Abstract
Studies of mouse monoclonal CD4+ T cell repertoires have revealed several mechanisms of self-tolerance, however, which mechanisms operate in normal repertoires is unclear. Here, polyclonal CD4+ T cells specific for green fluorescent protein expressed in different organs were studied, allowing determination of the effects of specific expression patterns on the same epitope-specific T cells. Peptides presented uniformly by thymic antigen-presenting cells were tolerated by clonal deletion, whereas thymus-excluded peptides were ignored. Peptides with limited thymic expression induced partial clonal deletion and impaired effector but enhanced regulatory T cell potential. These mechanisms were also active for T cell populations specific for endogenously expressed self-antigens. Thus, immune tolerance of polyclonal CD4+ T cells is maintained by distinct mechanisms, according to self-peptide expression patterns.
Keywords: CD4+ T lymphocyte, immunological tolerance, clonal deletion, immunological ignorance
Self-reactive CD4+ T cells are generated in the normal course of T cell receptor (TCR) rearrangement1. Low affinity interactions between TCR and self-peptide-major histocompatibility class II (self-peptide:MHCII) ligands are essential for positive selection of T cells in the thymus1. High affinity TCR-self-peptide:MHCII interactions, however, can cause autoimmunity. Therefore, tolerance mechanisms have evolved to limit T cells with TCRs with high affinities for self-peptide:MHCII ligands.
Different tolerance mechanisms may be engaged depending on where a self-peptide:MHCII epitope is displayed. A developing thymocyte may die by apoptosis or differentiate into a regulatory T cell (Treg cell) if its TCR binds strongly to a self-peptide:MHCII epitope displayed on particular antigen-presenting cells1, 2. Epitopes in this class may be derived from proteins expressed or taken up by MHCII+ thymic epithelial cells or dendritic cells. However, MHCII+ cells in the thymus might not display epitopes from all host proteins. Thus, T cells expressing TCRs with high affinities for certain self-epitopes may not be deleted in the thymus and exit to secondary lymphoid organs2, 3, 4, 5, 6. Some T cells in this class may be specific for epitopes from proteins that are not secreted or are expressed in locations with poor lymphatic drainage. Since these proteins would not reach antigen-presenting cells in secondary lymphoid organs, specific T cells would be retained in the repertoire in a fully responsive state of ignorance7, 8. Other tissue-restricted proteins that are secreted or found in dying cells may reach secondary lymphoid organs where they would be processed by antigen-presenting cells to form self-peptide:MHCII epitopes. CD4+ T cells that encounter and recognize these epitopes may differentiate into Treg cells2 or become anergic due to a lack of innate immune system-derived costimulation1, 9.
Much of the evidence cited above has come from studies of transgenic mice expressing monoclonal TCRs specific for self-peptide:MHCII epitopes7, 8, 10. These mice could reveal tolerance mechanisms because of the large numbers of T cells available for study. However, this approach has several limitations. Because a TCR transgenic mouse contains a single TCR from a larger repertoire, it can only reveal tolerance mechanisms that apply to TCRs with that affinity for a self-peptide:MHCII epitope11. Further, it is clear that an abundance of T cells with the same TCR can create abnormal outcomes12, 13, 14. This issue has come to the fore in several studies showing that clonal deletion is not as prominent a mechanism in polyclonal T cell repertoires as has been reported for certain monoclonal T cell populations 4, 5, 15. These studies, however, have been limited to epitopes with ubiquitous patterns of expression.
A need therefore exists for a systematic study of tolerance within normal polyclonal CD4+ T cell repertoires specific for self-peptides with defined patterns of expression. Here, we carried out such a study using peptide:MHCII tetramer-based cell enrichment. Intrathymic deletion was the mechanism of tolerance for T cells specific for peptides from proteins expressed uniformly by thymic dendritic cells and/or medullary thymic epithelial cells, whereas ignorance was the mechanism for peptides from cytosolic or nuclear proteins expressed only outside the thymus. Partial intrathymic deletion and impaired effector but enhanced Treg cell potential among remaining clones was the mechanism for many peptides from tissue-restricted proteins that were also expressed sparsely in the thymus. Thus, CD4+ T cells tolerate self-peptides with different expression patterns via different mechanisms.
RESULTS
eGFP transgenic mice model CD4+ T cell tolerance to self
Enhanced green, yellow, or cyan fluorescent proteins (eGFP, eYFP, CFP) were used as model self-antigens in transgenic mouse strains expressing these proteins and the I-Ab MHCII molecule. These proteins contain an I-Ab-binding peptide (HDFFKSAMPEGYVQE, eGFPp), which was incorporated into an I-Ab tetramer that was used to detect CD4+ T cells with TCRs specific for this epitope (Fig. 1a,b) 16. This experimental system allowed determination of the effects of different patterns of self-antigen expression on the same epitope-specific T cell repertoire. Another advantage of this approach was that wild-type C57BL/6 (B6) mice, which express the I-Ab molecule but not eGFP, could be used as controls to study the relevant T cell repertoire in the absence of the self-peptide.
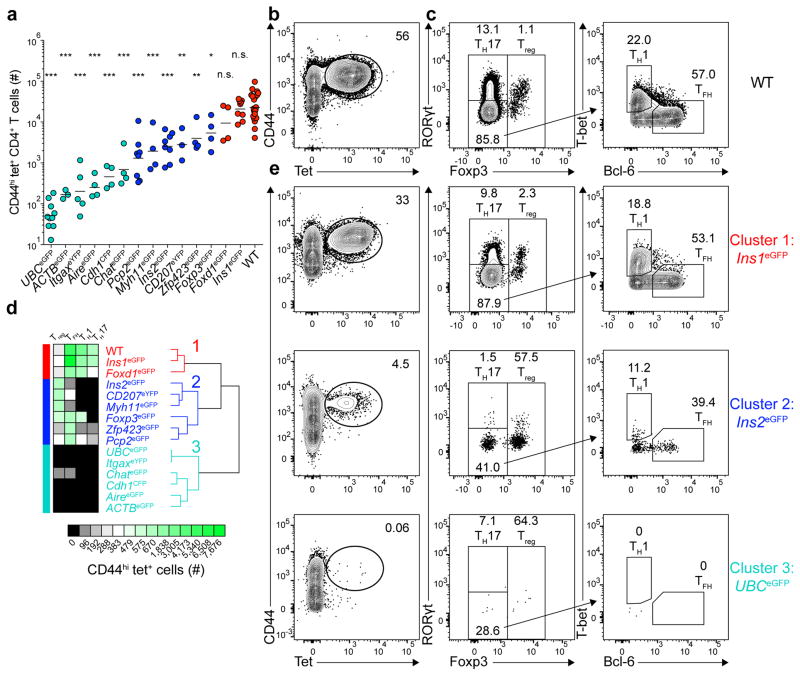
Figure 1. Three major patterns of tolerance to self-antigens.
(a) Total number of CD44hi tet+ (eGFPp:I-Ab-tetramer-binding) CD4+ T cells from pooled secondary lymphoid organs of mice 13–14 days after immunization with 100 μg of eGFPp emulsified in CFA. Horizontal bars indicate geometric mean values. Depicted results were pooled from 12 independent experiments, n=3–23 total mice per group. Circles represent individual mice and are colored according to the clusters identified in (d). (* P<0.05, ** P<0.005, *** P<0.0001, n.s. = not significant by one-way ANOVA of log10 transformed data relative to wild-type (WT) B6 mice).
(b) Contour plot showing CD44 and tetramer staining from tetramer-enriched CD4+ T cells from pooled secondary lymphoid organs of WT mice primed 14 days earlier with eGFPp in CFA. CD4+CD44hi tet+ cells are identified in the elliptical gate.
(c) Plots depicting transcription factor expression in the CD4+CD44hi tet+ cells identified in (b). Left, Foxp3 and RORγt expression by tet+ cells. Right, T-bet and Bcl-6 expression by Foxp3−RORγt− tet+ cells.
(d) Unsupervised hierarchical clustering of CD4+CD44hi tet+ T cells (geometric mean cell numbers and phenotypes) in the 15 indicated mouse strains using the HierarchicalClustering module from GenePattern. Three major clusters are indicated.
(e) Plots showing CD44 and tetramer staining of tetramer-enriched CD4+ T cells from pooled secondary lymphoid organs of three different eGFP expressing mouse strains that are representative examples of Clusters 1–3, identified in (d) (left panel) and transcription factor expression by the CD4+CD44hi tet+ cells identified in the elliptical gates in the left panels.
Numbers on each plot in (b–c) and (e) indicate the percent of cells in each gate.
