Abstract
Germline copy-number variants (CNVs) involving quadruplications are rare and the mechanisms generating them are largely unknown. Previously, we reported a 20-week gestation fetus with split-hand malformation; clinical microarray detected two maternally inherited triplications separated by a copy-number neutral region at 17p13.3, involving BHLHA9 and part of YWHAE. Here, we describe an 18-month-old male sibling of the previously described fetus with split-hand malformation. Custom high-density array and digital droplet PCR revealed the copy-number gains were actually quadruplications in the mother, the fetus and her later born son. This quadruplication-normal-quadruplication pattern was shown to be expanded from the triplication-normal-triplication CNV at the same loci in the maternal grandmother. We mapped two breakpoint junctions and demonstrated that both are mediated by Alu repetitive elements and identical in these four individuals. We propose a three-step process combining Alu-mediated replicative-repair-based mechanism(s) and intergenerational, intrachromosomal non-allelic homologous recombination to generate the quadruplications in this family.
Keywords: quadruplication, complex genomic rearrangement, Alu repetitive elements, split-hand malformation, digital droplet PCR
Germline copy-number variants (CNVs) involving quadruplications are rarely observed. Compared to duplications with one copy gained (three copies including the unaltered allele) and triplications with two copies gained (four copies in total), quadruplications are CNVs with three gained copies of a genomic region, and thus five copies of that locus in total in the diploid genome. Mechanisms responsible for the generation of these types of CNVs in humans are rarely discussed, although previous reports have suggested two potential models including a rolling circle mechanism (Beck et al., 2015) or two cycles of chromosome breakage followed by a U-type sister chromatid fusion (Recalcati et al., 2010).
Previously, we reported a prenatal case (BAB7218) with split-hand malformation detected by fetal ultrasound (Luk et al., 2014). Clinical microarray (NimbleGen CGX-135K array) showed two maternally inherited triplications separated by a copy-number neutral region at 17p13.3 in the fetus. The copy number gain regions included the gene BHLHA9 and a part of YWHAE. Due to the involvement of the YWHAE gene, gain of which has been associated with neurodevelopmental delay (MIM# 613215), the couple opted to terminate the pregnancy. The couple subsequently had a second pregnancy that resulted in an affected male child (BAB7221). Investigation of the 17p13.3 region by high-density array comparative genomic hybridization (aCGH) in this child led to the unexpected demonstration of two quadruplications – rather than triplications – at the same locus.
We observed this quadruplication-normal-quadruplication (QDR-NML-QDR) pattern in the mother, the aborted fetus, and her later born son. We further observed a triplication-normal-triplication (TRP-NML-TRP) pattern at the same loci in the maternal grandmother. Two identical breakpoint junctions were mapped in all four individuals and both are mediated by Alu repetitive elements; the Alu-Alu recombination resulting in a chimeric Alu element at the breakpoint junction (Gu et al., 2015; Mayle et al., 2015). We propose a model which consist of a three-step process involving replicative-repair-based mechanism(s) mediated via template switching between Alu elements plus two intergenerational non-allelic homologous recombination (NAHR) events for the generation of the end-product consisting of double quadruplications (QDR-NML-QDR) in this family.
The patient (BAB7221) was the second product of a non-consanguineous Chinese union. The couple’s first pregnancy (BAB7218) had bilateral split-hand-foot malformation, and was terminated at 23 weeks gestation. Clinical microarray (NimbleGen CGX-135K array) revealed two maternally-inherited triplications at chromosome 17p13.3. In the present case (BAB7221), a prenatal ultrasound at 20 weeks gestation demonstrated a split-hand malformation of the right hand, but no additional skeletal or soft tissue defects. After genetic counseling, the parents declined prenatal genetic testing. The patient was born by spontaneous vaginal delivery at 38 weeks and 3 days gestation with a birth weight of 3.72 kg (50–85th centile; length and head circumference not available). Right split-hand malformation was confirmed on neonatal examination, and no further skeletal abnormalities were noted (Supp. Figure S1). Posterior/anterior and lateral radiographs of the right hand confirmed absence of all three phalanges of the third digit, and normal formation of the third metacarpal (Supp. Figure S1). The patient is now 15 months of age with normal motor, fine motor, and speech development at 3 and 12 months of age. The mother has a history of mild right talipes equinovarus, but she has no additional skeletal anomalies and has a normal intellect (Luk et al., 2014).
