Abstract
The cerebral cortex controls our most distinguishing higher cognitive functions. Human-specific gene expression differences are abundant in the cerebral cortex, yet we have only begun to understand how these variations impact brain function. This review discusses the current evidence linking non-coding regulatory DNA changes, including enhancers, with neocortical evolution. Functional interrogation using animal models reveals converging roles for our genome in key aspects of cortical development including progenitor cell cycle and neuronal signaling. New technologies, including iPS cells and organoids, offer potential alternatives to modeling evolutionary modifications in a relevant species context. Several diseases rooted in the cerebral cortex uniquely manifest in humans compared to other primates, thus highlighting the importance of understanding human brain differences. Future studies of regulatory loci, including those implicated in disease, will collectively help elucidate key cellular and genetic mechanisms underlying our distinguishing cognitive traits.
Keywords: corticogenesis, enhancer, evolution, neocortex, stem cell
Introduction
A large six-layered neocortex is a unique feature of mammalian brains. This specialized outer covering of the brain controls our higher cognitive functions including abstract thought and language, which together help uniquely define us as humans. Our distinguishing cognitive capacities are specified within discrete cortical areas and are driven by dynamic communication between neurons of the neocortex and other brain regions, as well as glial cell populations (including oligodendrocytes, microglia, and astrocytes). Neurons are initially generated during human embryonic and early fetal development, where they migrate to appropriate regions and begin establishing functional connections during fetal and postnatal stages (Fig. 1). Disruptions to cerebral cortex function arising during either development or adulthood, can result in neurodevelopmental and neurode-generative disorders. Remarkably, several diseases affecting the cerebral cortex, such as autism spectrum disorder, primarily manifest in humans compared to other primates. Therefore, elucidating unique features of human neocortical development and function may help yield new insights into the inner workings of the brain and etiology of some neurological diseases.
Figure 1.
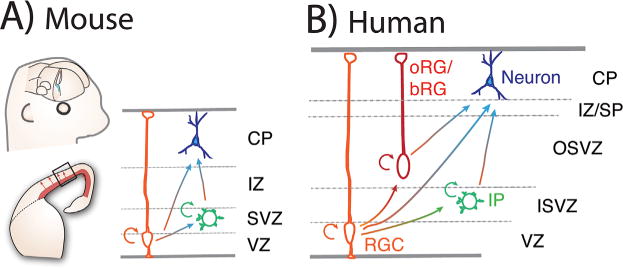
Overview of cortical neurogenesis in mice and humans. A: Cartoon representation of the embryonic mouse neocortex with a whole mount view (top left); 1/2 coronal section (bottom left) and high magnification view (right, boxed region from left). B: Simplified cartoon representation of neocortical section from human. For both A and B, the major cell populations and primary cortical layers are indicated. Major progenitors include: radial glial cell (orange, RGC); intermediate progenitor (green, IP); and outer radial glia/basal radial glia (red, oRG/bRG). RGCs undergo symmetric divisions to self-renew (shown as a half circle arrow). RGCs also undergo asymmetric divisions to generate new RGCs, IPs, and neurons (blue). Humans and other nonhuman primate neocortices contain abundant oRG/bRGs. In addition to RGCs, oRGs and IPs also produce neurons, as indicated by arrows. Major cortical layers of the mouse include cortical plate (CP), intermediate zone (IZ), sub-ventricular zone (SVZ), and ventricular zone (VZ). Major cortical layers of primate brains, including humans include the VZ, CP, IZ/sub-plate (IZ/SP). Primates also contain an expanded basal ventricular zone called the outer sub-ventricular zone (OSVZ) as well as an inner sub-ventricular zone (ISVZ). The mouse is shown for comparison with human as many functional studies have been performed in mice.
The human neocortex has many characteristic anatomical features [1–3]. Our neocortex, like that of some primates and mammals, is gyrencephalic, containing folds called gyri and sulci. Gyrencephaly is notably absent in rodents and some mammals such as manatees, which instead have a smooth neocortex [4]. These convolutions provide an expanded surface area and increased neuronal capacity. In addition, human neocortices are enlarged in volume, up to 2–3 times larger than chimpanzee neocortices [5]. Expansions also occur in the radial neocortical dimension such that primates, including humans, have enlarged supragranular layers (layers I–III), where neuronal connections are established between different cortical regions. Our neocortices also contain significantly more neurons than that of non-human primates (and approximately 1,000 times more than mice) [1]. Neurons of big-brained species, including humans and great apes, include von Economo neurons, which are large cells located in restricted cortical regions and postulated to promote rapid neuronal communication [6]. Beyond species-specific distribution in number and type, human neocortical neurons have a higher density of synapses and larger dendritic spines, which enhance neuronal communication [3].
Much of the structural distinctions in human and primate neocortices has been attributed to qualitative and quantitative differences in neural progenitor populations and proliferative behavior. Progenitors of humans, nonhuman primates, and rodents exhibit different cell cycle dynamics [1, 7–9], which vary across cortical areas [10]. Notably, humans have abundant and diverse basal progenitor populations, including basal/outer radial glia (termed bRGs or oRGs), located within an expanded outer sub-ventricular zone (OSVZ) [11–14] (Fig. 1). Progenitors of the OSVZ are thought to help expand the neuronal population and contribute to enlarged supragranular layers [10, 15, 16]. Live imaging of macaque and human oRGs reveal these progenitors are remarkably proliferative [14, 17]. These data increasingly lend support to the radial unit hypothesis, first posited in 1988 by Pasko Rakic, which predicted that the number and proliferative capacity of progenitor cells contributes to evolutionary neocortical differences, including distinct cytoarchitecture and size [7]. Prolonged duration of both fetal development and the neurogenic period, together with altered cell cycle dynamics, are thus posited to explain human-specific differences in neuron number and overall size of the neocortex [15, 18]. There is increasing empirical support for this hypothesis, as noted in this review.
Beyond defining anatomical and cellular differences, recent research has begun to unearth the genetic basis for our distinctively human cognitive faculties. These advances have been made possible with the advent of new genomic technologies, and collection of DNA sequences enabling comparative analyses between humans and non-human primates. Diverse genomic changes implicated in brain evolution include species-specific alterations to coding sequences, regulatory DNA, non-coding RNAs, and gene gains or losses [1, 2, 19]. Identification of these differences is no longer the limiting step in understanding the genetic basis of human neocortical evolution. Rather, the major challenge is our collective ability to assess function of these differences. This review will discuss examples of human genetic modifications, starting with an overview of gene expression differences, many of which define human oRG/bRG progenitors. The bulk of this discussion will focus on non-coding regulatory elements, emphasizing human accelerated enhancers that have been functionally implicated in human neocortical development and evolution. In many cases, these examples support the model that genetic modifications have converging influential roles in neuronal signaling and cell cycle, that together help explain unique aspects of our brains.