Tolerance in the eGFPp:I-Ab-specific CD4+ T cell repertoire was examined by immunizing 14 different transgenic strains expressing eGFP, eYFP, or CFP (Supplementary table 1) with eGFPp in complete Freunds adjuvant (CFA). Single cell suspensions of secondary lymphoid organs from immunized mice were stained with the eGFPp:I-Ab tetramer labeled with allophycocyanin, then with anti-allophycocyanin-coated magnetic beads. Tetramer-bound cells were enriched on magnetized columns, eluted, and stained with antibodies designed to identify T cells. TH1, TH17, follicular helper (TFH), and Treg cell differentiation by the eGFPp:I-Ab-specific CD4+ T cells was assessed by intracellular staining of the lineage-defining transcription factors T-bet (Tbx21), RORγt (Rorc), Bcl-6, and Foxp3, respectively (Fig. 1c).
Wild-type B6 mice, for which eGFPp is a foreign peptide, had on average 23,000 eGFPp:I-Ab-specific CD44hi CD4+ T cells in their secondary lymphoid organs 14 days after immunization (Fig. 1a). Most of these cells were Foxp3− effector T cells, although a small subpopulation of Foxp3+ Treg cells was present (Fig. 1c). The Foxp3− population consisted of 10% RORγt+ TH17 cells, 22% T-bet+ TH1 cells, 30% Bcl-6+ TFH cells, and 35% could not be assigned to a known lineage (Fig. 1c).
The eGFP-, eYFP-, or CFP-transgenic mice generated a spectrum of eGFPp:I-Ab-specific CD44hi CD4+ effector T cell numbers in secondary lymphoid organs following immunization (Fig. 1a). At one end, insulin 1 (Ins1)eGFP mice produced the same number of cells as wild-type mice, indicating a complete lack of tolerance. At the other end, ubiquitin (UBC)eGFP mice generated fewer than 100 eGFPp:I-Ab-specific CD44hi T cells, indicating a profound state of tolerance. The other 12 transgenic strains produced 200–10,000 eGFPp:I-Ab-specific CD44hi T cells (Fig. 1a).
The eGFPp:I-Ab-specific CD44hi T cells in the transgenic mice also displayed a range of phenotypes. Treg, TH1, TFH, and TH17 cells generated in each strain after priming with eGFPp in CFA were analyzed using an unsupervised hierarchical clustering algorithm17 to search for patterns of tolerance induction (Fig. 1d). Absolute cell numbers were used for this clustering to capture information about clonal proliferation and differentiation. This analysis revealed three major clusters. Cluster 1 was typified by Ins1eGFP mice and was characterized by wild-type numbers of TFH, TH1, and TH17 cells and relatively few Treg cells (Fig. 1d,e). Cluster 2 was exemplified by Ins2eGFP mice and contained low numbers of TFH, TH1, and TH17 cells and relatively large numbers of Treg cells (Fig. 1d,e). Cluster 3 was typified by UBCeGFP mice and characterized by profound reduction of all subsets (Fig. 1d,e). These results suggest that three major mechanisms account for CD4+ T cell tolerance to self-antigens with different patterns of expression.
Ignorance is the tolerance mechanism for Cluster 1
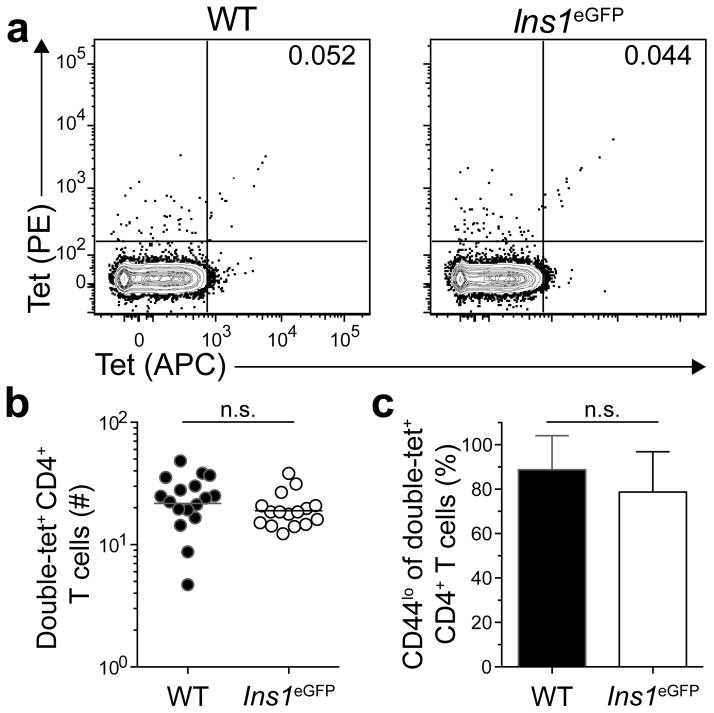
Cluster 1 contained wild-type mice and mice with patterns of very limited eGFP expression: Ins1eGFP mice express eGFP exclusively in pancreatic beta cells18, 19, whereas Forkhead box D1 (Foxd1)eGFP mice express eGFP during kidney and eye development20, 21. Because Ins1eGFP and Foxd1eGFP mice responded to eGFPp immunization like mice that did not express eGFP, it is likely that their eGFPp:I-Ab-specific T cell repertoires never encountered this epitope and were in a naive state. We tested this hypothesis in Ins1eGFP mice by enumerating eGFPp:I-Ab-specific CD4+ T cells in unimmunized mice (Fig. 2a,b). Single cell suspensions of secondary lymphoid organs were stained with eGFPp:I-Ab tetramers labeled with allophycocyanin or phycoerythrin, then with anti-phycoerythrin and anti-allophycocyanin magnetic beads before magnetic enrichment. Cells were stained with the same tetramer labeled with two different fluorochromes to maximize the TCR specificity of the assay16. Ins1eGFP mice contained an average of 19 eGFPp:I-Ab-specific CD4+ T cells in secondary lymphoid organs, like wild-type mice (Fig. 2b). These populations also had similar frequencies of naive-phenotype (CD44lo) cells (Fig. 2c). Together with the observation that Ins1eGFP and wild-type mice mounted comparable responses following immunization, these results indicated that tolerance in Cluster 1 was due to ignorance.
Figure 2. Ins1eGFP mice ignore the eGFPp:I-Ab epitope.
(a) Contour plots of eGFPp:I-Ab-streptavidin-phycoerythrin (tet-PE) versus eGFPp:I-Ab-streptavidin-allophycocyanin (tet-APC) staining of tetramer-enriched CD4+ T cells from pooled spleens and lymph nodes of unimmunized WT or Ins1eGFP mice. Numbers indicate the percent of double tetramer-positive eGFPp:I-Ab-specific (double-tet+) cells.
(b) Numbers of double-tet+ CD4+ T cells from pooled spleens and lymph nodes of unimmunized WT and Ins1eGFP mice. Horizontal bars indicate geometric mean values and circles represent individual mice. Results are pooled from 5 experiments, n=16–17 total mice per group. (n.s. = not significant by unpaired t-test of log10 transformed data).
(c) Mean percent of double-tet+ CD4+ T cells from pooled spleens and lymph nodes of unimmunized WT or Ins1eGFP mice that were CD44lo. Data are pooled from 5 experiments, n=16–17 total mice per group. Error bars indicate standard deviations. (n.s. = not significant by unpaired t-test).
Partial clonal deletion and Treg induction shape Cluster 2
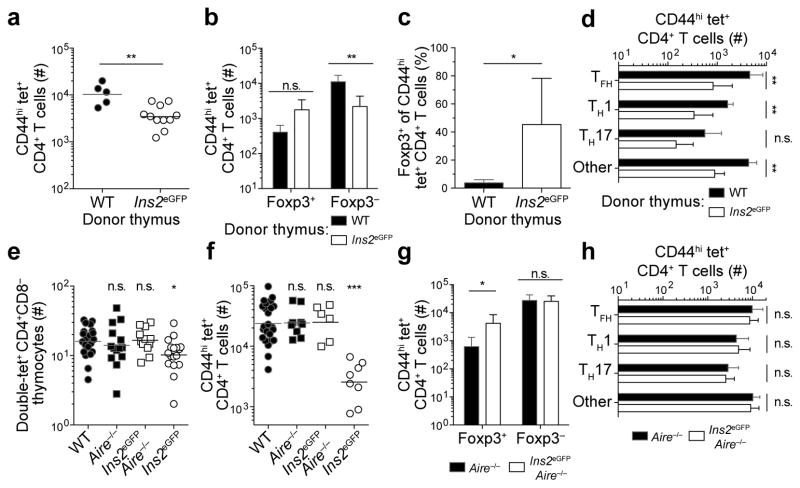
Cluster 2 was characterized by low numbers of effector cells and relatively large numbers of Treg cells in mice primed with eGFPp. It consisted of several mice with tissue-restricted promoters that are also expressed sparsely in the thymus. Ins2eGFP mice express eGFP fused to the C peptide of pro-insulin in pancreatic beta cells22. They also express eGFP in a small number of medullary thymic epithelial cells19. Forkhead box P3 (Foxp3)eGFP mice express eGFP in developing Treg cells in the thymus, whereas CD207eYFP mice23 express eYFP in epidermal Langerhans cells and perhaps thymic dendritic cells. Smooth muscle myosin heavy chain 11 (Myh11)eGFP and zinc finger protein 423 (Zfp423)eGFP mice are expected to express eGFP at moderate levels in medullary thymic epithelial cells24. Purkinje cell protein 2 (Pcp2)eGFP mice, which express eGFP in cerebellar Purkinje neurons in the brain25 were also in this cluster. Ins2eGFP mice were selected for further analysis of the tolerance mechanisms operating in Cluster 2.
