Abstract
Background & Aims
Glycine N-methyltransferase (GNMT) expression is decreased in some patients with severe NAFLD. Gnmt deficiency in mice (Gnmt-KO) results in abnormally elevated serum levels of methionine and its metabolite S-adenosylmethionine (SAMe), and this leads to rapid liver steatosis development. Autophagy plays a critical role in lipid catabolism (lipophagy), and defects in autophagy have been related to liver steatosis development. Since methionine and its metabolite SAMe are well known inactivators of autophagy, we aimed to examine whether high levels of both metabolites could block autophagy-mediated lipid catabolism.
Methods
We examined methionine levels in a cohort of 358 serum samples from steatotic patients. We used hepatocytes cultured with methionine and SAMe, and hepatocytes and livers from Gnmt-KO mice.
Results
We detected a significant increase in serum methionine levels in steatotic patients. We observed that autophagy and lipophagy were impaired in hepatocytes cultured with high methionine and SAMe, and that Gnmt-KO livers were characterized by an impairment in autophagy functionality, likely caused by defects at the lysosomal level. Elevated levels of methionine and SAMe activated PP2A by methylation, while blocking PP2A activity restored autophagy flux in Gnmt-KO hepatocytes, and in hepatocytes treated with SAMe and Methionine. Finally, normalization of methionine and SAMe levels in Gnmt-KO mice using a methionine deficient diet normalized the methylation capacity, PP2A methylation, autophagy, and ameloriated liver steatosis.
Conclusions
These data suggest that elevated levels of methionine and SAMe can inhibit autophagic catabolism of lipids contributing to liver steatosis.
Keywords: Autophagy, SAMe, methionine, liver, steatosis, Gnmt
Introduction
Liver steatosis results from an excessive delivery of free fatty acids from adipose tissue into the liver, and from a misbalance in de novo lipid synthesis and catabolism [1]. During autophagy, cellular proteins and organelles are sequestered by autophagosomes and degraded after fusion with lysosomes in response to a lack of nutrients or to stress[2]. Autophagy can also hydrolyze triglycerides (TG) in a process known as lipophagy[3], and inhibition of autophagy in hepatocytes leads to lipid accumulation[3]. Conversely, genetical[4] or pharmacological[5] promotion of autophagy alleviates hepatic steatosis in non-alcoholic fatty liver disease (NAFLD) in mice. These studies suggest that defects in autophagy might be involved in the development of fatty liver[3, 6], and that modulation of autophagy could be beneficial in NAFLD [5].
S-adenosylmethionine (SAMe), the principal biological methyl donor, is synthesized from methionine and ATP in a reaction catalyzed by the enzyme methionine adenosyltransferase (MAT). SAMe, upon transfer of its activated methyl group to an acceptor molecule such as glycine, is converted to S-adenosylhomocysteine (SAH)[7]. Glycine N-methyltransferase (GNMT) is one of the key enzymes involved in methionine and SAMe metabolism[8], and it has been proposed that GNMT maintains intracellular concentrations of SAMe within a narrow range[9]. There are several genetic conditions that lead to abnormally elevated plasma concentrations of methionine and SAMe that have been related with liver steatosis, such as GNMT, S-adenosylhomocysteine hydrolase and cystathionine β-synthase deficiency[10]. For example, Gnmt-KO mice, characterized by highly elevated SAMe and methionine levels, develop steatosis, fibrosis and hepatocelullar carcinoma[11].
Recently, it was shown that GNMT mRNA levels were often repressed in advanced NAFLD patients[12]. Hypermethioninemias has been associated with liver steatosis[13], and dietary methionine restriction can reverse liver steatosis in the ob/ob mice[14]. In a recent study in yeast, it was shown that methionine-induced inhibition of autophagy could be due to its conversion into SAMe[15]. It is an interesting posibility that high levels of methionine and/or its metabolite SAMe could also contribute to hepatic steatosis development by blocking lipid degradation through inhibition of lipophagy.
We found that the abnormally elevated levels of methionine and SAMe in the absence of Gnmt induced PP2A methylation that led to an inhibition of autophagy flux, contributing to lipid accumulation. Strikingly, we found that this effect was independent of MTOR, one of the critical inactivators of the autophagosome formation[16]. Altogether, these findings indicate that Gnmt deficiency, and/or hypermethioninemia and elevated SAMe levels could play a role in the pathogenesis of NAFLD by inhibiting lipophagy through PP2A methylation.
Material and Methods
Human Samples
Serum samples from non-steatotic and NAFLD patients were examined (non-steatotic=92, NAFLD=263). Clinical data were collected retrospectively using patient records described in a previous report [13]. Informed consent for all clinical investigations was obtained in accordance with the principles in the Declaration of Helsinki. The institutional review board of the Hospitals involved approved the protocol.
Animals
3-month-old male Gnmt-KO (n=10) or WT (n=10) mice were maintained on a methionine deficient diet (MDD) (S8946-E020 EF AIN 76A 0,15% L-methionine, from SSNIFF) for 21 days prior to analysis[17]. 3-month-old male WT (n=10) and Gnmt-KO (n=10) mice were euthanized after 8h and 24h of food starvation. Experiments were performed following the guidelines of the CIC bioGUNE institutional review committee on animal use.
Hepatocyte Isolation
Hepatocytes were isolated from mice livers by collagenase perfusion[18]. SAMe (Samyr®) was from Abbott. Purity of hepatocytes was determined by comparing their mRNA expression levels of Desmin (a hepatic stellate cell marker) and F4/80 (a macrophage marker) with the expression levels in purified cultures of hepatic stellate cells and Kupffer cells (each set as 100% value)[19]. We observed a maximum of 2.4% of contamination with Kupffer cells and 0.1% of contamination with hepatic stellate cells (data not shown).
Methionine and SAMe measurements
SAMe and methionine were determined as described in previous reports[11].
Western blot analysis
Immunoblotting was performed with specific antibodies (Supplementary Table 2). In selected figures, images of gels were constructed by splicing non-contiguous portions of the same gel (indicated by dashed lines).
Autophagic flux
Autophagic flux was determined by analyzing LC3-II turnover by preventing lysosomal degradation, using leupeptin (100 μM) and ammonium chloride treatment (20 mM) (N/L)][20] or using chloroquine (60 μM) (Chlo). LC3-flux was calculated by subtracting the densitometry value of normalized LC3-II in the sample treated with N/L or Chlo, by the value in the control sample (untreated). LC3-II flux was expressed relative to their respective controls.
