Abstract
Depression in adults is heritable with about 40 % of the phenotypic variance due to additive genetic effects and the remaining phenotypic variance due to unique (unshared) environmental effects. Common environmental effects shared by family members are rarely found in adults. One possible explanation for this finding is that there is an interaction between genes and the environment which may mask effects of the common environment. To test this hypothesis, we investigated genotype by environment interaction in a large sample of female and male adult twins aged 18–70 years. The anxious depression subscale of the Adult Self Report from the Achenbach System of Empirically Based Assessment (Achenbach and Rescorla in Manual for the ASEBA adult: forms and profiles, 2003) was completed by 13,022 twins who participate in longitudinal studies of the Netherlands Twin Register. In a single group analysis, we found genotype by unique environment interaction, but no genotype by common environment interaction. However, when conditioning on gender, we observed genotype by common environment interaction in men, with larger common environmental variance in men who are genetically less at risk to develop depression. Although the effect size of the interaction is characterized by large uncertainty, the results show that there is at least some variance due to the common environment in adult depression in men.
Keywords: Adult depression, Common environment, Genotype-by-environment interaction, Heritability, Heterogeneity
The heritability of depression in adults is estimated at around 40 % (Sullivan et al. 2000; Nivard et al. 2015). Interestingly, in adults the remaining phenotypic variance is consistently found to be solely due to the unique environment. In adolescents, however, at age 12 years variation in anxious depression is explained also by shared environmental factors, while at ages 14 and 16 these shared environmental effects were absent (Lamb et al. 2010). The absence of evidence for common environmental influences on depression after age 12 is remarkable, as it has been argued that, theoretically, at least some phenotypic variance in depression is expected to be due to the familial effects in childhood that persist into adulthood (Gatz et al. 1992). For instance, cognitive styles related to depression may be learned in the family (Monroe and Simons 1991; Mezulis et al. 2006; Ingram 2003), and familial traumatic events in childhood, such as divorce, affect children similarly (Bowlby 1977; Kessler et al. 1997; Silberg et al. 2001). Therefore, it has been argued that the recurring finding of no common environmental effects on adult depression may be spurious.
Duncan et al. (2014) hypothesized that the true effects of the common environment underlying depression are masked by non-linear effects. Specifically, the effects of the common environment (C) may depend on the genotype of the subject (A) that is an A×C interaction. If not explicitly modeled, such an interaction effect is included in the estimate of the genetic variance (Molenaar et al. 1990). The question arises whether common environmental effects on adult depression can be revealed by taking into account such non-linear effects.
Another question is why some individuals develop a depression after an adverse environmental event and others do not. Both linear and non-linear effects could explain this phenomenon. Given the ongoing debate of the usefulness of genetic variant by environment interaction studies, either in a candidate gene study or in a genome-wide association study, it is important to know whether non-linear effects are present for unique environmental effects (see e.g., Dick et al. 2015).
Therefore we investigated whether common and unique environmental variance influencing the vulnerability for adult depression can be detected by taking into account the non-linear effects of genotype by environment interaction. We also tested whether interaction effects differ between males and females. Gender differences in the prevalence of depression arise in adolescence and remain until older age (Kessler et al. 1993). The exact mechanism underlying the higher prevalence of depression in adult females is generally unknown (Piccinelli and Wilkinson 2000), although studies have indicated that environmental factors associated with depression are different for males than for females (Kendler et al. 2011; Klose and Jacobi 2004).
We analyzed data from a large sample of twins between the ages of 18 and 70 years from the Adult Netherlands Twin Register (Nivard et al. 2015). The twins completed the anxious depression subscale of the Adult Self-Report from the Achenbach System of Empirically Based Assessment (Achenbach and Rescorla 2003). We tested for unmeasured genotype by unmeasured environment interaction effects in these data using the heteroscedastic ACE model (Jinks and Fulker 1970; Molenaar et al. 2012). Using the item level data methodology (see Molenaar and Dolan 2014; Schwabe and van den Berg 2014), we modeled a latent depression factor as a function of additive genetic (A), common environment (C), unique environment (E), and non-linear effects (A×E and A×C). In addition, we extended the approach to enable tests on gender differences in these interaction effects. In the resulting model adopted here both the genetic and the environment effects are treated as latent factors. By studying the interaction at the level of the latent genetic and environmental variance, we did not require measured candidate genes and measured candidate environments. In addition, the modeling approach is insensitive to the scale properties of the data, which may otherwise result in spurious non-additive effects (Eaves et al. 1977; Molenaar and Dolan 2014; Schwabe and Van den Berg 2014).
Method
Participants and measures
The Netherlands Twin Register (NTR; see Nivard et al. 2015) includes the Young NTR (YNTR; van Beijsterveldt et al. 2013) and the Adult NTR (ANTR; Willemsen et al. 2013). In the YNTR, twins have been registered at birth by their parents since 1987 (Bartels et al. 2007). When twins reach the age of 18, they are enrolled in the ANTR. The ANTR originally included adolescent and adult twins who were recruited through city councils or who volunteered through the NTR website. Here we analyze the data from all twins aged 18 years and older. The dataset comprises 6511 twin pairs (no missing: 5923) between the age of 18 and 70 with information on depression and zygosity. These pairs consist of 3146 (no missing: 2895) are MZ twins and 3365 (no missing: 3028) are DZ twins.
The twin pairs completed the anxious depression subscale of the Adult Self Report (ASR), which is part of the Achenbach System of Empirically Based Assessment (Achenbach and Rescorla 2003). The twins were asked to indicate to what degree various statements concerning anxious depressive behavior and attitudes apply to them on a 3-point scale (‘not true’, somewhat or sometimes true’, ‘very or often true’). The ASR anxious depression subscale was included in 8 of the 11 surveys that have been collected for the NTR since 1991 (respectively in 1991, 1995, 1997, 2000, 2002, 2009, 2011, and 2013). The different surveys contained slightly different versions of the ASR, as over the years, the ASR has changed. However, for anxious depression there was a common set of 13 items included in all surveys that was analyzed in this project (see Appendix 1). Cronbach’s alpha for these items on the various measurement occasions and twin samples ranged between 0.83 and 0.90 which is an indication for good reliability. In addition, the correlations between the sum scores based on these 13 items and the sum scores based on all items at each measurement occasion are between 0.95 and 0.98. The validity of the ASR has been established by Achenbach and Rescorla (2003).
Not all twins participated at each measurement occasion (see Table 1). For instance, 2131 twin-1 members have data available at only one occasion and 1273 twin-2 members have data available at three occasions. We selected the data vector from the first occasion that has the least missing values for each twin. This data vector contains missing values due to twins not completing all items of the questionnaire. Here, we assumed that these missing values are missing at random so that we can take all missing values in the data into account in the model fitting approach described below.
Table 1.
The number of twin pairs that have data available on none, 1, 2, …, or all measurement occasions
| None | 1 | 2 | 3 | 4 | 5 | 6 | 7 | All | |
|---|---|---|---|---|---|---|---|---|---|
| Twin 1 | 382 | 2131 | 1675 | 1334 | 491 | 444 | 242 | 158 | 41 |
| Twin 2 | 374 | 2184 | 1642 | 1273 | 526 | 451 | 262 | 144 | 42 |
‘None’ means that only the co-twin has data available on 1 or more measurement occasions
Analysis
Our main objective is to test for genotype by environment interaction for males and females in the complete sample of twin pairs. However, we first needed to establish that the data are homogeneous with respect to measurement occasion and age. Below we discuss a standard measurement model and the biometric model. Next, within these models, we discuss how we assessed homogeneity of the data with respect to measurement occasion and age. Then, we introduce the gender dependent genotype by environment interactions in the biometric model.
