Abstract
Changes in cellular gene expression in response to small-molecule or genetic perturbations have yielded signatures that can connect unknown mechanisms of action (MoA) to ones previously established. We hypothesized that differential basal gene expression could be correlated with patterns of small-molecule sensitivity across many cell lines to illuminate the actions of compounds whose MoA are unknown. To test this idea, we correlated the sensitivity patterns of 481 compounds with ~19,000 basal transcript levels across 823 different human cancer cell lines and identified selective outlier transcripts. This process yielded many novel mechanistic insights, including the identification of activation mechanisms, cellular transporters, and direct protein targets. We found that ML239, originally identified in a phenotypic screen for selective cytotoxicity in breast cancer stem-like cells, most likely acts through activation of fatty acid desaturase 2 (FADS2). These data and analytical tools are available to the research community through the Cancer Therapeutics Response Portal.
Identifying the mechanisms by which small molecules affect cellular physiology is critical to their development into tools for research and effective medicines. Experiments using small molecules with clearly defined binding partners can yield new insights into biological mechanisms, illuminate novel therapeutic targets, identify appropriate cellular contexts for treatment, inform approaches for decoupling negative effects from beneficial effects, and suggest directions to improve efficacy1. In many cases, however, the relevant cellular targets of small molecules, including probes and drugs discovered through phenotypic screening, are unknown. Even for molecules with well-established primary targets, additional cellular interactions and mechanisms of metabolic processing, which may contribute to efficacy, toxicity, or drug resistance, are often difficult to predict. As such, systematic, unbiased approaches to identify mechanisms of action (MoA) are in demand2,3.
Measurements of genome-wide changes in mRNA expression following small-molecule treatments can provide insights into cellular processes (e.g., using gene-set enrichment analysis4) and make connections between perturbations acting through similar mechanisms (e.g., using connectivity mapping)5,6. However, the latter approach is limited by the need to connect a gene-expression response to a reference set of existing perturbation signatures. Additionally, signatures must be generated in an appropriate experimental context (e.g., a cell line with a relevant signaling pathway activated). Leveraging differences in basal (i.e., unperturbed) gene expression across cell lines may represent an alternative approach to identifying MoA. Examples of direct relationships between gene expression and compound action, such as the requirement of NQO1 expression for activation of the HSP90 inhibitor tanespimycin, suggest that correlating basal gene expression with small-molecule sensitivity profiles across cancer cell lines (CCLs) can yield insights into MoA7–12. However, the types of mechanisms and compounds suited for sensitivity profiling, as well as the reproducibility and reliability of profiling data, remain in question13.
Here, we report a new computational tool capable of identifying small-molecule MoA. We hypothesized that correlating sensitivity data across hundreds of CCLs with basal gene-expression profiles could illuminate novel small-molecule mechanisms. We also hypothesized that measuring hundreds of compounds would inform the uniqueness of implicated mechanisms and allow us to investigate differences between compounds sharing annotated targets. In this study, we used correlation-based analyses, based on the response of 860 human CCLs to 481 compounds, to investigate the relationships between small-molecule sensitivity profiles and basal gene expression. The inclusion of 115 small molecules with no annotated protein target allowed us to investigate for the first time whether this approach would generate novel insights into MoA. Our results demonstrate how outlier transcripts uniquely correlated with small-molecule response provide novel insights into small-molecule mechanisms, including metabolic processing targets, cellular import and export mechanisms, and direct protein targets. We have made these correlation methods available through the Cancer Therapeutics Response Portal (www.broadinstitute.org/ctrp), a public, interactive resource to enable the scientific community to explore genes and small molecules of interest.
RESULTS
Correlating chemical sensitivity to basal gene expression
To investigate whether differences in basal gene-expression profiles across hundreds of CCLs could be used to identify new MoA, we analyzed sensitivity measurements collected using an Informer Set of 481 tool compounds, probes, and drugs, including FDA-approved cancer therapeutics. We measured the response of 860 CCLs to each member of the Informer Set over a 16-point concentration range using an automated, high-throughput workflow, fit concentration–response curves, and calculated the area under the curve (AUC) as a measure of sensitivity (Supplementary Results, Supplementary Data Sets 1–3; see Methods). Overall, 823 unique CCLs profiled, spanning 23 lineages, were characterized genomically as part of the Cancer Cell Line Encyclopedia (CCLE) project7. Using basal genome-wide expression data previously collected from shared stocks of these CCLs (www.broadinstitute.org/ccle/)7, we calculated Pearson correlation coefficients between AUC values and expression of each of 18,543 transcripts, either across all CCLs or within subsets of CCL lineages. We applied Fisher’s z-transformation to the correlation coefficients to adjust for variation in CCL number across small molecules and contexts (Fig. 1a)14. This transformation allows comparison of lineage-specific correlation z-scores with the corresponding z-score across all CCLs (Supplementary Data Sets 4–5).
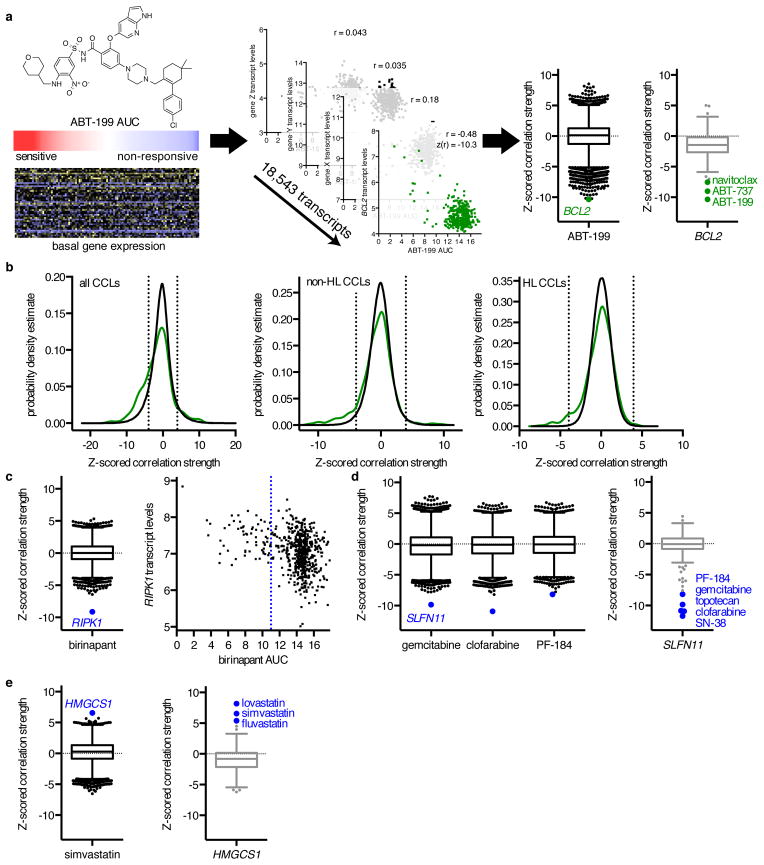
Figure 1. Correlating Gene Expression and CCL Sensitivity Data Illuminates Known Small-Molecule Mechanisms of Action.
(a) Calculation of z-scored Pearson correlation coefficients between small-molecule sensitivity data, expressed as areas under concentration-response curves (AUCs), with basal gene-expression measurements, expressed as log2 robust-multi-array-average values. We examined 18,543 correlation coefficients of transcript levels to ABT-199 sensitivity (black), and 481 correlation coefficients of small molecules to BCL2 expression (gray) across non-hematopoietic and lymphoid (non-HL) CCLs. Box-and-whisker plot outlier points represent Tukey outliers (1.5 × interquartile range). (b) Distribution of z-scored Pearson correlation coefficients between 660 annotated small-molecule–target pairs (green) across all CCLs, non-HL CCLs, and HL CCLs compared to random sampling of correlation coefficients (black). Dashed lines represent two-tailed Bonferroni-corrected significance (|z| = 3.96). (c–e) Expression–sensitivity correlations for target–pathway connections (blue), including (c) the Smac mimetic birinapant; (d) the nucleoside analogues clofarabine and gemcitabine, the IKKβ inhibitor PF-184, and the transcript SLFN11; and (e) the statin simvastatin and the transcript HMGCS1. The blue dashed line represents the low-AUC (left) cutoff for the robust z-score of birinapant AUCs.
To determine whether small-molecule sensitivity was associated with differential gene expression, we first focused on 358 small molecules in our Informer Set with annotated targets for which basal gene-expression measurements were available (660 total compound-target pairs; Supplementary Data Set 2). High expression of target transcripts was correlated significantly with sensitivity for 153/660 compound-target pairs (23%), while low expression was correlated significantly with sensitivity for 46/660 pairs (7%; Fig. 1b, Supplementary Data Set 6). Overall, expression of an annotated target was correlated significantly with compound sensitivity for 146/358 (41%) distinct compounds and 95/309 (31%) distinct targets.