Immunocytochemistry
Paraformaldehyde-fixed hepatocytes were incubated with antibodies against LC3 overnight (Supplementary Table 2). Then, the cells were incubated with CY3 or FITC-conjugated secondary goat antibodies and the DNA-binding fluorochrome DAPI (D9542, SIGMA). Samples were examined under Leica TCS-SP (UV) confocal laser microscope (60X objective). Quantification of LC3 puncta staining was performed using ImageJ software as detailed elsewhere[21]. LC3-flux was calculated by subtracting the number of LC3 puncta per cell in the sample treated with Chlo by the value of LC3 puncta per cell in the control sample (untreated with Chlo).
BODIPY staining
Hepatocytes in culture were incubated with BODIPY 493/503 (Molecular Probes) at a concentration of 1 mg/mL during 30 minutes prior to fixation (4% paraformaldehyde). Quantification of of lipid bodies was performed using Frida Software as detailed elsewhere[22].
Histological Analysis
Formalin-fixed liver sections were stained with hematoxylineosin or Periodic acid-Schiff. For sudan red staining, frozen liver tissue sections were used[11].
Quantification of lipids
Lysosome-enriched fraction was isolated from liver pieces (500 mg)[23]. Lipids were extracted from tissue, cell[24] and lysosome[25] homogenates, and TG were quantified using a commercial kit (37481 and 37484 from Menarini Diagnostics). Liver free cholesterol and cholesteryl esters, were separated by thin layer chromatography and quantified by optical densitometry as detailed elsewhere[25].
Lysosomal enzymatic activity
1.25 million hepatocytes were collected in PBS and dispensed in filter paper (13.5 mm). A 3 mm diameter circle from dried hepatocytes spots was used to analyze the lysosomal enzymatic activity. This method was adapted from a previously described report [26].
PP2A C Subunit Methylation Assay
The steady-state level of PP2Ac methylation was determined using an antibody specific for unmethylated PP2Ac as described previously[27]. A detailed description of methodology is found in supplementary material.
Immunoprecipitation
500 μg of total protein extract were immunoprecipitated with 5μg of IgG2a (BD Pharmigen), anti-MTOR antibody (Cell Signaling) and protein A Sepharose beads (Sigma-Aldrich), and immunoprecipitated proteins were analyzed by Western blot and incubated with anti-PP2A A subunit (Upstate).
Results
Methionine serum levels in NAFLD
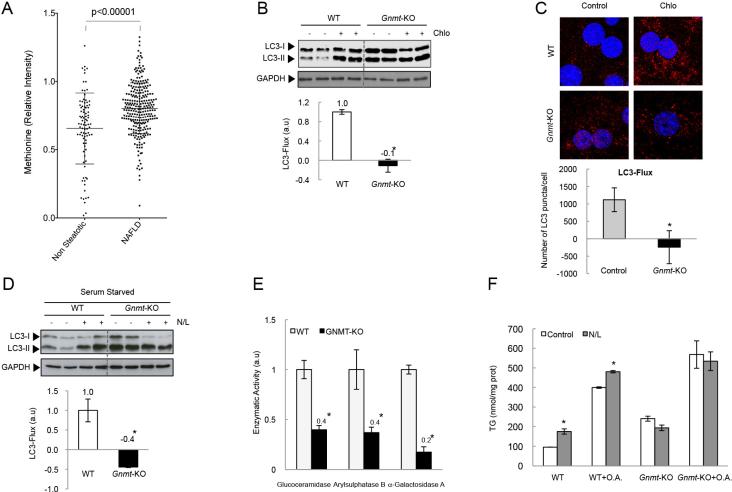
We examined whether methionine and SAMe were elevated in serum samples from NAFLD patients. We observed a significant increase of serum methionine levels in NAFLD patients compared to non-steatotic patients (Fig. 1A), suggesting a possible role for elevated methionine levels in the pathogenesis of NAFLD. Unfortunately, due to low SAMe stability, and the different origin and date of the set of serum samples, we were not able to obtain consistent data of SAMe levels.
Figure 1. Autophagy in Gnmt-KO hepatocytes.
(A) Methionine levels in serum samples from NAFLD and non-steatotic patients (mean +/− SD (*p<0.05) (Mann-Whitney t-test). (B,C) After 24h of culture, hepatocytes were incubated with Chloroquine (Chlo) during the last 4h. (D) Hepatocytes were serum-starved during 12h, and incubated with N/L during the last 4h. (E) Lysosomal enzymatic activity in hepatocytes. (F) TG in Gnmt-KO hepatocytes in absence or presence of O.A (1mM). Data are represented as mean +/− SEM (*p<0.05 N/L Vs. Control).
Autophagy flux in Gnmt-KO hepatocytes
To examine the role of a chronic excess of methionine and SAMe on autophagy flux, Gnmt-KO hepatocytes from 3-month old mice, characterized by abnormally elevated methionine and SAMe levels (Table 1), were used. We monitored autophagic flux by analyzing LC3-II turnover by Western blot or LC3 puncta immunolabelling, in the presence or absence of the inhibitor of lysosome-mediated proteolysis Chloroquine (Chlo), or in the presence or absence of the protease inhibitor Leupeptin and ammonium chloride (N/L)[20]. We observed reduced LC3-flux in basal (Fig. 1B and C) and serum-starved hepatocytes from Gnmt-KO mice (Fig. 1D), characterized by elevated LC3-II levels in untreated hepatocytes that do not increase after lysosomal blockage (Fig. 1B and D), suggesting that the impairment in autophagic flux was probably due to defects at the lysosomal compartment, as described previously [28]. To further confirm this, we analyzed the activity of three lysosomal hydrolases in Gnmt-KO hepatocytes. We observed a reduced activity of Glucoceramidase, Arylsulphatase B and α-Galactosidase A in Gnmt-KO hepatocytes (Fig. 1E) suggesting that Gnmt-KO hepatocytes could have an impairment in autophagy due to an inhibition of lysosomal functionality.
Table 1.
