Abstract
Hedgehog (Hh) pathway activation in R26-SmoM2;CAGGS-CreER mice, which carry a tamoxifen-inducible activated Smoothened allele (SmoM2), results in numerous microscopic tumor foci in mouse skeletal muscle. These tumors exhibit a highly differentiated myogenic phenotype and resemble human fetal rhabdomyomas. This study sought to apply previously established strategies to isolate lineally distinct populations of normal mouse myofiber-associated cells in order to examine cellular heterogeneity in SmoM2 tumors. We demonstrate that established SmoM2 tumors are composed of cells expressing myogenic, adipocytic and hematopoietic lineage markers and differentiation capacity. SmoM2 tumors thus recapitulate the phenotypic and functional hetereogeneity observed in normal mouse skeletal muscle. SmoM2 tumors also contain an expanded population of PAX7+ and MyoD+ satellite-like cells with extremely low clonogenic activity. Selective activation of Hh signaling in freshly isolated muscle satellite cells enhanced terminal myogenic differentiation without stimulating proliferation. Our findings support the conclusion that SmoM2 tumors represent an aberrant skeletal muscle state and demonstrate that, similar to normal muscle, myogenic tumors contain functionally distinct cell subsets, including cells lacking myogenic differentiation potential.
Keywords: Skeletal muscle, differentiation, Hedgehog signaling, intratumoral cellular heterogeneity
Introduction
Adult striated muscle is composed of highly organized bundles of multinucleated myofibers and a variety of functionally heterogeneous mononuclear cells [1–3], including myogenic (muscle-forming) and non-myogenic elements such as fibroadipogenic precursors (FAPs) and immune/ inflammatory cells of hematopoietic lineage. Within the myogenic cell compartment, cytoplasmic filaments such as Desmin, Actin and Myosin mark terminal myogenic differentiation, whereas the transcription factor PAX7 identifies satellite cells within the heterogenous pool of myofiber-associated mononuclear cells [2]. Upon injury, satellite cells proliferate, differentiate and fuse to generate new myofibers in a process that is governed by sequential expression of a series of myogenic regulatory factors including MyoD and Myogenin [4, 5]. These myogenic regulatory factors (MRFs) are generally silent in mature, resting muscle.
Skeletal muscle differentiation features can be found in a number of neoplastic conditions, including rhabdomyosarcomas, a varied group of soft-tissue sarcomas, and rhabdomyomas, benign tumors of striated muscle. These conditions have previously been linked to activation of certain oncogenic pathways, including activating mutations in Hedgehog (Hh) pathway genes, detected in fusion-negative human rhabdomyosarcomas [6, 7] and fetal rhabdomyomas [8, 7]. These tumors exhibit both terminal muscle differentiation markers (e.g. Actin) and myogenic regulatory factors (e.g. MyoD), and they represent an abnormal state of muscle differentiation [8, 9]. This study sought to examine cellular heterogeneity in myogenic tumors. We demonstrate that tumors arising in mouse skeletal muscle following induction of hyperactive Hh signaling [8, 9] recapitulate normal skeletal muscle cellular heterogeneity and contain an expanded pool of PAX7+, MyoD+ satellite-like cells.
Material and methods
Mice
R26-SmoM2(+/−) and R26-SmoM2(+/+) (mixed genetic background including 129/Sv and Swiss Webster as main components) [9] and R26-SmoM2(+/−);CAGGS-CreER [9] were bred at the Joslin Diabetes Center Animal Facility. Throughout this manuscript, R26-SmoM2(+/−) or R26-SmoM2(+/+) skeletal muscle is referred to as “control” muscle, and R26-SmoM2(+/−);CAGGS-CreER skeletal muscle as “SmoM2” muscle. C57BL6 mice were purchased from the Jackson Laboratory. Tamoxifen (Sigma, St Louis, MO) at a dose of 1 mg/40 g body weight was administered to R26-SmoM2(+/−);CAGGS-CreER intraperitoneally on postnatal day 10 (P10) to activate CreER-mediated recombination at transgene-encoded loxP sites. High rates of recombination in skeletal muscle were previously documented [9]. R26-SmoM2;CAGGS-CreER mice were monitored once weekly for the onset of soft-tissue tumors or other health problems, and they were sacrificed once they were ill. All animal experiments were approved by the Joslin Diabetes Center Institutional Animal Care and Use Committee.
Histopathological evaluation of skeletal muscle and tumors
Skeletal muscle and tumor tissue was dissected, fixed in 4% paraformaldehyde for 2 hours, and embedded in paraffin. Standard H&E stained sections were prepared. Staining for Myogenin (Dako, M3559, 1:100), MyoD1 (Dako, M3512, 1:50), Desmin (Dako, M0760, 1:50), FABP4 (Cell Signaling, D25B3, 1 :200), CD45 (Abcam, ab10558, 1:4000) and PAX7 (DSHB, 1:5) was performed as previously described [2].
Muscle and tumor dissociation
Upper extremity, lower extremity and pectoralis muscles from 4–8 week-old C57BL6/J wild-type, 4–9 week-old R26-SmoM2 mice and 3–9 week-old, tamoxifen-induced R26-SmoM2;CAGGS-CreER mice were harvested. Isolation of myofiber-associated cells was performed by two-step enzymatic digestion and mechanical dissociation as previously described [1]. Isolation of SmoM2 tumor cells was performed by one-step enzymatic digestion and mechanical dissociation as follows: Tumors were harvested, digested in DMEM + 0.2% collagenase type II (Invitrogen) + 0.05% dispase (Invitrogen) for 90 minutes at 37°C in a shaking waterbath, triturated to disrupt the remaining tumor pieces and filtered through a 70µm cell strainer. Red blood cells were lysed from tumor cell preparations by 3 min incubation in 0.15M ammonium chloride, 0.01M potassium bicarbonate solution on ice.
Fluorescence activated cell sorting (FACS) of myofiber-associated and tumor cells
Phenotypically distinct muscle and tumor cell subsets were sorted according to protocols that were previously established to isolate functionally discrete subsets of myofiber-associated cells [10, 11, 1]. In brief, cells were suspended in HBSS supplemented with 2% FBS. Antibody staining was performed for 20 minutes on ice. The following primary and secondary antibodies were used: APC-CY7-conjugated anti-mouse CD11b (1 in 200, BD Pharmingen, 557657), APC-CY7-conjugated anti-mouse CD45 (1 in 200, eBioscience, 557659), APC-CY7-conjugated anti-mouse TER119 (1 in 200, BioLegend, 116223), APC-conjugated anti-mouse Sca1 (1 in 200, eBioscience, 17-5981-82), PE-conjugated anti-mouse/ rat CD29 (1 in 400, BioLegend, 102207), biotin rat anti-mouse CD184 (1 in 100, BD Pharmingen, 551968), PECY7-conjugated Streptavidin (1 in 200, eBioscience, 25-4317-82). Antibody staining was performed for 20 minutes on ice. Prior to FACS sorting, cells were suspended in 1µg/ml propidium iodide and 10µM calcein blue (Invitrogen) to identify viable cells (Pi−Ca+). Cells were sorted twice to maximize purity.
RT PCR
Tumor samples were FACS sorted to isolate GFP+ cells. mRNA was isolated by TRIzol extraction and reverse transcribed using Superscript III First-Strand Synthesis System for RT-PCR (Invitrogen). Quantitative RT-PCR was performed using Taqman PCR reagents and the following primers: Taqman Gene Expression Gli1 (Mm00494654_m1), Ptch1 (Mm00436026_m1), Pax7 (Mm01354484_m1), 18s rRNA (Mm03928990_g1). For qRT-PCR, relative expression of Gli1, Ptch1 and Pax7 per sample was determined by normalization against the quantity of 18s rRNA within each sample.
