Abstract
Uranium has radiological and non-radiological effects within biological systems and there is increasing evidence for genotoxic and carcinogenic properties attributable to uranium through its heavy metal properties. In this study, we report that low concentrations of uranium (as uranyl acetate; <10 μM) is not cytotoxic to human embryonic kidney cells or normal human keratinocytes; however, uranium exacerbates DNA damage and cytotoxicity induced by hydrogen peroxide, suggesting that uranium may inhibit DNA repair processes. Concentrations of uranyl acetate in the low micromolar range inhibited the zinc finger DNA repair protein poly(ADP-ribose) polymerase (PARP)-1 and caused zinc loss from PARP-1 protein. Uranyl acetate exposure also led to zinc loss from the zinc finger DNA repair proteins Xeroderma Pigmentosum, Complementation Group A (XPA) and aprataxin (APTX). In keeping with the observed inhibition of zinc finger function of DNA repair proteins, exposure to uranyl acetate enhanced retention of induced DNA damage. Co-incubation of uranyl acetate with zinc largely overcame the impact of uranium on PARP-1 activity and DNA damage. These findings present evidence that low concentrations of uranium can inhibit DNA repair through disruption of zinc finger domains of specific target DNA repair proteins. This may provide a mechanistic basis to account for the published observations that uranium exposure is associated with DNA repair deficiency in exposed human populations.
Keywords: Uranium, Poly(ADP-ribose) polymerase-1 (PARP-1), Zinc finger, DNA damage, DNA repair
1. Introduction
Human exposure to uranium is often linked to occupational exposures such as mining activities, living in proximity to mine waste sites, or increased use of depleted uranium for industrial or military purposes (Craft et al., 2004; Brugge et al., 2005; Miller and McClain, 2007; deLemos et al., 2009; Briner, 2010; Brugge and Buchner, 2011; Hund et al., 2015). Natural uranium is a heavy metal predominantly comprised of two isotopes, 235U and 238U. The radiologic toxicity and carcinogenicity of uranium is well-established, but less is known regarding the toxic mechanisms of uranium that are associated with its heavy metal properties (Craft et al., 2004; Brugge et al., 2005; Miller and McClain, 2007; Briner, 2010; Brugge and Buchner, 2011). Uranium exposure in cell culture causes oxidative stress, DNA damage, and genotoxicity (Periyakaruppan et al., 2007; Periyakaruppan et al., 2009; Garmash et al., 2014). In addition, there is evidence that uranium may disrupt DNA repair. Several studies report that uranium exposure is associated with DNA repair deficiency in exposed populations (Au et al., 1995; Au et al., 1998; Au et al., 2010), but the mechanism is not established.
Proteins with zinc-finger structures are the most abundant class of zinc metalloproteins, representing an estimated 2–3% of human genes whose protein products regulate diverse functions in cells (Maret, 2012; Hudson et al., 2015). Certain metals can interact with zinc finger structures of proteins and thereby disrupt protein function (Witkiewicz-Kucharczyk and Bal, 2006; Beyersmann and Hartwig, 2008; Ding et al., 2009; Bal et al., 2011; Zhou et al., 2011; Hartwig, 2013; Sun et al., 2014; Zhou et al., 2014; Hudson et al., 2015). Metal inhibition of proteins involved in DNA repair has led to the hypothesis that low concentrations of certain metals may serve as co-carcinogens by preventing effective repair of DNA damage caused by another insult. Experimental evidence supports this hypothesis for metals such as arsenic where co-carcinogenic actions have been well established (Burns et al., 2004; Rossman et al., 2004; Beyersmann and Hartwig, 2008; Qin et al., 2008; Cooper et al., 2013), leading to questions regarding the co-carcinogenic potential of other metals.
The potential for uranium to act as a co-carcinogen through inhibition of DNA repair proteins has not been widely explored. There is limited evidence that uranium may interact with zinc finger proteins. One study demonstrated that at concentrations equal to or greater than 10 μM, uranyl acetate (UA) disrupted the DNA binding activity of two purified zinc finger proteins, Aart and specificity protein (Sp)1 in vitro (Hartsock et al., 2007). Furthermore, poly(ADP-ribose) polymerase-1 (PARP-1) was identified as a uranium binding protein using affinity chromatography and mass spectrometry approaches (Dedieu et al., 2009). Although these studies suggest that uranium may interfere with zinc finger DNA repair protein targets, the experiments did not investigate the impact of uranium on a relevant target protein isolated from exposed cells or direct inhibition of DNA repair activity.
In this study we provide evidence that low, non-cytotoxic concentrations of uranium (as uranyl acetate) enhance the cytotoxicity of another DNA damaging agent such as ultraviolet radiation (UVR), promote the retention of DNA damage, and disrupt the zinc binding functions of zinc finger DNA repair proteins including PARP-1. In addition, zinc largely overcomes the impact of uranium on PARP-activity and DNA damage. These findings suggest that uranium is capable of interfering with DNA repair processes through disruption of zinc finger DNA repair protein function and may act as a co-carcinogen through its inhibition of DNA repair.
2. Experimental procedures
2.1. Cell culture and treatment
Human embryonic kidney 293 (HEK293) cells were generously provided by Dr. H. Ward (University of New Mexico) and cultured in Dulbecco's Modified Eagle's Medium (DMEM). DMEM was supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine and antibiotics (penicillin, 100 U/ml and streptomycin, 100 μg/ml). Normal human neonatal epidermal keratinocytes (HEKn) and DermaLife K culture medium with supplements were purchased from Lifeline Cell Technologies (Oceanside, CA). The zinc content of DermaLife K culture medium was 500 nM as measured by inductively coupled plasma mass spectrometry (ICP-MS). Cells were cultured at 37 °C in 95% air/ 5% CO2 humidified incubator.
Uranyl acetate (99.6%) was purchased from Electron Microscopy Science (Hatfield, PA) and was comprised of 99.9% 238U and 0.1% 235U according to the product's technical bulletin. Uranyl acetate had a radioactive activity of 0.51 μCi g−1 and was handled according to the regulations set forth by the Radiation Safety office at the University of New Mexico. Sodium arsenite (99%) and zinc chloride (99%) were purchased from Fluka Chemie (Buchs, Germany). Other chemicals were obtained from Sigma-Aldrich unless otherwise indicated. Ten millimolar stock solutions of uranyl acetate (UA), sodium arsenite (As), zinc chloride (Zn), zinc sulfate (ZnSO4, 99%) and 100 mM stock solution of H2O2 were prepared in double-distilled water and sterilized using a 0.22-μm syringe filter. Working solutions were prepared by diluting the stock with complete cell growth medium. Etoposide solutions were prepared in absolute ethanol. For experiments involving cell exposures, cells were rinsed and placed in complete medium containing, UA, As, Zn, etoposide or H2O2 as indicated in the figure legends.
