Abstract
Background & Aims
During liver regeneration, hepatocytes are derived from pre-existing hepatocytes. However, if hepatocyte proliferation is compromised, biliary epithelial cells (BECs) become the source of new hepatocytes. We recently reported on a zebrafish liver regeneration model in which BECs extensively contribute to hepatocytes. Using this model, we performed a targeted chemical screen to identify important factors that regulate BEC-driven liver regeneration, the mechanisms of which remain largely unknown.
Methods
Using Tg(fabp10a:CFP-NTR) zebrafish, we examined the effects of 44 selected compounds on BEC-driven liver regeneration. Liver size was assessed by fabp10a:DsRed expression; liver marker expression was analyzed by immunostaining, in situ hybridization and quantitative PCR. Proliferation and apoptosis were also examined. Moreover, we used a mouse liver injury model, choline-deficient, ethionine-supplemented (CDE) diet.
Results
We identified 10 compounds that affected regenerating liver size. Among them, only bromodomain and extraterminal domain (BET) inhibitors, JQ1 and iBET151, blocked both Prox1 and Hnf4a induction in BECs. BET inhibition during hepatocyte ablation blocked BEC dedifferentiation into hepatoblast-like cells (HB-LCs). Intriguingly, after JQ1 washout, liver regeneration resumed, indicating temporal, but not permanent, perturbation of liver regeneration by BET inhibition. BET inhibition after hepatocyte ablation suppressed the proliferation of newly generated hepatocytes and delayed hepatocyte maturation. Importantly, Myca overexpression, in part, rescued the proliferation defect. Furthermore, oval cell numbers in mice fed CDE diet were greatly reduced upon JQ1 administration, supporting the zebrafish findings.
Conclusions
BET proteins regulate BEC-driven liver regeneration at multiple steps: BEC dedifferentiation, HB-LC proliferation, the proliferation of newly generated hepatocytes, and hepatocyte maturation.
Keywords: liver progenitor cells, dedifferentiation, JQ1, oval cells, zebrafish, BET inhibitors, liver regeneration
Introduction
In terms of the origin of regenerated hepatocytes, there are two types of liver regeneration: hepatocyte- and biliary-driven liver regeneration. Upon liver injury or resection, hepatocytes proliferate to recover the lost liver mass. However, if hepatocyte proliferation is compromised, as observed in chronic liver diseases and certain liver toxin injury models, liver progenitor cells (LPCs), also called oval cells, are activated and these proliferative LPCs contribute to regenerated hepatocytes [1, 2]. These LPCs appear to be derived from biliary epithelial cells (BECs) in the canals of Hering [1, 2]. This BEC-driven liver regeneration has been postulated based on in vitro data and marker expression analyses of rodent and human samples. However, recent lineage tracing studies in mice reveal that pre-existing hepatocytes are the main source of regenerated hepatocytes in current oval cell activation models [3, 4], raising a controversy about BEC-driven liver regeneration. Detailed marker analyses of human livers with cirrhosis [5] and massive hepatic necrosis [6] strongly suggest BEC contribution to regenerated hepatocytes in the human livers. However, the origin of regenerated hepatocytes in humans is still inconclusive due to a lack of cell-lineage tracing data. Importantly, the Forbes group recently reported the first evidence of robust BEC-driven liver regeneration in mice [7]. Hepatocyte-specific deletion of Mdm2 causes hepatocyte senescence and subsequent apoptosis, which completely blocks hepatocyte proliferation. In these mice, oval cell activation occurs and LPCs derived from BECs give rise to hepatocytes, thereby resulting in liver recovery. We previously reported that injury severity influences the extent of BEC-driven liver regeneration and that upon extreme hepatocyte ablation, BECs extensively give rise to hepatocytes in zebrafish [8], a phenomenon further confirmed by two separate groups [9, 10].
Liver transplantation is the only definitive treatment for end-stage liver disease; however, the shortage of donor livers makes this therapy extremely limited. Thus, augmenting innate BEC-driven liver regeneration in chronic liver diseases may be an attractive therapeutic alternative for such patients. A better understanding of this process at the molecular level will provide mechanistic insights and may lead to future clinical therapies. Given the extensive contribution of BECs to hepatocytes in the zebrafish regeneration model, we used this model to identify critical players that regulate BEC-driven liver regeneration.
Here, we report a targeted chemical screen that identified two compounds, iBET151 and JQ1, which inhibit the function of bromodomain and extraterminal (BET) proteins, as potent inhibitors of BEC-driven liver regeneration. The BET protein family, consisting of BRDT, BRD2, BRD3, and BRD4 in mammals, shares two highly conserved N-terminal bromodomains and a C-terminal extraterminal domain [11]. The bromodomain is a chromatin interaction module that recognizes acetylated lysine residues on histone tails; the extraterminal domain interacts with other histone modifying proteins. By binding to the acetylated lysine residues and recruiting transcriptional regulator components, BET proteins regulate the transcription of target genes. The recent development of the highly potent, specific BET inhibitors, JQ1 [12], iBET762 [13] and iBET151 [14], led to the explosion of BET protein research in the cancer field because of its potent anti-cancer effect on various tumors in animal models [15–17]. As a result, some BET inhibitors are in clinical trials for patients with T-cell lymphoma and multiple myeloma. In contrast to the extensive research on BET proteins in cancers, there are few reports describing their roles in regeneration [18] or liver biology [19]. Moreover, their role in liver regeneration has not been reported yet. Here, using the BET-specific inhibitors, we investigated the roles of BET proteins in BEC-driven liver regeneration and revealed their essential roles in this process.
Materials and methods
Zebrafish studies
Experiments were performed with approval of the Institutional Animal Care and Use Committee (IACUC) at the University of Pittsburgh. Embryos and adult fish were raised and maintained under standard laboratory conditions [20]. We used the following transgenic lines: Tg(fabp10a:CFP-nfsB)s931 [8], Tg(fabp10a:DsRed,ela31:EGFP)gz15 [21], Tg(EPV.TP1-Mmu.Hbb:hist2h2l-mCherry)s939 [22], Tg(EPV.TP1-Mmu.Hbb:Venus-Mmu.Odc1)s940 [22], Tg(fabp10a:mAGFP-gmnn,cryaa:ECFP)pt608, Tg(fabp10a:CAAX-EGFP)s942 [23], and Tg(hsp70l:VP16-myca,cryaa:ECFP)pt605 [referred to here as Tg(fabp10a:CFP-NTR), Tg(fabp10a:DsRed), Tg(Tp1:H2B-mCherry), Tg(Tp1:Venus-PEST), Tg(fabp10a:mAG-zGem), Tg(fabp10a:rasGFP), and Tg(hs:VP16-myca), respectively].
Hepatocyte ablation was performed by treating Tg(fabp10a:CFP-NTR) larvae with 10 mM metronidazole (Mtz) in egg water supplemented with 0.2% dimethyl sulfoxide (DMSO). For chemical screening, the larvae were treated with 44 compounds from A0h to R24h or from R0h to R48h, and fabp10a:DsRed expression at R24h or R48h was imaged to assess liver size using the Leica M205 FA microscope. For BET inhibition, larvae were treated with 3 μM JQ1 or 50 μM iBET151.