Standard measurement and biometric model
As advocated by Van den Berg et al. (2007), we analyzed the data at item level by separating between a measurement model for the items and a biometric model for the genetic and environmental variance. In the present study, the so-called graded response measurement model for ordinal item responses was used (Samejima 1969). Using this model, we separated the measurement properties of the item scores, Xi, from the underlying latent phenotypic factor (anxious-depression), denoted θ. Here, p = 1, …, N is used to index the twin pairs, j = 1, 2 is used to index the twin members, and i = 1, …, n is used to index the items.
In the graded response measurement model, the observed item scores, Xi, are regressed on the latent phenotypic factor, θ (using a multinomial probit regression function). The intercept and slope parameters in this regression are respectively referred to by threshold and discrimination parameters. These parameters are purported to capture the measurement characteristics of the item scores. In the present case, where we have a three point scale, we have 2 threshold parameters, τi1, and τi2. These parameters model the relative attractiveness of the answer options, that is, the degree to which the subjects use the different answer options. In an extreme depression item, for instance “I often think of suicide”, τi1 will likely be large, reflecting that the first answer option is attractive (most subjects score in the first answer category indicating that they do not think of suicide) and the second option is not. As the item scores are assumed to be ordered, the thresholds are also ordered, that is, τi2 should always be larger than τi1.
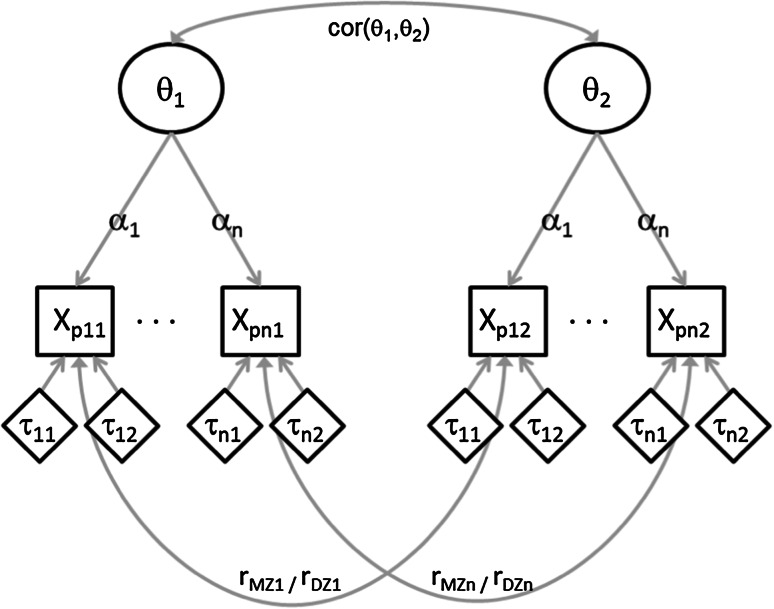
The slope, or discrimination parameter αi, in the regression of the item scores on the latent phenotypic variable, models the degree to which the item scores can distinguish between subjects with different levels of θ. The higher the value of αi, the better indicator the item is for θ. Besides the threshold and discrimination parameters, measurement models for twin data often also include the residual correlations between the item scores of the twin 1 and twin 2 members in the MZ sample (rMZ,i) and the DZ sample(rDZ,i). Such a correlation may indicate shared item specific genetic and/or environmental variance or it may indicate measurement problems resulting from filling in the questionnaire by two twins together. See Fig. 1 for a graphical representation of the measurement model including the parameters, and see Appendix 2 for a more technical discussion of the measurement model.
Fig. 1.
Graphical representation of the measurement model including the parameters
As the measurement model above captures the measurement properties of the item data in the αi, τi1, τi2, rMZi, and rDZi parameters, the latent phenotypic factor, θ, is unaffected by the scale properties in principle.1 In the standard phenotypic model, the phenotypic variable, θ, is decomposed into an additive genetic (A), common environmental (C), and unique environmental (E) variance component and an intercept, ν, that is
where COR(A1, A2) = 1 for MZ twins and COR(A1, A2) = 0.5 for DZ twins. In addition, COR(C1, C2) = 1 and COR(E1, E2) = 0, and E(A) = E(C) = E(E) = 0 for all twins. Under the assumption that the genetic and environmental variance components are mutually uncorrelated, the variance of θ can be decomposed as follows:
where the standardized estimate of VAR(A) is referred to as the heritability, h2.
Homogeneity with respect to measurement occasion
To investigate whether the eight measurement occasions differed in their biometric properties, we relied on estimates of the variance components (A, C, and E) within each occasion. We thus assumed that there are no important differences between the measurement models at each occasion. We fitted the measurement model measurement model and the biometric model simultaneously to the data of each occasion. Note that this was thus a standard ACE model at the level of the latent depression factor (biometric model). On basis of the 99 % highest posterior density regions (HPD) of these variance components, we judged whether the variance components differed importantly over measurement occasion. If not, we concluded that the data collected at the separate occasions were homogenous and could be aggregated.
Full homogeneity with respect to age
As the age range of our sample is wide (18–70), we also established homogeneity of the data with respect to age. As age is an important moderator in the literature, we wanted a more explicit test on homogeneity than the one discussed above. Here, we followed Nivard et al. (2015), and created age subsamples. We used the following groups: 18–19, 20–21, 22–24, 25–34, and 35–70 years. A major consideration in creating these subsamples was that the sample sizes within each age group need to be large enough to have sufficient power to detect heterogeneity. The resulting number of twin pairs within each age category that were in the analysis (i.e., twin pairs with a full or incomplete data record) is given in Table 2. In the case that the twin members have data in different age categories (due to a twin completing the questionnaire at a different age than his/her co-twin) we omitted their data from the present analysis as it requires independent groups. However, we included their data in the aggregated data analysis.
Table 2.