Global analysis of expression–sensitivity relationships
We then sought to identify global patterns in sensitivity profiles that could confound interpretation of genetic features associated with sensitivity. Consistent with our prior observations, CCLs from the hematopoietic and lymphoid (HL) lineage generally were more sensitive to certain compounds than CCLs from solid tumors8. Thus, marker genes for HL CCLs show significant correlation with many compounds; these HL-representative transcripts and HL-selective compounds emerged as the first principal component (PC) from an analysis of the full matrix of correlation coefficients (Supplementary Fig. 1a). However, we believe that these correlations reflect general sensitivity of the HL lineage, rather than a mechanistic connection of biomarker genes to compounds. For example, TNFRSF12A, a transcript almost exclusively expressed in non-HL CCLs, was the transcript most correlated with 36 small molecules (low expression correlated with sensitivity, p < 10−6; Supplementary Fig. 1b, Supplementary Data Sets 7–8). Small-molecule classes enriched for HL sensitivity include microtubule modulators, chromatin modifiers, topoisomerase inhibitors, and nucleotide analogs (Supplementary Data Set 9; Supplementary Fig. 1c). Nonetheless, many compounds were not selective for HL versus non-HL CCLs, including several kinase inhibitors (Supplementary Fig. 1d).
We then separated our data into HL and non-HL subsets, and analyzed each subset separately. We observed an 8.6-fold (p < 0.0001) and 1.5-fold (p = 0.007) increase in the frequency of significant target connections in HL and non-HL CCLs, respectively, including 15 subset-specific target–sensitivity relationships (Fig. 1b, Supplementary Fig. 1e, Supplementary Data Set 6). For example, ABL1 expression emerged as uniquely associated with response to imatinib in HL CCLs (Supplementary Data Set 6).
Because we included compounds that share annotated targets, we compared similarities of all 18,543 expression–sensitivity correlations for small molecules sharing 1) no annotated protein targets, 2) some, but not all, protein targets, and 3) all protein targets. While similarities among small molecules sharing some targets (median correlation coefficient ρm ~ 0.84; p < 2.7×10−67) or all targets (ρm ~ 0.88; p < 5.7×10−28) were significantly higher than those sharing no targets, we did observe a high degree of similarity (ρm ~ 0.53) among pairs of compounds with no shared targets across all CCLs (Supplementary Fig. 1f). When we restricted analysis to HL CCLs, the background similarity among compounds sharing no targets was largely eliminated (ρm ~ 0.10), while the enrichment of similarities among compounds sharing some (p < 7.6×10−152) or all (p < 1.4×10−29) targets improved (Supplementary Fig. 1g). For non-HL CCLs, we observed a similar effect on similarities among compounds sharing targets (some: p < 5.7×10−92; all: p < 3.2×10−43), but only partial reduction of background similarity (ρm ~ 0.22; Supplementary Fig. 1h).
PC analysis of non-HL CCLs revealed additional global patterns of similarity, including ‘fragile’ CCLs that were sensitive to many of the small molecules to which HL CCLs were sensitive (e.g., CCLs from the neuroblastoma lineage; PC_B1), and CCLs whose expression features reflect differences between epithelial and mesenchymal cell states (PC_B2) (Supplementary Fig. 1i–k, Supplementary Data Sets 8, 10). Accounting for these global patterns, by excluding HL CCLs and PC_B1 and PC_B2, further improved similarity enrichment among compounds with shared targets (some: p < 3.7×10−128; all: p < 4.5×10−44; background ρm ~ −0.01; Supplementary Fig. 1l). Collectively, these global analyses show that compelling similarities between small molecules with similar MoA emerge upon controlling for large-scale patterns in sensitivity profiling data.
Expression–sensitivity correlations illuminate MoA
The enrichment we observed between annotated compound–target pairs suggested that expression–sensitivity correlations could be used to identify novel target mechanisms (Fig. 1b, Supplementary Data Set 6). We took a three-step approach to identifying MoA, focusing on non-HL CCLs. We first identified the small molecules with the strongest connection to a single gene, using the cumulative distribution function for 18,543 correlations with each small molecule, considering positive and negative correlations separately (Supplementary Fig. 2a). Second, we scored transcripts for uniqueness, based on the number of small molecules to which that gene was significantly correlated (Supplementary Data Set 11). Finally, we investigated correlation distributions for evidence of multiple, distinct biological mechanisms associated with a single compound, by adjusting for the effects of the most correlated transcript. We used this approach to examine NQO1, a gene known to be involved in metabolic activation of tanespimycin10. Our results demonstrate that NQO1 expression is a key distinguishing feature between the response to tanespimycin and the response to other HSP90 inhibitors (Supplementary Fig. 2b–c).
Overall, this general MoA analysis yielded insights in three categories: 1) the encoded protein is a direct target of the small molecule; 2) the encoded protein is in a pathway including the small-molecule target; and 3) the encoded protein metabolically processes the small molecule (Supplementary Table 1). While we focus here on biological insights gained from the single transcripts most correlated with sensitivity, the approach can be tuned to prioritize multi-gene features (Supplementary Fig. 2d).
Direct target connections
We sought to validate our MoA method by first identifying direct target connections between transcripts and small molecules. A subset of annotated target transcripts, including PDGFRA, BCL2, and NAMPT (Fig. 1a, Supplementary Fig. 3a), correlated most strongly with small molecules known to target these proteins, informing comparative analyses across related compounds15. For example, sensitivity to four BCL2 inhibitors (obatoclax, TW-37, BRD-M00053801, and gossypol) did not correlate as strongly with high BCL2 expression as other BCL2 inhibitors (ABT-199, ABT-737, and navitoclax), suggesting that other mechanisms may be contributing to their cytotoxicity (Supplementary Fig. 3b). Indeed, our MoA analysis revealed that sensitivity to obatoclax was most associated with low expression of biliverdin reductase B (BLVRB; r = 0.33, z(r) = 8.9, p = 4.2×10−19; Supplementary Fig. 3c). Based on these data, and the structural similarities between obatoclax and biliverdins, we hypothesized that obatoclax might be reduced by BLVRB. We observed significant conversion of obatoclax by recombinant BLVRB in vitro, with comparable affinity to that reported for BCL2-family members in protein–protein interaction assays (Supplementary Fig. 3d)16. Binding and modification of obatoclax by BLVRB may therefore influence its ability to interact with BCL2-family proteins in cells.
Target pathway connections
We identified several instances in which transcript–small molecule correlation uncovered proteins involved in pathways relevant to the annotated target (e.g., proteins that interact directly with the target(s), using the STRING database17). First, high expression of receptor-interacting serine/threonine-protein kinase 1 (RIPK1) was associated with sensitivity to birinapant, a Smac mimetic under clinical investigation (r = −0.34, z(r) = −9.1, p = 3.7×10−20; Fig. 1c, Supplementary Fig. 3e). Birinapant binds inhibitor of apoptosis protein (IAP) family members, which prevent RIPK1-mediated pro-death signaling by targeting RIPK1 for ubiquitin-mediated degradation18. Second, we recapitulated the association between high expression of SLFN11 and response to topoisomerase-I inhibitors, and extended this relationship to additional compounds, including the nucleoside analogues gemcitabine (r = −0.39, z(r) = −9.8, p = 3.6×10−23) and clofarabine (r = −0.40, z(r) = −10.9, p = 4.6×10−28), and the IKKβ inhibitor PF-184 (r = −0.32, z(r) = −8.2, p = 1.4×10−16; Fig. 1d, Supplementary Fig. 3f)7,19. Finally, we observed pathway-relevant associations where low transcript expression correlated with sensitivity, such as between HMGCS1 (HMG-CoA synthase 1), which generates the substrate for the statin target HMG-CoA reductase, and response to statins (e.g., simvastatin; r = 0.26, z(r) = 6.6, p = 2.5×10−11; Fig. 1e).
