Abstract
PKR is a key component of the interferon-induced antiviral pathway in higher eukaryotes. Upon recognition of viral dsRNA, PKR is activated via dimerization and autophosphorylation. PKR contains two N-terminal dsRNA binding domains (dsRBD) and a C-terminal kinase domain. The dsRBDs and the kinase are separated by a long, unstructured ~80 amino acid linker in the human enzyme. The length of the N-terminal portion of the linker varies among PKR sequences and it is completely absent in one ortholog. Here, we characterize the effects of deleting the variable region from the human enzyme to produce PKRΔV. The linker deletion results in quantitative but not qualitative changes in catalytic activity, RNA binding and conformation. PKRΔV is somewhat more active and exhibits more cooperative RNA binding. As we previously observed for the full-length enzyme, PKRΔV is flexible in solution and adopts a range of compact and extended conformations. The conformational ensemble is biased towards compact states which may be related to weak interactions between the dsRBD and kinase domains. PKR retains RNA-induced autophosphorylation upon complete removal of the linker, indicating that the C-terminal, basic region is also not required for activity.
PKR is a key component of the interferon-induced antiviral pathway in higher eukaryotes.1,2 Upon recognition of intracellular dsRNA of 30 bp or longer it is activated via dimerization and autophophorylation.3 Activated PKR phosphorylates eIF2α, which locks the eIF2A heterotrimer in a GDP bound complex incapable of binding Met-tRNA and initiating translation. Consequently, protein synthesis is halted, leading to apoptosis and containment of the viral infection. In addition, PKR is involved in a plethora of signaling pathways involving stress response and inflammation and regulates cellular growth and proliferation, nutrient signaling and metabolism.4–6
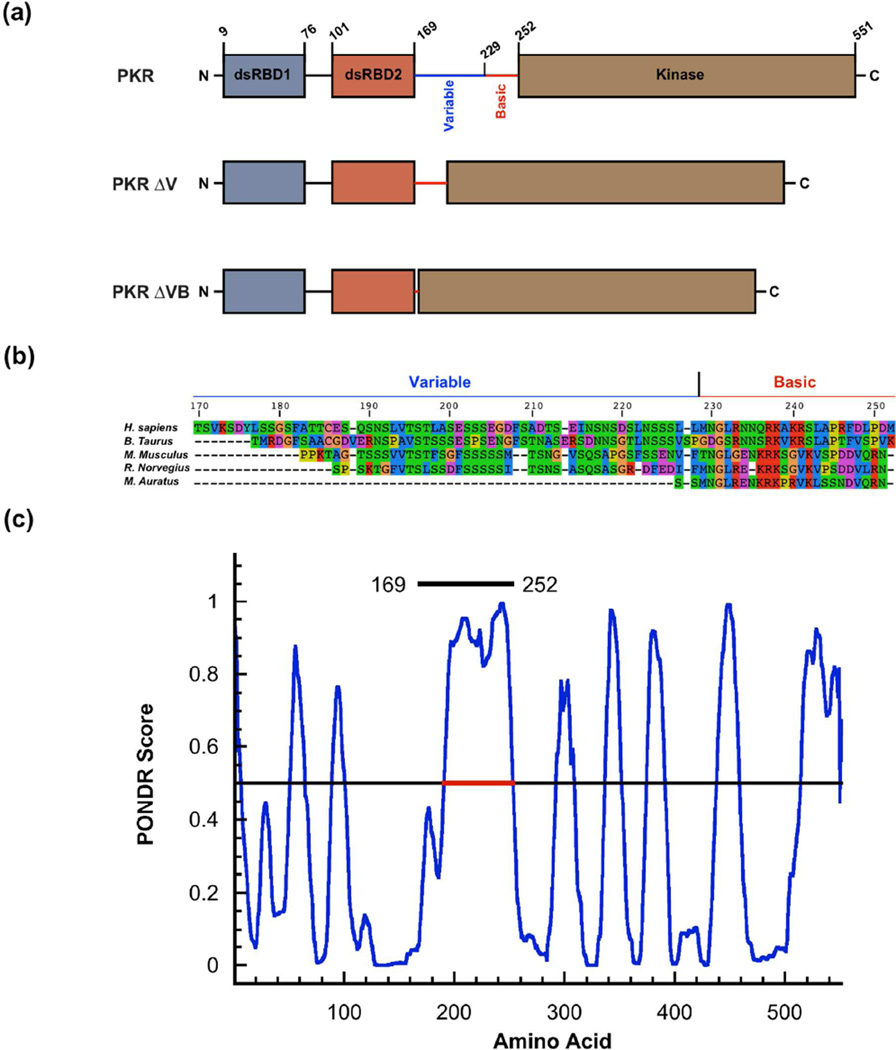
PKR is comprised of two N-terminal dsRNA binding domains (dsRBD) and a C-terminal kinase domain (Fig. 1a). The dsRBDs are separated by a short (~20 amino acid) linker. NMR analysis indicates that this region is disordered.7 In the human enzyme (hPKR), the regulatory and catalytic domains of the protein are separated by a long, ~80 amino acid linker. Residues 170–229 at the N-terminal end of the linker have the signatures of an intrinsically disordered region (Fig. 1b) with low sequence complexity and high content of polar amino acids interspersed with acidic residues. The C-terminal portion (230–252) is enriched in basic residues. An extended region within the linker from 191–252 has a PONDR score > 0.5 (Fig. 1c) and is thus predicted to be disordered. AFM,8 NMR,9 and SAXS10 measurements indicate that the linker is flexible and PKR can adopt a range of closed and open conformations. The PKR kinase domain crystalizes as a dimer1,11 and biochemical and biophysical analyses support a pivotal role for dimerization in RNA-mediated activation.3 PKR dimerizes only weakly (Kd ~ 500 µM); however, this process is capable of inducing autophosphorylation in the absence of dsRNA.12 The isolated kinase domain is not autoactivated; however, constructs including the adjoining basic region (228–551)13 or the entire linker (169–551)14 are activated in the absence of dsRNA, implying that the basic domain may contribute to dimerization or regulate activation.
Figure 1.
PKR domains and linker regions. (a) Domains of PKR and linker deleted constructs. The interdomain linker contains a variable region (169–229) and a basic region (230–252) depicted as blue and red solid lines respectively. (b) Linker sequence alignment for PKR orthologs prepared using SeaView.42 (c) Prediction of disordered regions in hPKR by PONDR.43 The region indicated by the red line (191–253) is predicted to be disordered. The entire linker (169–252) is indicated by the black line.
