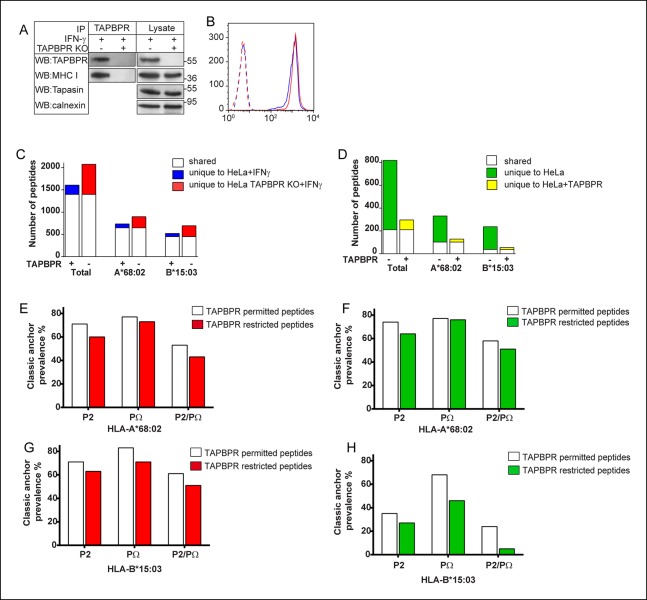
Figure 5. TAPBPR expression alters the peptide repertoire presented by MHC class I on cells.
(A) The TAPBPR:MHC class I complex was immunoprecipitated from IFN-γ treated HeLa and HeLa-TAPBPR KO cells. Western blot analysis was performed for TAPBPR (mouse anti-TAPBPR), tapasin (Rgp48N), MHC class I (HC10) and calnexin on lysates and TAPBPR immunoprecipitates as indicated. The data is representative of three independent experiments. (B) Cytofluorometric analysis of MHC class I detected with W6/32 on IFN-γ treated HeLa (Blue line) and HeLa-TAPBPR KO cells (Red line). Staining with an isotype control on both cell lines (dashed lines) is included as a control. (C,D) Peptide-MHC class I complexes were isolated by affinity chromatography using W6/32 from (C) IFN-γ treated HeLa and HeLa-TAPBPR KO cells or (D) HeLa and HeLa overexpressing WT-TAPBPR. Eluted peptides were analysed using LC-MS/MS. Graphs show the total number of peptides and also the number of peptides assigned as HLA-A*68:02 and HLA-B*15:03 binders based on their peptide motifs using the online programme NetMHC. The number of peptides shared between the cell lines (white bar) and the number of peptides unique to each cell line (coloured bars) is shown. The data was generated from tandem MS analysis performed five times on one immunoprecipitate. Two independent biological repeats have been performed in two different cell lines (HeLa-S cells shown in Figure 5—figure supplement 1, and KBM-7 cells shown in Figure 6) in which a similar pattern of increased peptide diversity in TAPBPR depleted cells was observed. (E–H) Conservation of P2 and C-terminal anchor residues (PΩ). Plots show prevalence of classic peptide anchors on (E,F) HLA-A*68:02 or (G, H) HLA-B*15:03. Bars show classic anchor conservation at P2, PΩ and P2/PΩ combined for (E,G) shared peptides found in both IFN-γ treated HeLa and HeLa TAPBPR KO cells (white bar) (presumably permitted expression in the presence of TAPBPR) and peptides unique to HeLa TAPBPR KO + IFNγ cells (red bar) (presumably restricted in the presence of TAPBPR), (F,H) shared peptides found in both HeLa and HeLa over-expressing TAPBPR (white bar) (presumably permitted expression in the presence of TAPBPR) and peptides unique to HeLa (green bar) (presumably restricted in the presence of TAPBPR).