Detailed screening procedures, analytic methods, and the methods for mouse studies are described in the Supplementary material.
Results
BET inhibitor treatment impairs BEC-driven liver regeneration
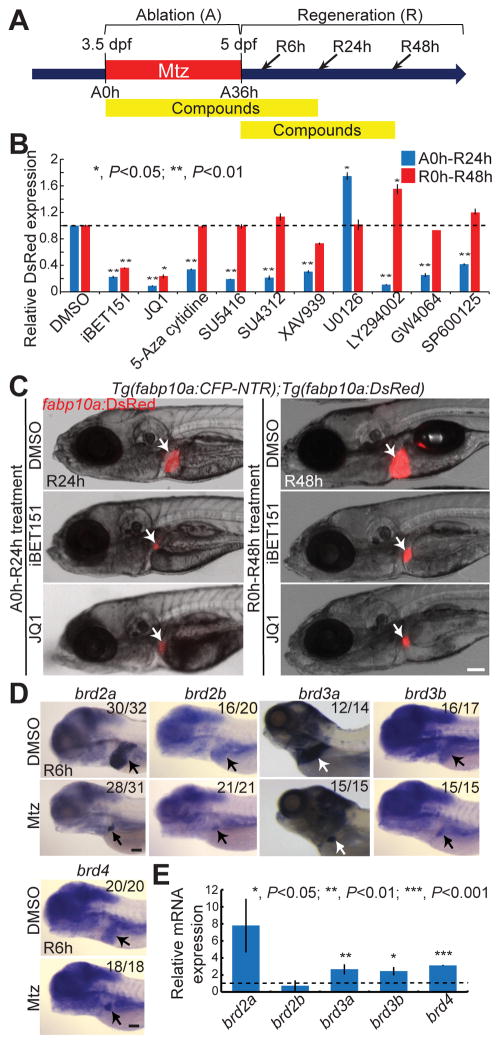
We [8] and others [9, 10] recently reported on a zebrafish liver regeneration model in which upon severe hepatocyte loss, regenerating hepatocytes are derived from BECs. In this model, transgenic fish that express nitroreductase (NTR) under the hepatocyte-specific fabp10a promoter were used. NTR converts the nontoxic prodrug, Mtz, into a cytotoxic drug; therefore, Mtz treatment results in hepatocyte ablation in the transgenic fish. Using this model, we performed a targeted chemical screen to identify pathways or factors that regulate BEC-driven liver regeneration (Supplementary Fig. 1). We used two transgenic lines for this screen: 1) Tg(fabp10a:CFP-NTR) that expresses NTR fused with cyan fluorescent protein (CFP) in hepatocytes, allowing for hepatocyte-specific ablation upon Mtz treatment, and 2) Tg(fabp10a:DsRed) that expresses DsRed in hepatocytes, allowing for easy assessment of liver size under an epifluorescence microscope. The double transgenic larvae were treated with Mtz from 3.5 to 5 days post-fertilization (dpf) for 36 hours (ablation, A36h). After Mtz washout (regeneration, R0h), which is scored as the start of regeneration, the liver size was analyzed at R24h or R48h. BEC-driven liver regeneration progresses through several steps, including BEC dedifferentiation into hepatoblast-like cells (HB-LCs), their proliferation and subsequent differentiation into hepatocytes, and the proliferation of newly generated hepatocytes (Supplementary Fig. 2) [8]. To identify the effect of compounds on both early and late steps of the regeneration, each compound was applied during two different time-windows: from A0h to R24h and from R0h to R48h (Fig. 1A). We screened 44 compounds with known targets to determine if they could regulate liver regeneration; of these, 10 compounds significantly affected liver size (Fig. 1B and Supplementary Table 1). Among these 10 compounds, only JQ1 and iBET151 significantly affected liver size, as assessed by fabp10a:DsRed expression, in both A0h-R24h and R0h-R48h treatments (Fig. 1B–1C).
Fig. 1. BET inhibitor treatment impairs BEC-driven liver regeneration.
(A) Scheme illustrating the periods of Mtz treatment (A, ablation), compound treatment, and liver regeneration (R). Arrows indicate analysis stages. (B) Graph showing the quantification of fabp10a:DsRed expression in the regenerating larvae treated with compounds from A0h to R24h (blue) or from R0h to R48h (red) (n=5–12). (C) Epifluorescence images showing fabp10a:DsRed expression in the regenerating larvae treated with iBET151 and JQ1. Arrows point to the liver. (D) WISH images showing the expression of brd2a, brd2b, brd3a, brd3b, and brd4 in the control and regenerating liver at R6h. Numbers indicate the proportion of larvae exhibiting the representative expression shown. Arrows point to the liver. (E) qPCR data showing the relative expression levels of brd2a, brd2b, brd3a, brd3b, and brd4 between control and regenerating liver at R6h. Error bars, ±SEM; scale bars, 100 μm.
To determine whether the hit compounds could block BEC dedifferentiation into HB-LCs, we examined the expression of both Prox1 and Hnf4a, used as HB-LC markers in combination [8], together with the expression of Tp1:H2B-mCherry, driven by Notch-responsive elements [22], which marks BECs and BEC-derived cells. The prolonged stability of H2B-mCherry allows one to trace cell lineages over several cell divisions [22]. JQ1 or iBET151 treatment blocked Prox1 and Hnf4a expression in Tp1:H2B-mCherry+ cells; GW4064, a farnesoid X receptor (FXR) agonist, reduced Hnf4a, but not Prox1, expression (Supplementary Table 1). Since the two BET inhibitors blocked both Prox1 and Hnf4a expression and BET proteins have not yet been implicated in liver regeneration, we selected these inhibitors for further detailed analyses. In the following analyses, JQ1 was preferably used over iBET151 due to the former’s superior efficacy and lower working concentration (Supplementary Fig. 3).
JQ1 specifically inhibits BET protein family members: BRDT, BRD2, BRD3, and BRD4. While Brdt is specifically expressed in testis, BRD2-4 are broadly expressed in mice [24, 25]. We examined the expression of their zebrafish orthologs, brd2a, brd2b, brd3a, brd3b and brd4, in control and regenerating larvae. All of the five genes are expressed in the normal liver and the regenerating liver at R6h (Fig. 1D). The expression of brd2a, brd3a, brd3b and brd4 was upregulated in the regenerating livers compared with the controls, as assessed by quantitative PCR (qPCR) (Fig. 1E). Altogether, these data suggest a critical role for BET proteins in BEC-driven liver regeneration.
