clones. Unfortunately it is not that straightforward. Some phenotypes such as large birth weight in cattle are not transmitted to offspring (Conway, 1996), neither is the old age obesity observed in some lines of cloned mice (Tamashiro et al., 2002). These observations provide evidence to suggest that some sort of epigenetic reprogramming can correct the abnormality. However, even cloning an animal with an abnormal phenotype can sometimes result in that phenotype disappearing, as in pigs with abnormal facial development (Kolber-Simonds et al., 2004; Cho et al., 2007), or contracted tendons (Randall S. Prather unpublished observations), and we have observed at least two clonal cell lines result in pigs with or without atresia ani (Randall S. Prather unpublished observations). Thus, some phenotypes are corrected while others are transmitted to offspring or clones.
METHODS TO IMPROVE NUCLEAR REMODELING AND REPROGRAMMING
The focus of this review is nuclear remodeling and reprogramming. Clearly some remodeling and reprogramming in clones is not complete. Since reprogramming is dependent upon remodeling the question becomes what can be done to improve the remodeling? Clearly the factors present in the cytoplasm of the oocyte, while they have the ability to facilitate a degree of remodeling, are not capable of sufficient remodeling in all nuclei. So what can be done to facilitate remodeling?
Protein Exchange
One of the main things that should happen for remodeling to occur is to have a free exchange of proteins between the cytoplasm of the oocyte and the transferred nucleus. This exchange probably needs to be at the level of the DNA. Since chromatin is packaged and there are physical constraints on the ability of proteins to exchange with chromatin, any treatments that would open up the chromatin to make it more accessible may improve nuclear remodeling. We should keep in mind that the three-dimensional structure of the nucleus prevents a given gene from undergoing both transcription and DNA synthesis at the same time, as both the DNA polymerases and RNA polymerases cannot occupy the same physical location at the same time. For example, transcription generally occurs in G1 and G2 phases of the cell cycle, while DNA synthesis occurs during S phase. Early embryos have very short, if nonexistent, G1 and G2 phases. So the proteins responsible for remodeling are competing with a very tight packaging that is responsible for chromosome condensation during mitosis, and if additional remodeling occurs during interphase these proteins would be competing with DNA polymerases during S phase of the first cell cycle.
Identification of the proteins that are responsible for determining the architecture of the nucleus may be helpful. Numerous studies have shown that fusing cells together and creating hybrids or simply adding a transcription factor(s) can result in a dramatic change in transcription (Tada et al., 1997). Generally this is called transdifferentiation (Eguchi and Kodama, 1993; Hakelien and Collas, 2002). A similar approach by using a cell free system has resulted in the uptake and assembly of T-cell-specific factors into fibroblast nuclei. This was correlated with chromatin remodeling and expression of T-cell-specific genes and proteins (Collas, 2003). Similar cell-free systems may aid in elucidating the mechanisms responsible for the remodeling properties found in oocyte cytoplasm (Novak et al., 2004).
Inhibition of Histone Deacetylases
While the structure of the transferred nucleus is not always adequately modified by the oocyte cytoplasm (Moreira et al., 2003), some treatments can help facilitate that change. One method is to inhibit histone deacetylases (HDACi). Compounds such as trichostatin A (TSA) and 6-(1,3-dioxo-1H, 3H-benzo[de]isoquinolin-2-yl)-hexanoic acid hydroxyamide (Scriptaid) are potent HDACis. Inhibition of the deacetylases results in an increase in the global acetylation of histones. Increase acetylation results in a change in the chromatin structure such that proteins like RNA polymerases can gain access to the DNA and begin transcription (Van Thuan et al., 2009). Trichostatin A or sodium butyrate treatment improves development of cloned embryos to the blastocyst stage (Li et al., 2008; Das et al., 2010), and in mice TSA improves both the nuclear remodeling (Maalouf et al., 2009) and development to term (Kishigami et al., 2006, 2007; Ding et al., 2008). Although TSA treatment has shown to improve cloning efficiency in mice, other groups reported neonatal death after the TSA treatment of rabbit (Meng et al., 2009) and pig embryos (Zhao et al., 2010a). TSA treatment can also result in an increase in the severity of placentomegaly (Kishigami et al., 2006). Treating reconstructed cloned pig zygotes (but not the donor cell) with Scriptaid (500 nM for 14 hr) results in an increase of histone acetylation intensity (AcH4K8) in the one-cell stage SCNT embryo to a level that is similar to IVF embryos at the same stage (Zhao et al., 2010a). Scriptaid treatment improved the production of cloned mice (Van Thuan et al., 2009) and pigs (Zhao et al., 2009, 2010a). In a report cloning inbred National Institutes of Health miniature pigs, Scriptaid increased the cloning efficiency from 0% to 1.3%, which is an increase in live piglet number from 0 to 14. Scriptaid has also been used to improve cloning efficiency in the large white breed from 0.4% to 1.6% for fetal fibroblast donor cell lines and 0% to 3.7% for more difficult to clone adult ear fibroblasts (Zhao et al., 2009). Minnesota mini piglets have also been successfully cloned by treating reconstructed zygotes with Scriptaid (Fig. 2F). It appears that increased acetylation after HDACi treatment allows for a more normal remodeling/reprogramming event. Studies to determine how Scriptaid is affecting gene expression after SCNT are currently being conducted.
Altering DNA Methylation
Not only can the three-dimensional structure of chromatin be changed by altering histone acetylation, it can be altered by changing DNA methylation. The most widely used chemical that alters DNA methylation is 5-aza-20-deoxycytidine (5-aza-dC). The net result of 5-aza-dC is a reduction in DNA methylation in donor cells. Unfortunately treatment of donor cells does not result in an increase in development in vitro or in vivo (Enright et al., 2005; Tsuji et al., 2009). Ding et al. (2008) treated bovine donor cells and embryos with both TSA and 5-aza-dC and showed increased histone acetylation, decreased DNA methylation and improved blastocyst development (both percentage and cell number) but the authors did not evaluate development beyond the blastocyst stage.
Inhibition of Proteasomal Machinery
Proteasomal inhibition was found to be necessary to clone rat embryos (Zhou et al., 2003) and has been thought to result from maintaining the recipient oocyte in metaphase by preventing the degradation of Cyclin B (Josefsberg et al., 2000). Rat oocytes will precociously activate when flushed from the oviduct; however, treatment with the specific and reversible proteasomal inhibitor MG132 prevents cyclin B degradation and spontaneous activation (Zhou et al., 2003). In this study, oocytes were pretreated with 5 μM MG132 and SCNT was performed within 30 min of the removal of the inhibitor resulting in fertile cloned rat offspring. There is evidence that delayed activation can assist in the remodeling of SCNT embryos that occurs to the nucleus (discussed above). Development of mouse SCNT embryos to the blastocyst stage (but not to term) has been improved with MG132 (Gao et al., 2005). In addition, proteasome inhibitors such as MG132 will prevent degradation of other oocyte-specific proteins beyond Cyclin B. Some of these proteins likely facilitate the nuclear remodeling that is observed (Prather et al., 2004). Are there other compounds that may inhibit the proteasomal pathway that may also enhance nuclear remodeling?
Chromosome Transfer Rather Than Nuclear Transfer
If we assume that it is important to remove as many of the proteins associated with the donor nucleus as possible prior to transfer to the oocyte, so that the only factors affecting the nuclear architecture of the clone are from the cytoplasm of the oocyte, then it might be beneficial to transfer chromosomes in metaphase or telophase rather that those in interphase (Prather, 2000). Theoretically, this should result in packaging of the SCNT nucleus with as few factors from the donor cell as possible and as many factors from the oocyte cytoplasm as possible. While these procedures successfully create clones, the efficiencies are not greatly improved and the same LOS phenotypes are encountered (Egli et al., 2007, 2009).