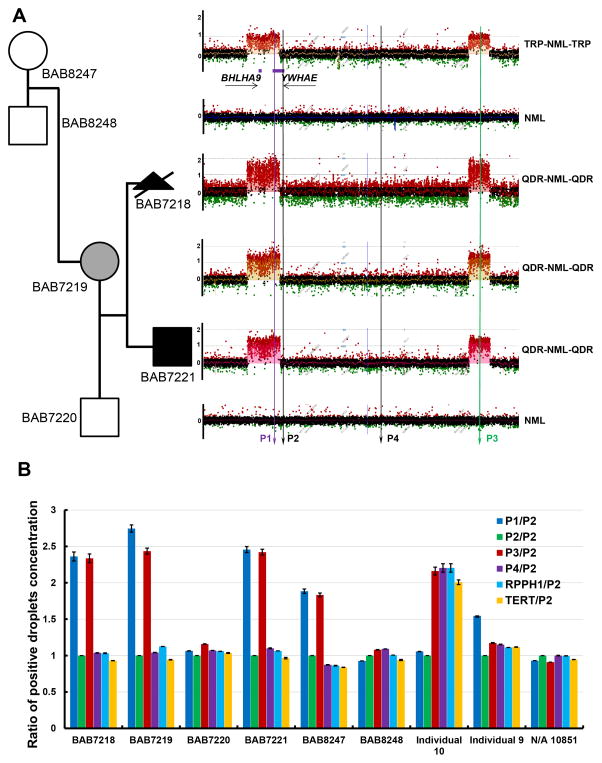
High-density aCGH detected two quadruplications separated by a copy-number neutral region in the mother (BAB7219), the aborted fetus (BAB7218) and her later born son (BAB7221) (log2 ratio>1, see Material and Methods), and detected double triplications (log2 ratio around 0.8) in the maternal grandmother (BAB8247) (Figure 1A, Supp. Figure S2, S3, Supp. Table S1). The two gained regions are 0.179 Mb and 0.115 Mb in size, respectively, and the middle unaltered region is 1.024 Mb in all four of these individuals. No copy-number variant was observed in the father (BAB7220) or maternal grandfather (BAB8248) in the 17p13.3 region. Microarray raw data was deposited to GEO (http://www.ncbi.nlm.nih.gov/geo/). Quadruplications and triplications were further verified by an independent orthogonal experimental approach for determining copy-number - digital droplet PCR (ddPCR) (Figure 1B, Supp. Figure S4, Supp. Table S2).
Figure 1. Pedigree, high-density aCGH plots and ddPCR results of all the family members.