SAMe and methionine levels in WT and Gnmt-KO hepatocytes cultured 42h in medium control, supplemented with SAMe and methionine, or medium deficient in methionine (n=3)
| Mouse Genotype | Methionine (pmol/μg) | SAMe (pmol/μg) |
|---|---|---|
| WT | 0.08 ± 0.01 | 0.25 ± 0.02 |
| WT+SAMe/Met | 0.94±0.04* | 2.79 ± 0.03* |
| Gnmt-KO | 0.14 ± 0.01* | 4.67 ± 0.39* |
| Gnmt-KO + MDM | 0.08 ± 0.03** | 0.07 ± 0.03** |
p<0.05 Group Vs. WT
p<0.05 Group Vs. Gnmt-KO
Next, we examined the impact of lysosomal inhibitors (N/L) on TG content. In WT hepatocytes, inhibition of lysosomal degradation significantly increased hepatocyte TG content both in absence or presence of lipid supplementation with oleic acid (O.A.) (Fig. 1F). In contrast, no such effect was seen in Gnmt-KO hepatocytes, suggesting that the decrease in autophagic flux observed in Gnmt-KO hepatocytes could impair lipophagy.
Autophagy in Gnmt-KO mice
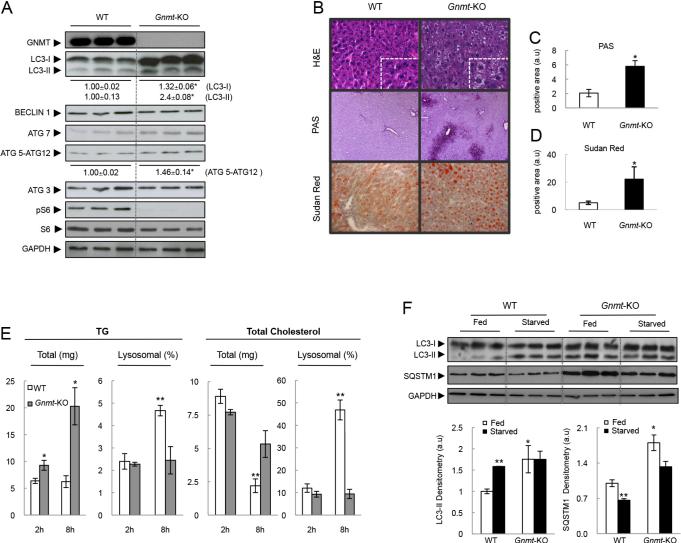
We examined autophagy functionality in Gnmt-KO mice in vivo. We found that LC3-II levels were basally elevated in livers from Gnmt-KO mice, similar to what we described in cultured Gnmt-KO hepatocytes above (Fig. 1B). Livers from Gnmt-KO mice were also characterized by elevated levels of ATG5 conjugated with ATG12 (ATG5-ATG12), and normal levels of ATG3, ATG7 and BECLIN1 proteins (Fig. 2A), suggesting an impairment in autophagic functionality.
Figure 2. Autophagy in Gnmt-KO mice.
(A) Western blot and densitometry analysis from 3-month-old Gnmt-KO livers. (B) Hematoxylin/Eosin, Sudan red, and PAS staining. (C, D) Quantification of positive areas of (C) PAS staining and (D) Sudan red staining of Gnmt-KO mice 24h after starvation. (E) TG and total cholesterol levels in total and lysosomal liver extracts from mice starved during 2 and 8 hours (t-test, *p<0.05 Gnmt-KO Vs. WT; **p<0.05 WT (2h) Vs. WT (8h)). (F) Western blot from WT and Gnmt-KO livers, 24h after starvation (t-test, *p<0.05 Gnmt-KO Vs. WT; **p<0.05 Starved Vs. Fed). Mean +/− SEM.
Next, we examined the response of Gnmt-KO mice to starvation, which is a well-characterized model of autophagy activation. Elevated levels of SAMe and methionine in Gnmt-KO mice were maintained after 8h of starvation (Table 2). Gnmt-KO livers showed higher glycogen accumulation than WT mice after 24h of starvation, as shown by PAS staining (Fig. 2B and C). This suggests an impairment in autophagy functionality since functional autophagy is required for glucose release from glycogen[29].
Table 2.
Hepatic SAMe and methionine levels from fed or starved mice (n=5)
| Mouse Genotype | Methionine (pmol/mg) | SAMe (pmol/mg) |
|---|---|---|
| Fed WT | 74.9 ± 9.5 | 70.4 ± 5.1 |
| Starved WT | 72.2 ± 6.7 | 52.1 ± 3.8* |
| Fed Gnmt-KO | 308.9 ± 30.2* | 1983.1 ± 90.9* |
| Starved Gnmt-KO | 177.0 ± 85.9** | 1412.6 ± 546.1* |
p<0.05 Group Vs. WT
p<0.05 Group Vs. Gnmt-KO
In hepatocytes, lipid droplets can associate with autophagic components during nutrient deprivation, and inhibition of autophagy in liver increases TG storage in lipid droplets[3]. Gnmt-KO livers showed higher lipid accumulation than WT mice after 24h of starvation, as shown by H&E and Sudan red staining (Fig. 2B and D), and by increased TG content in total liver 8h after starvation (Fig. 2E). Importantly, the increase in TG and total cholesterol levels seen in lysosomes during starvation in WT mice (Fig. 2E) was not observed in lysosomes from Gnmt-KO livers, suggesting an impaired delivery of TG and cholesterol to lysosomes.
Decreased SQSTM1/p62 levels are also associated with autophagy activation[20]. We found that the levels of SQSTM1 were strongly elevated in Gnmt-KO mice compared to WT mice both under fed and starved conditions. This correlated with a lack of response in LC3-II levels, i.e. the increase in LC3-II levels seen normally in starved WT mice compared to fed WT mice was not observed in the Gnmt-KO mice (Fig. 2F). These data suggest that autophagy is impaired in Gnmt-KO hepatocytes after starvation.
Altogether, our data show that Gnmt-KO mice have impairment in autophagic functionality both in vitro and in vivo, likely due to a defect at the lysosomal level.
