Abstract
Rationale
The liver-specific natural killer (NK) cell population is critical for local innate immune responses, but the mechanisms that lead to their selective homing and the definition of their functionally relevance remain enigmatic.
Objectives
We took advantage of the availability of healthy human liver to rigorously define the mechanisms regulating the homing of NK cells to liver and the repertoire of receptors that distinguish liver-resident NK (lr-NK) cells from circulating counterparts.
Findings
Nearly 50% of the entire liver NK cell population is composed of functionally relevant CD56bright lr-NK cells that localize within hepatic sinusoids. Further, CD56bright lr-NK cells express CD69, CCR5 and CXCR6 and this unique repertoire of chemokine receptors is functionally critical as it determines selective migration in response to the chemotactic stimuli exerted by CCL3, CCL5 and CXCL16. In addition, hepatic sinusoids express CCL3pos Kupffer cells, CXCL16pos endothelial cells and CCL5pos T and NK lymphocytes. The selective presence of these chemokines in sinusoidal spaces creates a tissue niche for lr-CD56bright NK cells that constitutively express CCR5 and CXCR6. CD56bright lr-NK cells co-exist with CD56dim conventional NK (c-NK) cells that are, interestingly, transcriptionally and phenotypically similar to their peripheral circulating counterparts. Indeed, CD56dim c-NK cells lack expression of CD69, CCR5, and CXCR6 but express selectins, integrins and CX3CR1.
Conclusion
Our findings disclosing the phenotypic and functional differences between lr-Nk cells and c-NK cells are critical to distinguish liver-specific innate immune responses. Hence, any therapeutic attempts at modifying the large population of CD56bright lr-NK cells will require modification of hepatic CCR5 and CXCR6.
Keywords: Liver immunology, Homing of hepatic NK cells, Chemokine Receptors
Introduction
Natural killer (NK) cells are important effectors of the innate immune system that can lyse tumor-transformed or virus-infected cells in the absence of prior antigen sensitization. NK cells are also endowed with immune-regulatory functions at tissue sites of inflammation through the establishment of cellular interactions and the production of pro-inflammatory cytokines including IFN-γ, TNF-α, CCL3 (Mip1-α, CCL4 (Mip1-α and CCL5 (RANTES) [1, 2]. Human peripheral blood NK (PB-NK) cells are divided into two functionally distinct subsets characterized by a different distribution of CD56 and CD16 surface markers. CD56bright/CD16neg-low (CD56bright) NK cells account for 5–15% of all PB-NK cells and, while poorly cytotoxic, can produce large amounts of cytokines. CD56dim/CD16pos (CD56dim) NK cells represent the majority of PB-NK cells (up to 95%) and serve primarily as cytotoxic effectors [3]. In order to spare autologous cells from cytotoxicity and ensure tolerance to self, NK cells receive inhibitory signals from a large family of inhibitory NK cell receptors (iNKRs), that include Killer cell immunoglobulin-like receptors (KIRs) and C-type lectins recognizing specific alleles of self MHC-class-I molecules (MHC-I). NK cell effector-functions are generally induced by the engagement of another family of activating NK cell receptors (aNKRs) that binds their ligands expressed on stressed, infected or tumor-transformed target cells that either lack or have a decreased expression of self-MHC-I [1, 4–6].
NK cells can account up to 50% of the total lymphocyte population in the human liver, which is in contrast to their lower frequency in blood (5–15%) or other peripheral tissues such as lymph nodes. Hence, great efforts have been placed over the recent years to understand the role of hepatic NK cells in the pathogenesis of liver disorders including fibrosis, viral infections, tumors and autoimmune diseases [7–9]. In this regard, a distinct subset of hepatic NK cells endowed with adaptive immune properties and exhibiting antigen-specific recall responses to viruses and haptens has been recently described in mice [10, 11]. This memory-like response is exerted by a unique subset of CD49apos/DX5neg liver-resident NK (lr-NK) cells that are phenotypically and functionally distinct from peripheral blood CD49aneg/DX5pos conventional NK (c-NK) cells circulating throughout spleen and liver. Likewise, it has been reported that NK cells in human liver are different from their circulating counterparts [7, 12–16], thus suggesting that a unique lr-NK cell subset also exists in the human liver under homeostatic conditions. However, the overall frequency and the precise phenotype of human lr-NK cells are still being debated, while the distribution and the homing mechanisms regulating their retention in the liver are unknown. The present study characterizes CD56bright lr-NK cells selectively located within hepatic sinusoids and accounting for half of the entire hepatic NK cell population. We demonstrate that CD56bright lr-NK cells are phenotypically and transcriptionally distinct from PB-CD56bright NK cells and constitutively express high levels of markers associated with tissue residency including CD69, CCR5 and CXCR6. This unique phenotype is functionally relevant as CD56bright lr-NK cells migrate in response to CCL3, CCL5 and CXCL16, the CCR5 and CXCR6 ligands highly expressed within liver sinusoids on Kupffer cells, T and NK lymphocytes as well as endothelial cells.
Material and Methods
Human Donors
Human peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats of healthy donors obtained in accordance with clinical protocols approved by the Institutional Review Board of Desio Hospital, Milan, Italy. Liver specimens were obtained from patients undergoing liver resection to remove liver metastases of colorectal carcinoma. Fragments of hepatic tissues used for our experiments were macroscopically and microscopically free of any diseases and considered healthy, as assessed by the Unit of Pathology of the Humanitas Research Hospital, Milan, Italy (Supplemental Figure 1). Specimens not meeting these criteria were excluded from our study. Liver specimens were obtained in accordance with clinical protocols approved by the Institutional Review Board (IRB) of Istituto Clinico Humanitas, Milan, Italy. Liver perfusates were obtained from Singapore Institute for Clinical Sciences as a part of the scientific collaboration with Dr. Antonio Bertoletti. The Gleneagles Hospital Ethics Committee, Singapore, Singapore approved the study and each patient gave written informed consent.
Cell preparation
PBMCs were obtained by Ficoll-Hypaque density gradient centrifugation (GE Healthcare Biosciences), as previously described [17, 18]. Liver mononuclear cells (LMNCs) were isolated by digestion of fresh liver samples using 2mg/ml of collagenese D (Roche) in HEPES Buffered Saline for 45 minutes followed by a brief mechanical digestion using the GentleMACS Dissociator (Miltenyi). Cells were layered over a 70%/30% discontinuous Percoll (GE-Healthcare) gradient. Cells between the 70%/30% layer contained LMNCs. Perfusate mononuclear cells from were isolated from healthy healthy liver before transplant using Ficoll-Paque PREMIUM (GE-Healthcare), as previously described [19].
Flow cytometry and in vitro functional assays
For multicolor flow cytofluorimetric analysis, PBMCs, LMNCs and PMNCs were stained with the following conjugated mAb as previously described: CD56-PE-Cy5 and NKp46-PE (Beckman Coulter, clone BAB281), CD16-PE-Cy7, CD19-APCH7, IFN-γ-PE, DNAM-1 PE, CD49e-PE, CD11c-PE, CCR5-PE, CD69-PE, CCR4-PE and CXCR3-PE (BD-Pharmigen), CD45-PB, CD62L-PE-Cy7, CCL5-AF647, CD161-PerCpCy5.5, CCR3-PE, CXCR2-PE and CXCR4-PerCpCy5.5 (Biolegend), CCR7-FITC and CXCR6-PE (R&D System), CX3CR1 PE (MBL). Aqua LIVE/DEAD (Life Technologies) was used to eliminate dead cells from the analysis. Intracellular staining was performed using Cytofix/Cytoperm (Beckton Dickinson), according to the manufacturers instructions. The gating strategy used to select NK cells from both PBMC and LMNC is depicted in Supplemental Figure 1. For measurement of IFN-γ production, whole PBMC and LMNC were stimulated with 18 hours with 20ng/ml rhIL-12 (R&D Systems) and 200U/ml rhIL-2 (Peprotech). For the final 4 hours, GolgiStop was added (Beckton Dickenson). Flow cytometry data was acquired using an LSR Fortessa (Beckton Dickinson) and data was analyzed using FlowJo Software (Tree Star).
Immunohistochemistry
Paraffin-embedded liver specimens were assessed for chemokine expression using the following mAbs: Anti-CCL3 (Mip1-α anti-CCL4 (Mip1-α (R&D) and anti-CCL5 (RANTES) (Abeam). Antigen retrieval was performed for 5 minutes at 125°C and for 3 minutes at 90°C in a pressure cooker using Diva Decloaker antigen retrieval solution (Biocare Medical). Endogenous peroxidase activity was blocked with Peroxidased I (Biocare Medical) for 5 minutes and non-specific proteins were blocked for 15 min with Background Sniper (Biocare Medical). The primary antibody was incubated for 1 hour at room temperature. For CCL5, MACH 4 HRP Polymer was used as the secondary antibody (Biocare Medical). For CCL3 and CCL4, Goat-on-Rodent HRP Polymer (Biocare Medical) was used as the secondary antibody. For identification of NK cells, a mAb anti-NKp46 (R&D Systems) was used. Samples were prepared by flash-freezing pieces of fresh liver in isopentane-cooled dry ice in OCT (Tissue-Tek). Sections were fixed in freshly prepared 4% paraformaldehyde and incubated with H202 in methanol. Blocking of non-specific proteins was performed using 0.04% horse serum followed by overnight incubation with the primary antibody at 4C. MACH 4 HRP Polymer was used as the secondary antibody. (Biocare Medical). All antigen detection was performed using 3,3-diaminobenzidine (DAB chromogen) and counterstaining of nuclei was performed using hemotoxylin. Images were obtained using optical microscopy (Olympus BX53).