Genomic studies expose gene expression differences distinct to the human neocortex
Comparative genomic studies using human, chimpanzee, rhesus macaque, and mice have uncovered human-specific spatial and temporal differences in neocortical gene expression [20–24]. Surveys of early postnatal human, chimpanzee, and rhesus macaque prefrontal cortices reveal subsets of human genes with delayed expression relative to other primates [21]. Pletikos et al. [22] noted differential human gene expression during prenatal stages and between cortical areas, including human-specific differences in gene expression relative to the macaque. In an effort to increase the spatial resolution of evolutionary gene expression differences specifically in the cortex, several groups have analyzed cortical tissue sections and layers. Using laser capture microscopy and RNA sequencing of cortical layers of human and mouse fetuses, Fietz et al. [24] discovered species-specific gene expression differences in inner and outer germinal zones. Likewise, Lui et al. [25] used differential gene co-expression analyses of serial sections to discover genes highly expressed in human, but not mouse, radial glia (Fig. 1). Among 18 genes discovered, the growth factor PDGFD is particularly interesting given that along with its receptor, PDGFRβ, it is required for radial glia cell cycle progression [25].
This exemplifies how human-specific gene expression changes may influence proliferation of key neocortical progenitors. Collectively, these genomic studies uncovered new temporal and spatial patterns of human-specific gene expression in the neocortex.
A potentially confounding feature of aforementioned analyses is the use of neocortical tissue that contains heterogeneous cell populations. This is particularly important given that one distinguishing characteristic of human brains relative to some species is abundant oRG/bRG progenitors. Studies by three independent groups recently tackled this issue by assessing gene expression in isolated human neural progenitors. Two studies by the same group used microfluidics and RNA sequencing to discover a repertoire of genes expressed in human but not mouse radial glia [26], and in human oRG/bRGs [17]. Using RNA sequencing of FAC-sorted individual progenitors from human, ferret, and mouse progenitors, Johnson et al. [27] also identified genes enriched in human oRG/bRGs, including target genes of the proneurogenic transcription factor Neurogenin. Likewise, Fietz et al. identified 56 genes preferentially expressed in human oRG/bRG [28]. Altogether, these analyses at a single progenitor level have uncovered genes expressed in human oRG/bRGs. Given the abundance of these progenitors in human brains, it is of interest to better understand functions of these genes.
Many observed gene expression differences are likely due to human-specific gene duplications [28–31] and gene deletions [32]. Notably, several human-specific duplicate genes are expressed in the neocortex [30, 31]. In a seminal study, Charrier et al. [29] used mouse models to show that human-specific duplications in SRGAP2 inhibit neuronal spine maturation and delay neuronal migration. Thus, in humans, this gene family may help prolong neurodevelopment and increase signal processing. In the Fietz et al. study described above, gene expression differences in ARHGAP11B are due to a human-specific duplication after our divergence from chimpanzees [28]. The authors demonstrated that transient over-expression of this gene by in utero electroporation of mouse embryonic brains, led to increased basal progenitor proliferation. Thus, it is posited that human-specific ARHGAP11B expression in the neocortex promotes amplification of the neuronal population, brain folding, and potentially brain size. This notion conforms with the idea that having more basal progenitors in humans impacts brain differences. Altogether these genome-wide analyses, along with detailed functional analysis of individual loci, indicate that gene expression differences are widespread and relevant for human brain evolution (also see Box 1).
Box 1. Genetic modifications of trans-regulators impact neocortical functions.
Point mutations in coding regions of transcription factors have long been hypothesized to promote human-specific neocortical function. Transcription factors are ideally suited to influence brain traits by modulating expression of downstream genes. Notably, comparison of human and chimpanzee brain tissue reveal differential expression of 90 transcription factors [82]]. Among these are 33 members of the KRAB-ZNF family, which is particular interesting as these transcription factors have accumulated a disproportionate high number of amino acid substitutions in humans compared to chimpanzees. Another transcription factor strongly implicated in human brain traits is FOXP2. Relative to non-human primates, human FOXP2 uniquely contains two amino acid substitutions [83, 84]. Foxp2 is enriched in excitatory neurons of the cerebral cortex [85] and FOXP2 mutations cause speech and language disorders [86, 87]. These observations led to the hypothesis that FOXP2 has an important role in the emergence of language in humans. Interestingly, studies of humanized knock-in mice (Foxp2hum/hum) demonstrate that evolutionary modifications in FOXP2 sequence alter neuronal dendrite morphology [85]. In pups, this is associated with altered vocalization patterns, however, a recent study indicates this phenotype is transient and absent in Foxp2hum/hum adults [88]. These coding substitutions impact downstream transcriptional gene expression as shown by Konopka et al. [89]. Thus, evolutionary changes in FOXP2 protein function alter neuronal behavior and in doing so may contribute to human language and vocalization capacities. FOXP2 also contains regulatory non-coding mutations, though these have not yet been shown to impact in vivo function [90]. Beyond FOXP2, there are just a few other examples of evolutionarily divergent coding changes that affect brain function. ASPM and CDK5RAP2 are outstanding candidates because both human genes are mutated in microcephaly (a disorder of reduced brain size), and genetic variation in these loci correlates with brain size [91]. Beyond transcription factors, changes in non-coding regulatory RNAs that influence gene expression are also strongly implicated in brain evolution. Both human-specific and primate-specific miRNAs are expressed in the developing brain [15, 92, 93]. Of note, there is an enrichment of primate-specific miRNAs that target cell cycle genes, consistent with the notion that cell cycle modifications play an instrumental role in brain evolution [27]. Another class of non-coding RNAs are long non-coding RNAs (lncRNAs). A recent single-cell analysis of human and mouse brains showed that many lncRNAs are differentially expressed between species and between progenitor and neuronal populations [93]. Thus, lncRNAs have emerged as an important class of non-coding transcripts highly relevant for human cortical evolution.
Human-specific non-coding regulatory DNA changes influence neocortical functions
Many species-specific changes in gene expression are likely explained by differential cis-transcriptional regulation. King and Wilson [33] first hypothesized that molecular alterations affecting non-coding regulatory DNA are especially critical for species-specific differences between humans and chimpanzees. The ability to sequence genomes from humans and nonhuman primates has now provided some empirical support for this hypothesis, revealing modifications to promoters and enhancers over the course of evolution [34, 35].