Methionine and SAMe levels affect autophagy
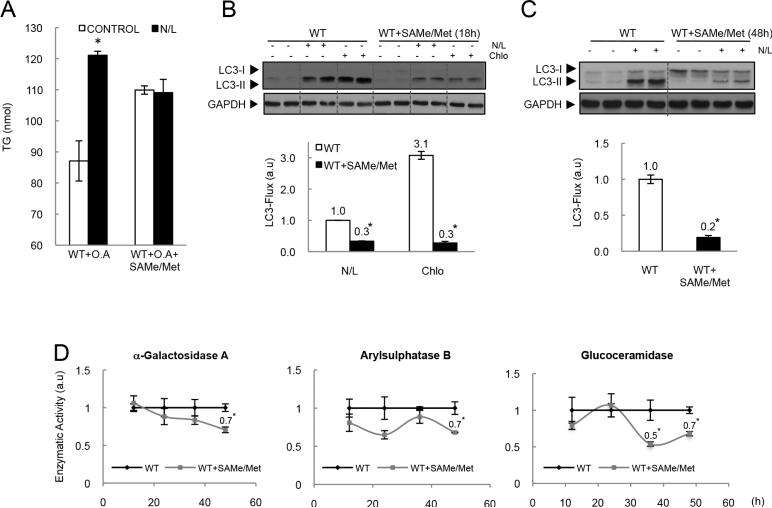
Autophagy can be compromised when the cells are exposed to a high lipid load due to a decrease in the convergence of autophagosomes and lysomes[30]. It is thus possible that the altered lipophagy found in Gnmt-KO mice could be due to the already established steatosis in these mice. Alternatively, it is also possible that the defect in lipophagy in these mice could be due to the high levels of methionine and SAMe. To further confirm this, we analyzed the effect of N/L on the TG content in WT hepatocytes treated with SAMe and methionine (4mM/1mM) and cultured in the presence of lipid supplementation with oleic acid (O.A.). In WT hepatocytes, inhibition of lysosomal degradation significantly increased hepatocyte TG content in presence of lipid supplementation with O.A. In contrast, no such effect was seen in SAMe and methionine treated hepatocytes (Fig. 3A), suggesting that SAMe and methionine decreased lipid degradation through autophagy, even under conditions of lipid overload. In addition, we found that treatment of WT hepatocytes with SAMe (4mM) and methionine (1mM) significantly prevented autophagy flux in the presence of lysosomal inhibitors after 18 or 48h (Fig. 3B and C) of treatment, whereas treatment with SAMe or methionine alone at the assayed doses, or lower doses of a combined treatment had no effect on LC3-flux (Supplementary Fig. 1A and B).
Figure 3. Methionine, SAMe and autophagy flux.
(A) Hepatocytes cultured 4h with oleic acid, where treated with SAMe and methionine during 12h, and incubated with N/L during the last 4h. (B) Hepatocytes were treated with SAMe and methionine during 18h (B) or 48h (C) and when indicated, were incubated in the presence or absence of N/L or Chlo for the last 4 hours of culture. (D) Hepatocytes were treated with SAMe and methionine during the indicated times and then lysosomal enzymatic activity was analyzed. (t-test, *p<0.05 Group Vs. WT).
Although autophagic flux was blocked both in SAMe- and methionine-treated cultures, and in Gnmt-KO hepatocytes, this seemed to be driven by different mechanisms in each case. In the case of Gnmt-KO hepatocytes, the impaired autophagic flux was likely due to an impaired lysosomal function, as shown by the elevated LC3-II levels in untreated hepatocytes that do not increase after lysosomal blockage (Fig. 1B). In the case of SAMe and methionine-treated cultures, on the other hand, this was likely to be due to an inhibition of upstream autophagosome formation, as shown by the reduced accumulation of LC3-II levels after lysosomal blockage[28] (Fig. 3B and C). These results could be due to the time of exposure to SAMe and methionine; Gnmt-KO hepatocytes are exposed for months to high doses of SAMe and methionine, whereas acute treatments with SAMe and methionine are typically performed for a maximum of 48 hours. In line with this, we observed a small reduction in the activity of three lysosomal hydrolases after 48h of treatment with SAMe and methionine (Fig. 3D), suggesting that longer periods of incubation with SAMe and methionine could be necessary to see impaired autophagy due to lysosomal dysfunction. Unfortunately, we were unable to mimic chronic conditions in vitro, since SAMe and methionine treatment for periods of time longer than 48h induced a decrease in hepatocyte viability (Supplementary Fig. 1C).
SAMe, methionine, MTOR and PP2A methylation
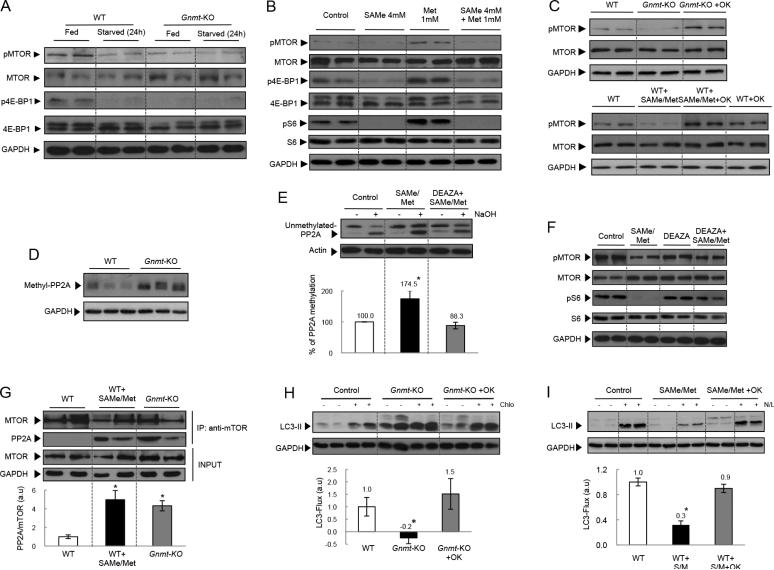
We next examined the mechanisms by which elevated levels of SAMe and methionine inhibited autophagy. Mammalian target of rapamycin (MTOR) is a serinethreonine kinase that can sense the nutrition status of cells and is one of the critical inactivators of the autophagosome formation [16]. Diverse hormones, growth factors, and amino acids stimulate the activation of MTOR complex (MTORC)[31, 32]. We found that the phosphorylation of MTOR and MTOR substrates 4EBP (Fig. 4A) and S6 (Fig. 2A) was significantly reduced in Gnmt-KO livers. This was an unexpected result since the block in autophagy in these mice should have coincided with an activation of MTOR, instead of the reduced phosphorylation of MTOR we actually observe. This suggests that SAMe and methionine inhibit autophagy by MTOR-independent pathways.
Figure 4. Methionine, SAMe, PP2A and MTOR.
(A) Gnmt-KO livers 24h after starvation. (B) SAMe and methionine treated hepatocytes during 16h. (C) Hepatocytes were pre-treated (2h) with okadaic acid (1 nM), and then cultured 18h. (D) Methylated PP2A in 3 months-old Gnmt-KO livers. (E) PP2A C subunit methylation assay. (F) Hepatocytes pre-treated with 3-Deaza (6h), and then cultured 24h. (G) MTOR-immunoprecipitated protein. (H) After 36h of culture hepatocytes were treated (2h) with okadaic acid (1 nM), and then incubated 4h with Chlo. (I) Hepatocytes were pre-treated 2h with okadaic acid and then cultured during 16h. Mean +/− SEM (t-test *p<0.05).