Immunofluorescence
For visualization of CCR5pos NK cells in liver sinusoids, samples of fresh liver were fixed with fresh 2% PFA for 2 hours, washed in PBS for 4 hours and then embedded in 30% sucrose overnight. Samples were then frozen in OCT. Sections were incubated with primary mAbs against NKp46 (R&D Systems), CCR5 (BD) and Lyve-1 (Abcam) to mark hepatic sinusoids for 1 hour at room temperature, washed and then incubated with the following secondary antibodies raised in donkey for 30 minutes: anti-mouse 488, anti-goat 647, anti-rabbit 594 (Life Technologies). Nuclei were counterstained with DAPI at a concentration of 1:50000 in distilled water. For visualization of CCL3pos/CD68pos cells, slides of paraffin-embedded healthy liver were exposed to ultraviolet light for 48 hours to remove background staining. Staining was performed as described above for immunohistochemical staining using anti-CCL3. For identification of Kupffer cells, anti CD68 (Dako) was used. After incubation with primary antibodies, the following secondary antibodies raised in donkey for 30 minutes: anti-mouse 488 and anti-goat 647 (Life Technologies). Nuclei were counterstained with DAPI at a concentration of 1:50000 in distilled water. Imaging was obtained by confocal microscopy (Olympus, Tokyo, Japan).
Chemotaxis assay
CD56pos cells were isolated from LMNC using CD56 positive selection kit (Stem Cell), and serum starved for 2 hours. Cells were then placed in serum-free medium in the upper well of transwell chambers (Costar, 5 µm insert, 24 well plate). Recombinant chemokines CCL3, CCL4, CCL5, CXCL16 and fractalkine (R&D Systems) were placed in the lower chamber. After 2 hours of migration at 37°C, the cells in the lower compartment were harvested, stained using antibodies specific for NK cells and analyzed by flow cytometry. Due to low recovery of purified NK cells from the liver, optimal concentrations of chemokines were determined by performing a titration of 4 concentrations on peripheral blood NK cells. From these data, 2 concentrations (1nM and 100nM) were selected based on the percentage of NK cells migrated. Each assay using hepatic NK cells was performed using these two concentrations and data displayed represents the chemokine concentration that gave the best results. The optimal concentrations for each chemokine were the following: 1nM for CCL3, CLL4, CCL5 and CXCL16, 100nM for fractalkine. The frequency of migration of hepatic NK cell subsets was determined as previously described [20]. Briefly, the percentage of hepatic NK cell populations was determined by dividing the number of cells migrated by the total number of cells in the input control well. The number of cells migrated was calculated by subtracting the background (from negative control) from the number of cells migrated to chemokine. Cells placed in the lower portion of the trans-well chamber only were defined as input control. Negative control is defined as the frequency of cells present in upper portion of the chamber in the absence of chemotactic stimulus in the lower portion of the chamber. Each chemokine concentration was performed in duplicate while the input control and negative control were performed in 4 different wells each. Each chemokine concentration was performed in duplicate while the input control and negative control were performed in 4 different wells each.
Gene expression
NK cells from liver perfusates and peripheral blood were FACS-sorted on the basis of their expression of CD56 and CD16 into the two subsets of CD56bright/CD16neg and CD56dim/CD16pos NK cell subsets (FACS Aria III, BD). All cells were sorted using a BD FACS Aria. Lysates from a minimum of 10,000 cells were analyzed using the preassembled nCounter GX Human Immunology Kit and the nCounter system (NanoString Technologies, Seattle, WA), a cut-off of 2 times the mean of the negative controls supplied in the kit was used to discriminate against nonspecific probe binding (noise). Samples were then normalized based on the geometric means of both the supplied positive controls and the panel of housekeeping genes, as recommended by the manufacturer. All data analyses were performed in R (version 3.0.2) using Bioconductor libraries (BioC 2.13) and R statistical packages on log2 expression values. Global unsupervised clustering was performed using the function hclust of R stats package with Pearson correlation as distance metric and average agglomeration method. Gene expression heatmaps were generated using the software dChip (http://www.hsph.harvard.edu/cli/complab/dchip/) after row-wise standardization of the expression values. To assess cluster-specific reproducibility, we calculated p-values for sample clusters via the multiscale bootstrap resampling method coded in the R pvclust package [21]. Then, p-values were computed for all clusters of the original data as the frequency that any cluster appears in the bootstrap replicates (Bootstrap Probability).
Statistical analysis
Statistical calculations were performed using the Student’s t test. Details of each calculation appear in the figure legends.
Results
CD56bright hepatic NK cells are enriched at high frequencies in the healthy human liver
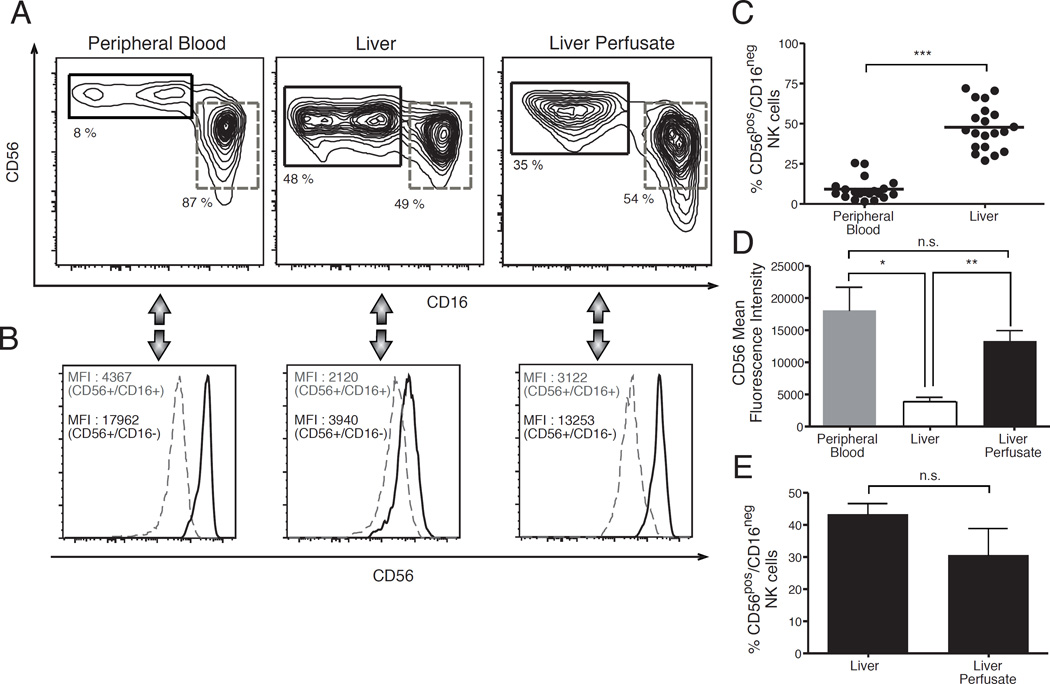
Similar to their circulating counterparts, human hepatic NK cells can be distinguished into two CD56pos/CD16neg and CD56pos/CD16pos cell subsets under homeostatic conditions [3, 19]. However, the frequency of CD56pos/CD16neg hepatic NK cells is significantly higher compared to that of CD56pos/CD16neg PB-NK cells in matched donors [7, 22] (Figures 1 A and 1C). CD56pos/CD16neg PB-NK cells are conventionally defined as CD56bright NK cells due to the higher mean fluorescence intensity (MFI) of CD56 compared to that of CD56pos/CD16pos PB-NK lymphocytes. Indeed, this latter population is defined as CD56dim NK cells. In freshly purified liver mononuclear cells (LMNCs) the MFI of CD56 on CD16neg NK cells is significantly lower compared to that of their circulating counterparts and is similar to that of CD16pos NK cells from both peripheral blood mononuclear cells (PBMCs) and LMNCs (Figures 1A, 1B and 1D). In this regard, it has been demonstrated that collagenase, the enzyme conventionally used to disrupt liver tissue for isolating LMNCs, induces a decrease in the surface expression of CD56 on NK cells [23]. To assess whether the lower MFI of CD56 on CD56pos/CD16neg hepatic NK is indeed an artifact associated with the use of collagenase, we analyzed the degree of CD56 expression on NK cells from liver perfusate (perf-NK cells). This biological specimen is conventionally obtained by flushing the donor’s healthy organ before transplantation with the cold University of Wisconsin solution, which lacks enzymes capable of cleaving or lowering the cellular expression of surface molecules [24]. We found that the subset distribution of perf-NK cells within perfusate mononuclear cells (PMNCs) recapitulates the one observed in LMNCs, as the frequency of CD56pos/CD16neg NK cells was similar in both specimens (Figures 1A and 1E). These results are line with previous data showing that PMNCs flushed out from hepatic sinusoids share with LMNCs a similar lymphocyte distribution [24, 25]. Moreover, we observed that the MFI of CD56 on CD56pos/CD16neg perf-NK cells is significantly higher compared to that of their LMNC counterparts and similar to that of CD56bright PB-NK cells (Figures 1B and 1D). Taken together, these results reveal that the degree of CD56 expression on CD56pos/CD16neg hepatic NK cells is indeed lowered by the enzymatic process of liver digestion. Therefore and in line with the nomenclature used for their circulating counterparts, CD56pos/CD16neg hepatic NK cells will be referred to as CD56bright NK cells henceforth.
Figure 1. Distribution of NK cell subsets in peripheral blood, liver tissues and liver perfusates.
(A,B) Flow cytometric contour plots (A) and histogram (B) graphs from a representative example showing the phenotypic distribution of CD56 and CD16 (A) and the mean fluorescence intensity (MFI) of CD56 (B) on CD56pos/CD16neg (black line) and CD56pos/CD16pos (dashed gray line) NK cell subsets freshly purified from peripheral blood (left), healthy liver tissue (middle) and healthy liver perfusates (right). (C) Summary graphs of statistical dot plots with p values and means showing the relative frequencies of CD56pos/CD16neg NK cell subset within total NK cell populations purified from peripheral blood and healthy liver tissue of matching donors. Data are presented as percentages and are representative of 20 donors. (D) Summary graphs of statistical histogram bars with p values and SD showing the MFI of CD56 on CD56pos/CD16neg NK cells freshly purified from peripheral blood (left), healthy liver tissues (middle) and healthy liver perfusates (right). Data are representative of 3 donors. (E) Summary graphs of statistical histogram bars with p values and SD showing the relative frequencies of CD56pos/CD16neg NK cell subset within total NK cell populations purified from healthy liver tissues (left) and healthy liver perfusates (right). Data are presented as percentages and are representative of 3 donors.