Clonal myogenesis assays
Cells were sorted at 1 cell per well into 96 well plates coated with 1µg/ml rat-tail collagen (Sigma) and 10µg/ml natural mouse laminin (Invitrogen). Cells were expanded for 5–7 days in growth medium (GM) composed of Ham’s F10 + 20% horse serum + 1% penicillin/streptomycin + 5ng/ml bFGF (Sigma). bFGF was replaced daily. Wells were screened for the presence of cell clones at 3, 5 and 8 days post sorting. The number of cells per individual clone was counted at the same time points.
Chemical modulation of Hh signaling
Chemical modulators of Hh signaling, including Cyclopamine (Toronto Research Chemicals) at a final concentration of 1000nM and Hh-conditioned medium (kindly supplied by Dr. J Mao, University of Massachusetts), were added to culture medium. Medium and chemicals were exchanged every 3 days in proliferation assays and every 2 days in differentiation assays.
Myogenic differentiation assays
Freshly sorted cells were expanded as described above. After 5–7 days, cells were passaged by aspiration of medium, re-plated onto 0.2% Matrigel (Fisher) coated chamber slides in growth medium for 2 days, and transitioned into differentiation medium (DM) consisting of Ham’s F10 + 2% horse serum + 1% penicillin/streptomycin. Alternatively, after 5–7 days of expansion, cells were transitioned into DM in 96 well plates. After 4 days in DM, medium was aspirated and cells were fixed with 4% paraformaldehyde and processed for immunofluorescence.
Adipogenic differentiation assay
Freshly sorted cells were expanded as described above. After 5–7 days, medium was changed to adipogenic induction medium consisting of DMEM + 10% FBS + 1% penicillin/streptomycin + 1mM Dexamethasone (Sigma) + 100nM Insulin (Sigma) + 1mM Roziglitazone (Cayman Chemical) + 0.5 mM 3-isobutyl-1-methylxanthine (Sigma) for 2 days, and then replaced with adipogenic differentiation medium consisting of DMEM + 10% FBS + 1% penicillin/streptomycin + 100nM Insulin. Cells were kept in differentiation medium for 6 days, and then fixed with 4% Paraformaldehyde and stained with Oil Red O (Sigma) for one hour at room temperature [11]. Oil Red O staining of lipid droplets within adipocytes was analyzed by standard microscopy using an Olympus IX51 inverted microscope.
PAX7 immunocytochemistry
Freshly sorted cells were fixed, blocked and stained for Pax7 (1:50, Developmental Studies Hybridoma Bank) as previously described [10].
Proliferation assays
Freshly sorted cells (1000–5000 cells/well) were plated in 96 well plates coated with 1 µg/ml rat-tail collagen (Sigma) and 10 µg/ml natural mouse laminin (Invitrogen). Cells were expanded in growth medium (GM) composed of Ham’s F10 + 20% horse serum + 1% penicillin/streptomycin + 5ng/ml bFGF (Sigma). bFGF was replaced daily. Cell growth was evaluated by MTT assay (Cayman Chemicals).
Statistics
Differences in cell composition, gene expression, clonal efficiency and differentiation indices were tested for statistical significance using Bonferroni’s multiple comparison testing.
Results
SmoM2 myogenic tumors contain a markedly heterogeneous cell pool
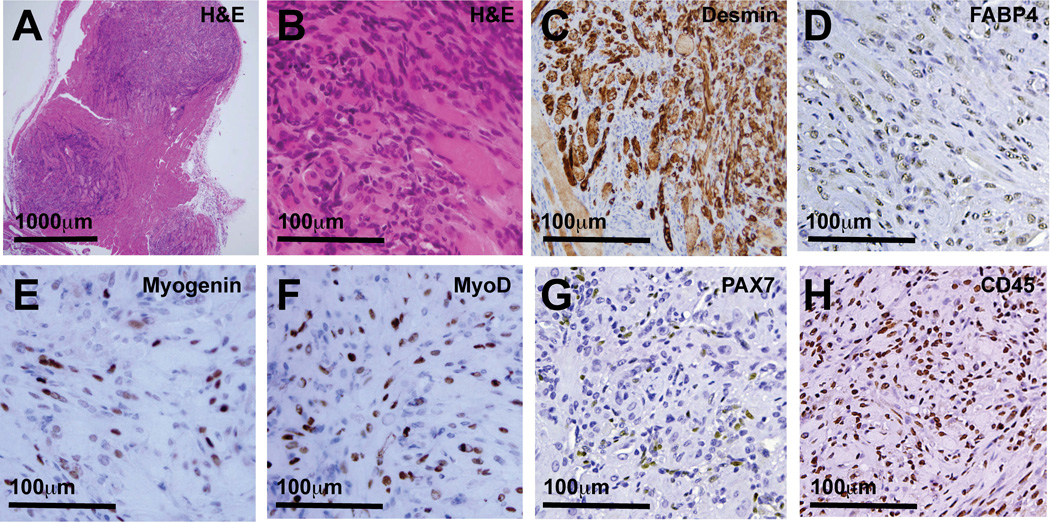
To activate Hh signaling in mouse skeletal muscle, expression of a mutant Smo allele (SmoM2) was activated via a Tamoxifen-inducible, ubiquitously expressed Cre recombinase (CAGGS-CreER) [9]. As previously reported, by 2 months of age, more than 90% of these SmoM2 mice developed large, multifocal tumors and numerous microscopic intramuscular tumor foci ([8] and Figure 1A). Tumors were composed of a mix of small, blue mononuclear cells and larger, often multinucleated cells with abundant eosinophilic cytoplasm (Figure 1B). The differentiation status of tumor cells was evaluated by immunohistochemical (IHC) staining for the myofilament Desmin, myogenic regulatory factors MyoD and Myogenin, the canonical satellite cell marker PAX7, the hematopoietic lineage marker CD45 and the adipocytic lineage marker FABP4. Desmin expression mostly localized to larger multinucleated cells with abundant cytoplasm and was notably absent in the mononuclear cells that infiltrated and often surrounded tumors (Figure 1C; >90% of cytoplasm-rich cells were Desmin-positive). Myogenin+, MyoD+ and PAX7+ nuclei were interspersed with Myogenin−, MyoD− and PAX7− nuclei throughout the tumors (Figures 1E–G). In tumor tissue, approximately 25% of nuclei were Myogenin-positive, 25–50% of nuclei were MyoD-positive, up to 10% of nuclei were PAX7-positive, 25–50% were FABP4-positive and 50–75% were CD45-positive (3 to 6 tumor evaluated for each antigen). Finally, tumors also contained numerous cells expressing either adipocytic (FABP4; Figure 1D) or hematopoietic (CD45; Figure 1H) lineage markers. These observations confirm a highly differentiated myogenic phenotype in SmoM2 tumors and reveal substantial heterogeneity within the SmoM2 tumor cell pool, including cells expressing markers of myogenic and non-myogenic lineages.
Figure 1.
Microscopic tumor foci in R26-SmoM2(+/−);CAGGS-CreER mouse skeletal muscle contain phenotypically heterogeneous cells. (A) Random muscle sections obtained from obtained from R26-SmoM2(+/−);CAGGS-CreER mice contained numerous microscopic tumor foci. (B) Tumors contained large, multinucleated cells with abundant cytoplasm interspersed with small round blue cells. (C) Desmin-expressing, (D) FABP4-expressing, (E) Myogenin-expressing, (F) MyoD-expressing, (G) PAX7-expressing, (H) CD45-expressing tumor cells were interspersed with many Desmin-, FABP4-, Myogenin-, MyoD-, Pax7- and CD45-negative tumor cells. (C) Desmin expression in tumors often localized to larger cells with abundant cytoplasm. Tumors contained numerous cells expressing (D) adipocytic cell lineage markers (FABP4) and (H) hematopoietic-lineage markers (CD45).