A). The mother (BAB7291) and her two offspring (BAB7281 and BAB7221) have double quadruplications separated by a copy-number neutral region, while the maternal grandmother (BAB8247) has double triplications separated by a copy-number neutral region. Maternal grandfather (BAB8248) and father (BAB7220) have no copy-number variants in the 17p13.3 region. TRP, triplication; NML, normal; QDR, quadruplication. B). Graph showing ratio of positive droplet concentrations (number of positive droplets per uL of PCR reaction) in each individual tested. Within each individual, positive droplet concentrations from PCR using primer pairs P1, P3, P4, RPPH1 and TERT were compared with that from P2. P1 (position indicated by purple vertical line in Figure 1A) and P3 (green line) are located in the two gained regions in BAB7218, 7219, 7221 and 8247. P2 and P4 (both black lines) are located in the unaltered copy-number region in between. Primer loci are also shown in Supp. Figure S4. RHHP1 and TERT are two known genes with normal, diploid copy-number in the human genome. Note that in BAB7218, 7219 and 7221 interpreted as having experimental evidence for quadruplications by aCGH, ratios of P1/P2 and P3/P2 are approximately 2.5 as expected (five copies in quadruplicated regions versus two copies in copy-number neutral region), and in BAB8247 with triplications, the ratios of P1/P2 and P3/P2 are approximately 2 (four copies in triplicated regions versus two copies in copy-number neutral region). In Individual 9 with duplication, P1/P2 is around 1.5 (three copies in duplicated region versus two copies in copy-number neutral region), and in Individual 10 with deletion, P3/P2, P4/P2, RPPH1/P2 and TERT/P2 are approximately 2 (two copies in copy-number neutral region versus one copy in deleted region). All three additional control primer pairs, P4, RPPH1 and TERT showed comparable ddPCR results to that of P2. Raw data of ddPCR is shown in Supp. Table S2, and primer sequences are shown in Supp. Table S3.
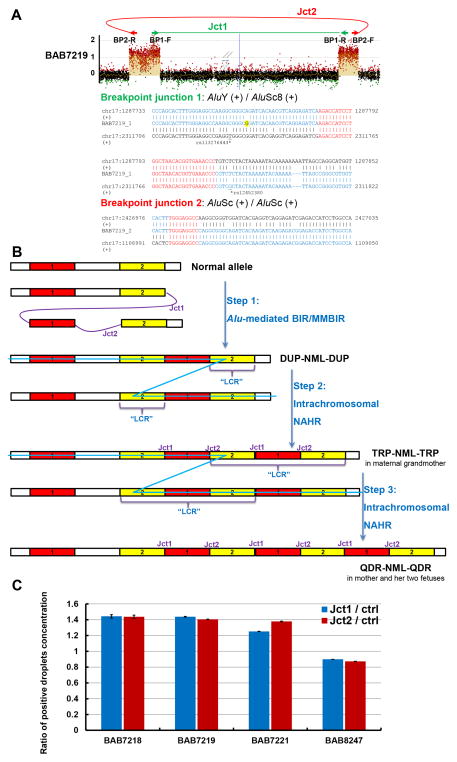
Two identical breakpoint junctions were mapped to nucleotide resolution by long-range PCR followed by Sanger sequencing in BAB7218, BAB7219, BAB7221 and BAB8247; the four individuals with copy-number gains. Breakpoint junction 1 (Jct1) joins the proximal side of the first gain to the distal side of the second gain, whereas breakpoint junction 2 (Jct2) joins the proximal side of the second gain to the distal side of the first gain (Figure 2A, Supp. Figure S5). Upon careful examination, the transitions from copy-number gain to copy-number neutral were direct in all four boundaries for the two gained regions, without any shoulders observed from aCGH (Supp. Figure S2). Multiple inward- and outward-facing primers were designed near each boundary; however, long-range PCRs did not detect additional breakpoint junctions. Since our custom designed high-density aCGH has an average genome coverage of ~150 bp per probe at 17p13.3 and 100–500 bp per probe around breakpoints, it is likely that Jct1 and Jct2 are the only two breakpoint junctions present. Both Jct1 and Jct2 are mediated by directly-oriented Alu pairs (Figure 2A).
Figure 2. Proposed three-step generation for the double quadruplications.