BET inhibition blocks BEC dedifferentiation into HB-LCs and their proliferation
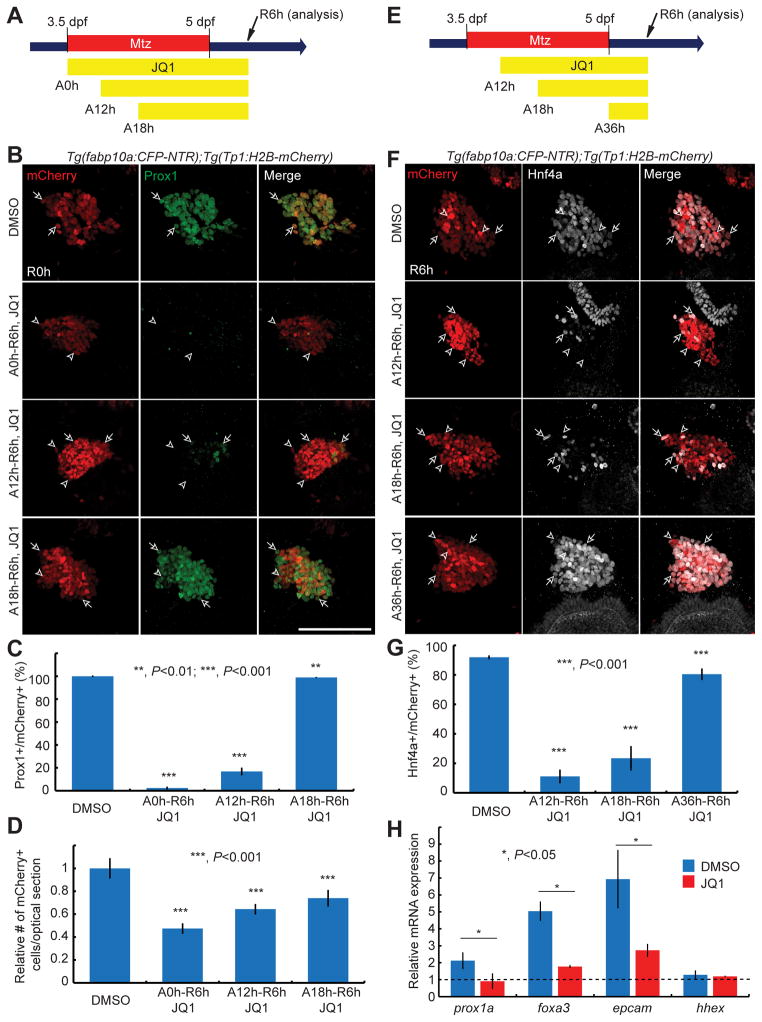
The finding that Prox1 and Hnf4a expression was greatly reduced in the regenerating iBET151 or JQ1-treated livers prompted us to investigate the role of BET proteins in BEC dedifferentiation. By treating hepatocyte-ablated larvae with JQ1 from several different stages (A0h, A12h, A18h, and A36h) to R6h (Fig. 2), we first defined the time-window during which BET proteins were required for the induction of Prox1 and Hnf4a in BECs. In the control regenerating liver at R6h, over 90% of Tp1:H2B-mCherry+ cells expressed Prox1 and Hnf4a. JQ1 treatment from A0h or A12h greatly blocked both Prox1 and Hnf4a expression in the regenerating liver, whereas JQ1 treatment from A18h greatly blocked Hnf4a, but not Prox1, expression (Fig. 2A–2G). JQ1 treatment from A36h modestly reduced the number of Hnf4a+ cells (Fig. 2E–2G). These data delineate A12h-A18h and A18h-A36h periods as the critical time-windows for Prox1 and Hnf4a induction in BECs, respectively.
Fig. 2. BET inhibition during hepatocyte ablation blocks BEC dedifferentiation into HB-LCs.
(A, E) Schemes illustrating the periods of Mtz and JQ1 treatment. (B, F) Confocal images showing the expression of Prox1 (B, green) or Hnf4a (F, grey) and Tp1:H2B-mCherry (red) in the regenerating liver. Tp1:H2B-mCherry expression reveals BECs and HB-LCs. Arrows point to the double-positive cells; arrowheads, mCherry single-positive cells. (C, G) Graphs showing the percentage of Prox1+ (C) or Hnf4a+ (G) cells among H2B-mCherry+ cells as shown in (B) (n=8–10) and (F) (n=5–18). (D) Graph showing the relative number of H2B-mCherry+ cells per optical section as shown in (B). (H) qPCR data showing the relative expression levels of prox1a, foxa3, epcam, and hhex in the liver at R6h between the control and regenerating larvae. The regenerating larvae were treated with DMSO (blue) or JQ1 (red) from A12h. Error bars, ±SEM; scale bars, 100 μm.
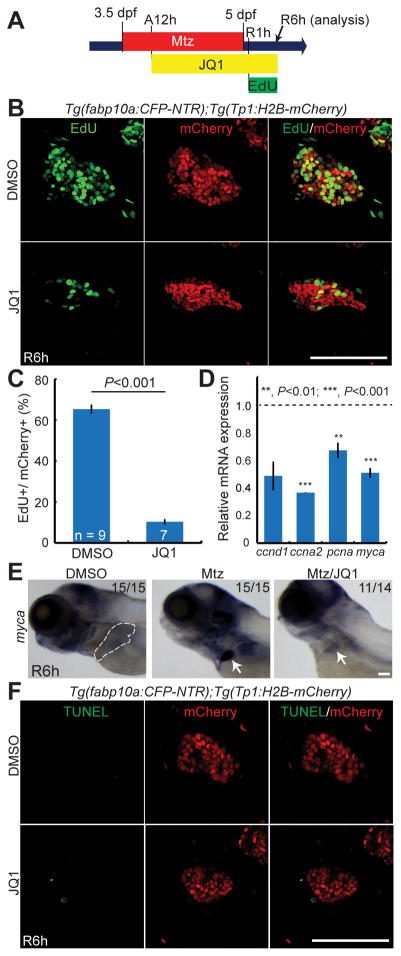
We next examined the expression levels of other hepatic markers expressed in hepatoblasts or LPCs, including foxa3, expressed in the endoderm even before hepatoblast specification [26, 27], prox1a and hhex, used as the early hepatoblast markers in zebrafish [26, 28], and epcam, used as a LPC marker in mammals [29, 30]. Non-ablated livers and regenerating livers treated with JQ1 or DMSO from A12h were dissected at R6h and used for qPCR analyses. foxa3, prox1a, and epcam, but not hhex, expression increased in the regenerating livers compared with the non-ablated livers; this increased expression was significantly reduced in the JQ1-treated regenerating livers (Fig. 2H). Since the number of Tp1:H2B-mCherry+ cells at R6h was reduced in larvae treated with JQ1 compared to controls (Fig. 2D), we also investigated cell proliferation and death. 5-ethynyl-2′-deoxyuridine (EdU) labeling revealed that the proliferation of Tp1:H2B-mCherry+ cells was greatly reduced in JQ1-treated larvae compared with controls at R6h (Fig. 3A–3C). Moreover, qPCR analyses revealed that the expression of proliferation-related genes, including ccnd1, ccna2, pcna, and myca, was reduced in the JQ1-treated regenerating livers compared with controls (Fig. 3D). Whole-mount in situ hybridization (WISH) further validated the reduced expression of myca in the JQ1-treated regenerating liver (Fig. 3E). However, there were no TUNEL+ cells among Tp1:H2B-mCherry+ cells in the control and the JQ1-treated regenerating liver at R6h (Fig. 3F), indicating that JQ1 treatment did not induce cell death in BECs or HB-LCs. Altogether, these data reveal the role of BET proteins in BEC dedifferentiation into HB-LCs and their proliferation.