SmoM2 tumors recapitulate skeletal muscle phenotypic heterogeneity
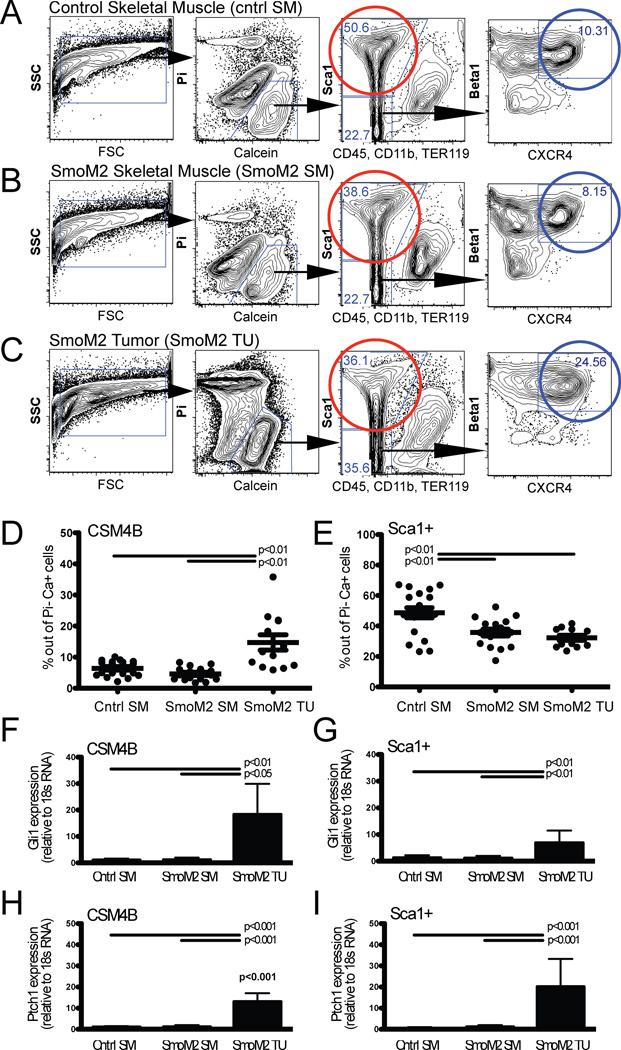
Normal skeletal muscle contains a variety of different cell types. Combinatorial staining for specific combinations of cell surface markers [10–13, 1], followed by FACS sorting, allows prospective isolation of phenotypically and functionally discrete subsets of cells from the myofiber-associated cell pool in muscle. For example, combinatorial staining for CD45, CD11b, TER119, Sca1, β1-integrin and CXCR4 distinguishes CD45−CD11b−TER119−Sca1−CXCR4+β1-integrin+ (CSM4B) myogenic precursors/ satellite cells (>90% PAX7-positive) [14, 10, 11, 15], CD45−CD11b−TER119−Sca1+ fibro-adipogenic precursors (Sca1+ cells) [10, 11, 3, 15, 16] and CD45+CD11b+TER119+ hematopoietic lineage cells [11, 1, 15] in skeletal muscle, including control mouse skeletal muscle (cntrl SM, isolated from 3–9 week-old R26-SmoM2(+/+) or R26-SmoM2(+/−) mice, Figure 2A). Interestingly, flow cytometry analyses demonstrated that analogous cell populations could be identified not only within tumor-predisposed SmoM2 skeletal muscle (isolated from 3–9 week-old tamoxifen-induced R26-SmoM2(+/−);CAGGS-CreER skeletal muscle that was free of macroscopic tumors; Figure 2B), but also from SmoM2 tumor tissue (isolated from macroscopic tumors arising in 3–9 week-old tamoxifen-induced R26-SmoM2(+/−);CAGGS-CreER mice; Figure 2C). The relative proportion of CSM4B cells was significantly increased in SmoM2 tumor tissue compared to tumor-free SmoM2 and control skeletal muscle (14.7±8.9% versus 4.6±2.0% and 6.4±2.4%, p<0.001, tumor and muscle tissue from 13–19 mice evaluated, Figure 2D). In contrast, the relative proportion of Sca1+ cells was reduced in SmoM2 tumor and tumor-free muscle tissue compared to control muscle (32.2±6% and 35.7±9.2% versus 48.7±14.6%, p<0.001, tumor and muscle tissue from 13–19 mice evaluated, Figure 2E). The proportion of hematopoietic lineage cells, marked by expression of CD45, CD11b, and/or TER119, was similar in control muscle, SmoM2 muscle and SmoM2 tumors (34.2±17% versus 48.7±17.2% and 42.4±13.5%, p>0.05, tumor and muscle tissue from 10–11 mice evaluated, data not shown).
Figure 2.
SmoM2 tumor cells exhibit immunophenotypic heterogeneity similar to normal mouse skeletal muscle. (A–C) Combinatorial staining for cell surface markers followed by fluorescence-activated cell sorting allows prospective isolation of phenotypically distinct cell populations from (A) control muscle (cntrl SM, isolated from R26-SmoM2(+/+) or R26-SmoM2(+/−) muscle), (B) SmoM2 muscle (SmoM2 SM, isolated from tamoxifen-induced R26-SmoM2(+/−);CAGGS-CreER muscle that was free of macroscopic tumors) and (C) SmoM2 tumor tissue (SmoM2 TU; isolated from R26-SmoM2(+/−);CAGGS-CreER muscle tumors). Data are presented as two-dimensional contour plots with nested gating as indicated by the black arrows. In normal muscle, CD45-Sca1-Mac1-CXCR4+β1-integrin+ (CSM4B) cells (blue gate) are enriched for PAX7-expressing satellite cells with myogenic precursor function [11, 1], and CD45-Sca1+ cells (red gate) are enriched for non-myogenic, adipogenic precursors [10, 3, 31]. Plots depict representative flow plots and are quantified in (D, E). (D, E) The frequency of CSM4B and Sca1+ cells was quantified by flow cytometry as percentage of Pi−Ca+ live cells in A–C. Individual data points, representing 12–14 individual mice, are presented as dots and are overlaid with mean +/− SD. P values indicate statistically significant differences and were calculated using Bonferroni’s correction for multiple comparisons. (D) The relative proportion of CSM4B cells was significantly increased, whereas (E) the relative proportion of Sca1+ fibroadipogenic stromal cells was reduced in SmoM2 tumor tissue compared to tumor-free SmoM2 and control muscle. (F–I) Increased expression of Hh targets Gli1 (F–G) and Ptch1 (H–I) in freshly sorted CSM4B and Sca1+ tumor cells was evaluated by qPCR. Data are presented as mean +/− SD and represent 7–9 independent experiments with 2–4 individual mice analyzed per experimental group in each experiment. P values indicate statistically significant differences and were calculated using Bonferroni’s correction for multiple comparisons.
Hedgehog pathway activation in SmoM2 tumor cell subsets
Expression of CAGGS-CreER is induced in many organs, and recombination in skeletal muscle was previously demonstrated to be efficient, yet incomplete [9]. Hh pathway activation in CSM4B and Sca1+ SmoM2 muscle and tumor cell subsets was evaluated by determining the expression of Hh target genes Gli1 (Figure 2F–G) and Ptch1 (Figure 2H–I) in freshly sorted cells (sorted from 2–4 mice per cell subset) by qPCR. Increased Gli1 and Ptch1 expression in CSM4B and Sca1+ cells from SmoM2 tumor tissue was consistent with activation of Hh signaling in both tumor cell subsets as compared to normal, tumor-free muscle from SmoM2 or wild-type mice (Figure 2F–I).