* = p < 0,05 – ** = p < 0,01 – *** = p < 0,001
CD56bright hepatic NK cells display unique transcriptional and phenotypic profiles
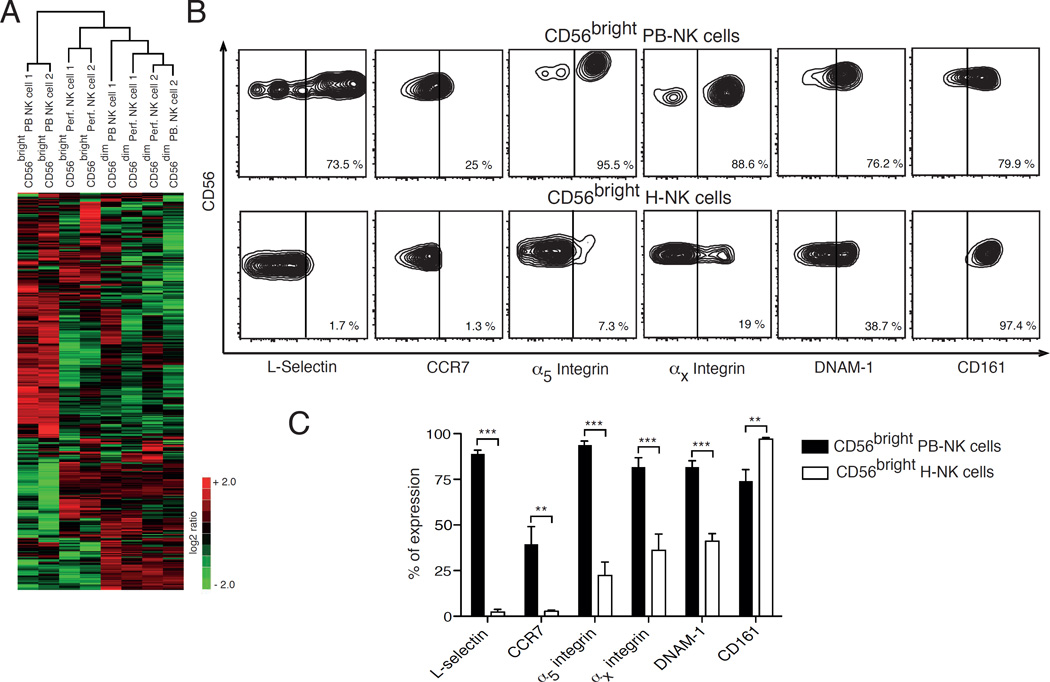
We then analyzed the gene expression profile of sorted PB- and perf-NK cell subsets by using the Nanostring technology. As mentioned previously, perf-NK cells were analyzed as a surrogate of hepatic NK cells since PMNCs recapitulate the overall lymphocyte distribution present in LMNCs under homeostatic conditions [24, 25]. An unsupervised cluster analysis performed on the entire dataset revealed striking differences between CD56bright PB- and perf-NK cell subsets that display different transcriptional profiles and separate into two main clades, thus suggesting that they represent two distinct NK cell populations. In contrast, the genetic profiles of CD56dim PB- and perf-NK cell subsets clustered together into a single clade, hence indicating a high similarity between these two populations (Figure 2A).
Figure 2. Transcriptional and phenotypic profiles of CD56bright hepatic NK cells.
(A) Heat map showing the unsupervised clustering of 441 genes differently expressed in the Nanostring dataset on CD56bright NK cell subset freshly purified from perfusates (Perf.) of healthy liver compared to that of CD56bright NK cell from peripheral blood (PB) and to CD56dim Perf- and PB-NK cells. (B) Flow cytometric contour plots from a representative example showing the percentages of L-selectin, CCR7, α5 integrin, αx integrin, DNAM-1 and CD161 on CD56bright NK cells from peripheral blood (PB-NK cells in the upper line) and healthy liver (H-NK cells in the lower line) of the same donor. (C) Summary graphs of statistical histogram bars with p values and SD showing the surface expression of L-selectin, CCR7, α5 integrin, αx integrin, DNAM-1 and CD161 on CD56bright NK cells from peripheral blood (black bars) and healthy livers (white bars) of matching donors. Data are presented as percentages and are representative of 5 human donors.
** = p < 0,01 – *** = p < 0,001
A more in-depth analysis of the 441 genes contained in the Nanostring dataset identified 50 genes as being significantly differentially regulated between CD56bright PB- and perf-NK cells (Supplemental tables 1 and 2). Among these genes, ccr7 and sell mRNA copies were decreased by respectively 32 and 10.85 fold in CD56bright perf-NK cells compared to their circulating counterparts. These two mRNAs encode for the surface receptors CCR7 and L-selectin, two important homing receptors known to be constitutively present on CD56bright PB-NK cells and regulating their migration to secondary lymphoid tissues (SLT) [26]. In line with their gene expression, we observed by flow cytometry that CD56bright hepatic NK cells lack the expression of CCR7 and L-selectin which, in contrast, are constitutively present at high levels on CD56bright PB-NK cells from matching donors (Figures 2B and 2C). Moreover, we found significantly lower levels of the mRNAs encoding for α5 (itga5) and αx (itgax) integrins on CD56bright perf-NK cells compared to CD56bright PB-NK cells. The same distribution pattern of these two integrins was also confirmed by flow cytometry on the surface of CD56bright hepatic and PB-NK cells from the same donors. Finally, we found that both the transcript level and surface expression of CD161 (klrb1) is significantly higher on CD56bright hepatic NK cells compared to CD56bright PB-NK cells (Figures 2B and 2C). These findings are in line with the reported high frequency of CD161-expressing lymphocytes in the human liver [19]. The phenotypic differences between CD56bright hepatic and PB-NK cells were not induced by the use of collagenase, as the phenotype of CD56bright perf-NK cells is identical to that of CD56bright hepatic NK cells (Supplemental Figure 2).
In contrast, perf- and PB-NK CD56dim cells were found to share a similar transcriptional profile (Supplemental Table 3). This homology in gene expression is corroborated by the identical surface expression of CCR7, L-selectin, α5 and αx integrins on CD56dim NK cell subsets from both LMNCs and PBMCs (data not shown). Instead, we found that these homing receptors are differently expressed on CD56bright hepatic NK cells compared to CD56bright PB-NK cells (Figures 2B and 2C). Among all aNKRs and iNKRs regulating the effector-functions of NK cells, we observed that only the expression of DNAM-1 was significantly decreased on CD56bright hepatic NK cells compared to their circulating counterparts (Figures 2B and C), while the repertoire of all remaining NKRs was nearly identical on both CD56bright and CD56dim hepatic NK cell subsets compared to their circulating counterparts (data not shown). We also measured the expression of Tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) that is constitutively expressed on a subset of murine hepatic NK cells [27]. In line with previous reports with human hepatic NK cells [7, 13], we did not find any detectable levels of TRAIL on both CD56bright and CD56dim H hepatic NK cell subsets (data not shown).
CD56bright and CD56dim hepatic NK cells express a different repertoire of chemokine receptors and markers associated with tissue-residency
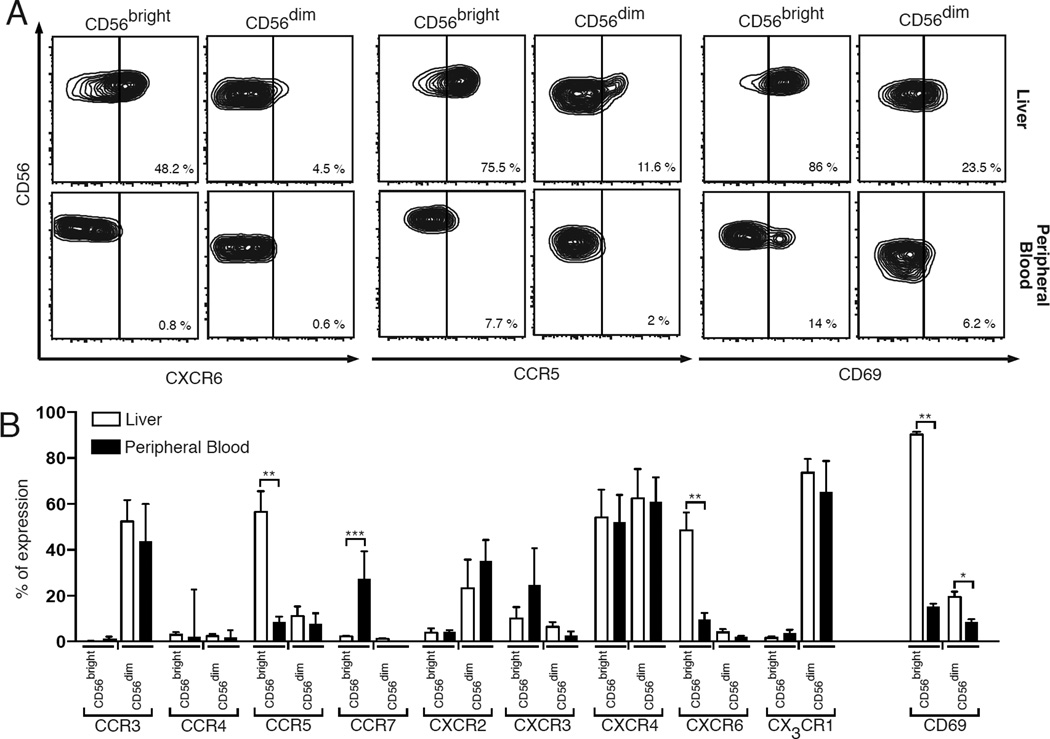
We then assessed whether CD56bright NK cells in liver also display a distinct homing pattern by analyzing on both hepatic and PB-NK cells from matching donors the expression of an extensive panel of chemokine receptors known to regulate the migration of circulating lymphocytes to peripheral tissues [26, 28]. First, we found that low transcript levels and surface expressions of CCR4, CCR5, CCR7, CXCR3 and CXCR6 characterize CD56dim hepatic and PB-NK cells. Moreover, both these subsets express similar high constitutive amounts of CCR3, CXCR2 and CX3CR1 that, in contrast, were found not to be present in CD56bright hepatic and PB-NK cells (Figure 3 and Supplemental Table 1). Interestingly, it has been previously reported that freshly purified PB-CD56dim NK cells are characterized by the high expression of CX3CR1 [3, 26], thus indicating that CX3CR1pos/CD56dim hepatic NK cells likely derives from their circulating counterparts flowing in liver parenchyma.