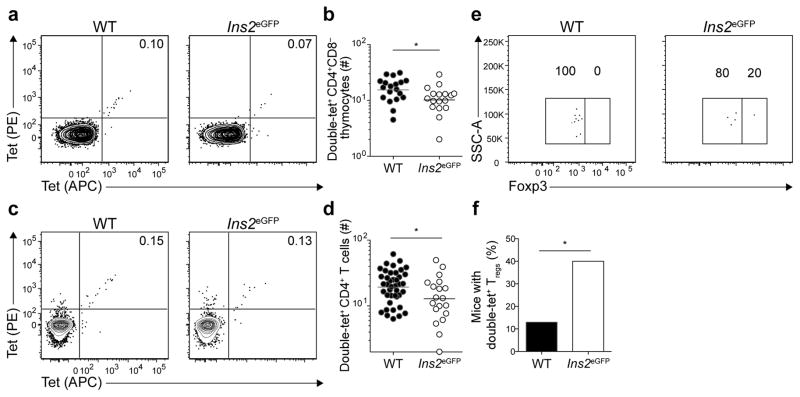
Evidence for intrathymic clonal deletion was sought by enumerating eGFPp:I-Ab-specific CD4+CD8− thymocytes (Fig. 3). Wild-type mice contained an average of 16 eGFPp:I-Ab-specific CD4+CD8− thymocytes and 20 peripheral CD4+ T cells, whereas Ins2eGFP mice had an average of 10 and 13 such cells (Fig. 3a–d). These results indicated that about 30% of eGFPp:I-Ab-specific T cell clones were deleted in Ins2eGFP mice. Foxp3+ eGFPp:I-Ab-specific CD4+CD8− thymocytes were detected in wild-type and Ins2eGFP thymuses, although not in all mice due to the small size of this population. However, significantly more Ins2eGFP thymuses (40%) contained eGFPp:I-Ab-specific Foxp3+ CD4+CD8− thymocytes than wild-type thymuses (Fig. 3e,f). Thus, Treg cells were spared from deletion or even increased in the presence of this self-antigen.
Figure 3. eGFPp:I-Ab-specific CD4+ T cells undergo limited clonal deletion in the thymus.
(a) Contour plots of tet-PE versus tet-APC staining of tetramer-enriched CD4+CD8− thymocytes from WT or Ins2eGFP mice.
(b) Numbers of double-tet+ CD4+CD8− thymocytes from WT and Ins2eGFP mice. Results are pooled from 5 experiments, n=17–19 total mice per group. (* P<0.05 by unpaired t-test of log10 transformed data). Horizontal bars indicate geometric mean values and circles represent individual mice.
(c) Contour plots of tet-PE versus tet-APC staining of tetramer-enriched CD4+ T cells from pooled spleens and lymph nodes of unimmunized WT or Ins2eGFP mice.
(d) Numbers of double-tet+ CD4+ T cells from pooled spleens and lymph nodes of WT or Ins2eGFP mice. Results are pooled from 8 experiments, n=20–41 total mice per group. Horizontal bars indicate geometric mean values and circles represent individual mice. (* P<0.05 by unpaired t-test of log10 transformed data).
(e) Dot plots of Foxp3 expression by double-tet+ CD4+CD8− thymocytes from WT and Ins2eGFP mice. Data are representative of samples from 5 independent experiments with n = 23 for WT mice and 20 mice for Ins2eGFP mice.
(f) Frequency of mice with double-tet+ Treg cells detected in the thymus. Data are pooled from 5 independent experiments with n = 23 for WT mice and 20 mice for Ins2eGFP mice. (*P<0.05 by Chi-square test).
Numbers on each plot in (a), (c), and (e) indicate the percent of cells in the quadrant or gate.
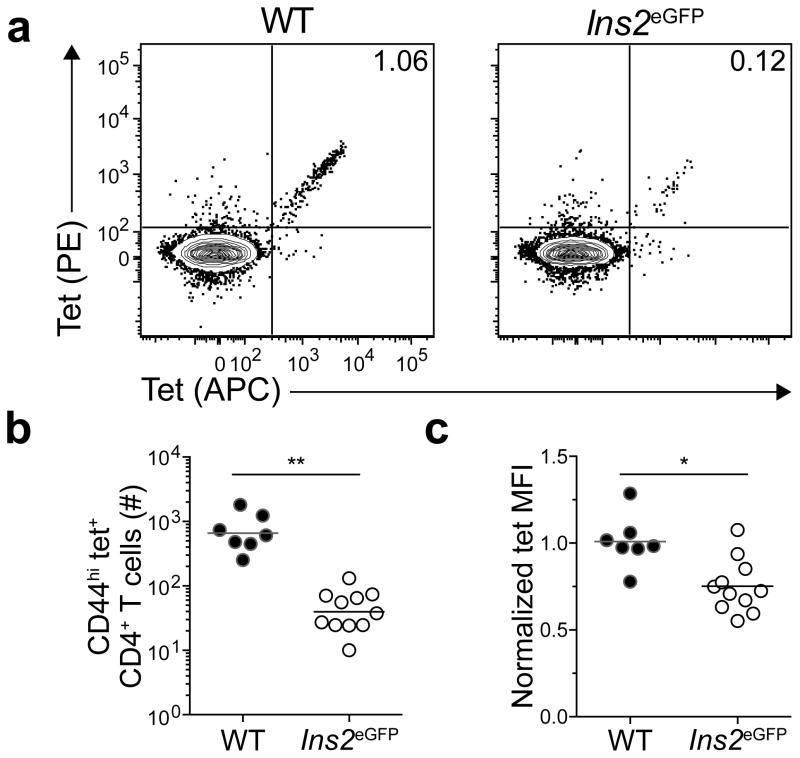
The clones that were deleted in Ins2eGFP mice might have had TCRs with the highest affinities for the eGFPp:I-Ab epitope. This possibility was addressed by measuring the mean fluorescence intensity of tetramer binding, which is a direct correlate of TCR affinity for peptide:MHCII26. Thymocytes could not be used for this purpose because they were too rare for an accurate measurement. The analysis was therefore performed on expanded populations of eGFPp:I-Ab-specific cells in immunized mice. No significant difference was found in the mean fluorescence intensity of eGFPp:I-Ab tetramer staining on cells from wild-type or Ins2eGFP mice immunized with eGFPp in CFA. Because CFA forms a long-lasting depot at the injection site27, persistent presentation of the eGFPp:I-Ab epitope may have given low affinity T cells an advantage thereby reducing the capacity to detect a subtle difference. The analysis was therefore repeated using an eGFP-expressing strain of attenuated (ΔactAΔinlB) Listeria monocytogenes (Lm-eGFP) that causes a very transient infection28. Lm-eGFP-induced clonal proliferation of eGFPp:I-Ab-specific T cells was significantly impaired in Ins2eGFP mice compared to wild-type mice (Fig. 4 a,b). Furthermore, the mean fluorescence intensity of eGFPp:I-Ab tetramer staining on cells from Ins2eGFP mice was lower than that of cells from wild-type mice (Fig. 4c). These results indicate that some clones with TCRs with high affinity for the eGFPp:I-Ab epitope are deleted in Ins2eGFP mice.
Figure 4. High affinity T cells are deleted in Ins2eGFP mice.
(a) Contour plots of tet-PE versus tet-APC staining of tetramer-enriched CD4+ T cells from pooled spleens and lymph nodes of WT or Ins2eGFP mice 6–7 days after infection with the ΔactAΔinlB strain of L. monocytogenes (Lm-eGFP). Numbers indicate the percent of double-tet+ cells.
(b) Numbers of CD44hi tet+ CD4+ T cells from pooled spleens and lymph nodes of mice 6–7 days after infection with Lm-eGFP. Horizontal bars indicate geometric mean values and circles represent individual mice. Data are pooled from 3 independent experiments, n=7–11 total mice per group. (** P<0.0001 by unpaired t-test of log10 transformed data).