Fig. 3. BET inhibition during hepatocyte ablation blocks HB-LC proliferation.
(A) Scheme illustrating the periods of Mtz, JQ1, and EdU treatment. (B, F) Confocal images of the regenerating liver at R6h processed for EdU (B) or TUNEL (F) labeling (green) and fluorescence detection of Tp1:H2B-mCherry (red). (C) Graph showing the percentage of EdU+ cells among H2B-mCherry+ cells as shown in (B). (D) qPCR data showing the relative expression levels of ccnd1, ccna2, pcna, and myca in the liver at R6h between the DMSO- or JQ1-treated larvae. (E) WISH images showing myca expression in the control and regenerating liver. Dashed lines outline the normal liver; arrows point to the regenerating liver. Error bars, ±SEM; scale bars, 100 μm.
BEC dedifferentiation resumes once BET inhibition is terminated
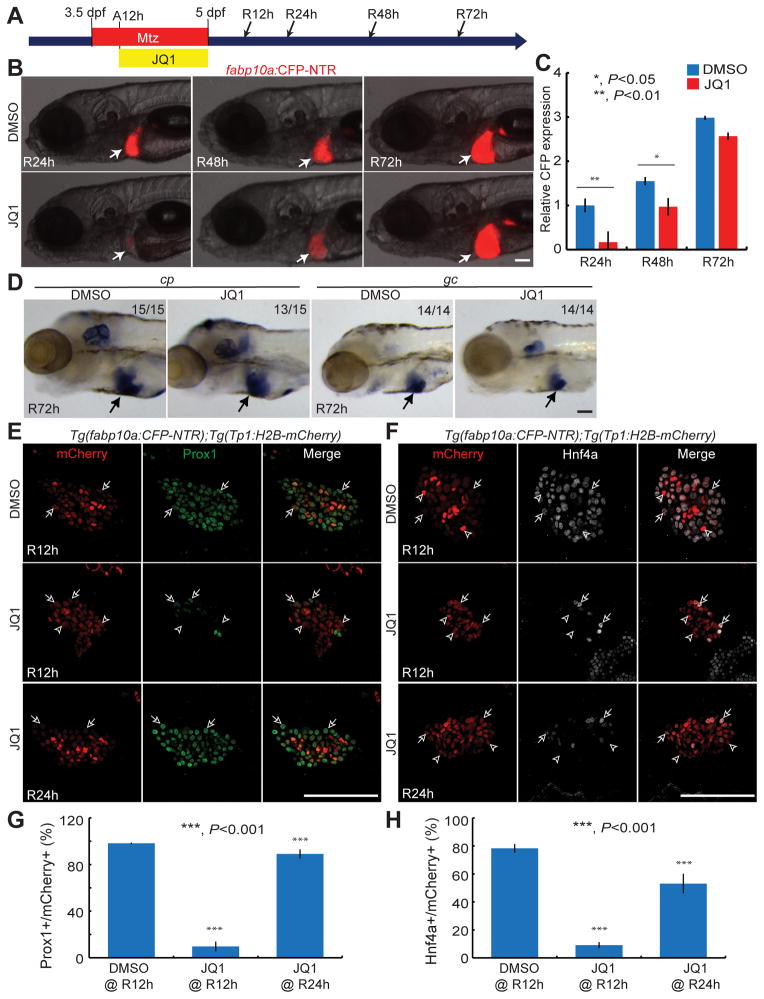
To examine whether BET inhibition blocked dedifferentiation permanently or whether BEC dedifferentiation could resume after JQ1 washout, larvae were treated with JQ1 only from A12h to R0h (Fig. 4A). In the JQ1-treated larvae at R24h, the liver size was smaller compared to controls; however, later at R48h, it greatly increased and finally reached the size of the control regenerating liver at R72h (Fig. 4B–4C). Moreover, hepatocyte markers, including ceruloplasmin (cp) and group-specific component (gc), were expressed in the JQ1-treated livers at R72h (Fig. 4D). Given the recovery of liver regeneration after JQ1 washout, we examined how early Prox1 and Hnf4a were induced after JQ1 washout. Prox1 and Hnf4a expression was detected in most Tp1:H2B-mCherry+ cells from R24h (Fig. 4E–4H). Altogether, these data indicate that temporal BET inhibition does not permanently impair BEC-driven liver regeneration.
Fig. 4. BEC-driven liver regeneration resumes after JQ1 washout.
(A) Scheme illustrating the periods of Mtz and JQ1 treatment and analysis stages (arrows). (B) Epifluorescence images showing fabp10a:CFP-NTR expression in the regenerating liver (arrows). (C) Graph showing the quantification of liver size shown in (B) (n=9–12). (D) WISH images showing cp and gc expression in the regenerating liver at R72h (arrows). (E, F) Single confocal optical section images of the regenerating liver immunostained for Prox1 (E, green) or Hnf4a (F, grey) and processed for Tp1:H2B-mCherry detection (red). Arrows point to the double-positive cells; arrowheads, mCherry single-positive cells. (G, H) Graphs showing the percentage of Prox1+ (G) or Hnf4a+ (H) cells among H2B-mCherry+ cells as shown in (E) (n=4–9) and (F) (n=5–10). Error bars, ±SEM; scale bars, 100 μm.