The total number of twin pairs within each age group that has been selected for the homogeneity analysis
| Age | MZ | DZ | Total | ||||
|---|---|---|---|---|---|---|---|
| Males | Females | Males | Females | Opposite-sex | MZ | DZ | |
| 18–19 | 298 (262) | 596 (552) | 230 (204) | 491 (431) | 355 (325) | 894 (814) | 1076 (960) |
| 20–21 | 219 (210) | 392 (369) | 160 (146) | 371 (336) | 238 (225) | 611 (579) | 769 (707) |
| 22–24 | 92 (86) | 173 (161) | 49 (44) | 123 (105) | 104 (97) | 265 (247) | 276 (246) |
| 25–34 | 77 (70) | 248 (229) | 46 (38) | 108 (94) | 114 (103) | 325 (299) | 268 (235) |
| 35–70 | 178 (163) | 512 (478) | 74 (66) | 174 (164) | 195 (188) | 690 (641) | 443 (418) |
| Agg | 981 (891) | 2165 (2004) | 645 (576) | 1525 (1349) | 1195 (1103) | 3146 (2895) | 3365 (3028) |
The number of twin pairs with a full data record (i.e., with data available for all 13 items in both twins) are in brackets
‘Agg’ denotes the data aggregated over age. If the twin members of the same pair have data in two separate age categories, this pair is omitted from the age grouping to enable multi-group analysis (which requires independent groups). However, this pair is not omitted from the interaction analysis in the aggregated data, leading to data in 6511 pairs for interaction analyses
We tested if some age groups differed importantly from other age groups (e.g., more variance in the phenotypic factor due to heterogeneity). As the age groups are independent (the members of a twin pair are always in the same age group), we could conduct a multi-group analysis and test for homogeneity of the measurement model (i.e., invariant αi, τi1, τi2, rMZ,i, rDZ,I; also referred to as measurement invariance, see Millsap and Yun-Tein 2004) and homogeneity of θ across ages [i.e., invariant MEAN(θ), VAR(θ), and COR(θ1,θ2)]. First, we tested for invariance of the αi, τi1, τi2, rMZ,i, and rDZ,i parameters in the five age groups. To this end we fitted the measurement model without the ACE decomposition, but with a correlation between θ1 and θ2, in the five age groups. We did this separately for the MZ and DZ twins. We started with an unconstrained model (step 0) in which all parameters were free to vary over age groups. Next, step-by-step, we constrained αi (step 1a), rMZ,i, and rDZ,i (step 1b), and τi1 and τi2 (step 1c) to be equal across groups. In step 1a we allowed for differences in the variance of θ between the age groups (i.e., we allowed for the possibility that older subjects have a higher/lower variance on the phenotypic depression variable as compared to the younger subjects). To do so, we constrained VAR(θ) = 1 in group 1, and estimated it freely in the remaining groups. In step 1c we allowed for a mean difference in θ between the age groups (i.e., we allowed for the possibility that older subjects have on average higher/lower levels on the phenotypic depression variable as compared to the younger subjects). To do so, we constrained ν = 0 in group 1, and estimated it freely in the remaining groups. If homogeneity with respect to the measurement model is tenable (i.e., step 1c), we could subsequently test for homogeneity of the population model with respect to age. To this end we tested whether the mean and variance of θ (step 2a), and COR(θ1,θ2) were equal across age groups (step 2b).
Measurement invariance with respect to gender
As we compared males and females in the interaction models, we analyzed whether homogeneity of the measurement model (measurement invariance) holds for males and females. We did not need to establish homogeneity of θ as we explicitly took possible differences in θ into account in the interaction model. Thus, we assessed whether the parameters αi, τic, rMZi, and rDZi were invariant over gender using the procedure from step 1a to 1c as described above. For the MZ subsample this was thus a two group analysis (males–females), and for the DZ subsample this was a three group analysis (male, female, and opposite-sex pairs).
Testing for interactions in a multi-group model
Recently, within the heteroscedastic ACE methodology (Jinks and Fulker 1970; Molenaar et al. 2012) an approach was presented to enable tests on genotype by environment interactions using the model discussed above (Molenaar and Dolan 2014; Schwabe and Van den Berg 2014). Specifically, retaining the measurement model for θ as discussed above, the biometric model can also be formulated as a conditional model. To this end, we condition θ on A, denoted θ | A. This results in
with
for the variance decomposition. Now, a genotype by environment interaction is operationalized as an AxC interaction and an A×E interaction:
that is, the variance of C and E are made a function of A. Within this function, γ0 and β0 are the intercept parameters for log[VAR(C)] and log[VAR(E)] respectively which model the size of VAR(C) and VAR(E) at A = 0. In addition, γ1 and β1 are the interaction parameters, which model the increase or decrease of VAR(C) and VAR(E) across A. The presence of A×C and/or A×E is established by testing whether γ1 and/or β1 depart from 0. If so, the parameter estimates can be used to infer the direction of the interaction effect. For instance, a β1 > 0 denotes that the unique environmental variance is larger for subjects with a higher genetic predisposition, A. In addition, β1 < 0 denotes that the unique environmental variance is smaller for subjects with a higher genetic predisposition, A. The same holds for the A×C parameter, γ1. This conceptualization of genotype by environment interaction is inspired by Jinks and Fulker (1970), who treated a genotype by environment interaction as an environmental variance that is heteroscedastic across the additive genetic factor. This conceptualization is somewhat different from that of Purcell (2002), who models genotype by environment interactions by making the variance of A a function of a measured moderator (which is not necessarily purely environmental). For a more technical discussion of the biometric model, see Appendix 2.
Gender effects
As we wanted to allow for gender effects in the aggregated data analysis, an extension of the model by Molenaar and Dolan (2014) was necessary. To account for gender differences in VAR(C|A), VAR(E|A), we used the following parameterization
with
where GENDERpj is coded 0 if twin j from twin pair p is a male and 1 if it is a female. In this way, the new parameters γ0,female and β0,female account for differences in the intercept parameters γ0,overall and β0,overall in the female group as compared to the male group. Similarly, the new parameters γ1,female and β1,female account for differences in the A×C and A×E parameters, γ1,overall and β1,overall, in the female group as compared to the male group. Thus, the AxE parameter β1 in the male group is equal to β1,overall, and the A×E parameter β1 in the female group is equal to β1,overall + β1,female. The same holds for the AxC parameter. In the model above, the intercept parameter, ν, captures a possible mean difference in θ between males and females.
To account for gender differences in VAR(A) we analogously defined:
that is, exp(ω0,overall) is the variance of A in the male group, and exp(ω0,overall + ωfemale) is the variance of A in the female group.
Identification and estimation
To identify the model, traditional scale and location constraints were imposed on θ (see Molenaar and Dolan 2014). We identified the scale of θ by fixing α1 = 1 for the MZ and DZ twin samples. In single group applications, the location of θ was fixed by imposing ν = 0. As discussed above, in the multi-group model including gender, ν was a free parameter in the female group and fixed to 0 in the male group.
We used a Bayesian approach to model fitting (Eaves and Erkanli 2003). Specifically, we implemented the model in the open-source OpenBUGS software package (Lunn et al. 2009). To this end, we extended the implementation by Molenaar and Dolan (2014) to include the multi-group components as discussed above. The adapted script is available from the website of the first author. Using this script, one can draw samples from the posterior distribution of the parameters using Markov Chain Monte Carlo (MCMC) sampling. From these samples one can determine the parameter means and HPD regions which can be used for statistical inference. The parameters of interest to be estimated were: αi, τic. rMZi, rDZ,i β0,overall, β1,overall, γ0,overall, γ1,overall, and ω0,overall in the single group analysis. For the multi-group gender analysis we additionally estimated ω0,female, β0,female, γ0,female, β1,female, γ1,female, and ν. As we used a Bayesian model fitting approach, we specified prior distributions for the parameters. Following Molenaar and Dolan (2014), we used an uniform distribution between −5 and 5 for all parameter except τic, rMZi, and rDZ,i. For τi1 we used a uniform distribution between −∞ and τi2; and for τi2 we used an uniform distribution between τi2 and ∞. These priors were meant to ensure that τi2 is larger than τi1 as discussed above. For rMZi and rDZi we specified a uniform prior between 0 and 1 for to prevent sign switching. Note that the missing data in our dataset provide no problem for parameter estimation: In the MCMC procedure these values are considered parameters and are included in the sampling routine. For more technical details concerning the implementation of the model see Molenaar and Dolan (2014).