A). Sequences of the two mapped breakpoint junctions. The same two breakpoint junctions were identified in BAB7218, BAB7219, BAB7221 and BAB8247, and array plot of BAB7219 was used for demonstration (upper panel). Breakpoint junction 1 (Jct 1) joins the proximal end of the first gained region to the distal end of the second gain, while breakpoint junction 2 (Jct 2) joins the other two ends of the two gains. Sequences of the two primer pairs (BP1-F/R and BP2-F/R) used for long-range PCR are shown in Supp. Table S3. Both breakpoint junctions are Alu-Alu mediated, and all the four CNV-mediated Alu elements are on the plus strand according to the reference genome (GRCh37/hg19 assembly) as indicated by the “+” sign. Breakpoint junction sequence is aligned to the proximal and distal genomic references (GRCh37/hg19 assembly) and color-matched. Microhomology at the breakpoint is indicated in red. A SNP near the microhomology of Jct 1 (“G” highlighted in yellow) was not found in any SNP databases, suggesting a potential de novo SNV (single nucleotide variant) mutation was generated during CNV formation; consistent with a replicative repair mechanism(Carvalho et al., 2013). B).Red blocks represent one of the gained regions (Region 1), while yellow blocks represent the other gained region (Region 2). During the first step, a DUP-NML-DUP rearrangement pattern is formed through Alu-mediated replicative based mechanism, joining the two gained regions together. During the second step, the duplicated Region 2 is used as an “LCR” for intrachromosomal NAHR, resulting in the TRP-NML-TRP rearrangement pattern observed in maternal grandmother. Blue lines represent the non-allelic crossover between two sister chromatids. During the third step, similar to Step 2, intrachromosomal NAHR resulted in the QDR-NML-QDR pattern observed in the mother. Note that the crossover in Step 3 could occur anywhere within the “Region 1+Region 2+Region1” segment formed in Step 2. Jct1, breakpoint junction1; Jct2, breakpoint junction 2. BIR, break-induced replication; MMBIR, microhomology-mediated BIR; LCR, low-copy repeats; NAHR, non-allelic homologous recombination; DUP, duplication; NML, normal; TRP, triplication; QDR, quadruplication. C). Graph showing ratio of positive droplet concentrations (number of positive droplets per uL of PCR reaction) for Jct1 and Jct2. Control primer pair amplifies the same locus of primer pair P4 in Figure 1B with a 1.5 kb product, consistent with the product length of Jct1 and Jct2. Note that in BAB7218, 7219 and 7221, ratios of Jct1/ctrl and Jct2/ctrl are approximately 1.5 as expected (three copies of Jct1 and Jct2 as shown in Figure 2B), and in BAB8247, these two ratios are approximately 1 (two copies). Raw data of ddPCR is shown in Supp. Table S2, and primer sequences are shown in Supp. Table S3.
This study demonstrates the limitation of clinical microarrays (NimbleGen CGX-135K array, average resolution of 140 kb across the genome and 40 kb or less in regions of clinical relevance (Luk et al., 2014)) in differentiating higher order copy-number changes, such as triplications and quadruplications. Higher copy-number changes (comparing triplication to duplication) are known to be associated with more severe phenotypes for certain dosage-sensitive genes, including YWHAE and BHLHA9 (Bi et al., 2009; Fuchs et al., 2007; Liu et al., 2014; Nagata et al., 2014). Therefore, it is of clinical importance to accurately detect the exact copy-number at a given locus. Both high-density aCGH and ddPCR allow accurate differentiation between quadruplications and triplications; however, the design and implementation of a high-density aCGH for every CNV would be expensive and time-consuming. The ddPCR technique, on the other hand, is fast, reliable and cost-effective, and therefore could be considered as a routine confirmation step for copy-number gains when more than 3 copies of a region (duplication) are potentially indicated by clinical testing.
Duplications and triplications containing the BHLHA9 gene are known to be associated with split-hand/foot malformation with long bone deficiency 3 (SHFLD3; MIM# 612576). Due to the low penetrance level (less than 50%) and the markedly variable inter-individual and intra-individual expressivity, increased copy-number of BHLHA9 was proposed to constitute a strong susceptibility factor rather than a causative gene for development of limb malformations (Klopocki et al., 2012). Alternative hypotheses to entertain regarding penetrance might include somatic mosaicism for a higher order copy number gain or reversion of a gain during development (Campbell et al., 2015).