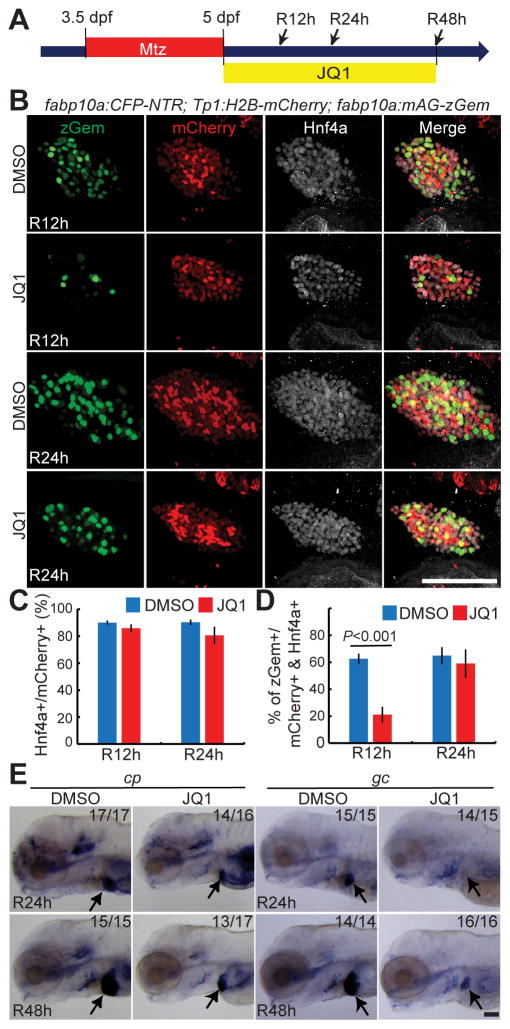
BET inhibition during the regeneration stage represses hepatocyte proliferation and delays its maturation
As BET inhibition from A36h/R0h reduced the liver size of the regenerating larvae at R48h (Fig. 1C), we explored the role of BET proteins during the regeneration period by examining 1) BEC dedifferentiation, 2) proliferation, 3) cell death, and 4) hepatocyte differentiation in the regenerating larvae treated with JQ1 from R0h (Fig. 5A). Hnf4a expression in the JQ1-treated liver was similar to that in the control regenerating liver (Fig. 5B), indicating that JQ1 treatment from R0h did not affect BEC dedifferentiation. To examine proliferation, we used the Tg(fabp10a:mAG-zGmn) line that expresses geminin fused with monomeric Azami green fluorescent proteins in hepatocytes. As geminin is degraded in G0 and G1 phases [31], this line reveals proliferating hepatocyte in S/G2/M phases. Intriguingly, JQ1 treatment from R0h reduced hepatocyte proliferation in the regenerating liver at R12h, but not R24h (Fig. 5B–5D). These data suggest that BET proteins regulate the proliferation of newly generated hepatocytes at an earlier, but not later, stage, which was further supported by the lack of a JQ1 effect on hepatocyte proliferation in the non-ablated liver at 5 dpf (Supplementary Fig. 4). TUNEL labeling revealed no dying cells among Tp1:H2B-mCherry+ cells in either the control or the JQ1-treated regenerating liver at R12h (Supplementary Fig. 5). Last, we examined cp and gc expression to determine the effect of JQ1 on hepatocyte differentiation. Both cp and gc were expressed in the control regenerating liver at R24h, whereas cp, but not gc, was expressed in the JQ1-treated liver at this stage (Fig. 5E). Later at R48h, weak gc expression was detected (Fig. 5E). Intriguingly, the order of cp and gc expression in the JQ1-treated regenerating liver is the same as that in the embryonic developing liver [32]. Collectively, the detailed analyses of the regenerating liver treated with JQ1 from R0h revealed two significant roles of BET proteins in BEC-driven liver regeneration: proliferation of newly generated hepatocytes and their maturation.
Fig. 5. BET inhibition from R0h reduces hepatocyte proliferation and delays its maturation.
(A) Scheme illustrating the periods of Mtz and JQ1 treatment and analysis stages (arrows). (B) Confocal images of the regenerating liver at R12h and R24h immunostained for Hnf4a (grey) and processed for Tp1:H2B-mCherry (red) and fabp10a:mAG-zGem (green) detection. Strong and weak H2B-mCherry+ cells are BECs and BEC-derived hepatocytes, respectively, at these stages. mAG-zGem+ cells are proliferating hepatocytes. (C, D) Graphs showing the percentage of Hnf4a+ (C) and mAG-zGem+ (D) cells among H2B-mCherry+ cells as shown in (B) (n=4–10). (E) WISH images showing cp and gc expression in the regenerating liver at R24h and R48h. Arrows point to the regenerating liver. Error bars, ±SEM; scale bars, 100 μm.
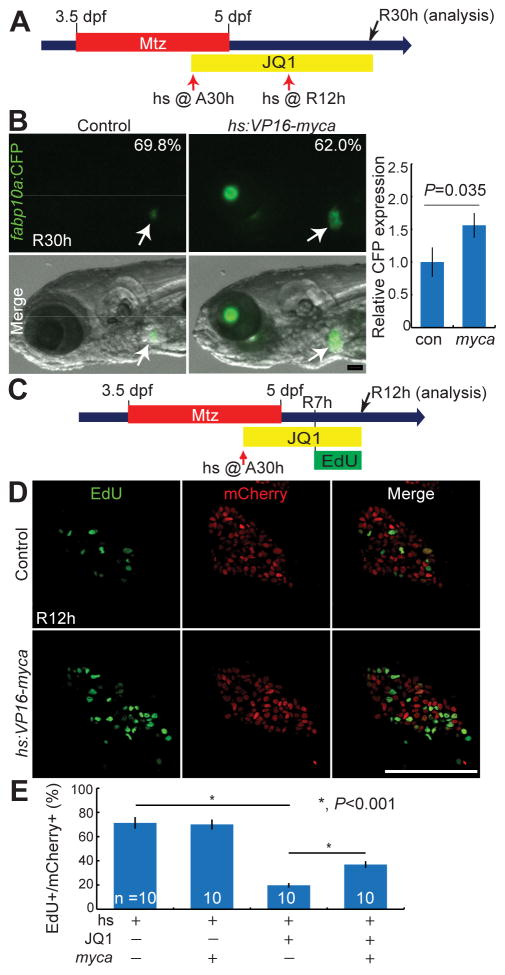
Myca overexpression partially rescues the hepatocyte proliferation defect observed in JQ1-treated regenerating larvae
BET inhibitors have a potent anti-cancer effect on multiple cancers via repressing the expression of key oncogenes, such as MYC [15] and BCL2 [14]. Importantly, MYC overexpression in multiple tumor cell lines partially rescued a proliferation defect elicited by BET inhibition [14, 15, 17]. Since myca was also greatly reduced in the JQ1-treated regenerating liver (Fig. 3E), we attempted to rescue the proliferation defect by overexpressing Myca. We used the Tg(hs:VP16-myca) line that expresses Myca fused with the VP16 transactivation domain upon heat-shock (Supplementary Fig. 6). As heat-shock alone between A33h and R6h killed most of the regenerating larvae (over 80%), we heat-shocked the larvae at A30h, immediately followed by JQ1 treatment, which increased larval survival over 90%. The second heat-shock was performed at R12h to sustain Myca overexpression. Myca overexpression significantly increased liver size, as assessed by fabp10a:CFP-NTR expression, in the JQ1-treated larvae (62%, n=163) compared with controls (Fig. 6A–6B). Since JQ1 treatment reduced liver size via reduced proliferation, we next examined whether Myca overexpression also rescued proliferation defects observed in JQ1-treated larvae. EdU labeling revealed that Myca overexpression significantly increased the proliferation of Tp1:H2B-mCherry+ cells in the JQ1-treated regenerating liver at R12h (Fig. 6C–6D), albeit not to the level observed in the regenerating liver without JQ1 treatment (Fig. 6E). Altogether, these rescue data suggest that BET proteins regulate proliferation in the regenerating liver, in part, via regulating myca expression.
Fig. 6. Myca overexpression partially rescues the proliferation defect observed in JQ1-treated larvae.