As mentioned above, SAMe levels are also markedly elevated in Gnmt-KO livers (Table 2). We thus examined if the high levels of SAMe could be responsible for blocking MTOR activation. First, we confirmed that the culture of hepatocytes with SAMe and methionine (4mM/1mM) increased their intracellular levels (Table 1). We found that methionine was not able to induce the phosphorylation of MTOR (Ser 2448), or 4E-BP1 (Ser 65) or S6 (Ser 235/236) in the presence of SAMe in WT hepatocytes (Fig. 4B). Similarly, insulin, another known MTOR activator, was not able to induce MTOR, and 4E-BP1 in Gnmt-KO hepatocytes or in WT hepatocytes cultured in the presence of SAMe and methionine (Supplementary Fig. 1D). The decrease in S6 phosphorylation in the presence of high SAMe and methionine levels was observed in treated cultures from 12 to 24h (Supplementary Fig. 1E). We also observed a decrease in MTOR (Ser 2448) and 4E-BP1 (Ser 65) phosphorylations in Gnmt-KO livers after 24 hours of starvation (Fig. 4A). Treatment of WT hepatocytes with low doses of SAMe and methionine was also less efficient at decreasing the levels of S6 (Ser 235/236) phosphorylation (Supplementary Fig. 1F).
PP2A is known to inhibit MTORC1[33]. Here, we found that the PP2A inhibitor, okadaic acid (OK), was able to restore MTOR phosphorylation in Gnmt-KO, and SAMe and methionine-treated hepatocytes (Fig. 4C). Recent studies in yeast have demonstrated that methionine inhibits autophagy by inducing SAMe-mediated methylation of PP2A[15]. We found that Gnmt-KO livers had increased methyl-PP2A levels (Fig. 4D), and SAMe and methionine-treated hepatocytes showed increased PP2A methylation (Fig. 4E). Treatment with Deaza, a strong inhibitor of methylation reactions[34, 35], prevented SAMe- and methionine-induced methylation of PP2A (Fig. 4E), and restored MTOR and S6 phosphorylation (Fig. 4F). The studies in yeast mentioned above, suggested that the methylation status of PP2A could influence TORC1 activity through modulating the phosphorylation state of Npr2p, which is a negative regulator of TORC1[15]. Here, instead, we observed a direct interaction of MTOR with PP2A in Gnmt-KO hepatocytes, and in SAMe- and methionine- treated hepatocytes that was completely absent in WT hepatocytes (Fig. 4G, Supplementary Fig. 1G). This suggests that the methylation status of PP2A could directly influence its interaction with TORC1 and therefore MTOR phosphorylation levels.
SAMe, methionine, PP2A and autophagy flux
We next examined if the impaired autophagy due to elevated SAMe and methionine levels in Gnmt-KO mice could be mediated by PP2A. We found that the PP2A inhibitor, okadaic acid, was able to restore LC3 flux in Gnmt-KO hepatocytes (Fig. 4H), and in SAMe and methionine treated hepatocytes (Fig. 4I). PP2A is composed of catalytic (PP2AC), scaffold (PP2AA) and regulatory (PP2AB) subunits. PP2AC exists in two isoforms, and [27]. We found that co-transfection of mouse liver progenitor cells (MLP-29) and Gnmt-KO hepatocytes with small interfering RNA (siRNA) constructs for PP2AC isoforms α and β, was able to restore LC3-flux in MLP-29 cells treated with SAMe and Met (Supplementary Fig. 2A and B), and in Gnmt-KO hepatocytes (Supplementary Fig. 2C and D). Taken together, our data suggest that SAMe and methionine inhibit autophagy in hepatocytes via PP2A methylation.
Methionine deficient diet in Gnmt-KO mice
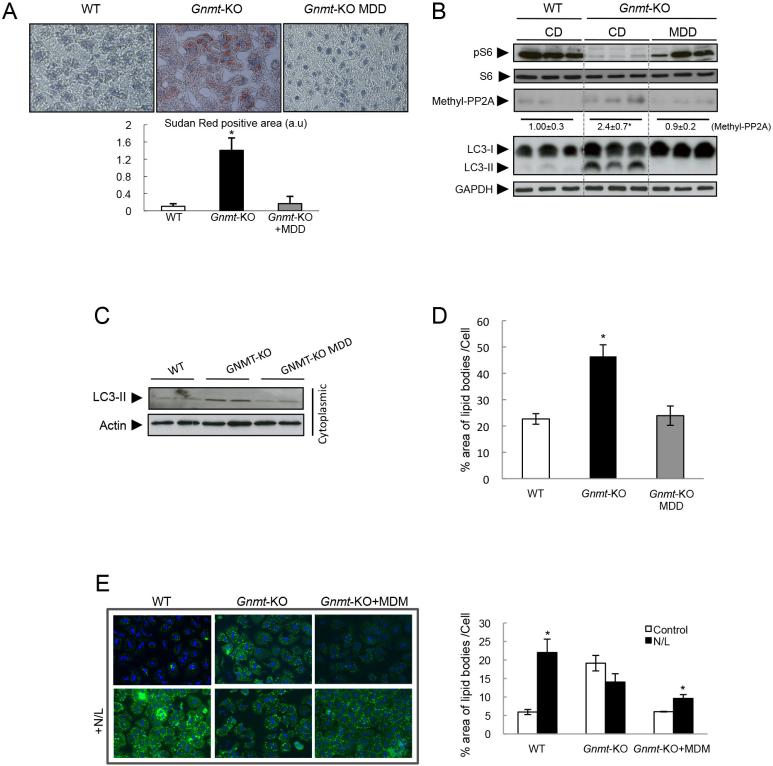
We have previously shown that feeding Gnmt-KO mice during 21 days with a methionine deficient diet (MDD) restored normal SAMe levels[17], recovered VLDL metabolism[36], decreased liver TG content, and ameliorated liver steatosis[17].
Here, we found that MDD in Gnmt-KO mice normalized methionine and SAMe hepatic content to WT levels, and decreased liver damage (see aminotransferases levels) (Table 3). MDD also restored lipid content (Fig. 5A) and decreased LC3-II levels (Fig. 5B) in liver. MDD ameliorated autophagy, as seen by cytoplasmic LC3-II levels in hepatocytes[20] isolated from livers from Gnmt-KO fed with a MDD (Fig. 5C). Importantly, we found that the elevated lipid levels in hepatocytes from Gnmt-KO mice were normalized by MDD (Fig. 5D). Notably, culture of Gnmt-KO hepatocytes in medium deficient in methionine (MDM) also normalized the intracellular levels of SAMe and methionine (Table 1), and the ability to catabolise lipids through autophagy, as seen by the effect of the inhibitor N/L on lipid accumulation (Fig. 5E).