Nonnuclear Remodeling and Reprogramming Improvements
While this review has focused on nuclear remodeling and reprogramming there are other areas for improvement. (1) Increase the quality of the in vitro matured oocytes. It is established that oocytes from sexually mature animals result in better development than from in vitro matured oocytes (discussed above). (2) Increase the efficiency of enucleation of the oocyte, such as using a microtubule polymerization inhibitor (Ibanez et al., 2003), or a polscope (Moon et al., 2003). (3) Develop methods that improve oocyte activation (discussed above or the use of ultrasound; Sato et al., 2005). (4) Improve the culture conditions for the 1–7 days that the embryos are cultured prior to transfer to a surrogate as these conditions may be different from a normal embryo (Gao et al., 2003) and may even be dependent upon the type of donor cell used. (5) Improve the ability of the chromosomes to segregate properly. Chromosome segregation may be enhanced by culturing with actin polymerization inhibitors compounds such as latrunculin A that have been shown to improve SCNT pig embryo development when used after cell fusion and activation (Himaki et al., 2010). Cell-cycle synchronization may be important as it is thought that donor cells in G1 or G0 of the cell cycle would be the most likely to undergo normal replication (Campbell et al., 1993; Prather, 1996). And (6) treat the surrogates so that they are more likely to carry what may be a compromised fetus to term (Tsuji et al., 2010).
CONCLUSIONS AND FUTURE DEVELOPMENTS
In conclusion, the nucleus that is transferred to the oocyte during SCNT is not structured or packaged as the maternal or paternal pronucleus is packaged. Thus, the cytoplasm of the oocyte is capable of remodeling the transferred nucleus only to a certain degree. In order to facilitate nuclear remodeling and nuclear reprogramming to increase cloning efficiency, additional protocols need to be developed to promote the restructuring that is necessary for normal development of the clone. Possibilities that have been discussed above include transferring chromosomes rather that a nucleus (or perhaps naked DNA) as the factors that are associated with the donor nucleus affect remodeling and transcription. Additional drug treatments that may delay activation of the oocyte such as MG132, open up the chromatin such as Scriptaid, or alter the DNA methylation status, may permit a change in the structure of the donor nucleus so that it is more zygote-like rather than somatic cell-like. Alternatively, there may be components associated with the metaphase chromosomes of the oocyte that would have a positive influence on remodeling the donor chromatin if the oocyte was treated so that these components remained in the cytoplasm at the time of enucleation. Clearly there is room for improvement and nuclear remodeling may be one of things that can be altered to increase the efficiency of cloning by nuclear transfer.
Acknowledgments
This study was funded by the National Institutes of Health (R01 RR013438, U42 RR018877, U42 RR018877-07S2, R21 NS059519, P01 HL051670), National Research Initiative Competitive Grant no. 2006-35203-17282 from the USDA CSREES, and Food for the 21st Century at the University of Missouri.
Abbreviations
- dpc
days post-coitis
- DTT
dithiothreitol
- EGA
embryonic genome activation
- ICM
inner cell mass
- LOS
large offspring syndrome
- SCNT
somatic cell nuclear transfer
- TE
trophectoderm
References
- Akagi S, Kaneyama K, Adachi N, Tsuneishi B, Matsukawa K, Watanabe S, Kubo M, Takahashi S. Bovine nuclear transfer using fresh cumulus cell nuclei and in vivo- or in vitro-matured cytoplasts. Cloning Stem Cells. 2008;10:173–180. doi: 10.1089/clo.2007.0047. [DOI] [PubMed] [Google Scholar]
- Antelman J, Manandhar G, Yi YJ, Li R, Whitworth KM, Sutovsky M, Agca C, Prather RS, Sutovsky P. Expression of mitochondrial transcription factor A (TFAM) during porcine gametogenesis and preimplantation embryo development. J Cell Physiol. 2008;217:529–543. doi: 10.1002/jcp.21528. [DOI] [PubMed] [Google Scholar]
- Aston KI, Li GP, Hicks BA, Sessions BR, Davis AP, Rickords LF, Stevens JR, White KL. Abnormal levels of transcript abundance of developmentally important genes in various stages of preimplantation bovine somatic cell nuclear transfer embryos. Cell Reprogram. 2010;12:23–32. doi: 10.1089/cell.2009.0042. [DOI] [PubMed] [Google Scholar]
- Betts D, Bordignon V, Hill J, Winger Q, Westhusin M, Smith L, King W. Reprogramming of telomerase activity and rebuilding of telomere length in cloned cattle. Proc Natl Acad Sci USA. 2001;98:1077–1082. doi: 10.1073/pnas.031559298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiani M, Eckardt S, Scholer HR, McLaughlin KJ. Oct4 distribution and level in mouse clones: Consequences for pluripotency. Genes Dev. 2002;16:1209–1219. doi: 10.1101/gad.966002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonk AJ, Cheong HT, Li R, Lai L, Hao Y, Liu Z, Samuel M, Fergason EA, Whitworth KM, Murphy CN, Antoniou E, Prather RS. Correlation of developmental differences to the methylation profiles of nuclear transfer donor cells. Epigenetics. 2007;2:179–186. doi: 10.4161/epi.2.3.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonk AJ, Li R, Lai L, Hao Y, Liu Z, Samuel M, Fergason EA, Whitworth KM, Murphy CN, Antoniou E, Prather RS. Aberrant DNA methylation in porcine in vitro-, parthenogenetic-, and somatic cell nuclear transfer-produced embryos. Mol Reprod Dev. 2008;75:250–264. doi: 10.1002/mrd.20786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortvin A, Eggan K, Skaletsky H, Akutsu H, Berry DL, Yanagimachi R, Page DC, Jaenisch R. Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development. 2003;130:1673–1680. doi: 10.1242/dev.00366. [DOI] [PubMed] [Google Scholar]
- Bourc’his D, Le Bourhis D, Patin D, Niveleau A, Comizzoli P, Renard JP, Viegas-Pequignot E. Delayed and incomplete reprogramming of chromosome methylation patterns in bovine cloned embryos. Curr Biol. 2001;11:1542–1546. doi: 10.1016/s0960-9822(01)00480-8. [DOI] [PubMed] [Google Scholar]
- Braude P, Bolton V, Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature. 1988;332:459–461. doi: 10.1038/332459a0. [DOI] [PubMed] [Google Scholar]
- Campbell KHS, Ritchie WA, Wilmut I. Nuclear-cytoplasmic interactions during the 1st cell cycle of nuclear transfer reconstructed bovine embryos—Implications for deoxyribonucleic acid replication and development. Biol Reprod. 1993;49:933–942. doi: 10.1095/biolreprod49.5.933. [DOI] [PubMed] [Google Scholar]