SmoM2 tumors recapitulate skeletal muscle functional heterogeneity
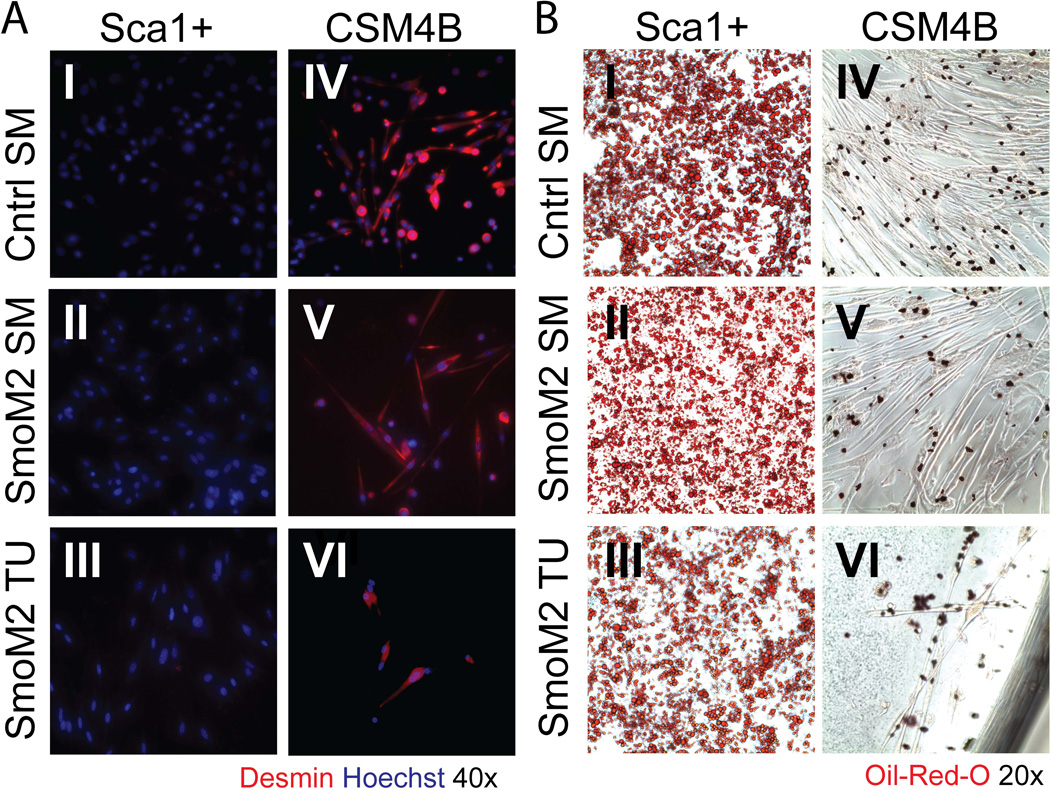
Previously published studies have established that, in normal mouse muscle, CSM4B and Sca1+ cells represent functionally discrete cell populations with distinct myogenic and adipogenic differentiation capacities [10, 3, 1]. To examine the differentiation potential of CSM4B and Sca1+ cells isolated from SmoM2 muscle and tumor tissue, cells were sorted at 1000–5000 cells per well, expanded in growth medium for 5 days and then transitioned to adipogenic or myogenic differentiation medium, respectively. Consistent with published findings [11], CSM4B myogenic precursors from control and SmoM2 muscle differentiated into Desmin-positive multinucleated myotubes in both myogenic and adipogenic culture conditions and exhibited no adipogenic differentiation capacity (Figure 3AIV–V and 3BIV–V). Conversely, Sca1+ cells obtained from control and SmoM2 muscle lacked myogenic activity and differentiated robustly into adipocytes [11] (Figure 3AI–II and 3BI–II). These same distinctions in myogenic and adipogenic differentiation capacity were observed in CSM4B and Sca1+ cells isolated from SmoM2 tumor tissue: SmoM2 tumor CSM4B cells differentiated into Desmin+ myotubes and lacked adipogenic capacity (Figure 3AVI and 3BVI), whereas tumor Sca1+ cells lacked myogenic activity and differentiated into adipocytes (Figure 3AIII and 3BIII). These data indicate that SmoM2 tumor cells recapitulate the phenotypic and functional heterogeneity of skeletal muscle and that these tumors contain distinct subsets of cells with and without myogenic differentiation potential.
Figure 3.
Freshly sorted SmoM2 tumor cells recapitulate the functional heterogeneity of mouse skeletal muscle. The adipogenic and myogenic differentiation potential of CSM4B and Sca1+ cells isolated from control muscle (top panels), SmoM2 muscle (middle panels) and SmoM2 tumor tissue (bottom panels) was determined by in vitro assays. Cells were freshly sorted at 1000 cells/well, expanded in growth medium and transitioned into myogenic (A) or adipogenic (B) differentiation medium. (A) Under myogenic conditions, muscle satellite cells and tumor CSM4B cells differentiated into Desmin-expressing myocytes (AIV–VI), while muscle and tumor Sca1+ cells lacked myogenic potential (AI–III). See Figure S1 for terminal differentiation of tumor CSM4B cells plated at 10,000 cells/well. (B) Under adipogenic conditions, muscle and tumor Sca1+ cells differentiated into Oil-Red-O-positive lipid droplet-containing adipocytes (BI–III). Muscle satellite cells and tumor CSM4B cells lacked adipogenic potential (BIV–VI). Differentiation assays were replicated in 2 independent experiments, with 1–2 mice analyzed in each experiment.
SmoM2 tumors contain an expanded pool of satellite-like cells
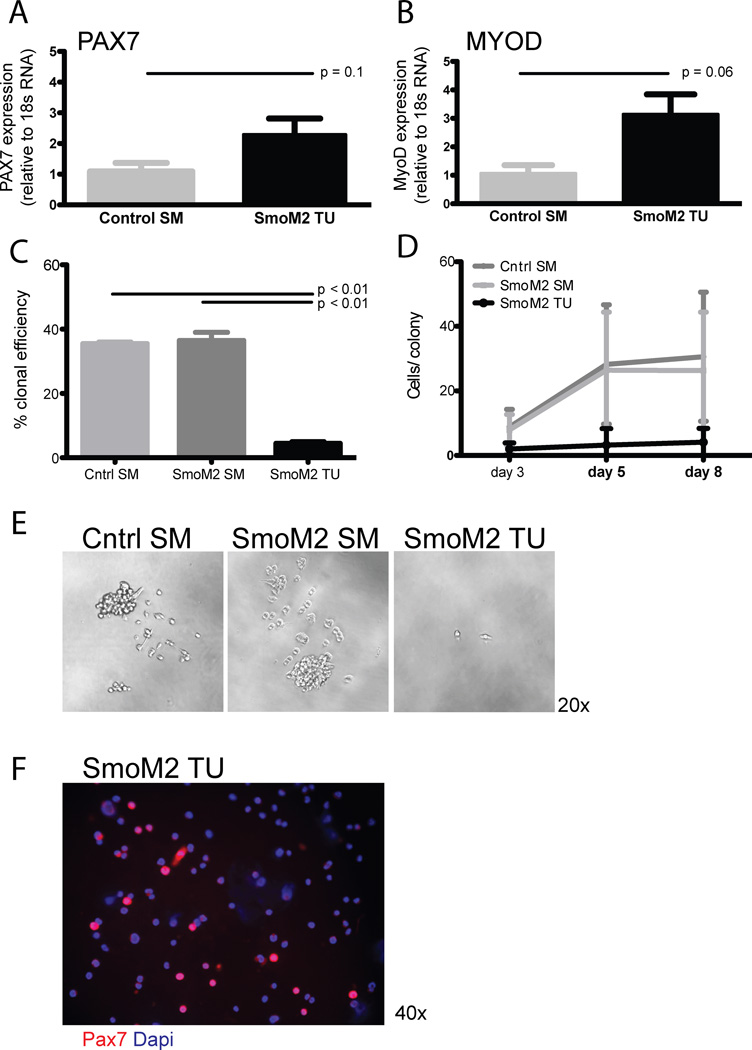
Compared to control and SmoM2 skeletal muscle, SmoM2 tumors contain a significantly increased frequency of CSM4B cells with selective myogenic activity (Figure 2D, AVI and S1). In normal skeletal muscle, CSM4B cells have been shown to be highly enriched for PAX7+ muscle satellite cells [10]. PAX7 is a widely-utilized, canonical marker of these cells, expressed by both quiescent and activated muscle satellite cells [17]. Up to 25% of resting satellite cells also express the myogenic regulatory factor MyoD [13], which is further induced upon satellite cell activation. MyoD expression is maintained as cells differentiate into early myoblasts (which lose Pax7 reactivity) [4, 5]. We analyzed Pax7 and MyoD expression in muscle and tumor CSM4B cells by qPCR. Both Pax7 and MyoD showed similar levels of expression in CSM4B cells from tumors as compared to CSM4B cells from control muscle (Figure 4A–B). PAX7 expression in freshly sorted tumor CSM4B cells was also confirmed by immunocytochemical staining of freshly sorted cells (Figure 4F). These data indicate that SmoM2 tumor CSM4B cells exhibit a satellite cell-like PAX7+, MyoD+ phenotype.
