Summary
Cyclophilin A (CypA) is over-expressed in a number of human cancer types, but the mechanisms by which CypA promotes oncogenic properties of cells are not understood. Here we demonstrate that CypA binds to and prevents the CrkII adaptor protein from switching to the inhibited state. CrkII is involved in cell motility and invasion by mediating signaling through its SH2 and SH3 domains. CrkII Tyr221 phosphorylation by the Abl or EGFR kinases induces an inhibited state of CrkII, by means of an intramolecular SH2-pTyr221 interaction, causing signaling interruption. We show that the CrkII phosphorylation site constitutes a binging site for CypA. Recruitment of CypA sterically restricts the accessibility of Tyr221 to kinases, thereby suppressing CrkII phosphorylation and promoting the active state. Structural, biophysical, and in vivo data show that CypA augments CrkII-mediated signaling. A strong stimulation of cell migration is observed in cancer cells wherein both CypA and CrkII are greatly up-regulated.
INTRODUCTION
Cyclophilin A (CypA) is a highly abundant protein, accounting for up to ~0.6% of the total cytosolic protein content1. CypA is involved in a growing number of biological processes, including protein folding, signal transduction, viral infection, trafficking, receptor assembly, immune response, and transcription regulation2. Although several proteins have been identified to interact with CypA3–6, the underlying mechanism of the CypA action and the physiological implications of the interactions remain in most cases unknown. CypA exhibits peptidyl–prolyl cis–trans isomerase (PPIase) activity by catalyzing cis–trans isomerization of peptide bonds preceding proline residues7. CypA can in principle act as an enzyme or a binding partner8 in mediating the biological processes.
Over the recent years a number of studies have consistently demonstrated that CypA is significantly up-regulated in various cancers and cancer lines9,10. The mRNA expression profile of different tumors shows an elevated level of CypA mRNA11 and proteomic studies of cells treated with anticancer drugs revealed a decrease in the level of CypA10. CypA has been directly linked to cell proliferation and tumor growth and treatment of cells with cyclosporin (CsA) or the downregulation of CypA by small interfering RNA (si-RNA) reduces the proliferative effects of CypA12,13. Although there is ample evidence to suggest that the presence of CypA confers a proliferative and anti-apoptotic effect, the molecular mechanisms of CypA function in oncogenesis remain elusive.
The Crk (CT-10 Regulation of Kinase) family of adaptor proteins are ubiquitously expressed in tissues and mediate the formation of protein complexes elicited by various extracellular stimuli, including growth and differentiation factors14. Crk proteins are overexpressed in many human cancers including lung and breast cancer, glioblastoma and synovial sarcoma15–19. The Crk expression levels positively correlate with the disease stage and poor survival outcome20. Crk knockdown results in attenuated invasion and migration of cancer cell lines21. Crk proteins stimulate the activity of Abl22, a kinase whose fusion to Bcr causes chronic myelogenous leukemia23, and interact strongly with the epidermal growth factor receptor (EGFR) kinase thereby mediating oncogenic EGFR signaling24.
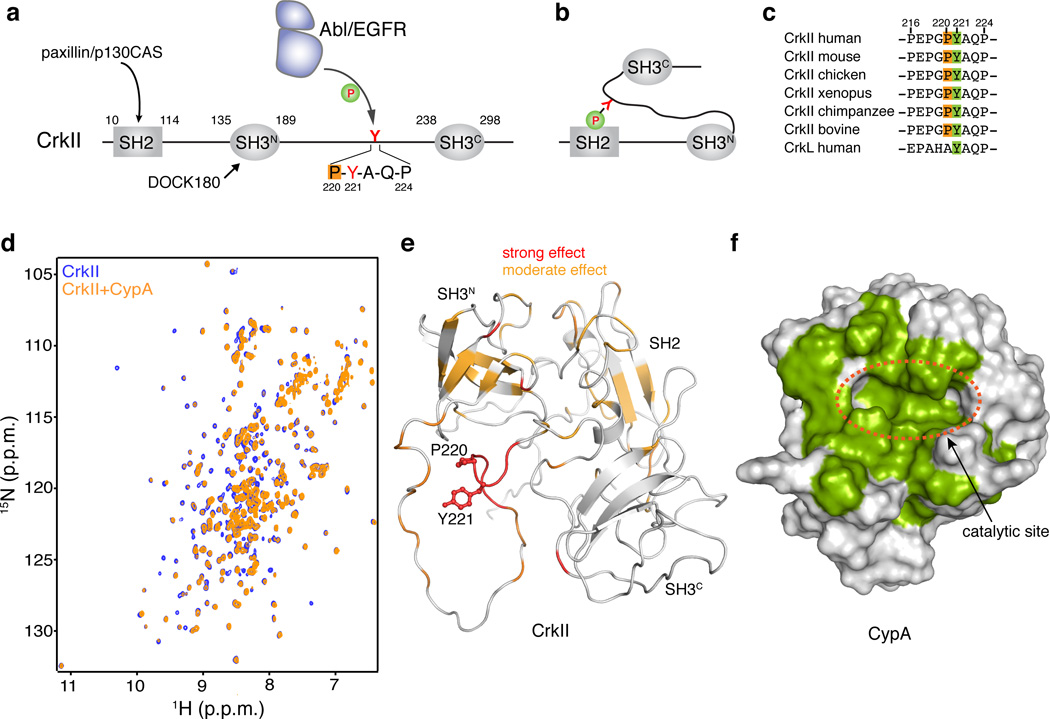
Cellular Crk (CrkII; 304 amino acids) consists of an SH2 domain, an N-terminal SH3 (SH3N) domain and a C-terminal SH3 domain (SH3C) (Fig. 1a). Previous NMR studies showed that a proline residue (Pro237, human numbering) located at the SH3C domain of chicken CrkII undergoes cis–trans isomerization25,26. The isomerization process regulated the intramolecular interaction between the two SH3 domains and the rate of the interconversion was shown to be accelerated catalytically by CypA25,26. In contrast, Pro237 does not exhibit isomerization in human or other mammalian CrkII proteins27 because of variations in the amino acids flanking Pro23728. The SH3N and SH3C domains are tethered by a 50 residue–long linker, which contains a tyrosine residue (Tyr221) that becomes phosphorylated in vivo by Abl29 and EGFR kinases30. Tyr221 phosphorylation results in intramolecular association between the SH2 domain and this region of the linker (pY221-A-Q-P)27,31 (Fig. 1b). This intramolecular association engages the SH2 domain and prevents its binding to focal adhesion scaffold proteins such as paxillin and p130CAS (Fig. 1a and 1b), thereby reducing drastically cell migration and motility32. This negative regulatory mechanism is absent in CrkI, an alternatively spliced isoform of CrkII that is only 204 residues long14. Thus, CrkI adopts a constitutively active form and for this reason CrkI is associated with more aggressive phenotypes19,27.
Figure 1. Binding between CypA and CrkII.
(a) Domain organization of human CrkII. Tyr221 and the Pro220 residues are highlighted. (b) Phosphorylation of Tyr2211 in CrkII results in intramolecular interaction between pTyr221 and the SH2 domain of CrkII. (c) Sequence alignment of the region Pro216-Pro231 of CrkII from different species. (d) Overlaid 1H-15N HSQC NMR spectra of labeled CrkII (blue) and equimolar unlabeled CypA (orange). (e) Chemical shift mapping of the effect of CypA binding to CrkII. A stretch of ~6 residues flanking Pro220 and encompassing Tyr221 is most affected. CrkII is shown in a ribbon representation (PDB ID 2EYZ). Tyr221 and Pro220 side chains are shown as sticks. (f) Chemical shift mapping of the effect of CrkII binding to CypA. CypA residues strongly affected by CrkII binding are colored green. CypA is shown as a solvent-accessible surface
Here we show that the amino acid sequence flanking Tyr221 in CrkII, a sequence that is universally conserved in all species (Fig. 1c), constitutes a recognition site for CypA. CypA is recruited by CrkII Pro220, which undergoes cis–trans isomerization, and complex formation cuases modulation of the level of CrkII phosphorylation by the Abl and EGFR kinases. This specific interaction between CypA and CrkII occurs both in vitro and in vivo. Suppression of Tyr221 phosphorylation leads to CrkII remaining in the active state wherein the SH2 domain is available to interact with paxillin or p130CAS thereby stimulating cell migration.
RESULTS
Proline isomerization at the CrkII phosphorylation site
Tyr221, the primary phosphorylation site in CrkII (Fig. 1a), is universally conserved among all CrkII proteins (Fig. 1c). Interestingly, the preceding residue, Pro220, is also universally conserved (Fig. 1c). NMR characterization of human CrkII shows that the amide signals of the residues flanking Pro220 (Glu217-Gln223) are duplicated (Supplementary Results, Supplementary Fig. 1a), indicating the presence of two conformations in slow exchange. Characteristic NOE correlations between Gly219 and Pro220, as well as 13Cβ and 13Cγ chemical shift analysis of Pro220 (Supplementary Fig. 1b), indicate that the Gly219–Pro220 prolyl bond exists both in the cis and trans conformation (Supplementary Fig. 1a and c). We conclude that the source of the conformational heterogeneity in the phosphorylation site of CrkII is the presence of cis–trans isomerization about the Gly220–Pro221 prolyl bond.