McClean et al. [32] showed that 510 human-specific gene deletions, each on average about 3 Kb, are found almost entirely in non-coding regions, often nearby genes involved in neural function. Notably, one human-specific deletion on chromosome 9 removes an enhancer located adjacent to GADD45G, a gene expressed in the developing neocortex known to repress cell proliferation. The authors speculate that human-specific loss of this enhancer promotes progenitor proliferation and neuron production. This tantalizing model fits with the idea that modifications to cell cycle influence brain evolution, however, it remains to be formally tested what the impact is of this or any enhancer deletion upon neocortical function.
Genome-wide studies have also exposed evolutionary differences in epigenetic features, which can demarcate enhancers [36]. Remarkably, DNA methylation marks can distinguish Neanderthals and Denisovans, both Homo species [37]. Gokhman et al. [37] assessed DNA methylation by taking advantage of the observation that over time there is natural decay of methylated cytosines to thymines, and unmethylated cytosines to uracils. Thus, the C:T ratio is used as a proxy for DNA methylation in ancient samples. Other studies have observed enhancer mark differences distinguishing humans from other species. Enhancers predicted by p300 ChIP-seq analysis of the developing mouse and human fetal neocortex revealed that one-third of these are poorly conserved between mice and humans [38] (Table 1). A recent study by Reilly et al. [39] further supported this finding, by profiling genome-wide histone acetylation and methylation marks in human, rhesus macaque, and mouse neocortices. They identified promoters and enhancers with gains of activity in humans, a large fraction of which are nearby genes associated with aspects of cortical development including cell proliferation. These epigenetic studies support the concept that global changes in promoters and enhancers influence brain evolution.
Table 1.
Glossary.
| Term | General definition |
|---|---|
| P300 ChIP-Seq | A technique in which the transcription factor, P300, is first bound to DNA by immunoprecipitation (IP), specific binding sites are isolated and the DNA is then sequenced. This enables discovery of putative enhancers. |
| Transgenic mouse | Mice are generated by injection of exogenous DNA transgenes into a fertilized zygote, which is grown to a blastocyst stage, and then implanted into a pseudo-pregnant mother. This is distinct from a knock-in mouse, which has endogenous DNA altered at a specific locus. |
| Symmetric divisions | Divisions of stem cell populations which produce identical progeny. Proliferative symmetric divisions are self-renewing, enabling them to produce two new stem cells. |
| Trans regulatory element | Molecules which affect gene expression by interacting with the DNA encoding the gene. These can be proteins (such as transcription factors) or non-coding RNAs. |
| Cis regulatory element | DNA modifications which affect expression of genes on the same DNA molecule (via intramolecular interactions). These can be enhancers. |
As genome-wide analyses enable the discovery of new loci relevant for brain evolution, an important next step is to assess the functional relevance of specific regulatory DNA changes. In one of the first studies to do so, Bae et al. [40] demonstrated that a 15 nt deletion within the 5′ promoter of human GPR56, a G protein coupled receptor, causes polymicrogyria in the cortical area controlling language. Compared to the mouse locus, human GPR56 has many additional promoters and spliceoforms. Using mouse transgenics, the authors assessed how promoter element differences impact gene expression. The mouse element promoted broad expression in the developing neocortex, recapitulating the endogenous gene expression. In contrast, the same promoter from humans, or three other mammals, drove activity in a localized region of the lateral neocortex, suggesting that broad GPR56 expression in human brains is dependent on additional newly acquired promoter elements. Use of knockout and transgenic mice showed that GPR56 levels influence progenitor proliferation and gyral patterning. The authors postulate that evolutionary differences in GPR56 expression and splicing influence brain shape and folding relevant for mammals, including humans. Taken together, these aforementioned studies implicate modifications of non-coding loci, specifically promoters and enhancers, in human brain evolution. Clearly, these studies are just the beginning, given the breadth of epigenetic differences now known to distinguish humans from other species.
Human-accelerated enhancers influence gene expression in the developing brain
One class of non-coding regulatory modifications that is increasingly implicated in cortical evolution are non-coding human-accelerated regions, termed ncHARs [41–45]. In general, HARs show rapid sequence changes along the human lineage in an otherwise ultra-conserved sequence, suggestive that these changes were fixed in humans for functional reasons [46] (see Fig. 2A). A recent review by Hubisz and Pollard [46] summarizes the discovery and unique features of HARs in detail. Importantly, different criteria have been used for defining HARs. Capra et al. [47] report that approximately 2,700 ncHARs have been identified to date, with the majority found within non-coding regions, at least 30 active in development, and about 10% predicted to function as developmental brain enhancers. This observation that ncHARs are frequently located nearby genes implicated in brain development and function has also been supported by independent studies [47–49].
Figure 2.
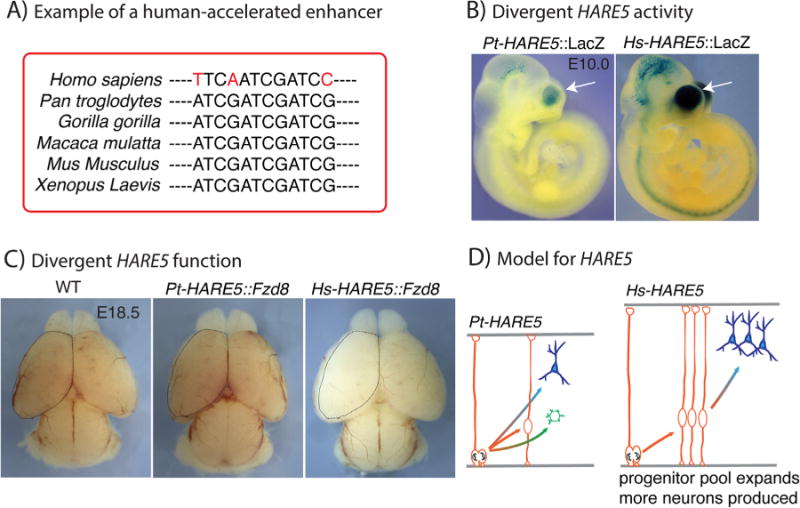
An example of a human-accelerated enhancer influencing corticogenesis. A: A mock example of a human-accelerated locus showing extremely high evolutionary conservation up until the Homo sapiens divergence, when there is rapid accumulation of changes. B: E10.0 mouse transgenic embryos depicting β-galactosidase enhancer activity (blue) driven by either chimpanzee or human HARE5. Hs-HARE5 has stronger enhancer activity in the developing neocortex. C: E18.5 mouse brains from indicated genotypes, with dotted line drawn from WT (control) overlaid on other two genotypes. Hs-HARE5 activation of Fzd8 induces larger embryonic brains with more Foxp1-positive neurons. This phenotype was evident in multiple independent transgenic lines. D: A schematic model for how HARE5 may influence human brain size, by promoting expansion of neural progenitors that eventually leads to production of more neurons and a larger brain. Panel B is reproduced from [59] with permission form Elsevier Publishing.