(A) Scheme illustrating the periods of Mtz and JQ1 treatment and the stages of heat-shock (red arrows). (B) Epifluorescence images showing fabp10a:CFP-NTR expression in the regenerating larvae at R30h (arrows) and their quantification. Numbers indicate the percentage of larvae showing the corresponding liver size (controls, n=63; hs:VP16-myca larvae, n=163). A graph shows the relative liver size, as assessed by hepatic CFP expression, between control and VP16-Myca-overexpressing larvae (controls, n=14; hs:VP16-myca larvae, n=20). (C) Scheme illustrating the periods of Mtz, JQ1, and EdU treatment and the stages of heat-shock (red arrow) and analysis (black arrow). (D) Single confocal optical section images of the regenerating liver at R12h processed for EdU labeling (green) and fluorescence detection of Tp1:H2B-mCherry (red). (E) Graph showing the percentage of EdU+ cells among H2B-mCherry+ cells as shown in (D). Error bars, ±SEM; scale bars, 100 μm.
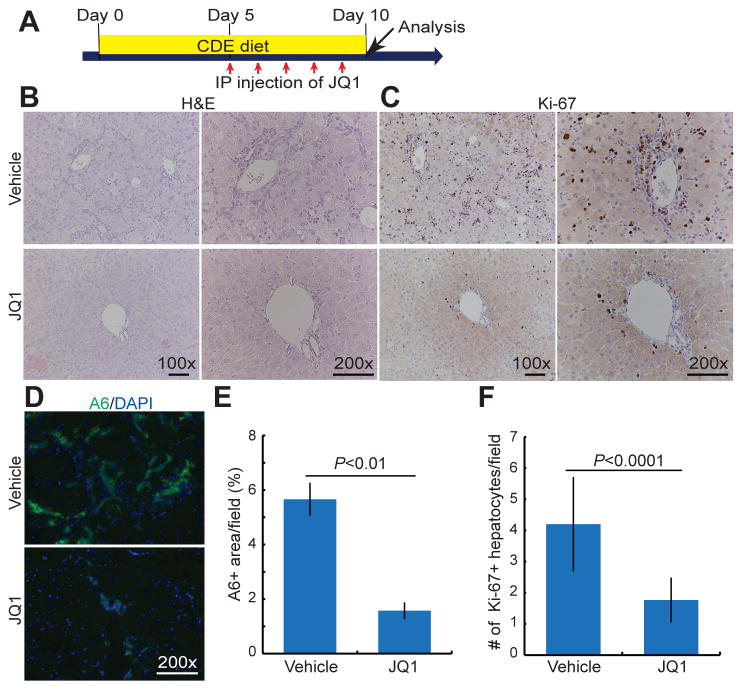
BET inhibition greatly reduces oval cell number in mice fed a CDE diet
The critical roles of BET proteins in BEC-driven liver regeneration in zebrafish prompted us to investigate its role in oval cell activation in mice. We selected a choline-deficient, ethionine-supplemented (CDE) diet model for this investigation because in this liver injury model, oval cells are mainly derived from BECs [4, 33]. Mice were fed a CDE diet for 10 days; JQ1 or vehicle control was injected daily into the mice from day 5 to day 9 (Fig. 7A). H&E staining on hepatic sections from the vehicle-treated group shows a clear expansion of the portal area with a large number of small cells with high nuclear to cytoplasmic ratio expanding into the hepatic lobule (Fig. 7B). These oval cells were notably reduced in the JQ1-injected group (Fig. 7B). To verify the identity of these cells, we next examined immunofluorescence staining for A6, a well-known oval cell marker [34]. Greatly reduced A6-positive oval cells were identified in the JQ1-injected mice compared with controls (Fig. 7D and 7E). Proliferation, as assessed by the expression of Ki-67, a marker for the S phase of the cell cycle, was also greatly reduced in the JQ1-injected mice as compared with controls (Fig. 7C). The quantification of Ki-67+ hepatocytes also revealed reduced hepatocyte proliferation (Fig. 7F). To rule out that the observed reduction in oval cell number is not due to reduced liver damage in the JQ1-injected mice, we compared liver damage between the control and JQ1-injected groups. Serum alanine aminotransferase (ALT), total bilirubin, and the number of TUNEL+ hepatocytes, denoting cell death, were comparable between the two groups (Supplementary Fig. 7D–7G). Likewise, the number of macrophages, as assessed by F4/80 expression, was not reduced in the livers of JQ1-injected animals as compared with the controls (Supplementary Fig. 7C). Interestingly, fewer CD45+ inflammatory cells were observed in the livers of the JQ1-injected group (Supplementary Fig. 7B), and may be secondary to reduced oval cells, which are an important source of pro-inflammatory cytokines [35]. Altogether, these mouse data indicate that BET proteins regulate oval cell activation in mice as well as in zebrafish.
Fig. 7. BET inhibition greatly reduces oval cell number and its proliferation in mice fed a CDE diet.
(A) Scheme illustrating the period of a CDE diet and JQ1 injection stages. (B, C) Representative images of the liver sections processed for H&E staining (B) or immunostained for Ki-67 (C). (D, E) Epifluorescence images of the liver sections processed for A6 immunostaining and their quantification (n=3). (F) Quantification of the number of Ki-67+ hepatocytes as shown in (C). Error bars, ±SEM; scale bars, 100 μm.
Discussion
In this study, we performed a targeted chemical screen using the zebrafish hepatocyte ablation model. Among the 10 identified compounds that affected BEC-driven liver regeneration, we focused on BET inhibitors because they blocked both Prox1 and Hnf4a expression in BECs, implicating BET proteins in the BEC dedifferentiation step of BEC-driven liver regeneration. Besides the initial step of BEC-driven liver regeneration, BET proteins regulate HB-LC proliferation and the proliferation and maturation of newly generated hepatocytes. Furthermore, we show that BET proteins are important for mouse oval cell activation, consistent with their roles in zebrafish BEC-driven liver regeneration.
Given the small size and optical transparency of zebrafish embryos/larvae and their growth in water, zebrafish embryos/larvae have widely been used for in vivo chemical screening [36, 37]. Our liver regeneration model, which simultaneously results in severe and synchronous hepatocyte ablation in hundreds of larvae, made it possible for us to complete the targeted chemical screen reported here, which will be very challenging in any rodent liver regeneration models.
BET proteins bind to acetylated lysine on histone tails, and function as a scaffold protein to assemble diverse chromatin protein complexes [11, 38, 39]. By recruiting transcriptional regulatory complexes, they regulate the transcription of target genes [11, 39]. BET-specific inhibition greatly reduces proliferation and enhances apoptosis in various cancer cells by repressing key oncogenes, such as Myc, Bcl2, and Fosl1 [15, 16, 40]. Importantly, MYC overexpression rescues the proliferation defect elicited by BET inhibition in multiple cancer cells [15, 17]. We observed that in the regenerating liver, not only was myca expression greatly increased, but this increase was also suppressed by JQ1 treatment. Importantly, Myca overexpression rescued the hepatocyte proliferation defect observed in JQ1-treated larvae at R12h, implicating BET proteins in the regulation of hepatocyte proliferation during BEC-driven liver regeneration via myca activation. However, this regulation was not observed in the non-ablated normal liver or in the regenerating liver at R24h (Fig. 5D), suggesting that BET proteins regulate the proliferation of newly generated, immature, but not mature, hepatocytes. This regulation of immature hepatocyte proliferation is supported by a mouse study showing that Brd4+/− mice displayed reduced hepatic proliferation at E15 [19], when the liver consists of hepatoblasts and immature hepatocytes [41].