The effect of Pro220 cis–trans isomerization is local, with only 7 residues (Glu217-Gln223) showing duplicate peaks. This is expected since Pro220 is located in an unstructured region of the linker26,27. A synthetic peptide encompassing the Pro216-Ser225 region exhibits cis–trans isomerization at Pro220 demonstrating that this process is an intrinsic property of this sequence (Supplementary Fig. 2a, b). The sequence of the region around Pro220 is absolutely conserved among all CrkII proteins (Fig. 1c) and thus this isomerization process appears to be evolutionary conserved.
CypA is recruited at the CrkII phosphorylation site
The presence of a heterogeneous proline residue undergoing cis–trans isomerization provides a potential binding site for PPIase enzymes. In fact, the Gly219–Pro220 motif in CrkII presents a preferred recognition site for CypA33. To determine whether CypA interacts with this region in CrkII, we titrated CypA (human) to 15N-labeled CrkII (human) (Fig. 1d). Increased amounts of CypA resulted in specific chemical shift perturbations, with the residues close to the Pro220 binding site being most affected (Fig. 1e). Weaker effect was observed for several other residues in CrkII, presumably because of minor rearrangements in its assembly and domain interactions27. The reverse titration, where unlabeled CrkII was titrated to 15N-labeled CypA, further corroborated the specific interaction between the two proteins (Supplementary Fig 3a). Chemical shift and intensity perturbation analysis shows that CrkII binds to the catalytic site of CypA (Fig. 1f). Indeed, addition of CsA, an analog that binds with high affinity to the CypA catalytic site34, abolishes binding between CypA and CrkII (Supplementary Fig. 3b, c). NMR characterization of the interaction between CypA and CrkII showed that the dissociation constant (Kd) of the complex is ~15±4 µM (Supplementary Fig. 3d), an affinity typical of CypA–ligand interactions33. Substitution of Pro220 by Ala in CrkII abrogates binding to CypA (Supplementary Fig. 3e) further corroborating the observation that the heterogeneous Pro220 provides the only CypA-binding site in human CrkII (Supplementary Fig. 2b). Taken together, the data demonstrate that CypA and CrkII form a stoichiometric and specific complex.
Structural basis for the interaction of CypA with CrkII
To understand the basis for the specific interaction between CypA and CrkII, we determined the solution structure of CypA in complex with a peptide encompassing the CypA-binding region of CrkII (Pro216-Ser225) (Supplementary Table 1). NMR analysis demonstrates that the chemical shift effect induced on the binding site of CypA by the peptide is very similar to the effect induced by full-length CrkII suggesting that the contacts between the CrkII peptide or full-length CrkII and CypA are essentially identical (Supplementary Fig. 3a).
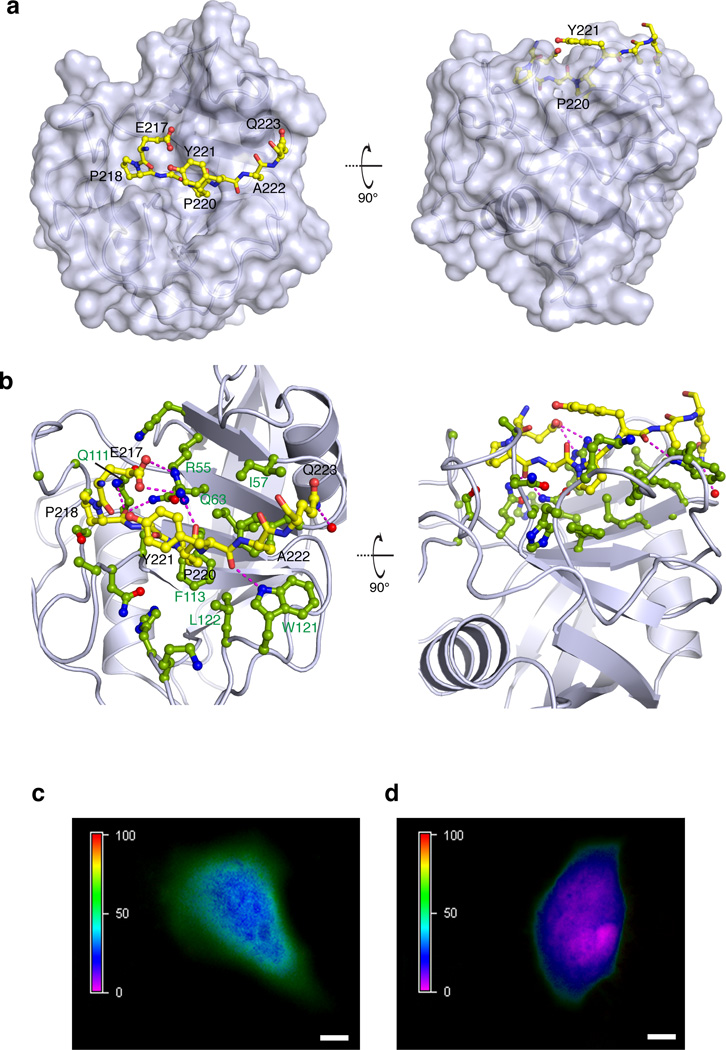
The structural data show that the Gly219–Pro220 region of CrkII binds deep into the active site cleft of CypA (Fig. 2a), in an overall structural arrangement that is reminiscent of the binding of the Gly–Pro motif of the HIV-capsid protein (CA) to CypA6 (Supplementary Fig. 4b). Pro220 fits snugly into a hydrophobic pocket formed by CypA residues Phe60, Met61, Ala101, Phe113, and Leu122 (Fig. 2b). Several hydrogen bonds between polar side chains in CypA and the backbone of CrkII are also present (Fig. 2b). The side chain of Tyr221 is exposed to the solvent (Fig. 2a) and appears to form no contacts with CypA. The complex is suitably arranged for the catalysis of the cis–trans isomerization in CrkII by CypA. Indeed, 15N ZZ-exchange NMR spectroscopy4 shows that the addition of catalytic amounts of CypA accelerates the cis–trans isomerization process (Supplementary Fig. 3f). Thus, CypA uses its catalytic site to interact specifically with the CrkII linker region that encompasses the tyrosine-phosphorylation site.
Figure 2. Specific interaction between CypA and CrkII in vitro and in vivo.
(a) Structure of CypA in complex with the CrkII peptide consisting of residues Ser216-Pro225. CypA is shown in a solvent-accessible surface representation whereas CrkII is shown as a ball-and-stick model. CrkII residues Glu217-Gln223 are shown (CrkII residues Pro216, Pro224 and Pro225 do not interact with CypA). The backbone of the CrkII region interacting with CypA is buried inside the catalytic cleft of CypA. Two views related by a 90° rotation about the x axis are shown. CrkII is in the trans conformation. (b) Interacting interface between CypA and CrkII. CypA is shown in light blue ribbon and the side chains of the residues interacting with CrkII are shown in ball-and-stick (green). CrkII is shown in yellow ball-and-stick. The magenta lines denote inter-molecular polar contacts (hydrogen bonds and/or salt bridges). (c) FRET analysis of HeLa cells transiently co-transfected with CrkII-YFP and CypA-CFP. The FRET efficiency is color coded from 0–100% on the left bar. Images were acquired on live cells after confirming comparable expression of both proteins. An average FRET efficiency of 70% is observed between the two fluorophores. Scale bar: 10 µm. (d) FRET analysis of HeLa cells transiently co-transfected with CrkII-YFP and CypA-CFP and treated with 25 µM of CsA before imaging. The average FRET efficiency drops to ~15% indicating that the complex is disrupted in the cell. Scale bar: 10 µm.
CypA and CrkII form a specific complex in the cell
To further corroborate that CrkII is indeed a CypA substrate, we tested their interaction in live cells using fluorescence resonance energy transfer (FRET) microscopy imaging35. We transiently transfected HeLa cells with CrkII tagged with yellow fluorescent protein (CrkII-YFP) and CypA tagged with a cyan fluorescent protein (CypA-CFP) and monitored the expression levels and distribution of the two proteins. Signal from the fluorescent tags indicated that both proteins reside in the cytoplasm and for CrkII-YFP the distribution agrees with previously published results35 (Supplementary Fig. 5a). In the cells cotransfected with both plasmids, maximum FRET was observed near the membrane (~70±3.3%) (Fig. 2c and Supplementary Fig. 5b). Addition of CsA resulted in a dramatic decrease in FRET efficiency to ~15% (Fig. 2d). Thus, the FRET imaging data demonstrate that CrkII and CypA form a specific complex in vivo mediated by the CypA catalytic site.