- Campbell KH, McWhir J, Ritchie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64–66. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- Carroll JA, Carter DB, Korte SW, Prather RS. Evaluation of the acute phase response in cloned pigs following a lipopolysaccharide challenge. Domest Anim Endocrinol. 2005;29:564–572. doi: 10.1016/j.domaniend.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Cassar-Malek I, Picard B, Jurie C, Listrat A, Guillomot M, Chavatte-Palmer P, Heyman Y. Myogenesis is delayed in bovine fetal clones. Cell Reprogram. 2010;12:191–202. doi: 10.1089/cell.2009.0065. [DOI] [PubMed] [Google Scholar]
- Chae JI, Lee KS, Kim DJ, Han YM, Lee DS, Lee KK, Koo DB. Abnormal gene expression in extraembryonic tissue from cloned porcine embryos. Theriogenology. 2009;71:323–333. doi: 10.1016/j.theriogenology.2008.07.025. [DOI] [PubMed] [Google Scholar]
- Chang CC, Gao S, Sung LY, Corry GN, Ma Y, Nagy ZP, Tian XC, Rasmussen TP. Rapid elimination of the histone variant MacroH2A from somatic cell heterochromatin after nuclear transfer. Cell Reprogram. 2010;12:43–53. doi: 10.1089/cell.2009.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavatte-Palmer P, de Sousa N, Laigre P, Camous S, Ponter AA, Beckers JF, Heyman Y. Ultrasound fetal measurements and pregnancy associated glycoprotein secretion in early pregnancy in cattle recipients carrying somatic clones. Theriogenology. 2006;66:829–840. doi: 10.1016/j.theriogenology.2006.01.061. [DOI] [PubMed] [Google Scholar]
- Chavatte-Palmer P, Guillomot M, Roiz J, Heyman Y, Laigre P, Servely JL, Constant F, Hue I, Ellis SA. Placental expression of major histocompatibility complex class I in bovine somatic clones. Cloning Stem Cells. 2007;9:346–356. doi: 10.1089/clo.2006.0086. [DOI] [PubMed] [Google Scholar]
- Cho SK, Kim JH, Park JY, Choi YJ, Jae B, Hwang KC, Cho EJ, Sohn SH, Uhm SJ, Koo DB, Lee KK, Kim T. Serial cloning of pigs by somatic cell nuclear transfer: Restoration of phenotypic normality during serial cloning. Dev Dyn. 2007;236:3369–3382. doi: 10.1002/dvdy.21308. [DOI] [PubMed] [Google Scholar]
- Collas P. Nuclear reprogramming in cell-free extracts. Philosophical Transactions of the Royal Society of London—Series B Biol Sci. 2003;358:1389–1395. doi: 10.1098/rstb.2003.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constant F, Guillomot M, Heyman Y, Vignon X, Laigre P, Servely JL, Renard JP, Chavatte-Palmer P. Large offspring or large placenta syndrome? Morphometric analysis of late gestation bovine placentomes from somatic nuclear transfer pregnancies complicated by hydrallantois. Biol Reprod. 2006;75:122–130. doi: 10.1095/biolreprod.106.051581. [DOI] [PubMed] [Google Scholar]
- Conway KL. Birth weight of bovine calves produced by nuclear transfer (cloning) and their offspring (embryo transfer) Diss Abstr Int. 1996;57:3462. [Google Scholar]
- Daniels R, Hall V, Trounson AO. Analysis of gene transcription in bovine nuclear transfer embryos reconstructed with granulosa cell nuclei. Biol Reprod. 2000;63:1034–1040. doi: 10.1095/biolreprod63.4.1034. [DOI] [PubMed] [Google Scholar]
- Das ZC, Gupta MK, Uhm SJ, Lee HT. Increasing histone acetylation of cloned embryos, but not donor cells, by sodium butyrate improves their in vitro development in pigs. Cell Reprogram. 2010;12:95–104. doi: 10.1089/cell.2009.0068. [DOI] [PubMed] [Google Scholar]
- de Montera B, El Zeihery D, Muller S, Jammes H, Brem G, Reichenbach H-D, Scheipl F, Chavatte-Palmer P, Zakhartchenko V, Schmitz OJ, Wolf E, Renard J-P, Hiendleder S. Quantification of leukocyte genomic 5-methylcytosine levels reveals epigenetic plasticity in healthy adult cloned cattle. Cell Reprogram. 2010;12:175–181. doi: 10.1089/cell.2009.0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sousa PA, King T, Harkness L, Young LE, Walker SK, Wilmut I. Evaluation of gestational deficiencies in cloned sheep fetuses and placentae. Biol Reprod. 2001;65:23–30. doi: 10.1095/biolreprod65.1.23. [DOI] [PubMed] [Google Scholar]
- Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, Wolf E, Reik W. Conservation of methylation reprogramming in mammalian development: Aberrant reprogramming in cloned embryos. Proc Natl Acad Sci USA. 2001;98:13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dementyeva EV, Shevchenko AI, Zakian SM. X-chromosome upregulation and inactivation: Two sides of the dosage compensation mechanism in mammals. Bioessays. 2009;31:21–28. doi: 10.1002/bies.080149. [DOI] [PubMed] [Google Scholar]
- Ding X, Wang Y, Zhang D, Wang Y, Guo Z, Zhang Y. Increased pre-implantation development of cloned bovine embryos treated with 5-aza-2′-deoxycytidine and trichostatin A. Theriogenology. 2008;70:622–630. doi: 10.1016/j.theriogenology.2008.04.042. [DOI] [PubMed] [Google Scholar]
- Eggan K, Akutsu H, Hochedlinger K, Rideout W, Yanagimachi R, Jaenisch R. X-chromosomes inactivation in cloned mouse embryos. Science. 2000;290:1578–1581. doi: 10.1126/science.290.5496.1578. [DOI] [PubMed] [Google Scholar]
- Egli D, Rosains J, Birkhoff G, Eggan K. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature. 2007;447:679–685. doi: 10.1038/nature05879. [DOI] [PubMed] [Google Scholar]
- Egli D, Sandler VM, Shinohara ML, Cantor H, Eggan K. Reprogramming after chromosome transfer into mouse blastomeres. Curr Biol. 2009;19:1403–1409. doi: 10.1016/j.cub.2009.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi G, Kodama R. Transdifferentiation. Curr Opin Cell Biol. 1993;5:1023–1028. doi: 10.1016/0955-0674(93)90087-7. [DOI] [PubMed] [Google Scholar]
- Enright BP, Sung LY, Chang CC, Yang X, Tian XC. Methylation and acetylation characteristics of cloned bovine embryos from donor cells treated with 5-aza-2′-deoxycytidine. Biol Reprod. 2005;72:944–948. doi: 10.1095/biolreprod.104.033225. [DOI] [PubMed] [Google Scholar]
- Estrada J, Sommer J, Collins B, Mir B, Martin A, York A, Petters RM, Piedrahita JA. Swine generated by somatic cell nuclear transfer have increased incidence of intrauterine growth restriction (IUGR) Cloning Stem Cells. 2007;9:229–236. doi: 10.1089/clo.2006.0079. [DOI] [PubMed] [Google Scholar]
- Fuks F. DNA methylation and histone modifications: Teaming up to silence genes. Curr Opin Genet Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Gao SR, Chung YG, Williams JW, Riley J, Moley K, Latham KE. Somatic cell-like features of cloned mouse embryos prepared with cultured myoblast nuclei. Biol Reprod. 2003;69:48–56. doi: 10.1095/biolreprod.102.014522. [DOI] [PubMed] [Google Scholar]