In contrast to the proliferation defect, we did not observe any cell death in BECs or BEC-derived cells in the JQ1-treated regenerating liver. BET inhibition results in both cell cycle arrest and apoptosis in many cancers [15, 40, 42]. These two phenotypes appear to result from the repression of distinct genes, because MYC overexpression rescues the proliferation, but not apoptosis, phenotype in acute myeloid leukemia [17] and BCL2 overexpression rescues the apoptosis, but not proliferation, phenotype in MLL-fusion leukemia [14]. This independent regulation of proliferation and apoptosis might explain why BET inhibition did not induce apoptosis, but did block proliferation in the regenerating liver.
Since JQ1 washout restored BEC dedifferentiation and subsequent hepatocyte differentiation, we wondered about the identity of BECs in the presence of JQ1. Since these cells failed to express Prox1 or Hnf4a, they cannot be classified as HB-LCs. Moreover, they differ from BECs in non-ablated livers because a subset of genes, which are upregulated during BEC-driven liver regeneration, remained upregulated in the JQ1-treated regenerating liver (e.g., mmp15b and atf7ip; Supplementary Fig. 8). Given the role of BET proteins as a reader of chromatin modifications [39], it is likely that chromatin modifications necessary for BEC dedifferentiation occurred and stalled in the JQ1-treated regenerating liver. Therefore, after JQ1 washout, BEC dedifferentiation and subsequent hepatocyte differentiation resumed. Interestingly, deleterious phenotypes observed in the mouse skin and intestine following the inducible overexpression of Brd4 shRNA completely resolve after Brd4 shRNA expression is turned off [18], consistent with the resumption of BEC-driven liver regeneration after JQ1 washout.
Since five brd genes were expressed in the regenerating liver and four of them were upregulated during liver regeneration, our studies using pan-BET inhibitors cannot determine which brd genes play a key role in BEC-driven liver regeneration. As Brd4 knockdown results in the same defects elicited by BET inhibition in multiple cancer cells [17, 42, 43] and the effects of JQ1 on cardiomyocyte hypertrophy appear to be mediated by altering BRD4-dependent transcriptional program [44], Brd4 might be a key BET protein that regulates BEC-driven liver regeneration. However, it is still possible that other BET proteins are involved. Given the early embryonic lethality of Brd2 [25] and Brd4 [19] homozygous mutant mice, liver-specific conditional knockout animals will help to determine which BET proteins, individually or in combination, regulate BEC-driven liver regeneration.
Using the zebrafish liver injury model, we discovered multiple roles of BET proteins in BEC-driven liver regeneration. However, in mice, we only observed their role in the initial step of this process, mainly due to the limitation of current mouse liver injury models in which BECs barely or minimally contribute to hepatocytes [3, 4, 33]. Although we could not determine the role of BET proteins in later steps of BEC-driven liver regeneration in mice, we did observe that BET proteins play a crucial role in oval cell activation, the first step of LPC-driven liver regeneration in rodents. This step includes the change of BECs or LPCs into oval cells and subsequent oval cell proliferation [1, 2], which are similar to BEC dedifferentiation into HB-LCs and their proliferation in the zebrafish model. Given the recent advent of a mouse model in which robust BEC-driven liver regeneration occurs [7], it will be interesting to determine the effects of JQ1 on the entire process of BEC-driven liver regeneration in mice as shown here in zebrafish.
Supplementary Material
Acknowledgments
Financial support: The work was supported in part by grants from the March of Dimes Foundation (5-FY12-39) and the NIH (DK101426) to D.S. and from the NIH (DK62277 and DK100287) and by Endowed Chair for Experimental Pathology to S.P.S.M. J.O.R was funded on an NIH Training grant (T32EB001026).
We thank James Bradner for JQ1 compounds, Tanaka Minoru for the CDE protocol, Hirohisa Okabe for the initial setup of the CDE liver injury model, Isla Cheung and Didier Stainier for the Tg(fabp10a:rasGFP) line, Neil Hukriede and Michael Tsang for discussions, and Mehwish Khaliq for critical reading of the manuscript. The work was supported in part by grants from the March of Dimes Foundation (5-FY12-39) and the NIH (DK101426) to D.S. and from the NIH (DK62277 and DK100287) and by Endowed Chair for Experimental Pathology to S.P.S.M. J.O.R is funded on a Training grant (T32EB001026) entitled Cellular Approaches to Tissue Engineering and Regeneration.
List of abbreviations
- BEC
biliary epithelial cell
- CDE
choline-deficient, ethionine-supplemented
- BET
bromodomain and extraterminal domain
- HB-LCs
hepatoblast-like cells
- LPC
liver progenitor cell
- Mtz
metronidazole
- DMSO
dimethyl sulfoxide
- NTR
nitroreductase
- CFP
cyan fluorescent protein
- dpf
days post-fertilization
- FXR
farnesoid X receptor
- qPCR
quantitative polymerase chain reaction
- EdU
5-ethynyl-2′-deoxyuridine
- WISH
whole-mount in situ hybridization
- ALT
alanine aminotransferase
Footnotes
Author names in bold designate shared co-first authorship.
Conflict of interest: The authors declare no conflict of interest.