Figure 4.
SmoM2 tumors contain an expanded pool of activated satellite cells. (A–B) Similar expression of (A) PAX7 and (B) MyoD in muscle and tumor CSM4B cells. Data are presented as mean +/− SD for analysis of 3–4 independent cell samples for each experimental group. (C–E) Myogenic activity of CSM4B satellite cells sorted from control or SmoM2 muscle as well as CSM4B cells sorted from SmoM2 tumors was evaluated by sorting 1 cell per well in Collagen/Laminin coated 96-well-plates. The myogenic colony-initiating efficiency of tumor CSM4B cells was significantly lower than that of normal muscle CSM4B satellite cells. Data are presented as (C) ratio of number of colonies out of total number of CSM4B cells plated, (D) number of cells per colony and (E) representative brightfield images of CSM4B cell colonies from wild-type or SmoM2 muscle or tumor. Muscle CSM4B cell colonies (E; left and middle panels), but not tumor CSM4B cell colonies (E; right panel), quickly expanded in culture under myogenic growth conditions. Clonal assays were replicated in 2 independent experiments, representing 1–2 mice per experimental group in each experiment. Data are presented as mean +/− SD. (F) Immunocytochemical staining confirmed PAX7 expression in tumor CSM4B cells.
The myogenic function of tumor-derived CSM4B cells was also evaluated using ex vivo colony forming assays (Figure 4C–E). This well-established experimental approach [10, 18–20] determines the frequency of cells capable of activating a muscle regenerative program by counting the number of individual satellite cells that proliferate to form a colony of muscle cells after isolation from muscle and the number of cells per colony after in vitro culture. Our experiments revealed significantly impaired colony-initiating potential of SmoM2 tumor CSM4B cells as compared to that of muscle CSM4B cells (p<0.001, Figure 4C). Consistent with previously published findings in normal skeletal muscle [10], 69 of 192 clonally sorted control muscle CSM4B cells (36%) and 70 of 192 clonally sorted SmoM2 muscle CSM4B cells (36%) gave rise to rapidly expanding myogenic cell colonies (Figure 4C–E, data pooled from 2 independent experiments in which CSM4B cells from the indicated muscle type were sorted at 1 cell/well). However, only 9 of 192 clonally sorted SmoM2 tumor CSM4B cells (5%) initiated detectable cell growth in these assays, and these colonies did not expand over 8 days in culture (Figure 4D–E). These studies suggest that ectopic activation of the Hh signaling pathway in SmoM2 tumors may impair the ability of CSM4B cells exhibiting a satellite cell-like phenotype (Figure 2C, F, H) to survive and proliferate in response to stimuli that induce a regenerative myogenic program in normal satellite cells.
Hh pathway activation enhances terminal differentiation of muscle satellite cells
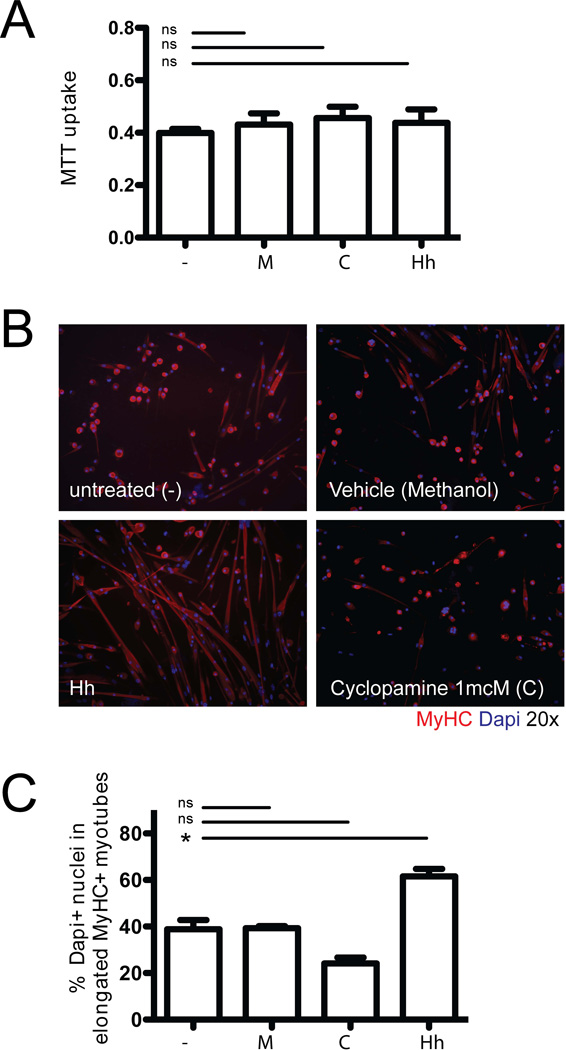
To complement these studies in SmoM2 tumor cells, we also assayed directly the effects of Hh ligand exposure in muscle satellite cells. We exposed sorted CSM4B cells from wild-type C57BL6 mice to conditioned medium (CM) containing Hh ligand or to cyclopamine, a Hh antagonist. Cells were assayed for ex-vivo growth and differentiation under myogenic culture conditions. Cell growth in the presence of Hh CM or cyclopamine was evaluated by MTT assay after 6 days, and revealed no differences in the growth of Hh- or cyclopamine-treated cells compared to control cells (Figure 5A). Next, to evaluate Hh effects on satellite cell differentiation, CSM4B cells from wild-type mice were expanded in growth medium for 5–7 days, transferred into chamber slides, and then exposed to either Hh CM or cyclopamine under myogenic differentiation conditions (Figure 5B–C). Myogenic differentiation was quantified as the percentage of Dapi+ nuclei in myosin heavy-chain (MyHC)-positive elongated myotubes (out of all Dapi+ nuclei). Exposure to Hh CM enhanced myogenic differentiation in these assays (58.3±4.6% vs 34.8±5.6 Dapi+ nuclei in MyHC+ myotubes in Hh treated cultures compared to control cultures; p<0.05; Figure 5B–C). In contrast, cyclopamine appeared to reduce myogenic differentiation, but these effects did not reach statistical significance (21.6±3.6% vs 34.8±5.6% Dapi+ nuclei in MyHC+ myotubes in Cyclopamine treated cultures compared to control cultures; p>0.05; Figure 5B–C). These data suggest that activation of Hh signaling via Hh ligand exposure in primary, purified muscle satellite cells may enhance terminal myogenic differentiation and that it is unlikely that Hh pathway activation intrinsically, in satellite cells themselves, drives the expansion of CSM4B cells noted in SmoM2 tumors (Figure 2D).