While some HARs function as non-coding RNA molecules, others act as enhancers. A seminal finding in this field was the discovery of HAR1, a long non-coding RNA specifically expressed in Cajal-Retzius neurons of the developing human neocortex and thought to potentially influence cortical structure [44]. In subsequent years, many HARs have been implicated as developmental enhancers, based on analysis in transgenic embryos or the presence of epigenetic marks [47, 50–52]. The first study to demonstrate that HARs could impact gene expression in an evolutionary divergent fashion came from Prabhakar et al. [53], which used transgenic reporter mice to discover a HAR, termed HACNS1, with human-specific gain of activity in the limb. Following this influential study, several additional HARs have now been empirically demonstrated to function as developmental enhancers in an evolutionarily divergent fashion in vivo [46, 47]. Below, I consider those HARs active in the developing neocortex.
Capra et al. [47] used mouse transient transgenic assays to test the developmental activity of 29 HARs, noting several with evolutionary divergent activity in the developing neocortex. Among these, human 2xHAR.238 had weaker reporter activity than chimpanzee 2xHAR.238 in the developing E11.5 forebrain [47]. Although the gene(s) regulated by 2XHAR.238 have not been formally identified, one of its closest genes on the chromosome, GLI2, has a similar developmental expression profile [47]. It is of interest to determine if 2XHAR.238 regulates GLI2 expression and how this relationship impacts neocortical function.
Several genes relevant for neocortical development contain a high density of HARs. AUTS2, a gene associated with autism spectrum disorder (ASD) and intellectual disability, contains three intronic HARs [51, 54]. In addition to these HARs, the N-terminus of AUTS2 shows the most significant statistical sequence differences between the genomes of humans and Neanderthals [55], further supporting the potential evolutionary significance of this gene. Oksenberg et al. [51] used zebrafish and mouse transgenic assays to assess enhancer activity for two HARs within AUTS2. They noted one, HACNS369, is active in the mid-brain tectum, which is thought to be important for auditory and visual reflexes. As a first step toward understanding the role of HACNS369 in brain function, it will be important to determine if human and nonhuman primate HACNS369 enhancer activity differs in the brain. Such identification of human-specific enhancer activity may help expose how AUTS2 impacts disease.
With 14 human-accelerated elements, NPAS3 is among those genes with the highest density of HARs [56]. NPAS3 encodes a transcription factor that is highly expressed in the developing brain and is implicated in schizophrenia [56, 57]. Npas3-deficient mice have altered brain development including reduced hippocampal size, corpus callosum hypoplasia, and enlarged ventricles, as well as behavioral deficits consistent with schizophrenia like behaviors [57, 58]. Given these findings, the study of NAPS3 is of particular interest. Kamm et al. [56] used transgenic zebrafish assays to show that 11 human NPAS3 HARs function as enhancers, with the majority active in the developing forebrain and other neural structures. Interestingly, one human HAR, HAR202, was not active in the developing forebrain, whereas the chimpanzee orthologous enhancer was. In a follow up study, this same group used E12.5 transgenic reporter mouse embryos to describe another HAR showing the opposite outcome, with the human ortholog driving more robust forebrain expression compared to the chimpanzee and mouse enhancers [52]. Taken together, these two studies highlight evolutionarily relevant HARs of NPAS3 in the developing brain and expose many exciting questions for the future. For example, do other NPAS3 HARs exhibit evolutionarily divergent enhancer activity and/or impact neocortical development/function? This study and others described above succeeded in establishing a critical framework for identifying and studying HARs in the brain. Given the strong association of many of these loci with disease, functional studies could be clinically relevant.
A human-accelerated enhancer of Fzd8 is relevant for neural progenitor proliferation and brain size
A recent study published from our collaborative group in 2015 was the first to demonstrate the functional relevance of a HAR, with the discovery of a human-accelerated enhancer of FZD8 [59], a receptor of the Wnt pathway implicated in brain development and size [60, 61]. This HAR was termed HARE5, because of its ability to activate enhancer activity in vivo (hence the “E” in HARE). Bird et al. [43] originally identified HARE5 as a human-accelerated locus called ANC516, but did not denote it as an enhancer. The presence of P300 binding sites in E11.5 mouse neocortices indicated HARE5 may be an enhancer [62]. We tested this prediction by generating stable transgenic reporter mice containing either the human 1.2 Kb Hs-HARE5 sequence or the chimpanzee Pt-HARE5 sequence, which is identical except for 16 differences. Hs-HARE5 enhancer activity was detectable throughout the embryonic neocortex beginning at E10.5, coincident with the onset of cortical development [16]. In contrast, reporter activity driven by Pt-HARE5 lagged behind that of Hs-HARE5 in the developing neocortex (Fig. 2B). For example, at E10.5 Pt-HARE5 was limited to only the most lateral ventral domains, whereas Hs-HARE5 activity was evident throughout the neocortex, with the exception of the mid-line. This enhancer activity mimics the spatial timing of corticogenesis that occurs along a gradient, initiating in lateral ventral domains and extending medially as development proceeds [16]. For these comparative studies, both β-galactosidase and fluorescent-destabilized reporter mice were used (see Box 2). Fluorescent analyses further demonstrated that both enhancers were active in neural progenitors. A potential caveat of these experiments is that reporter proteins could mature differently, thus we used quantitative PCR of the reporter transcripts to independently demonstrate expression differences.
Box 2. Considerations for transgenic analyses.