(c) Normalized tetramer mean fluorescence intensity (MFI) of cells identified in (b). MFIs within each experiment were normalized to those of WT controls. Horizontal bars indicate mean values and circles represent individual mice. Data are pooled from 3 independent experiments, n=7–11 total mice per group. (* P<0.005 by unpaired t-test).
The fact that eGFP was likely expressed in the thymuses of Ins2eGFP mice under the control of the transcriptional regulator Aire led to an investigation of whether tolerance occurred in this organ in an Aire-dependent fashion. This question was first addressed by transplanting wild-type or Ins2eGFP thymuses into athymic B6-nude mice29. Eight-twelve weeks later, mice were primed subcutaneously with eGFPp in CFA. B6-nude recipients of wild-type thymus grafts generated large numbers of CD44hi eGFPp:I-Ab-specific CD4+ T cells as seen in intact wild-type mice, whereas B6-nude mice that received Ins2eGFP thymus grafts produced a much smaller population (Fig. 5a). B6-nude recipients of Ins2eGFP thymus grafts produced higher numbers and frequencies of CD44hi eGFPp:I-Ab-specific Treg cells than the recipients of wild-type thymus grafts (Fig. 5b,c). Foxp3− effector cell differentiation was also impaired in Ins2eGFP thymus graft recipients (Fig. 5d). Thus, much of the tolerance observed in Ins2eGFP mice was induced in the thymus.
Figure 5. Aire-mediated thymic expression of eGFP promotes clonal deletion and Treg induction in Ins2eGFP mice.
(a) Numbers of CD44hi tet+ CD4+ T cells per B6-nude mouse 14 days after immunization with 100 μg of eGFPp in CFA. Mice received WT or Ins2eGFP thymus grafts eight-twelve weeks before immunization, n=5–11 total mice per group.
(b) Numbers of CD44hi tet+ CD4+ Foxp3+ Treg cells and Foxp3− effector T cells from mice in (a).
(c) Frequency of Foxp3+ T cells among cells identified in (a).
(d) Numbers of each Foxp3− T effector cell subset in populations from (a).
(e) Numbers of double-tet+ CD4+CD8− thymocytes in WT, Aire−/−, Ins2eGFPAire−/−, and Ins2eGFP mice. Cells were double-tetramer stained and analyzed as in Figure 2a. Data for WT and Ins2eGFP mice are replotted from Figure 3b, n=12–21 total mice per group.
(f) Numbers of CD44hi tet+ CD4+ T cells per mouse 14 days after immunization of WT, Aire−/−, Ins2eGFPAire−/−, and Ins2eGFP mice with eGFPp in CFA. Data for WT and Ins2eGFP mice are replotted from Figure 1a, n=6–23 total mice per group.
(g) Numbers of CD44hi tet+ Foxp3+ Treg cells and Foxp3− effector T cells from mice in (f).
(h) Numbers of each Foxp3− T effector cell subset among cells enumerated in (f). Data are from 2 (a–d), 6 (e), and 5 (f–h) independent experiments. Data are from pooled spleens and lymph nodes (a–d) and (f–h) or thymuses (e) of indicated mice. Horizontal bars indicate geometric means and symbols represent individual mice (a) and (e–f). Error bars indicate standard deviations in (b–d) and (g–h). * P<0.05, ** P<0.005, *** P<0.0001, n.s. = not significant by unpaired t-test of log10 transformed data (a–b), (d), (g–h); unpaired t-test (c); or one-way ANOVA of log10 transformed data (e–f).
The involvement of Aire was tested using Ins2eGFPAire−/− mice. These mice had comparable numbers of eGFPp:I-Ab-specific CD4+CD8− thymocytes as wild-type and Aire−/− mice (Fig. 5e), demonstrating that clonal deletion in Ins2eGFP mice was Aire-dependent. Furthermore, thymic Treg cell numbers in Ins2eGFPAire−/− mice were similar to wild-type levels (data not shown) and clonal proliferation and differentiation of Foxp3− effector cells was very similar in Ins2eGFPAire−/− and Aire−/− mice following immunization (Fig. 5f–h). Therefore, much of the tolerance in Ins2eGFP mice was induced in the thymus by an Aire-dependent mechanism. Total numbers of Foxp3+ Treg cells, however, were still elevated in some immunized Ins2eGFPAire−/− mice compared to wild-type mice, suggesting that a non-Aire-dependent mechanism could play a role in this aspect of tolerance (Fig. 5g).
Overall, these studies indicate that tolerance in CD4+ T cell repertoires specific for self-epitopes expressed in a limited fashion in the thymus is associated with intrathymic deletion of some clones and impaired effector, but enhanced Treg cell generation from the remaining clones.
Clonal deletion regulates self-antigens in Cluster 3
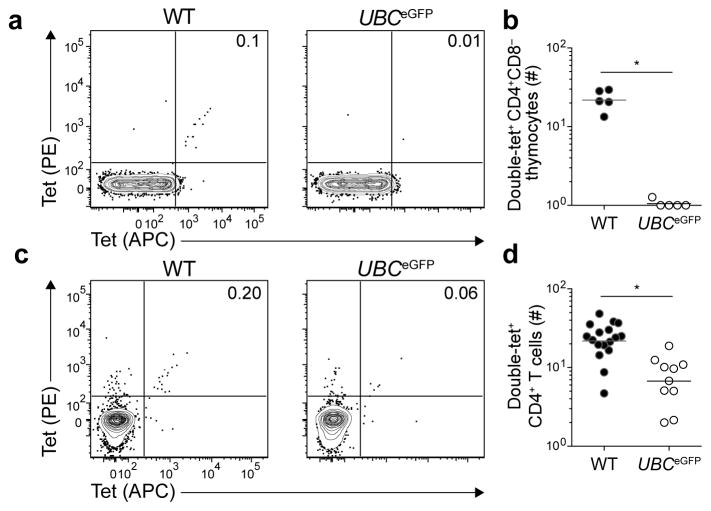
Cluster 3 was characterized by a profound clonal proliferation defect following immunization. This cluster contained mice that express eGFP under the control of non-Aire-regulated promoters in MHCII+ cells in the thymus. UBCeGFP and beta actin (ACTB)eGFP mice express eGFP in thymic dendritic cells, medullary thymic epithelial cells, and all other nucleated cells30, 31. CD11c (Itgax)eYFP mice express eYFP in thymic dendritic cells32, whereas choline acetyltransferase (Chat)eGFP 33, 34, cadherin 1 (Cdh1)CFP 35 and AireeGFP 36 mice express eGFP in medullary thymic epithelial cells. UBCeGFP mice were chosen for further analysis as a representative of Cluster 3.
eGFPp:I-Ab-specific CD4+CD8− thymocytes were enumerated to determine whether intrathymic clonal deletion was responsible for the proliferation defect observed in UBCeGFP mice (Fig. 6). Wild-type mice contained an average of 20 eGFPp:I-Ab-specific CD4+CD8− thymocytes, while UBCeGFP mice had less than one. A few eGFPp:I-Ab-specific CD4+ T cells were, however, detected in the secondary lymphoid organs of UBCeGFP mice (Fig. 6c,d). These results suggest that most eGFPp:I-Ab-specific CD4+CD8− thymocytes are deleted in the thymus, although a few escape and accumulate in the secondary lymphoid organs. The profound lack of proliferation following immunization indicated that these few cells were unresponsive. These results indicate that MHCII-bound peptides from proteins that are abundantly expressed in MHCII+ cells in the thymus induce a profound state of immune tolerance mediated in large part by intrathymic clonal deletion.
Figure 6. Expression of a self-epitope by thymic antigen-presenting cells induces intrathymic clonal deletion.
(a) Contour plots of tet-PE versus tet-APC staining of tetramer-enriched CD4+CD8− thymocytes from WT or UBCeGFP mice.
(b) Number of CD4+CD8− double-tet+ cells in the thymuses of WT or UBCeGFP mice. Horizontal bars indicate geometric mean values and circles represent individual mice. Data were pooled from two independent experiments, n=5 total mice per group. (* P < 0.0001 by unpaired t-test of log10 transformed data).
(c) Contour plots of tet-PE versus tet-APC staining of tetramer-enriched CD4+ T cells from pooled spleens and lymph nodes of WT or UBCeGFP mice.
(d) Number of CD4+ double-tet+ cells in pooled spleens and lymph nodes of WT or UBCeGFP mice. Horizontal bars indicate geometric mean values and circles represent individual mice. Data were pooled from 4 independent experiments, n=10–17 total mice per group. Data for WT mice are replotted from Figure 2b. (* P < 0.0001 by unpaired t-test of log10 transformed data).
Numbers on each plot in (a) and (c) indicate the percent of double-tet+ cells.