2.2. Cell viability
HEKn and HEK293 cells (4 × 103 cells per well) were cultured in 96-well plates for 24 h, then treated with UA (0–30 μM) and incubated for an additional 24 h. HEKn and HEK293 cells were then exposed to 100 μM H2O2 for 10 min, washed twice with phosphate-buffered saline (PBS), placed in UA containing medium and returned to the incubator for an additional 24 h. PrestoBlue (Life Technologies) was added to each well at a final concentration of 10% and plates returned to the incubator for an additional 2 h. Fluorescence was measured using a microplate reader (SpectraMax M2e, Molecular devices) with excitation 555 nm and emission 585 nm. Values were normalized to untreated controls. For certain experiments as indicated in the figure legends, cell viability was measured using the CellTiter 96 Non-radioactive cell proliferation assay kit following the manufacturer's instructions (Promega, Madison, WI). HEK293 cells were cultured as above except plating density was adjusted so cells were either confluent or approximately 50% (sub-confluent) at time of UA treatment (0–100 μM) for 24, 48 and 72 h. In addition, images were obtained for each UA treatment condition after staining with Trypan blue (0.02%, Invitrogen, Carlsbad, CA) for 5 min. Viable and non-viable cells were counted and values normalized to number of viable cells per 106 cells.
2.3. DNA damage assessment and quantification
HEK293 cells were cultured on chamber slides and treated with concentrations of UA as indicated in the figure legends for 24 h, then DNA damage was induced by exposure to either H2O2 (100 μM), ssUVR (3 kJ/m2) or etoposide (as indicated in the figure legends). Select experiments included treatment with Zn (2 μM) and UA plus Zn. Slides were fixed at 0, 1, and 6 h post-induction of DNA damage. DNA damage was assessed by indirect immunofluorescence using anti-phospho-H2Ax (1:250; Cell Signaling Technologies, Danvers, MA), and anti-cyclobutane pyrimidine dimers (1:200; CPDs, Thymine clone KTM53, Kamiya Laboratories, Seattle, WA) antibodies in combination with anti-rabbit FITC conjugated secondary antibodies (1:1000; Abcam, Cambridge, MA) diluted in blocking buffer (PBS containing 2% bovine serum albumin (BSA), 5% horse serum and 0.05% triton X-100). Cover glass was mounted with VectaShield + 4′,6-diamidino-2-phenylindole (DAPI, Vector Laboratories, Burlingame, CA). DNA damage markers were visualized with an Olympus IX70 fluorescence microscope equipped with a DP72 digital camera and cellSens Dimension 9.1 (Olympus America, Center Valley, PA) software. A minimum of 10 images per slide were obtained per treatment and average fluorescence intensity of nuclei was quantified using cellSens Dimension 9.1 Count & Measure image analysis software.
2.4. UV source
UVR exposures were performed using an Oriel 1000 W Watt Solar Ultraviolet Simulator (Oriel Corp., Stratford, CT). This solar simulator produces a high intensity UVR beam in both the UVA (320–400 nm) and UVB (280–320 nm) spectrum with an emission ratio of 14:1 (UVA: UVB). The proportion and intensity of UVA/UVB was measured using a radiospectrometer (Optronics Laboratories, Inc.; Orlando, FL) and exposure times were calculated to give the desired dose of approximately 1.1 minimum erythema dose (MED). Measurements were made with Erythema UV and UVA intensity meter (Solar Light Co., Inc., Philadelphia, PA) in order to estimate MED.
2.5. PARP activity assay
Cells were treated with UA (0–100 μM), Zn (2 μM) or both for 24 h, then PARP-1 activated by exposure to either H2O2 (100 μM) for 10 min or etoposide (80 μM) for 4 h. Whole cell extracts were collected in cell lysis buffer (10 mM Tris, pH 7.4, 100 mM sodium chloride, 1 mM EDTA, 1 mM EGTA, 1 mM sodium fluoride, 20 mM sodium pyrophosphate, 2 mM sodium vanadate, 1% Triton X-100, 10% glycerol, 0.1% SDS, 0.5% deoxycholate and 10 μl/ml protease inhibitor cocktail (Thermo Scientific) and total protein content determined via the BCA assay kit (Pierce, Rockford, IL). Cell lysates were assayed for PARP activity via the PAR ELISA assay as described by Liu et al. (2008). Briefly, PARP standards and total protein (5 μg) were placed in triplicate in antibody (PAR monoclonal; #4335-MC-100) coated plates and incubated overnight at 4 °C. Plates were then incubated with polyclonal PAR antibody (#4336-BPC-100) and incubated for 2 h at room temperature followed by an additional 1 h incubation with goat anti-rabbit HRP conjugated antibody at room temperature. Total PAR content was determined by the addition of SuperSignal ELISA pico substrate (Pierce, Rockford, IL) and luminescence measured on a SpectraMax M5 plate reader. Luminescence was compared to the standard curve and PAR content reported as picograms per mg of protein. Reagents for this protocol were purchased from Trevigen (Gaithersburg, MD) unless otherwise noted.
2.6. Zinc release assay
HEK293 and HEKn cells were treated with UA (0–30 μM), Zn (2 μM) or both for 24 h and whole cell lysates collected in lysis buffer as described above without the addition of EGTA or EDTA. PARP-1, XPA, APTX and SP1 were isolated from cells by immunoprecipitation. Cells were nomore than 75% confluent at the time of collection. Immunoprecipitation and measurement of zinc content was performed as described previously (Zhou et al., 2011; Cooper et al., 2013). Briefly, total protein was collected and desired proteins isolated by immunoprecipitation (500 μg in 500 μl) with primary antibodies (1:100 PARP-1, Cell Signaling Tech; 1:100 XPA, Abcam; 1:100 APTX, Abcam; and 1:50 SP1, Cell Signaling Tech). Protein samples and zinc standards (100 μl) were transferred to 96 well plates and zinc content measured by adding 10 μl of 1 mM 4, (2-pyridylazo)-resorcinol (PAR). Absorbance was measured at 493 nm using a SpectraMax M2 plate reader (Molecular Devices; Sunnyvale, CA). Wavelength scans for PAR absorbance of zinc standards and standards containing UA concentrations up to 50 μM overlapped indicating that the presence of experimental concentrations of UA did not alter the absorbance shift of PAR due to zinc. (data not shown). Immunoprecipitated protein concentrations were determined by the Bradford assay (Amresco, Solon, OH) and zinc content was normalized to sample protein concentration.
2.7. Statistical analyses
Data from a minimum of 3 separate experiments were pooled and statistical significance assessed by one-way ANOVA with Tukey's multiple comparison tests or two-way ANOVA with Bonferroni posttests conducted using GraphPad Prism 5.03 (San Diego, CA).