Most studies of HARs have used β-galactoside reporters, which rely on enzyme activity of the β-galactosidase gene of the X-gal substrate, which yields a blue precipitate wherever β-gal is actively expressed. This reporter is a traditional tool for measuring enhancer activity and is useful for whole-embryo studies because it is permanent, robust, and easy to use. As an alternative, one can use fluorescent reporters. These reporters have standard requirements and challenges associated with fluorescent proteins, including necessity of a fluorescent dissecting microscope and detection system, and the problem of photobleaching and fading, which can make long-term analyses problematic. In contrast to β-galactosidase, which have a long persistence, destabilized fluorescent reporters containing PEST sequences are only stable for two hours post-translation due to rapid proteasome-mediated degradation [94]. Hence, they can be useful for revealing temporal activity of enhancers, which could be masked with other approaches in which an enhancer would appear to be active longer than it actually is. In addition, fluorescent reporters enable the more facile spatial comparison of enhancers to each other or to other cell specific markers, particularly in tissue sections. An important consideration in the use of transgenics is to perform analyses across several independent lines in order to limit concerns about position effect due to DNA integration site. Observation of comparable phenotypes in multiple independent transgenic lines increases confidence in transgenic phenotypes.
Next our group pursued the question of determining what, if any, is the functional impact of these genomic differences. We used a genomics approach called chromosome conformation capture (3C) to demonstrate specific neocortical DNA interactions between HARE5 and the promoter of Frizzled 8 (Fzd8) [63]. Relative to HARE5, FZD8 is the closest gene on the chromosome and shares similar neocortical expression patterns [64, 65]. This implicated HARE5 acts as a Fzd8 enhancer. With this information in hand, we used another set of transgenic mice to assess if expression of mouse Fzd8 protein by either Pt-HARE5 or Hs-HARE5 affects neocortical development. Measurement of neural progenitor cell cycle duration at E12.5 revealed Hs-HARE5 accelerated the cell cycle by 23%. The seminal studies of Takahashi et al. [66] demonstrated that as neurogenesis proceeds, progenitor cell cycle lengthens, and predicts that progenitors undergo about 11 total cell divisions. Following this line of thought, if the results for Hs-HARE5 were extrapolated over the course of 4 more days of mouse development, we predict progenitors would undergo 3–4 extra cell divisions. This prediction is interesting in light of our observation that compared to Pt-HARE5 and control brains, E18.5 Hs-HARE5 transgenic cortices are larger (Fig. 2C). The surface enlarged area of these brains was likely due in part as to longer tangential dimensions, and more mid/upper layer neurons, but relatively normal cortical thickness [59] (Fig. 2C). Indeed increased symmetric divisions of progenitors are predicted to drive expansion tangentially. In conclusion, the faster cell cycle is consistent with the larger neocortical size and production of more neurons. But what do these changes mean in the context of brain evolution? Genetic modifications that promote expansion of the founder population (i.e. progenitors) could be evolutionarily beneficial for brain development and function [67] (Fig. 2D). Expansion of progenitors over a longer human gestational period could ultimately generate significantly more neurons and ultimately lead to an enlarged brain [67].
These analyses illuminated important functions of HARE5, yet also exposed additional questions. (i) What is the impact of HARE5 upon adult brain size and function? It is exciting to consider that HARE5 modifications may impact mouse behaviors controlled by the cerebral cortex. Before pursuing such experiments, it is prudent to first assess which cortical area(s) in the adult brain are impacted by HARE5 activity; (ii) given the role for HARE5 in brain size and progenitor proliferation, are human HARE5 mutations associated with cortical neurodevelopmental disorders, such as autism or macrocephaly? (iii) does HARE5 regulate expression of additional genes beyond Fzd8? The 3C approach used in our study was not designed to detect interactions in an unbiased fashion. However, 4C approaches, which detect interchromosomal interactions, have been performed in fibroblasts. Among over one million long-range interactions identified in a study by Jin et al. [68], the chromosomal region containing HARE5 is predicted to interact with additional loci beyond Fzd8. Another approach to address this question is to measure the impact upon gene expression of replacing the mouse HARE5 locus with Hs- or Pt-HARE5. We attempted to do this experiment; however, the Fzd8 location nearby the centromere made homologous recombination events unsuccessful. Analysis of mice with targeted HARE5 replacement is also desirable as it would help assuage remaining concerns about the presence of the endogenous locus. However, it is important to point out that we noted no significant impact of the endogenous mouse locus, as evidenced with multiple independent transgenic lines and analysis of control littermates; (iv) how does HARE5 influence neural progenitor proliferation? We predict it modulates canonical WNT signaling, given that overexpression of stabilized β-catenin, a canonical downstream WNT signaling protein, induces a larger gyrencephalic brain [60]; (v) which of the 16 human–chimpanzee nt difference(s) are relevant for the observed phenotypes? Transcription factor predictions suggest specific changes that may be especially relevant, but this remains to be empirically tested; (vi) finally – and perhaps most challenging – how do HARE5 differences manifest in the context of human or chimpanzee tissue? Below, I discuss approaches that may be utilized going forward to address this relevant question.
Future challenges and emerging directions: Discovery and functional validation of human-accelerated enhancers
Identification of evolutionary important enhancers
We now have virtually complete genomes for many primate species, and this has made it possible to identify thousands of human-accelerated enhancers. However, a big challenge is selecting which HARs to prioritize for follow up functional studies. One approach is to identify HARs that distinguish different Homo species. Recent sequence builds of the Neanderthal and Denisovan genomes will be useful for this purpose [69]. Another interesting approach will be to apply comparative epigenetic analyses – which has been fruitful for identifying enhancers active in tissue – to specific progenitor populations and across developmental time points. An especially important future direction will be the identification of HARs relevant for disease pathogenesis such as autism. As described above, recent studies have begun to do this. Kamm et al. [56] identified several HAR-enriched genes such as NPAS4, CNTNAP2, and RBFOX1, each associated with schizophrenia and autism. Xu et al. [70] also recently discovered HARs nearby genes associated with schizophrenia. In addition to focusing on HARs nearby or within disease genes, it will also be of interest to identify pathological mutations within human-specific HAR sequences. Given their high level of conservation, mutations affecting HARs are predicted to have roles in disease pathogenesis.
Suitable assays for testing enhancer function
As highlighted in this review, to date most HARs have been functionally assessed using mouse and zebrafish transgenics. These approaches, particularly mouse models, are fruitful for studying enhancers important for many aspects of cortical development [71]. Mouse and human corticogenesis share many commonalities, and mice provide an in vivo context for studies that cannot be performed in humans. Mice also enable researchers to take full advantage of available genetic and cell biological tools for studying cortical function and development. However, there are some biological disadvantages associated with use of mice, as they do not accurately model all human features. Unlike human brains, which are gyrencephalic (with folds), mouse brains are smooth (lissencephalic). Additionally, as highlighted in this review, humans have distinct and more diverse progenitor populations, some of which are only sparingly present in mice [11–13, 72]. Thus, for studies of evolutionary important enhancers, use of mice can be limiting because they may not necessarily contain the appropriate trans machinery, such as key transcription factors, found in humans.