Thymic self-antigen expression correlates with tolerance
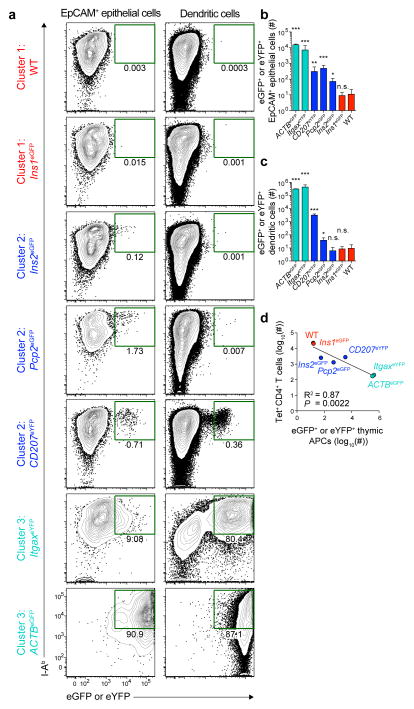
The specific mechanisms of tolerance associated with self-epitopes in Clusters 1–3 appeared to depend on self-antigen expression by MHCII+ cells in the thymus. To test this hypothesis, representatives from these clusters were examined for expression of eGFP and eYFP by EpCAM+ thymic epithelial cells and thymic dendritic cells (Supplementary Fig. 1). Thymuses from wild-type and Ins1eGFP mice, members of Cluster 1, contained fewer than 10 I-Ab+ eGFP+ or eYFP+ EpCAM+ epithelial cells or dendritic cells, consistent with the ignorant phenotype of eGFPp:I-Ab-specific CD4+ T cells in these mice (Fig. 7a–c). At the other end of the spectrum, ItgaxeYFP and ACTBeGFP mice, members of Cluster 3, contained more than 300,000 I-Ab+ eYFP+ or eGFP+ dendritic cells and EpCAM+ epithelial cells in their thymuses, likely accounting for the substantial clonal deletion observed in these mice (Fig. 7a–c). Finally, Ins2eGFP, Pcp2eGFP, and CD207eYFP thymuses contained several hundred I-Ab+ eGFP+ or eYFP+ EpCAM+ epithelial cells, and CD207eYFP thymuses also contained ~3,000 I-Ab+ eYFP+ dendritic cells (Fig. 7a–c). A significant inverse correlation was observed between the number of eGFP+ or eYFP+ thymic antigen-presenting cells and the number of eGFPp:I-Ab-specific CD4+ T cells present 14 days after immunization (Fig. 7d). These results are consistent with the hypothesis that the number of MHCII+ thymic antigen-presenting cells expressing a particular self-antigen determines the responsiveness of the corresponding CD4+ T cell population.
Figure 7. Frequency of self-antigen+ thymic antigen-presenting cells correlates with the responsiveness of the corresponding CD4+ T cell population.
(a) Contour plots of MHCII (I-Ab) and eGFP+ or eYFP+ expression by EpCAM+ thymic epithelial cells (left) and thymic dendritic cells (right) from the indicated members of Clusters 1–3. Plots are from 5 experiments. Numbers indicate the percent of cells that are I-Ab+ and either eGFP+ or eYFP+.
(b) Numbers of eGFP+ or eYFP+ EpCAM+ thymic epithelial cells in the indicated mouse strains, identified as in (a). Bars are colored according to the clusters identified in Figure 1d. Error bars represent the standard deviations. (n=3–6 total mice per group, * P<0.05, ** P<0.0005, *** P<0.0001, n.s. = not significant by one-way ANOVA of log10 transformed data).
(c) Numbers of eGFP+ or eYFP+ thymic dendritic cells in the indicated mouse strains, identified as in (a). Bars are colored according to the clusters identified in Figure 1d. Error bars represent the standard deviations. (n=3–6 total mice per group, * P<0.05, *** P<0.0001, n.s. = not significant by one-way ANOVA of log10 transformed data).
(d) Linear regression analysis of the log10(number of eGFP+ or eYFP+ thymic antigen-presenting cells) versus log10(number of tet+ CD4+ T cells 14 days after immunization). Each point represents values for an individual mouse strain as indicated. For each point, the y-axis value represents the log10(geometric mean from Figure 1a), and the x-axis value represents the log10(geometric mean for the total number of I-Ab+ eGFP+ or eYFP+ EpCAM+ thymic epithelial cells and dendritic cells for each strain). Circles and labels are colored according to the clusters identified in Figure 1d.
Tolerance to true self-antigens occurs by these mechanisms
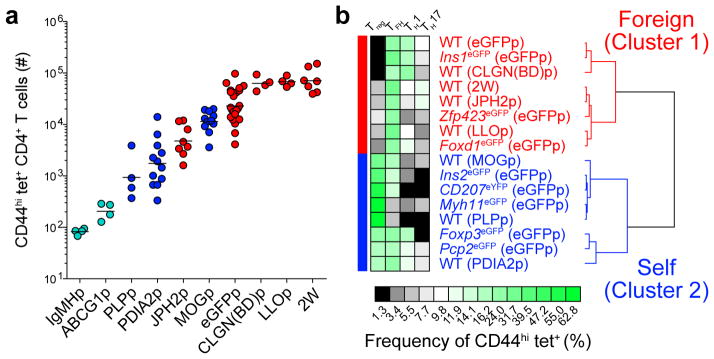
Information from the eGFP system was then used to identify the tolerance mechanisms for CD4+ T cell repertoires for 6 genuine self-peptide:I-Ab epitopes. B6 mice were immunized with the self-peptides or foreign peptides as controls, and the compositions of the resulting Foxp3− effector and Foxp3+ Treg cell populations were determined. The percentages of TFH, TH1, TH17, and Treg cells were analyzed using an unsupervised hierarchical clustering algorithm to determine if the responses to the self-peptides aligned with the clusters defined in the eGFP system. Percentages of cells in each subset, rather than absolute numbers, were used for this comparison because the sizes of the pre-immunization repertoires for the natural epitopes were unknown. The use of absolute numbers of cells in each subset would have been problematic for epitopes with populations that had the same differentiation potential but simply varied in size.
Peptides from the constant region of IgM (IgMHp), which is very abundant in the blood, or ATP-binding cassette sub-family G, member 1 (ABCG1p), which is highly expressed in thymic dendritic cells and medullary thymic epithelial cells24, elicited fewer than 200 CD44hi cells that bound the relevant peptide:I-Ab tetramers (Fig. 8a). These peptides elicited too few CD44hi cells to accurately determine the percentages of each subset and thus were not analyzed by the clustering algorithm. The minimal responses to these self-epitopes, however, indicated that these CD4+ T cell repertoires underwent extensive intrathymic clonal deletion as in the eGFP transgenic mice in Cluster 3.
Figure 8. Tolerance mechanisms identified in eGFP expressing mouse strains govern tolerance to true self-antigens.
(a) Numbers of CD44hi peptide:I-Ab+ (tet+) CD4+ T cells from pooled spleens and lymph nodes of mice 13–14 days after immunization with 100 μg of each indicated peptide emulsified in CFA. Horizontal bars indicate geometric mean values. Data were pooled from 17 experiments, n=4–23 total mice per epitope. Circles represent individual mice and are colored according to the clusters identified in (b).
(b) Unsupervised hierarchical clustering of 16 self- and foreign- tet+ CD4+ T cell populations from (a). Profiles were clustered by percentage of cells of each subset as displayed in the heatmap. Two major clusters are indicated.
The unsupervised hierarchical clustering analysis produced 2 clusters. The first (‘foreign’) cluster contained responses to foreign peptides from listeriolysin O protein of Listeria monocytogenes (LLOp)37, 38, calnexin from Blastomyces dermatitidis (CLGN(BD)p), and a model peptide called 2W39, as well as the Cluster 1 (Fig. 1d) responses to eGFPp in wild-type, Ins1eGFP, and Foxd1eGFP mice (Fig. 8b). The second (‘self’) cluster consisted of responses to the eGFP epitope in the eGFP transgenic mice that comprised Cluster 2 (Fig. 1d).
A peptide from junctophilin 2 (JPH2p), which is expressed exclusively in skeletal muscle and the heart40, elicited 5,000 CD44hi cells that bound the relevant peptide:I-Ab tetramer (Fig. 8a). This population produced a Treg and effector cell composition similar to that of populations specific for foreign peptides, as evidenced by its assignment to the ‘foreign’ cluster (Fig. 8b). These results indicate that CD4+ T cell tolerance to the junctophilin 2 epitope in B6 mice is maintained by ignorance.