CypA inhibits CrkII phosphorylation in vitro
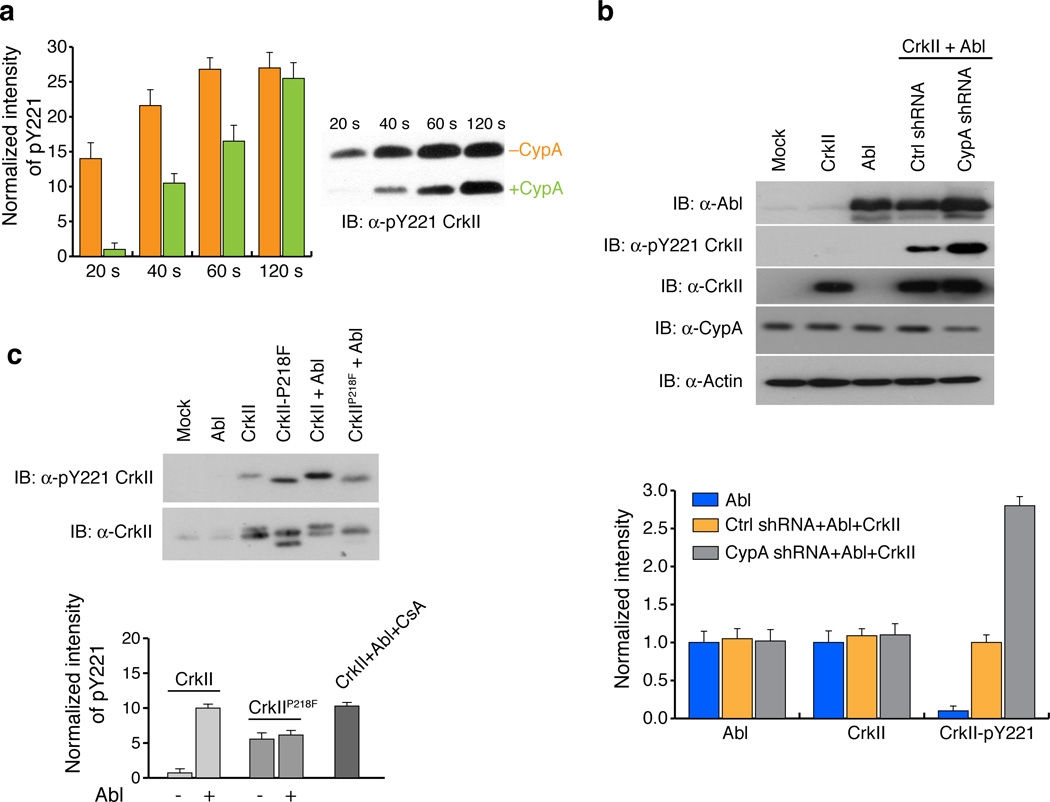
When CypA is bound to CrkII, the backbone of the residues flanking the phosphorylation site is buried in the catalytic cleft (Fig. 2a). Therefore the CypA-CrkII complex could prevent phosphorylation of CrkII (replace with original). To test the hypothesis that CrkII–CypA complex formation results in suppression of phosphorylation of CrkII, we used a kinase assay to measure the efficiency of CrkII Tyr221 phosphorylation by Abl in the presence and absence of CypA. The data show that the level of CrkII Tyr221 phosphorylation is significantly lower in the presence of CypA (up to a factor of ~12) (Fig. 3a and Supplementary Fig. 6). As expected, phosphorylation increases with time because of the moderate affinity and the dynamic nature of the CypA–CrkII complex.
Figure 3. CypA attenuates CrkII Tyr221 phosphorylation in vitro and in vivo.
(a) Effect of CypA on CrkII Tyr221 phosphorylation by Abl kinase was investigated using an in vitro kinase assay (left). The intensity of the pTyr221 bands were quantified and plotted (right). Full gels are shown in Supplementary Fig. 13. Without CypA (orange) and with CypA (green) (b) Effect of CypA on CrkII Tyr221 phosphorylation in 293T cells was investigated by stably expressing scrambled shRNA or CypA shRNA and transiently transfected with Abl kinase and CrkII plasmids for 48 hours. Lysates were prepared and analyzed by western blotting (top) with the antibodies indicated. Intensity of bands were quantified and plotted (bottom) Color code is as displayed in the figure panel. Full gels are shown in Supplementary Fig. 13. (c) Effect of CrkII Tyr221 phosphorylation on CrkIIP218F in 293T cells transfected with the plasmids or treatment indicated for 48 hours followed by immunoprecipitation of CrkII from lysate. Quantification of CrkII Tyr221 phosphorylation normalized to total CrkII (bottom) was performed after western blotting (top). Full gels are shown in Supplementary Fig. 13 For all graphs data represents the results from 3 independent experiments. (±SEM)
CypA-mediated CrkII phosphorylation suppression in vivo
Next we investigated whether CypA regulates CrkII Tyr221 phosphorylation in cells. To test the effect of CypA on CrkII phosphorylation, we prepared 293T cells where CypA was knocked-down using short hairpin RNA (shRNA). Consistent with the in vitro kinase assay, transient CypA knockdown (~50% efficiency) enhanced phosphorylation of CrkII at Tyr221 ~3 fold (Fig. 3b). Phosphorylation of CrkII was achieved by co-expression of CrkII and Abl in 293T cells. To determine the effect that increased amounts of CypA, as is the case in several cancer lines, have on CrkII phosphorylation, we used various approaches to ectopically overexpress CypA. However, all our efforts were unsuccessful and we could not achieve overexpression to a level greater than ~20% of the endogenous CypA level. For this reason, in an effort to increase the population of the CypA–CrkII complex in the cell we sought to design a CrkII variant with higher affinity for CypA. Analysis of the CypA–CrkII structure suggested that substitution of Pro218 (Supplementary Fig. 7a) by a bulkier hydrophobic residue would optimize juxtaposition of the two proteins. Molecular modeling indicated that a CrkII variant with a Phe residue in place of Pro218 (CrkIIP218F) optimized the surface complementarity in the complex with CypA (Supplementary Fig. 7b). Indeed, the CrkIIP218F variant binds CypA with a ~20-fold higher affinity than wild-type CrkII (Supplementary Fig. 7c). Interestingly, when 293T cells were transfected with CrkIIP218F a significant decrease in Tyr221 phosphorylation was observed when compared with cells transfected with wild type CrkII (Fig. 3c).
CypA suppresses CrkII phosphorylation in Cancer Cells
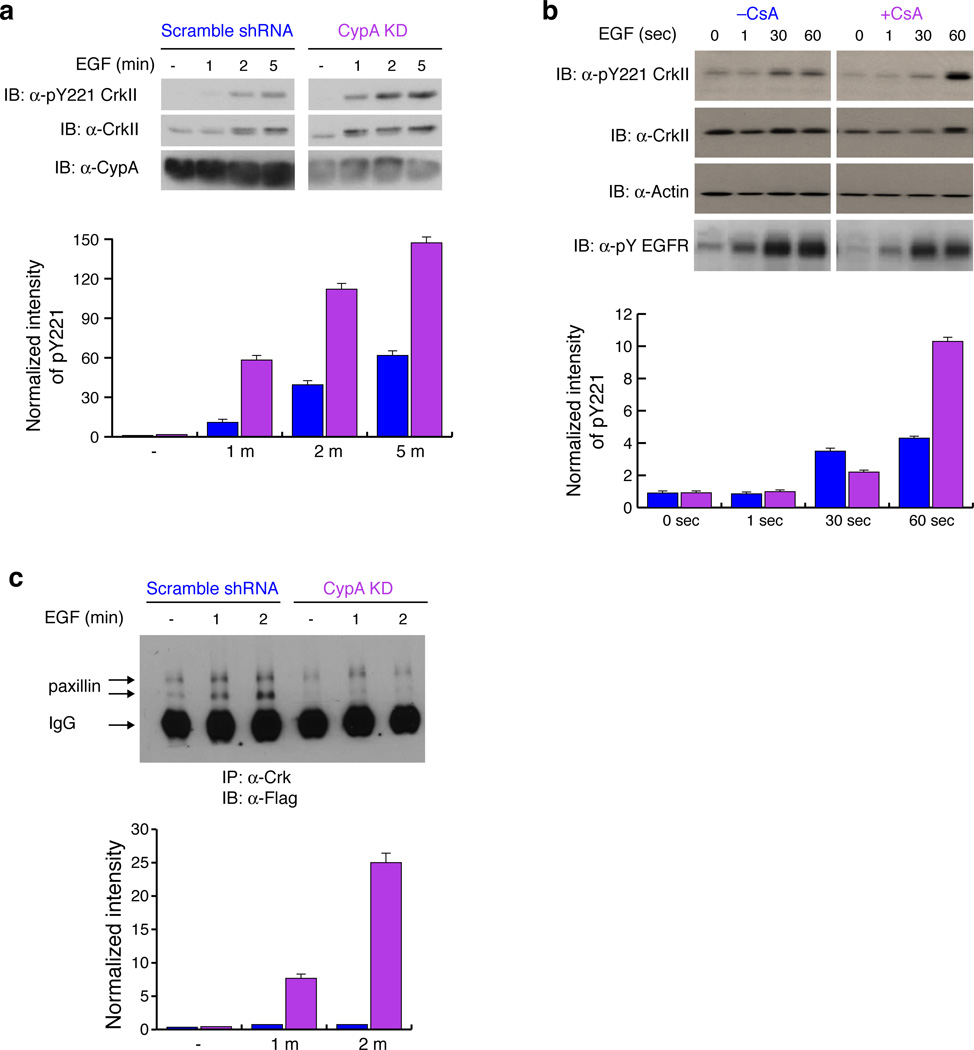
CypA is overexpressed in many cancers2. We screened several cancer cell lines and found that the MDA-MB-468 breast-cancer cell line showed one of the highest levels of CypA expression among them (Supplementary Fig. 8a). Incidentally this particular cell line overexpresses EGFR36, which also phosphorylates CrkII at Tyr22130, a reaction that can be stimulated by the addition of EGF. CypA knock-down MDA-MB-468 cells displayed up to ~10-fold higher phosphorylation at CrkII Tyr221 when compared to cells that were treated with a scrambled shRNA (at 1min) (Fig. 4a). In order to further investigate this inhibitory property of CypA, we treated serum starved MDA-MB-468 cells with CsA. As expected, the addition of CsA significantly increases phosphorylation at CrkII Tyr221 because of the disruption of the CypA–CrkII complex (Fig. 4b).
