Abstract
The performance of complex networks, like the brain, depends on how effectively their elements communicate. Despite the importance of communication, it is virtually unknown how information is transferred in local cortical networks, consisting of hundreds of closely spaced neurons. To address this, it is important to record simultaneously from hundreds of neurons at a spacing that matches typical axonal connection distances, and at a temporal resolution that matches synaptic delays. We used a 512-electrode array (60 μm spacing) to record spontaneous activity at 20 kHz from up to 500 neurons simultaneously in slice cultures of mouse somatosensory cortex for 1 h at a time. We applied a previously validated version of transfer entropy to quantify information transfer. Similar to in vivo reports, we found an approximately lognormal distribution of firing rates. Pairwise information transfer strengths also were nearly lognormally distributed, similar to reports of synaptic strengths. Some neurons transferred and received much more information than others, which is consistent with previous predictions. Neurons with the highest outgoing and incoming information transfer were more strongly connected to each other than chance, thus forming a “rich club.” We found similar results in networks recorded in vivo from rodent cortex, suggesting the generality of these findings. A rich-club structure has been found previously in large-scale human brain networks and is thought to facilitate communication between cortical regions. The discovery of a small, but information-rich, subset of neurons within cortical regions suggests that this population will play a vital role in communication, learning, and memory.
SIGNIFICANCE STATEMENT Many studies have focused on communication networks between cortical brain regions. In contrast, very few studies have examined communication networks within a cortical region. This is the first study to combine such a large number of neurons (several hundred at a time) with such high temporal resolution (so we can know the direction of communication between neurons) for mapping networks within cortex. We found that information was not transferred equally through all neurons. Instead, ∼70% of the information passed through only 20% of the neurons. Network models suggest that this highly concentrated pattern of information transfer would be both efficient and robust to damage. Therefore, this work may help in understanding how the cortex processes information and responds to neurodegenerative diseases.
Keywords: effective connectivity, information transfer, microcircuits, rich club, transfer entropy
Introduction
The performance of complex networks depends crucially on how they route their traffic, whether it is passengers, supplies, or phone calls (Barrat et al., 2008). Despite the importance of this, we have only a few clues about how information is distributed in local cortical networks, consisting of several hundred closely spaced neurons.
Patch-clamp recordings in cortex have shown that the distributions of synaptic strengths (Song et al., 2005) and firing rates (Hromádka et al., 2008) are long tailed and approximately lognormal (Buzsáki and Mizuseki, 2014). Moreover, cortical layer 2/3 pyramidal neurons with high firing rates are more likely to share synaptic connections than neurons with low firing rates (Yassin et al., 2010). Do the above results indicate that most information transfer occurs within a small percentage of strongly connected neurons? To draw this conclusion at the population level, it is necessary to record simultaneously, preferably from hundreds of closely spaced neurons. To quantify information transfer at typical synaptic delays of 1–20 ms (Mason et al., 1991; Swadlow, 1994), it is essential to measure spike trains with millisecond precision (Panzeri et al., 2001; Wehr and Zador, 2003; Ito et al., 2011, 2014).
As it is currently difficult to achieve all of these requirements in vivo, we primarily used a high-density (60 μm spacing) 512-electrode array to record spontaneous spike trains at submillisecond resolution from up to 500 neurons [mean (±SD), 347 ± 119 neurons] simultaneously in slice cultures of somatosensory cortex (N = 15). To check the generality of our findings, we also analyzed some multisite recording data from rodent cortex in vivo. For all data, we applied a previously validated algorithm for transfer entropy (TE; Schreiber, 2000; Ito et al., 2011) to extract directed networks of information transfer (IT). TE has been used widely in neuroscience (Gourévitch and Eggermont, 2007; Garofalo et al., 2009; Lindner et al., 2011; Vicente et al., 2011; Orlandi et al., 2014; Timme et al., 2014) because it can accommodate nonlinear relationships, and it compares favorably to other measures (Lungarella et al., 2007). Following Schreiber (2000), we defined TE as predictive information. If the spike train of neuron A contains information about the future spiking of neuron B not given by the past of neuron B, then TE detects information transfer from neuron A to neuron B. We have used TE before on cortical networks and found that it reveals nonrandom structures in the unweighted connectivity maps (Shimono and Beggs, 2015).