Results
Homogeneity with respect to measurement occasion
Estimates for the contributions of heritability, common and unique environment are given in Table 3. Note that these estimates are based on data from the same that were selected for the interaction analyses as described above. Hence the number of participants in Table 3 is smaller than the total number of twins who took part at each measurement occasion. As can be seen, estimates for heritability tend to be higher than the estimates of 0.4 that are commonly found. Also, there are no differences across measurement occasion in the scaled contributions of the A, C, and E factors to the latent phenotypic depression factor, that is, all 99 % HPD regions overlap. We therefore aggregated the data over the measurement occasions, to obtain a sufficiently large sample size for the heteroscedastic ACE model fitting.
Table 3.
The proportion of variance explained in the latent depression phenotype by the additive genetic factor (heritability; h2), the unique environment (e2), and the common environment (c2) at each occasion (year of data collection)
| Occasion | MZ | DZ | h2 | e2 | c2 | ||
|---|---|---|---|---|---|---|---|
| Twin 1 | Twin 2 | Twin 1 | Twin 2 | ||||
| 1991 | 290 | 297 | 456 | 450 | 0.52 (0.37; 0.61) | 0.46 (0.38; 0.57) | 0.03 (0.01; 0.15) |
| 1995 | 300 | 306 | 426 | 432 | 0.62 (0.51; 0.70) | 0.37 (0.29; 0.45) | 0.02 (0.01; 0.14) |
| 1997 | 342 | 335 | 334 | 343 | 0.63 (0.48; 0.71) | 0.35 (0.28; 0.45) | 0.04 (0.01; 0.21) |
| 2000 | 472 | 457 | 409 | 420 | 0.52 (0.35; 0.61) | 0.45 (0.38; 0.54) | 0.04 (0.01; 0.20) |
| 2002 | 221 | 227 | 178 | 170 | 0.51 (0.40; 0.59) | 0.48 (0.40; 0.55) | 0.03 (0.01; 0.15) |
| 2009 | 891 | 896 | 874 | 835 | 0.51 (0.41; 0.58) | 0.47 (0.42; 0.53) | 0.02 (0.00; 0.11) |
| 2011 | 144 | 149 | 171 | 172 | 0.52 (0.37; 0.60) | 0.46 (0.40; 0.54) | 0.04 (0.01; 0.21) |
| 2013 | 486 | 479 | 517 | 543 | 0.50 (0.40; 0.57) | 0.49 (0.43; 0.56) | 0.02 (0.01; 0.10) |
The 99 % Highest Posterior Density regions are in brackets for h2, e2, and c2
Full homogeneity with respect to age
The RMSEA model fit statistic for the MZ and DZ twin samples is depicted in Table 4 for the different models. As can be seen, all RMSEA values were well below the 0.05 criterion of good model fit (Schermelleh-Engel et al. 2003). Although in both the MZ and DZ twin samples the invariance of τi1 and τi2 was associated with a small deterioration of model fit by 0.002 RSMEA points, there was no obvious source of misfit as indicated by the modification indices (the largest modification index was 11.68 for τi1 of item 4 in the first age group). We therefore concluded that measurement invariance is tenable.
Table 4.
RMSEA fit statistic for the multi-group models fit to test measurement invariance across the age groups
| Step | MZ | DZ | |
|---|---|---|---|
| 0 | Baseline | 0.027 | 0.022 |
| 1a | Invariance of αi | 0.027 | 0.021 |
| 1b | + Invariance of rMzi and rDZi | 0.026 | 0.020 |
| 1c | + Invariance of τic | 0.028 | 0.022 |
| 2a | No differences in θ | 0.028 | 0.022 |
| 2b | No differences in COR(θ1,θ2) | 0.024 | 0.019 |
Table 5 contains the estimated means and variances of θ in the different age groups in step 1c. As can be seen, only the mean in the 25–34 age group of the DZ sample was significantly different from zero at a 0.01 significance level. For the variances, only the variance of θ in the 20–21 age group of the MZ sample departed significantly from 1. In addition, restricting the means of θ in all age groups to equal 0 and all variances of θ to be equal to 1 (step 1d) did not deteriorate the model fit, see Table 4. Finally, we tested the latent phenotypic twin correlation, COR(θ1, θ2) to be equal across age groups (step 1e). As can be seen from Table 4, a model with equal latent phenotypic correlations across age groups improved the RMSEA in both the MZ and DZ twin samples. We therefore concluded that there was no overall age effect detectable. That is, either there is no age effect in the data or the age effect is very small. In both cases we can safely conclude that age did not confound the analysis on the aggregated data as reported below.
Table 5.
Estimated means and variances of the latent phenotypic factor, θ, in the different age groups
| Age | MEAN(θ) | VAR(θ) | ||
|---|---|---|---|---|
| Estimate | se | Estimate | se | |
| DZ | ||||
| 18–19 | 0a | – | 1a | – |
| 20–21 | −0.06 | 0.04 | 0.92 | 0.03 |
| 22–24 | 0.14 | 0.06 | 0.95 | 0.04 |
| 25–34 | 0.13 | 0.06 | 1.05 | 0.04 |
| 35–70 | 0.02 | 0.05 | 1.01 | 0.03 |
| MZ | ||||
| 18–19 | 0a | – | 1a | – |
| 20–21 | 0.045 | 0.053 | 1.013 | 0.037 |
| 22–24 | 0.095 | 0.074 | 1.118 | 0.054 |
| 25–34 | 0.217 | 0.066 | 1.076 | 0.046 |
| 35–70 | −0.116 | 0.050 | 1.017 | 0.037 |
aThese parameters are constrained for identification purposes
Measurement invariance with respect to gender
We started with a baseline model (step 0) in which all measurement model parameters αi, τi1, τi2, rMZ,i, and rDZ,I were allowed to differ across males and females. Next, we fitted the models from step 1a, 1b, and 1c to the data as discussed above. The results are in Table 6. As can be seen all models fitted well according to the 0.05 criterion. However, in step 1c, the model fit deteriorated notable in both the MZ and DZ twin samples. The modification indices suggested that τi1 of item 3 (‘I cry a lot’) accounts for this misfit (the modification index equaled 112.20 in the male MZ sample). Indeed, as can be seen from the table in step 1c′, freeing this parameter improved the model fit. Results showed that for both the MZ and DZ twins, the threshold parameter τi1 of item 3 was estimated to be much larger for the males as compared to the females indicating that the males tend to use the lower category too often as compared to the females (or similarly, females use the lower category too little as compared to the males). In the final model (step 1c′), the mean difference on θ between males and females (i.e., parameter ν) was estimated to be 0.47 (se 0.04) in the MZ sample and 0.47 (se 0.03) in the DZ sample. In addition, the variance in the female group was estimated to be 1.02 (se 0.03) in the MZ sample and 1.06 (se 0.03) in the DZ sample indicating that there was no variance difference between males and females (the male variance was fixed to 1).
Table 6.