Authors’ contributions:
S.K.: study concept and design; acquisition, analysis and interpretation of data; writing the manuscript
T.Y.C.: study concept and design; acquisition, analysis and interpretation of data
J.O.R.: acquisition, analysis and interpretation of data; writing the manuscript
J.S.: material support
S.S.M.: study design; analysis and interpretation of data; writing the manuscript
D.S.: study concept and design; analysis and interpretation of data; study supervision; writing the manuscript
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Duncan AW, Dorrell C, Grompe M. Stem Cells and Liver Regeneration. Gastroenterology. 2009;137:466–481. doi: 10.1053/j.gastro.2009.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fausto N, Campbell JS. The role of hepatocytes and oval cells in liver regeneration and repopulation. Mechanisms of Development. 2003;120:117–130. doi: 10.1016/s0925-4773(02)00338-6. [DOI] [PubMed] [Google Scholar]
- 3.Yanger K, Knigin D, Zong YW, Maggs L, Gu GQ, Akiyama H, et al. Adult Hepatocytes Are Generated by Self-Duplication Rather than Stem Cell Differentiation. Cell Stem Cell. 2014;15:340–349. doi: 10.1016/j.stem.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaub JR, Malato Y, Gormond C, Willenbring H. Evidence against a Stem Cell Origin of New Hepatocytes in a Common Mouse Model of Chronic Liver Injury. Cell Rep. 2014;8:933–939. doi: 10.1016/j.celrep.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stueck AE, Wanless IR. Hepatocyte buds derived from progenitor cells repopulate regions of parenchymal extinction in human cirrhosis. Hepatology. 2015 doi: 10.1002/hep.27706. [DOI] [PubMed] [Google Scholar]
- 6.Craig CEH, Quaglia A, Selden C, Lowdell M, Hodgson H, Dhillon AP. The histopathology of regeneration in massive hepatic necrosis. Semin Liver Dis. 2004;24:49–64. doi: 10.1055/s-2004-823101. [DOI] [PubMed] [Google Scholar]
- 7.Lu WY, Bird TG, Boulter L, Tsuchiya A, Cole AM, Hay T, et al. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat Cell Biol. 2015;17:971–U943. doi: 10.1038/ncb3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi TY, Ninov N, Stainier DY, Shin D. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology. 2014;146:776–788. doi: 10.1053/j.gastro.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang M, Chang A, Choi M, Zhou D, Anania FA, Shin CH. Antagonistic interaction between Wnt and Notch activity modulates the regenerative capacity of a zebrafish fibrotic liver model. Hepatology. 2014 doi: 10.1002/hep.27285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He J, Lu H, Zou Q, Luo L. Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology. 2014;146:789–800. e788. doi: 10.1053/j.gastro.2013.11.045. [DOI] [PubMed] [Google Scholar]
- 11.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci U S A. 2012;109:19408–19413. doi: 10.1073/pnas.1216363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolden JE, Tasdemir N, Dow LE, van Es JH, Wilkinson JE, Zhao Z, et al. Inducible in vivo silencing of Brd4 identifies potential toxicities of sustained BET protein inhibition. Cell Rep. 2014;8:1919–1929. doi: 10.1016/j.celrep.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houzelstein D, Bullock SL, Lynch DE, Grigorieva EF, Wilson VA, Beddington RS. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Molecular and cellular biology. 2002;22:3794–3802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) University of Oregon Press; 2000. [Google Scholar]
- 21.Korzh S, Pan X, Garcia-Lecea M, Winata CL, Pan X, Wohland T, et al. Requirement of vasculogenesis and blood circulation in late stages of liver growth in zebrafish. Bmc Dev Biol. 2008;8:84. doi: 10.1186/1471-213X-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ninov N, Borius M, Stainier DYR. Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development. 2012;139:1557–1567. doi: 10.1242/dev.076000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung ID, Bagnat M, Ma TP, Datta A, Evason K, Moore JC, et al. Regulation of intrahepatic biliary duct morphogenesis by Claudin 15-like b. Dev Biol. 2012;361:68–78. doi: 10.1016/j.ydbio.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paillisson A, Levasseur A, Gouret P, Callebaut I, Bontoux M, Pontarotti P, et al. Bromodomain testis-specific protein is expressed in mouse oocyte and evolves faster than its ubiquitously expressed paralogs BRD2, -3, and -4. Genomics. 2007;89:215–223. doi: 10.1016/j.ygeno.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Shang E, Wang X, Wen D, Greenberg DA, Wolgemuth DJ. Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse. Dev Dyn. 2009;238:908–917. doi: 10.1002/dvdy.21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Field HA, Ober EA, Roeser T, Stainier DY. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev Biol. 2003;253:279–290. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- 27.Odenthal J, Nusslein-Volhard C. fork head domain genes in zebrafish. Dev Genes Evol. 1998;208:245–258. doi: 10.1007/s004270050179. [DOI] [PubMed] [Google Scholar]
- 28.Ober EA, Verkade H, Field HA, Stainier DY. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006;442:688–691. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- 29.Yoon SM, Gerasimidou D, Kuwahara R, Hytiroglou P, Yoo JE, Park YN, et al. Epithelial Cell Adhesion Molecule (EpCAM) Marks Hepatocytes Newly Derived from Stem/Progenitor Cells in Humans. Hepatology. 2011;53:964–973. doi: 10.1002/hep.24122. [DOI] [PubMed] [Google Scholar]
- 30.Okabe M, Tsukahara Y, Tanaka M, Suzuki K, Saito S, Kamiya Y, et al. Potential hepatic stem cells reside in EpCAM(+) cells of normal and injured mouse liver. Development. 2009;136:1951–1960. doi: 10.1242/dev.031369. [DOI] [PubMed] [Google Scholar]
- 31.Sugiyama M, Sakaue-Sawano A, Iimura T, Fukami K, Kitaguchi T, Kawakami K, et al. Illuminating cell-cycle progression in the developing zebrafish embryo. Proc Natl Acad Sci U S A. 2009;106:20812–20817. doi: 10.1073/pnas.0906464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noel ES, dos Reis M, Arain Z, Ober EA. Analysis of the Albumin/alpha-Fetoprotein/Afamin/Group specific component gene family in the context of zebrafish liver differentiation. Gene Expression Patterns. 2010;10:237–243. doi: 10.1016/j.gep.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Espanol-Suner R, Carpentier R, Van Hul N, Legry V, Achouri Y, Cordi S, et al. Liver Progenitor Cells Yield Functional Hepatocytes in Response to Chronic Liver Injury in Mice. Gastroenterology. 2012;143:1564–1575. doi: 10.1053/j.gastro.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 34.Preisegger KH, Factor VM, Fuchsbichler A, Stumptner C, Denk H, Thorgeirsson SS. Atypical ductular proliferation and its inhibition by transforming growth factor beta1 in the 3,5-diethoxycarbonyl-1,4-dihydrocollidine mouse model for chronic alcoholic liver disease. Lab Invest. 1999;79:103–109. [PubMed] [Google Scholar]
- 35.O’Hara SP, Tabibian JH, Splinter PL, LaRusso NF. The dynamic biliary epithelia: Molecules, pathways, and disease. J Hepatol. 2013;58:575–582. doi: 10.1016/j.jhep.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rennekamp AJ, Peterson RT. 15 years of zebrafish chemical screening. Curr Opin Chem Biol. 2015;24C:58–70. doi: 10.1016/j.cbpa.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaufman CK, White RM, Zon L. Chemical genetic screening in the zebrafish embryo. Nat Protoc. 2009;4:1422–1432. doi: 10.1038/nprot.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prinjha RK, Witherington J, Lee K. Place your BETs: the therapeutic potential of bromodomains. Trends Pharmacol Sci. 2012;33:146–153. doi: 10.1016/j.tips.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Bandopadhayay P, Bergthold G, Nguyen B, Schubert S, Gholamin S, Tang Y, et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin Cancer Res. 2014;20:912–925. doi: 10.1158/1078-0432.CCR-13-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zorn AM. Liver development. StemBook; Cambridge (MA): 2008. [Google Scholar]
- 42.Tolani B, Gopalakrishnan R, Punj V, Matta H, Chaudhary PM. Targeting Myc in KSHV-associated primary effusion lymphoma with BET bromodomain inhibitors. Oncogene. 2014;33:2928–2937. doi: 10.1038/onc.2013.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fiskus W, Sharma S, Qi J, Valenta JA, Schaub LJ, Shah B, et al. Highly active combination of BRD4 antagonist and histone deacetylase inhibitor against human acute myelogenous leukemia cells. Mol Cancer Ther. 2014;13:1142–1154. doi: 10.1158/1535-7163.MCT-13-0770. [DOI] [PubMed] [Google Scholar]
- 44.Anand P, Brown JD, Lin CY, Qi J, Zhang R, Artero PC, et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154:569–582. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.