3. Results
3.1. Cytotoxicity of uranyl acetate
Uranium has been reported to be cytotoxic at high concentrations (>100 μM) in a number of cell lines (Stearns et al., 2005; Coryell and Stearns, 2006; Knobel et al., 2006; Periyakaruppan et al., 2007; Thiebault et al., 2007; Milgram et al., 2008; Periyakaruppan et al., 2009; Darolles et al., 2010; Rouas et al., 2010; Heintze et al., 2011; Orona and Tasat, 2012; Garmash et al., 2014), but there is less published information regarding lower concentrations of uranium. We compared uranyl acetate (UA) toxicity in human embryonic kidney cells (HEK293) and normal human keratinocytes (HEKn). Cell viability was measured in cells treated with increasing concentrations of UA (0– 100 μM) for 48 h. In keeping with the published findings, UA concentrations of up to 10 μM displayed limited cytotoxicity in HEK 293 or HEKn cells (Fig. 1A, B) and Jurkat cells were resistant to UA toxicity (not shown). At 100 μM UA, cell viability was 52.4% and 66.5% for HEK293 and HEKn cells, respectively. Density of HEK293 cells affected sensitivity to UA; subconfluent cells showed a decrease in tolerance to UA (Suppl. Fig. 1). Trypan blue staining confirmed the toxicity of UA at 30 and 100 μM (Suppl. Fig. 2).
Fig. 1.
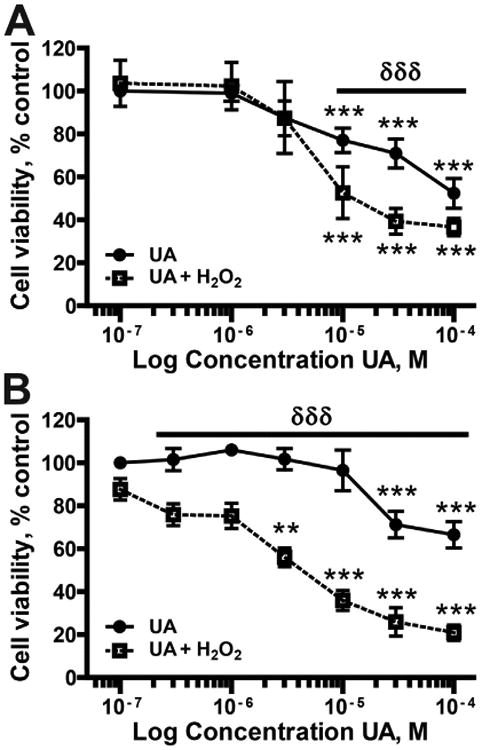
Cell viability following UA exposure. (A) HEK 293 cells or (B) normal keratinocytes (HEKn) were exposed to the indicated concentrations of UA for 48 h (closed circles, solid line). H2O2 (100 μM; 10 min: closed squares, dashed line) was added to a subset of samples at 24 h post-UA treatment and incubated for an additional 24 h. Cellular viability was determined using PrestoBlue Reagent. The data are presented as the means ± SEM, n = 3–4, **p < 0.01, ***p < 0.001 significantly different from exposure matched untreated controls; δδδp < 0.001 significantly different between UA and UA + H2O2 groups.
3.2. Uranyl acetate enhances cytotoxicity and DNA damage by hydrogen peroxide
UA pretreatment for 24 h enhances the cytotoxicity of H2O2 in both HEK293 and HEKn cells (Fig. 1A, B). UA treatment after H2O2 exposure had no effect on cell viability (Suppl. Fig. 3). Inhibition of DNA repair by UA is one possible mechanism to account for the increased DNA damage following UA pretreatment when compared to H2O2 alone. UA causes strand breaks as detected by increased pH2AX in a dose dependent manner (Fig 2A). Exposure to 10 μM UA for 24 h caused a 1.85 fold increase in pH2AX staining increasing to 2.95 fold at 100 μM UA. UA also increased the magnitude and persistence of DNA damage induced by ssUVR (Fig. 2B). Six h post-ssUVR exposure pH2AX staining was at control (no ssUVR) levels in cells that did not receive UA. In contrast, pH2AX staining was elevated 3.55-fold over initial values in cells incubated with 10 μM UA for 24 h before exposure to ssUVR. This finding suggests that UA interferes with DNA repair processes in the cells.
Fig. 2.
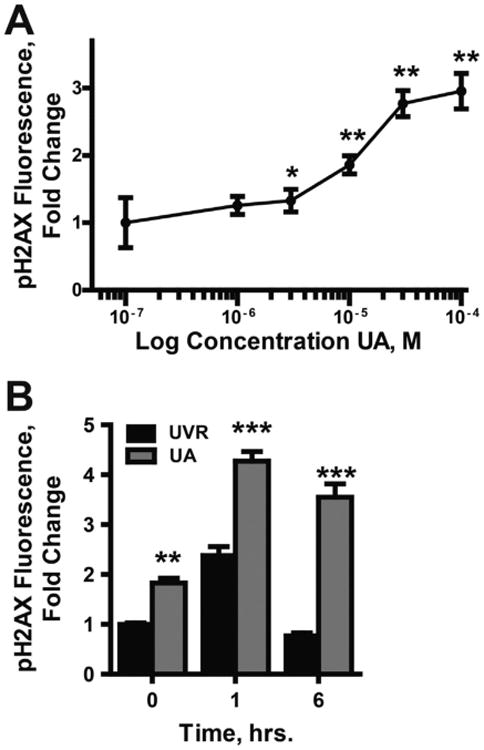
Enhanced DNA damage due to UA exposure. (A) HEK293 cells were treated with increasing concentrations of UA (0–100 μM) for 24 h. DNA damage was assessed via indirect immunofluorescence with anti-pH2AX antibodies. Images were collected with an Olympus IX70 equipped with a DP72 digital camera (Olympus America). Fluorescence intensity was quantified using cellSens Dimension 1.9 Count & Measure software (Olympus America). *p < 0.05, **p < 0.01 significantly different from untreated control; n = 3. (B) Cells were exposed to UA (10 μM) for 24 h then exposed to ssUVR (3 kJ/m2). Cell were fixed at zero, 1, and 6 h post-ssUVR exposure, then stained and fluorescence quantified as described in (A). Data is reported as fluorescence per nuclei normalized to the untreated control. Graphs represent a minimum of 10 images per treatment from 3 independent experiments. **p < 0.01; ***p < 0.001 significantly different from treatment matched t = 0; n = 3.
3.3. UA disrupts the zinc finger function of DNA repair proteins
Zinc finger proteins are molecular targets for certain metals and PARP-1 is one such protein. PARP-1 binds to DNA through two C3H1 zinc finger motifs and binding to damaged DNA activates PARP-1 enzymatic activity (Gibson and Kraus, 2012; Dantzer and Santoro, 2013; Langelier and Pascal, 2013). UA exposure alone does not decrease basal PARP activity and a modest increase in PARP activity is detected at 100 μM UA (Fig. 3A). In contrast, lower levels of UA inhibit PARP-1 activation (Fig. 3B). HEKn cells were pre-treated with UA for 24 h then exposed to etoposide (80 μM, 4 h) to induce DNA damage. UA treatment decreased PARP activation in a concentration-dependent manner with an inhibitory concentration (IC)50 value of 4.95 μM (Fig. 3B).
Fig. 3.