Figure 4. CypA attenuates CrkII Tyr221 phosphorylation cancer cell line MDA-MB-468.
(a) Effect of CypA on CrkII Tyr221 phosphorylation was measured upon EGF stimulation in MDA-MB-468 cells. Lysates were analyzed by western blotting with the antibodies indicated. The intensity of the bands (top) was quantified and pTyr221 CrkII phosphorylation levels were normalized to overall CrkII expression and plotted (bottom). Full gels are shown in Supplementary Fig. 13. Scrambled shRNA (blue) and CypA-KD (magenta). (b) Effect of CsA on CrkII Tyr221 phosphorylation was investigated in MDA-MB-468 cells pretreated with DMSO or CsA and stimulated with EGF for the times indicated, and lysates analyzed for CrkII Tyr221 phosphorylation. The intensity of the bands (top) was quantified and pY221 CrkII phosphorylation levels were normalized to overall CrkII expression and plotted (bottom). Full gels are shown in Supplementary Fig. 13. Without CsA (blue) and with CsA (magenta). (c) Effect of CypA on paxillin-CrkII complex formation was investigated in MDA-MB-468 cells stably expressing scrambled shRNA or CypA shRNA transiently transfected with flag-paxillin and stimulated with EGF for 1 and 2 minutes. Immunoprecipitation of CrkII from lysates was analyzed with anti-flag antibody. The intensity of the bands (top) representing paxillin were quantified, normalized against the paxillin level in untreated cell line expressing CypA shRNA and plotted (bottom). Scrambled shRNA(blue) and CypA-KD (magenta). The two bands of paxillin represent differentially phosphorylated forms of paxillin. All experiments were analyzed by western blotting. Full gels are shown in Supplementary Fig. 13. For all graphs data represents the results from 3 independent experiments. (±SEM)
CypA binding to CrkII increases paxillin-CrkII complex
To determine the effect of CypA-mediated suppression of CrkII Tyr211 phosphorylation on CrkII association with signaling proteins, we probed changes in the interaction between CrkII and paxillin. The paxillin–CrkII complex forms when the activation of integrin receptor, in the context of cell adhesion and cell migration, results in the phosphorylation of paxillin creating a docking site for the CrkII SH2 domain37. Examination of the complex formation between paxillin and CrkII in CypA knock-down MDA-MB-468 cells revealed a significant decrease (~10-fold) of paxillin association with CrkII when compared to MDA-MB-468 cells treated with scrambled shRNA. (Fig. 4c). In addition the CypA knockdown cells were passaged until the CypA levels were restored and the complex formation between CrkII and paxillin were reexamined. As expected the association of paxillin and CrkII increases to that of the control cell line. (Supplementary Fig. 8b). Thus, the specific interaction of CypA with CrkII results in a significant decrease in CrkII Tyr221 phosphorylation, which leads to an increase in the amount of CrkII bound to paxillin or other proteins interacting with the SH2 domain of CrkII such as p130CAS.
CypA–CrkII complex stimulates cell migration
To determine whether the CypA-effected suppression in CrkII phosphorylation has an impact on the physiology of the cell, we investigated the migratory properties of cells as a function of the CypA-CrkII interaction. Crk has been shown to enhance the migration of several patient-derived cancer cell lines including breast cancer38. Phosphorylation of Tyr221 has been shown to inhibit Crk-mediated motility in breast cancer cells39,40. Migration is a complex process that involves an intricate balance between several protein complexes that eventually leads to a reorganization of the cytoskeleton39,40. We argued that a straightforward comparison of the migratory properties of cells in the presence and absence of CypA would provide direct evidence of whether the CypA–CrkII complex and the subsequent suppression of Tyr221 phosphorylation could significantly perturb the dynamic cell motility equilibrium.
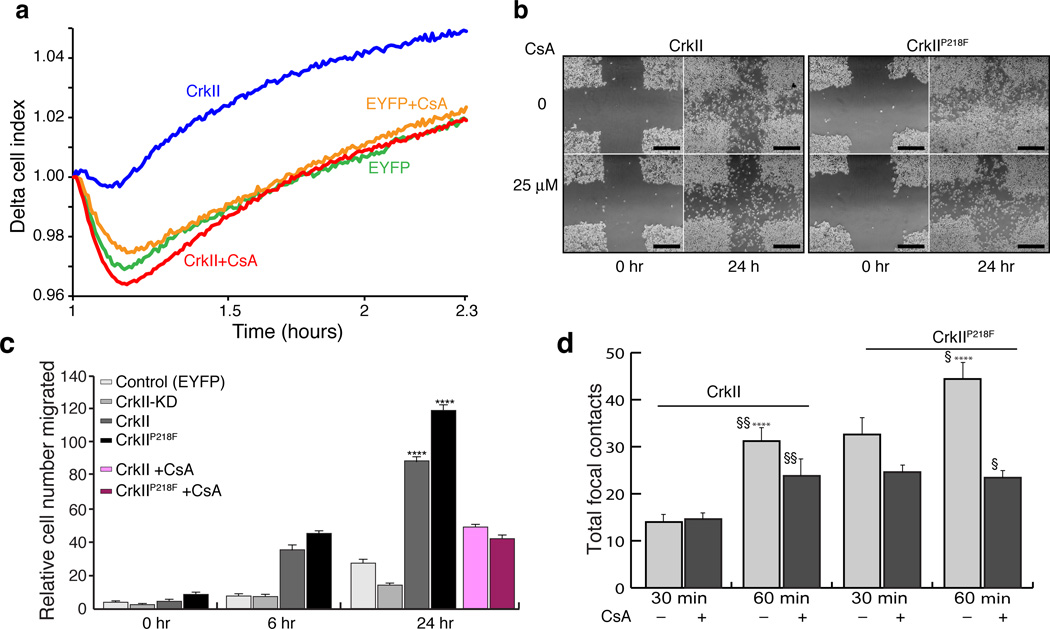
We tested the effect of the interaction between CrkII and CypA on cell motility since CrkII is a key molecule in regulating cell motility. We prepared three cell lines of MDA-MB-468 cells (i) overexpressed wild type CrkII (CrkII); (ii) overexpressed CrkIIP218F; and, (iii) CrkII knock-down (CrkII-KD). A control cell line expressing EYFP was used in parallel for comparison. To obtain quantitative information about the cell migration properties, we used the xCELLIgence system (Roche) that measures migration by detecting the electrical impedance using microelectronic biosensor technology41. We followed the migration of MDA-MB-468 cells collecting data at 15 minute intervals. Both CrkII and CrkIIP218F greatly enhanced cell motility compared to the control cells while CrkII-KD cells had no significant motility, in agreement with the crucial role of CrkII in cell motility (Supplementary Fig. 9a). Migration in CsA treated cells were markedly lower at the 4 hour time point. (Supplementary Fig. 9b). In a separate set of experiments, we also quantified the effects of CsA at earlier time points and increased the data collection frequency from 15 minutes to 15 seconds. Addition of CsA caused a significant decrease in migration (Fig. 5a). Taken together, the results indicate that formation of the CypA-CrkII complex stimulates the migratory properties of the cells and that disruption of this complex by CsA results in suppression of cell migration.
Figure 5. Effect of the CypA–CrkII complex on cell migration.
(a) MDA-MB-468 cells stably expressing EYFP or CrkII were serum starved and motility was analyzed towards EGF (25 ng ml−1) in real-time using xCelligence technology (Roche). Delta Cell Index is plotted on the y-axis as an average of duplicate samples versus time on the x-axis (b) Wound healing assay of wild type CrkII (left) and CrkIIP218F (right) in the presence and absence of CsA. Cells were cultured to near confluence, scratched using a pipette and imaged after 24 hours. Scale bars: 500µm(c) Summary of the measurement of cell migration from the wound healing assay in panel b and Supplementary Fig. 9c was quantified for each condition indicated. Data represents mean of 3 independent experiments ±SEM. Two-tailed t-test, **** p≤0.001. (d) Focal contacts based on signal from GFP-Paxillin were quantified for each cell line and treatment using ImageJ and plotted. Light grey without CsA, dark grey with CsA. (Supplementary Fig. 10a, b). Data represents a mean of 6 cells ±SEM (n=6; two-tailed t-test **p and, §§≤0.05. § and ****≤0.001.