Figure 3. Repertoire of chemokine receptors and CD69 expression on hepatic NK cell subsets compared to their circulating counterparts.
(A) Flow cytometric contour plots graphs showing the surface expression of the chemokine receptors CXCR6, CCR5 and CD69 on CD56bright (first column) and CD56dim (second column) purified from liver (upper line) and peripheral blood (lower line) of matching donors. (B) Summary graphs of statistical histogram bars with p values and SD showing the surface expression of CCR3, CCR4, CCR5, CCR7, CXCR2, CXCR3, CXCR4, CXCR6, CX3CR1 and CD69 on CD56bright and CD56dim NK cells from healthy livers (white bars) and peripheral blood (black bars) of matching donors. Data are presented as percentages and are representative of 5 donors.
* = p < 0,05 – ** = p < 0,01 – *** = p< 0,001
While CD56dim hepatic and PB- NK cells share a similar repertoire of chemokine receptors, CD56bright hepatic NK cells possess a unique pattern of homing receptors that differs from that of CD56bright PB-NK cells (Figure 3 and Supplemental Table 1). Indeed, CXCR6 is highly expressed only on the surface of hepatic CD56bright NK cells. This was also confirmed at transcriptional level as CD56bright perf-NK cells showed a nearly 18-fold increase of mRNA encoding for CXCR6 compared to CD56bright PB-NK cells. We also found that CD56bright hepatic NK cells, but not their circulating counterparts, express high surface levels of CCR5, a chemokine receptor present on lymphocytes in non-lymphoid tissues under homeostatic conditions [29, 30]. These latter results were not confirmed at the mRNA level, likely indicating the presence of a post-transcriptional mechanism associated with the high levels of CCR5 selectively on CD56bright hepatic NK cells.
The high amounts of chemokine receptors associated with tissue residency and the lack of adhesion molecules as well as chemokine receptors involved in the homing of circulating immune cells to peripheral tissues prompted us to postulate that CD56bright hepatic NK cells might represent a subset of liver-resident lymphocytes in humans. To confirm this hypothesis, we analyzed the surface expression of CD69, a molecule known to characterize tissue-resident lymphocytes [31]. We found that almost all CD56bright hepatic NK cells constitutively express CD69, while CD56bright PB-NK as well as CD56dim hepatic NK cells and CD56dim PB-NK cells lack the expression of this tissue-residency marker (Figure 3). We also analyzed on CD56dim hepatic NK cells the repertoire of KIRs, which are known to distinguish CD56dim from CD56bright PB-NK cells [3]. We observed that both transcript levels and surface expressions of KIRs on CD56dim hepatic NK cells are nearly identical to those of their circulating counterpart (Supplemental Table 1 and data not shown), thus further indicating that CD69neg/CD56dim hepatic NK cells are not tissue-resident but retain the homing features and phenotype of CD56dim PB-NK. Indeed, CD56dim perf- and PB-NK cell subsets share a highly similar transcriptional profile as assessed by the Nanostring dataset showing that only 7 out of 441 genes are differentially expressed between these two populations (Supplemental Table 3).
CD56bright hepatic NK cells are functional and migrate to CCR5 and CXCR6 ligands
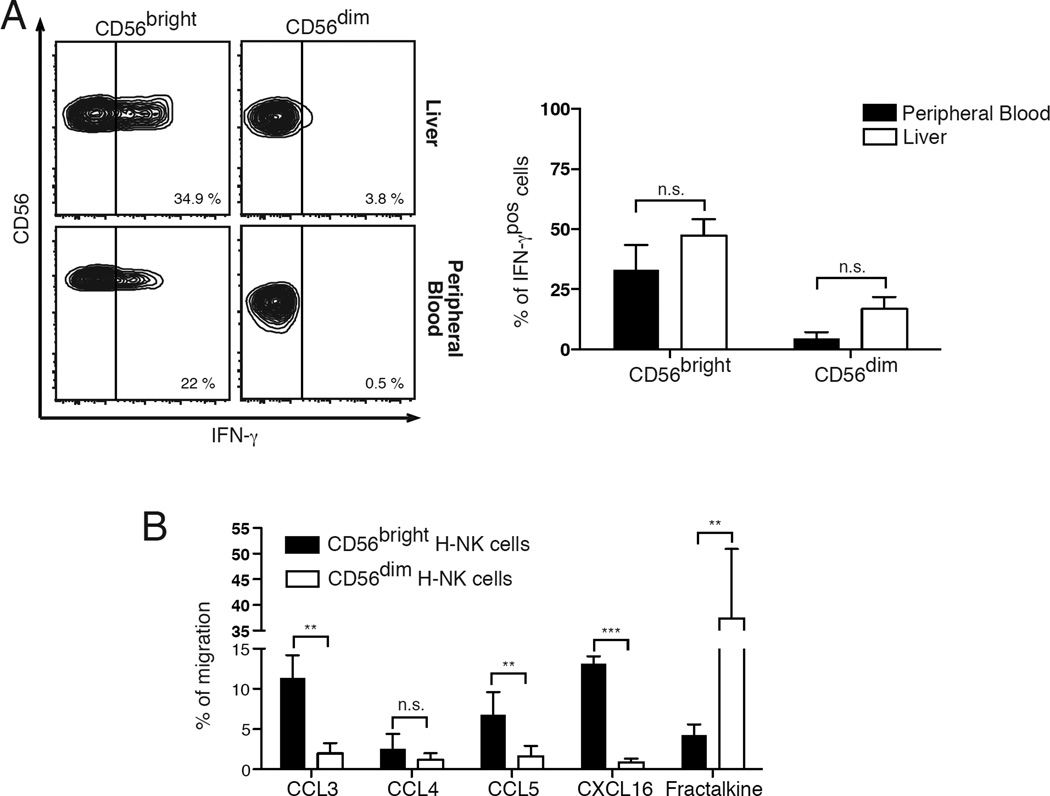
Similar to CD56bright PB-NK cells [3], we found that CD56bright hepatic NK cells are able to produce large quantities of IFN-γ following stimulation with IL-2 and IL-12. This is in contrast with what we observed for CD56dim hepatic NK cells, as their degree of IFN-γ production was significantly lower compared to that of CD56bright hepatic NK and equivalent to that of CD56dim PB-NK (Figure 4A). These results indicate that CD56bright and CD56dim hepatic NK cell subsets mirror the same dichotomy in immune-regulatory functions compared to their circulating counterparts.
Figure 4. Functional relevance and migration patterns of hepatic NK cell subsets.
(A) Flow cytometric contour plots graphs from the same representative donor showing the percentages of IFN-γpos/CD56bright (left column) and IFN-γpos/CD56dim (right column) NK cells purified from liver (upper line) and peripheral blood (lower line)
(B) Summary graphs of statistical histogram bars with p values and SD from matching donors showing the percentages of IFN-γpos/CD56bright and IFN-γpos/CD56dim NK cells purified from healthy livers (white bars) and peripheral blood (black bars). Data are presented as percentages and are representative of 6 human donors.
(C) Summary graphs of statistical histogram bars with p values and SD showing the percentages of CD56bright (black bars) and CD56dim (white bars) H-NK cells from matching donors migrating in response to CCL3, CCL4, CCL5, CXCL16 and fractalkine. Data are representative of 5 human donors.
* = p < 0,05 – ** = p < 0,01 – *** = p < 0,001
We then proceeded to determine if the constitutive expression of CXCR6 and CCR5 on CD56bright hepatic NK cells is associated with the ability of this subset to migrate to their natural ligands (Figure 4B). By using an in vitro chemotaxis assay, we found that CCL3 and CCL5, but not CCL4, induced a significantly higher migration of CCR5pos/CD56bright hepatic NK cells compared to that of CCR5neg/CD56dim hepatic NK cells that, as expected, did not respond to CCR5 ligands. Similarly, CXCL16 was able to induce the migration of CXCR6pos/CD56bright but not CXCR6neg/CD56dim hepatic NK cells. We then measured the chemotactic function of CX3CR1, the chemokine receptor constitutively expressed on CD56dim but not on CD56bright hepatic NK cells. As expected, its ligand fractalkine induced the chemotaxis of CX3CR1pos/CD56dim hepatic NK cells but not of CX3CR1neg/CD56bright hepatic NK cells. Hence, the different repertoires of chemokine receptors on CD56bright and CD56dim hepatic NK cell subsets are functionally relevant and also associated with distinct migratory patterns of these two populations.