Metabolic mechanisms of small-molecule activation
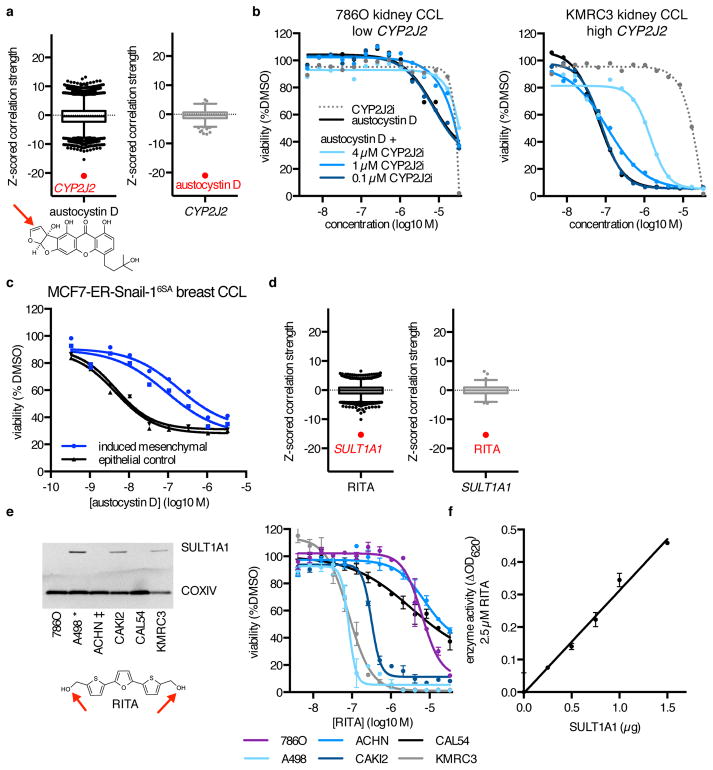
A majority of transcripts implicated by MoA analysis were neither annotated small-molecule targets nor involved in target-relevant pathways (Supplementary Fig. 2a). The two strongest of these single-gene outliers suggested a role for metabolic activation in small-molecule sensitivity. Differential cytotoxicity of the DNA-damaging agent austocystin D is thought to result from modification by cytochrome P450 enzymes, but the specific CYP enzyme involved has not been identified20. Sensitivity to austocystin D was uniquely correlated with high expression of CYP2J2, but not other CYP enzymes (r = −0.70, z(r) = −21.0, p = 4.7×10−98; Fig. 2a, Supplementary Fig. 4a–b). Accordingly, austocystin D cytotoxicity was attenuated by co-treatment with a selective CYP2J2 inhibitor (Fig. 2b, Supplementary Fig. 4c)21. We noted that another strong outlier for austocystin D, HOOK1, is adjacent to CYP2J2 on chromosome 1p32. HOOK1, and other transcripts most correlated with austocystin D sensitivity, are highly co-expressed with CYP2J2 across CCLs. Their associations with sensitivity are diminished upon adjustment for CYP2J2 expression, suggestive of a single coherent signal underlying response (Supplementary Fig. 4d). Both gene-set enrichment analysis (GSEA) and our PC analysis suggest that CYP2J2 is expressed predominantly in epithelial cells (Supplementary Data Sets 8, 10, 12). Consistent with this notion of epithelial selectivity, conversion of the MCF7 breast epithelial CCL to a more mesenchymal cell state, by overexpressing the transcription factor SNAI1, resulted in decreased response to austocystin D (Fig. 2c)22.
Figure 2. MoA Analysis Reveals New Mechanisms of Small-Molecule Metabolism.
(a) Expression–sensitivity correlations for the compound austocystin D and CYP2J2 expression. Reactive functionalities are depicted with a red arrow. (b) Cytotoxicity of austocystin D and a selective CYP2J2 inhibitor (CYP2J2i) across renal CCLs with different expression levels of CYP2J2, and effects of co-treatment of austocystin D with CYP2J2i. Cell viability values are normalized to vehicle-only (DMSO) treatment, with each point representing the mean of n = 2 independent experiments with 2 technical replicates each. (c) Cytotoxicity of austocystin D in MCF7-ER-Snail-16SA cells either induced to undergo epithelial-to-mesenchymal transition (blue) or vehicle-treated (black). Each point is the mean of n = 2 technical replicates. Two independent inductions are shown. (d) Expression–sensitivity correlations for the compound RITA and SULT1A1 expression. (e) Protein expression of SULT1A1 and sensitivity to RITA of six renal CCLs. CCLs described in ref. 23 as capable (*) or incapable (‡) of metabolizing RITA into a cytotoxic form are indicated. Each point is mean ± s.d. for n = 3 independent experiments. For uncropped gel image, see Supplementary Figure 8b. (f) Sulfotransferase enzyme activity of varying amounts of recombinant SULT1A1 at a fixed concentration of RITA in the presence of the sulfate donor 3′-phosphoadenosine-5′-phosphosulfate. Each point is mean ± s.d. for n = 3 independent experiments.
The second strongest correlation involved RITA (NSC652887), a putative modulator of p53 signaling, and SULT1A1, which encodes a phenol-preferring sulfotransferase (r = −0.54, z(r) = −15.3, p = 2.1×10−53; Fig. 2d, Supplementary Fig. 4e)23. Although previous studies suggested that differential cytotoxicity of RITA across CCLs depends on its conversion to an active metabolite capable of forming DNA–protein and DNA–DNA crosslinks, the actual mechanism of activation remained unclear12,23. Sulfotransferases transfer sulfo groups to hormones, xenobiotics, and other nucleophilic small molecules, which yield electrophilic sulfate species capable of reacting with or binding to macromolecules24. We found that SULT1A1 protein expression in six renal CCLs correlated with the reported ability of RITA to form cross-links across these CCLs (Fig. 2e, Supplementary Fig. 4f)23. Further, RITA is sulfated by recombinant SULT1A1 in vitro, suggesting that sulfate modification of RITA primes the compound for cross-linking and underlies its selective cytotoxicity (Fig. 2f, Supplementary Fig. 4g).
Small-molecule import and export mechanisms
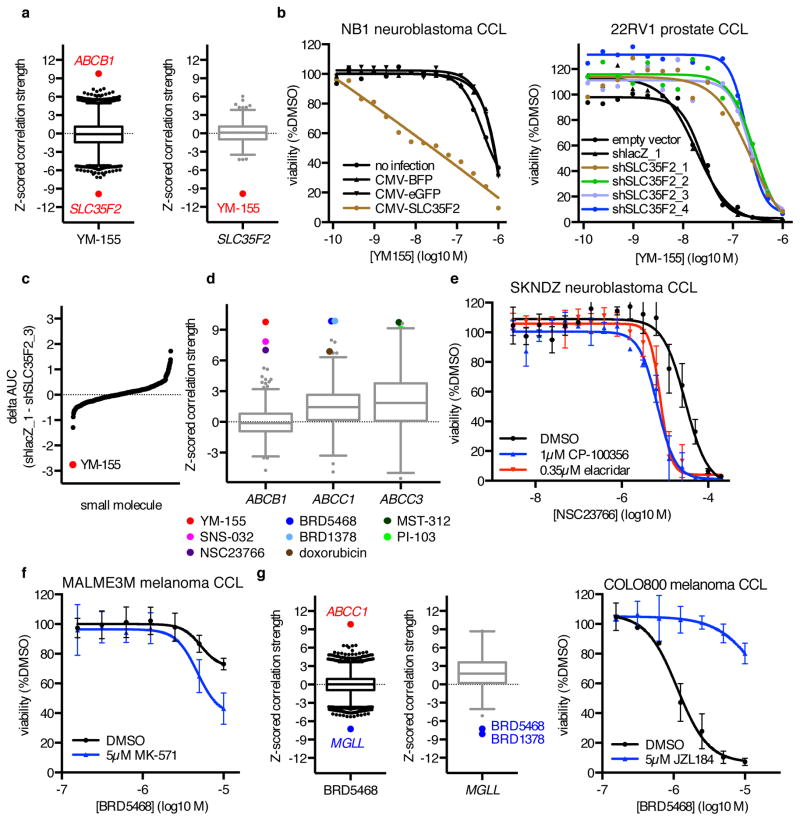
Another outlier relationship unique to one small molecule was the correlation between high expression of SLC35F2, a transmembrane solute carrier, and response to YM-155 (r = −0.38, z(r) = −9.9, p = 3.4×10−23; Fig. 3a)25,26. YM-155, a clinical candidate that induces DNA damage and inhibits survivin expression, is positively charged and requires active transport into cells. SLC35F2 is highly expressed in PC3 cells, the cell line in which YM-155 was initially characterized (Supplementary Fig. 5a). We found that overexpression of SLC35F2 in the NB1 neuroblastoma CCL, which does not express SLC35F2, increased sensitivity to YM-155, while knockdown of SLC35F2 reduced sensitivity of both PC3 and 22RV1 prostate CCLs to YM-155 (Fig. 3b, Supplementary Fig. 5b–d). Further, knockdown of SLC35F2 reduced YM-155 cytotoxicity to a much greater extent than for 439 other small molecules from our Informer Set (Fig. 3c). The limited expression of SLC35F2 in normal tissues (Supplementary Fig. 5e) suggests a potential therapeutic avenue for targeting SLC35F2-expressing cancer cells relative to normal tissue, particularly for cancer lineages with consistently high expression (e.g., prostate and pancreas). Our results are consistent with previous reports showing the importance of SLC35F2 expression for YM-155 activity by an insertional mutagenesis approach27.