The role of the long interdomain linker in PKR is not well understood. In the context of the dimerization model for PKR activation,3 the linker functions as a simple tether that brings two kinase domains into close proximity upon binding of two PKRs to a single dsRNA lattice. However, it has been suggested that the linker mediates communication between the RNA binding domain and the kinase.9 Recently, we demonstrated that dsRNAs capable of activating PKR induce PKR kinase domain dimerization, but dimerization is inefficient on nonactivating short (20 bp) dsRNAs capable of binding two PKRs at low salt.15 Thus, the dsRNA binding mode affects kinase dimerization, implying some degree of interdomain communication. Although the long linker appears to be unstructured in the free enzyme it may become ordered upon binding RNA. Interestingly, the sequence and length of the N-terminal portion of the linker are not conserved among mammalian PKR sequences and it is virtually absent in the Syrian golden hamster (M. auratus) (Fig. 1b). This natural variability suggests that this region might not play a critical role in the activation mechanism. Indeed, it was recently reported that deletions of the linker region from hPKR do not affect dsRNA binding, activation or inhibition by noncoding viral RNA inhibitors.16
In order to gain insight into the contribution of the linker to the functional and conformational properties of PKR we initially decided to compare the human enzyme with that from M. auratus (maPKR), which contains the shortest known linker region. Because maPKR is not amenable to biophysical investigations, we prepared a human deletion construct that is analogous to maPKR. Removal of the variable region results in quantitative but not qualitative changes in PKR activation and dsRNA binding affinity. SAXS measurements indicate that this construct is flexible in solution and exists in a range of extended and compact conformations. Although the conformational heterogeneity is greatly reduced upon complete removal of the linker, dsRNA induced activation is retained, indicating that a flexible linker is not required for function.
Materials and Methods
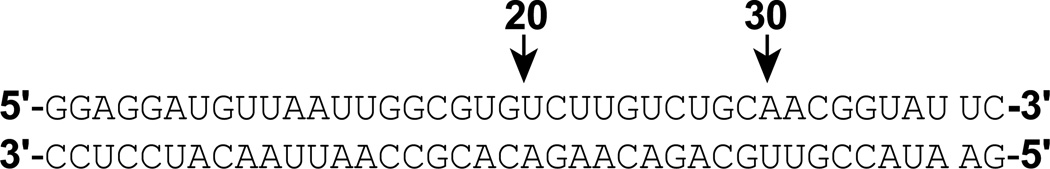
All buffers were made from reagent grade chemicals with deionized, distilled water (Mili-Q, Millipore, MA). Linker-deleted hPKR constructs PKRΔV and PKRΔVB were created by deleting regions 167–228 and 167–253 respectively from the full-length PKR plasmid using PCR. The dsRBD (1–169) was created by deleting the 170–551 region. These proteins were expressed and purified as described previously for hPKR.17 The maPKR sequence was cloned into the pET-PKR-PPase vector previously used to express the human enzyme.12 This vector encodes phage λ phosphatase to produce fully dephosphorylated PKR. maPKR was expressed in Arctic Express RIL DE3 cells (Agilent, CA) which supply cold shock protein. The cultures were grown to an OD600 of 0.8 and were induced with 1 mM IPTG at 10°C for 16 hours. The cells were harvested, lysed and maPKR was purified using the protocol for the human enzyme. All proteins were purified by size exclusion chromatography immediately prior to use on either Superdex 75 or 200 (GE Healthcare) in AU200 buffer (20 mM HEPES, pH 7.5, 200 mM NaCl, 0.1 mM EDTA, 0.1 mM TCEP) RNAs of 20 – 40 nt were obtained from GE Dharmacon (Lafayette, CO) and were purified on denaturing 8M urea gels followed by electroelution. Complementary strands were annealed by heating to 80°C and slowly cooling to room temperature. The sequences are depicted below: Autophosphorylation assays were performed using [γ-32P] ATP as described previously.12,18 RNA-dependent activation was measured using at 0.1 µM of PKR and RNA-independent assays were performed using 0.05 µM – 2 µM PKR. Autophosphorylation reactions were allowed to proceed for 15 minutes at 32°C. Sedimentation velocity analysis of protein-dsRNA interactions was performed as previously described19 using a Beckman Coulter XL-I analytical ultracentrifuge with absorbance optics. Normalized g(s*) distributions were produced using DCDT+20 and global analysis was performed using SEDANAL.21 The binding stoichiometries were determined as previously described.19 Sedimentation equilibrium analysis of PKR dimerization was performed using interference optics as previously described12. Data at multiple loading concentrations were obtained at 18,000 and 22,000 rpm and were globally fit using HeteroAnalysis22 after subtraction of buffer blanks and trimming the gradients to a maximum concentration difference of 4mg/ml between the meniscus and the top of the gradient.
SAXS measurements were performed in AU200 buffer at 4°C using a Nanostar instrument (Bruker, Madison, WI) equipped with a rotating anode source and 550 µm scatter less pinholes. Proteins were centrifuged at 14,000 rpm for 10 minutes prior to measurements. Data were collected in four 1 h frames at a sample-detector distance of 67.2 cm providing a range of q from 0.007 to 0.37 Å−1, defined as q = (4π sinθ)/λ, where 2θ is the scattering angle and λ is the radiation wavelength. Data were reduced using Bruker SAXS software. The four scans overlaid, indicating the absence of radiation damage. Buffer background scattering (15 1h frames) was subtracted from the data prior to analysis. Guinier analysis was performed using the low-q portion of the data where Rg•q<1.3. The p(r) distributions were produced using GNOM23 and the maximum dimension (Dmax) was manually adjusted to the distance where the p(r) distribution approaches zero. Structural modeling was performing with EOM24,25 as previously described10 over a range of q ~0.01 – 0.3 Å−1. Default parameters were used unless otherwise indicated. SAXS data were simulated in IGOR Pro (Wavemetrics, OR) using scattering curves corresponding to an ensemble of 10 structures derived from the pool of 10,000 structures. The Rgs were spaced about equally along the pool distribution and the amplitudes were derived from the pool. The normalized standard deviation corresponding to the level of the 5 mg/ml PKRΔV data set was obtained by fitting a plot of standard deviation/intensity vs. q to a fifth-order polynomial. Gaussian noise corresponding the amplitude of the standard deviation was added to the simulated data.