CCL3, CCL5 and CXCL16 are highly expressed in the sinusoids of the healthy liver
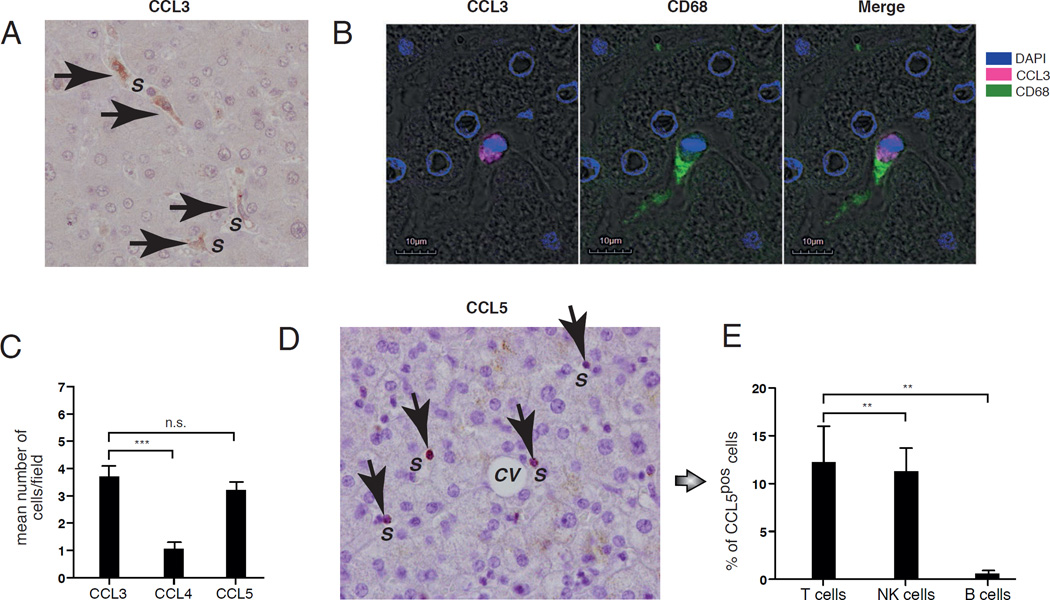
We then assessed by immunohistochemistry the expression and distribution of CCR5 and CXCR6 ligands in healthy human liver. We found that CCL3 is expressed within hepatic sinusoids on the surface of cells characterized by an amoeboid shape and adherent to neighboring cells. These irregular morphologic features are known to characterize Kupffer cells, which are the liver macrophages residing within the hepatic sinusoids [32]. To confirm that these CCL3pos cells are indeed Kupffer cells, we labeled by confocal microscopy CD68, a marker constitutively expressed on liver-resident macrophages [33, 34]. Our data showed that CCL3 co-localizes with CD68 on irregular shaped cells within hepatic sinusoids, thereby confirming that they are indeed Kupffer cells (Figure 5A and 5B).
Figure 5. Distribution of CCR5 ligands in healthy human liver.
(A) Representative immunohistochemistry image (magnification of 20X) showing-CCL3pos (brown indicated with arrows) cells in healthy liver. The hepatic sinusoids are indicated with S. (B) Representative fluorescence microscopic image showing the expression of CCL3 (purple on the left) and CD68 (green in the middle) alone or co-localized on the same cells (right). DAPI in blue labels cell nuclei. The hepatic sinusoids are indicated with S (C) Summary graphs of statistical histogram bars with p values and SD showing the mean number for field of CCL3pos, CCL4pos and CCL5pos cell in the in parenchyma of healthy liver determined by immunohistochemistry experiments. Cells were counted at 20X magnification and a least 10 fields per specimen were analyzed. Data are representative of 5 donors. (D) Representative immunohistochemistry image showing the CCL5pos (brown indicated with arrows) cells in parenchyma of healthy liver. The hepatic sinusoids are indicated with S. (e) Summary graphs of statistical histogram bars with p values and SD showing the percentage of CCL5pos/CD3pos T cells, CCL5pos/CD3neg/CD56pos NK cells and CCL5pos/CD19pos B cells freshly purified from healthy liver and analyzed by flow cytometry. Data are representative of 5 donors.
** = p < 0,01 – *** = p < 0,001
Regarding the other two CCR5 ligands, we found that the frequency of CCL4pos cells in the liver is very low and significantly smaller compared to that of CCL3pos cells. In contrast, CCL5pos appeared to also be localized on cells within hepatic sinusoids at a frequency similar to that of CCL3pos Kupffer cells. However, CCL5pos cells were small in size, circular in shape and displayed a large nucleus surrounded by little cytoplasm. These features resemble the morphology of a lymphocyte rather than a macrophage. To confirm this and to identify the lymphocyte subset expressing CCL5, we performed flow cytometry on freshly purified LMNCs. Our results showed that CCL5 is indeed expressed on CD3pos hepatic T cells and CD3neg/CD56pos hepatic NK cells, but not on CD19pos hepatic B cells (Figures 5C, 5D and 5E). We also found a similar constitutive expression and distribution of CCL5 in liver perfusates (data not shown), thus confirming that this chemokine is located within hepatic sinusoids.
As for CXCL16, it has been previously reported that liver sinusoidal endothelial cells constitutively express this chemokine under homeostatic conditions [35].
CD56bright hepatic NK cells are localized within the hepatic sinusoids
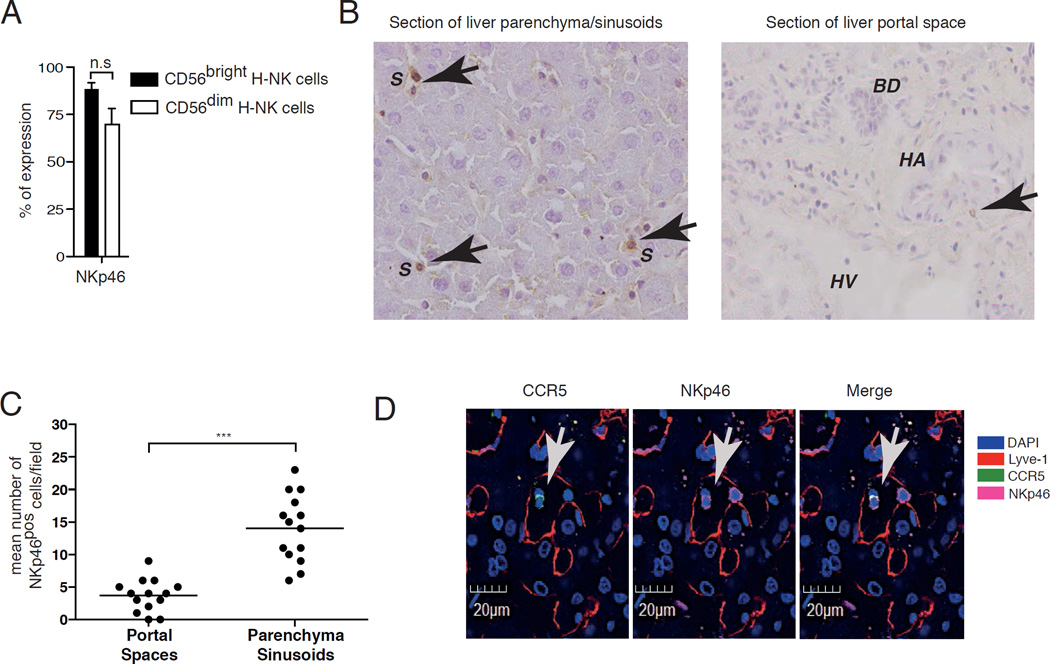
We then analyzed whether the high frequency of CCL3, CCL5 and CXCL16 in hepatic sinusoids is associated with the retention of CD56bright hepatic NK cells expressing both CCR5 and CXCR6 in liver sinusoidal spaces. Given that NKp46 is constitutively present on both CD56bright and CD56dim hepatic NK cell subsets and is also widely used to identify NK cells in tissues [36], we determined by immunohistochemistry the expression of this natural cytotocixity receptor (NCR) in sections of healthy liver. Our results showed that NKp46pos NK cells are preferentially localized within liver sinusoids, while their frequency is very low in portal spaces (Figures 6A, 6B and 6C).
Figure 6. CD56bright NK cells are preferentially located in hepatic sinusoids.
(A) Summary graphs of statistical histogram bars with p values and SD showing the percentage of expression of NKp46 on CD56bright and CD56dim H-NK cells from healthy liver and analyzed by flow cytometry. Data are representative of 5 donors. (B) Representative immunohistochemistry image showing NKp46pos H-NK cells (brown indicated with arrows) cells in sections of parenchyma (left) and portal spaces (right) of healthy liver. The hepatic sinusoids are indicated with S, bile ducts with BD, hepatic arteries with HA and hepatic veins with HV. (C) Summary graphs of statistical dot plots with means and P value showing the mean number for field of NKp46pos H-NK cells in the in parenchyma of healthy liver determined in immunohistochemistry experiments. Cells were counted at 40X magnification and a least 10 fields per specimen were analyzed. Data are representative of 3 donors. (D) Representative fluorescence microscopic image showing the expression of CCR5 (green on the left) and NKp46 (green in the middle) alone or co-localized on the same cells (right) within hepatic sinusoids labeled with Lyve-1 (red). DAPI in blue labels cell nuclei.
To assess if NKp46pos hepatic NK cells found in hepatic sinusoids under homeostatic conditions belong to the CD56bright hepatic NK cell subset, we performed confocal microscopy on frozen sections of healthy liver using an anti-CCR5 monoclonal antibody (mAb) in combination with another mAb specific for Lyve-1, a marker specific for liver sinusoidal endothelial cells delineating sinusoidal spaces [37]. We found that the co-localization of NKp46 with CCR5 on hepatic NK cells is confined within the areas defined by Lyve-1, thus demonstrating that the subset of CCR5pos/CD56bright hepatic NK cells is contained within sinusoidal spaces (Figure 6D).
Finally, we analyzed by flow cytometry the expression of chemokine receptors on perf-NK cells. Our results showed that, similar to what we observed in liver specimens, liver perfusates contain both CCR5pos/CD56bright and CX3CR1pos/CD56dim NK cells (Supplemental Figure 3), thus further confirming that that CD56bright NK cells expressing markers associated with tissue residency are present in the healthy liver and are located in the hepatic sinusoids.
Discussion
The present study characterizes a novel subset of CD56bright lr-NK cells, whose transcriptional and phenotypic profiles differ from those of its circulating counterparts. We show that CD56bright lr-NK cells constitutively express markers of tissue-residency including CD69, CCR5 and CXCR6. The unique repertoire of chemokine receptors on CD56bright lr-NK cells is functionally relevant as this subset selectively migrates in response to the chemotactic stimuli given by CCR5 (i.e. CCL3 and CCL5) and CXCR6 (i.e. CXCL16) ligands. Under homeostatic conditions, CD56bright lr-NK cells are preferentially located within hepatic sinusoids containing CCL3pos Kupffer cells as well as CCL5pos T and NK lymphocytes. Moreover, it has been reported that hepatic sinusoidal endothelial cells also express high constitutive levels of CXCL16 [35]. Hence, the abundance of these chemokines in liver sinusoidal spaces creates a unique tissue niche hosting CD56bright lr-NK cells constitutively expressing CCR5 and CXCR6.