Figure 3. MoA Analysis Reveals Small-Molecule Transport Mechanisms.
(a) Expression–sensitivity correlations for YM-155 and SLC35F2. (b) Effects of SLC35F2 overexpression in NB1 cells, or of SLC35F2 knockdown in 22RV1 cells, on YM-155 cytotoxicity. Each point represents the mean of n = 2 independent experiments with 3 (22RV1) –4 (NB1) technical replicates each. (c) Difference in 8-point AUC values between 22RV1-shlacZ_1 and 22RV1-shSLC35F2_3 cells for 439 small molecules tested in duplicate. (d) Expression–sensitivity correlations for multidrug resistance genes implicated by MoA analysis. (e) Effects of co-treatment with DMSO, the ABCB1 inhibitor CP-100356, or the ABCB1 inhibitor elacridar on NSC23766 cytotoxicity in SKNDZ cells. (f) Effects of co-treatment with DMSO or the ABCC1 inhibitor MK-571 on BRD5468 cytotoxicity in MALME3M cells. (g) Expression–sensitivity correlations for BRD5468 and MGLL, and effects of co-treatment with DMSO or the MGLL inhibitor JZL184 on BRD5468 cytotoxicity in COLO800 cells. For (e–g), each point is mean ± s.d. for n = 3 independent experiments.
We also observed an outlier correlation for YM-155 with low expression of ABCB1, encoding ATP-Binding Cassette, sub-family B (MDR/TAP), member 1 (r = 0.38, z(r) = 9.8, p = 7.4×10−23; Supplementary Fig. 5f–g). In fact, several other highly correlated low-expression outliers implicated basal expression of efflux pumps, a common mechanism of drug resistance. These connections included both known and unreported small-molecule substrates, and we obtained experimental support for two such novel predicted interactions (Fig. 3d; Supplementary Table 1)28,29. Co-treatment with the RAC1 inhibitor NSC23766 and either of the ABCB1 inhibitors elacridar or CP-100356 enhanced NSC23766 cytotoxicity in SKNDZ cells (Fig. 3e). Similarly, co-treatment with BRD5468, a differentially cytotoxic reactive-oxygen-species–enhancing tool compound of unknown mechanism, and the ABCC1 inhibitor MK-571 enhanced BRD5468 cytotoxicity in MALME3M cells (Fig. 3f)30. The observation that expression of distinct transporters correlates with sensitivity to different small molecules suggests that CTRP can be used to identify compound-specific resistance mechanisms.
Novel targets uncovered by MoA analysis
Several instances of outlier correlations between single genes and a small molecule could not readily be explained by prior knowledge about the compound, metabolic processing, or general resistance or toxicity mechanisms. For example, BRD5468, along with its correlation to ABCC1, was independently associated with MGLL, encoding monoglyceride lipase (r = −0.28, z(r) = −7.2, p = 2.2×10−13; Fig. 3g). MGLL regulates cellular monoglyceride and free fatty acid levels, and has been implicated in tumorigenesis and metastasis31,32. CCLs with high expression of MGLL were most sensitive to BRD5468 and the structurally related molecule BRD1378 (Supplementary Fig. 5h). Treatment of sensitive CCLs with the MGLL inhibitor JZL184, which was relatively nontoxic across a panel of 240 CCLs, attenuated cytotoxicity of BRD5468 and BRD1378 in COLO800 cells (Fig. 3g, Supplementary Fig. 5i)8. Knockdown of MGLL with four separate shRNA hairpins also reduced cytotoxicity (Supplementary Fig. 5j). These observations suggest that MGLL activity is required for the cytotoxic effects of these compounds.
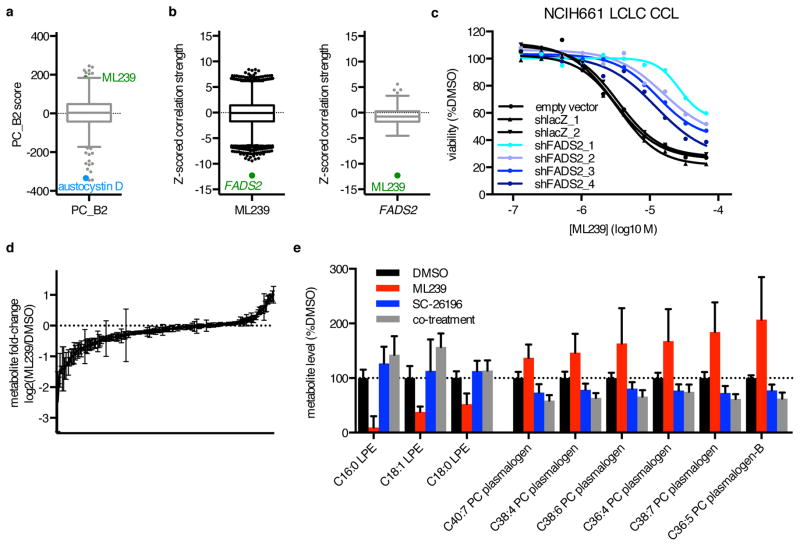
Motivated by the ability of this method to identify novel small-molecule targets, we applied the approach to study ML239, a probe discovered for selective toxicity to breast epithelial cells induced to undergo epithelial-mesenchymal transition (EMT) by knockdown of CDH1, encoding E-cadherin (Supplementary Fig. 6a)33. EMT has been implicated in increased cell motility and metastasis, and in drug resistance and cancer stem cells34. Consistent with these findings, sensitivity to ML239 was correlated with both the mesenchymal PC signature PC_B2 and low expression of CDH1 (r = 0.22, z(r) = 5.6, p = 9.1×10−9; Fig. 4a, Supplementary Fig. 6b). An unbiased analysis showed that sensitivity to ML239 was correlated with high expression of FADS2, which encodes delta(6) fatty acid desaturase (r = −0.45, z(r) = −12.3, p = 5.1×10−35; Fig. 4b, Supplementary Fig. 6c). Consistent with this result, we found that FADS2 expression was higher in shECad cells relative to shGFP cells (Supplementary Fig. 6d), and that FADS2 is a component of an orthogonal EMT expression signature derived from HMLE cells35. FADS2 is involved in polyunsaturated fatty acid (PUFA) metabolism important for oxidative generation of lipid signaling molecules such as arachidonic acid and arachidonic acid-derived oxidized lipid mediators36.
Figure 4. MoA Analysis Identifies and Illuminates the Basis for a Requirement for FADS2 Activity in ML239 Cytotoxicity.
(a) Correlation of PC2 score excluding HL CCLs (PC_B2) with sensitivity to all small molecules. (b) Expression–sensitivity correlations for ML239 and FADS2. (c) Effects of FADS2 knockdown on ML239 cytotoxicity in NCIH661 LCLC cells. Each point represents the mean of n = 2 independent experiments. (d) Change in levels of 183 cellular lipids in NCIH661 cells upon 24-hour treatment with 2 μM ML239. Bars represent mean ± s.e.m. for n=9. (e) Significantly changed lipid species from (d) upon 24-hour treatment with ML239, 2 μM SC-26196, or both (p < 0.001, two-way ANOVA with Bonferroni correction). Bars represent mean ± s.e.m. for n=9 for DMSO, ML239, and SC-26196, and n=3 for co-treatment.
We selected large-cell lung carcinoma (LCLC) for further investigation of ML239, since these CCLs demonstrate a wide range of FADS2 expression and response to ML239 (Supplementary Fig. 6e–f). Co-treatment with the selective FADS2 inhibitor SC-26196 reduced ML239 cytotoxicity in sensitive CCLs (Supplementary Fig. 6g–h)37. FADS2 knockdown reduced ML239 cytotoxicity in NCIH661 cells in a manner dependent upon knockdown efficiency (Fig. 4c, Supplementary Fig. 6i). These data are consistent with a model in which FADS2 activity is required for sensitivity to ML239.