In this family, the maternal grandmother (BAB8247) with BHLHA9 triplication showed no apparent phenotype, while the mother (BAB7219) with BHLHA9 quadruplication had mild talipes equinovarus as a child. The phenotypic differences between these two individuals could potentially be attributed to the increased copy-number of BHLHA9. Compared to the mother (BAB7219), the aborted fetus (BAB7218) and her later born son (BAB7221) shared identical BHLHA9 quadruplication, however, exhibited the more severe phenotype of split-hand malformation. This could potentially be explained by the presence of a paternally inherited deleterious variant(s) or haplotype on the other allele in BAB7218 and BAB7221 (Wu et al., 2015). A similar hypothesis regarding a role for genetic modifiers has been proposed in the Japanese population with BHLHA9 duplications and triplications (Nagata et al., 2014).
By utilizing directly oriented paralogous low-copy repeats (DP-LCRs), reciprocal deletions and duplications can be generated by an ectopic crossover via the NAHR mechanism (Liu et al., 2012). Two important findings support a model in which the transition from TRP-NML-TRP to QDR-NML-QDR was NAHR-mediated using already replicated Region 1 and/or Region 2 as LCRs: (1) the TRP-NML-TRP CNV present in the maternal grandmother becomes a QDR-NML-QDR structure in the mother, and this copy number change results from a combined gain of both “Region 1” and “Region 2” in the absence of other copy number changes (Figure 1A, Figure 2B), and (2) Region 1 and Region 2 were joined by Jct1, demonstrating that these regions are directly adjacent. We propose a three-step process for generation of this CNV (Figure 2B): Step 1, a duplication-normal-duplication (DUP-NML-DUP) rearrangement pattern is formed from the normal allele via a replication-based mechanism(s), e.g. break-induced replication (BIR) (Malkova and Ira, 2013) or microhomology-mediated BIR (MMBIR) (Hastings et al., 2009), and by virtue of template switches between Alu repetitive elements (Mayle et al., 2015), generating Alu-Alu mediated Jct1 and Jct2. Similar Alu-mediated rearrangements with DUP-NML-DUP have been observed in our previous studies on 17p13.3 complex genomic rearrangements (Individual 2) (Gu et al., 2015). Step 2, by utilizing the duplicated Region 2 as potential LCRs, Region 1 and Region 2 are further amplified through intrachromosomal (interchromatidal) NAHR, resulting in the TRP-NML-TRP pattern observed in the maternal grandmother (BAB8247). Step 3, another intergenerational intrachromosomal NAHR results in the QDR-NML-QDR pattern in the mother and her two offspring. Note that in this step, the ectopic crossover could happen anywhere within the “Region 1 + Region 2 + Region 1” segment generated in Step 2 (Figure 2B). ddPCR of the two breakpoint junctions demonstrated the presence of three Jct1 and Jct2 fragments in BAB7218, 7219 and 7221 versus two of each in BAB8247, consistent with the proposed end-product structure and model (Figure 2C).
Previous studies have demonstrated that the frequency of NAHR is positively correlated with the degree of sequence homology of the NAHR-mediating LCRs and LCR length, and negatively correlated with the distance between the DP-LCRs (Dittwald et al., 2013; Liu et al., 2011). When LCRs share > ~97% sequence identity and are located less than ~ 10 Mb apart, they could lead to ectopic synapsis and mediate NAHR (Stankiewicz and Lupski, 2010). The model we propose to generate QDR-NML-QDR from TRP-NML-TRP (and similarly TRP-NML-TRP from DUP-NML-DUP) utilizes identical Region 1 and/or Region 2 as LCRs, and as such they share 100% sequence homology (assuming no point mutation has occurred). This degree of homology cannot be achieved by any natural DP-LCRs in the human genome. In addition, comparing to NPHP1 duplications/deletions, which are the most commonly observed recurrent CNVs utilizing ~45 kb DP-LCRs about 290 kb apart (Dittwald et al., 2013), the Region 1 and/or Region 2 proposed as LCRs in this family are longer (~179 kb for Region 1 and ~115 kb for Region 2) and more closely located. These factors may explain the repeated identical copy-number gain at the same locus potentially through NAHR in this family.