The key roles of these two chemokine receptors in regulating the homeostasis of CD56bright lr-NK cells are further supported by previous studies performed in murine models. Indeed, CCR5-deficient mice have a drastic reduction of NK cell frequency [38], while CXCR6 regulates the survival, the trafficking and the effector-function of murine hepatic NK cells [11]. Our experimental evidence demonstrates, for the first time, that the engagement of these two chemokine receptors is required to ensure the constitutive presence of a large subset of lr-NK cells in human hepatic sinusoids.
CD56bright lr-NK cells coexist with another subset of CD56dim c-NK cells that, instead, are transcriptionally and phenotypically similar to their circulating counterparts. In particular, CD56dim c-NK cells are characterized by a CD69neg/CCR5neg/CXCR6neg phenotype and express adhesion molecules as well as functionally relevant chemokine receptors such as CX3CR1 associated with the homing of circulating immune cells to peripheral tissues. These results indicate that CD56dim c-NK cells are capable of circulating throughout the liver without being retained in hepatic sinusoids. Notably, it has been reported that, similar to human CD56dim c-NK cells, CX3CR1 is also expressed on murine NK cells able to circulate in the periphery but not on liver-resident NK cells [10]. In line with previous studies [39, 40], we also observed that fractalkine is not present within hepatic sinusoids but is preferentially expressed by bile ducts under homeostatic conditions (data not shown). Nevertheless, we found very few NK cells in the portal areas surrounding the bile ducts of healthy liver. In this regard, it has been reported that the frequency of NK cells increases during the course of autoimmune hepatic disorders such as primary biliary sclerosis. This phenomenon is associated with the establishment of a pathologic inflammatory milieu at injured liver sites inducing a higher expression of fractalkine and several adhesion molecules such as ICAM-1 and VCAM-1 on biliary epithelial cells [40, 41]. We also show that CD56dim c-NK cells constitutively express CCR3, CXCR2, and CXCR4, but the potential binding of these chemokine receptors with their ligands in a redundant chemokine network does not appear be relevant in the homing of this subset in liver sinusoids under homeostatic conditions. Indeed, many studies have shown the lack of expression of the ligands to CXCR4 (CXCL12), CXCR2 (CXCL1 and CXCL8) and CCR3 (CCL7, CCL15, CCL28) in hepatic sinusoids of healthy human liver [15, 22, 42–45].
A unique subset of lr-NK cells expressing markers of tissue-residency and located within hepatic sinusoids has recently been reported in mice [10, 11, 46]. These NK cells are characterized by a CD49apos/DX5neg phenotype and are endowed with memory-like features in contrast to CD49aneg/DX5pos c-NK cells found in liver, blood, bone marrow and spleen. The direct translation of this knowledge gained in mice into the human setting is precluded by the fact that DX5 is not conserved in humans and CD56 does not define murine NK cells. Nonetheless, a population of human CD56pos/CD49apos hepatic NK cells displaying some similarities with the murine CD49apos/DX5neg lr-NK cells has been recently described [16]. In this study, human CD56pos/CD49apos hepatic NK cells were detected at low frequencies (mean of 2.3 % of the entire hepatic NK cell population) in the healthy liver of only a fraction (41 %) of all donors tested. Our experimental evidence indicates that this subset of CD49apos/DX5neg hepatic NK cells represents only a small fraction of the large human subset of lr-NK cells. In fact, we show here that human CD69pos/CD56bright lr-NK cells expressing constitutively high levels of CXCR6 and CCR5 (means 48,5 % and 56,5%, respectively) account for half of the entire hepatic NK cell population (mean of 47.8 %) and are consistently found in all donors tested. Furthermore, we show here that the frequencies of both CD56bright and CD56dim NK cells in liver perfusates differ from those normally detected in peripheral blood PBMCs [3], but are similar to those observed in liver [7, 47]. Indeed, draining immune cells from liver before transplantation flushes out from hepatic sinusoids high frequencies of CD69pos/CD56bright lr-NK normally retained within this anatomic compartment and not circulating in the blood. On the other side, CD69neg/CD56bright PB-NK cells do not express CCR5 and CXCR6 but were positive for CCR7 and L-selectin, a phenotype classically associated with the homing of circulating NK cells to secondary lymphoid tissues [26]. These data demonstrate that CD56bright NK cells in blood and liver represent two distinct populations present in different anatomic compartments. The fact that the liver harbors a large population of tissue-resident CD56bright NK cell is not unique to this organ, as high frequencies of unique CD56bright NK cell subsets have been found in lymph nodes, decidua and intestinal mucosa [3, 48, 49]. These NK cells generally display a phenotype and function specific to the organ of occupancy, while they are either absent or present at very low frequencies in peripheral blood [50–52]. Similar to their counterparts in other tissues, the existence of lr-NK cells certainly plays a key role in the immune-surveillance against tumor transformation and pathogens. Moreover, it is conceivable to hypothesize an important contribution exerted by CD56bright lr-NK in setting a threshold for immunologic tolerance in the liver given the large numbers of foreign antigens drained daily from the gut. Indeed, NK cell dysfunctions have been reported to be greatly involved in the pathogenesis of autoimmune liver disease [7].
Our data showing the enrichment of CD56bright lr-NK cells in the hepatic sinusoids alongside the abundant Kupffer cell population reveals that the sinusoidal spaces represent an important tissue niche where macrophages and NK cells interact. Indeed, many lines of evidence clearly demonstrated over the past decade that NK cells are able to engage bi-directional interplays with other members of innate immunity, such as dendritic cells (DCs), macrophages and neutrophils. The final outcome of these synergic interactions mediated by both cell-to-cell contacts and soluble mediators is the coordination and optimization of both innate and adaptive immune responses [2, 53–63]. In particular, the NK cell-macrophage cross-talk is highly relevant in the context of host-pathogen interactions and tumor biology as both these innate immune cells are deeply involved in the pathogenesis of microbial infection and cancer. Disclosing the mechanisms regulating the homing and the synergic interactions between hepatic NK and Kupffer cells under homeostatic conditions is key to better understand the physiopathology of liver diseases, including viral infections, autoimmune diseases and tumor.
The characterization of lr-NK cells has also prompted immunologists to hypothesize that certain tissues may replace the bone marrow as a site for NK cell development and/or differentiation in order to generate organ-specific NK cells [10, 64]. This idea is supported by data showing that CD56bright NK cells can develop not only from CD34pos hematopoietic stem cell (HSC) precursors present in the bone marrow, but also from HSCs found in the SLT, thymus, fetal liver, uterus and intestine [65–71]. Moreover, all five NK cell developmental intermediates (NKDIs) have been identified in the adult liver and it has been also reported that differentiation stages 1–3 can give rise to CD56bright NK cells in vitro, similar to what was shown for SLT [72]. Preliminary data developed in our laboratory have confirmed that healthy human liver contains all NKDIs (data not shown). Additional studies are required to determine if these NKDIs can give rise in vitro to the unique subset of human CD56bright lr-NK cells we have identified and characterized in the present study. Finally, we are delighted that this manuscript will be part of an issue, which recognizes the enormous contributions of Diego and Giorgina Vergani in autoimmunity. It is part of the special series of the Journal of Autoimmunity that devotes topics and issues to important figures and critical events that we believe will greatly advance improved care of patients [73–75].
Supplementary Material
Highlights.
Characterization of the mechanisms regulating the homing of NK cells to liver and the repertoire of receptors that distinguish liver-resident NK (lr-NK) cells from circulating counterparts (c.NK cells).
Nearly 50% of the entire liver NK cell population is composed of functionally relevant CD56bright lr-NK cells that localize within hepatic sinusoids.
CD56bright lr-NK cells express CD69, CCR5 and CXCR6 and this unique repertoire of chemokine receptors is functionally for their migration to hepatic sinusoids expressing their putative ligands.
The selective presence of these chemokines in sinusoidal spaces creates a tissue niche for lr-CD56bright NK cells that constitutively express CCR5 and CXCR6.
This study releases the phenotypic and functional differences that distinguish lr-Nk cells from c-NK cells.
Acknowledgments
We are deeply grateful to all human donors and patients participating to this study for their generosity and support. This work was supported by the Italian Ministry of Health (Bando Giovani Ricercatori, GR-2008-1135082 to D.M.), Italian Association for Cancer Research (AIRC IG 14687 to D.M.), the European Union (Marie Curie International Reintegration Grant 322093 to E.L.), the National Institutes of Health (DK39588 to M.E.G.) and by the intramural research program of Humanitas Clinical and Research Center to D.M. K.H. is the recipient of a fellowship from the “Fondazione Umberto Veronesi”. E.P. is a recipient of “Luigi Tocco and Liliana Mirizio” fellowship from the Italian Foundation for Cancer Research (FIRC).
List of abbreviations
- lr-NK cells
liver resident Natural Killer cells
- c-NK cells
conventional Natural Killer cells
- perf-NK cells
perfusate Natural Killer cells
- iNKRs
inhibitory NK cell receptors
- aNKRs
activating NK cell receptors
- KIRs
Killer cell immunoglobulin-like receptors
- MHC-I
MHC-class-I molecules
- MAIT
mucosal-associated invariant T cells
- PBMCs
peripheral blood mononuclear cells
- LMNCs
liver mononuclear cells
- PMNCs
perfusate mononuclear cells
- MFI
mean fluorescence intensity
- SLT
secondary lymphoid tissues
- TRAIL
Tumor necrosis factor (TNF)-related apoptosis-inducing ligand
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have declared that no conflict of interest exists.
Author Contributions
Conception and design: K.H. and D.M; Contributed unpublished, essential data, or reagents (recruited patients for liver specimens): M.D., M.C. and G.T.; Contributed unpublished, essential data, or reagents (recruited patients for liver-perfusate specimens): M.H. and A.B; Acquisition of data, analysis and interpretation of data: K.H., E.P., P.T., M.P., S.B., P.I., E.L., M.E.G., D.M. K.H. and D.M.; Drafting the article: K.H. and D.M.