To examine the effects of ML239 on cellular lipid metabolism, we isolated lipids from NCIH661 cells treated with DMSO, ML239, SC-26196, or both compounds, and subjected the samples to mass spectrometry (Fig. 4d, Supplementary Data Set 14). Upon treatment with ML239, we observed significant depletion of saturated and monounsaturated lysophosphatidylethanolamines and increases in polyunsaturated plasmalogens, particularly phosphatidylcholines, relative to treatment with SC-26196 or co-treatment with ML239 and SC-26196 (Fig. 4e). While the functions of individual plasmalogen species are not well defined, plasmalogens are associated with oxidative environments, and are thought to act as antioxidants in membranes by protecting unsaturated fatty acids and membrane proteins from oxidation38,39. We hypothesized that accumulation of polyunsaturated plasmalogens reflects increased oxidative lipid stress due to increased flux through FADS2, and thus investigated whether antioxidants could rescue the effects of ML239. ML239 cytotoxicity was attenuated by quercetin and α-tocopherol, but not by N-acetylcysteine or the ferroptosis inhibitor ferrostatin-1, suggesting that the oxidative effects of ML239 are distinct from other small molecules that induce cell death via modulation of lipid pathways through glutathione depletion or ferroptosis (e.g., ML210; Supplementary Fig. 6j–k)40.
To explore the potential relationship between FADS2 and E-cadherin, we knocked down CDH1 in MCF7 cells, an epithelial breast CCL commonly used as an EMT model and known to have low basal FADS2 function41. We observed neither sensitization to ML239 nor changes in FADS2 expression upon CDH1 knockdown, or indirect transcriptional repression of CDH1 through inducible overexpression of SNAI1 (Supplementary Fig. 6l)22. Similarly, FADS2 expression was not significantly altered in several other publicly available EMT-related expression signatures derived from other cell lines (e.g., MCF10A22, NCIH35842) and/or projected across tumor samples (e.g., non-small cell lung carcinoma43). These findings suggest that increased FADS2 expression is not a universal feature of mesenchymal transition models, but rather that FADS2 expression and function, and not CDH1 levels or a mesenchymal cell state, are the critical determinants of sensitivity to ML239.
Profiling requirements for MoA discovery
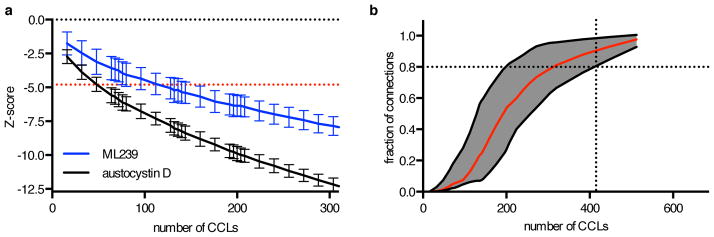
Given the time and resources necessary to run these profiling experiments, we investigated the number CCLs required to identify the mechanisms in Supplementary Table 1 by randomly sampling CCLs, focusing on novel MoA. To establish a significant connection between ML239 and FADS2, more than 160 CCLs were required; for austocystin D and CYP2J2, the strongest pair in our dataset, 68 CCLs were required (Fig. 5a). Overall, we found that to establish 80% of the 43 connections highlighted (Supplementary Table 1), more than 400 CCLs were needed (Fig. 5b, Supplementary Fig. 7), supporting the importance of profiling at large scale.
Figure 5. Large numbers of CCLs are required to identify MoA.
(a) Significance of connection between austocystin D AUC and CYP2J2 expression (black), and ML239 AUC and FADS2 expression (blue) as a function of CCL number in non-HL CCLs. Red, Bonferroni-corrected z-score cutoff (|z| > 4.79) for random sampling of CCLs. (b) Simulation of whether connections to 43 small molecules listed in Supplementary Table 1 could be identified using smaller numbers of CCLs. Depicted are the fraction (red trace) of connections that were statistically significant relative to the null distribution after Bonferroni correction for multiple hypothesis testing. Correlations one standard deviation above or below the mean are in gray.
DISCUSSION
Our results demonstrate the value of systematic correlation of sensitivity data with basal gene-expression data to identify cellular targets and MoA of small molecules. We observed instances where high expression of a cellular target correlated with sensitivity, which may reflect a dependency of sensitive CCLs on the target, enhanced metabolic activation or import of the small molecule, or a dependency on the presence or activity of target–drug complexes7,44. We also observed instances where low expression correlated with sensitivity, which may reflect a buffering effect, where high expression of the target protein titrated away the effects of the small molecule, or drug inactivation or efflux mechanisms.
To explore the generality of this approach, we investigated correlations between chemical sensitivity and expression of previously annotated targets, observing significant correlation with at least one target for 154/358 (43%) small molecules (Supplementary Data Set 6). Reasons contributing to lack of correlation for other small-molecule–target pairs may include lack of differential expression of targets across CCLs, domination of correlation profiles by other features (e.g., mechanisms of metabolism or engagement of un-annotated cellular targets), small molecules not exhibiting widespread toxicity in this assay format, or a need to test larger numbers or specific subsets of CCLs. In particular, the global patterns of expression across CCLs highlighted by our PC analysis support further investigation of correlations specific to cellular lineages or states. For example, the combination of global and lineage-specific analyses highlighted the poly-pharmacology of the dual BCR-ABL/PDGFRA inhibitor imatinib, as high PDGFRA and ABL1 expression were correlated significantly with imatinib sensitivity in solid tumor and HL CCLs, respectively (Supplementary Fig. 3a, Supplementary Data Set 6).
To identify novel targets, we focused on single-transcript correlation outliers, which frequently pointed to interpretable biology. Comparing the correlations of transcripts across many small molecules is a critical feature of this analysis, as it can inform specificity of associations, reveal global confounding factors, or highlight transcripts that actually contribute to differences in small-molecule response. For example, we identified novel mechanisms of selective CCL toxicity dependent on the activity of the proteins MGLL and FADS2, both of which promote generation of reactive oxidized lipid species and have been suggested as potential therapeutic targets for inhibition in cancer32,41.
However, correlations produce lists of associated transcripts that can be challenging to interpret, even where outliers exist. An advantage of testing across many cell lines with a range of expression features is the ability to distinguish between co-regulated and distinctly regulated transcripts correlated with compound sensitivity. For several novel MoA compounds (e.g., RITA, austocystin D, and ML239), adjusting for the expression of the most-correlated transcript diminished all other associations (Supplementary Fig. 4, Supplementary Fig. 6). For other small molecules (e.g., BRD5468), we observed evidence for multiple independent associations that prompted further biological investigation (Supplementary Fig. 5). Using CTRP to interrogate transcripts beyond those most associated with small-molecule response may reveal additional targets, while querying multi-gene signatures, or signatures reporting on pathway activity and cancer-cell states, may identify new types of dependencies and therapeutic vulnerabilities (Supplementary Fig. 2d). For example, the association of a polyunsaturated fatty acid desaturase (FADS2) and arachidonic acid epoxygenase (CYP2J2) with mesenchymal and epithelial CCLs suggests that the role of oxidative lipid metabolism in these cellular states may warrant further investigation.
There are potential limitations to this approach in identifying novel MoA, such as when a relevant target is not differentially expressed, or when differential expression of a target does not affect toxicity (e.g., as for certain inhibitors of metabolic pathways45). However, this approach is complementary to existing target-identification methods, with particular value for compounds with pre-existing evidence of differential cytotoxicity in subsets of CCLs (e.g., molecules profiled in the NCI-6012). Indeed, ML239, BRD1378, and BRD5468 were identified for their differential cytotoxicity in isogenic or near-isogenic phenotypic screens, and elicited strong differential responses in this dataset30,33. Similarly, for RITA and austocystin D, previous studies had suggested a differential mode of metabolic activation across CCLs, but in both cases, the precise molecular mechanism remained elusive12,20,23. Notably, the ability to identify specific metabolic connections is a particular strength of this approach relative to profile-matching methods, where the output of differential drug metabolism is either an ‘active’ or ‘inactive’ profile, without reporting on the underlying metabolic mechanism.
In addition to providing the scientific community with the tools and dataset to apply basal gene-expression correlation widely in the CTRP (www.broadinstitute.org/ctrp), the approach presented here also lays the framework for future experimental and computational investigations. Importantly, we found that hundreds of CCLs were required to capture the expression–sensitivity connections reported here. The strength of association did not plateau at the maximum number of CCLs tested (Supplementary Table 1, Supplementary Fig. 7), suggesting that testing even more CCLs across different cancer types may capture novel biological connections. While testing large numbers of CCLs is a significant undertaking, new technical advances (e.g., the ability to test small molecules against multiplexed pools of CCLs46) may increase the feasibility of this approach. Further, these correlation methods can be applied to orthologous datasets collected across shared CCLs as they become publicly available (e.g., metabolite data, shRNA data, or additional small-molecule sensitivity data). We plan to incorporate such analyses into subsequent versions of CTRP, enabling identification of additional features associated with small-molecule response. Such applications of CTRP data will likely accelerate progress in characterization of the cellular mechanisms of small molecules, and may enable translation of these insights into treatment of disease.