Beyond these biological differences, there are also some technical disadvantages to using transgenic mice for HAR studies. Mouse generation and analyses are time-consuming, expensive, and low-throughput. Traditional transgenics allow integration of DNA randomly in the genome, and therefore copy number and integration sites can vary between transgenics, both of which may influence expression of a given transgene. Transgenics therefore require investigators to perform analyses across multiple independent lines, as we did for analysis of HARE5 activity and function [59]. One suitable work-around is the use of so-called “Safe Harbor Integration” mice, which enable one to target a transgene to a specific locus such as Rosa26 [73]. An optimal approach for functional analysis is targeted replacement of a mouse enhancer with an orthologous enhancer using either traditional homologous recombination or CRISPR/Cas9.
Independent of mouse models, new model systems have emerged for study of human brain function. With the ability to apply CRISPR/Cas9, there is increasing interest in using primate models that can model important aspects of human brain function. Relative to macaques, marmosets reach reproductive age earlier and are born with immature brains, making them amenable for studies of brain development [74]. Another animal model that is also useful for modeling human cortical development is the gyrencephalic ferret [75].
Induced pluripotent stem cells (iPSCs) provide an alternative model that does not require generation of transgenic animals. iPSCs can be produced from both human and non-human primate fibroblasts, and provide a relevant species background for studying enhancers [76, 77]. Prescott et al. [78] recently demonstrated that iPSCs can be used to discover evolutionary enhancer differences, including HARs, from human and chimpanzee IPSC-derived neural crest cells. Additionally, species-specific iPS cells may be utilized to assay enhancer activity via luciferase assays. Toward performing functional studies, iPS cells can induced toward a neural lineage, and genetically modified using CRISPR/Cas9. Hence, we now have the ability to replace a human enhancer with a chimpanzee enhancer in human-induced neural progenitors, and then assess the impact of specific nucleotide differences upon neuronal proliferation, differentiation, and synaptogenesis. Beyond 2D cell culture, an especially exciting application of iPS cells for studying HARs is the generation of mini-brains [79] or organoids [80, 81]. Use of these approaches will enable researchers to assess HAR functions in a species-specific tissue context. The initial reports of mini-brains do not necessarily recapitulate all features of human brains, such as the presence of glia. Another critical limitation of human iPSC-derived mini-brains is their marked variation in size and shape. However, as this technology improves, mini-brains will be an extremely powerful approach for dissecting HAR functions. Use of iPS cells is not without its challenges, because these too can be time-consuming and require much effort to maintain and characterize. Thus, it may be necessary to combine approaches to balance the proper biological context with high-throughput, in vivo, and affordable experimental paradigms.
Conclusions and perspectives
Understanding the genetic basis for human brain differences is an important, albeit challenging, endeavor. These discoveries will yield the most fundamental insights into our distinguishing cognitive traits and have important ramifications for elucidating the basis for neurological disease. As we learn more about human-specific loci, we can begin to uncover processes especially relevant for human brain function. In this review, I have highlighted several human-specific non-coding loci that impact progenitor cell cycle, supporting the central influence of cell cycle regulation for differentially shaping species brain differences. However, genetic modifications also influence processes that do not rely upon proliferation, such as neuronal morphology. This review has discussed a number of genetic differences unique to humans, differences spanning genomic rearrangements to point mutations to gene regulation. One clear theme that emerges is that there are many loci yet to be functionally examined. As new technologies interface with new discoveries, it is an exciting time to study the genetic regulation of human neocortical evolution.
Acknowledgments
DLS acknowledges members of the Silver lab for helpful discussions, LJ Pilaz for assistance with the graphical abstract, and funding from the NIH (R01NS803897), Broad Foundation, Holland-Trices, and Whitehead (to DLS). The author regrets that not all of the relevant work of other authors could be discussed, because of space limitations.
Footnotes
The author has declared no conflicts of interest.
References
- 1.Geschwind DH, Rakic P. Perspective. Neuron. 2013;80:633–47. doi: 10.1016/j.neuron.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Somel M, Liu X, Khaitovich P. Human brain evolution: transcripts, metabolites and their regulators. Nat Rev Neurosci. 2013;14:112–27. doi: 10.1038/nrn3372. [DOI] [PubMed] [Google Scholar]
- 3.DeFelipe J. The evolution of the brain, the human nature of cortical circuits, and intellectual creativity. Front Neuroanat. 2011;5:29. doi: 10.3389/fnana.2011.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Florio M, Huttner WB. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141:2182–94. doi: 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- 5.Herculano-Houzel S. The remarkable, yet not extraordinary, human brain as a scaled-up primate brain and its associated cost. Proc Natl Acad Sci USA. 2012;109:10661–8. doi: 10.1073/pnas.1201895109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nimchinsky EA, Gilissen E, Allman JM, Perl DP, et al. A neuronal morphologic type unique to humans and great apes. Proc Natl Acad Sci USA. 1999;96:5268–73. doi: 10.1073/pnas.96.9.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–6. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 8.Kornack DR, Rakic P. Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proc Natl Acad Sci USA. 1998;95:1242–6. doi: 10.1073/pnas.95.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dehay C, Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8:438–50. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- 10.Lukaszewicz A, Savatier P, Cortay V, Giroud P, et al. G1 phase regulation, area-specific cell cycle control, and cytoarchitectonics in the primate cortex. Neuron. 2005;47:353–64. doi: 10.1016/j.neuron.2005.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen DV, Lui JH, Parker PRL, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–61. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 12.Fietz SA, Kelava I, Vogt J, Wilsch-Bräuninger M, et al. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci. 2010;13:690–9. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- 13.Betizeau M, Cortay V, Patti D, Pfister S, et al. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron. 2013;80:442–57. doi: 10.1016/j.neuron.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 14.Pfeiffer M, Betizeau M, Waltispurger J, Pfister SS, et al. Unsupervised lineage-based characterization of primate precursors reveals high proliferative and morphological diversity in the OSVZ. J Comp Neurol. 2015 doi: 10.1002/cne.23820. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dehay C, Kennedy H, Kosik KS. The outer subventricular zone and primate-specific cortical complexification. Neuron. 2015;85:683–94. doi: 10.1016/j.neuron.2014.12.060. [DOI] [PubMed] [Google Scholar]