Figure 5.
Hh pathway activation enhances terminal differentiation of muscle satellite cells. (A) Freshly sorted CSM4B muscle satellite cells (from C57BL/6 wild-type muscle) were expanded in growth medium supplemented with Hh conditioned medium (Hh), cyclopamine (C) or methanol (M, vehicle control) for 6 days. Exposure to Hh pathway modulators did not change cell growth as measured by MTT assay after 6 days in culture (B–D). Freshly sorted satellite cells were expanded in GM for 5 days, passaged, re-plated onto Matrigel-coated chamber slides and transitioned into myogenic differentiation medium. (B) Selective activation of Hh signaling under these conditions enhanced terminal myogenic differentiation into MyHC-expressing myocytes. (C) Myogenic differentiation was quantified as percentage Dapi+ nuclei in MyHC-positive elongated myotubes (out of all Dapi+ nuclei) and showed a significant increase in terminally differentiated cells after Hh exposure. Assays were replicated in 2 independent experiments, representing 3–4 mice per group per experiment.
Discussion
Hedgehog (Hh) signaling is essential in embryonic development [21] and contributes to the formation of various tumors, including tumors with myogenic differentiation [22]. In mouse skeletal muscle, activation of Hh signaling via a ubiquitously expressed SmoM2 allele gives rise to multifocal myogenic tumors [8, 9]. Here, we demonstrate that these myogenic tumors contain a complex pool of cells that recapitulates normal skeletal muscle phenotypic and functional heterogeneity. The cluster of cells contained within myogenic tumors includes cells with terminal muscle differentiation features, distinct subsets of mononuclear cells with differential myogenic and adipogenic differentiation capacity, and an expanded pool of Pax7+ and MyoD+ myogenic satellite-like cells. This latter observation is consistent with published findings demonstrating that Hh-driven myogenic tumors display a gene expression profile similar to activated satellite cells [23].
The frequency of CSM4B satellite-like cells in SmoM2 tumors was significantly increased compared to the frequency of normal CSM4B satellite cells in skeletal muscle. Similar to CSM4B muscle satellite cells, SmoM2 tumor satellite-like cells were selectively myogenic and expressed PAX7 and MyoD. However, tumor CSM4B satellite-like cells exhibited extremely poor colony-forming activity and growth in vitro when evaluated in clonal assays, suggesting that, unlike freshly sorted CSM4B muscle satellite cells [10], they were unable to support a normal muscle regenerative program. The observation that the satellite-like cell pool in SmoM2 tumors is expanded, while, at the same time, their intrinsic colony-forming/ proliferative activity is substantially impaired, is somewhat surprising; yet, it has also been reported that SmoM2 tumor cells exhibit extremely poor tumor-propagating capacity in secondary recipients, even when transplanted at very high numbers [24]. The poor tumor-propagating capacity of SmoM2 myogenic tumors stands in marked contrast to the highly efficient induction of secondary tumors by Ras-driven rhabdomyosarcomas [24]. That Hh-driven mouse myogenic tumors exhibit differences in tumor behavior and phenotype compared to other types of mouse myogenic tumors that more closely resemble human fusion-negative rhabdomyosarcomas [24, 8, 25] emphasizes the need for further studies to investigate if a subset of tumors on the rhabdomyosarcoma spectrum may recapitulate the functional heterogeneity observed within the SmoM2 tumor cell pool in this study.
Regarding the role of Hh signaling in normal muscle maintenance and regeneration, findings to date have been somewhat conflicting. Hh signaling is activated in acutely injured mouse muscle [26, 27], and Elia et al showed that Hh stimulates myoblast differentiation [28], similar to the results reported here. Straface et al also documented Hh pathway activation in mouse muscle satellite cells following crush/ cardiotoxin injury, and showed that when Hh signaling was impeded by cyclopamine treatment of mice, satellite cell activation and muscle regeneration were impaired [27]. However, Koleva et al reported a differentiation-blocking effect of Hh on mouse myoblast cultures [29]. Similarly, overexpression of Gli1 or Gli2 in pluripotent mesenchymal cell lines has been linked to both inhibitory and stimulating effects on MyoD transcriptional activation [30, 31]. We evaluated the effects of Hh pathway activation on terminal differentiation of freshly sorted, highly purified satellite cells [10, 1] into MyHC-expressing myofibers in vitro and observed a small but statistically significant differentiation-promoting effect of Hh pathway activation, but no effect on satellite cell proliferation. These data, together with the observation that selective activation of SmoM2 in satellite cells fails to yield myogenic tumors [23], suggests that it is unlikely that direct effects of Hh signaling in satellite cells themselves contribute to the expansion of satellite-like cells in SmoM2 tumors and tumorigenesis in SmoM2 mice.
The cellular origins of Hh-driven myogenic tumors remain controversial. Published findings support the notion that certain fusion-negative rhabdomyosarcomas, driven by RAS or Hippo pathway activation, originate in activated satellite cells [11, 32]. However, selective activation of SmoM2 in postnatal, PAX7-expressing satellite cells via a Tamoxifen-inducible PAX7-CreER did not result in any tumors [23]. This observation is consistent with findings from our study showing that chemical stimulation of Hh signaling in freshly sorted satellite cells enhances differentiation but does not directly induce satellite cell proliferation and expansion. Prior studies in Ptch1+/− mice indicate that the development of Hh-driven myogenic tumors strictly depends on prenatal Ptch1 deletion [33] with the highest tumor incidence (around 80%) apparent when Ptch1 is deleted embryonically (at E9.5) [34]. Activation of SmoM2 during early muscle development via Myf5-Cre, Pax3-Cre or MyoD1-Cre resulted in embryonic lethality without tumor induction [23]. Myogenin-Cre activation of SmoM2 induced tongue tumors [23]. Finally, activation of SmoM2 in FABP4-expressing cells was shown to efficiently induce myogenic tumors in mice [23], thereby raising the possibility that SmoM2 myogenic tumors originate from Hh-mediated transdifferentiation events in adipocytic cells. Alternatively, it is conceivable that Hh-pathway activation in FABP4-expressing cells initiates a cascade of paracrine events resulting in satellite cell expansion, activation and differentiation. Such a paracrine mechanism, involving intermediary events initiated by Hh activation in other cell types, could also explain the apparent conflict between satellite cell expansion and activation in SmoM2 tumors and the direct, differentiating effects of Hh ligand on satellite cells described here. Interestingly, non-cell-autonomous influences of Hh signaling on tumorigenesis have been noted previously in epithelial malignancies, which exhibit a paracrine requirement for Hh ligand activation in tumor stromal cells [35].
Finally, findings from our study demonstrate that SmoM2 mouse myogenic tumors contain functionally heterogeneous cells, including a sizeable number of immune and inflammatory cells of hematopoietic lineage and a subset of stromal cells that lacks myogenic differentiation potential. CAGGS-CreER recombination in skeletal muscle is incomplete [9], and Hh pathway activation in sorted SmoM2 tumor cells was not evaluated at a single-cell level, which represents a limitation of this study. Yet, Hh pathway activation in freshly sorted myogenic and non-myogenic tumor cell subsets was confirmed at the population level by PCR detection of Hh pathway effectors Ptch1 and Gli1. Importantly, the presence of cells lacking myogenic differentiation capacity in Hh-driven myogenic tumors could be relevant in the context of therapeutic considerations aiming to induce terminal myodifferentiation in rhabdomyosarcoma [32, 36, 37], as the presence of a sizeable subset of tumor cells without myogenic differentiation potential could represent a potential obstacle, or novel target, in the development of differentiation-promoting rhabdomyosarcoma therapies.