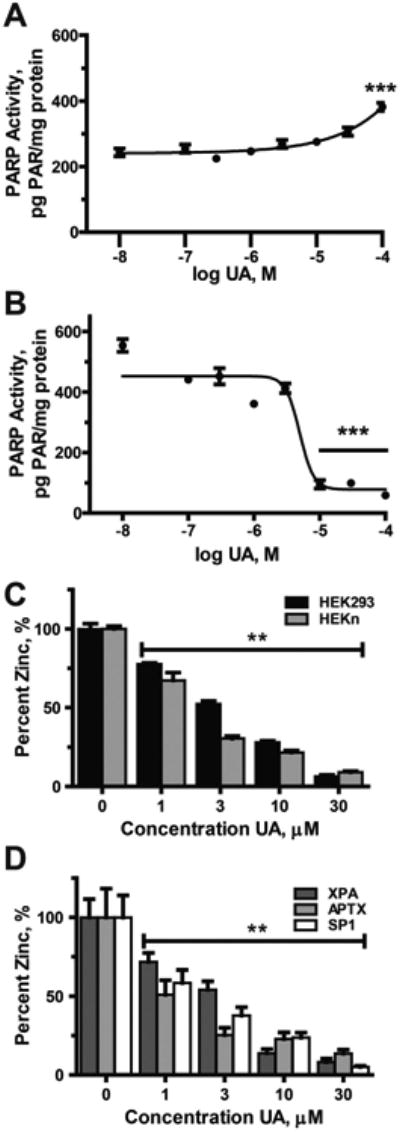
Inhibition of zinc finger-proteins by UA HEKn cells were treated with the indicated concentrations of UA for 24 h. A & B) Total protein was collected and PARP activity assessed via the PAR ELISA as described in “Experimental procedures”. A) PARP activity was stimulated by high concentration of UA alone. B) DNA damage-induced PARP activity (stimulated via the addition of etoposide (80 μM) for 4 h) inhibited by UA C & D). Cells were exposed to UA (0–30 μM) for 24 h. PARP-1 was immunoprecipitated from 500 μg of whole cell extract and zinc content of the protein determined via the Zinc Release Assay as described in “Experimental procedures”. C) UA induces loss of zinc from PARP-1 in both HEK293 (black bars) and HEKn (gray bars). D) Zinc assessment of immunoprecipitated XPA (dark gray bars), APTX (light gray bars) and SP1 (white bars) from HEKn cells. Graphs represent normalized means ± SEM of at least 3 independent experiments. **p < 0.01, ***p < 0.001 significantly different from untreated control.
PARP-1 DNA binding and activation requires zinc coordination within the zinc finger motifs (Langelier and Pascal, 2013). UA treatment of HEK 293 and HEKn cells for 24 h led to a concentration-dependent loss of zinc from PARP-1 protein isolated from exposed cells (Fig. 3C) suggesting that UA disrupts the zinc finger function of PARP-1 protein. We further investigated whether UA exposure led to zinc loss from other zinc finger proteins in HEKn cells (Fig. 3D). Xeroderma Pigmentosum, Complementation Group A (XPA) has a single C4 zinc finger that is sensitive to other carcinogenic metals such as arsenic (Piatek et al., 2008; Zhou et al., 2011; Hudson et al., 2015) and zinc content of XPA was reduced after treatment with UA. UA was previously reported to interfere with the DNA binding capacity of the C2H2 zinc finger protein specificity protein 1 (Sp1) (Hartsock et al., 2007) and UA treatment caused zinc loss from this protein as well as another C2H2 zinc finger protein, aprataxin (APTX). The IC50 values for zinc loss were between 1 and 3 μM for each of these zinc finger proteins suggesting that UA may interfere with DNA repair at much lower concentrations than required to cause DNA damage and cytotoxicity.
3.4. Zinc protects against the effects of UA on PARP activity and DNA damage
It has been reported that zinc is protective against uranium induced cytotoxicity (Hao et al., 2014) and since UA interfered with zinc finger function of several proteins (Fig. 3C&D), we tested whether zinc could offset the effects of UA on PARP-1 activity and DNA damage repair. Co-exposure of cells to UA and zinc largely counteracted the effect of UA on inhibition of PARP-1 activity (Fig. 4). Similarly, retention of DNA damage after exposure to solar simulated (ss) UVR (Fig. 5) was investigated. ssUVR was selected as an exposure because it causes both oxidative DNA damage and direct photo damage and these lesions are repaired by different DNA repair mechanisms. The 3 kJ/m2 dose used was experimentally determined to approximate 1.1 MED. The increase in pH2AX staining induced by ssUVR exposure alone was resolved within 6 h. Preincubation of cells with UA at 10 μM for 24 h significantly increased pH2AX staining above ssUVR alone 1 h post-ssUVR exposure (4 fold greater) and remained elevated at 6 h in contrast to the no UA treatment. ssUVR-induced DNA damage was equivalent to no UA treatment when cells were co-incubated with UA (10 μM) and zinc (2 μM) (Fig. 5A). Similar findings were obtained for detection of direct ssUVR DNA damage as measured by cyclobutane pyrimidine dimers (CPDs) (Fig 5B). DNA damage was retained at 6 h post-UVR exposure in cells pre-treated with UA (2 fold greater) and DNA damage was near UVR only levels in cells co-incubated with UA and zinc. As the chemical properties of UA and its interaction with H2O2 within the cellular environment are largely unknown, we investigated the impact of UA on DNA damage repair with an alternate inducer of DNA damage etoposide (Suppl. Fig. 4). Initial (1 h post-induction) and retained (6 h post-induction) levels of DNA damage assessed by ICC or by flow cytometry were comparable to those shown in Fig. 5. These findings suggest that UA inhibits DNA repair through disruption of zinc finger function and zinc is protective.
Fig. 4.
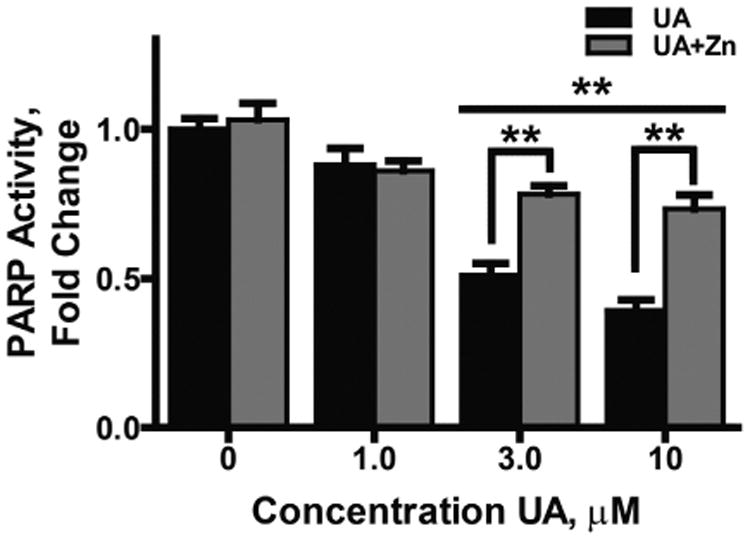
Supplemental zinc is protective for UA inhibition of PARP1 activity. HEKn cells were treated with increasing concentrations of UA (0–10 μM) or UA plus Zn (2 μM) for 24 h. A) PARP activity was stimulated by inducing DNA damage via the addition of etoposide (80 μM) for 4 h. Total protein was collected and PARP activity assessed via the PAR ELISA as described in “Experimental procedures.” Data was normalized to the untreated control and graph represents mean ± SEM of at least 3 independent experiments. **p<0.01; n = 3.