References
- 1.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lugli E, Marcenaro E, Mavilio D. NK Cell Subset Redistribution during the Course of Viral Infections. Frontiers in immunology. 2014;5:390. doi: 10.3389/fimmu.2014.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends in immunology. 2001;22:633–640. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 4.Lanier LL. NK cell recognition. Annual review of immunology. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 5.Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annual review of immunology. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 6.Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annual review of immunology. 2013;31:227–258. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hudspeth K, Pontarini E, Tentorio P, Cimino M, Donadon M, Torzilli G, et al. The role of natural killer cells in autoimmune liver disease: a comprehensive review. Journal of autoimmunity. 2013;46:55–65. doi: 10.1016/j.jaut.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Oliviero B, Varchetta S, Paudice E, Michelone G, Zaramella M, Mavilio D, et al. Natural killer cell functional dichotomy in chronic hepatitis B and chronic hepatitis C virus infections. Gastroenterology. 2009;137:1151–1160. doi: 10.1053/j.gastro.2009.05.047. 60 e1-7. [DOI] [PubMed] [Google Scholar]
- 9.Rehermann B. Pathogenesis of chronic viral hepatitis: differential roles of T cells and NK cells. Nature medicine. 2013;19:859–868. doi: 10.1038/nm.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng H, Jiang X, Chen Y, Sojka DK, Wei H, Gao X, et al. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. The Journal of clinical investigation. 2013;123:1444–1456. doi: 10.1172/JCI66381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paust S, Gill HS, Wang BZ, Flynn MP, Moseman EA, Senman B, et al. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nature immunology. 2010;11:1127–1135. doi: 10.1038/ni.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Zhang S, Zou Z, Shi J, Zhao J, Fan R, et al. Hypercytolytic activity of hepatic natural killer cells correlates with liver injury in chronic hepatitis B patients. Hepatology. 2011;53:73–85. doi: 10.1002/hep.23977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishiyama K, Ohdan H, Ohira M, Mitsuta H, Arihiro K, Asahara T. Difference in cytotoxicity against hepatocellular carcinoma between liver and periphery natural killer cells in humans. Hepatology. 2006;43:362–372. doi: 10.1002/hep.21035. [DOI] [PubMed] [Google Scholar]
- 14.Boisvert J, Kunkel EJ, Campbell JJ, Keeffe EB, Butcher EC, Greenberg HB. Liver-infiltrating lymphocytes in end-stage hepatitis C virus: subsets, activation status, and chemokine receptor phenotypes. Journal of hepatology. 2003;38:67–75. doi: 10.1016/s0168-8278(02)00328-8. [DOI] [PubMed] [Google Scholar]
- 15.Wald O, Pappo O, Safadi R, Dagan-Berger M, Beider K, Wald H, et al. Involvement of the CXCL12/CXCR4 pathway in the advanced liver disease that is associated with hepatitis C virus or hepatitis B virus. European journal of immunology. 2004;34:1164–1174. doi: 10.1002/eji.200324441. [DOI] [PubMed] [Google Scholar]
- 16.Marquardt N, Beziat V, Nystrom S, Hengst J, Ivarsson MA, Kekalainen E, et al. Cutting Edge: Identification and Characterization of Human Intrahepatic CD49a+ NK Cells. Journal of immunology. 2015 doi: 10.4049/jimmunol.1402756. [DOI] [PubMed] [Google Scholar]
- 17.Hudspeth K, Fogli M, Correia DV, Mikulak J, Roberto A, Della Bella S, et al. Engagement of NKp30 on Vdelta1 T cells induces the production of CCL3, CCL4, and CCL5 and suppresses HIV-1 replication. Blood. 2012;119:4013–4016. doi: 10.1182/blood-2011-11-390153. [DOI] [PubMed] [Google Scholar]
- 18.Gupta N, Arthos J, Khazanie P, Steenbeke TD, Censoplano NM, Chung EA, et al. Targeted lysis of HIV-infected cells by natural killer cells armed and triggered by a recombinant immunoglobulin fusion protein: implications for immunotherapy. Virology. 2005;332:491–497. doi: 10.1016/j.virol.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 19.Tang XZ, Jo J, Tan AT, Sandalova E, Chia A, Tan KC, et al. IL-7 licenses activation of human liver intrasinusoidal mucosal-associated invariant T cells. Journal of immunology. 2013;190:3142–3152. doi: 10.4049/jimmunol.1203218. [DOI] [PubMed] [Google Scholar]
- 20.Campbell JJ, Haraldsen G, Pan J, Rottman J, Qin S, Ponath P, et al. The chemokine receptor CCR4 in vascular recognition by cutaneous but not intestinal memory T cells. Nature. 1999;400:776–780. doi: 10.1038/23495. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 22.Tachibana Y, Nakamoto Y, Mukaida N, Kaneko S. Intrahepatic interleukin-8 production during disease progression of chronic hepatitis C. Cancer letters. 2007;251:36–42. doi: 10.1016/j.canlet.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 23.Curry MP, Norris S, Golden-Mason L, Doherty DG, Deignan T, Collins C, et al. Isolation of lymphocytes from normal adult human liver suitable for phenotypic and functional characterization. Journal of immunological methods. 2000;242:21–31. doi: 10.1016/s0022-1759(00)00204-0. [DOI] [PubMed] [Google Scholar]
- 24.Tu Z, Bozorgzadeh A, Crispe IN, Orloff MS. The activation state of human intrahepatic lymphocytes. Clinical and experimental immunology. 2007;149:186–193. doi: 10.1111/j.1365-2249.2007.03415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jonsson JR, Hogan PG, Balderson GA, Ooi LL, Lynch SV, Strong RW, et al. Human liver transplant perfusate: an abundant source of donor liver-associated leukocytes. Hepatology. 1997;26:1111–1114. doi: 10.1002/hep.510260504. [DOI] [PubMed] [Google Scholar]
- 26.Campbell JJ, Qin S, Unutmaz D, Soler D, Murphy KE, Hodge MR, et al. Unique subpopulations of CD56+ NK and NK-T peripheral blood lymphocytes identified by chemokine receptor expression repertoire. Journal of immunology. 2001;166:6477–6482. doi: 10.4049/jimmunol.166.11.6477. [DOI] [PubMed] [Google Scholar]
- 27.Takeda K, Cretney E, Hayakawa Y, Ota T, Akiba H, Ogasawara K, et al. TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood. 2005;105:2082–2089. doi: 10.1182/blood-2004-08-3262. [DOI] [PubMed] [Google Scholar]
- 28.Berahovich RD, Lai NL, Wei Z, Lanier LL, Schall TJ. Evidence for NK cell subsets based on chemokine receptor expression. Journal of immunology. 2006;177:7833–7840. doi: 10.4049/jimmunol.177.11.7833. [DOI] [PubMed] [Google Scholar]
- 29.Kunkel EJ, Boisvert J, Murphy K, Vierra MA, Genovese MC, Wardlaw AJ, et al. Expression of the chemokine receptors CCR4, CCR5, and CXCR3 by human tissue-infiltrating lymphocytes. The American journal of pathology. 2002;160:347–355. doi: 10.1016/S0002-9440(10)64378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heydtmann M, Hardie D, Shields PL, Faint J, Buckley CD, Campbell JJ, et al. Detailed analysis of intrahepatic CD8 T cells in the normal and hepatitis C-infected liver reveals differences in specific populations of memory cells with distinct homing phenotypes. Journal of immunology. 2006;177:729–738. doi: 10.4049/jimmunol.177.1.729. [DOI] [PubMed] [Google Scholar]
- 31.Sathaliyawala T, Kubota M, Yudanin N, Turner D, Camp P, Thome JJ, et al. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity. 2013;38:187–197. doi: 10.1016/j.immuni.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naito M, Hasegawa G, Ebe Y, Yamamoto T. Differentiation and function of Kupffer cells. Medical electron microscopy : official journal of the Clinical Electron Microscopy Society of Japan. 2004;37:16–28. doi: 10.1007/s00795-003-0228-x. [DOI] [PubMed] [Google Scholar]
- 33.Negash AA, Ramos HJ, Crochet N, Lau DT, Doehle B, Papic N, et al. IL-1beta production through the NLRP3 inflammasome by hepatic macrophages links hepatitis C virus infection with liver inflammation and disease. PLoS pathogens. 2013;9:e1003330. doi: 10.1371/journal.ppat.1003330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alabraba EB, Curbishley SM, Lai WK, Wigmore SJ, Adams DH, Afford SC. A new approach to isolation and culture of human Kupffer cells. Journal of immunological methods. 2007;326:139–144. doi: 10.1016/j.jim.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 35.Heydtmann M, Lalor PF, Eksteen JA, Hubscher SG, Briskin M, Adams DH. CXC chemokine ligand 16 promotes integrin-mediated adhesion of liver-infiltrating lymphocytes to cholangiocytes and hepatocytes within the inflamed human liver. Journal of immunology. 2005;174:1055–1062. doi: 10.4049/jimmunol.174.2.1055. [DOI] [PubMed] [Google Scholar]
- 36.Walzer T, Blery M, Chaix J, Fuseri N, Chasson L, Robbins SH, et al. Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3384–3389. doi: 10.1073/pnas.0609692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mouta Carreira C, Nasser SM, di Tomaso E, Padera TP, Boucher Y, Tomarev SI, et al. LYVE-1 is not restricted to the lymph vessels: expression in normal liver blood sinusoids and down-regulation in human liver cancer and cirrhosis. Cancer research. 2001;61:8079–8084. [PubMed] [Google Scholar]
- 38.Weiss ID, Shoham H, Wald O, Wald H, Beider K, Abraham M, et al. Ccr5 deficiency regulates the proliferation and trafficking of natural killer cells under physiological conditions. Cytokine. 2011;54:249–257. doi: 10.1016/j.cyto.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 39.Aspinall AI, Curbishley SM, Lalor PF, Weston CJ, Blahova M, Liaskou E, et al. CX(3)CR1 and vascular adhesion protein-1-dependent recruitment of CD16(+) monocytes across human liver sinusoidal endothelium. Hepatology. 2010;51:2030–2039. doi: 10.1002/hep.23591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isse K, Harada K, Zen Y, Kamihira T, Shimoda S, Harada M, et al. Fractalkine and CX3CR1 are involved in the recruitment of intraepithelial lymphocytes of intrahepatic bile ducts. Hepatology. 2005;41:506–516. doi: 10.1002/hep.20582. [DOI] [PubMed] [Google Scholar]