- Gao SR, Chung YG, Parseghian MH, King GJ, Adashi EY, Latham KE. Rapid H1 linker histone transitions following fertilization or somatic cell nuclear transfer: Evidence for a uniform developmental program in mice. Dev Biol. 2004;266:62–75. doi: 10.1016/j.ydbio.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Gao S, Han Z, Kihara M, Adashi E, Latham KE. Protease inhibitor MG132 in cloning: No end to the nightmare. Trends Biotechnol. 2005;23:66–68. doi: 10.1016/j.tibtech.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Brennan B, Fairman S, Mohun TJ. Transcription of muscle-specific actin genes in early Xenopus development: Nuclear transplantation and cell dissociation. Cell. 1984;38:691–700. doi: 10.1016/0092-8674(84)90264-2. [DOI] [PubMed] [Google Scholar]
- Hakelien AM, Collas P. Novel approaches to transdifferentiation. Cloning Stem Cells. 2002;4:379–387. doi: 10.1089/153623002321025050. [DOI] [PubMed] [Google Scholar]
- Heyman Y, Chavatte-Palmer P, LeBourhis D, Camous S, Vignon X, Renard JP. Frequency and occurrence of late-gestation losses from cattle cloned embryos. Biol Reprod. 2002;66:6–13. doi: 10.1095/biolreprod66.1.6. [DOI] [PubMed] [Google Scholar]
- Hiiragi T, Solter D. Reprogramming is essential in nuclear transfer. Mol Reprod Dev. 2005;70:417–421. doi: 10.1002/mrd.20126. [DOI] [PubMed] [Google Scholar]
- Hill JR, Burghardt RC, Jones K, Long CR, Looney CR, Shin T, Spencer TE, Thompson JA, Winger QA, Westhusin ME. Evidence for placental abnormality as the major cause of mortality in first-trimester somatic cell cloned bovine fetuses. Biol Reprod. 2000a;63:1787–1794. doi: 10.1095/biolreprod63.6.1787. [DOI] [PubMed] [Google Scholar]
- Hill JR, Winger QA, Long CR, Looney CR, Thompson JA, Westhusin ME. Development rates of male bovine nuclear transfer embryos derived from adult and fetal cells. Biol Reprod. 2000b;62:1135–1140. doi: 10.1095/biolreprod62.5.1135. [DOI] [PubMed] [Google Scholar]
- Himaki T, Mori H, Mizobe Y, Miyoshi K, Sato M, Takao S, Yoshida M. Latrunculin A dramatically improves the developmental capacity of nuclear transfer embryos derived from gene-modified clawn miniature pig cells. Cell Reprogram. 2010;12:127–132. doi: 10.1089/cell.2009.0066. [DOI] [PubMed] [Google Scholar]
- Hyun S, Lee G, Kim D, Kim H, Lee S, Nam D, Jeong Y, Kim S, Yeom S, Kang S, Han J, Lee B, Hwang W. Production of nuclear transfer-derived piglets using porcine fetal fibroblasts transfected with the enhanced green fluorescent protein. Biol Reprod. 2003;69:1060–1068. doi: 10.1095/biolreprod.102.014886. [DOI] [PubMed] [Google Scholar]
- Ibanez E, Albertini DF, Overstrom EW. Demecolcine-induced oocyte enucleation for somatic cell cloning: Coordination between cell-cycle egress, kinetics of cortical cytoskeletal interactions, and second polar body extrusion. Biol Reprod. 2003;68:1249–1258. doi: 10.1095/biolreprod.102.008292. [DOI] [PubMed] [Google Scholar]
- Inoue K, Ogonuki N, Miki H, Hirose M, Noda S, Kim JM, Aoki F, Miyoshi H, Ogura A. Inefficient reprogramming of the hematopoietic stem cell genome following nuclear transfer. J Cell Sci. 2006;119:1985–1991. doi: 10.1242/jcs.02913. [DOI] [PubMed] [Google Scholar]
- Jarrell VL, Day BN, Prather RS. The transition from maternal to zygotic control of development occurs during the 4-cell stage in the domestic pig, sus scrofa; quantitative and qualitative aspects of protein synthesis. Biol Reprod. 1991;44:62–68. doi: 10.1095/biolreprod44.1.62. [DOI] [PubMed] [Google Scholar]
- Jiang LA, Carter DB, Xu J, Yang X, Prather RS, Tian XC. Telomere lengths in cloned transgenic pigs. Biol Reprod. 2004;70:1589–1593. doi: 10.1095/biolreprod.103.022616. [DOI] [PubMed] [Google Scholar]
- Jiang L, Jobst P, Ayares D, Lai L, Samuel M, Prather RS, Tian XC. Expression of growth-regulating imprinted genes in decreased and surviving cloned pigs. Cloning Stem Cells. 2007a;9:97–106. doi: 10.1089/clo.2006.0041. [DOI] [PubMed] [Google Scholar]
- Jiang L, Jobst P, Prather RS, Ayares D, Yang X, Tian XC. Expression of X-linked genes in deceased neonates and surviving cloned female pigs. Mol Reprod Dev. 2007b;75:265–273. doi: 10.1002/mrd.20758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsberg LBY, Galiani D, Dantes A, Amsterdam A, Dekel N. The proteasome is involved in the first metaphase-to-anaphase transition of meiosis in rat oocytes. Biol Reprod. 2000;62:1270–1277. doi: 10.1095/biolreprod62.5.1270. [DOI] [PubMed] [Google Scholar]
- Jouneau A, Zhou Q, Camus A, Brochard V, Maulny L, Collignon J, Renard JP. Developmental abnormalities of NT mouse embryos appear early after implantation. Development. 2006;133:1597–1607. doi: 10.1242/dev.02317. [DOI] [PubMed] [Google Scholar]
- Kang YK, Koo DB, Park JS, Choi YH, Chung AS, Lee KK, Han YM. Aberrant methylation of donor genome in cloned bovine embryos. Nat Genet. 2001;28:173–177. doi: 10.1038/88903. [DOI] [PubMed] [Google Scholar]
- Katayama M, Zhong Z, Lai L, Sutovsky P, Prather RS, Schatten H. Mitochondrial distribution and microtubule organization in fertilized and cloned porcine embryos: Implications for developmental potential. Dev Biol. 2006;299:206–220. doi: 10.1016/j.ydbio.2006.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishigami S, Mizutani E, Ohta H, Hikichi T, Van Thuan N, Wakayama S, Bui HT, Wakayama T. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem Biophys Res Commun. 2006;340:183–189. doi: 10.1016/j.bbrc.2005.11.164. [DOI] [PubMed] [Google Scholar]
- Kishigami S, Bui HT, Wakayama S, Tokunaga K, Thuan NV, Hikichi T, Mizutani E, Ohta H, Suetsugu R, Sata T, Wakayama T. Successful mouse cloning of an outbred strain by trichostatin A treatment after somatic nuclear transfer. J Reprod Dev. 2007;53:165–170. doi: 10.1262/jrd.18098. [DOI] [PubMed] [Google Scholar]
- Kolber-Simonds D, Lai L, Watt SR, Denaro M, Arn S, Augenstein ML, Betthauser J, Carter DB, Greenstein JL, Hao Y, Im G-S, Liu Z, Mell GD, Murphy CN, Park K-W, Rieke A, Ryan D, Sachs DH, Forsberg EJ, Prather RS, Hawley RJ. Alpha-1,3-galactosyltransferase null pigs via nuclear transfer with fibroblasts bearing loss of heterozygosity mutations. Proc Natl Acad Sci USA. 2004;101:7335–7340. doi: 10.1073/pnas.0307819101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubiak JZ, Prather RS, Maul GG, Schatten G. Cytoplasmic modification of the nuclear lamina during pronuclear-like transformation of mouse blastomere nuclei. Mech Dev. 1991;35:103–111. doi: 10.1016/0925-4773(91)90061-a. [DOI] [PubMed] [Google Scholar]
- Kühholzer B, Hawley RJ, Lai L, Kolber-Simonds D, Prather RS. Clonal lines of transgenic fibroblast cells derived from the same fetus result in different development when used for nuclear transfer in pigs. Biol Reprod. 2001;64:1695–1698. doi: 10.1095/biolreprod64.6.1695. [DOI] [PubMed] [Google Scholar]