- 16.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollen AA, Nowakowski TJ, Chen J, Retallack H, et al. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewitus E, Kelava I, Kalinka AT, Tomancak P, et al. An adaptive threshold in mammalian neocortical evolution. PLoS Biol. 2014;12:e1002000. doi: 10.1371/journal.pbio.1002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bae B-I, Jayaraman D, Walsh CA. Genetic changes shaping the human brain. Dev Cell. 2015;32:423–34. doi: 10.1016/j.devcel.2015.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konopka G, Friedrich T, Davis-Turak J, Winden K, et al. Human-specific transcriptional networks in the brain. Neuron. 2012;75:601–17. doi: 10.1016/j.neuron.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Somel M, Franz H, Yan Z, Lorenc A, et al. Transcriptional neoteny in the human brain. Proc Natl Acad Sci USA. 2009;106:5743–8. doi: 10.1073/pnas.0900544106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pletikos M, Sousa AMM, Sedmak G, Meyer KA, et al. Temporal specification and bilaterality of human neocortical topographic gene expression. Neuron. 2014;81:321–32. doi: 10.1016/j.neuron.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernard A, Lubbers LS, Tanis KQ, Luo R, et al. Transcriptional architecture of the primate neocortex. Neuron. 2012;73:1083–99. doi: 10.1016/j.neuron.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fietz SA, Lachmann R, Brandl H, Kircher M, et al. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proc Natl Acad Sci USA. 2012;109:11836–41. doi: 10.1073/pnas.1209647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lui JH, Nowakowski TJ, Pollen AA, Javaherian A, et al. Radial glia require PDGFD-PDGFRβ signalling in human but not mouse neocortex. Nature. 2014;515:264–8. doi: 10.1038/nature13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollen AA, Nowakowski TJ, Shuga J, Wang X, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol. 2014;32:1053–8. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson MB, Wang PP, Atabay KD, Murphy EA, et al. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat Neurosci. 2015;18:637–46. doi: 10.1038/nn.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Florio M, Albert M, Taverna E, Namba T, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347:1465–70. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- 29.Charrier C, Joshi K, Coutinho-Budd J, Kim J-E, et al. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell. 2012;149:923–35. doi: 10.1016/j.cell.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dennis MY, Nuttle X, Sudmant PH, Antonacci F, et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell. 2012;149:912–22. doi: 10.1016/j.cell.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sudmant PH, Kitzman JO, Antonacci F, Alkan C, et al. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–6. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mclean CY, Reno PL, Pollen AA, Bassan AI, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471:216–9. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King M-C, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–16. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 34.Nord AS, Blow MJ, Attanasio C, Akiyama JA, et al. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell. 2013;155:1521–31. doi: 10.1016/j.cell.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shim S, Kwan KY, Li M, Lefebvre V, et al. Cis-regulatory control of corticospinal system development and evolution. Nature. 2012;486:74–9. doi: 10.1038/nature11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nord AS, Pattabiraman K, Visel A, Rubenstein JLR. Genomic perspectives of transcriptional regulation in forebrain development. Neuron. 2015;85:27–47. doi: 10.1016/j.neuron.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gokhman D, Lavi E, Prüfer K, Fraga MF, et al. Reconstructing the DNA methylation maps of the Neandertal and the Denisovan. Science. 2014;344:523–7. doi: 10.1126/science.1250368. [DOI] [PubMed] [Google Scholar]
- 38.Visel A, Taher L, Girgis H, May D, et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reilly SK, Yin J, Ayoub AE, Emera D, et al. Evolutionary genomics. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science. 2015;347:1155–9. doi: 10.1126/science.1260943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bae B-I, Tietjen I, Atabay KD, Evrony GD, et al. Evolutionarily dynamic alternative splicing of GPR56 regulates regional cerebral cortical patterning. Science. 2014;343:764–8. doi: 10.1126/science.1244392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindblad-Toh K, Garber M, Zuk O, Lin MF, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–82. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prabhakar S, Noonan JP, Pääbo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- 43.Bird CP, Stranger BE, Liu M, Thomas DJ, et al. Fast-evolving noncoding sequences in the human genome. Genome Biol. 2007;8:R118. doi: 10.1186/gb-2007-8-6-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pollard KS, Salama SR, Lambert N, Lambot M-A, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–72. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- 45.Bush EC, Lahn BT. A genome-wide screen for noncoding elements important in primate evolution. BMC Evol Biol. 2008;8:17. doi: 10.1186/1471-2148-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hubisz MJ, Pollard KS. Exploring the genesis and functions of human accelerated regions sheds light on their role in human evolution. Curr Opin Genet Dev. 2014;29C:15–21. doi: 10.1016/j.gde.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 47.Capra JA, Erwin GD, McKinsey G, Rubenstein JLR, et al. Many human accelerated regions are developmental enhancers. Philos Trans R SocLond B Biol Sci. 2013;368:20130025. doi: 10.1098/rstb.2013.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haygood R, Babbitt CC, Fedrigo O, Wray GA. Contrasts between adaptive coding and noncoding changes during human evolution. Proc Natl Acad Sci USA. 2010;107:7853–7. doi: 10.1073/pnas.0911249107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cotney J, Leng J, Yin J, Reilly SK, et al. The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell. 2013;154:185–96. doi: 10.1016/j.cell.2013.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oksenberg N, Stevison L, Wall JD, Ahituv N. Function and regulation of AUTS2, a gene implicated in autism and human evolution. PLoS Genet. 2013;9:e1003221. doi: 10.1371/journal.pgen.1003221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamm GB, López-Leal R, Lorenzo JR, Franchini LF. A fastevolving human NPAS3 enhancer gained reporter expression in the developing forebrain of transgenic mice. Philos Trans R Soc Lond B Biol Sci. 2013;368:20130019–9. doi: 10.1098/rstb.2013.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prabhakar S, Visel A, Akiyama JA, Shoukry M, et al. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–50. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oksenberg N, Ahituv N. The role of AUTS2 in neurodevelopment and human evolution. Trends Genet. 2013;29:600–8. doi: 10.1016/j.tig.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Green RE, Krause J, Briggs AW, Maricic T, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–22. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamm GB, Pisciottano F, Kliger R, Franchini LF. The developmental brain gene NPAS3 contains the largest number of accelerated regulatory sequences in the human genome. Mol Biol Evol. 2013;30:1088–102. doi: 10.1093/molbev/mst023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Erbel-Sieler C, Dudley C, Zhou Y, Wu X, et al. Behavioral and regulatory abnormalities in mice deficient in the NPAS1 and NPAS3 transcription factors. Proc Natl Acad Sci USA. 2004;101:13648–53. doi: 10.1073/pnas.0405310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brunskill EW, Ehrman LA, Williams MT, Klanke J, et al. Abnormal neurodevelopment, neurosignaling and behaviour in Npas3-deficient mice. Eur J Neurosci. 2005;22:1265–76. doi: 10.1111/j.1460-9568.2005.04291.x. [DOI] [PubMed] [Google Scholar]