- 41.Gershwin ME, Ansari AA, Mackay IR, Nakanuma Y, Nishio A, Rowley MJ, et al. Primary biliary cirrhosis: an orchestrated immune response against epithelial cells. Immunological reviews. 2000;174:210–225. doi: 10.1034/j.1600-0528.2002.017402.x. [DOI] [PubMed] [Google Scholar]
- 42.Isse K, Harada K, Nakanuma Y. IL-8 expression by biliary epithelial cells is associated with neutrophilic infiltration and reactive bile ductules. Liver international : official journal of the International Association for the Study of the Liver. 2007;27:672–680. doi: 10.1111/j.1478-3231.2007.01465.x. [DOI] [PubMed] [Google Scholar]
- 43.Eksteen B, Miles A, Curbishley SM, Tselepis C, Grant AJ, Walker LS, et al. Epithelial inflammation is associated with CCL28 production and the recruitment of regulatory T cells expressing CCR10. Journal of immunology. 2006;177:593–603. doi: 10.4049/jimmunol.177.1.593. [DOI] [PubMed] [Google Scholar]
- 44.Tsuneyama K, Harada K, Yasoshima M, Hiramatsu K, Mackay CR, Mackay IR, et al. Monocyte chemotactic protein-1, -2, and -3 are distinctively expressed in portal tracts and granulomata in primary biliary cirrhosis: implications for pathogenesis. The Journal of pathology. 2001;193:102–109. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH725>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Wu J, Zhang W, Zhang N, Guo H. Identification of serum CCL15 in hepatocellular carcinoma. British journal of cancer. 2013;108:99–106. doi: 10.1038/bjc.2012.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Y, Kuang DM, Pan WD, Wan YL, Lao XM, Wang D, et al. Monocyte/macrophage-elicited natural killer cell dysfunction in hepatocellular carcinoma is mediated by CD48/2B4 interactions. Hepatology. 2013;57:1107–1116. doi: 10.1002/hep.26192. [DOI] [PubMed] [Google Scholar]
- 48.Freud AG, Becknell B, Roychowdhury S, Mao HC, Ferketich AK, Nuovo GJ, et al. A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity. 2005;22:295–304. doi: 10.1016/j.immuni.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 49.Carrega P, Bonaccorsi I, Di Carlo E, Morandi B, Paul P, Rizzello V, et al. CD56(bright)perforin(low) noncytotoxic human NK cells are abundant in both healthy and neoplastic solid tissues and recirculate to secondary lymphoid organs via afferent lymph. Journal of immunology. 2014;192:3805–3815. doi: 10.4049/jimmunol.1301889. [DOI] [PubMed] [Google Scholar]
- 50.Koopman LA, Kopcow HD, Rybalov B, Boyson JE, Orange JS, Schatz F, et al. Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. The Journal of experimental medicine. 2003;198:1201–1212. doi: 10.1084/jem.20030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanna J, Goldman-Wohl D, Hamani Y, Avraham I, Greenfield C, Natanson-Yaron S, et al. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nature medicine. 2006;12:1065–1074. doi: 10.1038/nm1452. [DOI] [PubMed] [Google Scholar]
- 52.Ferlazzo G, Thomas D, Lin SL, Goodman K, Morandi B, Muller WA, et al. The abundant NK cells in human secondary lymphoid tissues require activation to express killer cell Ig-like receptors and become cytolytic. Journal of immunology. 2004;172:1455–1462. doi: 10.4049/jimmunol.172.3.1455. [DOI] [PubMed] [Google Scholar]
- 53.Degli-Esposti MA, Smyth MJ. Close encounters of different kinds: dendritic cells and NK cells take centre stage. Nat Rev Immunol. 2005;5:112–124. doi: 10.1038/nri1549. [DOI] [PubMed] [Google Scholar]
- 54.Zitvogel L. Dendritic and natural killer cells cooperate in the control/switch of innate immunity. J Exp Med. 2002;195:F9–F14. doi: 10.1084/jem.20012040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moretta A. Natural killer cells and dendritic cells: rendezvous in abused tissues. Nature reviews Immunology. 2002;2:957–964. doi: 10.1038/nri956. [DOI] [PubMed] [Google Scholar]
- 56.Cooper MA, Fehniger TA, Fuchs A, Colonna M, Caligiuri MA. NK cell and DC interactions. Trends Immunol. 2004;25:47–52. doi: 10.1016/j.it.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 57.Bellora F, Castriconi R, Dondero A, Reggiardo G, Moretta L, Mantovani A, et al. The interaction of human natural killer cells with either unpolarized or polarized macrophages results in different functional outcomes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21659–21664. doi: 10.1073/pnas.1007654108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nedvetzki S, Sowinski S, Eagle RA, Harris J, Vely F, Pende D, et al. Reciprocal regulation of human natural killer cells and macrophages associated with distinct immune synapses. Blood. 2007;109:3776–3785. doi: 10.1182/blood-2006-10-052977. [DOI] [PubMed] [Google Scholar]
- 59.Michel T, Hentges F, Zimmer J. Consequences of the crosstalk between monocytes/macrophages and natural killer cells. Frontiers in immunology. 2012;3:403. doi: 10.3389/fimmu.2012.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Costantini C, Calzetti F, Perbellini O, Micheletti A, Scarponi C, Lonardi S, et al. Human neutrophils interact with both 6-sulfo LacNAc+ DC and NK cells to amplify NK-derived IFN{gamma}: role of CD18, ICAM-1, and ICAM-3. Blood. 2011;117:1677–1686. doi: 10.1182/blood-2010-06-287243. [DOI] [PubMed] [Google Scholar]
- 61.Lapaque N, Walzer T, Meresse S, Vivier E, Trowsdale J. Interactions between human NK cells and macrophages in response to Salmonella infection. Journal of immunology. 2009;182:4339–4348. doi: 10.4049/jimmunol.0803329. [DOI] [PubMed] [Google Scholar]
- 62.Romo N, Magri G, Muntasell A, Heredia G, Baia D, Angulo A, et al. Natural killer cell-mediated response to human cytomegalovirus-infected macrophages is modulated by their functional polarization. Journal of leukocyte biology. 2011;90:717–726. doi: 10.1189/jlb.0311171. [DOI] [PubMed] [Google Scholar]
- 63.Mattiola I, Pesant M, Tentorio P, Molgora M, Marcenaro E, Lugli E, Locati M, Mavilio D. Priming of human resting NK cells by autologous M1 macrophages via the engagement of IL-1β, IFN-β and IL-15 pathways. Journal of immunology. 2015 doi: 10.4049/jimmunol.1500325. In press. [DOI] [PubMed] [Google Scholar]
- 64.Yu J, Freud AG, Caligiuri MA. Location and cellular stages of natural killer cell development. Trends in immunology. 2013;34:573–582. doi: 10.1016/j.it.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fathman JW, Bhattacharya D, Inlay MA, Seita J, Karsunky H, Weissman IL. Identification of the earliest natural killer cell-committed progenitor in murine bone marrow. Blood. 2011;118:5439–5447. doi: 10.1182/blood-2011-04-348912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Freud AG, Yokohama A, Becknell B, Lee MT, Mao HC, Ferketich AK, et al. Evidence for discrete stages of human natural killer cell differentiation in vivo. The Journal of experimental medicine. 2006;203:1033–1043. doi: 10.1084/jem.20052507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mingari MC, Poggi A, Biassoni R, Bellomo R, Ciccone E, Pella N, et al. In vitro proliferation and cloning of CD3- CD16+ cells from human thymocyte precursors. The Journal of experimental medicine. 1991;174:21–26. doi: 10.1084/jem.174.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodewald HR, Moingeon P, Lucich JL, Dosiou C, Lopez P, Reinherz EL. A population of early fetal thymocytes expressing Fc gamma RII/III contains precursors of T lymphocytes and natural killer cells. Cell. 1992;69:139–150. doi: 10.1016/0092-8674(92)90125-v. [DOI] [PubMed] [Google Scholar]
- 69.Jaleco AC, Blom B, Res P, Weijer K, Lanier LL, Phillips JH, et al. Fetal liver contains committed NK progenitors, but is not a site for development of CD34+ cells into T cells. Journal of immunology. 1997;159:694–702. [PubMed] [Google Scholar]
- 70.Vacca P, Vitale C, Montaldo E, Conte R, Cantoni C, Fulcheri E, et al. CD34+ hematopoietic precursors are present in human decidua and differentiate into natural killer cells upon interaction with stromal cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2402–2407. doi: 10.1073/pnas.1016257108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chinen H, Matsuoka K, Sato T, Kamada N, Okamoto S, Hisamatsu T, et al. Lamina propria c-kit+ immune precursors reside in human adult intestine and differentiate into natural killer cells. Gastroenterology. 2007;133:559–573. doi: 10.1053/j.gastro.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 72.Moroso V, Famili F, Papazian N, Cupedo T, van der Laan LJ, Kazemier G, et al. NK cells can generate from precursors in the adult human liver. European journal of immunology. 2011;41:3340–3350. doi: 10.1002/eji.201141760. [DOI] [PubMed] [Google Scholar]
- 73.Ansari AA. Clinical features and pathobiology of Ebolavirus infection. J Autoimmun. 2014;55:1–9. doi: 10.1016/j.jaut.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 74.Berrih-Aknin S. Myasthenia Gravis: paradox versus paradigm in autoimmunity. J Autoimmun. 2014;52:1–28. doi: 10.1016/j.jaut.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 75.Steinman L, Shoenfeld Y. From defining antigens to new therapies in multiple sclerosis: honoring the contributions of Ruth Arnon and Michael Sela. J Autoimmun. 2014;54:1–7. doi: 10.1016/j.jaut.2014.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.