RMSEA fit statistic for the multi-group models fit to test measurement invariance across gender
| Step | MZ | DZ | |
|---|---|---|---|
| 0 | Baseline | 0.022 | 0.018 |
| 1a | Invariance of αi | 0.023 | 0.020 |
| 1b | + Invariance of rMzi and rDZi | 0.022 | 0.019 |
| 1c | + Invariance of τic | 0.028 | 0.026 |
| 1c’ | Free τi1 for i = 3 | 0.024 | 0.021 |
This final model without τi1 for item 3 fitted acceptable as compared to the other models. In addition, there was no obvious source of misfit as judged by the modification indices (the largest modification index equaled 12.17 for τi1 of item 4 in the male MZ sample). We concluded that measurement invariance was tenable for all items except item 3. As item 3 was not associated with the same measurement properties for males and females, we omitted this item from the remaining analysis to ensure a meaningful comparison.
Results of the interaction model
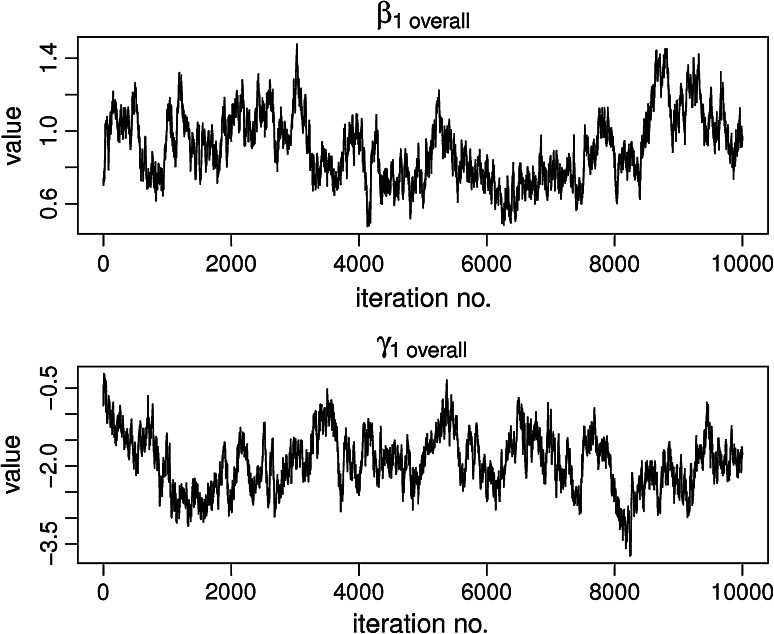
We drew 20,000 samples from the posterior parameter distribution of which we discarded the first 10,000 as burn-in. From the Gelman and Rubin (1992) diagnostic (based on two chains) and trace plots of the parameters this number of samples appeared to be sufficient to ensure that the chains converged to their stationary distribution. See Fig. 2 for example trace plots of the interaction parameters, β1,overall and γ1,overall of the full interaction model including gender.
Fig. 2.
Trace plots of the interaction parameters β1 and γ1 in the full interaction model in age group 18–19
We fitted a model without gender differences (i.e., model M1) and a model with gender differences (i.e., model M2, the full gender interaction model). The parameter estimates of the measurement model parameters for model M2 are in Table 7. As can be seen, these correlations were notably smaller in the DZ twin group. This indicates that some item specific genetic and/or shared environmental variance may underlie the scores. By means of the residual correlation, we accounted for this common variance.
Table 7.
Item parameter estimates (99 % HPD) in the full gender interaction model
| Item | αi | τi1 | τi2 | rDZ,i | rMZ,i |
|---|---|---|---|---|---|
| 1 | 1.00a | 0.96 (0.89; 1.02) | 3.10 (3.00; 3.21) | 0.12 (0.01; 0.22) | 0.22 (0.12; 0.32) |
| 2 | 1.00 (0.94; 1.07) | 1.69 (1.61; 1.78) | 3.37 (3.24; 3.52) | 0.04 (0.00; 0.16) | 0.31 (0.19; 0.42) |
| 3 | – | – | – | – | – |
| 4 | 0.71 (0.66; 0.76) | 1.32 (1.25; 1.39) | 2.85 (2.74; 2.96) | 0.09 (0.01; 0.19) | 0.26 (0.16; 0.35) |
| 5 | 0.52 (0.49; 0.56) | 0.09 (0.05; 0.14) | 1.47 (1.42; 1.53) | 0.12 (0.05; 0.18) | 0.31 (0.25; 0.37) |
| 6 | 0.89 (0.83; 0.96) | 1.87 (1.78; 1.97) | 3.35 (3.21; 3.50) | 0.09 (0.00; 0.22) | 0.35 (0.21; 0.48) |
| 7 | 1.46 (1.36; 1.56) | 2.08 (1.96; 2.20) | 4.27 (4.06; 4.47) | 0.12 (0.00; 0.31) | 0.26 (0.12; 0.40) |
| 8 | 1.00 (0.93; 1.06) | 0.35 (0.29; 0.42) | 2.68 (2.57; 2.78) | 0.04 (0.00; 0.13) | 0.26 (0.18; 0.35) |
| 9 | 1.04 (0.98; 1.11) | 1.40 (1.32; 1.49) | 3.31 (3.17; 3.44) | 0.03 (0.00; 0.13) | 0.36 (0.26; 0.46) |
| 10 | 0.88 (0.83; 0.94) | 1.33 (1.26; 1.40) | 3.03 (2.92; 3.16) | 0.07 (0.00; 0.18) | 0.22 (0.11; 0.32) |
| 11 | 0.85 (0.80; 0.91) | 0.57 (0.52; 0.63) | 2.44 (2.36; 2.53) | 0.14 (0.05; 0.22) | 0.32 (0.25; 0.40) |
| 12 | 1.35 (1.27; 1.45) | 1.53 (1.43; 1.64) | 4.02 (3.84; 4.22) | 0.08 (0.00; 0.21) | 0.20 (0.07; 0.33) |
| 13 | 1.15 (1.09; 1.23) | 0.18 (0.11; 0.25) | 2.42 (2.31; 2.52) | 0.09 (0.00; 0.17) | 0.23 (0.14; 0.31) |
aThis parameter has been constrained for identification purposes. In addition, item 3 was omitted from the analysis as it violated measurement invariance across gender
In Table 8, the parameter estimates of the interaction parameters in model M1 and M2 are depicted. As can be seen in model M1 without gender differences, β1 departed from 0 and was positive indicating the presence of A×E the variance of E increasing for increasing levels of A. In addition, the γ1 parameter did not depart from 0, indicating the absence of A×C in the full sample.
Table 8.
Parameter estimates (99 % highest posterior density region) of the A×E and A×C parameters in the aggregated data analysis using a model without (M1) and a model with (M2) gender differences in the parameters
| Group | VAR(A) | β0 | β1 | γ0 | γ1 | |
|---|---|---|---|---|---|---|
| M1 | – | 0.64 (0.53; 0.74) | −0.68 (−0.81; −0.53) | 0.34 (0.18; 0.53) | −4.04 (−4.99; −2.43) | −1.38 (−3.05; 1.61) |
| M2 | Males | 0.40 (0.25; 0.57) | −0.87 (−1.07; −0.66) | 0.91 (0.53; 1.41) | −2.24 (−3.63; −1.45) | −1.93 (−3.26; −0.61) |
| Females | 0.64 (0.57; 0.73) | −0.65 (−0.78; −0.51) | 0.21 (0.003; 0.37) | −5.60 (−7.85; −2.87) | 0.54 (−2.90; 3.25) |
VAR(A) is calculated as exp(ω0,overall) for the males and as exp(ω0,overall + ω0,female) for the females. Similar applies to β0, β1, γ0, and γ1, see the paragraph on the parametrization of the gender effects
As can be seen in Table 8, when gender differences were taken into account (model M2), a different pattern of results emerged. That is, for both males and females, A×E was present with positive β1, but for males, there was evidence for A×C as the HPD region of γ1 did not include 0, while for females there was no evidence for A×C. The mean difference between males and females in the latent phenotypic factor, θ, was hardly affected by taking the A×E and A×E interactions into account. That is, ν in the female sample was estimated to be 0.51 (99 % HPD 0.45; 0.59) which was about equal to the estimate reported above in the case of no interactions. It can also be seen from Table 8 that the results from the females follow the results from the entire sample (i.e., M1), while the results from the males are different from the entire sample. We will return to this point in the discussion.