Fig. 5.
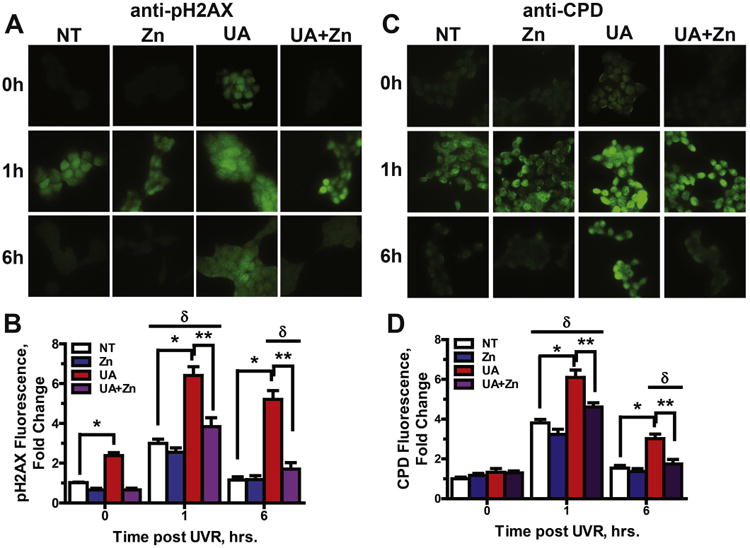
Retention of DNA damage following UA exposure is rescued by zinc. Immunocytochemistry of HEK293 cells was used to illustrate oxidative (A, pH2AX) and direct (B, CPDs) DNA damage. Cells were cultured on chamber slides and treated with UA (10 μM), Zn(2 μM), both or neither for 24 h then DNA damage was induced by exposure to ssUVR (3 kJ/m2). Cells were fixed at 0,1 and 6 h post-ssUVR exposure and stained for the DNA damage markers pH2AX (A) and CPDs (C). Images were collected with an Olympus IX70 equipped with a digital camera and fluorescence intensity was quantified using cellSens Dimension 1.9 Count & Measure software (Olympus America) and graphs shown (B, pH2AX and D, CPDs) are mean ± SEM of the intensity per nuclei of at least 10 images from each treatment group from 3 independent experiments. *p < 0.05; **p < 0.01 significantly different from indicated groups; δp < 0.01 significantly different from treatment matched t = 0 group.
4. Discussion
There is mounting evidence that the metal properties of uranium may contribute to adverse human health effects. Our results both support previous reports that uranium exposure leads to DNA damage both in vitro and in human populations, and provides a plausible mechanism for those observations through inhibition of DNA repair proteins. UA alone displayed limited cytotoxicity at levels below 10 μM in keeping with other reports in the literature where cytotoxicity is detected at uranium levels at or greater than 100 μM (Stearns et al., 2005; Periyakaruppan et al., 2007; Thiebault et al., 2007; Milgram et al., 2008; Rouas et al., 2010; Heintze et al., 2011; Orona and Tasat, 2012; Garmash et al., 2014). However, there is evidence that elevated blood uranium levels are associated with increased DNA damage (Popp et al., 2000; Guimaraes et al., 2010; Lourenco et al., 2013) and decreased DNA repair capacity in human populations (Au et al., 1995; Au et al., 1998; Au et al., 2010). In cells, uranium causes DNA damage and mutations (Stearns et al., 2005; Coryell and Stearns, 2006; Knobel et al., 2006; Wilson et al., 2014) and we find evidence for strand break in response to 10 μM UA in normal human keratinocytes. Uranium has been shown to cause DNA adducts and strand breaks in the presence of ascorbate (Wilson et al., 2014) and potentiate UV-induced strand breaks through a mechanism involving photoactivation of the uranyl ion (George et al., 2011; Wilson et al., 2015). In addition to these mechanisms, we find that low, non-cytotoxic concentrations of UA inhibit DNA repair and enhance UVR-induced DNA damage indicating that uranium may act as a co-carcinogen when combined with other DNA damaging insults. The potential of uranium to act as a co-carcinogen is further supported by population-based findings that uranium-exposed smokers had increased frequency of chromosomal aberrations when compared to non-exposed smokers (Prabhavathi et al., 2000). It is also important to note that the concentration of UA required to significantly disrupt zinc finger function and increase DNA damage is more than 10-fold lower than required to detect minimal cytotoxicity in this cell system. This indicates that significant impact of uranium on DNA damage and repair processes can occur at low micromolar concentrations.
Zinc fingers are protein structures where there is tetrahedral coordination of the zinc ion by cysteine (C) and histidine (H) residues. Our findings indicate that uranium is capable of disrupting zinc finger DNA repair proteins similar to what has been reported for other carcinogenic and co-carcinogenic metals such as arsenic (Beyersmann and Hartwig, 2008; Ding et al., 2009; Bal et al., 2011; Zhou et al., 2011; Hartwig, 2013; Sun et al., 2014; Zhou et al., 2014; Hudson et al., 2015). Past work on interactions of uranium with zinc finger proteins is limited, but proteomics analysis of uranium-interacting proteins identified PARP-1 (Dedieu et al., 2009), and uranium inhibits DNA binding of certain transcription factors including the C2H2 zinc finger proteins Sp1 and Aart (Hartsock et al., 2007). We find that UA inhibits PARP-1 activity and causes zinc loss from the protein. Furthermore, in contrast to arsenite, UA displays non-selective disruption of zinc fingers in DNA repair proteins; UA-dependent loss of zinc occurs in all three zinc finger configurations and there are C2H2, C3H1 and C4 ZF in DNA repair pathways. We previously reported that arsenite causes zinc loss from C3H1 and C4, but not C2H2, zinc finger proteins (Ding et al., 2009; Zhou et al., 2011; Zhou et al., 2014). The lack of zinc finger motif selectivity for uranium disruption suggests that uranium may be more deleterious to DNA repair inhibition than arsenite based on an increased number of potential targets in the DNA repair pathway. However, the evidence that uranium also inhibits non-zinc finger DNA binding proteins (Hartsock et al., 2007) indicates that UA has other mechanisms of action in addition to the disruption of zinc finger containing DNA repair proteins. Inhibition of DNA binding lacking zinc fingers may be due to binding of uranyl ions to carboxylate sites within protein structures (Huang et al., 2005).