To further investigate the effect of CypA-CrkII interaction on cell migration, we used a wound healing assay (Fig. 5b and Supplementary Fig. 9c). Using the control as a basis for the comparison, the CrkII-KD cells, even at the 24-hr time point displayed the least migratory tendency including a lack of cell spreading and absence of lamellipodea (Fig. 5c and Supplementary Fig. 9c, d). In contrast, overexpression of CrkII promoted cell spreading and numerous lamellipodia were observed at the leading edges of the culture (Supplementary Fig. 9d). The cell lines overexpressing CrkII and CrkIIP218F displayed increased cell proliferation and migration (Fig. 5c and Supplementary Fig. 9e) especially at the edges of the monoculture (Fig. 5b and Supplementary Fig. 9c). The CrkIIP218F mutant presented a more aggressive migratory phenotype and greater cell-spreading tendency when compared to the cells overexpressing CrkII (Fig. 5b, c and Supplementary Fig. 9c). Treatment of either CrkII or CrkP218F cells with CsA resulted in significant decrease in migration (Fig. 5b,c and Supplementary Fig. 9e), further establishing CrkII Tyr221 phosphorylation as a key event in cell migration. This role is consistent with the observation that expression of CrkY221F nullifies Ephrin-B2 induced inhibition of cell migration in breast cancer cells39. Taken together, these results suggest that the CypA-CrkII interaction delays phosphorylation of CrkII at Tyr221 thereby decreasing Crk-mediated cell motility.
Finally we followed the distribution of paxillin using fluorescent microscopy. Paxillin is integral to the formation of focal contacts and is recruited to nascent focal adhesions. It is also important for the focal adhesion turnover, which is essential for cell migration42,43. We reasoned that the increased migratory behavior we observed in the CrkIIP218F cells would translate to an increase in the number of focal contacts that are observed. Using GFP-Paxillin as a marker for focal adhesion we stably reconstituted CrkII or CrkIIP218F plasmid in Crk(−/−) MEFs and observed focal adhesion formation at early time points after seeding (15 min, 30 min and 60 min) (Supplementary Fig. 10a). At the 60-min time point the CrkIIP218F cell line had significantly more focal contacts (by ~50%) than the cells expressing CrkII (Fig. 5d and Supplementary Fig. 10b). Treatment of CsA caused a marked decrease on focal contacts in the cells expressing CrkII or CrkIIP218F. Interestingly, we also observed that by 60 mins, a greater number of the CrkIIP218F cells were spread out with robust paxillin signal from the leading edges (Supplementary Fig. 10a). In the Crk knockout cells paxillin was diffused throughout the cell with few focal contacts. Taken together, these results support the coimmunoprecipitation data that the increased affinity for CrkII to CypA has a direct effect on the complex formation between CrkII and paxillin and as a consequence the distribution of paxillin in the cell.
Abl-mediated CrkL phosphorylation is not affected by CypA
Crk-like (CrkL), encoded by a distinct gene, exhibits high sequence identity (~60%) to CrkII (Supplementary Fig. 11a). Similarly to CrkII, CrkL also features a highly conserved Tyr residue (Tyr207) that becomes phosphorylated by Abl44. CrkL is a preferred substrate of the Bcr-Abl kinase and its tyrosine-phosphorylation is used as a diagnostic tool for Philadelphia-positive leukemia45. Notably, the residue preceding Tyr207 in CrkL is Ala, as opposed to Pro in CrkII (Fig. 1c), and thus, CrkL does not undergo cis–trans isomerization as shown by NMR analysis. (Supplementary Fig. 11b). As a result, CypA is not recruited to this region in CrkL and no binding was detected between the two proteins (Supplementary Fig. 11c). Kinase assay performed with Abl kinase and CrkL as a substrate also show no inhibition of phosphorylation in the presence of CypA (Supplementary Fig. 11d). This provides another important difference between CrkII and CrkL in addition to the structural differences identified previously46. Thus, whereas CypA regulates CrkII phosphorylation by Abl, it does not regulate CrkL phosphorylation. This may be one of the factors that contribute to the preferential phosphorylation of CrkL by Abl.
DISCUSSION
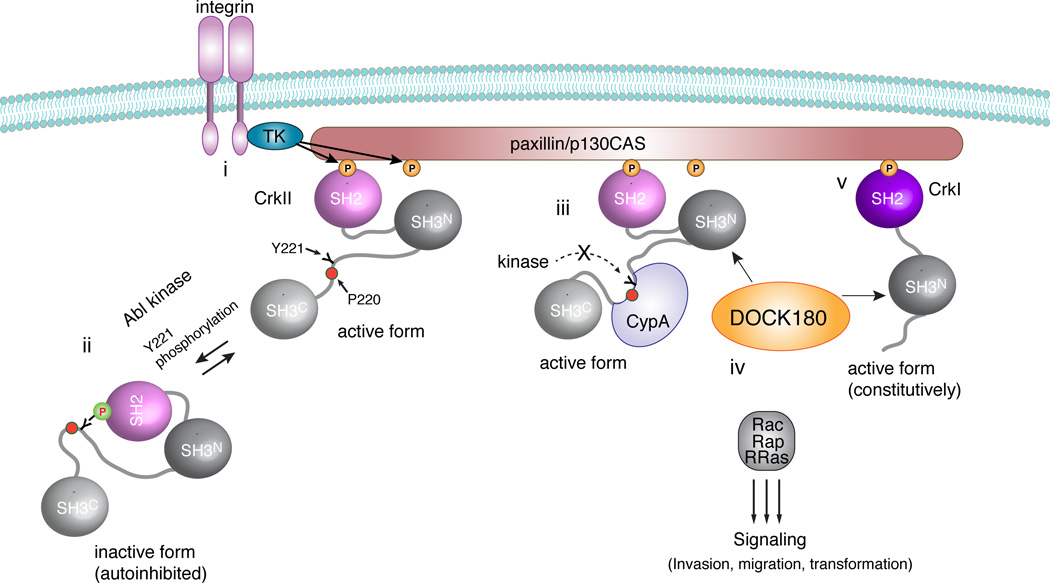
PPIases have been implicated in an ever-growing number of cellular processes47. Their ubiquitous presence in cells suggests that their full repertoire remains to be explored. Recent studies have identified CypA as one of the proteins upregulated in many human cancers. However, the molecular mechanisms underlying CypA action and the signaling pathways through which it exerts its function are poorly understood. Here we show that CypA binds to the CrkII adaptor protein and inhibits a key phosphorylation process that shuts down signaling by CrkII. As a result, CypA augments CrkII-mediated signaling giving rise to enhanced cell motility and invasion (Fig. 6).
Figure 6. Effect of CypA binding to CrkII in integrin signaling.
(i) Integrin activation elicits paxillin/p130CAS phosphorylation by tyrosine kinases (TK), and, as a result, Crk proteins are recruited. (ii) Abl-induced phosphorylation of CrkII forces its dissociation from paxillin/p130CAS and thus results in signaling suppression. (iii) Binding of CypA to Pro220 sterically inhibits CrkII Tyr221 phosphorylation and CrkII remains in the active form. (iv) GEFs (for example, DOCK180 and C3G) associate with CrkII via its SH3N domain, giving rise to efficient localized activation of small GTPases at the membrane. (v) CrkI is a short alternatively spliced form of Crk that lacks the phosphorylation site and thus forms the active state constitutively.
Integrin-mediated cell adhesion and migration are important processes both in normal cell activities as well as in pathological conditions, such as tumorigenesis and metastasis48. Tyrosine phosphorylation of the paxillin or p130CAS scaffold proteins, which are important players in the formation of focal adhesions, creates binding sites for the SH2 domain of Crk proteins49. Crk proteins bind to Rho-GTPase exchange factors DOCK180, SOS1 and C3G via their SH3N domain enabling efficient localized activation of small GTPases, such as Rac1 or Rap1 at the cell membrane. These events induce cell migration through actin cytoskeleton remodeling, pseudopodia extension and focal adhesion turnover. Because the Crk family of proteins form a bridge between the focal adhesion proteins and the GTPases, dysregulation of Crk signaling either by mutation or expression levels negatively regulates cell migration. Phosphorylation of CrkII Tyr221 by Abl or EGFR kinases is a key event35 that induces intra-molecular binding of pTyr221 to the SH2 domain giving rise to an autoinhibited form of CrkII. In this form, CrkII cannot associate with paxillin or p130CAS scaffolding proteins causing the signaling cascade to be interrupted and cell movement to decrease (Fig. 6).