Conclusions
Hedgehog (Hh) pathway activation in R26-SmoM2;CAGGS-CreER mice, expressing a mutant activated Smoothened allele (SmoM2) via a ubiquitously expressed Cre-recombinase induction system, results in the formation of tumors with a highly differentiated myogenic phenotype. These tumors contain a mixture of cells expressing myogenic, adipocytic and hematopoietic lineage markers, recapitulate the phenotypic and functional hetereogeneity observed in normal mouse skeletal muscle and include an expanded population of satellite-like cells. Moreover, similar to normal skeletal muscle, SmoM2 tumors contain cells with differential myogenic and adipogenic differentiation capacity, including cells lacking myogenic potential.
Supplementary Material
Freshly sorted SmoM2 tumor CSM4B cells differentiate into Desmin-expressing myocytes. The myogenic differentiation potential of SmoM2 tumor CSM4B cells was determined by in vitro assays. 10,000 double-sorted cells per well were plated.
Highlights.
Hedgehog activation induces multifocal tumors in skeletal muscle.
Hedgehog-driven tumors in muscle recapitulate skeletal muscle phenotypic and functional heterogeneity.
Hedgehog-driven tumors in muscle contain cells with differential myogenic and adipogenic capacity.
Hedgehog activation enhances satellite cell differentiation.
Acknowledgements
We thank G. Buruzula, J. LaVecchio, and A. Wakabayashi at Joslin’s DRC Flow Cytometry Core for excellent flow cytometry support, A. Pinkhasov and T. Genest at Joslin’s DRC Histology Core for help with tumor sectioning, and C.L. Unitt and T. Bowman at the DF/HCC Histopathology Core for help with immunohistochemistry. This work was funded in part by grants from the NIH (DK036836), the Burroughs-Wellcome Fund and the Harvard Stem Cell Institute (to AJW), from Hope Street Kids, P.A.L.S. Bermuda/St. Baldrick’s, ALSF and Bear Necessities (to SH) and from NINDS (NS033642; to APM). Content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or other funding agencies.
List of abbreviations
- FAP
fibroadipogenic progenitors
- PAX7
paired box 7
- MRF
myogenic regulatory factor
- Hh
Hedgehog
- MyoD1
myogenic differentiation 1
- Smo
Smoothened
- Gli1
Gli family zinc finger 1
- Ptch1
Patched 1
- rRNA
ribosomal ribonucleic acid
- CAGGS
CMV early enhancer/chicken beta actin
- ER
estrogen receptor
- FABP4
fatty acid binding protein 4
- DMEM
Dulbecco’s Modified Eagle Medium
- DM
differentiation medium
- CM
conditioned medium
- Hank’s
Balanced Salt Solution
- FBS
fetal bovine serum
- bFGF
basic fibroblastic growth factor
- FACS
fluorescence activated cell sorting
- HBSS
APC, allophycocyanin
- Cy7
cyanine 7
- PE
phycoerythrin
- PI
propidium iodide
- Ca
Calcein
- GFP
green fluorescent protein
- PCR
polymerase chain reaction
- RT-PCR
real time polymerase chain reaction
- qRT-PCR
quantitative real-time polymerase chain reaction
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest disclosures
The authors declare that they have no competing interests.
References
- 1.Sherwood RI, Christensen JL, Conboy IM, Conboy MJ, Rando TA, Weissman IL, et al. Isolation of adult mouse myogenic progenitors: functional heterogeneity of cells within and engrafting skeletal muscle. Cell. 2004;119(4):543–554. doi: 10.1016/j.cell.2004.10.021. doi:S0092867404010360 [pii] 10.1016/j.cell.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 2.Wagers AJ, Conboy IM. Cellular and molecular signatures of muscle regeneration: current concepts and controversies in adult myogenesis. Cell. 2005;122(5):659–667. doi: 10.1016/j.cell.2005.08.021. doi:S0092-8674(05)00858-5 [pii] 10.1016/j.cell.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 3.Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J, et al. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12(2):153–163. doi: 10.1038/ncb2015. doi:ncb2015 [pii] 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornelison DD, Wold BJ. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev Biol. 1997;191(2):270–283. doi: 10.1006/dbio.1997.8721. doi:S0012-1606(97)98721-2 [pii] 10.1006/dbio.1997.8721. [DOI] [PubMed] [Google Scholar]
- 5.Cooper RN, Tajbakhsh S, Mouly V, Cossu G, Buckingham M, Butler-Browne GS. In vivo satellite cell activation via Myf5 and MyoD in regenerating mouse skeletal muscle. J Cell Sci. 1999;112(Pt 17):2895–2901. doi: 10.1242/jcs.112.17.2895. [DOI] [PubMed] [Google Scholar]
- 6.Bridge JA, Liu J, Weibolt V, Baker KS, Perry D, Kruger R, et al. Novel genomic imbalances in embryonal rhabdomyosarcoma revealed by comparative genomic hybridization and fluorescence in situ hybridization: an intergroup rhabdomyosarcoma study. Genes Chromosomes Cancer. 2000;27(4):337–344. doi: 10.1002/(sici)1098-2264(200004)27:4<337::aid-gcc1>3.0.co;2-1. doi:10.1002/(SICI)1098-2264(200004)27:4<337::AID-GCC1>3.0.CO;2-1 [pii]. [DOI] [PubMed] [Google Scholar]
- 7.Tostar U, Malm CJ, Meis-Kindblom JM, Kindblom LG, Toftgard R, Unden AB. Deregulation of the hedgehog signalling pathway: a possible role for the PTCH and SUFU genes in human rhabdomyoma and rhabdomyosarcoma development. J Pathol. 2006;208(1):17–25. doi: 10.1002/path.1882. [DOI] [PubMed] [Google Scholar]
- 8.Hettmer S, Teot LA, van Hummelen P, Macconaill L, Bronson RT, Dall'osso C, et al. Mutations in Hedgehog pathway genes in fetal rhabdomyomas. J Pathol. 2013 doi: 10.1002/path.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mao J, Ligon KL, Rakhlin EY, Thayer SP, Bronson RT, Rowitch D, et al. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 2006;66(20):10171–10178. doi: 10.1158/0008-5472.CAN-06-0657. doi:66/20/10171 [pii] 10.1158/0008-5472.CAN-06-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cerletti M, Jurga S, Witczak CA, Hirshman MF, Shadrach JL, Goodyear LJ, et al. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell. 2008;134(1):37–47. doi: 10.1016/j.cell.2008.05.049. doi:S0092-8674(08)00755-1 [pii] 10.1016/j.cell.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hettmer S, Liu J, Miller CM, Lindsay MC, Sparks CA, Guertin DA, et al. Sarcomas induced in discrete subsets of prospectively isolated skeletal muscle cells. Proc Natl Acad Sci U S A. 2011;108(50):20002–20007. doi: 10.1073/pnas.1111733108. doi:1111733108 [pii] 10.1073/pnas.1111733108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, et al. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309(5743):2064–2067. doi: 10.1126/science.1114758. doi:1114758 [pii] 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- 13.Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456(7221):502–506. doi: 10.1038/nature07384. doi:nature07384 [pii] 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerletti M, Jang YC, Finley LW, Haigis MC, Wagers AJ. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell Stem Cell. 2012;10(5):515–519. doi: 10.1016/j.stem.2012.04.002. doi:S1934- 5909(12)00167-1 [pii] 10.1016/j.stem.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan KY, Eminli S, Hettmer S, Hochedlinger K, Wagers AJ. Efficient generation of iPS cells from skeletal muscle stem cells. PLoS One. 2011;6(10):e26406. doi: 10.1371/journal.pone.0026406. doi:10.1371/journal.pone.0026406 PONE-D-11-08506 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol. 2010;12(2):143–152. doi: 10.1038/ncb2014. doi:ncb2014 [pii] 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- 17.Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102(6):777–786. doi: 10.1016/s0092-8674(00)00066-0. doi:S0092-8674(00)00066-0 [pii]. [DOI] [PubMed] [Google Scholar]