Zinc is the critical ion within zinc finger structures, so we tested the potential for zinc to offset the actions of uranium. Our previous work demonstrated that supplemental zinc is protective against the effects of arsenite on PARP-1 function, DNA damage, and mutagenesis (Ding et al., 2009; Zhou et al., 2011; Cooper et al., 2013). Furthermore, zinc abolished arsenite enhancement of UV-induced DNA damage in mouse skin in vivo (Cooper et al., 2013). Similarly, co-treatment of cells with UA (10 μM) and Zn (2 μM) largely blocked UA inhibition of PARP-1 activity and Zn was protective for the uranium enhancement of DNA damage. These findings illustrate that modest levels of zinc are effective in counteracting these toxic responses to uranium and may offer an avenue for protection in at risk populations.
Our results may be relevant for populations vulnerable to chronic low-dose exposures to uranium and other metals from living in proximity to some of more than four thousand abandoned uranium mines in the Western US (USEPA, 2008a; USEPA, 2008b). In addition to uranium, these mine wastes include numerous other heavy metals and metalloids including arsenic (USEPA, 1995). These mine wastes create a strong potential for adjacent communities to be exposed to metal mixtures that may share mechanisms of toxicity as we have found for uranium and arsenic inhibition of DNA repair. Many of these abandoned waste sites are located on or in proximity to Native American lands in the West where existing disparities in health raise additional concern of vulnerability to environmental toxicant insults. Studies by our group and others have indicated that community members living in proximity to uranium contaminated sites have an increased likelihood of developing chronic diseases including hypertension, kidney disease, and diabetes (Wagner et al., 2010; Hund etal., 2015); as well as immune dysfunction and autoimmunity (Lewis, 2013; Lu-Fritts et al., 2014). Our research group has also found that urine uranium concentrations within the Navajo population are greater than the national norms, underscoring the need for additional studies within these exposed communities (Lewis, 2013). Further studies to understand the mechanisms of action of metals and metal mixtures will help to expand our knowledge of toxicity and linkage to adverse health effects in exposed communities.
Supplementary Material
Acknowledgments
This project was supported in part by the National Center for Research Resources and the National Center for Advancing Translational Sciences of the National Institutes of Health through Grant Number 8UL1TR000041, the University of New Mexico Clinical and Translational Science Center and the New Mexico Center for the Advancement of Research Engagement through grant number UL1TR001449 and Science on Health Disparities (NM CARES Health Disparities Center) through grant number NIMHD/USEPA P20 MD004811. EJD received support from The Academic Science Education and Research Training (ASERT) program (2K12GM088021) and RT received support from the Initiative for Minority Student Development (IMSD) program (5R25GM060201-15).
Abbreviations
- DAPI
4′,6-diamidino-2-phenylindole
- APTX
aprataxin
- As
arsenite
- CPDs
cyclobutane pyrimidine dimers
- C
cysteine
- DMEM
Dulbecco's modified Eagle's medium
- H
histidine
- HEK293
human embryonic kidney 293
- ICP-MS
inductively coupled plasma mass spectrometry
- HEKn
normal human neonatal epidermal keratinocytes
- PBS
phosphate-buffered saline
- pH2AX
phospho-histone H2A.X
- PARP-1
poly(ADP-ribose) polymerase-1
- SP-1
specificity protein-1
- ssUVR
solar-simulated ultraviolet radiation
- UA
uranyl acetate
- XPA
Xeroderma Pigmentosum, Complementation Group A
- Zn
zinc
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.taap.2015.11.017.
Conflict of interest: The authors declare that there are no conflicts of interest.
Transparency document: The Transparency document associated with this article can be found, in the online version.
References
- Au WW, Lane RG, Legator MS, Whorton EB, Wilkinson GS, Gabehart GJ. Biomarker monitoring of a population residing near uranium mining activities. Environ Health Perspect. 1995;103:466–470. doi: 10.1289/ehp.95103466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au WW, McConnell MA, Wilkinson GS, Ramanujam VM, Alcock N. Population monitoring: experience with residents exposed to uranium mining/milling waste. Mutat Res. 1998;405:237–245. doi: 10.1016/s0027-5107(98)00141-9. [DOI] [PubMed] [Google Scholar]
- Au WW, Giri AK, Ruchirawat M. Challenge assay: a functional biomarker for exposure-induced DNA repair deficiency and for risk of cancer. Int J Hyg Environ Health. 2010;213:32–39. doi: 10.1016/j.ijheh.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Bal W, Protas AM, Kasprzak KS. Genotoxicity of metal ions: chemical insights. Met Ions Life Sci. 2011;8:319–373. [PubMed] [Google Scholar]
- Beyersmann D, Hartwig A. Carcinogenic metal compounds: recent insight into molecular and cellular mechanisms. Arch Toxicol. 2008;82:493–512. doi: 10.1007/s00204-008-0313-y. [DOI] [PubMed] [Google Scholar]
- Briner W. The toxicity of depleted uranium. Int J Environ Res Public Health. 2010;7:303–313. doi: 10.3390/ijerph7010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugge D, Buchner V. Health effects of uranium: new research findings. Rev Environ Health. 2011;26:231–249. doi: 10.1515/reveh.2011.032. [DOI] [PubMed] [Google Scholar]
- Brugge D, de Lemos JL, Oldmixon B. Exposure pathways and health effects associated with chemical and radiological toxicity of natural uranium: a review. Rev Environ Health. 2005;20:177–193. doi: 10.1515/reveh.2005.20.3.177. [DOI] [PubMed] [Google Scholar]