Interestingly, it has been shown that the SH3N domain of CrkII, which binds to Rho-GTPase exchange factors, is not accessible for binding in the phosphorylated form of CrkII27. Thus, suppression of Tyr221 phosphorylation not only extends the lifetime of the paxillin-CrkII complex but it also allows CrkII to continue mediating signaling through its SH3N domain50.
The Pro residue preceding Tyr221 is conserved in all CrkII proteins (Fig. 1c). Using NMR spectroscopy we demonstrate that Pro220 undergoes cis–trans isomerization and creates a binding site for CypA. The structure of the CypA–CrkII complex shows that the CrkII region encompassing Tyr221 is buried in the binding cleft of CypA and thus cannot be accessed by kinases (Fig. 2a). As a result, CypA binding to CrkII suppresses Tyr221 phosphorylation and CrkII remains in the active form that mediates cell motility (Fig. 6).
In principle, CypA can function as a binding partner (in stoichiometric amounts) or as an enzyme (in catalytic, sub-stoichiometric amounts). Pro220 is located in an unstructured region of the 50-residue long linker that tethers the two SH3 domains. The NMR data show that Pro220 isomerization has a very local and limited effect since only about seven residues exhibit duplicated peaks. Furthermore, the NMR data showed that both the cis and trans conformers of the Pro220–Tyr221 prolyl bond in CrkII are phosphorylated by Abl and both the cis and trans conformers of pTyr221 bind to the SH2 domain (Supplementary Fig. 12). Thus, there seems to be no conformer-specific binding selectivity. Moreover, the effect of catalytic amounts of CypA on inhibiting CrkII phosphorylation is negligible further corroborating that CypA exerts its inhibitory effect as a binding partner and not as an enzyme. Because the affinity of CrkII for CypA is relative low (Kd ~15 µM), the effect of CypA on suppressing Tyr221 phosphorylation becomes significant and noticeable only when the proteins are present in significant amounts. This is especially the case in cancer cells wherein both CypA and CrkII are vastly upregulated.
Binding of CypA to CrkII is expected to have a similar, albeit likely diminished, phenotype to the CrkIIY221F mutant, which cannot be phosphorylated. Indeed, motility of cells expressing CrkIIY221F is not inhibited downstream of extracellular stimuli that induce Tyr221 phosphorylation such as Ephrin B239. Moreover, dephosphorylation of pCrkII by phosphatase PTB1B enhances motility of cells expressing CrkII40. CrkI, an alternatively spliced Crk isoform that consists only of the first 204 residues of CrkII, and thus lacks the Tyr221 phosphorylation site, remains always in the active form (Fig. 6) and is associated with a particularly aggressive phenotype19. The lack in CrkI of the segment encompassing Pro220 explains why CrkI does not associate with CypA50.
In contrast to CrkII, CrkL showed no interaction with CypA. CrkL is missing the critical heterogeneous Pro residue next to the Tyr phosphorylation site (Tyr207 in human CrkL; Fig. 1c) and is unable to recruit CypA. This highlights a potential mechanism by which CypA can influence signaling by discriminating between two similar proteins and may account, to some extent, for the differential behavior of the CrkII and CrkL proteins.
ONLINE METHODS
Protein preparation and purification
The following constructs were used for the experiments: human CrkII (1–304), human CrkII linker-SH3C (193–304), chicken CrkII (1–305), chicken CrkII linker-SH3C (193–305), human CrkL (1–303), human CrkL linker-SH3C (191–304) and human CypA (1–165). Crk and CrkL proteins were cloned into pet42a using NcoI and XhoI restriction sites. A TEV protease cleavage site was introduced between the histidine tag and the protein. Isotopically labeled samples for NMR studies were prepared by growing the cells in M9 minimal media supplemented with 1g l−1 of 15NH4Cl and 2 g l−1 of 13C6 glucose. All CrkII and CypA constructs were grown at 37°C, and protein synthesis was induced by the addition of 0.25 mM IPTG at OD600 ~0.4. Cells were lysed by sonication and the cytosolic fraction was separated by centrifugation at 50,000 × g. The lysate was loaded onto Ni-NTA agarose resin (GE) equilibrated with Tris buffer and 1M NaCl, pH 8. Elution was performed with 400 mM imidazole and after TEV cleavage the sample was concentrated and applied to a Superdex 75 size – exclusion column (GE). For NMR studies the samples were dialyzed in NMR buffer (50mM KPi, pH 6.5, 150 mM NaCl and 1 mM β-mercaptoethanol) and concentrated using Amicon cell units (Millipore). All samples were monomeric in solution at concentrations used for the NMR studies (0.3mM–0.5mM) as confirmed by multi angle light scattering (MALS).
Kinase assay
Kinase assay was carried out in the NMR buffer with either recombinant Crk or CrkL supplemented with 5 mM MgCl2 and 5 mM ATP. The kinase reaction was started by addition of enzymatic quantity of recombinant Abl kinase and the reaction was stopped using 2× Laemmli buffer. The samples were analyzed using western blotting with the indicated antibody. Quantitation was performed using imageJ.
FRET assay and imaging
CFP-CypA and YFP-CrkII were constructed by inserting the respective genes into pcDNA3.1 already containing either the CFP or YFP genes. Each plasmid was sequenced for verification. Fluorescence imaging to measure FRET was conducted 24 hrs post-transfection using the Nikon TE2000 Fluorescence Microscope equipped with a motorized barrier filter wheel. Images were acquired using the CFP channel (excitation: 430 nm, emission: 470/24nm), YFP channel (excitation: 500 nm, emission: 535/30nm) and CFP-YFP channel (excitation 430 nm, emission: 535/30nm). Cells transfected with only CFP-CypA and only YFP-CrkII were used to calibrate the FRET measurements for spectral bleed-through. The NIS Elements Imaging Software was used to measure the FRET efficiency using the sensitized emission method.
Cell culture and plasmid transfection
HeLa cell lines were seeded in 24-well polystyrene-coated culture plates in Dulbecco’s modified eagle medium (DMEM). The medium was supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 µg/ml streptomycin, and 2 mM L-glutamine. Cells were seeded to achieve 70% confluency at the time of transfection. The cells are transfected with the fusion plasmids using the commercial available transfection reagent X-tremeGENE (Roche). Briefly, equal amounts of the CFP-CypA and YFP-CrkII plasmids were mixed together and complexed with the recommended volume of XtremeGENE for 20 min. The complexes were then added dropwise to the cells in serum-free OptiMEM media. After 6 hr the cells were washed two times with fresh cell culture medium. For the inhibition studies, CsA was dissolved in DMSO and added to the cell culture medium to achieve a final concentration of 25 µM CsA and 0.1% DMSO. CsA was added 30 min prior to conducting fluorescence imaging.
Isothermal Titration Calorimetry
All calorimetric titrations were performed on an iTC200 microcalorimeter (GE). Protein samples were extensively dialyzed against the ITC buffer containing 50 mM KPi, pH 6.8,150 mM NaCl and 1 mM TCEP at 8 °C. The sample cell was typically filled with ~40 µM of CypA and the injection syringe with ~400 µM of Crk proteins or peptides. Crk peptide solutions were prepared by dissolving the peptide in the flow through of the last exchange buffer. Each titration typically consisted of a preliminary injection followed by 7–8 subsequent injections. Data for the preliminary injection, which are affected by diffusion of the solution from and into the injection syringe during the initial equilibration period, were discarded. The data were analyzed with Origin 7.0.
Reagents
The following antibodies were used: anti-pY221 CrkII (Sigma SAB4503813), anti-CrkII (Sigma SAB1405658), anti-actin (Sigma A2066), anti-Flag (Sigma F1804) and anti-Abl (Sigma SAB4501043). Protein G agarose beads were purchased from Santa Cruz Biotechnology, HRP-conjugated goat anti-mouse and goat anti-rabbit were purchased from Jackson Immunologicals and Type I Collagen was purchased from BD. Anti-Crk (RF-51) anti-sera was prepared as described previously51. Transfections were performed with the Xtremegene HP reagent according to the manufacturers’ protocol (Roche). Human EGF was purchased from Life Technologies.
Cell culture and growth factor stimulation
MCF10A, MDA-MB-468, MCF7, MDA-MB-231, T47D and 293T cells were obtained from ATCC. Cells were cultured in DMEM with 10% Fetal Bovine Serum (FBS) and 1% Penicillin/Streptomycin. Cells were starved in DMEM containing 0.5% FBS over-night and then stimulated with EGF at 100 ng ml−1. Where indicated, cells were pre-treated before stimulation with CsA (Cell Signaling Technology).
Generation of stable cell lines
For stable over-expression, EYFP or Crk proteins in the pMSCV vector backbone, pCL-Eco and pMD2.G (encoding the VSV glycoprotein) were transiently transfected into the BOSC23 packaging cell line. Viral supernatants were collected 48 hours post-transfection and transferred to MDA-MB-468 cells in the presence of Polybrene (8 µg ml−1). Stable polyclonal populations were selected in the presence of puromycin (1 µg ml−1). For stable knockdowns, scrambled shRNA, CypA shRNA or Crk shRNA in the pLKO.1 vector backbone, psPAX2 and pMD2.G were co-transfected into HEK 293T cells. Viral supernatants were collected 60 hours post-transfection, virus was concentrated by ultra-centrifugation and transferred to MDA-MB-468 cells. Stable polyclonal populations were selected in the presence of puromycin (1 µg/ml).