- Lai LX, Kolber-Simonds D, Park KW, Cheong HT, Greenstein JL, Im GS, Samuel M, Bonk A, Rieke A, Day BN, Murphy CN, Carter DB, Hawley RJ, Prather RS. Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science. 2002;295:1089–1092. doi: 10.1126/science.1068228. [DOI] [PubMed] [Google Scholar]
- Lanza RP, Cibelli JB, Blackwell C, Cristofalo VJ, Francis MK, Baerlocher GM, Mak J, Schertzer M, Chavez EA, Sawyer N, Lansdorp PM, West MD. Extension of cell life-span and telomere length in animals cloned from senescent somatic cells. Science. 2000;288:665–669. doi: 10.1126/science.288.5466.665. [DOI] [PubMed] [Google Scholar]
- Latham KE. Mechanisms and control of embryonic genome activation in mammalian embryos. Int Rev Cytol. 1999;193:71–124. doi: 10.1016/s0074-7696(08)61779-9. [DOI] [PubMed] [Google Scholar]
- Lee J, Inoue K, Ono R, Ogonuki N, Kohda T, Kaneko-Ishino T, Ogura A, Ishino F. Erasing genomic imprinting memory in mouse clone embryo produced from day 11.5 primoridal germ cells. Development. 2002;129:1807–1817. doi: 10.1242/dev.129.8.1807. [DOI] [PubMed] [Google Scholar]
- Lee G-S, Hyun S-H, Kim H-S, Kim D-Y, Lee S-H, Lim J-M, Lee E-S, Hwang W-S. Improvement of a porcine somatic cell nuclear transfer technique by optimizing donor cell and recipient oocyte preparations. Theriogenology. 2003;59:1949–1957. doi: 10.1016/s0093-691x(02)01294-3. [DOI] [PubMed] [Google Scholar]
- Lee RSF, Peterson AJ, Donnison MJ, Ravelich S, Ledgard AM, Li N, Oliver JE, Miller AL, Tucker FC, Breier B, Wells DN. Cloned cattle fetuses with the same nuclear genetics are more variable than contemporary half-siblings resulting from artificial insemination and exhibit fetal and placental growth deregulation even in the first trimester. Biol Reprod. 2004;70:1–11. doi: 10.1095/biolreprod.103.020982. [DOI] [PubMed] [Google Scholar]
- Li X, Kato Y, Tsunoda Y. Comparative analysis of development-related gene expression in mouse preimplantation embryos with different developmental potential. Mol Reprod Dev. 2005;72:152–160. doi: 10.1002/mrd.20346. [DOI] [PubMed] [Google Scholar]
- Li J, Svarcova O, Villemoes K, Kragh PM, Schmidt M, Bogh IB, Zhang Y, Du Y, Lin L, Purup S, Xue Q, Bolund L, Yang H, Maddox-Hyttel P, Vajta G. High in vitro development after somatic cell nuclear transfer and trichostatin A treatment of reconstructed porcine embryos. Theriogenology. 2008;70:800–808. doi: 10.1016/j.theriogenology.2008.05.046. [DOI] [PubMed] [Google Scholar]
- Lin H, Gupta V, Vermilyea MD, Falciani F, Lee JT, O’Neill LP, Turner BM. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol. 2007;5:e326. doi: 10.1371/journal.pbio.0050326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Ju JC, Yang X. Differential inactivation of maturation-promoting factor and mitogen-activated protein kinase following parthenogenetic activation of bovine oocytes. Biol Reprod. 1998a;59:537–545. doi: 10.1095/biolreprod59.3.537. [DOI] [PubMed] [Google Scholar]
- Liu L, Ju JC, Yang X. Parthenogenetic development and protein patterns of newly matured bovine oocytes after chemical activation. Mol Reprod Dev. 1998b;49:298–307. doi: 10.1002/(SICI)1098-2795(199803)49:3<298::AID-MRD10>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Liu ZH, Schatten H, Hao YH, Lai LX, Wax D, Samuel M, Zhong ZS, Sun QY, Prather RS. The nuclear mitotic apparatus (NuMA) protein is contributed by the donor cell nucleus in cloned porcine embryos. Front Biosci. 2006;11:1945–1957. doi: 10.2741/1937. [DOI] [PubMed] [Google Scholar]
- Maalouf WE, Liu Z, Brochard V, Renard JP, Debey P, Beaujean N, Zink D. Trichostatin A treatment of cloned mouse embryos improves constitutive heterochromatin remodeling as well as developmental potential to term. BMC Dev Biol. 2009;9:11. doi: 10.1186/1471-213X-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machaty Z, Wang WH, Day BN, Prather RS. Complete activation of porcine oocytes induced by the sulfhydryl reagent, thimerosal. Biol Reprod. 1997;57:1123–1127. doi: 10.1095/biolreprod57.5.1123. [DOI] [PubMed] [Google Scholar]
- Machaty Z, Rickords LF, Prather RS. Parthenogenetic activation of porcine oocytes after nuclear transfer. Cloning. 1999a;1:101–110. doi: 10.1089/15204559950019988. [DOI] [PubMed] [Google Scholar]
- Machaty Z, Wang WH, Day BN, Prather RS. Calcium release and subsequent development induced by modification of sulfhydryl groups in porcine oocytes. Biol Reprod. 1999b;60:1384–1391. doi: 10.1095/biolreprod60.6.1384. [DOI] [PubMed] [Google Scholar]
- Majewska M, Panasiewicz G, Majewski M, Szafranska B. Localization of chorionic pregnancy-associated glycoprotein family in the pig. Reprod Biol. 2006;6:205–230. [PubMed] [Google Scholar]
- Mananadhar G, Schatten H, Lai L, Ezashi T, Letko J, Laurincik J, Caamano JN, Sutovsky M, Day BN, Sutovsky P. Centrosomal protein centrin is not detectable during early cleavages but reappears during late blastocyst stage in porcine embryos. Biol Reprod. 2004;72:2–13. doi: 10.1530/rep.1.00983. [DOI] [PubMed] [Google Scholar]
- Mann MRW, Chung YG, Nolen LD, Verona RI, Latham KE, Bartolomei MS. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol Reprod. 2003;69:902–914. doi: 10.1095/biolreprod.103.017293. [DOI] [PubMed] [Google Scholar]
- Martin C, Brochard V, Migne C, Zink D, Debey P, Latham KE. Architectural reorganization of the nuclei upon transfer into oocytes accompanies genome reprogramming. Mol Reprod Dev. 2006;73:1102–1111. doi: 10.1002/mrd.20506. [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Embryogenesis—Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Meng Q, Polgar Z, Liu J, Dinnyes A. Live birth of somatic cell-cloned rabbits following trichostatin A treatment and co-transfer of parthenogenetic embryos. Cloning Stem Cells. 2009;11:203–208. doi: 10.1089/clo.2008.0072. [DOI] [PubMed] [Google Scholar]
- Miyashita N, Shiga K, Yonai M, Kaneyama K, Kobayashi S, Kojima T, Goto Y, Kishi M, Aso H, Suzuki T, Sakaguchi M, Nagai T. Remarkable differences in telomere lengths among cloned cattle derived from different cell types. Biol Reprod. 2002;66:1649–1655. doi: 10.1095/biolreprod66.6.1649. [DOI] [PubMed] [Google Scholar]
- Miyashita N, Shiga K, Fujita T, Umeki H, Sato W, Suzuki T, Nagai T. Normal telomere lengths of spermatozoa in somatic cell-cloned bulls. Theriogenology. 2003;59:1557–1565. doi: 10.1016/s0093-691x(02)01195-0. [DOI] [PubMed] [Google Scholar]
- Moon JH, Hyun CS, Lee SW, Son WY, Yoon SH, Lim JH. Visualization of the metaphase II meiotic spindle in living human oocytes using the Polscope enables the prediction of embryonic developmental competence after ICSI. Hum Reprod. 2003;18:817–820. doi: 10.1093/humrep/deg165. [DOI] [PubMed] [Google Scholar]