- Burns FJ, Uddin AN, Wu F, Nadas A, Rossman TG. Arsenic-induced enhancement of ultraviolet radiation carcinogenesis in mouse skin: a dose–response study. Environ Health Perspect. 2004;112:599–603. doi: 10.1289/ehp.6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KL, King BS, Sandoval MM, Liu KJ, Hudson LG. Reduction of arsenite-enhanced ultraviolet radiation-induced DNA damage by supplemental zinc. Toxicol Appl Pharmacol. 2013;269:81–88. doi: 10.1016/j.taap.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coryell VH, Stearns DM. Molecular analysis of hprt mutations generated in Chinese hamster ovary EM9 cells by uranyl acetate, by hydrogen peroxide, and spontaneously. Mol Carcinog. 2006;45:60–72. doi: 10.1002/mc.20155. [DOI] [PubMed] [Google Scholar]
- Craft E, Abu-Qare A, Flaherty M, Garofolo M, Rincavage H, Abou-Donia M. Depleted and natural uranium: chemistry and toxicological effects. J Toxicol Environ Health B Crit Rev. 2004;7:297–317. doi: 10.1080/10937400490452714. [DOI] [PubMed] [Google Scholar]
- Dantzer F, Santoro R. The expanding role of PARPs in the establishment and maintenance of heterochromatin. FEBS J. 2013;280:3508–3518. doi: 10.1111/febs.12368. [DOI] [PubMed] [Google Scholar]
- Darolles C, Broggio D, Feugier A, Frelon S, Dublineau I, De Meo M, Petitot F. Different genotoxic profiles between depleted and enriched uranium. Toxicol Lett. 2010;192:337–348. doi: 10.1016/j.toxlet.2009.11.009. [DOI] [PubMed] [Google Scholar]
- Dedieu A, Berenguer F, Basset C, Prat O, Quemeneur E, Pible O, Vidaud C. Identification of uranyl binding proteins from human kidney-2 cell extracts by immobilized uranyl affinity chromatography and mass spectrometry. J Chromatogr A. 2009;1216:5365–5376. doi: 10.1016/j.chroma.2009.05.023. [DOI] [PubMed] [Google Scholar]
- deLemos JL, Brugge D, Cajero M, Downs M, Durant JL, George CM, Henio-Adeky S, Nez T, Manning T, Rock T, Seschillie B, Shuey C, Lewis J. Development of risk maps to minimize uranium exposures in the Navajo Churchrock mining district. Environ Heal. 2009;8:29. doi: 10.1186/1476-069X-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W, Liu W, Cooper KL, Qin XJ, de Souza Bergo PL, Hudson LG, Liu KJ. Inhibition of poly(ADP-ribose) polymerase-1 by arsenite interferes with repair of oxidative DNA damage. J Biol Chem. 2009;284:6809–6817. doi: 10.1074/jbc.M805566200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmash SA, Smirnova VS, Karp OE, Usacheva AM, Berezhnov AV, Ivanov VE, Chernikov AV, Bruskov VI, Gudkov SV. Pro-oxidative, genotoxic and cytotoxic properties of uranyl ions. J Environ Radioact. 2014;127:163–170. doi: 10.1016/j.jenvrad.2012.12.009. [DOI] [PubMed] [Google Scholar]
- George SA, Whittaker AM, Stearns DM. Photoactivated uranyl ion produces single strand breaks in plasmid DNA. Chem Res Toxicol. 2011;24:1830–1832. doi: 10.1021/tx200410x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- Guimaraes AC, Antunes LM, Ribeiro HF, dos Santos AK, Cardoso PC, de Lima PL, Seabra AD, Pontes TB, Pessoa C, de Moraes MO, Cavalcanti BC, Sombra CM, Bahia Mde O, Burbano RR. Cytogenetic biomonitoring of inhabitants of a large uranium mineralization area: the municipalities of Monte Alegre, Prainha, and Alenquer, in the State of Para, Brazil. Cell Biol Toxicol. 2010;26:403–419. doi: 10.1007/s10565-010-9152-8. [DOI] [PubMed] [Google Scholar]
- Hao Y, Ren J, Liu C, Li H, Liu J, Yang Z, Li R, Su Y. Zinc protects human kidney cells from depleted uranium-induced apoptosis. Basic Clin Pharmacol Toxicol. 2014;114:271–280. doi: 10.1111/bcpt.12167. [DOI] [PubMed] [Google Scholar]
- Hartsock WJ, Cohen JD, Segal DJ. Uranyl acetate as a direct inhibitor of DNA-binding proteins. Chem Res Toxicol. 2007;20:784–789. doi: 10.1021/tx600347k. [DOI] [PubMed] [Google Scholar]
- Hartwig A. Metal interaction with redox regulation: an integrating concept in metal carcinogenesis? Free Radic Biol Med. 2013;55:63–72. doi: 10.1016/j.freeradbiomed.2012.11.009. [DOI] [PubMed] [Google Scholar]
- Heintze E, Aguilera C, Davis M, Fricker A, Li Q, Martinez J, Gage MJ. Toxicity of depleted uranium complexes is independent of p53 activity. J Inorg Biochem. 2011;105:142–148. doi: 10.1016/j.jinorgbio.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Chaudhary S, Van Horn JD. Uranyl-peptide interactions in carbonate solution with DAHK and derivatives. Inorg Chem. 2005;44:813–815. doi: 10.1021/ic049528l. [DOI] [PubMed] [Google Scholar]
- Hudson LG, Cooper KL, Atlas SR, King BK, Liu KJ. Arsenic interaction with zinc finger motifs. In: States JC, editor. Arsenic: Exposure Sources, Health Risks and Mechanisms of Toxicity. Wiley & Sons Inc.; 2015. pp. 291–314. [Google Scholar]
- Hund L, Bedrick E, Miller C, Huerta G, Nez T, Ramone S, Shuey C, Cajero M, Lewis J. A Bayesian framework for estimating disease risk due to exposure to uranium mine and mill waste on the Navajo Nation. J R Stat Soc Ser A (Stat Soc) 2015 [Google Scholar]
- Knobel Y, Glei M, Weise A, Osswald K, Schaferhenrich A, Richter KK, Claussen U, Pool-Zobel BL. Uranyl nitrilotriacetate, a stabilized salt of uranium, is genotoxic in nontransformed human colon cells and in the human colon adenoma cell line LT97. Toxicol Sci. 2006;93:286–297. doi: 10.1093/toxsci/kfl060. [DOI] [PubMed] [Google Scholar]
- Langelier MF, Pascal JM. PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Curr Opin Struct Biol. 2013;23:134–143. doi: 10.1016/j.sbi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JL. DiNEH Project Update: Survey → Clinic → Mechanisms. USEPA Stakeholder Workshop 2013. 2013 http://www.epa.gov/region9/superfund/navajo-nation/pdf/stakeholder/2013/dr-lewis-di-neh-project-update2013.pdf.