Results
Analysis of maPKR
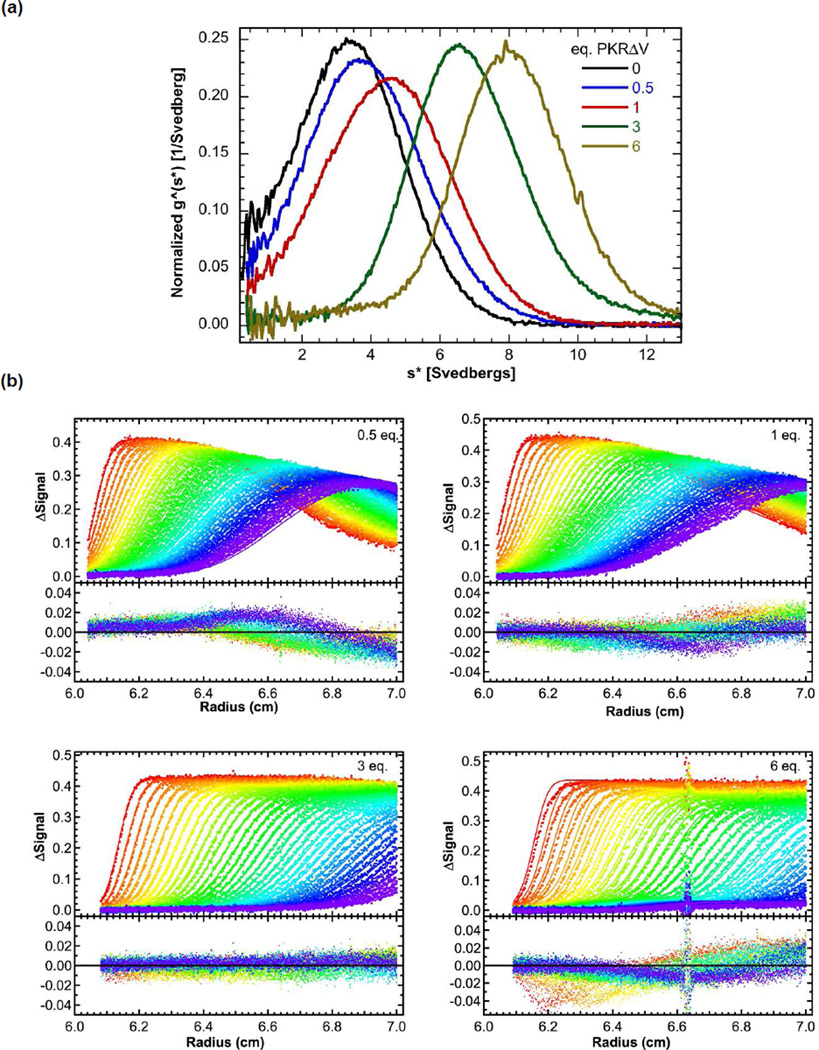
In order to explore the importance of the variable region of the linker, we set out to characterize maPKR, a naturally occurring PKR variant in which this region is largely absent. maPKR does not yield soluble protein when it is expressed in E. coli using conditions optimized for the human enzyme.12 Expression at lower temperature in Arctic Express RIL DE3 cells that supply cold shock chaperones results in increased solubility. However, the Cpn-10 chaperone binds strongly to maPKR and it was not feasible to produce large amounts of homogenous enzyme for biophysical measurements. Sedimentation velocity measurements indicate that maPKR binds to a 40 bp dsRNA (Fig. S1A). The g(s*) distributions shift to the right with increasing protein concentration. Addition of 6 eq. of maPKR induces a large shift from ~ 3.5 S to ~ 9 S (Fig. 4a), which is higher than the fitted sedimentation coefficient for the 2:1 complex formed with hPKR (7.6 S).18 Thus, the magnitude of the shift indicates that three maPKRs bind to the 40 bp RNA. In contrast only two hPKRs bind to this RNA under identical conditions.18,26 maPKR is activated by 40 bp dsRNA and inhibited at high concentrations in a manner analogous to hPKR (Fig S1B). However, the maximum extent of phosphorylation is ~8 fold higher for the hamster enzyme.
Figure 4.
Effect of the variable linker region on RNA binding. PKRΔV binding to 40 bp dsRNA measured by sedimentation velocity. Data were obtained at 0.75 uM RNA and variable protein concentrations. (a) Normalized g(s*) distributions for samples containing 0 (black), 0.5 (blue), 1 (red), 3 (green) and 6 (tan) equivalents of PKRΔV. (b) Global analysis of difference curves for PKRΔV binding to 40 bp dsRNA using a model of three sequential binding events. The top panels show the data (points) and fit (lines) and the bottom panels show the residuals (points). The best fit parameters are presented in Table 1.
Analysis of PKRΔV
Outside of the linker region missing in the hamster enzyme, human and hamster PKR are very closely related, with about 60% sequence identity. Due to the practical limitations of obtaining pure maPKR we examined an analogous construct derived from hPKR in which the variable region is deleted. This construct, denoted PKRΔV (Fig. 1A), displayed an appreciable improvement in solubility and stability relative to hPKR. The activation and RNA binding properties of this construct are similar to the hamster enzyme (vide infra). A construct lacking the linker sequence, PKRΔVB was also created to probe the importance of the C-terminal, basic region of the linker.
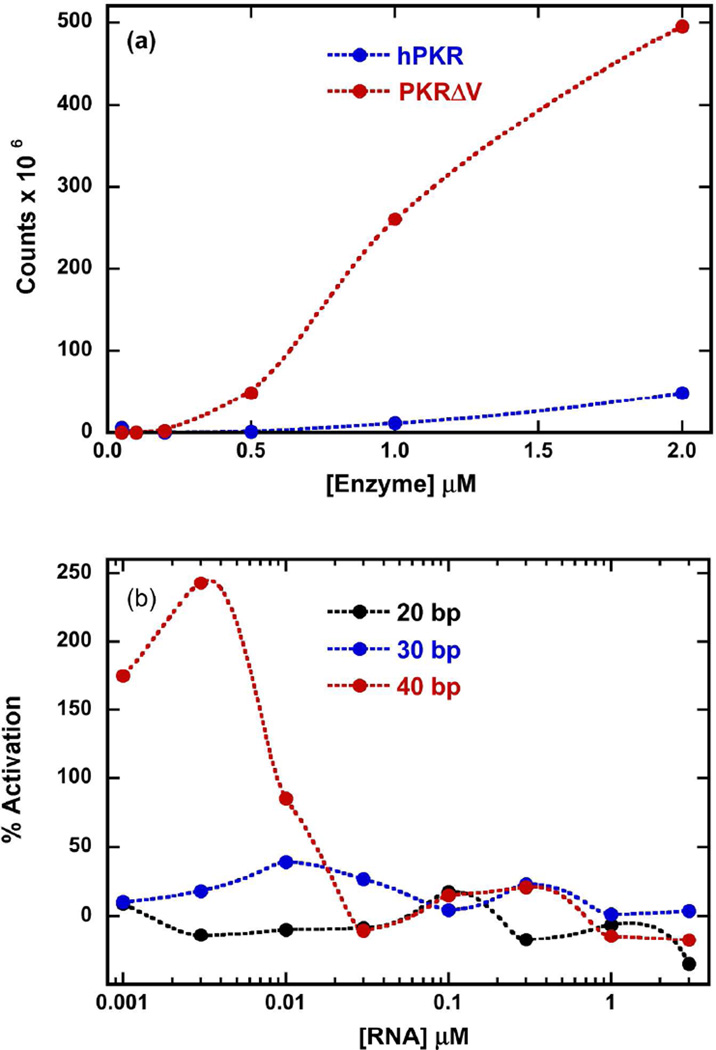
hPKR is capable of being activated in an RNA-independent fashion at higher protein concentrations due to weak dimerization.12 Similarly, PKRΔV also undergoes RNA-independent autophosphorylation (Fig. 2a). The degree of autophosphorylation increases with protein concentrations and is significantly enhanced relative to hPKR. At lower protein concentrations where autoactivation is not present, PKRΔV is activated by dsRNAs with a characteristic bell-shaped activation curve (Fig. 2b). The longest dsRNA examined (40 bp) activates PKRΔV ~2.5 fold more potently than hPKR and the maximum occurs at a low dsRNA concentration of 3 nM compared to about 100 nM for hPKR.26 In contrast to hPKR, the 30 bp dsRNA is a very poor activator of PKRΔV and no activation is detected for the 20 bp duplex.