ONLINE METHODS
Cancer cell-line (CCL) sensitivity profiling
An Informer Set of 481 small molecules was measured against 860 publicly available human CCLs encompassing 25 lineages. Small-molecule and cell-line information, including CCL contexts and growth conditions, is provided in Supplementary Data Sets 1–2 and the CTRP website (www.broadinstitute.org/ctrp) and downloadable from the NCI-CTD2 Data Portal (ctd2.nci.nih.gov/). To verify that CCLs tested in sensitivity profiling uniquely matched those used to generate microarray expression measurements, genomic DNA was extracted from 803/860 CCLs tested (93%) and used to genotype 96 SNPs using the Fluidigm 96.96 system; SNP calls were matched to a database of 1045 CCLs7,47. 771 CCLs positively matched to a CCLE reference genotype; 32 (4%) did not match any reference CCL (Supplementary Data Set 1). As further information becomes available (e.g., matching of unconfirmed samples), we will provide updated information and analyses reflecting any changes at the NCI-CTD2 Data Portal and in the CTRP.
Small molecules were selected individually to interrogate important targets and/or cellular processes in cancer with high reported selectivity, and collectively to target diverse nodes in cancer cell circuitry, from sources including FDA-approved drugs, clinical candidates, previous screening and sensitivity profiling experiments, scientific literature and patents, bioactives, and collaborator contributions (Supplementary Data Set 2)8,12. CCLs were plated at a density of 500 cells/well in white opaque tissue-culture-treated Aurora 1536-well MaKO plates (Brooks Automation) in the provider-recommended growth media using a highly automated platform. Compounds were added by acoustic transfer using a Labcyte Echo 555 (Labcyte Inc.) 24 hours after plating. The effects of small molecules were measured over a 16-point concentration range (two-fold dilution) in duplicate. DMSO was used at a constant concentration of 0.33%, including vehicle-only control wells. As a surrogate for viability, cellular ATP levels were assessed 72 hours after compound transfer by addition of CellTiterGlo (Promega) followed by luminescence measurement using a ViewLux Microplate Imager (PerkinElmer). Duplicates were averaged and luminescence values normalized to vehicle (DMSO) treatment and background (media-only) wells.
Area under curve (AUC) calculation
Sixteen-point concentration-response curves were generated using nonlinear fits to 3-parameter sigmoid functions with the low-concentration asymptote set to the DMSO-normalized value (100% viability) using the equation:
When such a fit resulted in a predicted EC50 higher than the top concentration tested, curves were re-fit using a 2-parameter sigmoid function with the value of the lower asymptote set equal to the value of complete killing (0% viability):
The area under curve (AUC) for each compound-CCL pair was calculated by integration under the 16-point curve. Calculations were performed using MATLAB (MathWorks).
Correlation of AUC with gene expression
Gene-centric robust multi-array average (RMA)-normalized basal mRNA gene expression data measured on the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array were downloaded from the Cancer Cell Line Encyclopedia (CCLE_Expression_Entrez_2012-10-18.res; www.broadinstitute.org/ccle)7. Accession numbers were merged with Entrez gene identifiers; 445/18988 rows that did not map to a gene symbol were removed (Supplementary Data Set 14). Pearson correlation coefficients were calculated between gene expression and AUCs across all overlapping CCLs (n = 842) and for up to 306 additional CCL sub-contexts including lineage, histology, growth mode, and gender in MATLAB. Correlations were normalized using Fisher’s Z-transformation14. Small-molecule target annotations were used to correlate known targets with response and compared to correlations generated by randomly sampling correlations (1,024 permutations), with significance corresponding to a Bonferroni-corrected, two-tailed distribution with family-wise error-rate α < 0.025 in each tail for 660 compound-target pairs (|z| > 3.96). The frequency of significant compound–target pairs was compared to all compound–transcript pairs exceeding this |z| threshold using a chi-squared test.
The normal cumulative distribution function of correlation coefficients was used to calculate the p-value difference [−log10(ptranscript 1/ptranscript 2)] between the 1st-ranked transcript and the 2nd-ranked transcript to identify outliers using MATLAB. Positive and negative correlation distributions were calculated separately. Multi-transcript outliers were prioritized using the p-value difference between the 2nd-ranked and Tth-ranked transcript [−log10(ptranscript 2/ptranscript T)], where T = the number of transcripts that are Tukey outliers, i.e., 1.5 times the interquartile range of the correlation coefficient distribution (see Supplementary Fig. 2d). The number of small molecules to which each top-ranked transcript was significantly associated was calculated with Bonferroni correction using an estimated p-value (if 16,384 randomly-permuted correlations between small-molecule sensitivity and transcript levels passed a Kolmogorov-Smirnov test for normality) or otherwise an empirical p-value generated from randomly permuting CCL labels (n ≥ 16,384).
Estimated adjusted correlation coefficients from semipartial correlation controlling for the expression of individual transcripts were calculated using the ppcor R package. Box-and-whisker plots were generated from correlation coefficients using GraphPad Prism.
Principal component analysis and expression–sensitivity correlation similarities
Principal component analysis of the correlation matrix of 18,543 transcripts across 481 small molecules was performed in MATLAB for 1) all CCLs and 2) non-HL CCLs. 18,543 transcript loadings per principal component were then correlated with basal gene expression of these 18,543 transcripts across CCLs to generate a PC score for each CCL using Pearson correlation (Supplementary Fig. 1a). The number of small molecules to which each individual transcript was associated (Supplementary Fig. 1j) was calculated using a Bonferroni-corrected, two-tailed distribution with family-wise error-rate α < 0.025 in each tail. We compared patterns of correlations among compounds annotated to share no, some, or all targets by calculating median correlation coefficients between small-molecule correlation distributions (18,543 transcripts) and comparing correlations using the Wilcoxon rank-sum test.
STRING database queries
The complete list of annotated small-molecule targets for 481 compounds was used to query the STRING v9.1 database for human proteins (string-db.org)17. All resulting putative interactions were intersected with the list of transcripts most correlated with small-molecule sensitivity; these were manually curated for published support for interactions.
Gene set enrichment analysis (GSEA)
18,543 transcript loadings for each PC were used as inputs for pathway analysis in gene set enrichment analysis (GSEA) querying the C2 and C5 gene sets using the default parameters4. Analogously, 18,543 z-scored correlations between expression and small-molecule sensitivity were used to identify pathways enriched for the transcripts most correlated with austocystin D sensitivity.
CCL subsampling analysis
CCLs were sampled randomly 28 times at each of 31 numbers of CCLs (16, 32, 48, 64, 128, 132, 136, 140, 144, 160, 176, 192, 196, 200, 204, 208, 224, 240, 256, 272, 288, 304, 320, 384, 448, 512, 528, 544, 560, 576, 640). For each number of CCLs, average correlation coefficients across all samplings were compared to null distributions of 210 correlation coefficients obtained by permuting CCL labels. The z-score cutoff for statistical significance relative to these null distributions corresponds to a Bonferroni-corrected, two-tailed distribution with family-wise error-rate α < 0.025 in each tail. Correlations one standard deviation above or below the mean were used as a measure of the noise in the simulation. Calculations were carried out in MATLAB.
Cellular validation and follow-up assays
CCLs were plated in white, opaque tissue-culture-treated 384-well plates (Corning) in the provider-recommended growth media at 1000 cells/well. Small molecules were transferred using a CyBi-Well vario pin-transfer machine 24 hours after plating (CyBio). DMSO was used at a constant concentration of 0.33%, including for vehicle-control wells, except for compound co-treatment experiments where DMSO was used at a constant concentration of 0.66%. Sensitivity was measured using CellTiterGlo 72 hours after addition of small molecules as previously described7. DMSO-normalized concentration-response curves were generated using four-parameter nonlinear regression in GraphPad Prism for visual presentation:
For comparison of 22RV1-shlacZ_1 and 22RV1-SLC35F2_3, 440 small molecules were tested over an 8-point concentration range in duplicate. Duplicates were averaged and curves fit as previously described with successful fits for 439/440 compounds7.
SNAI1-inducible MCF7 cells
Retroviral generation of MCF7 cells with inducible Snail-1 under the control of an estrogen response element (MCF7-ER-Snail-16SA) has been described previously22. MCF7-ER-Snail-16SA cells were treated with 1 μM 4-hydroxytamoxifen (4-OHT; Sigma) or vehicle (ethanol) for 120 hours, with 4-OHT replenished every 48 hours. 4-OHT was then withdrawn and cells were plated in 384-well plates (800 cells/well), allowed to adhere for 24 hours, and treated with compound for 72 hours before sensitivity measurements using CellTiterGlo. Expression changes (induced versus vehicle) in EMT-related transcripts were measured using a Fluidigm Real-Time PCR system to confirm induction of EMT as described previously22.