Cell migration assays using xCelligence
CIM plates (Acea Biosciences) were coated on the underside with 10 µg/ml collagen. 40,000 viable cells were seeded into the top chamber while EGF was added only to the bottom chamber. In this system, the bottom of the transwell insert is lined with electrodes that send a pulse of current periodically to record impedance. As cells move through the membrane and make contact with the electrode, they contribute to impedance which is used to generate a cell index (y-axis) that is directly proportional to the number of migrating cells. Data are plotted as Delta cell index versus time where delta cell index refers to cell index after background subtraction from each well.
Wound healing migration assay
Cell migration for the four MDA-MB-468 cell lines (control EYFP, CrkIIP218F, CrkII and knock-down CrkII) was evaluated by a scratched wound healing assay. All four cell lines were seeded in 6-well polystyrene-coated culture plates in Dulbecco’s modified eagle medium (DMEM). The medium was supplemented with 10% fetal bovine serum (FBS), 100 U ml−1 penicillin, 100 µg ml−1 streptomycin, and 2 mM L-glutamine. Cells were grown to confluence and then wounded using a pipette tip. Two wounds were made for each sample, and all were imaged at 0 hr, 6 hr and 24 hrs at the same position to compare the degree of migration. For the inhibition studies, CsA was dissolved in DMSO and added to the cell culture medium to achieve a final concentration of 25 µM CsA and 0.1% DMSO. CsA was added immediately after wounding.
Western blotting and immunoprecipitation
Samples were analyzed by SDS PAGE and then transferred to a PVDF membrane (Millipore). After blocking, the membrane was incubated with primary and secondary antibodies according to the manufacturer’s protocols. Antibody dilutions used were Anti-CypA: 1:1000, anti-CrkII: 1:5000, anti-CrkII-pY221: 1:2000, anti-Flag: 1:1000, anti-Abl: 1:1000, anti-actin: 1:1000. Binding was detected by chemiluminescence and then quantified using ImageJ software. Immunoprecipitation was performed as described previously52.
Image acquisition and processing
Images were taken on an inverted wide field Zeiss microscope equipped with a Plan-Apo 63× oil immersion objective with NA 1.4 using Axiovision 4.8.2 software (Carl Zeiss, Inc). For each field binning was set to 2×2 using a CoolSNAP HQ2 (Photometrics) camera and 60 z-stacks with a step size of 0.240 µm. Deconvolution was iterative with autolinear normalization for all images. Focal contacts were quantified using ImageJ. (Rasband, W.S)
Statistics
Bar graphs represent mean ±SEM. Data were analyzed using Excel. Statistical significance was tested using two-tailed t-tests.
NMR spectroscopy
NMR experiments were conducted at 25 or 32 °C on Bruker 700 MHz spectrometer equipped with a TCI cryoprobe. Spectra were processed with NMRPipe53 and visualized using Sparky. Complete backbone and sidechain assignment for the [U-13C,15N]-CypA was achieved using standard 3D triple-resonance, 3D-1H-13C -TOCSY, and COSY experiments. Assignments were extended, verified and distance restraints were extracted from 3D 15N,13C-edited NOESY spectra acquired with 120 ms mixing time. The 1H assignment for the CypA-bound CrkII peptide and intermolecular protein-protein NOE contacts were obtained using a double half-filtered (3D F1-13C/15N-filtered, F3-13C-edited) NOESY experiment. 1H-15N ZZ-heteronuclear exchange NMR spectra of 15N-CrkII in the absence and presence of catalytic amounts (2–10%) of CypA were performed as detailed before25.
Structure calculation and refinement
Structures for CypA–CrkII complex were calculated using CYANA 3.054. Initial restraints for the backbone torsion angles Φ and Ψ were obtained from chemical shifts using TALOS-N55. Resonance assignments, NOESY peak lists from all NOESY experiments, and TALOS-derived dihedral angles were used as CYANA input. Structural refinement was performed using restrained molecular dynamics in explicit water with CNS 1.354. The ensemble of 20 lowest energy structures and restraints for CypA–CrkII complex have been deposited in PDB (PDB ID 2MS4). Chemical shifts were deposited in BMRB (BMRB ID 25104). The summary of NMR restraints and structure refinement statistics is presented in Supplementary Table 1. HADDOCK 2.156 was used to model the CypA-CrkIIP218F structure.
Supplementary Material
Acknowledgements
We thank Nakul Sheth for helping with the cell biological experiments. This work was supported by the US National Institutes of Health (GM80308 to C.G.K and CA165077 to R.B.B). The Rutgers University Busch Biomedical Research Grant to A.J.R. and NIH Director’s Innovator Award (1DP20D006462-01 to K.-B.L).
Footnotes
Accession codes
Protein Data Bank (PDB): coordinates for the CypA-CrkII complex have been deposited with accession code 2MS4.
Author Contributions
T.S. and C.G.K. designed the study; T.S. and W.J. recorded and analyzed the NMR data; W.J. and P.R. determined the structure of the complex; T.S., G.S. and R.B.B. collected and analyzed the kinase phosphorylation and cell migration data; T.S., S.S. and K.B.L. collected and analyzed the FRET and wound healing data; T.S., L.A.C. and A.J.R. collected and analyzed the fluorescence microscopy data; T.S. and C.G.K. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests
References
- 1.Ryffel B, et al. Distribution of the cyclosporine binding protein cyclophilin in human tissues. Immunology. 1991;72:399–404. [PMC free article] [PubMed] [Google Scholar]
- 2.Nigro P, Pompilio G, Capogrossi MC. Cyclophilin A: a key player for human disease. Cell Death & Disease. 2013;4:e888. doi: 10.1038/cddis.2013.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonfils C, et al. Cyclophilin A as negative regulator of apoptosis by sequestering cytochrome c. Biochem Biophys Res Commun. 2010;393:325–330. doi: 10.1016/j.bbrc.2010.01.135. [DOI] [PubMed] [Google Scholar]
- 4.Bosco DA, Eisenmesser EZ, Pochapsky S, Sundquist WI, Kern D. Catalysis of cis/trans isomerization in native HIV-1 capsid by human cyclophilin A. Proc Natl Acad Sci U S A. 2002;99:5247–5252. doi: 10.1073/pnas.082100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brazin KN, Mallis RJ, Fulton DB, Andreotti AH. Regulation of the tyrosine kinase Itk by the peptidyl-prolyl isomerase cyclophilin A. Proc Natl Acad Sci U S A. 2002;99:1899–1904. doi: 10.1073/pnas.042529199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howard BR, Vajdos FF, Li S, Sundquist WI, Hill CP. Structural insights into the catalytic mechanism of cyclophilin A. Nature Structural Biology. 2003;10:475–481. doi: 10.1038/nsb927. [DOI] [PubMed] [Google Scholar]
- 7.Gothel SF, Marahiel MA. Peptidyl-prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. Cell Mol Life Sci. 1999;55:423–436. doi: 10.1007/s000180050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer G, Tradler T, Zarnt T. The mode of action of peptidyl prolyl cis/trans isomerases in vivo: binding vs. catalysis. FEBS Letters. 1998;426:17–20. doi: 10.1016/s0014-5793(98)00242-7. [DOI] [PubMed] [Google Scholar]
- 9.Lee J, Kim SS. An overview of cyclophilins in human cancers. J Int Med Res. 2010;38:1561–1574. doi: 10.1177/147323001003800501. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, et al. Proteomics identification of cyclophilin a as a potential prognostic factor and therapeutic target in endometrial carcinoma. Mol Cell Proteomics. 2008;7:1810–1823. doi: 10.1074/mcp.M700544-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howard BA, et al. Stable RNA interference-mediated suppression of cyclophilin A diminishes non-small-cell lung tumor growth in vivo. Cancer Res. 2005;65:8853–8860. doi: 10.1158/0008-5472.CAN-05-1219. [DOI] [PubMed] [Google Scholar]
- 12.Obchoei S, et al. Cyclophilin A enhances cell proliferation and tumor growth of liver fluke-associated cholangiocarcinoma. Mol Cancer. 2011;10:102. doi: 10.1186/1476-4598-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart T, Tsai SC, Grayson H, Henderson R, Opelz G. Incidence of de-novo breast cancer in women chronically immunosuppressed after organ transplantation. Lancet. 1995;346:796–798. doi: 10.1016/s0140-6736(95)91618-0. [DOI] [PubMed] [Google Scholar]