Peptides from myelin oligodendrocyte glycoprotein (MOG) and proteolipid protein (PLP), which are expressed in the nervous system, and protein disulfide associated isomerase 2 (PDIA2), which is expressed in the pancreas and digestive tract41, elicited 1,000–10,000 peptide:I-Ab-specific CD44hi cells (Fig. 8a). These populations contained reduced Foxp3− effector and increased Treg cell frequencies compared to populations specific for non-mouse peptides (Fig. 8b). The responses to these self-epitopes were placed in the ‘self’ cluster, which also contained responses to the eGFP epitope in the eGFP transgenic mice that comprised Cluster 2 in Figure 1d. Thus, tolerance to these endogenously expressed peptides resembled that associated with limited thymic eGFP expression.
DISCUSSION
Here we demonstrate that tolerance in the normally diverse CD4+ T cell repertoire is attributed to one of three mechanisms depending on the expression pattern of the self-epitope.
A profound state of tolerance existed in CD4+ T cell repertoires specific for epitopes from proteins that are easily accessed or expressed by many MHCII+ cells in the thymus. This tolerance manifested as nearly complete deletion of self-epitope-specific T cells in the thymus, which likely resulted from abundant self-epitope display on MHCII+ thymic dendritic cells and/or medullary thymic epithelial cells. Although substantial clonal deletion was reported for human and murine T cells specific for self-epitopes from the Y-chromosome or driven by the ACTB promoter4, 5, 15, it was not as extensive as that observed here for an epitope expressed from the UBC promoter. This promoter may have led to an extraordinarily large number of self-epitopes on thymic antigen-presenting cells, causing deletion of nearly all self-epitope-specific T cells. Alternatively, deletion within the small eGFPp:I-Ab-specific CD4+ T cell repertoire might have been more efficient because of limited inter-clonal competition.
A state of immunological ignorance existed within CD4+ T cell repertoires specific for epitopes from non-Aire-regulated proteins that are expressed exclusively outside of the thymus. In addition, these epitopes are probably not displayed in secondary lymphoid organs because their parent proteins are cytosolic and expressed by cells that do not turn over in a way that allows local dendritic cells to produce the epitopes or access draining lymph nodes. The complete lack of presentation of these self-epitopes dictates that corresponding CD4+ T cells exist in a naive and fully responsive state of ignorance. Self-epitopes in this cluster are essentially foreign to the relevant T cell repertoires.
A complex form of tolerance controlled CD4+ T cell repertoires specific for epitopes from proteins with tissue-restricted patterns of expression, but that are also expressed by small numbers of MHCII+ thymic antigen-presenting cells. Analysis of the Ins2eGFP member of this group revealed that tolerance was associated with intrathymic deletion of ~30% of the epitope-specific clones. Partial clonal deletion was also observed in studies of a polyclonal CD8+ T cell population specific for a tissue-restricted epitope11. In our studies, the deleted clones had TCRs with the highest affinities for the eGFPp:I-Ab ligand. Thymocytes with high affinity TCRs might be especially sensitive to deletion because they can garner sufficient signal from single encounters with rare thymic antigen-presenting cells. In contrast, deletion of thymocytes with lower affinity TCRs may require numerous encounters42, 43, which are unlikely to occur when relevant thymic antigen-presenting cells are rare, as in Cluster 2.
The partial deletion of the epitope-specific polyclonal population studied in Ins2eGFP mice depended on Aire, indicating involvement of medullary thymic epithelial cells. Furthermore, we found that expression of eGFP from the Ins2, Pcp2, and CD207 loci occurred in only a small number of thymic epithelial cells and dendritic cells, likely allowing eGFPp:I-Ab-specific T cells with lower affinity TCRs to escape deletion. In agreement with this possibility, eGFPp:I-Ab-specific T cells were nearly completely deleted in AireeGFP mice, which express eGFP in all medullary thymic epithelial cells36. Although medullary thymic epithelial cells were a likely source of eGFP in these models, antigen transfer to neighboring dendritic cells could have mediated deletion44.
The non-deleted T cells specific for Cluster 2 epitopes generated Foxp3− effector T cells less well than corresponding wild-type T cells. This defect extended to TFH cells but was particularly striking for TH1 and TH17 cells. Other work has shown that TH1 clones tend to have higher affinity TCRs than TFH cells45. Thus, it is possible that the lower affinity TCRs found on epitope-specific T cells in this cluster were incapable of generating enough signals to form TH1 and TH17 cells. Alternatively, these cells might have been induced into a partially anergic state due to persistent self-epitope recognition in a non-inflammatory environment as suggested by previous TCR transgenic studies46, 47, 48. If so, the thymic transplant experiments presented here suggest that this anergy induction occurs mainly in the thymus.
Enhanced Treg cell proliferation after immunization was another component of tolerance to the epitopes in Cluster 2. One explanation for this feature is that the pre-immunization repertoires of T cells specific for these epitopes are enriched for thymus-derived Treg cells, which might occur when cells are shunted into the Treg cell lineage following moderately high affinity self-epitope recognition in the thymus49. The observation that Treg cells were detected more frequently in the pre-immunization eGFPp:I-Ab-specific population in Ins2eGFP mice than in wild-type mice is consistent with this possibility. This is only a tentative conclusion however, because the eGFPp:I-Ab-specific populations were very small in both situations. It is also possible that Foxp3− cells in the eGFPp:I-Ab-specific population had enhanced potential to differentiate into induced Treg cells after immunization. In any case, our results suggest that Treg cells are especially important for tolerance to epitopes from tissue-restricted antigens that also exhibit limited thymic expression.
The mechanisms of tolerance identified for the epitopes studied here all appeared to be induced in the thymus. It is possible that other epitopes are tolerated by mechanisms that operate in the secondary lymphoid organs, for example, epitopes from paternal fetal proteins that appear in pregnant females. Peripheral deletion and anergy appear to be the tolerance mechanisms for these antigens50.
ONLINE METHODS
Mice
Four- to 8-week old C57BL/6 (B6), B6.Cg-Foxn1nu/J (B6-nude), B6.PL-Thy1a/CyJ (CD90.1), C57BL/6-Tg(UBC-GFP)30Scha/J (UBCeGFP), B6.Cg-Tg(Ins1-EGFP)1Hara/J (Ins1eGFP), B6.Cg-Tg(RP23-268L19-EGFP)2Mik/J (ChateGFP), B6;FVB-Tg(Zfp423-EGFP)7Brsp/J (Zfp423eGFP)51, B6.Cg-Tg(Myh11-cre,-EGFP)2Mik/J (Myh11eGFP)52, B6.129P2(Cg)-Cdh1tm1Cle/J (Cdh1eGFP), B6;129S4-Foxd1tm1(GFP/cre)Amc/J (Foxd1eGFP), and C57BL/6-Tg(CAG-EGFP)131Osb/LeySopJ (ACTBeGFP) mice were purchased from the Jackson Laboratory or the National Cancer Institute Mouse Repository (Frederick, MD, USA). Mice were used at 6–12 weeks of age. (Ins2eGFP x B6), (Ins2eGFP x CD90.1), (Ins1eGFP x B6), (Ins2eGFPAire+/− x Aire+/−), STOCK Tg(Pcp2-EGFP)BT153Gsat/Mmmh, (backcrossed >8 generations onto the B6 background) (Pcp2eGFP), B6.Cg-Tg(Itgax-Venus)1Mnz/J (ItgaxeYFP), CD207cre x B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J (CD207eYFP)23, and Foxp3eGFP mice53 were bred and housed under specific-pathogen free conditions as per the guidelines of the University of Minnesota Institutional Animal Care and Use Committee and the National Institutes of Health. AireeGFP mice36 were bred and housed under specific-pathogen free conditions in accordance with the National Institutes of Health and American Association of Laboratory Animal Care standards, and with the animal care and use regulations of the University of California, San Francisco. Analysis of thymus cell numbers was carried out between 3 and 5 weeks of age.
Immunizations
Mice were injected subcutaneously with 100 μl (split over 2 sites) of CFA (Sigma-Aldrich) emulsion containing 100 μg of peptide (GenScript) in DPBS (Life Technologies). Mice were immunized with the specific peptides contained in the tetramers below with the following exceptions: MOGp (GWYRSPFSRVVHL) and CLGN(BD)p (LVVKNPAAHHAI).
Infections
The ΔactAΔinlB strain of Lm-eGFP (PL1113) was provided by Peter Lauer. Briefly, the eGFP construct (hyperSPO1 promoter-eGFP)54 was cloned into the pPL1 vector55 and integrated at the comK locus in the actA-inlB-double mutant56. Mice were injected i.v. with 1×107 Lm-eGFP bacteria and analyzed 6–7 days post-infection.