Table 3.
Hepatic SAMe, methionine, and SAH levels and serum aminotransferases from WT and Gnmt-KO mice, fed with control or Methionine Deficient Diet. (n=5)
| Mouse Genotype | Methionine (pmol/mg) | SAMe (pmol/mg) | SAH (pmol/mg) | SAMe/SAH | AST (U/L) | ALT (U/L) |
|---|---|---|---|---|---|---|
| WT | 73.2 ± 10.2 | 75.2 ± 2.5 | 8.6 ± 3.1 | 8.7 ± 0.8 | 55 ± 4 | 26 ± 3 |
| Gnmt-KO | 335.3 ± 1.6* | 1909.5± 92.5* | 73.4 ± 11.6* | 26.0 ± 8.0* | 240 ± 9* | 169 ± 10* |
| Gnmt-KO + MDD | 57.5 ± 10.0 | 76.2± 13.9 | 12.1 ± 9.9 | 6.3± 1.4 | 52 ± 3 | 27 ± 2 |
p<0.05 Group Vs. WT.
Figure 5. Methionine deficient diet in Gnmt-KO mice.
(A) Sudan red staining and quantification in liver from WT and Gnmt-KO mice fed with normal or methionine deficient diet (MDD). (B,C) Western blot from total protein from livers (B) and cytoplasmic proteins from hepatocytes (C) of WT and Gnmt-KO mice fed with MDD. (D) Percentage area of lipid bodies per hepatocyte in culture after O.A. treatment (1mM) for 8h. (E) BODIPY staining and percentage area of lipid bodies per cell in WT, Gnmt-KO hepatocytes, and Gnmt-KO hepatocytes cultured for 24h in methionine deficient medium (MDM). (Mean +/− SEM, t-test *p<0.05).
An increase in the ratio of SAMe to SAH predicts high methylation capacity[37] Here, we found that MDD restored the SAMe/SAH ratio, decreasing the methylation capacity of Gnmt-KO livers (Table 3). Importantly, we observed that PP2A methylation and S6 phosphorylation were also restored in livers from Gnmt-KO mice after MDD (Fig. 5B).
As mentioned before, MDD led to a normalization of the supra-physiological levels of hepatic SAMe and methionine in Gnmt-KO livers to near WT levels. In WT mice, MDD also significantly reduced hepatic SAMe and methionine, and in this case, led to a decrease to below-physiological levels (Supplementary Fig. 3A). This decrease in methionine and SAMe levels did not affect the lipid content (Supplementary Fig. 3A and B) and induced an increased LC3 flux (Supplementary Fig. 3C).
Discussion
Recent studies have shown that autophagy can hydrolyze TG in a process known as lipophagy[3], and that defects in autophagy might be involved in the development of fatty liver[3, 6]. Here, in our study, we have shown that one possible cause of deficient lipophagy during liver steatosis development could be elevated levels of methionine and SAMe arising from low GNMT expression, which we have modeled in vivo using the Gnmt-KO mice. This study is relevant to human pathology as GNMT expression is repressed in at least a subset of advanced NAFLD patients[12], and that we have observed a significant increase of serum methionine levels in NAFLD patients compared to non-steatotic patients.
Several data suggest that lipid degradation by autophagy is impaired in conditions of GNMT deficiency due to a failure in the lysosomal compartment, as including (1) elevated basal LC3-II levels in untreated Gnmt-KO hepatocytes that do not increase after lysosomal blockage, (2) impaired delivery of TG and cholesterol to lysosomes, and (3) the decrease in lysosomal enzymatic activity in Gnmt-KO hepatocytes. It has been suggested that the chronic steatotic condition in diseased liver could be responsible for lipophagy impairment[3, 30] by changing the membrane lipid composition and by reducing autophagosome/lysosome fusion[30]. This suggests that the impaired lipophagy observed in Gnmt-KO mice could be due to the steatotic condition. However, in our study, we have observed that the blockage of autophagy in steatotic hepatocytes from Gnmt-KO mice could be reversed in vitro, either by a normalization of the supra-physiological levels of hepatic SAMe and methionine during 24h, or by inhibition of PP2A activity. This suggests that the impaired autophagy in the Gnmt-KO mice is mediated by PP2A activity on one hand, and is more than a consequence of the steatotic process itself on the other hand.
In the case of SAMe and methionine-treated hepatocytes, the observation of reduced accumulation of LC3-II levels after lysosomal blockage, suggested that SAMe and methionine-induced inhibition of autophagy is likely due to an impaired upstream autophagosome formation. The observation of a small reduction in the activity of three lysosomal hydrolases after 48h of treatment with SAMe and methionine lead us to propose that the chronic exposure to high SAMe and methionine in vivo would progressively alter lysosomal enzymatic activity leading to the lysosomal dysfunctions observed in Gnmt-KO hepatocytes. Importantly, as shown for Gnmt-KO hepatocytes, blockage of PP2A activity similarly restored autophagy flux in SAMe- and methionine-treated cells, suggesting that impaired autophagy due to elevated SAMe and methionine levels in Gnmt-KO mice could be mediated by PP2A.
MTOR regulates cell growth and coordinates the response to amino acid availability[16]. We found that the excess of SAMe in Gnmt-KO hepatocytes was responsible for preventing MTOR activation, by inducing methylation of the PP2A catalytic subunit, which increased the interaction of PP2A with MTOR, hence blocking its activation. Importantly, we have observed that the normalization of methionine and SAMe levels in Gnmt-KO mice using the methionine deficient diet decreased the methylation capacity of Gnmt-KO livers, reduced PP2A methylation, and restored MTOR phosphorylation and autophagy.
Previous studies have suggested that MTOR reactivation is an autophagy-dependent process, and requires the degradation of autolysosomal products[38]. For example, fibroblasts lacking lysosomal α-galactosidase activity have impaired MTOR reactivation and defective lysosome reformation[38]. Thus, it is a distinct possibility that the decrease in lysosomal hydrolases activity observed in Gnmt-KO and in SAMe- and methionine-treated hepatocytes could contribute, together with the SAMe and methionine-mediated PP2A activation, to the observed impairment in MTOR reactivation.