- 14.Birge RB, Kalodimos C, Inagaki F, Tanaka S. Crk and CrkL adaptor proteins: networks for physiological and pathological signaling. Cell Commun Signal. 2009;7:13. doi: 10.1186/1478-811X-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linghu H, et al. Involvement of adaptor protein Crk in malignant feature of human ovarian cancer cell line MCAS. Oncogene. 2006;25:3547–3556. doi: 10.1038/sj.onc.1209398. [DOI] [PubMed] [Google Scholar]
- 16.Nishihara H, et al. Molecular and immunohistochemical analysis of signaling adaptor protein Crk in human cancers. Cancer Lett. 2002;180:55–61. doi: 10.1016/s0304-3835(01)00763-7. [DOI] [PubMed] [Google Scholar]
- 17.Miller CT, et al. Increased C-CRK proto-oncogene expression is associated with an aggressive phenotype in lung adenocarcinomas. Oncogene. 2003;22:7950–7957. doi: 10.1038/sj.onc.1206529. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe T, et al. Adaptor protein Crk induces Src-dependent activation of p38 MAPK in regulation of synovial sarcoma cell proliferation. Mol Cancer Res. 2009;7:1582–1592. doi: 10.1158/1541-7786.MCR-09-0064. [DOI] [PubMed] [Google Scholar]
- 19.Takino T, et al. CrkI adapter protein modulates cell migration and invasion in glioblastoma. Cancer research. 2003;63:2335–2337. [PubMed] [Google Scholar]
- 20.Kumar S, Fajardo JE, Birge RB, Sriram G. Crk at the quarter century mark: perspectives in signaling and cancer. Journal of Cellular Biochemistry. 2014;115:819–825. doi: 10.1002/jcb.24749. [DOI] [PubMed] [Google Scholar]
- 21.Fathers KE, et al. Crk adaptor proteins act as key signaling integrators for breast tumorigenesis. Breast Cancer Res. 2012;14:R74. doi: 10.1186/bcr3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shishido T, et al. Crk family adaptor proteins trans-activate c-Abl kinase. Genes to Cells. 2001;6:431–440. doi: 10.1046/j.1365-2443.2001.00431.x. [DOI] [PubMed] [Google Scholar]
- 23.Sirvent A, Benistant C, Roche S. Cytoplasmic signalling by the c-Abl tyrosine kinase in normal and cancer cells. Biol Cell. 2008;100:617–631. doi: 10.1042/BC20080020. [DOI] [PubMed] [Google Scholar]
- 24.Petschnigg J, et al. The mammalian-membrane two-hybrid assay (MaMTH) for probing membrane-protein interactions in human cells. Nat Methods. 2014;11:585–592. doi: 10.1038/nmeth.2895. [DOI] [PubMed] [Google Scholar]
- 25.Sarkar P, Reichman C, Saleh T, Birge RB, Kalodimos CG. Proline cis-trans isomerization controls autoinhibition of a signaling protein. Molecular Cell. 2007;25:413–426. doi: 10.1016/j.molcel.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkar P, Saleh T, Tzeng SR, Birge RB, Kalodimos CG. Structural basis for regulation of the Crk signaling protein by a proline switch. Nat Chem Biol. 2011;7:51–57. doi: 10.1038/nchembio.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobashigawa Y, et al. Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK. Nat Struct Mol Biol. 2007;14:503–510. doi: 10.1038/nsmb1241. [DOI] [PubMed] [Google Scholar]
- 28.Schmidpeter PA, Schmid FX. Molecular determinants of a regulatory prolyl isomerization in the signal adapter protein c-CrkII. ACS Chem Biol. 2014;9:1145–1152. doi: 10.1021/cb500001n. [DOI] [PubMed] [Google Scholar]
- 29.Feller SM, Knudsen B, Hanafusa H. c-Abl kinase regulates the protein binding activity of c-Crk. EMBO Journal. 1994;13:2341–2351. doi: 10.1002/j.1460-2075.1994.tb06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hashimoto Y, et al. Phosphorylation of CrkII adaptor protein at tyrosine 221 by epidermal growth factor receptor. The Journal of biological chemistry. 1998;273:17186–17191. doi: 10.1074/jbc.273.27.17186. [DOI] [PubMed] [Google Scholar]
- 31.Rosen MK, et al. Direct demonstration of an intramolecular SH2-phosphotyrosine interaction in the Crk protein. Nature. 1995;374:477–479. doi: 10.1038/374477a0. [DOI] [PubMed] [Google Scholar]
- 32.Chodniewicz D, Klemke RL. Regulation of integrin-mediated cellular responses through assembly of a CAS/Crk scaffold. Biochimica et biophysica acta. 2004;1692:63–76. doi: 10.1016/j.bbamcr.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Piotukh K, et al. Cyclophilin A binds to linear peptide motifs containing a consensus that is present in many human proteins. The Journal of biological chemistry. 2005;280:23668–23674. doi: 10.1074/jbc.M503405200. [DOI] [PubMed] [Google Scholar]
- 34.Handschumacher RE, Harding MW, Rice J, Drugge RJ, Speicher DW. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science. 1984;226:544–547. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- 35.Ting AY, Kain KH, Klemke RL, Tsien RY. Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc Natl Acad Sci U S A. 2001;98:15003–15008. doi: 10.1073/pnas.211564598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filmus J, Pollak MN, Cailleau R, Buick RN. MDA-468, a human breast cancer cell line with a high number of epidermal growth factor (EGF) receptors, has an amplified EGF receptor gene and is growth inhibited by EGF. Biochem Biophys Res Commun. 1985;128:898–905. doi: 10.1016/0006-291x(85)90131-7. [DOI] [PubMed] [Google Scholar]
- 37.Escalante M, et al. Phosphorylation of c-Crk II on the negative regulatory Tyr222 mediates nerve growth factor-induced cell spreading and morphogenesis. J Biol Chem. 2000;275:24787–24797. doi: 10.1074/jbc.M000711200. [DOI] [PubMed] [Google Scholar]
- 38.Yamada S-I, et al. Overexpression of CRKII increases migration and invasive potential in oral squamous cell carcinoma. Cancer Lett. 2011;303:84–91. doi: 10.1016/j.canlet.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 39.Noren NK, Foos G, Hauser CA, Pasquale EB. The EphB4 receptor suppresses breast cancer cell tumorigenicity through an Abl-Crk pathway. Nature Cell Biology. 2006;8:815–825. doi: 10.1038/ncb1438. [DOI] [PubMed] [Google Scholar]
- 40.Takino T, et al. Tyrosine phosphorylation of the CrkII adaptor protein modulates cell migration. J Cell Sci. 2003;116:3145–3155. doi: 10.1242/jcs.00632. [DOI] [PubMed] [Google Scholar]
- 41.Diemert S, et al. Impedance measurement for real time detection of neuronal cell death. J Neurosci Methods. 2012;203:69–77. doi: 10.1016/j.jneumeth.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 42.Deakin NO, Turner CE. Paxillin comes of age. J Cell Sci. 2008;121:2435–2444. doi: 10.1242/jcs.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu YL, et al. FAK and paxillin dynamics at focal adhesions in the protrusions of migrating cells. Sci Rep. 2014;4:6024. doi: 10.1038/srep06024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Jong R, ten Hoeve J, Heisterkamp N, Groffen J. Tyrosine 207 in CRKL is the BCR/ABL phosphorylation site. Oncogene. 1997;14:507–513. doi: 10.1038/sj.onc.1200885. [DOI] [PubMed] [Google Scholar]
- 45.Hemmeryckx B, et al. BCR/ABL P190 transgenic mice develop leukemia in the absence of Crkl. Oncogene. 2002;21:3225–3231. doi: 10.1038/sj.onc.1205452. [DOI] [PubMed] [Google Scholar]
- 46.Jankowski W, et al. Domain organization differences explain Bcr-Abl’s preference for CrkL over CrkII. Nature Chemical Biology. 2012;8:590–596. doi: 10.1038/nchembio.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bell ES, Park M. Models of crk adaptor proteins in cancer. Genes Cancer. 2012;3:341–352. doi: 10.1177/1947601912459951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bravo-Cordero JJ, Hodgson L, Condeelis J. Directed cell invasion and migration during metastasis. Curr Opin Cell Biol. 2012;24:277–283. doi: 10.1016/j.ceb.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klemke RL, et al. CAS/Crk coupling serves as a "molecular switch" for induction of cell migration. Journal of Cell Biology. 1998;140:961–972. doi: 10.1083/jcb.140.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nath PR, Dong G, Braiman A, Isakov N. Immunophilins Control T Lymphocyte Adhesion and Migration by Regulating CrkII Binding to C3G. Journal of Immunology. 2014;193:3966–3977. doi: 10.4049/jimmunol.1303485. [DOI] [PubMed] [Google Scholar]
- 51.Reichman C, et al. Transactivation of Abl by the Crk II adapter protein requires a PNAY sequence in the Crk C-terminal SH3 domain. Oncogene. 2005;24:8187–8199. doi: 10.1038/sj.onc.1208988. [DOI] [PubMed] [Google Scholar]
- 52.Sriram G, et al. Phosphorylation of Crk on tyrosine 251 in the RT loop of the SH3C domain promotes Abl kinase transactivation. Oncogene. 2011;30:4645–4655. doi: 10.1038/onc.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 54.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 55.Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.