Figure 2.
Effect of the variable linker region on PKR activation. (a) dsRNA-independent activation of hPKR and PKRΔV. (b) dsRNA-induced activation of PKRΔV. Activation was monitored at 0.1 µM PKRΔV with varying concentrations of 20 bp (black), 30 (blue) and 40 bp (red) dsRNA. The data are normalized relative to the extent of (wild-type) hPKR activation at 0.03 uM 40 bp dsRNA.
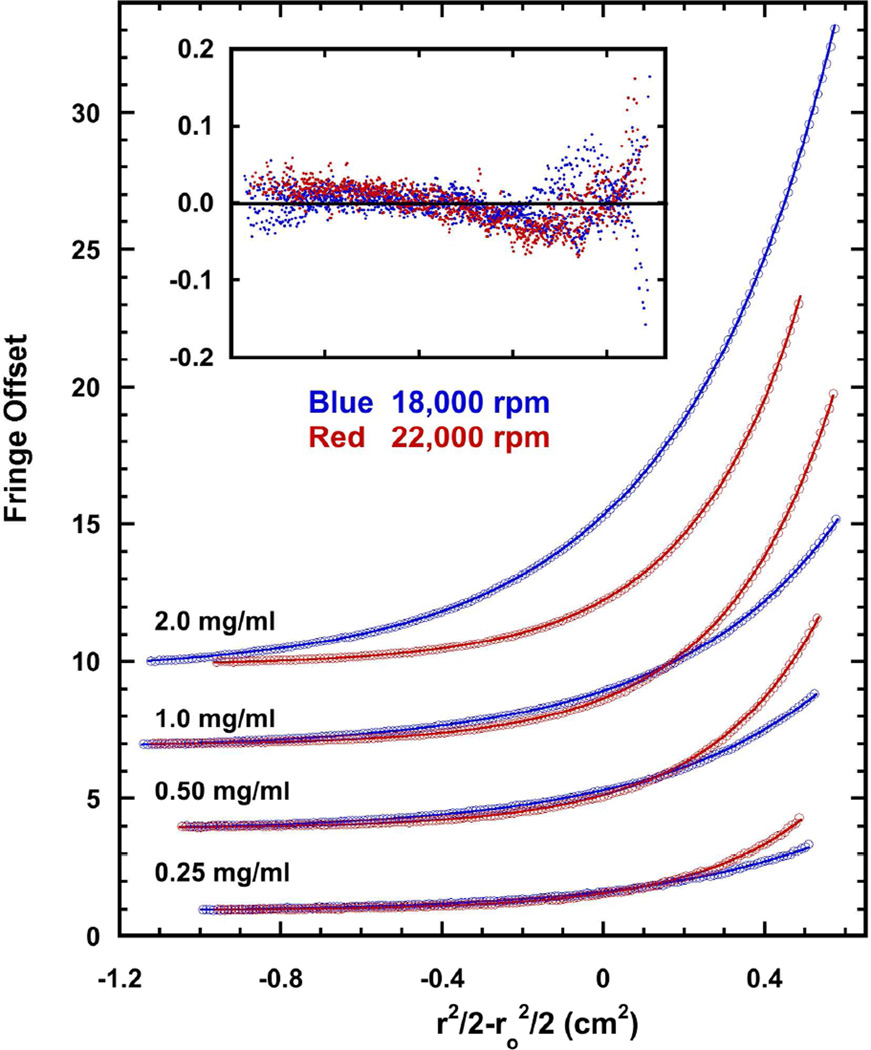
To test whether the increased activation of PKRΔV is due to enhanced dimerization, the dimer dissociation constant of PKRΔV was measured by sedimentation equilibrium (Fig. 3). The weight-average molecular weights increase with protein concentration, indicating that PKRΔV self-associates (data not shown). Data were collected over a protein concentration range of 0.25 – 2 mg/ml and two rotor speeds and were globally fit to a monomer-dimer equilibrium model. The best fit Kd = 1.105 (1.103, 1.210) mM, where the values in parentheses correspond to the 95% joint confidence intervals. Thus, deletion of the variable portion of the linker does not enhance dimerization affinity, indicating that the enhanced degree of RNA-independent autophosphorylation of PKRΔV must be due to other effects.
Figure 3.
Dimerization of PKRΔV. The Kd for self-association of PKRΔV was measured using sedimentation equilibrium. Data (open circles) were collected for 0.25, 0.5, 1, and 2 mg/mL PKRΔV at two rotor speeds: 18,000 rpm (blue) and 22,000 rpm (red). For clarity, only every other data point is shown and the curves are vertically offset. The solid lines indicate a global fit of the data to a monomer-dimer equilibrium model. The best-fit Kd = 1.105 (1.103, 1.210) mM with an rmsd of 0.0256 fringes Inset: residuals.
The stoichiometries and affinities for PKRΔV binding to dsRNA were measured for 20–40 bp sequences using sedimentation velocity and compared with hPKR. The stoichiometries were determined from the magnitude of the shift in the g(s*) sedimentation coefficient distribution function upon titration of the dsRNA with protein.19 For the 40 bp dsRNA, addition of 6 eq. of PKRΔV induces a large shift from ~ 3.5 S to ~8 S (Fig. 4a), which is greater than the fitted sedimentation coefficient for the 2:1 complex formed with hPKR (7.6 S).18 Thus, more than two PKRΔV bind to the 40 bp dsRNA in AU200 buffer whereas only two hPKR monomers bind under comparable conditions. The data obtained at four protein concentrations were globally fit a 3:1 binding model using SEDANAL (Fig. 4b). A good fit is obtained with a low rmsd of 0.0070 OD and small residuals. The well defined confidence intervals for binding of the third PKR support the 3:1 model (Table 1). The best-fit Kds for the first and second PKR binding events are 75 nM and 56 nM, respectively, with considerably weaker binding of the third monomer. For hPKR, Kd1 = 38 nM and Kd2 = 813 nM.18 Thus, the first PKRΔV binds to the 40 bp dsRNA with similar affinity as hPKR but the affinity for the second PKRΔV is enhanced by about 10-fold. Similarly, for the 30 bp dsRNA the affinity for the second PKRΔV is enhanced about 7-fold.
Table 1.