Tetramers
Biotin-labeled I-Ab monomers containing the eGFPp (HDFFKSAMPEGYVQE), 2W (EAWGALANWAVDSA), LLOp (NEKYAQAYPNVS), CLGN(BD)p (LVVKNPAAHHAIS), JPH2p (VYTWPSGNTFE), MOGp (GWYRSPFSRVV), PLPp (CLVGAPFASLVA), IgMHp (EKYVTSAPMPEPGAPG)57, I-Ab-binding-stabilized variant of ABCG1p (YYMTPQPSDVV), or PDIA2p (EYSKAAALLAA) peptides covalently linked to the I-Ab beta chain were grown in Drosophila melanogaster S2 cells also expressing the I-Ab alpha chain, purified and combined with streptavidin (SA)-phycoerythrin (PE) or (SA)-allophycocyanin (APC) (Prozyme, San Leandro, CA, USA) to produce fluorescently labeled I-Ab tetramers as previously described 16, 37, 58.
Cell enrichment and flow cytometry
Single cell suspensions were prepared from pooled murine spleens and lymph nodes (axillary, brachial, inguinal, cervical, mesenteric, pancreatic, and para-aortic) or thymuses by mechanical disruption. Cells were stained for 1 hour at room temperature with allophycocyanin-conjugated tetramers. In some experiments cells were also stained with anti-CXCR5 (2G8, BD) antibody or phycoerythrin-conjugated tetramers. Tetramer-binding cells were enriched on magnetized columns as described previously58. For thymic epithelial cell and dendritic cell analysis, thymuses were harvested and enzymatically digested as previously described59. Single cell suspensions were stained with a biotinylated antibody to CD11c (HL3, BD) and an allophycocyanin-labeled antibody to EpCAM (G8.8, Biolegend). Cells were incubated with anti-biotin and anti-allophycocyanin cocktails and magnetic beads (StemCell Technologies). Cells were enriched using the EasySep system according to the manufacturer’s instructions (StemCell Technologies).
Antibodies
Tetramer-enriched samples were stained for surface markers for 30 minutes on ice using antibodies to: CD4 (GK1.5, BD), CD8 (53–6.7, BD), CD90.1 (HIS51), CD90.2 (53–2.1, 30-H12), CD3e (145-2C11, BD), CD11b (M1/70), CD11c (N418), F4/80 (BM8), CD44 (IM7, BD), CD45.1 (A20, Biolegend), CD45.2 (104), B220 (RA3–6B2), and/or PD1 (J43). Cellular viability was confirmed using GhostDye Red 780 (Tonbo Biosciences). For transcription factor expression analysis, stained cells were fixed and permeablized using the Foxp3/Transcription Factor Staining Buffer Set (eBioscience) according to the manufacturer’s instructions. Cells were stained overnight at 4°C with antibodies to T-bet (4B10, Biolegend), Foxp3 (FJK-16s), RORγt (Q31-378, BD), and Bcl-6 (K112-91, BD). For thymic dendritic cell and thymic epithelial cell analyses cells, following enrichment cells were stained with antibodies to CD11c (N418), CD19 (6D5, Biolegend), I-A/I-E (M5/114.15.2, Biolegend), CD8 (53–6.7, BD), CD90.2 (53–2.1, 30-H12), CD11b (M1/70), and CD45.2 (104, BD). Antibodies were purchased from eBioscience unless otherwise indicated. Cells were analyzed by flow cytometry on a Fortessa (BD). Data were analyzed using FlowJo software (TreeStar).
Thymus transplants
Thymus transplants were performed as previously described 60. Briefly, thymus lobes were harvested from prenatal (post-conception day 18) pups generated by crossing Ins2eGFP and CD90.1+ B6 mice. An incision was made on the left abdomen of anesthetized 4–6 week old female B6-nude to expose the kidney and the donor thymic lobes were grafted beneath the kidney capsule. The muscle layer was sutured and the skin was closed using surgical clips. Thymic engraftment was assessed 6 weeks after transplantation by examining peripheral blood for the presence of mature αβ T cells by flow cytometry. Donor genotypes were determined post-transplantation.
Statistics
Two-tailed, unpaired Student’s t tests, one-way ANOVA, Chi-square test, and linear regression were performed using Prism (Graphpad) software.
Hierarchical Clustering
Unsupervised hierarchical clustering and heatmap generation was carried out using the ‘HierarchicalClustering’ and ‘HierarchicalClusteringViewer’ modules from GenePattern (http://genepattern.broadinstitute.org)17, 61, 62.
Supplementary Material
Acknowledgments
We thank Dan Mueller for reviewing the manuscript and Jennifer Walter and Orion Rainwater for technical assistance. We thank Peter Lauer (and Aduro Biotech) for providing the Lm-eGFP strain: PL1113. Supported by grants from the US National Institutes of Health (P01 AI35296 to M.K.J. and K.A.H.; F32 AI114050 to D.M.; T32 GM008244 and F30 DK093242 to R.W.N; T32 AI07313 to J.L.L; K99 AI114884 to Y.J.L), the Wallin Neuroscience Discovery Fund (to H.T.O. and M.K.J.), and the Juvenile Diabetes Research Foundation Grant 2-2011-662 (to B.T.F.).
Footnotes
AUTHOR CONTRIBUTIONS
D. M. designed the study, did experiments, analyzed data, and wrote the manuscript, J. L. L, T. D., Y. J. L., R.W.N., W.E.P., J.V.P., and M. S. A. did experiments, K. A. H., B.T.F., and H. T. O. provided discussion, M. K. J. designed the study, analyzed data, and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Xing Y, Hogquist KA. T-cell tolerance: central and peripheral. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wing K, Sakaguchi S. Regulatory T cells exert checks and balances on self tolerance and autoimmunity. Nat Immunol. 2010;11:7–13. doi: 10.1038/ni.1818. [DOI] [PubMed] [Google Scholar]
- 3.Semana G, Gausling R, Jackson RA, Hafler DA. T cell autoreactivity to proinsulin epitopes in diabetic patients and healthy subjects. J Autoimmun. 1999;12:259–267. doi: 10.1006/jaut.1999.0282. [DOI] [PubMed] [Google Scholar]
- 4.Moon JJ, et al. Quantitative impact of thymic selection on Foxp3+ and Foxp3- subsets of self-peptide/MHC class II-specific CD4+ T cells. Proc Natl Acad Sci USA. 2011;108:14602–14607. doi: 10.1073/pnas.1109806108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu W, et al. Clonal deletion prunes but does not eliminate self-specific alphabeta CD8(+) T lymphocytes. Immunity. 2015;42:929–941. doi: 10.1016/j.immuni.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeda Y, et al. Detection of self-reactive CD8(+) T cells with an anergic phenotype in healthy individuals. Science. 2014;346:1536–1540. doi: 10.1126/science.aaa1292. [DOI] [PubMed] [Google Scholar]
- 7.Ohashi PS, et al. Ablation of “tolerance” and induction of diabetes by virus infection in viral antigen transgenic mice. Cell. 1991;65:305–317. doi: 10.1016/0092-8674(91)90164-t. [DOI] [PubMed] [Google Scholar]
- 8.Oldstone MB, Nerenberg M, Southern P, Price J, Lewicki H. Virus infection triggers insulin-dependent diabetes mellitus in a transgenic model: role of anti-self (virus) immune response. Cell. 1991;65:319–331. doi: 10.1016/0092-8674(91)90165-u. [DOI] [PubMed] [Google Scholar]
- 9.Jenkins MK, Schwartz RH. Antigen presentation by chemically modified splenocytes induces antigen-specific T cell unresponsiveness in vitro and in vivo. J Exp Med. 1987;165:302–319. doi: 10.1084/jem.165.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akkaraju S, et al. A range of CD4 T cell tolerance: partial inactivation to organ-specific antigen allows nondestructive thyroiditis or insulitis. Immunity. 1997;7:255–271. doi: 10.1016/s1074-7613(00)80528-2. [DOI] [PubMed] [Google Scholar]
- 11.Zehn D, Bevan MJ. T cells with low avidity for a tissue-restricted antigen routinely evade central and peripheral tolerance and cause autoimmunity. Immunity. 2006;25:261–270. doi: 10.1016/j.immuni.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hataye J, Moon JJ, Khoruts A, Reilly C, Jenkins MK. Naive and memory CD4+ T cell survival controlled by clonal abundance. Science. 2006;312:114–116. doi: 10.1126/science.1124228. [DOI] [PubMed] [Google Scholar]
- 13.Marzo AL, et al. Initial T cell frequency dictates memory CD8+ T cell lineage commitment. Nat Immunol. 2005;6:793–799. doi: 10.1038/ni1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bautista JL, et al. Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nat Immunol. 2009;10:610–617. doi: 10.1038/ni.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouneaud C, Kourilsky P, Bousso P. Impact of negative selection on the T cell repertoire reactive to a self-peptide: a large fraction of T cell clones escapes clonal deletion. Immunity. 2000;13:829–840. doi: 10.1016/s1074-7613(00)00080-7. [DOI] [PubMed] [Google Scholar]
- 16.Nelson RW, et al. T cell receptor cross-reactivity between similar foreign and self peptides influences naive cell population size and autoimmunity. Immunity. 2015;42:95–107. doi: 10.1016/j.immuni.2014.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 18.Hara M, et al. Transgenic mice with green fluorescent protein-labeled pancreatic beta -cells. Am J Physiol Endocrinol Metab. 2003;284:E177–183. doi: 10.1152/ajpendo.00321.2002. [DOI] [PubMed] [Google Scholar]