On the other hand, it is well established that MTOR inhibition leads to an activation of autophagy[16], whereas, here, we observed that high levels of SAMe and methionine inhibited both MTOR and autophagy. These data suggest that SAMe and methionine-induced inhibition of autophagy could be mediated by MTOR-independent mechanisms. Several MTOR-independent pathways that regulate autophagy have been described in different systems, including increased intracellular levels of cAMP, lithium or ceramide[39, 40]. It would be important in future studies to identify the MTOR-independent mechanisms, by which SAMe and methionine block autophagy in hepatocytes.
In summary, we found that impaired lipophagy during steatosis could be due to elevated SAMe and methionine levels rather than being a consequence of the steatotic process itself and that normalization of their levels could be sufficient to restore autophagic functionality and reduce liver steatosis. Thus, our work provides evidence of a novel model of lipid metabolism regulation that could contribute to liver steatosis development. Based on this study and in previous work, we thus propose that excess methionine and its metabolite SAMe could induce liver steatosis by regulating lipid metabolism at several different levels, including increasing PP2A methylation that leads to a decrease in autophagy functionality and AMPK phosphorylation[41], and inducing PE methylation, increasing PEMT flux that leads to increased TG synthesis[17].
Based on our present data from the Gnmt-KO mice and on the observation that NAFLD patients have a significant increase of serum methionine levels, together with recent data showing that the expression of three hypermethioninemia-causative genes (Gnmt, Mat1a and Ahcy) was often repressed in advanced NAFLD patients[12], we suggest that in the cases of Gnmt deficiency, hypermethioninemia and elevated SAMe levels could play a role in the pathogenesis of NAFLD through the inhibition of autophagy-mediated degradation of lipids. It has been suggested that pharmacological agents that stimulate autophagy could be considered as a novel aproach for alleviating liver condition in NAFLD [5]. Here we suggest that NAFLD patients with elevated methionine and SAMe levels, arising from low GNMT expression could benefit from methionine restricted diets.
Supplementary Material
Acknowledgements
We thank the Metabolomics platform at CIC bioGUNE and OWL metabolomics for methionine and SAMe measurements.
Finantial Support: This work is supported by NIH grants RO1AT-1576 (to SCL, JMM and MLM-C), ETORTEK-2011, Sanidad Gobierno Vasco 2013, Educación Gobierno Vasco 2011 (PI2011/29) and FIS (PI11/01588) (to MLM-C), SAF 2011-29851 (to JMM), Sanidad Gobierno Vasco 2012 (to MVR), Basque Goverment IT-336-10, Unidad de formación e investigación UFI11/20, and University of Basque Country (to PA), FIS PS12/00005, Fundación Científica de la Asociación Española Contra el Cáncer (Cancer Infantil), Educación Gobierno Vasco (PI2013-46) (to AW), and the Program Ramón y Cajal, Ministry of Science and Innovation, Spain (to AW and NB), Asociación Española contra el Cáncer (PF-T and MLM-C), FIS PS12/00402 (to NB and MVR), FIS PI10/00067 and FIS PI 13/01299 (to CG-M). Ciberehd is funded by the Instituto de Salud Carlos III.
List of Abbreviations
- GNMT
Glycine N-methyltransferase
- SAMe
S-Adenosylmethionine
- MTOR
The mammalian target of rapamycin
- PP2A
protein phosphatase 2A
- LD
lipid droplets
- MAT
methionine adenosyltransferase
- SAH
S-adenosylhomocysteine
- DG
diglyceride
- TG
Triglycerides
- PEMT
phosphatidylethanolamine N-methyltransferase
- AHCY
S-adenosylhomocysteine hydrolase
- NAFLD
Non-alcoholic Fatty liver disease
- Deaza
Deazaadenosine
- LC3
microtubule-associated light chain 3
- AST
aspartate transaminase
- ALT
alanine transaminase
- H&E
Hematoxylin and eosin
- SQSTM1/p62
Sequestosome 1
- Chlo
Chloroquine
- N/L
Ammonium chloride and leupeptin
- LAMP2
lysosome-associated membrane protein type 2
- O.A
oleic acid
- 4EBP
Eukaryotic translation initiation factor 4E-binding protein 1
- PE
Phosphatidylethanolamine
- PC
to Phosphatidylcholine
- MDD
methionine deficient diet
- MDM
Medium deficient in methionine
- SAH
S-Adenosyl-L-homocysteine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: None of the authors have conflict of interest to declare.
Author Contributions
M.V-R., I.Z-F., J.L.G-R., M.M-U., N.M-L., S.L-M., F.A., M.L.F., L.A-E., N.B., A.W., P.F.-T., L.B.-T., D.F-R., J.M.F-P., F.L-O., and V.G.-J. performed the experiments, Z.L., C.W., gave technical support, A.W., C.G.M., S.C.L., P.A., and J.M.M. gave conceptual advice. M.V.R. and M.L.M.C. designed and supervised the study, wrote the manuscript and prepared figures.