We sought to answer three specific questions. First, do the TE strengths between cortical neuron pairs follow a lognormal distribution, as reported for synaptic strengths (Song et al., 2005)? Second, as predicted by Koulakov et al. (2009), do some neurons transfer and receive more information than others? Third, do the neurons that transfer the most information preferentially interact with each other, thus forming a “rich club” (van den Heuvel and Sporns, 2011; Towlson et al., 2013)? These questions are aimed at probing inhomogeneity at the level of connections, single neurons, and the network, respectively.
Several studies motivated us to investigate the potential existence of inhomogeneous patterns of information transfer. A model by Koulakov et al. (2009) suggested that lognormal firing rates arise because some neurons receive much stronger inputs, driving them to fire at much higher rates than the mean. Computational models have shown that highly concentrated connectivity patterns can facilitate bistability in cortical neurons, thus promoting Up and Down states, which are commonly seen in cortical recordings and are thought to be relevant for working memory (Klinshov et al., 2014). Computational models also have suggested that a rich-club structure can increase the number of distinct network activity states that could serve long-term memory (Senden et al., 2014). If a relatively small, but information-rich, population of neurons does exist in cortical networks, it seems likely that such neurons would play vital roles in communication, learning, and memory. In addition, we expect that knowledge about such neurons would help us to understand how cortical networks respond to neurodegenerative diseases.
Materials and Methods
The methods used here, particularly for assessing effective connectivity, are complex and involve many steps. For clarity, we first provide the following outline to give an overview of what will be described in more detail below: (1) in vitro preparation and recording; (2) in vivo preparation and recording; 3. estimating effective connectivity (transfer entropy measure, controlling for firing rate, controlling for network drive, controlling for common drive and transitive effects, and validation of effective connectivity); and (4) graph theory measures (cumulative information transfer, rich club, betweenness centrality, dynamic importance, and neuron diversity).
In vitro preparation and recording
Cultured tissue from animals was prepared according to guidelines from the National Institutes of Health, and all animal procedures were approved by the Animal Care and Use Committees at Indiana University and at the University of California, Santa Cruz. For these studies, we chose organotypic cultures because they preserve many of the features characteristic of cortex, including neuronal morphology (Klostermann and Wahle, 1999), cytoarchitecture (Caeser et al., 1989; Götz and Bolz, 1992), precise intracortical connectivity (Bolz et al., 1990), and intrinsic electrophysiological properties (Plenz and Aertsen, 1996). In addition, they produce a variety of emergent network activity patterns that have been found in vivo, including precisely timed responses (Buonomano, 2003), Up states (Johnson and Buonomano, 2007), oscillations (Baker et al., 2006; Gireesh and Plenz, 2008), synchrony (Beggs and Plenz, 2004; Baker et al., 2006), waves (Harris-White et al., 1998), repeating activity patterns (Beggs and Plenz, 2004; Ikegaya et al., 2004), and neuronal avalanches (Beggs and Plenz, 2003; Friedman et al., 2012).
When cortical slice cultures are grown with subcortical target structures, as was done here, it has been shown that they form appropriate connections that are not exuberant (Leiman and Seil, 1986; Baker and Van Pelt, 1997). However, organotypic cortical cultures have been reported to have disrupted layer structure (Staal et al., 2011). We prepared organotypic cultures using methods previously reported (Tang et al., 2008). Briefly, brains from postnatal day 6 (P6) to P7 black 6 mouse pups (RRID:Charles_River:24101632, Harlan) were removed under a sterile hood and placed in chilled Gey's balanced salt solution (Sigma-Aldrich) for 1 h at 0°C. After 30 min, half the solution was changed. Brains were next blocked into 5 mm3 sections containing somatosensory cortex (Paxinos and Watson, 1986). Blocks were then sliced with a thickness of 400 μm.