- 19.Anderson MS, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 20.Carreres MI, et al. Transcription factor Foxd1 is required for the specification of the temporal retina in mammals. J Neurosci. 2011;31:5673–5681. doi: 10.1523/JNEUROSCI.0394-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi A, et al. Identification of a multipotent self-renewing stromal progenitor population during mammalian kidney organogenesis. Stem Cell Reports. 2014;3:650–662. doi: 10.1016/j.stemcr.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ben-Yehudah A, et al. Specific dynamic and noninvasive labeling of pancreatic beta cells in reporter mice. Genesis. 2005;43:166–174. doi: 10.1002/gene.20165. [DOI] [PubMed] [Google Scholar]
- 23.Kaplan DH, et al. Autocrine/paracrine TGFbeta1 is required for the development of epidermal Langerhans cells. J Exp Med. 2007;204:2545–2552. doi: 10.1084/jem.20071401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heng TS, Painter MW Immunological Genome Project C. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 25.Gong S, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 26.Crawford F, Kozono H, White J, Marrack P, Kappler J. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity. 1998;8:675–682. doi: 10.1016/s1074-7613(00)80572-5. [DOI] [PubMed] [Google Scholar]
- 27.Warren HS, Vogel FR, Chedid LA. Current status of immunological adjuvants. Annu Rev Immunol. 1986;4:369–388. doi: 10.1146/annurev.iy.04.040186.002101. [DOI] [PubMed] [Google Scholar]
- 28.Portnoy DA, Auerbuch V, Glomski IJ. The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell-mediated immunity. J Cell Biol. 2002;158:409–414. doi: 10.1083/jcb.200205009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordier AC, Haumont SM. Development of thymus, parathyroids, and ultimo-branchial bodies in NMRI and nude mice. Am J Anat. 1980;157:227–263. doi: 10.1002/aja.1001570303. [DOI] [PubMed] [Google Scholar]
- 30.Schaefer BC, Schaefer ML, Kappler JW, Marrack P, Kedl RM. Observation of antigen-dependent CD8+ T-cell/dendritic cell interactions in vivo. Cell Immunol. 2001;214:110–122. doi: 10.1006/cimm.2001.1895. [DOI] [PubMed] [Google Scholar]
- 31.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 32.Lindquist RL, et al. Visualizing dendritic cell networks in vivo. Nat Immunol. 2004;5:1243–1250. doi: 10.1038/ni1139. [DOI] [PubMed] [Google Scholar]
- 33.Panneck AR, et al. Cholinergic epithelial cell with chemosensory traits in murine thymic medulla. Cell Tissue Res. 2014;358:737–748. doi: 10.1007/s00441-014-2002-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tallini YN, et al. BAC transgenic mice express enhanced green fluorescent protein in central and peripheral cholinergic neurons. Physiol Genomics. 2006;27:391–397. doi: 10.1152/physiolgenomics.00092.2006. [DOI] [PubMed] [Google Scholar]
- 35.Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 36.Gardner JM, et al. Deletional tolerance mediated by extrathymic Aire-expressing cells. Science. 2008;321:843–847. doi: 10.1126/science.1159407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pepper M, Pagan AJ, Igyarto BZ, Taylor JJ, Jenkins MK. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 2011;35:583–595. doi: 10.1016/j.immuni.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geginat G, Schenk S, Skoberne M, Goebel W, Hof H. A novel approach of direct ex vivo epitope mapping identifies dominant and subdominant CD4 and CD8 T cell epitopes from Listeria monocytogenes. J Immunol. 2001;166:1877–1884. doi: 10.4049/jimmunol.166.3.1877. [DOI] [PubMed] [Google Scholar]
- 39.Rees W, et al. An inverse relationship between T cell receptor affinity and antigen dose during CD4(+) T cell responses in vivo and in vitro. Proc Natl Acad Sci USA. 1999;96:9781–9786. doi: 10.1073/pnas.96.17.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K. Junctophilins: a novel family of junctional membrane complex proteins. Mol Cell. 2000;6:11–22. doi: 10.1016/s1097-2765(00)00003-4. [DOI] [PubMed] [Google Scholar]
- 41.Fu XM, Dai X, Ding J, Zhu BT. Pancreas-specific protein disulfide isomerase has a cell type-specific expression in various mouse tissues and is absent in human pancreatic adenocarcinoma cells: implications for its functions. J Mol Histol. 2009;40:189–199. doi: 10.1007/s10735-009-9230-5. [DOI] [PubMed] [Google Scholar]
- 42.Rosette C, et al. The impact of duration versus extent of TCR occupancy on T cell activation: a revision of the kinetic proofreading model. Immunity. 2001;15:59–70. doi: 10.1016/s1074-7613(01)00173-x. [DOI] [PubMed] [Google Scholar]
- 43.Au-Yeung BB, et al. A sharp T-cell antigen receptor signaling threshold for T-cell proliferation. Proc Natl Acad Sci USA. 2014;111:E3679–3688. doi: 10.1073/pnas.1413726111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see) Nat Rev Immunol. 2014;14:377–391. doi: 10.1038/nri3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tubo NJ, et al. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell. 2013;153:785–796. doi: 10.1016/j.cell.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Apostolou I, Sarukhan A, Klein L, von Boehmer H. Origin of regulatory T cells with known specificity for antigen. Nat Immunol. 2002;3:756–763. doi: 10.1038/ni816. [DOI] [PubMed] [Google Scholar]
- 47.Tanchot C, Barber DL, Chiodetti L, Schwartz RH. Adaptive tolerance of CD4+ T cells in vivo: multiple thresholds in response to a constant level of antigen presentation. J Immunol. 2001;167:2030–2039. doi: 10.4049/jimmunol.167.4.2030. [DOI] [PubMed] [Google Scholar]
- 48.Kearney ER, Pape KA, Loh DY, Jenkins MK. Visualization of peptide-specific T cell immunity and peripheral tolerance induction in vivo. Immunity. 1994;1:327–339. doi: 10.1016/1074-7613(94)90084-1. [DOI] [PubMed] [Google Scholar]
- 49.Lee HM, Bautista JL, Scott-Browne J, Mohan JF, Hsieh CS. A broad range of self-reactivity drives thymic regulatory T cell selection to limit responses to self. Immunity. 2012;37:475–486. doi: 10.1016/j.immuni.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.PrabhuDas M, et al. Immune mechanisms at the maternal-fetal interface: perspectives and challenges. Nat Immunol. 2015;16:328–334. doi: 10.1038/ni.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta RK, et al. Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell Metab. 2012;15:230–239. doi: 10.1016/j.cmet.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xin HB, Deng KY, Rishniw M, Ji G, Kotlikoff MI. Smooth muscle expression of Cre recombinase and eGFP in transgenic mice. Physiol Genomics. 2002;10:211–215. doi: 10.1152/physiolgenomics.00054.2002. [DOI] [PubMed] [Google Scholar]
- 53.Fontenot JD, et al. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 54.Shen A, Higgins DE. The 5′ untranslated region-mediated enhancement of intracellular listeriolysin O production is required for Listeria monocytogenes pathogenicity. Mol Microbiol. 2005;57:1460–1473. doi: 10.1111/j.1365-2958.2005.04780.x. [DOI] [PubMed] [Google Scholar]
- 55.Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J Bacteriol. 2002;184:4177–4186. doi: 10.1128/JB.184.15.4177-4186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brockstedt DG, et al. Listeria-based cancer vaccines that segregate immunogenicity from toxicity. Proc Natl Acad Sci USA. 2004;101:13832–13837. doi: 10.1073/pnas.0406035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chu HH, Moon JJ, Kruse AC, Pepper M, Jenkins MK. Negative selection and peptide chemistry determine the size of naive foreign peptide-MHC class II-specific CD4+ T cell populations. J Immunol. 2010;185:4705–4713. doi: 10.4049/jimmunol.1002276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moon JJ, et al. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malhotra D, et al. Transcriptional profiling of stroma from inflamed and resting lymph nodes defines immunological hallmarks. Nat Immunol. 2012;13:499–510. doi: 10.1038/ni.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berzins SP, Boyd RL, Miller JF. The role of the thymus and recent thymic migrants in the maintenance of the adult peripheral lymphocyte pool. J Exp Med. 1998;187:1839–1848. doi: 10.1084/jem.187.11.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 62.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.