Binding constants for PKRΔV and duplex dsRNAs
| RNA | Kd1 (nM) | Kd2 (nM) | Kd3 (nM) | rmsda |
|---|---|---|---|---|
| 20 bp | 235 (189, 286) | - | - | 0.0079 |
| 30 bp | 77 (24, 215) | 865 (480, 1700) | - | 0.0160 |
| 40 bp | 75 (43, 121) | 56 (27, 99) | 2,290 (1,380, 3.870) | 0.0070 |
Parameters obtained by global nonlinear least square analysis of sedimentation velocity experiments.
The values in parentheses represent the 95% joint confidence intervals obtained using the F-statistic.
Root-mean square deviation in absorbance units.
Analysis of PKRΔVB
Recently, it was reported that deletion of the entire linker (170–250) does not affect activation of hPKR by dsRNA or inhibition by a structured viral RNA, Adenovirus VAI.16 A similar deletion construct, PKRΔVB, is activated by 40 bp dsRNA (Fig. S2). However, the maximal activation level is only about 30% relative to hPKR. The maximal activation occurs at 0.3 µM RNA, which is similar to hPKR. Thus, the basic region is not necessary for RNA-induced activation but it does affect the extent of autophosphorylation. Sedimentation equilibrium analysis of PKRΔVB indicates weak dimerization, with Kd = 1.73 (1.53, 1.98) mM and a low rmsd of 0.0185 fringes for the global fit (data not shown).
SAXS analysis of PKR conformation
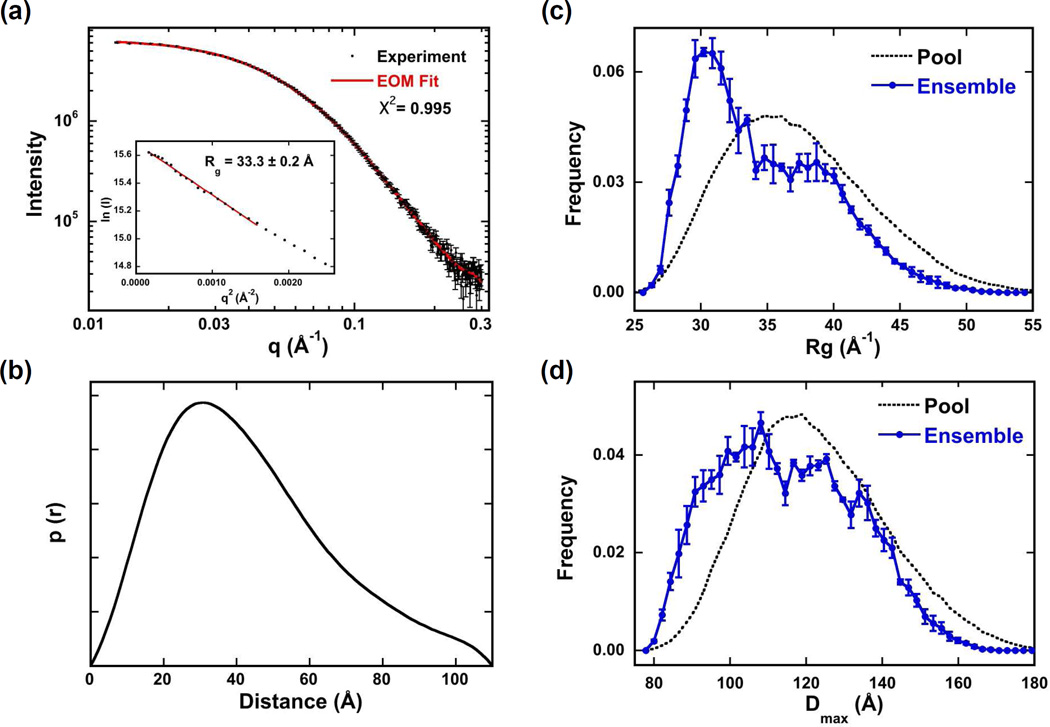
hPKR contains two flexible linker regions and adopts a range of compact and extended conformations.10 The contribution of the variable linker region of PKR to flexibility was determined by small angle scattering analysis of PKRΔV. Data were collected over a concentration range from 1 - 5 mg/ml. The parameters derived from Guinier and GNOM analysis did not vary substantially over this concentration range (Table S1) so the data at the highest concentration were used for further analysis. A linear Guinier plot (Fig. 5a) indicates the absence of aggregation. The radius of gyration (Rg) of 33.3 ± 0.2 Å is substantially smaller than for hPKR (~ 40 A),10 indicating that PKRΔV does not adopt as extended a conformation as the wild type protein. Similar to hPKR, the p(r) distribution of interatomic distances peaks at about 30 Å with a tail to longer distances, suggesting the presence of flexible regions (Fig. 5b). However, the Dmax for PKRΔV is smaller (110 Å) relative to hPKR (175 Å). A Kratky plot of the scattering data (Fig. S3) exhibits a peak at lower q and rises at higher q, which is characteristic of proteins containing both folded and disordered regions.27
Figure 5.
SAXS analysis of PKRΔV conformation. Data were collected at 4°C at a protein concentration of 5 mg/ml. (a) Scattering data (points) and EOM fit (solid red line). Inset: Guinier analysis yields Rg = 33.3 ± 0.2 Å. (b) p(r) distance distribution produced by transforming the data in part (a) using GNOM.23 Rg (c) and Dmax (d) distributions produced by fitting the data in part (a) using EOM.24,25 The distributions of the random pool of 10,000 structures are shown in black dotted lines and the distributions selected by the genetic algorithm to fit the experimental data are shown in solid blue. The error bars correspond to the standard deviations of the distributions produced from three runs of GAJOE.
Based on the evidence that PKRΔV retains flexibility we have pursued structural models consisting of three folded domains (dsRBD1, dsRBD2 and kinase) connected by linkers that can adopt multiple conformations. The EOM program24,25 was used to generate a large (10,000) pool of structures and select the minimal ensemble from this pool that best fits the SAXS data. Figures 5c and 5d show distribution of Rg and Dmax, respectively, for the initial pool and the ensembles selected by EOM. In both cases, the selected structures exhibit broad distributions, indicating the contribution of compact and extended conformations to the ensemble. However, the distributions are biased towards more compact conformations relative to the random pool, suggesting the presence of attractive interactions between domains. We have confirmed that PKRΔV is flexible by fitting the SAXS data using EOM with an ensemble size of one or alternatively, by rigid body modeling of single structure using BUNCH.28 The X2 of the fit increases from 0.996 in the default EOM analysis to 1.303 using an ensemble of one and 1.56 using BUNCH (Fig. S4). Thus, the SAXS data fit poorly to models incorporating only a single conformation.