- 59.Boyd JL, Skove SL, Rouanet JP, Pilaz L-J, et al. Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex. Curr Biol. 2015;25:772–9. doi: 10.1016/j.cub.2015.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–9. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- 61.Freese JL, Pino D, Pleasure SJ. Wnt signaling in development and disease. Neurobiol Dis. 2010;38:148–53. doi: 10.1016/j.nbd.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Visel A, Blow MJ, Li Z, Zhang T, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hagège H, Klous P, Braem C, Splinter E, et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2:1722–33. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 64.Fischer T, Guimera J, Wurst W, Prakash N. Distinct but redundant expression of the Frizzled Wnt receptor genes at signaling centers of the developing mouse brain. Neurosci. 2007;147:693–711. doi: 10.1016/j.neuroscience.2007.04.060. [DOI] [PubMed] [Google Scholar]
- 65.Liu C, Bakeri H, Li T, Swaroop A. Regulation of retinal progenitor expansion by Frizzled receptors: implications for microphthalmia and retinal coloboma. Hum Mol Genet. 2012;21:1848–60. doi: 10.1093/hmg/ddr616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takahashi T, Nowakowski RS, Caviness VS. The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J Neurosci. 1995;15:6046–57. doi: 10.1523/JNEUROSCI.15-09-06046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rakic P. A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci. 1995;18:383–8. doi: 10.1016/0166-2236(95)93934-p. [DOI] [PubMed] [Google Scholar]
- 68.Jin F, Li Y, Dixon JR, Selvaraj S, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–4. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prüfer K, Racimo F, Patterson N, Jay F, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–9. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu K, Schadt EE, Pollard KS, Roussos P, et al. Genomic and network patterns of schizophrenia genetic variation in human evolutionary accelerated regions. Mol Biol Evol. 2015;32:1148–60. doi: 10.1093/molbev/msv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nord AS. Learning about mammalian gene regulation from functional enhancer assays in the mouse. Genomics. 2015;106:178–84. doi: 10.1016/j.ygeno.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 72.Wang X, Tsai J-W, Lamonica B, Kriegstein AR. A new subtype of progenitor cell in the mouse embryonic neocortex. Nat Neurosci. 2011;14:555–61. doi: 10.1038/nn.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tasic B, Hippenmeyer S, Wang C, Gamboa M, et al. Site-specific integrase-mediated transgenesis in mice via pronuclear injection. Proc Natl Acad Sci US. 2011;A108:7902–7. doi: 10.1073/pnas.1019507108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Belmonte JCI, Callaway EM, Caddick SJ, Churchland P, et al. Brains, genes, and primates. Neuron. 2015;86:617–31. doi: 10.1016/j.neuron.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gertz CC, Kriegstein AR. Neuronal migration dynamics in the developing ferret cortex. J Neurosci. 2015;35:14307–15. doi: 10.1523/JNEUROSCI.2198-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wunderlich S, Kircher M, Vieth B, Haase A, et al. Primate iPS cells as tools for evolutionary analyses. Stem Cell Res. 2014;12:622–9. doi: 10.1016/j.scr.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 77.Pfefferle LW, Wray GA. Insights from a chimpanzee adipose stromal cell population: opportunities for adult stem cells to expand primate functional genomics. Genome Biol Evol. 2013;5:1995–2005. doi: 10.1093/gbe/evt148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Prescott SL, Srinivasan R, Marchetto MC, Grishina I, et al. Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell. 2015;163:68–83. doi: 10.1016/j.cell.2015.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lancaster MA, Renner M, Martin C-A, Wenzel D, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–9. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kadoshima T, Sakaguchi H, Nakano T, Soen M, et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci USA. 2013;110:20284–9. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mariani J, Coppola G, Zhang P, Abyzov A, et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell. 2015;162:375–90. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nowick K, Gernat T, Almaas E, Stubbs L. Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proc Natl Acad Sci USA. 2009;106:22358–63. doi: 10.1073/pnas.0911376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Enard W, Przeworski M, Fisher SE, Lai CSL, et al. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–72. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- 84.Zhang J, Webb DM, Podlaha O. Accelerated protein evolution and origins of human-specific features: Foxp2 as an example. Genetics. 2002;162:1825–35. doi: 10.1093/genetics/162.4.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Enard W, Gehre S, Hammerschmidt K, HOlter SM, et al. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell. 2009;137:961–71. doi: 10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 86.Lai S, Fisher SE, Hurst JA, Vargha-Khadem F, et al. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–23. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 87.MacDermot KD, Bonora E, Sykes N, Coupe A-M, et al. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet. 2005;76:1074–80. doi: 10.1086/430841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hammerschmidt K, Schreiweis C, Minge C, Paabo S, et al. A humanized version of Foxp2 does not affect ultrasonic vocalization in adult mice. Genes Brain Behav. 2015 doi: 10.1111/gbb.12237. [DOI] [PubMed] [Google Scholar]
- 89.Konopka G, Bomar JM, Winden K, Coppola G, et al. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462:213–7. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maricic T, Gunther V, Georgiev O, Gehre S, et al. A recent evolutionary change affects a regulatory element in the human FOXP2 gene. Mol Biol Evol. 2013;30:844–52. doi: 10.1093/molbev/mss271. [DOI] [PubMed] [Google Scholar]
- 91.Montgomery SH, Mundy NI. Microcephaly genes evolved adaptively throughout the evolution of eutherian mammals. BMC Evol Biol. 2014;14:120. doi: 10.1186/1471-2148-14-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Berezikov E, Thuemmler F, van Laake LW, Kondova I, et al. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006;38:1375–7. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- 93.Arcila ML, Betizeau M, Cambronne XA, Guzman E, et al. NeuroResource. Neuron. 2014;81:1255–62. doi: 10.1016/j.neuron.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–9. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]