In summary, we demonstrate robust detection of higher order copy-number gains using high-density aCGH and ddPCR in a family with triplications and quadruplications. In addition, we propose a three-step process combining one step of Alu-mediated replicative-repair-based mechanism(s) and two steps of intergenerational intrachromosomal NAHR for the generation of double quadruplications in this family. We speculate that the incidence of higher order copy-number gains such as quadruplications may be greater than anticipated and simply undetected by commonly used clinical arrays because of the reduced resolution and dynamic range for the comparative genomic analyses methods when trying to discern higher order copy number gains (e.g. quadruplication vs triplication). Moreover, the mechanism for the generation of such complex genomic rearrangements (CGR) may vary (Beck et al., 2015; Recalcati et al., 2010). Precise detection of copy-number gains in clinical diagnosis is essential for understanding the clinical relevance, potential recurrence risk, and mechanisms of generation of these CGR.
Supplementary Material
Acknowledgments
J.R.L. has stock ownership in 23 and Me, is a paid consultant for Regeneron Pharmaceuticals, has stock options in Lasergen, Inc. and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases and bacterial genomic fingerprinting. J.E.P. was supported by the Medical Genetics Research Fellowship Program NIH/NIGMS NIH T32 GM07526. This study was approved by the Institutional Review Board for Human Subject Research at BCM. This work was supported in part by grants from the US National Institute of Neurological Disorders and Stroke (R01NS058529), National Institute of General Medical Sciences (R01GM106373) and National Human Genome Research Institute (NHGRI)/National Heart Lung and Blood Institute (NHLBI) (U54 HG006542) to the Baylor-Hopkins Center for Mendelian Genomics.
References
- Beck CR, Carvalho CM, Banser L, Gambin T, Stubbolo D, Yuan B, Sperle K, McCahan SM, Henneke M, Seeman P, Garbern JY, Hobson GM, et al. Complex genomic rearrangements at the PLP1 locus include triplication and quadruplication. PLoS Genet. 2015;11:e1005050. doi: 10.1371/journal.pgen.1005050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Sapir T, Shchelochkov OA, Zhang F, Withers MA, Hunter JV, Levy T, Shinder V, Peiffer DA, Gunderson KL, Nezarati MM, Shotts VA, et al. Increased LIS1 expression affects human and mouse brain development. Nat Genet. 2009;41:168–177. doi: 10.1038/ng.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell IM, Shaw CA, Stankiewicz P, Lupski JR. Somatic mosaicism: implications for disease and transmission genetics. Trends Genet. 2015;31:382–392. doi: 10.1016/j.tig.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Pehlivan D, Ramocki MB, Fang P, Alleva B, Franco LM, Belmont JW, Hastings PJ, Lupski JR. Replicative mechanisms for CNV formation are error prone. Nat Genet. 2013;45:1319–1326. doi: 10.1038/ng.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittwald P, Gambin T, Szafranski P, Li J, Amato S, Divon MY, Rodriguez Rojas LX, Elton LE, Scott DA, Schaaf CP, Torres-Martinez W, Stevens AK, et al. NAHR-mediated copy-number variants in a clinical population: mechanistic insights into both genomic disorders and Mendelizing traits. Genome Res. 2013;23:1395–1409. doi: 10.1101/gr.152454.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs J, Nilsson C, Kachergus J, Munz M, Larsson EM, Schule B, Langston JW, Middleton FA, Ross OA, Hulihan M, Gasser T, Farrer MJ. Phenotypic variation in a large Swedish pedigree due to SNCA duplication and triplication. Neurology. 2007;68:916–922. doi: 10.1212/01.wnl.0000254458.17630.c5. [DOI] [PubMed] [Google Scholar]