Small molecules for validation assays
JZL184 (Cat. no. 13158; ≥97% purity) was purchased from Cayman Chemical. ML239 (SML0442; ≥98%), NSC23766 (SML0952; ≥97%), CP-100356 (PZ0171; ≥98%), elacridar (SML0486; ≥98%), ML210 (SML0521; ≥98%), ferrostatin-1 (SML0583; ≥95%), α-tocopherol (T3251; ≥99%), and N-acetyl-L-cysteine (A7250; ≥99%) were purchased from Sigma. YM-155 (S1130; ≥99%), obatoclax mesylate (S1057; ≥99%), and RITA (S2781; 99%) were purchased from SelleckChem. SC-26196 (4189; 99.2%), quercetin (1125; >98%), and MK-571 (2238; >96.9%) were purchased from Tocris. Austocystin D was purchased from eMolecules. The selective CYP2J2 inhibitor 1-(4-bromophenyl)-4-[4-(hydroxydiphenylmethyl)piperidin-1-yl]butan-1-one was synthesized in-house according to the published methodology (>95% purity)48. All small molecules were dissolved in DMSO, except N-acetylcysteine, which was dissolved in cell culture medium then adjusted to pH 7.5.
Western blotting
Whole-cell lysates were prepared by incubating cell pellets in radioimmunoprecipitation assay buffer (Pierce) containing Complete Mini EDTA-free protease inhibitor cocktail (Roche) for 15 minutes on ice. Following centrifugation (15000g, 10 min, 4°C), protein concentration of the supernatant was determined by BCA assay (Pierce). 50 μg of total protein was heated (95°C, 10 minutes) in 4X Bolt LDS sample buffer (Life Technologies) containing 5% 2-mercaptoethanol (Sigma-Aldrich) and loaded onto Bolt 4%–12% Bis-Tris Plus gels (Life Technologies) for electrophoresis. Proteins were then transferred to nitrocellulose membranes using the iBlot system (Life Technologies), blocked for 60 minutes in Tris-buffered saline + 0.1% Tween (TBST) containing 3% milk, and incubated with primary antibody overnight in TBST at 4°C. Primary antibodies and dilutions used were: mouse SULT1A1 (1:1000, with 5% milk) from Abcam (ab57849), rabbit CYP2J2 (H-60) (1:100) from Santa Cruz Biotechnology (sc-67276), rabbit MGLL (1:500) from Abcam (ab152002), and rabbit COX IV (3E11) (1:2000) from Cell Signaling (#5247). Blots were washed with TBST and incubated with HRP-conjugated secondary antibody [Anti-mouse IgG #7076 or Anti-rabbit IgG #7074 (Cell Signaling) diluted 1:1000 in TBST] for 60 minutes at room temperature, developed using SuperSignal West Pico Chemiluminescent Substrate (Pierce), and detected on a Kodak Image Station 4000MM Pro.
SULT1A1 and BLVRB kinetic assays
Recombinant human SULT1A1 (R&D Systems) activity in the presence of 0.2 mM 3′-phosphoadenosine-5′-phosphosulfate and the indicated concentrations of RITA was measured by a phosphatase-coupled assay using the Universal Sulfotransferase Activity Kit (R&D Systems) according to the manufacturer’s instructions. The assay was carried out in Corning clear 96-well plates and incubated (37°C, 1 hour) before addition of detection reagents and visualization at 620 nm using a SpectraMax M5 microplate reader using the SoftMax Pro software (Molecular Devices).
Recombinant human BLVRB was purchased from R&D Systems, Inc. BLVRB activity was measured in the presence of 20 ng/μL BLVRB, obatoclax, and 0.01 mM NADPH in 100 mM sodium phosphate (pH 7.5) in a 20 μL volume in white, opaque 384-well plates. The reaction was incubated for 3 hours at 37°C before detection using the NADPH-Glo assay (Promega) according to the manufacturer’s instructions. NADPH and sodium phosphate were purchased from Sigma.
Lentiviral knockdown and overexpression
High-titer virus preparation and viral packaging were performed by the Broad Institute Genetic Perturbation Platform according to the public protocol [www.broadinstitute.org/rnai/public/resources/protocols and ref. 49]. Cells (30000/well for COLO800, 100000/well for NCIH661 and PC3, 350000/well for 22RV1, and 200000/well for NB1) were seeded in 6-well dishes and allowed to adhere. Upon wells reaching ~50% confluence, media was then changed to regular growth media containing polybrene (8 μg/mL), and virus (approximately 6 × 105 viral particles/well) was added. Media was changed 24 hours later for selection with puromycin for shRNA (Life Technologies; 2 μg/mL for 22RV1, COLO800, and NCIH661; 5 μg/mL for PC3) or blasticidin S for overexpression vectors (Life Technologies; 4 μg/mL for NB1). Cells were maintained under continuous selection, passaged twice, and then assayed.
Real-time PCR
CCLs were seeded at 3000 cells/well in tissue-culture-treated 384-well plates in triplicate. After 24 hours, cells were lysed and RNA collected using the Cells-to-CT kit (Life Technologies) according to the manufacturer’s instructions but with a lysis volume of 20 μL. Following preparation of cDNA (total volume 25 μL), Real-time PCR was carried out in duplicate for each sample using 4 μL of cDNA mixture per reaction and 1 μL probe (target gene and ACTB for each sample) in a total reaction volume of 20 μL on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). Results were normalized to ACTB using the delta CT method, and to control shRNA using the delta-delta CT method. Probes used were Hs99999903_m1 (ACTB), Hs00213850_m1 (SLC35F2), Hs00200752_m1 (MGLL), and Hs00927433_m1 (FADS2) (Life Technologies).
Cellular lipid profiling
One hundred thousand NCIH661 cells were seeded in 6-well dishes. After 24 hours, cells were treated with 2 μM ML239, 2 μM SC-26196, 2 μM ML239 + 2 μM SC-26196, or DMSO as vehicle control and incubated for 24 hours before metabolite extraction. Media was vacuum-aspirated and then cells were immediately washed with cold PBS without calcium or magnesium (Life Technologies). Following PBS aspiration, 800 μL of 80% ice-cold LC-MS grade methanol was added and plates were transferred to −80°C for 15 minutes. Plates were then scraped on dry ice with a cell scraper and transferred to 1.5 mL tubes on dry ice. Following centrifugation (9000g, 10 min, 4°C), pellets were resuspended in 100 μL 80% methanol and centrifuged again (9000g, 5 min, 4°C). Supernatants were stored in fresh tubes until mass spectrometry analysis.