- 18.Hayhurst M, Wagner AK, Cerletti M, Wagers AJ, Rubin LL. A cell-autonomous defect in skeletal muscle satellite cells expressing low levels of survival of motor neuron protein. Dev Biol. 2012;368(2):323–334. doi: 10.1016/j.ydbio.2012.05.037. doi:S0012-1606(12)00307-7 [pii] 10.1016/j.ydbio.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shefer G, Rauner G, Yablonka-Reuveni Z, Benayahu D. Reduced satellite cell numbers and myogenic capacity in aging can be alleviated by endurance exercise. PLoS One. 2010;5(10):e13307. doi: 10.1371/journal.pone.0013307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shefer G, Van de Mark DP, Richardson JB, Yablonka-Reuveni Z. Satellite-cell pool size does matter: defining the myogenic potency of aging skeletal muscle. Dev Biol. 2006;294(1):50–66. doi: 10.1016/j.ydbio.2006.02.022. doi:S0012-1606(06)00119-9 [pii] 10.1016/j.ydbio.2006.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15(23):3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 22.Corcoran RB, Scott MP. A mouse model for medulloblastoma and basal cell nevus syndrome. J Neurooncol. 2001;53(3):307–318. doi: 10.1023/a:1012260318979. [DOI] [PubMed] [Google Scholar]
- 23.Hatley ME, Tang W, Garcia MR, Finkelstein D, Millay DP, Liu N, et al. A mouse model of rhabdomyosarcoma originating from the adipocyte lineage. Cancer Cell. 2012;22(4):536–546. doi: 10.1016/j.ccr.2012.09.004. doi:S1535-6108(12)00393-5 [pii] 10.1016/j.ccr.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hettmer S, Bronson RT, Wagers AJ. Distinct malignant behaviors of mouse myogenic tumors induced by different oncogenetic lesions. Front Oncol. 2015;5:50. doi: 10.3389/fonc.2015.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kappler R, Bauer R, Calzada-Wack J, Rosemann M, Hemmerlein B, Hahn H. Profiling the molecular difference between Patched- and p53-dependent rhabdomyosarcoma. Oncogene. 2004;23(54):8785–8795. doi: 10.1038/sj.onc.1208133. doi:1208133 [pii] 10.1038/sj.onc.1208133. [DOI] [PubMed] [Google Scholar]
- 26.Pola R, Ling LE, Silver M, Corbley MJ, Kearney M, Blake Pepinsky R, et al. The morphogen Sonic hedgehog is an indirect angiogenic agent upregulating two families of angiogenic growth factors. Nat Med. 2001;7(6):706–711. doi: 10.1038/89083. doi:10.1038/89083 89083 [pii]. [DOI] [PubMed] [Google Scholar]
- 27.Straface G, Aprahamian T, Flex A, Gaetani E, Biscetti F, Smith RC, et al. Sonic hedgehog regulates angiogenesis and myogenesis during post-natal skeletal muscle regeneration. J Cell Mol Med. 2009;13(8B):2424–2435. doi: 10.1111/j.1582-4934.2008.00440.x. doi:JCMM440 [pii] 10.1111/j.1582-4934.2008.00440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elia D, Madhala D, Ardon E, Reshef R, Halevy O. Sonic hedgehog promotes proliferation and differentiation of adult muscle cells: Involvement of MAPK/ERK and PI3K/Akt pathways. Biochim Biophys Acta. 2007;1773(9):1438–1446. doi: 10.1016/j.bbamcr.2007.06.006. doi:S0167-4889(07)00160-7 [pii] 10.1016/j.bbamcr.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Koleva M, Kappler R, Vogler M, Herwig A, Fulda S, Hahn H. Pleiotropic effects of sonic hedgehog on muscle satellite cells. Cell Mol Life Sci. 2005;62(16):1863–1870. doi: 10.1007/s00018-005-5072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerber AN, Wilson CW, Li YJ, Chuang PT. The hedgehog regulated oncogenes Gli1 and Gli2 block myoblast differentiation by inhibiting MyoD-mediated transcriptional activation. Oncogene. 2007;26(8):1122–1136. doi: 10.1038/sj.onc.1209891. doi:1209891 [pii] 10.1038/sj.onc.1209891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voronova A, Coyne E, Al Madhoun A, Fair JV, Bosiljcic N, St-Louis C, et al. Hedgehog signaling regulates MyoD expression and activity. J Biol Chem. 2013;288(6):4389–4404. doi: 10.1074/jbc.M112.400184. doi:M112.400184 [pii] 10.1074/jbc.M112.400184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tremblay AM, Missiaglia E, Galli GG, Hettmer S, Urcia R, Carrara M, et al. The Hippo transducer YAP1 transforms activated satellite cells and is a potent effector of embryonal rhabdomyosarcoma formation. Cancer Cell. 2014;26(2):273–287. doi: 10.1016/j.ccr.2014.05.029. doi:S1535-6108(14)00230-X [pii] 10.1016/j.ccr.2014.05.029. [DOI] [PubMed] [Google Scholar]
- 33.Zibat A, Uhmann A, Nitzki F, Wijgerde M, Frommhold A, Heller T, et al. Time-point and dosage of gene inactivation determine the tumor spectrum in conditional Ptch knockouts. Carcinogenesis. 2009;30(6):918–926. doi: 10.1093/carcin/bgp068. doi:bgp068 [pii] 10.1093/carcin/bgp068. [DOI] [PubMed] [Google Scholar]
- 34.Nitzki F, Zibat A, Frommhold A, Schneider A, Schulz-Schaeffer W, Braun T, et al. Uncommitted precursor cells might contribute to increased incidence of embryonal rhabdomyosarcoma in heterozygous Patched1-mutant mice. Oncogene. 2011;30(43):4428–4436. doi: 10.1038/onc.2011.157. doi:onc2011157 [pii] 10.1038/onc.2011.157. [DOI] [PubMed] [Google Scholar]
- 35.Yauch RL, Gould SE, Scales SJ, Tang T, Tian H, Ahn CP, et al. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455(7211):406–410. doi: 10.1038/nature07275. doi:nature07275 [pii] 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 36.Svalina MN, Keller C. YAPping about differentiation therapy in muscle cancer. Cancer Cell. 2014;26(2):154–155. doi: 10.1016/j.ccr.2014.07.011. doi:S1535-6108(14)00302-X [pii] 10.1016/j.ccr.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ciarapica R, De Salvo M, Carcarino E, Bracaglia G, Adesso L, Leoncini PP, et al. The Polycomb group (PcG) protein EZH2 supports the survival of PAX3-FOXO1 alveolar rhabdomyosarcoma by repressing FBXO32 (Atrogin1/MAFbx) Oncogene. 2013 doi: 10.1038/onc.2013.471. doi:onc2013471 [pii] 10.1038/onc.2013.471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Freshly sorted SmoM2 tumor CSM4B cells differentiate into Desmin-expressing myocytes. The myogenic differentiation potential of SmoM2 tumor CSM4B cells was determined by in vitro assays. 10,000 double-sorted cells per well were plated.