Results in terms of the contributions of heritability, common and unique environment are given in Table 9. Note that these estimates are based on the marginal variance of C and E, as the conditional variance differs across A. The marginal variance for C and E can be calculated using exp(β0 + 0.5 × β21) and exp(γ0 + 0.5 × γ21) respectively (see Hessen and Dolan 2009). As can be seen from the Table by taking into account the gender differences in the interactions (M2) the heritability (h2) drops from 0.54 in the full sample to 0.35 in the male group. In addition, the contribution of the common environment increases from 0.04 in the full sample to 0.22 in the male group. It should be noted however that the uncertainty in this estimate is relatively large, reflected by the wide 99 % HPD region which runs from 0.09 to 0.37. But at least we can conclude that there is some contribution of the common environment to depression for males.
Table 9.
The proportion of variance in the latent depression phenotype explained by the additive genetic factor (heritability; h2), unique environment (e2) and common environment (c2) in the full genotype-by-environment interaction model
| Group | a2 | e2 | c2 | |
|---|---|---|---|---|
| M1 | – | 0.52 (0.43; 0.58) | 0.43 (0.37; 0.48) | 0.04 (0.01; 0.16) |
| M2 | Males | 0.35 (0.21; 0.49) | 0.44 (0.33; 0.53) | 0.22 (0.09; 0.37) |
| Females | 0.54 (0.49; 0.59) | 0.45 (0.41; 0.50) | 0.01 (0.00; 0.06) |
Here, c2 and e2 are the standardized variance of C and E marginally over A
From the results in Table 9 it can be calculated that 33 % of the heritability in males is due to genotype by environment interaction (i.e., 1 − 0.35/0.52; see Molenaar et al. in press). It is clear that this percentage is due to A×C to a large extent, however the exact amount of A×C variance in the male group is difficult to assess. That is, theoretically, the distinction between the effects of A×C and A×E interactions is clear: when not included into the model, regular genetic covariance structure analysis cannot distinguish between an additive genetic factor A and an A×C interaction factor, or between an environmental factor E and an A×E interaction factor (Molenaar et al. 1990). In practical applications however, the parameter estimates for A×C and A×E are correlated which complicate quantification of the exact amount of A×C and A×E variance in the data (see Molenaar et al. 2012).
Discussion
We studied whether an additive genetic by unique environment interaction (A×E) and/or an additive genetic by common environment interaction (A×C) play any role in adult depression. In a measurement model for categorical item scores, the depression phenotype was operationalized as a latent variable. In a first set of analyses, omitting interaction effects, we found heritability estimates of around 0.5–0.6, which are somewhat larger than those commonly found using an MDD diagnosis or symptom count sum score (i.e., around 0.3–0.4; Sullivan et al. 2000; Nivard et al. 2015). This discrepancy is both of interest and expected, as latent variables always contain less measurement error as compared to observed measures (see Van den Berg et al. 2007).
The present undertaking was aimed at testing the hypothesis that common environmental variance in depression is masked by interaction effects (Duncan et al. 2014). In a single group analysis of the complete sample, we found that the unique environmental variance is larger for individuals with a higher predisposition to develop depression (i.e., higher A factor score). However, we obtained no evidence for A×C interaction in the single group analysis. Next, in a multi-group analysis, we took gender differences into account. We found A×E in both males and females similarly as in the single group analysis, but additionally, there was A×C for the male group. Specifically, in males, common environmental variance is smaller in twins with a higher genetic predisposition to develop depression. The marginal contribution of C increased from 0.04 in the full sample to 0.22 for males by taking gender differences in AxC into account. Although the uncertainty in this estimate is large, it can be concluded that in males, at least some common environment variance is masked by non-linearity due to an A×C interaction.
As mentioned above, the results of the female sample follow the results of the entire sample while the results of the males are different from the results in the entire sample. As interaction effects result in non-normality, the present method detects specific departures from bivariate normality in the latent phenotypic factor θ that are due to A×C (see Molenaar et al. 2012). In the entire sample (collapsing over gender), the non-normality due to A×C in the male sample is masked due to the females that score higher on θ. The distribution of θ in the entire sample is thus approximately normal. In the male sample, the distribution of θ departs from normality resulting in different estimates of the parameters as compared to the entire sample. As there is no A×C interaction in the female sample, the distribution of θ is approximately normal and the results follow the results from the entire sample.
As we argued in this paper, using a measurement model for the item data in testing for genotype by environment interactions may solve the scaling issues commonly encountered in genotype by environment interaction research (Eaves 2006). However, some common limitations of genotype by environment interaction research remain (see Molenaar et al. in press). That is, the presence of a non-linear genotype by environment correlation may conflate the genotype-by-environment interaction. In addition, a genotype-by-environment interaction may spuriously arise if the twin sample is unrepresentative of the population (e.g., the higher phenotypes are underrepresented). Note however that these shortcomings are not unique to the item level approach used here, as they are also problematic in, for instance, the popular genotype by measured environment approach (Purcell 2002).
It would be of interest to model the complete longitudinal dataset including all items at all measurement occasions. That is, the measurement model approach adopted here allows for so-called ‘item linking’ (Kolen and Brennan 2004). In addition, extending the genotype by environment model with a longitudinal component would allow inclusion of the data from all measurement occasions into the analysis, resulting in the largest power possible to detect an interaction effect. However, such an analysis is currently impossible as the required longitudinal genotype by environment models are not yet developed and because of the large sample size, the mathematically complex model, and the tremendous number of missing data in the complete dataset, a full longitudinal item linking approach is numerically intractable.
The present approach provides what may be considered an omnibus test of A×C and A×E, as the interactions are modeled at the level of the latent variables A, C, and E. We consider this an advantage as we do not need to include measured moderators (candidate genes, environmental variables). We emphasize that a failure to detect A×C or A×E using the present method should be interpreted as a result pertaining to A, C, and E. We do not consider the absence of say A×C in females necessarily incompatible with the presence of an interaction detected with a measured moderator, as the power to detect the effect of an interaction with a measured moderator may be greater than the power to detect A×C. The question of which mechanisms underlie gender differences in depression is important. With the present results we hope to have provided a point of departure for further research into the etiology of differences between males and females in the development of depression. Most importantly, we found some empirical evidence for the claim by Duncan et al. (2014) that effects of the common environment underlying depression are masked by non-linear effects. It is therefore advisable to account for these non-linearity when studying the genetic and environmental underpinning of depression.