Each slice was placed on a small circular cutout of permeable membrane (Millipore) that was then placed on top of a larger membrane that spanned a culture well. Culture medium consisted of HBSS (catalog #H9394, Sigma-Aldrich) 1:4, minimum essential medium (catalog #M4083, Sigma-Aldrich) 2:4, horse serum (catalog #H1270, Sigma-Aldrich) 1:4, and 4 ml of l-glutamine, with penicillin/streptomycin 1:1000 volume of media (catalog #P4083, Sigma-Aldrich). Slices were maintained at an interface between medium below and atmosphere above. The plates of wells were incubated at 37°C in humidified atmosphere with 5% CO2.
After 2–3 weeks, the cultures were then gently placed on a microelectrode recording array by lifting the small circular cutout of membrane with tweezers. Each culture was placed tissue side down, with the membrane facing up. We attempted to place the tissue such that somatosensory cortex was on the electrode array. During recording, cultures were perfused at 1 ml/min with culture medium that was saturated with 95% O2/5% CO2. Spontaneous activity was recorded for 1.5 h. The first 0.5 h was not used in analysis. Spikes were recorded with a 512-electrode array system that has been used previously for retinal (Litke et al., 2004; Shlens et al., 2006; Petrusca et al., 2007; Field et al., 2010) and cortical (Tang et al., 2008; Friedman et al., 2012) experiments. The outline of the array was rectangular (1.8 mm × 0.9 mm) and electrodes were arranged in a hexagonal lattice with an interelectrode distance of 60 μm. Spike sorting was performed off-line, as previously described (Litke et al., 2004; Tang et al., 2008). Briefly, signals that crossed a threshold of 8 SDs were marked, and the waveforms found on the marked electrode and its six adjacent neighbors were projected into five-dimensional principal component space. A mixture-of-Gaussians model was fit to the distribution of features based on maximum likelihood. Neurons with well separated clusters in principal components space were retained for further analysis; duplicate neurons and neurons with significant refractory period violations were excluded.
The quality of the data collected for this study had a unique combination of features that made them particularly useful for uncovering effective connectivity in neural networks at this scale. The high temporal resolution of the recordings allowed the sequence of spike activations to be identified in many cases, letting us measure directed, rather than undirected, connections. The close electrode spacing (∼60 μm) was within the radius of most synaptic contacts between cortical pyramidal neurons (Song et al., 2005) and enhanced the probability that neurons recorded on neighboring electrodes would share direct synaptic contacts. The large number of neurons simultaneously recorded, as well as the long recording durations (≥1 h) enhanced the statistical power, allowing many connections to be identified that would have otherwise gone undetected. Very few previous studies have been able to combine all of these features simultaneously in cortex.
In vivo preparation and recording
To check the generality of our results, we also analyzed data collected in vivo from rodent cortex where silicon-based microprobes, built in a fashion that was similar to those in the study by Shobe et al. (2015), were placed into orbitofrontal cortex of awake, behaving mice. All in vivo recording procedures were approved by the University of California, Los Angeles, Chancellor's Animal Research Committee. Singly housed male C57BL/6J mice (N = 7, 12–16 weeks old; The Jackson Laboratory) were used in the experiments. Animals underwent an initial surgery under isoflurane anesthesia in a stereotaxic apparatus to bilaterally fix stainless steel head restraint bars (10 × 7.5 mm, 0.6 g, laser cut at Fab2Order) on the skull in preparation for recording from awake head-fixed animals. They recovered for 7 d, and subsequently animals were anesthetized with isoflurane for a second surgery on the recording session day to make a rectangular craniotomy over the orbitofrontal cortex for silicon microprobe insertion. An additional craniotomy was made over the posterior cerebellum for placement of an electrical reference wire. After the craniotomy surgery, animals recovered from anesthesia in their home cage for ∼2 h before being head fixed while awake and free to move on a spherical uniaxial polystyrene treadmill. The silicon microprobes were slowly lowered to stereotaxically defined coordinates targeting the orbitofrontal cortex (2.2–2.3 mm anterior to bregma). The microprobes contained a total of 256 electrode recording sites that were densely distributed (hexagonal array geometry with 25 