Supplementary Material
Acknowledgments
This work was supported by the NCI’s Cancer Target Discovery and Development (CTD2) Network (grant number U01CA176152, awarded to S.L.S.). We acknowledge the following colleagues and centers for contributing compounds and for valuable critique: Boston University, J. Bradner, P. Brown, C. Chen, J. Clardy, CNIO, E.J. Corey, the Drug Synthesis and Chemistry Branch (Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute), Eutropics, J. Gutterman, E. Holson, Karyopharm, M. Meyerson, A. Myers, J. Porco, J. Qi, Sanford-Burnham, M. Serrano-Wu, M. Shair, B. Stockwell, L. Walensky, X. Wang, and D. Zaharevitz. We thank A. Deik for conducting lipid profiling measurements; S. Chattopadhay, J. Law, G. Schaefer, M. Stewart, V. Viswanathan, and other members of the Schreiber lab for advice and helpful discussions; S. Wang for helping curate the Informer Set; K. Emmith, J. Aseidu, and the CSofT informatics group for development and support of cell-line and data-tracking software; A. Vrcic and the Broad Compound Management team for handling the Informer Set; J. Boehm, A. Tsherniak, A. Aguirre, and the Broad Cancer Program for training and advice; the Broad Biological Samples Platform for providing CCLs; and Levi Garraway and the Broad-Novartis Cancer Cell Line Encyclopedia (CCLE) team. S.L.S. is an Investigator at the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
M.G.R. designed and executed validation experiments, analyzed and interpreted data, made the figures, and wrote the manuscript; B.S.L. designed and executed validation experiments, analyzed and interpreted data, and wrote the manuscript; J.H.C. designed and executed the primary cell-line screen; D.J.A. designed and executed validation experiments and analyzed data; E.V.P. executed the primary cell-line screen; S.G. executed validation experiments; S.J. executed validation experiments and analyzed data; M.E.C. executed the primary cell-line screen; V.L.J. executed the primary cell-line screen; N.E.B. analyzed and interpreted primary cell-line screen data; C.K.S. executed the primary cell-line screen; B.A. developed the CTRP; A.L. analyzed and interpreted data; P.M. developed the CTRP; J.D.K. wrote the manuscript; C.S.H. designed the primary cell-line screen; B.M. designed the primary cell-line screen; T.L. developed the CTRP; V.D. analyzed and interpreted primary cell-line screen data; D.A.H. designed validation experiments; C.B.C. designed and executed lipid profiling experiments and analyzed data; J.A.B. designed the primary cell-line screen; M.P. designed the primary cell-line screen; B.K.W. executed validation experiments and wrote the manuscript; P.A.C. designed, analyzed, and interpreted primary cell-line screening data, directed the study, and wrote the manuscript; A.F.S. designed the primary cell-line screen, interpreted data, directed the study, and wrote the manuscript; S.L.S. designed the primary cell-line screen, directed the study, and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Hughes JP, Rees S, Kalindjian SB, Philpott KL. Principles of early drug discovery. Br J Pharmacol. 2011;162:1239–49. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schenone M, Dančík V, Wagner BK, Clemons PA. Target identification and mechanism of action in chemical biology and drug discovery. Nat Chem Biol. 2013;9:232–40. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10:507–19. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 4.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–26. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 6.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 7.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basu A, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151–61. doi: 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo W, et al. Formation of 17-allylamino-demethoxygeldanamycin (17-AAG) hydroquinone by NAD(P)H:quinone oxidoreductase 1: role of 17-AAG hydroquinone in heat shock protein 90 inhibition. Cancer Res. 2005;65:10006–15. doi: 10.1158/0008-5472.CAN-05-2029. [DOI] [PubMed] [Google Scholar]
- 11.Papillon-Cavanagh S, et al. Comparison and validation of genomic predictors for anticancer drug sensitivity. J Am Med Inform Assoc. 2013;20:597–602. doi: 10.1136/amiajnl-2012-001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–23. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 13.Haibe-Kains B, et al. Inconsistency in large pharmacogenomic studies. Nature. 2013;504:389–93. doi: 10.1038/nature12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dančík V, et al. Connecting Small Molecules with Similar Assay Performance Profiles Leads to New Biological Hypotheses. J Biomol Screen. 2014;19:771–781. doi: 10.1177/1087057113520226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams DJ, et al. NAMPT is the cellular target of STF-31-like small-molecule probes. ACS Chem Biol. 2014 doi: 10.1021/cb500347p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhai D, Jin C, Satterthwait AC, Reed JC. Comparison of chemical inhibitors of antiapoptotic Bcl-2-family proteins. Cell Death Differ. 2006;13:1419–21. doi: 10.1038/sj.cdd.4401937. [DOI] [PubMed] [Google Scholar]
- 17.Franceschini A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–15. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benetatos CA, et al. Birinapant (TL32711), a bivalent SMAC mimetic, targets TRAF2-associated cIAPs, abrogates TNF-induced NF-κB activation, and is active in patient-derived xenograft models. Mol Cancer Ther. 2014;13:867–79. doi: 10.1158/1535-7163.MCT-13-0798. [DOI] [PubMed] [Google Scholar]
- 19.Zoppoli G, et al. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci U S A. 2012;109:15030–5. doi: 10.1073/pnas.1205943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marks KM, et al. The selectivity of austocystin D arises from cell-line-specific drug activation by cytochrome P450 enzymes. J Nat Prod. 2011;74:567–73. doi: 10.1021/np100429s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lafite P, Dijols S, Zeldin DC, Dansette PM, Mansuy D. Selective, competitive and mechanism-based inhibitors of human cytochrome P450 2J2. Arch Biochem Biophys. 2007;464:155–68. doi: 10.1016/j.abb.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Javaid S, et al. Dynamic chromatin modification sustains epithelial-mesenchymal transition following inducible expression of Snail-1. Cell Rep. 2013;5:1679–89. doi: 10.1016/j.celrep.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rivera MI, et al. Selective toxicity of the tricyclic thiophene NSC 652287 in renal carcinoma cell lines: differential accumulation and metabolism. Biochem Pharmacol. 1999;57:1283–95. doi: 10.1016/s0006-2952(99)00046-5. [DOI] [PubMed] [Google Scholar]
- 24.Gamage N, et al. Human sulfotransferases and their role in chemical metabolism. Toxicol Sci. 2006;90:5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- 25.Glaros TG, et al. The “survivin suppressants” NSC 80467 and YM155 induce a DNA damage response. Cancer Chemother Pharmacol. 2012;70:207–12. doi: 10.1007/s00280-012-1868-0. [DOI] [PubMed] [Google Scholar]
- 26.Nakahara T, et al. YM155, a novel small-molecule survivin suppressant, induces regression of established human hormone-refractory prostate tumor xenografts. Cancer Res. 2007;67:8014–21. doi: 10.1158/0008-5472.CAN-07-1343. [DOI] [PubMed] [Google Scholar]
- 27.Winter GE, et al. The solute carrier SLC35F2 enables YM155-mediated DNA damage toxicity. Nat Chem Biol. 2014 doi: 10.1038/nchembio.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamath AV, Chong S, Chang M, Marathe PH. P-glycoprotein plays a role in the oral absorption of BMS-387032, a potent cyclin-dependent kinase 2 inhibitor, in rats. Cancer Chemother Pharmacol. 2005;55:110–6. doi: 10.1007/s00280-004-0873-3. [DOI] [PubMed] [Google Scholar]
- 29.Young LC, et al. Expression of multidrug resistance protein-related genes in lung cancer: correlation with drug response. Clin Cancer Res. 1999;5:673–80. [PubMed] [Google Scholar]
- 30.Adams DJ, et al. Discovery of small-molecule enhancers of reactive oxygen species that are nontoxic or cause genotype-selective cell death. ACS Chem Biol. 2013;8:923–9. doi: 10.1021/cb300653v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long JZ, et al. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat Chem Biol. 2009;5:37–44. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nomura DK, et al. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Germain AR, et al. Identification of a selective small molecule inhibitor of breast cancer stem cells. Bioorg Med Chem Lett. 2012;22:3571–4. doi: 10.1016/j.bmcl.2012.01.035. [DOI] [PubMed] [Google Scholar]
- 34.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–73. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 35.Taube JH, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;107:15449–54. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sprecher H. Metabolism of highly unsaturated n-3 and n-6 fatty acids. Biochim Biophys Acta. 2000;1486:219–31. doi: 10.1016/s1388-1981(00)00077-9. [DOI] [PubMed] [Google Scholar]
- 37.Obukowicz MG, et al. Identification and characterization of a novel delta6/delta5 fatty acid desaturase inhibitor as a potential anti-inflammatory agent. Biochem Pharmacol. 1998;55:1045–58. doi: 10.1016/s0006-2952(97)00665-5. [DOI] [PubMed] [Google Scholar]
- 38.Catala A. Lipid peroxidation of membrane phospholipids generates hydroxy-alkenals and oxidized phospholipids active in physiological and/or pathological conditions. Chem Phys Lipids. 2009;157:1–11. doi: 10.1016/j.chemphyslip.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 39.Snyder F, Lee T, Wykle RL. Ether-linked lipids and their bioactive species. Biochemistry of Lipids, Lipoproteins, and Membranes. 2002;36:233–262. [Google Scholar]
- 40.Yang WS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–31. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park WJ, Kothapalli KS, Lawrence P, Brenna JT. FADS2 function loss at the cancer hotspot 11q13 locus diverts lipid signaling precursor synthesis to unusual eicosanoid fatty acids. PLoS One. 2011;6:e28186. doi: 10.1371/journal.pone.0028186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salt MB, Bandyopadhyay S, McCormick F. Epithelial-to-mesenchymal transition rewires the molecular path to PI3K-dependent proliferation. Cancer Discov. 2014;4:186–99. doi: 10.1158/2159-8290.CD-13-0520. [DOI] [PubMed] [Google Scholar]
- 43.Byers LA, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19:279–90. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fischer ES, et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature. 2014;512:49–53. doi: 10.1038/nature13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmer AC, Kishony R. Opposing effects of target overexpression reveal drug mechanisms. Nat Commun. 2014;5:4296. doi: 10.1038/ncomms5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu C, Golub TR. US Patent WO 2013138585 A1. Multiplex methods to assay mixed cell populations simultaneously. 2013
- 47.Cowley GS, et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data. 2014;1:140035. doi: 10.1038/sdata.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C, et al. Selective inhibitors of CYP2J2 related to terfenadine exhibit strong activity against human cancers in vitro and in vivo. J Pharmacol Exp Ther. 2009;329:908–18. doi: 10.1124/jpet.109.152017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X, et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8:659–61. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.