Acknowledgements
Funding was provided by the Netherlands Scientific Organization (NWO) (912-100-20): “Genetic influences on stability and change in psychopathology from childhood to young adulthood” and the European Research Council: “Genetics of Mental Illness” (ERC-230374) and “Beyond the genetics of Addiction” (ERC-284167). Data collection has been funded by multiple grants from NWO and ZONMW. Zygosity typing was done by the Avera Institute for Human Genetics in Sioux Falls South Dakota. OpenBUGS input files are available from www.dylanmolenaar.nl. We are grateful to two anonymous reviewers whose comments led to substantial improvements of this paper. We warmly thank all participants for their contributions to this study.
Appendix 1
The following 13 items of the anxious depression scale of the ASR were overlapping in all surveys and used in present analysis (translated from Dutch):
I feel lonely (‘Ik voel me eenzaam’),
I feel confused or in a fog (‘Ik voel me in de war of denk wazig’),
I cry a lot (‘Ik huil veel’),
I am afraid I might think or do something bad (‘Ik ben bang dat ik iets slechts zou kunnen doen of denken’),
I feel that I have to be perfect (‘Ik heb het gevoel dat ik perfect moet zijn’),
I feel that no one loves me (‘Ik heb het gevoel dat niemand van mij houdt’),
I feel worthless or inferior (‘Ik voel me waardeloos of minderwaardig’),
I am nervous or tense (‘Ik ben nerveus of gespannen’),
I am too fearful or anxious (‘Ik ben te angstig of bang’),
I feel too guilty (‘Ik voel me erg schuldig’),
I am self-conscious or easily embarrassed (‘Ik schaam me gauw of voel me niet op mijn gemak’),
I am unhappy, sad or depressed (‘Ik ben ongelukkig, verdrietig of depressief’), and
I worry a lot (‘Ik maak me vaak zorgen’).
Appendix 2
Here we provide the technical details of the model. As discussed, we distinguish between a measurement model for the item scores Xi, and a biometric model for the latent phenotypic factor, θ. First, as a measurement model we use the graded response model (Samejima, 1969). For the MZ twins, the model is given by:
where Φ(.) is the cumulative normal distribution function, Xpij denotes the score of twin member j = 1,2 of twin pair p = 1, …, N on item i, and θpj denotes the latent phenotypic factor. In addition, Q denotes the maximum possible item score (here Q = 2, as we code the data 0, 1, and 2). The parameter rMZi models the residual correlation between the twin 1 and twin 2 scores on the same item. In the DZ subsample this parameter is replaced by rDZi. As discussed in Molenaar and Dolan (2014), δi is a standard normally distributed latent variable that is common to Xi1 and Xi2 (the scores of the twin 1 and twin 2 members of a twin pair). The loadings of Xi1 on δi and Xi2 on δi are equal to in the MZ sample and to in the DZ sample. As the residual polychoric variance of Xi (i.e., the polychoric variance of Xi conditional on θ) is constrained to be equal to 1 - rMZi for MZ and to 1 - rDZi for DZ twins (see the equation above), the residual (polychoric) correlation between Xi1 and Xi2 (i.e., the polychoric correlation between Xi1 and Xi2 conditional on θ1 and θ2) is equal to rMZi and rDZi respectively, see Molenaar and Dolan (2014).
In the biometric model, θ is submitted to the ACE decomposition, that is,
where COR(A1, A2) = 1 for MZ twins and COR(A1, A2) = 0.5 for DZ twins. In addition, COR(C1, C2) = 1 and COR(E1, E2) = 0 for all twins. Under the assumption that the genetic and environmental variance components are mutually uncorrelated, the variance of θ can be decomposed as follows
As discussed in the main text, genotype by environment interactions are operationalized by conditioning θ on the additive genetic factor A. The conditional variance is given by
As the variance of C and E are now conditional on A, we can make them a function of A, that is,
The variance of C and E now depend on the level of A, that is, the variance due to the environment depends on the genotypic factor. Note that because COR(C1, C2) = 1 by definition, imposing
Thus, in the full model, the vector A = [A1, A2] is distributed as
with
and ρ = 1 for MZ twins and ρ = 0.5 for DZ twins.
By conditioning on A, the conditional distribution of the vector θ = [θ1, θ2] is given by
with
with
and
.
Now we can condition on θ and specify the distribution of the observed data:
where the probabilities in the categorical distribution can be determined using the graded response model above. See Molenaar and Dolan (2014) for a discussion on how these distributions are exactly implemented in OpenBUGS (Lunn et al. 2009).
Compliance with Ethical Standards
Conflicts of Interest
Dylan Molenaar, Christel M. Middeldorp, Gonneke Willemsen, Lannie Ligthart, Michel G. Nivard, & Dorret I. Boomsma have no conflict of interest.
Human and Animal Rights and Informed Consent
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Footnotes
Molenaar and Dolan (2014) show that even under severe measurement problems (severe floor effects), AxE and AxC tests are relatively unbiased. However, the power to detect an interaction is diminished.
References
- Achenbach TM, Rescorla LA. Manual for the ASEBA adult: forms and profiles. Burlington: University of Vermont, Research Center for Children, Youth and Families; 2003. [Google Scholar]
- Bartels M, Beijsterveldt VC, Derks EM, Stroet TM, Polderman TJ, Hudziak JJ, Boomsma DI. Young Netherlands Twin Register (Y-NTR): a longitudinal multiple informant study of problem behavior. Twin Res Human Genetics. 2007;10(01):3–11. doi: 10.1375/twin.10.1.3. [DOI] [PubMed] [Google Scholar]
- Bowlby J. The making and breaking of affectional bonds. II. Some principles of psychotherapy. The fiftieth Maudsley Lecture. Br J Psychiatry. 1977;130(5):421–431. doi: 10.1192/bjp.130.5.421. [DOI] [PubMed] [Google Scholar]
- Dick DM, Agrawal A, Keller MC, Adkins A, Aliev F, Monroe S, Hewitt JK, Kendler KS, Sher KJ. Candidate gene–environment interaction research reflections and recommendations. Perspect Psychol Sci. 2015;10(1):37–59. doi: 10.1177/1745691614556682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AE, Munn-Chernoff MA, Hudson DL, Eschenbacher MA, Agrawal A, Grant JD, Nelson EC, Waldron M, Glowinski AL, Sartor CE, Bucholz KK, Madden PA, Heath AC. Genetic and environmental risk for major depression in African-American and European-American women. Twin Res Human Genetics. 2014;17(04):244–253. doi: 10.1017/thg.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves LJ. Genotype x environment interaction in psychopathology: fact or Artifact? Twin Res Human Genet. 2006;9:1–8. doi: 10.1375/twin.9.1.1. [DOI] [PubMed] [Google Scholar]
- Eaves LJ, Erkanli A. Markov chain Monte Carlo approaches to analysis of genetic and environmental components of human developmental change and GxE interaction. Behav Genet. 2003;33:279–299. doi: 10.1023/A:1023446524917. [DOI] [PubMed] [Google Scholar]