- Moreira PN, Robl JM, Collas P. Architectural defects in pronuclei of mouse nuclear transplant embryos. J Cell Sci. 2003;116:3713–3720. doi: 10.1242/jcs.00692. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major transition in early Xenopus embryos. I. Characterization and timing of cellular changes at the midblastula stage. Cell. 1982a;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major transition in early Xenopus embryos. II. Control of the onset of transcription. Cell. 1982b;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- Niemann H, Wrenzycki C, Lucas-Hahn A, Brambrink T, Kues WA, Carnwath JW. Gene expression patterns in bovine in vitro-produced and nuclear transfer-derived embryos and their implications for early development. Cloning Stem Cells. 2002;4:29–38. doi: 10.1089/153623002753632020. [DOI] [PubMed] [Google Scholar]
- Niemann H, Carnwath JW, Herrmann D, Wieczorek G, Lemme E, Lucas-Hahn A, Olek S. DNA methylation patterns reflect epigenetic reprogramming in bovine embryos. Cell Reprogram. 2010;12:33–42. doi: 10.1089/cell.2009.0063. [DOI] [PubMed] [Google Scholar]
- Nolen LD, Gao SR, Han ZM, Mann MRW, Chung YG, Otte AP, Bartolomei MS, Latham KE. X chromosome reactivation and regulation in cloned embryos. Dev Biol. 2005;279:525–540. doi: 10.1016/j.ydbio.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Novak S, Paradis F, Savard C, Tremblay K, Sirard MA. Identification of porcine oocyte proteins that are associated with somatic cell nuclei after co-incubation. Biol Reprod. 2004;71:1279–1289. doi: 10.1095/biolreprod.103.027037. [DOI] [PubMed] [Google Scholar]
- Oback B, Wells DN. Donor cell differentiation, reprogramming, and cloning efficiency: Elusive or illusive correlation? Mol Reprod Dev. 2007;74:646–654. doi: 10.1002/mrd.20654. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Heard E. Lessons from comparative analysis of X-chromosome inactivation in mammals. Chromosome Res. 2009;17:659–669. doi: 10.1007/s10577-009-9057-7. [DOI] [PubMed] [Google Scholar]
- Ouhibi N, Fulka J, Kanka J, Moor RM. Nuclear transplantation of ectodermal cells in pig oocytes—Ultrastructure and radiography. Mol Reprod Dev. 1996;44:533–539. doi: 10.1002/(SICI)1098-2795(199608)44:4<533::AID-MRD13>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Park KW, Cheong HT, Lai L, Im GS, Kühholzer B, Bonk A, Samuel M, Rieke A, Day BN, Murphy CN, Carter DB, Prather RS. Production of nuclear transfer-derived swine that express the enhanced green fluorescent protein. Anim Biotechnol. 2001a;12:173–181. doi: 10.1081/abio-100108344. [DOI] [PubMed] [Google Scholar]
- Park KW, Lai L, Cheong HT, Im GS, Sun QY, Wu G, Day BN, Prather RS. Developmental potential of porcine nuclear transfer embryos derived from transgenic fetal fibroblasts infected with the gene for the green fluorescent protein: Comparison of different fusion/activation conditions. Biol Reprod. 2001b;65:1681–1685. doi: 10.1095/biolreprod65.6.1681. [DOI] [PubMed] [Google Scholar]
- Parry TW, Prather RS. Carry-over of mRNA during nuclear transfer in pigs. Reprod Nutr Dev. 1995;35:313–318. doi: 10.1051/rnd:19950307. [DOI] [PubMed] [Google Scholar]
- Patrat C, Okamoto I, Diabangouaya P, Vialon V, Le Baccon P, Chow J, Heard E. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc Natl Acad Sci USA. 2009;106:5198–5203. doi: 10.1073/pnas.0810683106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister-Genskow M, Myers C, Childs LA, Lacson JC, Patterson T, Betthauser JM, Goueleke PJ, Koppang RW, Lange G, Fisher P, Watt SR, Forsberg EJ, Zheng Y, Leno GH, Schultz RM, Liu B, Chetia C, Yang X, Hoeschele I, Eilertsen KJ. Identification of differentially expressed genes in individual bovine preimplantation embryos produced by nuclear transfer: Improper reprogramming of genes required for development. Biol Reprod. 2005;72:546–555. doi: 10.1095/biolreprod.104.031799. [DOI] [PubMed] [Google Scholar]
- Prather RS. Progress in cloning embryos from domesticated livestock [Review] Proc Soc Exp Biol Med. 1996;212:38–43. doi: 10.3181/00379727-212-43989. [DOI] [PubMed] [Google Scholar]
- Prather RS. Cloning mammals by nuclear transfer. Encycl Reprod I. 1999:638–644. [Google Scholar]
- Prather RS. Cloning—Pigs is pigs. Science. 2000;289:1886–1887. doi: 10.1126/science.289.5486.1886. [DOI] [PubMed] [Google Scholar]
- Prather RS, Rickords LF. Developmental regulation of a snRNP core protein epitope during pig embryogenesis and after nuclear transfer for cloning. Mol Reprod Dev. 1992;33:119–123. doi: 10.1002/mrd.1080330202. [DOI] [PubMed] [Google Scholar]
- Prather RS, Schatten G. Construction of the nuclear matrix at the transition from maternal to zygotic control of development in the mouse: An immunocytochemical study. Mol Reprod Dev. 1992;32:203–208. doi: 10.1002/mrd.1080320304. [DOI] [PubMed] [Google Scholar]
- Prather RS, Sims MM, Maul GG, First NL, Schatten G. Nuclear lamin antigens are developmentally regulated during porcine and bovine embryogenesis. Biol Reprod. 1989;41:123–132. doi: 10.1095/biolreprod41.1.123. [DOI] [PubMed] [Google Scholar]
- Prather RS, Sims MM, First NL. Nuclear transplantation in the pig embryo: Nuclear swelling. J Exp Zool. 1990;255:355–358. doi: 10.1002/jez.1402550312. [DOI] [PubMed] [Google Scholar]
- Prather RS, Kubiak J, Maul GG, First NL, Schatten G. The expression of nuclear lamin A and C epitopes is regualted by the developmental stage of the cytoplasm in mouse oocytes or embryos. J Exp Zool. 1991;257:110–114. doi: 10.1002/jez.1402570114. [DOI] [PubMed] [Google Scholar]
- Prather RS, Kühholzer B, Lai L, Park KW. Changes in the structure of nuclei after transfer to oocytes. Cloning. 2000;2:117–122. doi: 10.1089/152045500750039815. [DOI] [PubMed] [Google Scholar]
- Prather RS, Sutovsky P, Green JA. Nuclear remodeling and reprogramming in transgenic pig production. Proc Soc Exp Biol Med. 2004;229:1120–1126. doi: 10.1177/153537020422901106. [DOI] [PubMed] [Google Scholar]
- Ramsoondar JJ, Christopherson RL, Guilbert LJ, Dixon WT, Ghahary A, Ellis S, Wegmann TC, Piedrahita JA. Lack of class I major histocompatibility antigens on trophoblast of periimplantation blastocysts and term placenta in the pig. Biol Reprod. 1999;60:387–397. doi: 10.1095/biolreprod60.2.387. [DOI] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Roh S, Shim H, Hwang WS, Yoon JT. In vitro development of green fluorescent protein (GFP) transgenic bovine embryos after nuclear transfer using different cell cycles and passages of fetal fibroblasts. Reprod Fertil Dev. 2000;12:1–6. doi: 10.1071/rd00021. [DOI] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- Sato K, Yoshida M, Miyoshi K. Utility of ultrasound stimulation for activation of pig oocytes matured in vitro. Mol Reprod Dev. 2005;72:396–403. doi: 10.1002/mrd.20352. [DOI] [PubMed] [Google Scholar]