The Rg distribution and to a lesser extent, the Dmax distribution, derived from EOM analysis of PKRΔV appears bimodal. This observation implies that PKRΔV may cluster into two families of compact and extended conformations. Consistent with this picture, the X2 of the fit increases only slightly to 1.056 as the ensemble size is reduced to two. However, simulations indicate that such bimodal distributions may be an artifact (Fig. S5). A continuous distribution of structures was simulated by selecting ten conformations from the pool with amplitudes derived from the pool distribution. Noise equal to the experimental PKRΔV scattering was added and the resulting simulated data set was fit using EOM with the same pool. Interestingly, the selected distribution was also bimodal and reducing the ensemble size to two results in only a marginal increase in X2 from 1.023 to 1.040. Thus, the bimodal shape of the PKRΔV distribution does not necessarily imply the presence of two families of conformers. Repeating this process using noise-free data resulting in a selected distribution that reproduced the underlying pool more faithfully (data not shown), indicating that the noise level is an important determinant of the quality of the distributions produced by EOM.
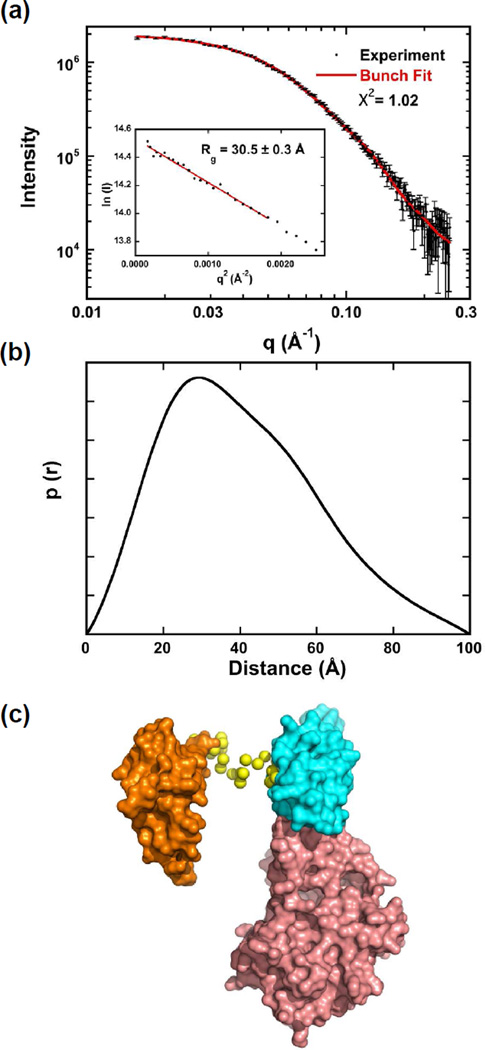
The flexibility of PKRΔVB was also analyzed by SAXS. As expected, the Rg of 30.5 ± 0.3 Å (Fig. 6a and Table S2) is slightly smaller than for PKRΔV. Like the other PKR constructs, the p(r) distribution also peaks near 30 Å but it is less extended, with a Dmax of 100 Å (Fig. 6b). The scattering data fit well to a model of a single structure generated using BUNCH with X2 = 1.02 (Fig. 6a). In this structure the dsRBD2 and kinase form an interface and dsRBD1 is extended away from dsRBD2 (Fig. 6c). The lack of interdomain contacts is a characteristic of dynamic systems.29 Presumably, the short (21 aa) region lying between dsRBD1 and dsRBD2 remains flexible in PKRΔVB. Fitting the data to a model allowing for an ensemble of conformations using EOM results in only a slight improvement in the fit quality (X2 = 0.937). Presumably the dsRBD1-dsRBD2 flexibility contributes weakly to the overall scattering curve so that the data fit reasonable well to a model of a single conformation.
Figure 6.
SAXS analysis of PKRΔVB conformation. Data were collected at 4°C at a protein concentration of 5 mg/ml. (a) Scattering data (points) and BUNCH fit (solid red line). Inset: Guinier analysis yields Rg = 30.5 ± 0.3 Å. (b) p(r) distance distribution produced by transforming the data in part (a) using GNOM.23 (c) Structural model produced by BUNCH.28 The domains are shown in surface representation: dsRBD1 (orange), dsRBD2 (cyan) and kinase (salmon). The linker residues are depicted as yellow spheres.
Discussion
It is noteworthy that the length of the interdomain linker in PKR orthologs is highly variable, leading to the question of how this variable region affects the structure and function of the enzyme. This region is virtually absent in maPKR and we initial sought to compare this construct to hPKR. However, maPKR is difficult to obtain in purified form. We created an analogous deletion construct of the human enzyme, PKRΔV, which is highly homologous to the M. auratus enzyme and was chosen as a suitable alternative to probe the role of the variable linker. Control experiments verify that PKRΔV behaves similarly to maPKR.
A key observation to emerge from our analyses is that the removal of the variable region of the linker results in quantitative but not qualitative changes in the catalytic activity, RNA binding and conformational properties of PKR. PKRΔV retains enzymatic activity and undergoes both RNA-independent and RNA induced phosphorylation. The enhanced autoactivation of PKRΔV indicates that the linker in hPKR negatively regulates the rate of PKR autophosphorylation. This enhancement is not simply due to higher affinity dimerization. PKR autophosphorylates at multiple sites lying within the kinase domain, the basic region and the dsRBD.30–33 Potentially the variable length linker separates the domains and limits the rate with which the kinase domain catalyzes cis phosphorylation of sites within the dsRBD. Deletion of the linker may allow more rapid phosphorylation of serines and threonines in the dsRBD due to induced proximity. Alternatively, the variable region may act as a steric barrier that limits access to the sites located in the dsRBD.
Like hPKR, PKRΔV exhibits a bell-shape curve for activation by dsRNA where high concentration inhibit. This effect has been attributed to dissociation of PKR dimers onto separate RNAs at high RNA concentrations.34,35 The length dependence of dsRNA activation can be correlated with binding stoichiometry: 30 bp is the minimal length capable of binding two PKRs; however, in contrast to full length hPKR, PKRΔV is barely activated by 30 bp dsRNA. However, this may not represent a fundamental difference in the dsRNA activation requirements for the two enzymes. Because of the potent activation of PKRΔV in the absence of dsRNA, it is necessary to reduce the protein concentration from 200 to 100 nM in the assays. Under these conditions, the concentration of the active species containing two PKRs bound to a single RNA is reduced and it may be difficult to detect weak activation.