- Liu X, Palma J, Kinders R, Shi Y, Donawho C, Ellis PA, Rodriguez LE, Colon-Lopez M, Saltarelli M, LeBlond D, Lin CT, Frost DJ, Luo Y, Giranda VL. An enzyme-linked immunosorbent poly(ADP-ribose) polymerase biomarker assay for clinical trials of PARP inhibitors. Anal Biochem. 2008;381:240–247. doi: 10.1016/j.ab.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Lourenco J, Pereira R, Pinto F, Caetano T, Silva A, Carvalheiro T, Guimaraes A, Goncalves F, Paiva A, Mendo S. Biomonitoring a human population inhabiting nearby a deactivated uranium mine. Toxicology. 2013;305:89–98. doi: 10.1016/j.tox.2013.01.011. [DOI] [PubMed] [Google Scholar]
- Lu-Fritts PY, Kottyan LC, James JA, Xie C, Buckholz JM, Pinney SM, Harley JB. Association of systemic lupus erythematosus with uranium exposure in a community living near a uranium-processing plant: a nested case–control study. Arthritis Rheum. 2014;66:3105–3112. doi: 10.1002/art.38786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maret W. New perspectives of zinc coordination environments in proteins. J Inorg Biochem. 2012;111:110–116. doi: 10.1016/j.jinorgbio.2011.11.018. [DOI] [PubMed] [Google Scholar]
- Milgram S, Carriere M, Thiebault C, Malaval L, Gouget B. Cytotoxic and phenotypic effects of uranium and lead on osteoblastic cells are highly dependent on metal speciation. Toxicology. 2008;250:62–69. doi: 10.1016/j.tox.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Miller AC, McClain D. A review of depleted uranium biological effects: in vitro and in vivo studies. Rev Environ Health. 2007;22:75–89. doi: 10.1515/reveh.2007.22.1.75. [DOI] [PubMed] [Google Scholar]
- Orona NS, Tasat DR. Uranyl nitrate-exposed rat alveolar macrophages cell death: influence of superoxide anion and TNF alpha mediators. Toxicol Appl Pharmacol. 2012;261:309–316. doi: 10.1016/j.taap.2012.04.022. [DOI] [PubMed] [Google Scholar]
- Periyakaruppan A, Kumar F, Sarkar S, Sharma CS, Ramesh GT. Uranium induces oxidative stress in lung epithelial cells. Arch Toxicol. 2007;81:389–395. doi: 10.1007/s00204-006-0167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periyakaruppan A, Sarkar S, Ravichandran P, Sadanandan B, Sharma CS, Ramesh V, Hall JC, Thomas R, Wilson BL, Ramesh GT. Uranium induces apoptosis in lung epithelial cells. Arch Toxicol. 2009;83:595–600. doi: 10.1007/s00204-008-0396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatek K, Schwerdtle T, Hartwig A, Bal W. Monomethylarsonous acid destroys a tetrathiolate zinc finger much more efficiently than inorganic arsenite: mechanistic considerations and consequences for DNA repair inhibition. Chem Res Toxicol. 2008;21:600–606. doi: 10.1021/tx7003135. [DOI] [PubMed] [Google Scholar]
- Popp W, Plappert U, Muller WU, Rehn B, Schneider J, Braun A, Bauer PC, Vahrenholz C, Presek P, Brauksiepe A, Enderle G, Wust T, Bruch J, Fliedner TM, Konietzko N, Streffer C, Woitowitz HJ, Norpoth K. Bio-markers of genetic damage and inflammation in blood and bronchoalveolar lavage fluid among former German uranium miners: a pilot study. Radiat Environ Biophys. 2000;39:275–282. doi: 10.1007/s004110000072. [DOI] [PubMed] [Google Scholar]
- Prabhavathi PA, Fatima SK, Rao MS, Reddy PP. Analysis of chromosomal aberration frequencies in the peripheral blood lymphocytes of smokers exposed to uranyl compounds. Mutat Res. 2000;466:37–41. doi: 10.1016/s1383-5718(99)00236-3. [DOI] [PubMed] [Google Scholar]
- Qin XJ, Hudson LG, Liu W, Timmins GS, Liu KJ. Low concentration of arsenite exacerbates UVR-induced DNA strand breaks by inhibiting PARP-1 activity. Toxicol Appl Pharmacol. 2008;232:41–50. doi: 10.1016/j.taap.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman TG, Uddin AN, Burns FJ. Evidence that arsenite acts as a cocarcinogen in skin cancer. Toxicol Appl Pharmacol. 2004;198:394–404. doi: 10.1016/j.taap.2003.10.016. [DOI] [PubMed] [Google Scholar]
- Rouas C, Bensoussan H, Suhard D, Tessier C, Grandcolas L, Rebiere F, Dublineau I, Taouis M, Pallardy M, Lestaevel P, Gueguen Y. Distribution of soluble uranium in the nuclear cell compartment at subtoxic concentrations. Chem Res Toxicol. 2010;23:1883–1889. doi: 10.1021/tx100168c. [DOI] [PubMed] [Google Scholar]
- Stearns DM, Yazzie M, Bradley AS, Coryell VH, Shelley JT, Ashby A, Asplund CS, Lantz RC. Uranyl acetate induces hprt mutations and uranium-DNA adducts in Chinese hamster ovary EM9 cells. Mutagenesis. 2005;20:417–423. doi: 10.1093/mutage/gei056. [DOI] [PubMed] [Google Scholar]
- Sun X, Zhou X, Du L, Liu W, Liu Y, Hudson LG, Liu KJ. Arsenite binding-induced zinc loss from PARP-1 is equivalent to zinc deficiency in reducing PARP-1 activity, leading to inhibition of DNA repair. Toxicol Appl Pharmacol. 2014;274:313–318. doi: 10.1016/j.taap.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebault C, Carriere M, Milgram S, Simon A, Avoscan L, Gouget B. Uranium induces apoptosis and is genotoxic to normal rat kidney (NRK-52E) proximal cells. Toxicol Sci. 2007;98:479–487. doi: 10.1093/toxsci/kfm130. [DOI] [PubMed] [Google Scholar]
- USEPA. Extraction and beneficiation of ores and minerals. In: Waste, O.o.S, editor. Volume 5, Uranium. USEPA Office of Solid Waste; Washington, DC: 1995. [Google Scholar]
- USEPA. Technical report on technologically enhanced naturally occurring radioactive materials from uranium mining. In: Air, O.o.R.a.I, editor. Volume 1: Mining and Reclaimation Background. USEPA; Washington, DC: 2008a. [Google Scholar]
- USEPA. Technical report on technologically enhanced naturally occurring radioactive materials from uranium mining. In: Air, O.o.R.a. I, editor. Volume 2: Investigation of Potential Health, Geographic, and Environmental Issues of Abandoned Uranium Mines. USEPA; Washington, DC: 2008b. [Google Scholar]
- Wagner SE, Burch JB, Bottai M, Pinney SM, Puett R, Porter D, Vena JE, Hebert JR. Hypertension and hematologic parameters in a community near a uranium processing facility. Environ Res. 2010;110:786–797. doi: 10.1016/j.envres.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J, Young A, Civitello ER, Stearns DM. Analysis of heat-labile sites generated by reactions of depleted uranium and ascorbate in plasmid DNA. J Biol Inorg Chem. 2014;19:45–57. doi: 10.1007/s00775-013-1057-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J, Zuniga MC, Yazzie F, Stearns DM. Synergistic cytotoxicity and DNA strand breaks in cells and plasmid DNA exposed to uranyl acetate and ultraviolet radiation. J Appl Toxicol. 2015;35:338–349. doi: 10.1002/jat.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkiewicz-Kucharczyk A, Bal W. Damage of zinc fingers in DNA repair proteins, a novel molecular mechanism in carcinogenesis. Toxicol Lett. 2006;162:29–42. doi: 10.1016/j.toxlet.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Zhou X, Sun X, Cooper KL, Wang F, Liu KJ, Hudson LG. Arsenite interacts selectively with zinc finger proteins containing C3H1 or C4 motifs. J Biol Chem. 2011;286:22855–22863. doi: 10.1074/jbc.M111.232926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Sun X, Mobarak C, Gandolfi AJ, Burchiel SW, Hudson LG, Liu KJ. Differential binding of monomethylarsonous acid compared to arsenite and arsenic trioxide with zinc finger peptides and proteins. Chem Res Toxicol. 2014;27:690–698. doi: 10.1021/tx500022j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