- Eaves LJ, Last K, Martin NG, Jinks JL. A progressive approach to non-additivity and genotype-environmental covariance in the analysis of human differences. Br J Math Stat Psychol. 1977;30:1–42. doi: 10.1111/j.2044-8317.1977.tb00722.x. [DOI] [Google Scholar]
- Gatz M, Pederson NL, Plomin R, Nesselroade JR. Importance of shared genes and shared environments for symptoms of depression in older adults. J Abnorm Psychol. 1992;101(4):701. doi: 10.1037/0021-843X.101.4.701. [DOI] [PubMed] [Google Scholar]
- Gelman A, Rubin DB. Inference from iterative simulation using multiple sequences. Stat Sci. 1992;7:457–511. doi: 10.1214/ss/1177011136. [DOI] [Google Scholar]
- Hessen DJ, Dolan CV. Heteroscedastic one-factor models and marginal maximum likelihood estimation. Br J Math Stat Psychol. 2009;62(1):57–77. doi: 10.1348/000711007X248884. [DOI] [PubMed] [Google Scholar]
- Ingram R. Origins of cognitive vulnerability to depression. Cogn Ther Res. 2003;27:77–88. doi: 10.1023/A:1022590730752. [DOI] [Google Scholar]
- Jinks JL, Fulker DW. Comparison of the biometrical genetical, mava, and classical approaches to the analysis of human behavior. Psychol Bull. 1970;73:311–349. doi: 10.1037/h0029135. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Thornton LM, Prescott CA. Gender differences in the rates of exposure to stressful life events and sensitivity to their depressogenic effects. Am J Psychiatry. 2011;158:587–593. doi: 10.1176/appi.ajp.158.4.587. [DOI] [PubMed] [Google Scholar]
- Kessler RC, McGonagle KA, Swartz M, Blazer DG, Nelson CB. Sex and depression in the National Comorbidity Survey I: lifetime prevalence, chronicity and recurrence. J Affect Disord. 1993;29(2):85–96. doi: 10.1016/0165-0327(93)90026-G. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Davis CG, Kendler KS. Childhood adversity and adult psychiatric disorder in the US National Comorbidity Survey. Psychol Med. 1997;27(05):1101–1119. doi: 10.1017/S0033291797005588. [DOI] [PubMed] [Google Scholar]
- Klose M, Jacobi F. Can gender differences in the prevalence of mental disorders be explained by sociodemographic factors? Arch Womens Mental Health. 2004;7(2):133–148. doi: 10.1007/s00737-004-0047-7. [DOI] [PubMed] [Google Scholar]
- Kolen MJ, Brennan RL. Test equating, scaling, and linking. New York: Springer; 2004. pp. 201–205. [Google Scholar]
- Lamb DJ, Middeldorp CM, van Beijsterveldt CE, Bartels M, van der Aa N, Polderman TJ, Boomsma DI. Heritability of anxious-depressive and withdrawn behavior: age related changes during adolescence. J Am Acad Child Adolesc Psychiatry. 2010;49(3):248–255. [PubMed] [Google Scholar]
- Lunn D, Spiegelhalter D, Thomas A, Best N. The BUGS project: evolution, critique, and future directions. Stat Med. 2009;28:3049–3067. doi: 10.1002/sim.3680. [DOI] [PubMed] [Google Scholar]
- Mezulis AH, Hyde JS, Abramson LY. The developmental origins of cognitive vulnerability to depression: temperament, parenting, and negative life events in childhood as contributors to negative cognitive style. Dev Psychol. 2006;42:1012–1025. doi: 10.1037/0012-1649.42.6.1012. [DOI] [PubMed] [Google Scholar]
- Millsap RE, Yun-Tein J. Assessing factorial invariance in ordered-categorical measures. Multivar Behav Res. 2004;39(3):479–515. doi: 10.1207/S15327906MBR3903_4. [DOI] [Google Scholar]
- Molenaar D, Dolan CV. Testing systematic genotype by environment interactions using item level data. Behav Genet. 2014;44:212–231. doi: 10.1007/s10519-014-9647-9. [DOI] [PubMed] [Google Scholar]
- Molenaar PCM, Boomsma DI, Neeleman D, Dolan CV, Rao DC, Vogler GP. Using factor scores to detect G × E Interactive Origin of “pure” genetic or environmental factors obtained in genetic covariance structure analysis. Genet Epidemiol. 1990;7(1):93–100. doi: 10.1002/gepi.1370070116. [DOI] [PubMed] [Google Scholar]
- Molenaar D, van der Sluis S, Boomsma DI, Dolan CV. Detecting specific genotype by environment interactions using marginal maximum likelihood estimation in the classical twin design. Behav Genet. 2012;42:483–499. doi: 10.1007/s10519-011-9522-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar D, Middeldorp CM, van Beijsterveldt CEM, Boomsma DI (in press). Analysis of behavioral and emotional problems in children highlights the role of genotype-by environment interaction. Child Develop [DOI] [PubMed]
- Monroe SM, Simons AD. Diathesis-stress theories in the context of life stress research: implications for the depressive disorders. Psychol Bull. 1991;110(3):406. doi: 10.1037/0033-2909.110.3.406. [DOI] [PubMed] [Google Scholar]
- Nivard MG, Dolan CV, Kendler KS, Kan KJ, Willemsen G, van Beijsterveldt CE, et al. Stability in symptoms of anxiety and depression as a function of genotype and environment: a longitudinal twin study from ages 3 to 63 years. Psychol Med. 2015;45:1039–1049. doi: 10.1017/S003329171400213X. [DOI] [PubMed] [Google Scholar]
- Piccinelli M, Wilkinson G. Gender differences in depression Critical review. Br J Psychiatry. 2000;177(6):486–492. doi: 10.1192/bjp.177.6.486. [DOI] [PubMed] [Google Scholar]
- Purcell S. Variance components models for gene–environment interaction in twin analysis. Twin Res. 2002;5(06):554–571. doi: 10.1375/136905202762342026. [DOI] [PubMed] [Google Scholar]
- Samejima F (1969) Estimation of ability using a response pattern of graded scores. Psychometrika Monograph, No. 17
- Schermelleh-Engel K, Moosbrugger H, Müller H. Evaluating the fit of structural equation models: tests of significance and descriptive goodness-of-fit measures. Methods Psychol Res Online. 2003;8(2):23–74. [Google Scholar]
- Schwabe I, van den Berg SM. Assessing genotype by environment interaction in case of heterogeneous measurement error. Behav Genet. 2014;44:394–406. doi: 10.1007/s10519-014-9649-7. [DOI] [PubMed] [Google Scholar]
- Silberg J, Rutter M, Neale M, Eaves L. Genetic moderation of environmental risk for depression and anxiety in adolescent girls. Br J Psychiatry. 2001;179(2):116–121. doi: 10.1192/bjp.179.2.116. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157(10):1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- van Beijsterveldt CEM, Groen-Blokhuis M, Hottenga JJ, Franić S, Hudziak JJ, Lamb D, et al. The Young Netherlands Twin Register (YNTR): longitudinal twin and family studies in over 70,000 children. Twin Res Human Genet. 2013;16:252–267. doi: 10.1017/thg.2012.118. [DOI] [PubMed] [Google Scholar]
- van den Berg SM, Glas CA, Boomsma DI. Variance decomposition using an IRT measurement model. Behav Genet. 2007;37(4):604–616. doi: 10.1007/s10519-007-9156-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen G, Vink JM, Abdellaoui A, den Braber A, van Beek JHDA, Draisma HHM, et al. The adult Netherlands twin register: twenty-five years of survey and biological data collection. Twin Res Human Genet. 2013;16:271–281. doi: 10.1017/thg.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]