The RNA affinities for binding of the first monomer PKRΔV are similar to those we previously reported for hPKR18 with some enhanced affinity for the second PKR binding to the 30 and 40 bp dsRNAs and detection of a third PKR binding to the 40 bp sequence. Previously, we only detected a third PKR binding to this sequence upon reducing the NaCl concentration from 200 to 75 mM.18 The fact that the first binding events occur with similar affinity for hPKR and PKRΔV suggest that intrinsic free energy of association is similar for the two constructs. The enhanced affinities for the second and third PKRΔV suggests that removal of the variable linker region enhances the cooperativity of assembly. The enhanced activation of PKRΔV by the 40 bp dsRNA is correlated with the increased binding affinity. Based on the measured dissociation constants18 and the PKR concentration used for the activation assays (100 nM), we have simulated the equilibria in the titration of PKR with the 40 bp to define the maximal fraction of PKR residing in the active species containing two PKRs bound to a single RNA. For hPKR, the maximum is 5.5% but for PKRΔV the maximum increases to 36%. In the latter case, the population of the species containing three bound PKRs is negligible due to the low protein concentration in the assay. Thus, the enhanced activation of PKRΔV, and possibly maPKR, by the 40 bp dsRNA is at least partially due to the increased affinity for binding of the second monomer.
hPKR retains RNA-induced activation upon complete removal of the linker in PKRΔVB, indicating that the basic region is not absolutely required for activity. Similarly, Conn and coworkers that reported that removal of the linker did not block activation of PKR by dsRNA or inhibition by VAI RNA.16 They found that the functional properties of hPKR and a linker-deleted construct are identical, whereas we detected some quantitative differences in RNA-independent and dependent activation and RNA binding affinities. Interestingly, rat PKR was reported to bind viral RNA inhibitor with the same affinity as the human enzyme but the inhibition potency was greatly reduced. Surprisingly, the apparent affinity is affected by reversal of the orientation of the titrant and titrand. The stoichiometry and affinity of PKR binding to VAI RNA is modulated by divalent ion,36 and these discrepancies may be due to differences in the experimental conditions: 100 mM NaCl with no divalent ion for the isothermal titration calorimetry binding measurements and 50 mM KCl and 2 mM Mg2+ for the activity assays.16
SAXS analysis reveals that PKRΔV retains significant conformational flexibility despite the removal of the bulk of the interdomain linker. The shapes of the Kratky plot and p(r) distribution are characteristic of proteins containing disordered regions. Structural modeling treating the regions lying between dsRBD1 and dsRBD2 and basic region as flexible linkers reveals that good fits to the scattering data require models that incorporate an ensemble of conformations. Although we cannot ascribe significance to the bimodal shapes of the Rg and Dmax distributions, they are clearly biased towards more compact distributions that than the random pool. Similar bias was observed in our previous SAXS studies of full length PKR.10 NMR chemical shift perturbation measurements37,38 indicate interactions between dsRBD2 and the kinase domain that may be responsible for the compaction of the ensemble. This interaction is weak and is expected to lead to population of both closed and open states.39 The persistence of this conformational bias in PKRΔV implies that the basic linker region remains flexible to allow the kinase and dsRBD2 to interact. Consistent with this interpretation, deletion of the basic region in PKRΔVB quenches the bulk of the flexibility in PKRΔV. However, further investigation is required to parse the contributions of the dsRBD1-dsRBD2 linker and the basic region to the overall flexibility of the PKR.
Our observation that the structural and functional properties of PKR are not strongly affected by the presence of the variable linker region leads to the questions: why does this region exist and why do PKR orthologs contain different length linkers? PKR is activated by a diverse array of RNAs, some of which contain extensive tertiary structure,40 and we have previously suggested that flexible tethers would permit dimerization of kinase domains and activation in cases where the dsRBDs of two PKR monomers are bound at more distal regions of RNA activators.10 In this case, the effects of linker length would become more pronounced for RNAs that contain duplex binding sites that are far apart. However, PKRΔV is not more susceptible than the full-length enzyme to the effects of insertion of 2'-O-Me dsRNA barriers in the middle of a 30 bp dsRNA (data not shown). Further investigation using a more diverse range of PKR ligands may reveal a linker dependence. Alternatively, the linker region may contribute to PKR function in vivo.16 Unstructured regions are often involved in protein-protein interactions41 and PKR is involved in a plethora of signaling pathways4–6 that may be dependent on interactions with the unstructured linker.
Supplementary Material
Scheme 1.
Table 2.
SAXS parameters for PKRΔV
| [Protein] mg/ml |
Guinier Rg(Å)a | GNOM Rg(Å)b | Porod Volume (Å3)b |
|---|---|---|---|
| 5 | 33.30 ± 0.21 | 34.10 ± 0.14 | 87,200 |
| 2 | 32.20 ± 0.41 | 33.62 ± 0.20 | 86,200 |
| 1 | 31.60 ± 0.67 | 33.46 ± 0.31 | 87,800 |
Rg determined from the slope of the Guinier plot.
Rg determined using GNOM.23
Acknowledgements
We thank Michael Stobart and Dr. David Knox (University of Manitoba) for supplying maPKR DNA.
Funding Source: This work was supported by grant number AI-53615 from the NIH to J.L.C. The SAXS instrument was acquired with support from NSF-MRI 1228817.
Abbreviations
- dsRBD
double-stranded RNA binding domain
- dsRNA
double-stranded RNA
- eIF2α
eukaryotic initiation factor 2α
- hPKR
human PKR
- maRNA
M. auratus PKR
- PKR
RNA activated protein kinase
- rmsd
root-mean square deviation
Footnotes
Supporting Information
Supporting Information Available. Figure S1. Comparison of hamster PKR and PKRΔV. Figure S2. Activation of PKRΔVB by dsRNA. Figure S3. Kratky plot for PKRΔV. Figure S4. BUNCH analysis of PKRΔV. Figure S5. Simulation of the PKRΔV ensemble. Table S1. SAXS parameters for PKRΔVB. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 2.Unterholzner L, Bowie AG. The interplay between viruses and innate immune signaling: recent insights and therapeutic opportunities. Biochem. Pharmacol. 2008;75:589–602. doi: 10.1016/j.bcp.2007.07.043. [DOI] [PubMed] [Google Scholar]
- 3.Cole JL. Activation of PKR: an open and shut case? Trends Biochem Sci. 2007;32:57–62. doi: 10.1016/j.tibs.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiology and Molecular Biology Reviews. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pindel A, Sadler A. The role of protein kinase R in the interferon response. J Interferon Cytokine Res. 2011;31:59–70. doi: 10.1089/jir.2010.0099. [DOI] [PubMed] [Google Scholar]