- Gu S, Yuan B, Campbell IM, Beck CR, Carvalho CM, Nagamani SC, Erez A, Patel A, Bacino CA, Shaw CA, Stankiewicz P, Cheung SW, et al. Alu-mediated diverse and complex pathogenic copy-number variants within human chromosome 17 at p13.3. Hum Mol Genet. 2015;24:4061–4077. doi: 10.1093/hmg/ddv146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopocki E, Lohan S, Doelken SC, Stricker S, Ockeloen CW, Soares Thiele de Aguiar R, Lezirovitz K, Mingroni Netto RC, Jamsheer A, Shah H, Kurth I, Habenicht R, et al. Duplications of BHLHA9 are associated with ectrodactyly and tibia hemimelia inherited in non-Mendelian fashion. J Med Genet. 2012;49:119–125. doi: 10.1136/jmedgenet-2011-100409. [DOI] [PubMed] [Google Scholar]
- Liu P, Carvalho CM, Hastings PJ, Lupski JR. Mechanisms for recurrent and complex human genomic rearrangements. Curr Opin Genet Dev. 2012;22:211–220. doi: 10.1016/j.gde.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Gelowani V, Zhang F, Drory VE, Ben-Shachar S, Roney E, Medeiros AC, Moore RJ, DiVincenzo C, Burnette WB, Higgins JJ, Li J, et al. Mechanism, prevalence, and more severe neuropathy phenotype of the Charcot-Marie-Tooth type 1A triplication. Am J Hum Genet. 2014;94:462–469. doi: 10.1016/j.ajhg.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Lacaria M, Zhang F, Withers M, Hastings PJ, Lupski JR. Frequency of nonallelic homologous recombination is correlated with length of homology: evidence that ectopic synapsis precedes ectopic crossing-over. Am J Hum Genet. 2011;89:580–588. doi: 10.1016/j.ajhg.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk HM, Wong VC, Lo IF, Chan KY, Lau ET, Kan AS, Tang MH, Tang WF, She WM, Chu YW, Sin WK, Chung BH. A prenatal case of split-hand malformation associated with 17p13.3 triplication - a dilemma in genetic counseling. Eur J Med Genet. 2014;57:81–84. doi: 10.1016/j.ejmg.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Malkova A, Ira G. Break-induced replication: functions and molecular mechanism. Curr Opin Genet Dev. 2013;23:271–279. doi: 10.1016/j.gde.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayle R, Campbell IM, Beck CR, Yu Y, Wilson M, Shaw CA, Bjergbaek L, Lupski JR, Ira G. DNA REPAIR. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science. 2015;349:742–747. doi: 10.1126/science.aaa8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata E, Kano H, Kato F, Yamaguchi R, Nakashima S, Takayama S, Kosaki R, Tonoki H, Mizuno S, Watanabe S, Yoshiura K, Kosho T, et al. Japanese founder duplications/triplications involving BHLHA9 are associated with split-hand/foot malformation with or without long bone deficiency and Gollop-Wolfgang complex. Orphanet J Rare Dis. 2014;9:125. doi: 10.1186/s13023-014-0125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recalcati MP, Valtorta E, Romitti L, Giardino D, Manfredini E, Vaccari R, Larizza L, Finelli P. Characterisation of complex chromosome 18p rearrangements in two syndromic patients with immunological deficits. Eur J Med Genet. 2010;53:186–191. doi: 10.1016/j.ejmg.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- Wu N, Ming X, Xiao J, Wu Z, Chen X, Shinawi M, Shen Y, Yu G, Liu J, Xie H, Gucev ZS, Liu S, et al. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N Engl J Med. 2015;372:341–350. doi: 10.1056/NEJMoa1406829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.