- Sawai K, Takahashi M, Moriyasu S, Hirayama H, Minamihashi A, Hashizume T, Onoe S. Changes in the DNA methylation status of bovine embryos from the blastocyst to elongated stage derived from somatic cell nuclear transfer. Cell Reprogram. 2010;12:15–22. doi: 10.1089/clo.2009.0039. [DOI] [PubMed] [Google Scholar]
- Senda S, Wakayama T, Yamazaki Y, Ohgane J, Hattori N, Tanaka S, Yanagimachi R, Shiota K. Skewed X-inactivation in cloned mice. Biochem Biophys Res Commun. 2004;321:38–44. doi: 10.1016/j.bbrc.2004.06.096. [DOI] [PubMed] [Google Scholar]
- Shen PC, Lee SN, Liu BT, Chu FH, Wang CH, Wu JS, Lin HH, Cheng WT. The effect of activation treatments on the development of reconstructed bovine oocytes. Anim Reprod Sci. 2008;106:1–12. doi: 10.1016/j.anireprosci.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Shiels PG, Kind AJ, Campbell KHS, Waddington D, Wilmut I, Colman A, Schnieke AE. Analysis of telomere lengths in cloned sheep. Nature. 1999;399:316–317. doi: 10.1038/20580. [DOI] [PubMed] [Google Scholar]
- Shimozawa N, Ono Y, Kimoto S, Hioki K, Araki Y, Shinkai Y, Kono T, Ito M. Abnormalities in cloned mice are not transmitted to the progeny. Genesis. 2002;34:203–207. doi: 10.1002/gene.10143. [DOI] [PubMed] [Google Scholar]
- Shin MR, Park SW, Shim H, Kim NH. Nuclear and microtubule reorganiztion in nuclear-transferred bovine embryos. Mol Reprod Dev. 2002;62:74–82. doi: 10.1002/mrd.10069. [DOI] [PubMed] [Google Scholar]
- Simerly C, Navara C, Hyun SH, Lee BC, Kang SK, Capuano S, Gosman G, Dominko T, Chong KY, Compton D, Hwang WS, Schatten G. Embryogenesis and blastocyst development after somatic cell nuclear transfer in nonhuman primates: Overcoming defects caused by meiotic spindle extraction. Dev Biol. 2004;276:237–252. doi: 10.1016/j.ydbio.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Sinclair KD, Young LE, Wilmut I, McEvoy TG. In-utero overgrowth in ruminants following embryo culture: Lessons from mice and a warning to men. Hum Reprod. 2000;15:68–86. doi: 10.1093/humrep/15.suppl_5.68. [DOI] [PubMed] [Google Scholar]
- Smith SL, Everts RE, Tian XC, Du FL, Sung LY, Rodriguez-Zas SL, Jeong BS, Renard JP, Lewin HA, Yang XZ. Global gene expression profiles reveal significant nuclear reprogramming by the blastocyst stage after cloning. Proc Natl Acad Sci USA. 2005;102:17582–17587. doi: 10.1073/pnas.0508952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers J, Smith C, Donnison M, Wells DN, Henderson H, McLeay L, Pfeffer PL. Gene expression profiling of individual bovine nuclear transfer blastocysts. Reproduction. 2006;131:1073–1084. doi: 10.1530/rep.1.00967. [DOI] [PubMed] [Google Scholar]
- Suemizu H, Aiba K, Yoshikawa T, Sharov AA, Shimozawa N, Tamaoki N, Ko MSH. Expression profiling of placentomegaly associated with nuclear transplantation of mouse ES cells. Dev Biol. 2003;253:36–53. doi: 10.1006/dbio.2002.0870. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Prather RS. Nuclear remodeling after SCNT: A contractor’s nightmare. Trends Biotechnol. 2004;22:205–208. doi: 10.1016/j.tibtech.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Manandhar G, Laurincik J, Letko J, Caamano JN, Day BN, Lai L, Prather RS, Sharpe-Timms KL, Zimmer R, Sutovsky M. Expression and proteasomal degradation of the major vault protein (MVP) in mammalian oocytes and zygotes. Reproduction. 2005;129:269–282. doi: 10.1530/rep.1.00291. [DOI] [PubMed] [Google Scholar]
- Suzuki J, Jr, Therrien J, Filion F, Lefebvre R, Goff AK, Smith LC. In vitro culture and somatic cell nuclear transfer affect imprinting of SNRPN gene in pre- and post-implantation stages of development in cattle. BMC Dev Biol. 2009;9:9. doi: 10.1186/1471-213X-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranska B, Miura R, Ghosh D, Ezashi T, Xie S, Roberts RM, Green JA. Gene for porcine pregnancy-associated glycoprotein 2 (poPAG2): Its structural organization and analysis of its promoter. Mol Reprod Dev. 2001;60:137–146. doi: 10.1002/mrd.1070. [DOI] [PubMed] [Google Scholar]
- Tada M, Tada T, Lefebvre L, Barton SC, Surani MA. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 1997;16:6510–6520. doi: 10.1093/emboj/16.21.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamashiro KLK, Wakayama T, Akutsu H, Yamazaki Y, Lachey JL, Wortman MD, Seeley RJ, D’Alessio DA, Woods SC, Yanagimachi R, Sakai RR. Cloned mice have an obese phenotype not transmitted to their offspring. Nat Med. 2002;8:262–267. doi: 10.1038/nm0302-262. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Oda M, Toyoshima Y, Wakayama T, Tanaka M, Yoshida N, Hattori N, Ohgane J, Yanagimachi R, Shiota K. Placentomegaly in cloned mouse concepti caused by expansion of the spongiotrophoblast layer. Biol Reprod. 2001;65:1813–1821. doi: 10.1095/biolreprod65.6.1813. [DOI] [PubMed] [Google Scholar]
- Telford NA, Watson AJ, Schultz GA. Transition from maternal to embryonic control in early mammalian development: A comparison of several species. Mol Reprod Dev. 1990;26:90–100. doi: 10.1002/mrd.1080260113. [DOI] [PubMed] [Google Scholar]
- Telugu BP, Green JA. Characterization of the peptidase activity of recombinant porcine pregnancy-associated glycoprotein-2. J Biochem. 2008;144:725–732. doi: 10.1093/jb/mvn127. [DOI] [PubMed] [Google Scholar]
- Telugu BP, Spate L, Prather RS, Green JA. Acid peptidase activity released from in vitro produced porcine embryos: A candidate marker to predict developmental competence. Mol Reprod Dev. 2009;76:417–428. doi: 10.1002/mrd.20965. [DOI] [PubMed] [Google Scholar]
- Teranishi T, Tanaka M, Kimoto S, Ono Y, Miyakoshi K, Kono T, Yoshimura Y. Rapid replacement of somatic linker histones with the oocyte-specific linker histone H1 foo in nuclear transfer. Dev Biol. 2004;266:76–86. doi: 10.1016/j.ydbio.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Tian XC, Xu J, Yang XZ. Normal telomere lengths found in cloned cattle. Nat Genet. 2000;26:272–273. doi: 10.1038/81559. [DOI] [PubMed] [Google Scholar]
- Tsuji Y, Kato Y, Tsunoda Y. The developmental potential of mouse somatic cell nuclear-transferred oocytes treated with trichostatin A and 5-aza-2′-deoxycytidine. Zygote. 2009;17:109–115. doi: 10.1017/S0967199408005133. [DOI] [PubMed] [Google Scholar]
- Tsuji Y, Kato Y, Tsunoda Y. Effect of human chorionic gonadotropin and progesterone administration on the developmental potential of mouse somatic cell nuclear-transferred oocytes. Cell Reprogram. 2010;12:183–189. doi: 10.1089/cell.2009.0064. [DOI] [PubMed] [Google Scholar]