Author names in bold designate shared co-first authorship
References
- 1.Varela-Rey M, Embade N, Ariz U, Lu SC, Mato JM, Martinez-Chantar ML. Non-alcoholic steatohepatitis and animal models: understanding the human disease. The international journal of biochemistry & cell biology. 2009;41:969–976. doi: 10.1016/j.biocel.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 2.Kaushik S, Singh R, Cuervo AM. Autophagic pathways and metabolic stress. Diabetes, obesity & metabolism. 2010;12(Suppl 2):4–14. doi: 10.1111/j.1463-1326.2010.01263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell metabolism. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin CW, Zhang H, Li M, Xiong X, Chen X, Chen X, et al. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. Journal of hepatology. 2013;58:993–999. doi: 10.1016/j.jhep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuervo AM. Preventing lysosomal fat indigestion. Nature cell biology. 2013;15:565–567. doi: 10.1038/ncb2778. [DOI] [PubMed] [Google Scholar]
- 7.Finkelstein JD. Methionine metabolism in mammals. The Journal of nutritional biochemistry. 1990;1:228–237. doi: 10.1016/0955-2863(90)90070-2. [DOI] [PubMed] [Google Scholar]
- 8.Luka Z. Methyltetrahydrofolate in folate-binding protein glycine N-methyltransferase. Vitamins and hormones. 2008;79:325–345. doi: 10.1016/S0083-6729(08)00411-1. [DOI] [PubMed] [Google Scholar]
- 9.Lu SC, Mato JM. S-adenosylmethionine in liver health, injury, and cancer. Physiological reviews. 2012;92:1515–1542. doi: 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mudd SH. Hypermethioninemias of genetic and non-genetic origin: A review. American journal of medical genetics Part C, Seminars in medical genetics. 2011;157:3–32. doi: 10.1002/ajmg.c.30293. [DOI] [PubMed] [Google Scholar]
- 11.Martinez-Chantar ML, Vazquez-Chantada M, Ariz U, Martinez N, Varela M, Luka Z, et al. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology. 2008;47:1191–1199. doi: 10.1002/hep.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy SK, Yang H, Moylan CA, Pang H, Dellinger A, Abdelmalek MF, et al. Relationship Between Methylome and Transcriptome in Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barr J, Caballeria J, Martinez-Arranz I, Dominguez-Diez A, Alonso C, Muntane J, et al. Obesity-dependent metabolic signatures associated with nonalcoholic fatty liver disease progression. Journal of proteome research. 2012;11:2521–2532. doi: 10.1021/pr201223p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malloy VL, Perrone CE, Mattocks DA, Ables GP, Caliendo NS, Orentreich DS, et al. Methionine restriction prevents the progression of hepatic steatosis in leptin-deficient obese mice. Metabolism: clinical and experimental. 2013 doi: 10.1016/j.metabol.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 15.Sutter BM, Wu X, Laxman S, Tu BP. Methionine Inhibits Autophagy and Promotes Growth by Inducing the SAM-Responsive Methylation of PP2A. Cell. 2013;154:403–415. doi: 10.1016/j.cell.2013.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Una M, Varela-Rey M, Cano A, Fernandez-Ares L, Beraza N, Aurrekoetxea I, et al. Excess S-adenosylmethionine reroutes phosphatidylethanolamine towards phosphatidylcholine and triglyceride synthesis. Hepatology. 2013 doi: 10.1002/hep.26399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leffert HL, Koch KS, Moran T, Williams M. Liver cells. Methods in enzymology. 1979;58:536–544. doi: 10.1016/s0076-6879(79)58168-3. [DOI] [PubMed] [Google Scholar]
- 19.Mederacke I, Dapito DH, Affo S, Uchinami H, Schwabe RF. High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nature protocols. 2015;10:305–315. doi: 10.1038/nprot.2015.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez-Lopez N, Athonvarangkul D, Mishall P, Sahu S, Singh R. Autophagy proteins regulate ERK phosphorylation. Nature communications. 2013;4:2799. doi: 10.1038/ncomms3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gold ES, Ramsey SA, Sartain MJ, Selinummi J, Podolsky I, Rodriguez DJ, et al. ATF3 protects against atherosclerosis by suppressing 25-hydroxycholesterol-induced lipid body formation. The Journal of experimental medicine. 2012;209:807–817. doi: 10.1084/jem.20111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez MJ, Ruiz JI, Lacort M, Ochoa B. Diurnal variations of rat liver enzymes catalyzing cholesterol ester hydrolysis. Biochimica et biophysica acta. 1991;1085:106–111. doi: 10.1016/0005-2760(91)90237-c. [DOI] [PubMed] [Google Scholar]
- 24.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Canadian journal of biochemistry and physiology. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 25.Ruiz JI, Ochoa B. Quantification in the subnanomolar range of phospholipids and neutral lipids by monodimensional thin-layer chromatography and image analysis. Journal of lipid research. 1997;38:1482–1489. [PubMed] [Google Scholar]
- 26.Gasparotto N, Tomanin R, Frigo AC, Niizawa G, Pasquini E, Blanco M, et al. Rapid diagnostic testing procedures for lysosomal storage disorders: alpha-glucosidase and beta-galactosidase assays on dried blood spots. Clinica chimica acta; international journal of clinical chemistry. 2009;402:38–41. doi: 10.1016/j.cca.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Jackson JB, Pallas DC. Circumventing cellular control of PP2A by methylation promotes transformation in an Akt-dependent manner. Neoplasia. 2012;14:585–599. doi: 10.1593/neo.12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korolchuk V, Kaushik S, et al. In search of an “autophagomometer”. Autophagy. 2009;5:585–589. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 29.Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy in glucose homeostasis. Pathology, research and practice. 2006;202:631–638. doi: 10.1016/j.prp.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weichhart T. Mammalian target of rapamycin: a signaling kinase for every aspect of cellular life. Methods Mol Biol. 2012;821:1–14. doi: 10.1007/978-1-61779-430-8_1. [DOI] [PubMed] [Google Scholar]
- 32.Sancak Y, Sabatini DM. Rag proteins regulate amino-acid-induced mTORC1 signalling. Biochemical Society transactions. 2009;37:289–290. doi: 10.1042/BST0370289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Findlay GM, Yan L, Procter J, Mieulet V, Lamb RF. A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. The Biochemical journal. 2007;403:13–20. doi: 10.1042/BJ20061881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mato JM, Alvarez L, Ortiz P, Pajares MA. S-adenosylmethionine synthesis: molecular mechanisms and clinical implications. Pharmacology & therapeutics. 1997;73:265–280. doi: 10.1016/s0163-7258(96)00197-0. [DOI] [PubMed] [Google Scholar]
- 35.Chiang PK. Biological effects of inhibitors of S-adenosylhomocysteine hydrolase. Pharmacology & therapeutics. 1998;77:115–134. doi: 10.1016/s0163-7258(97)00089-2. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Una M, Varela-Rey M, Mestre D, Fernandez-Ares L, Fresnedo O, Fernandez-Ramos D, et al. S-Adenosylmethionine increases circulating very-low density lipoprotein clearance in non-alcoholic fatty liver disease. Journal of hepatology. 2014 doi: 10.1016/j.jhep.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mato JM, Martinez-Chantar ML, Lu SC. S-adenosylmethionine metabolism and liver disease. Annals of hepatology. 2013;12:183–189. [PMC free article] [PubMed] [Google Scholar]
- 38.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomez-Sanchez JA, Carty L, Iruarrizaga-Lejarreta M, Palomo-Irigoyen M, Varela-Rey M, Griffith M, et al. Schwann cell autophagy, myelinophagy, initiates myelin clearance from injured nerves. The Journal of cell biology. 2015;210:153–168. doi: 10.1083/jcb.201503019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiological reviews. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 41.Martinez-Chantar ML, Vazquez-Chantada M, Garnacho M, Latasa MU, Varela-Rey M, Dotor J, et al. S-adenosylmethionine regulates cytoplasmic HuR via AMP-activated kinase. Gastroenterology. 2006;131:223–232. doi: 10.1053/j.gastro.2006.04.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.