- 6.Lu B, Nakamura T, Inouye K, Li J, Tang Y, Lundbäck P, Valdes-Ferrer SI, Olofsson PS, Kalb T, Roth J, Zou Y, Erlandsson-Harris H, Yang H, Ting JPY, Wang H, Andersson U, Antoine DJ, Chavan SS, Hotamisligil GS, Tracey KJ. Novel role of PKR in inflammasome activation and HMGB1 release. Nature. 2012;488:670–674. doi: 10.1038/nature11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nanduri S, Carpick BW, Yang Y, Williams BR, Qin J. Structure of the double-stranded RNA binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemaire PA, Tessmer I, Craig R, Erie DA, Cole JL. Unactivated PKR exists in an open conformation capable of binding nucleotides. Biochemistry. 2006;45:9074–9084. doi: 10.1021/bi060567d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKenna SA, Lindhout DA, Kim I, Liu CW, Gelev VM, Wagner G, Puglisi JD. Molecular Framework for the Activation of RNA-dependent Protein Kinase. Journal of Biological Chemistry. 2007;282:11474–11486. doi: 10.1074/jbc.M700301200. [DOI] [PubMed] [Google Scholar]
- 10.Vanoudenhove J, Anderson E, Krueger S, Cole JL. Analysis of PKR structure by small-angle scattering. J Mol Biol. 2009;387:910–920. doi: 10.1016/j.jmb.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F, Li S, Wang Z, Shen Y, Zhang T, Yang X. Structure of the kinase domain of human RNA-dependent protein kinase with K296R mutation reveals a face-to-face dimer. Chin. Sci. Bull. 2013;58:998–1002. [Google Scholar]
- 12.Lemaire PA, Lary J, Cole JL. Mechanism of PKR activation: dimerization and kinase activation in the absence of double-stranded RNA. J Mol Biol. 2005;345:81–90. doi: 10.1016/j.jmb.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 13.Wu S, Kaufman RJ. A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA-activated protein kinase PKR. J Biol Chem. 1997;272:1291–1296. doi: 10.1074/jbc.272.2.1291. [DOI] [PubMed] [Google Scholar]
- 14.McKenna SA, Lindhout DA, Shimoike T, Puglisi JD. Biophysical and biochemical investigations of dsRNA-activated kinase PKR. Meth Enzymol. 2007;430:373–396. doi: 10.1016/S0076-6879(07)30014-1. [DOI] [PubMed] [Google Scholar]
- 15.Husain B, Hesler S, Cole JL. Regulation of PKR by RNA: Formation of Active and Inactive Dimers. Biochemistry. 2015;54:6663–6672. doi: 10.1021/acs.biochem.5b01046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sunita S, Schwartz SL, Conn GL. The Regulatory and Kinase Domains but Not the Interdomain Linker Determine Human Double-stranded RNA-activated Kinase (PKR) Sensitivity to Inhibition by Viral Non-coding RNAs. Journal of Biological Chemistry. 2015;290:28156–28165. doi: 10.1074/jbc.M115.679738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson E, Pierre-Louis WS, Wong CJ, Lary JW, Cole JL. Heparin Activates PKR by Inducing Dimerization. J Mol Biol. 2011;413:973–984. doi: 10.1016/j.jmb.2011.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Husain B, Mukerji I, Cole JL. Analysis of high-affinity binding of protein kinase R to double-stranded RNA. Biochemistry. 2012;51:8764–8770. doi: 10.1021/bi301226h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong CJ, Launer-Felty K, Cole JL. Analysis of PKR-RNA interactions by sedimentation velocity. Meth Enzymol. 2011;488:59–79. doi: 10.1016/B978-0-12-381268-1.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 21.Stafford WF, Sherwood PJ. Analysis of heterologous interacting systems by sedimentation velocity: curve fitting algorithms for estimation of sedimentation coefficients, equilibrium and kinetic constants. Biophys Chem. 2004;108:231–243. doi: 10.1016/j.bpc.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 22.Cole JL. Analysis of heterogeneous interactions. Meth Enzymol. 2004;384:212–232. doi: 10.1016/S0076-6879(04)84013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Semenyuk AV, Svergun DI. GNOM – a program package for small-angle scattering data processing. J Appl Crystallogr. 1991;24:537–540. [Google Scholar]
- 24.Tria G, Mertens HDT, Kachala M, Svergun DI. Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ. 2015;2:207–217. doi: 10.1107/S205225251500202X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernadó P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 26.Lemaire PA, Anderson E, Lary J, Cole JL. Mechanism of PKR Activation by dsRNA. J Mol Biol. 2008;381:351–360. doi: 10.1016/j.jmb.2008.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kikhney AG, Svergun DI. A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett. 2015;589:2570–2577. doi: 10.1016/j.febslet.2015.08.027. [DOI] [PubMed] [Google Scholar]
- 28.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernadó P. Effect of interdomain dynamics on the structure determination of modular proteins by small-angle scattering. Eur Biophys J. 2010;39:769–780. doi: 10.1007/s00249-009-0549-3. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, Herring CJ, Romano PR, Szczepanowska J, Brzeska H, Hinnebusch AG, Qin J. Identification of phosphorylation sites in proteins separated by polyacrylamide gel electrophoresis. Anal Chem. 1998;70:2050–2059. doi: 10.1021/ac971207m. [DOI] [PubMed] [Google Scholar]
- 31.Zhang F, Romano PR, Nagamura-Inoue T, Tian B, Dever TE, Mathews MB, Ozato K, Hinnebusch AG. Binding of double-stranded RNA to protein kinase PKR is required for dimerization and promotes critical autophosphorylation events in the activation loop. J Biol Chem. 2001;276:24946–24958. doi: 10.1074/jbc.M102108200. [DOI] [PubMed] [Google Scholar]
- 32.Taylor DR, Tian B, Romano PR, Hinnebusch AG, Lai MM, Mathews MB. Hepatitis C virus envelope protein E2 does not inhibit PKR by simple competition with autophosphorylation sites in the RNA-binding domain. Journal of Virology. 2001;75:1265–1273. doi: 10.1128/JVI.75.3.1265-1273.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romano PR, Garcia-Barrio MT, Zhang X, Wang Q, Taylor DR, Zhang F, Herring C, Mathews MB, Qin J, Hinnebusch AG. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2alpha kinases PKR and GCN2. Mol Cell Biol. 1998;18:2282–2297. doi: 10.1128/mcb.18.4.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manche L, Green SR, Schmedt C, Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kostura M, Mathews MB. Purification and activation of the doublestranded RNA-dependent eIF-2 kinase DAI. Mol Cell Biol. 1989;9:1576–1586. doi: 10.1128/mcb.9.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Launer-Felty K, Wong CJ, Wahid AM, Conn GL, Cole JL. Magnesium-dependent interaction of PKR with adenovirus VAI. J Mol Biol. 2010;402:638–644. doi: 10.1016/j.jmb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gelev V, Aktas H, Marintchev A, Ito T, Frueh D, Hemond M, Rovnyak D, Debus M, Hyberts S, Usheva A, Halperin J, Wagner G. Mapping of the auto-inhibitory interactions of protein kinase R by nuclear magnetic resonance. J. Mol. Biol. 2006;364:352–363. doi: 10.1016/j.jmb.2006.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nanduri S, Rahman F, Williams BR, Qin J. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J. 2000;19:5567–5574. doi: 10.1093/emboj/19.20.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson E, Cole JL. Domain stabilities in protein kinase R (PKR): evidence for weak interdomain interactions. Biochemistry. 2008;47:4887–4897. doi: 10.1021/bi702211j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nallagatla SR, Toroney R, Bevilacqua PC. Regulation of innate immunity through RNA structure and the protein kinase PKR. Curr Opin Struct Biol. 2011;21:119–127. doi: 10.1016/j.sbi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 42.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 43.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.