- Van Thuan N, Bui HT, Kim JH, Hikichi T, Wakayama S, Kishigami S, Mizutani E, Wakayama T. The histone deacetylase inhibitor scriptaid enhances nascent mRNA production and rescues full-term development in cloned inbred mice. Reproduction. 2009;138:309–317. doi: 10.1530/REP-08-0299. [DOI] [PubMed] [Google Scholar]
- Wakayama T, Shinkai Y, Tamashiro KL, Niida H, Blanchard DC, Blanchard RJ, Ogura A, Tanemura K, Tachibana M, Perry AC, Colgan DF, Mombaerts P, Yanagimachi R. Cloning of mice to six generations. Nature. 2000;407:318–319. doi: 10.1038/35030301. [DOI] [PubMed] [Google Scholar]
- Wakefield L, Gurdon JB. Cytoplasmic resulation of 5S RNA genes in nuclear-transplant embryos. EMBO J. 1983;2:1613–1619. doi: 10.1002/j.1460-2075.1983.tb01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakisaka-Saito N, Kohda T, Inoue K, Ogonuki N, Miki H, Hikichi T, Mizutani E, Wakayama T, Kaneko-Ishino T, Ogura A, Ishino F. Chorioallantoic placenta defects in cloned mice. Biochem Biophys Res Commun. 2006;349:106–114. doi: 10.1016/j.bbrc.2006.08.057. [DOI] [PubMed] [Google Scholar]
- Wei Y, Zhu J, Huan Y, Liu Z, Yang C, Zhang X, Mu Y, Xia P, Liu Z. Aberrant expression and methylation status of putatively imprinted genes in placenta of cloned piglets. Cell Reprogram. 2010;12:213–222. doi: 10.1089/cell.2009.0090. [DOI] [PubMed] [Google Scholar]
- Wells DN, Misica PM, Day TA, Tervit HR. Production of cloned lambs from an established embryonic cell line: A comparison between in vivo- and in vitro-matured cytoplasts. Biol Reprod. 1997;57:385–393. doi: 10.1095/biolreprod57.2.385. [DOI] [PubMed] [Google Scholar]
- Whitaker M, Swann K. Lighting the fuse at fertilization. Development. 1993;117:1–12. [Google Scholar]
- Whitworth KM, Agca C, Kim JG, Patel RV, Springer GK, Bivens NJ, Forrester LJ, Mathialagan N, Green JA, Prather RS. Transcriptional profiling of pig embryogenesis by using a 15-K member unigene set specific for pig reproductive tissues and embryos. Biol Reprod. 2005;72:1437–1451. doi: 10.1095/biolreprod.104.037952. [DOI] [PubMed] [Google Scholar]
- Whitworth KM, Li R, Spate LD, Wax DM, Rieke A, Whyte JJ, Manandhar G, Sutovsky M, Green JA, Sutovsky P, Prather RS. Method of oocyte activation affects cloning efficiency in pigs. Mol Reprod Dev. 2009;76:490–500. doi: 10.1002/mrd.20987. [DOI] [PubMed] [Google Scholar]
- Whitworth KM, Spate LD, Li R, Rieke A, Sutovsky P, Green JA, Prather RS. Activation method does not alter abnormal placental gene expression and development in cloned pigs. Mol Reprod Dev. 2010;77:1016–1030. doi: 10.1002/mrd.21235. [DOI] [PubMed] [Google Scholar]
- Wilson JM, Williams JD, Bondioli KR, Looney CR, Westhusin ME, McCalla DF. Comparison of birth weight and growth characteristics of bovine calves produced by nuclear transfer (cloning), embryo transfer and natural mating. Anim Reprod Sci. 1995;38:73–83. [Google Scholar]
- Winger QA, Hill JR, Shin TY, Watson AJ, Kraemer DC, Westhusin ME. Genetic reprogramming of lactate dehydrogenase, citrate synthase, and phosphofructokinase mRNA in bovine nuclear transfer embryos produced using bovine fibroblast cell nuclei. Mol Reprod Dev. 2000;56:458–464. doi: 10.1002/1098-2795(200008)56:4<458::AID-MRD3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Wooding FB, Roberts RM, Green JA. Light and electron microscope immunocytochemical studies of the distribution of pregnancy associated glycoproteins (PAGs) throughout pregnancy in the cow: Possible functional implications. Placenta. 2005;26:807–827. doi: 10.1016/j.placenta.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Wrenzycki C, Wells D, Herrmann D, Miller A, Oliver J, Tervit R, Niemann H. Nuclear transfer protocol affects messenger RNA expression patterns in cloned bovine blastocysts. Biol Reprod. 2001;65:309–317. doi: 10.1095/biolreprod65.1.309. [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Fujita TC, Low EW, Alarcon VB, Yanagimachi R, Marikawa Y. Gradual DNA demethylation of the Oct4 promoter in cloned mouse embryos. Mol Reprod Dev. 2006;73:180–188. doi: 10.1002/mrd.20411. [DOI] [PubMed] [Google Scholar]
- Yang X, Smith SL, Tian XC, Lewin HA, Renard JP, Wakayama T. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat Genet. 2007;39:295–302. doi: 10.1038/ng1973. [DOI] [PubMed] [Google Scholar]
- Young LE, Beaujean N. DNA methylation in the preimplantation embryo: The differing stories of the mouse and sheep. Anim Reprod Sci. 2004;82–83:61–78. doi: 10.1016/j.anireprosci.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Young LE, Sinclair KD, Wilmut I. Large offspring syndrome in cattle and sheep. Rev Reprod. 1998;3:155–163. doi: 10.1530/ror.0.0030155. [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M. Activation of embryonic genes during preimplantation rat development. Mol Reprod Dev. 1994;38:30–35. doi: 10.1002/mrd.1080380106. [DOI] [PubMed] [Google Scholar]
- Zhao J, Ross JW, Hao Y, Spate LD, Walters EM, Samuel MS, Rieke A, Murphy CN, Prather RS. Significant improvement in cloning efficiency of an inbred miniature pig by histone deacetylase inhibitor treatment after somatic cell nuclear transfer. Biol Reprod. 2009;81:525–530. doi: 10.1095/biolreprod.109.077016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hao Y, Ross JW, Spate LD, Walters EM, Samuel MS, Rieke A, Murphy CN, Prather RS. Histone deacetylase inhibitors improve in vitro and in vivo developmental competence of somatic cell nuclear transfer porcine embryos. Cell Reprogram. 2010a;12:75–83. doi: 10.1089/cell.2009.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Whyte JJ, Prather RS. Effect of epigenetic regulation during swine embryogenesis and on cloning by nuclear transfer. Cell Tissue Res. 2010b;341:13–21. doi: 10.1007/s00441-010-1000-x. [DOI] [PubMed] [Google Scholar]
- Zhong ZS, Zhang G, Meng XQ, Zhang YL, Chen DY, Schatten H, Sun QY. Function of donor cell centrosome in intraspecies and interspecies nuclear transfer embryos. Exp Cell Res. 2005;306:35–46. doi: 10.1016/j.yexcr.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Zhong Z, Spate L, Hao Y, Li R, Lai L, Katayama M, Sun Q-Y, Prather RS, Schatten H. Remodeling of centrosomes in intraspecies and interspecies nuclear transfer porcine embryos. Cell Cycle. 2007;6:1510–1520. [PubMed] [Google Scholar]
- Zhou Q, Renard JP, Le Friec G, Brochard V, Beaujean N, Cherifi Y, Fraichard A, Cozzi J. Generation of fertile cloned rats by regulating oocyte activation. Science. 2003;302:1179. doi: 10.1126/science.1088313. [DOI] [PubMed] [Google Scholar]
- Zimmerman U, Vienken J. Electric field-induced cell-to-cell fusion. J Membr Biol. 1982;67:158–1174. doi: 10.1007/BF01868659. [DOI] [PubMed] [Google Scholar]