μm spacing) on five prongs (0.3–0.4 mm spacing) spanning the medial to lateral orbitofrontal cortex. Animals were killed after the recording session; and brain tissue was sectioned, immunostained for NeuN, and imaged to verify the silicon prong placement. Signals from all recording sites were simultaneously sampled at 25 kHz. Spike sorting was performed off-line using a semi-automated Matlab script. The data used for this analysis were prepared by concatenating resting period activity in between periods when the mouse was performing an odor discrimination task in the same recording session. Resting corresponded to periods of immobility (no treadmill movement and no licking) and a lack of explicitly presented stimuli. The firing rates were fairly constant across resting periods except for the last 2 s of each resting period, where a consistent increase in firing rate was detected. Hence, we deleted the last 2 s from each resting period and then concatenated the different resting periods identified across the entire recording session together to obtain the final spike trains for each recorded neuron. Although we use the resting period spiking activity, when the animal is immobile, to create the effective connectivity network, this does not necessarily mean that during the resting periods the animal did not receive any sensory input. This could result in the activity being nonstationary. The culture data on the other hand do not receive any external sensory input and could be thought of as approximately stationary. We looked at the coefficient of variation (SD over the mean) of the number of spikes per second per neuron for the entire recording periods for both the in vitro and the in vivo data. We found that the coefficient of variation for the in vivo data was smaller than that for the in vitro datasets. Thus, although the analysis method does not explicitly take into account the nonstationary nature of the data, this information suggests that the nonstationarity in the in vivo datasets is smaller than that observed in the in vitro datasets. A recent study (Wollstadt et al., 2014) looked into the issue of nonstationarity in multiple neuron recordings in greater detail and suggested new methods to analyze such data. Applying such methods here, however, was beyond the scope of this study.
Estimating effective connectivity
Broadly speaking, the neuroscience literature discusses the following three types of connections: structural, functional, and effective. At the level of neurons that we are dealing with here, a synapse or gap junction would constitute a structural connection; a correlation between spike trains would constitute a functional connection; and a predictive relationship from the spike train of neuron J to the spike train of neuron I would constitute an effective connection from neuron J to neuron I. Here we examine effective connectivity to infer the directed transfer of information among hundreds of neurons.
Transfer entropy measure.
There is a large and growing literature on metrics for effective connectivity (Okatan et al., 2005; Hlaváčková-Schindler et al., 2007; Pillow et al., 2008; Gerhard et al., 2011); we selected TE (Schreiber, 2000) because several previous studies (Lungarella et al., 2007; Garofalo et al., 2009; Ito et al., 2011; Vicente et al., 2011; Stetter et al., 2012) indicated that it compared quite favorably with other metrics in terms of detecting connections. As an information theoretic measure, TE is also capable of detecting nonlinear interactions and allows the quantification of information transfer in general units of bits that can be compared across systems.
In neuroscience terms, given the spike trains of neurons J and I, TE is nonzero if including information about the spiking activity of neuron J improves the prediction of the activity of neuron I beyond the prediction based on the past of neuron I alone. The original definition of TE (Schreiber, 2000) was given as follows:
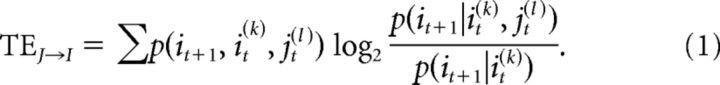 |
Here, it denoted the status of neuron I at time t, and could be either 1 or 0, indicating a spike or no spike, respectively; jt denoted the status of neuron J at time t; it+1 denoted the status of neuron I at time t + 1; p denoted the probability of having the status in the following parentheses; and the vertical bar in the parentheses denoted the conditional probability. The sum was over all possible combinations of it+1, it(k), and jt(l). Probability for a particular triplet state is calculated by dividing the number of occurrences of that state by the total number of observable states. The parameters k and l gave the order of TE, meaning the number of time bins in the past that were used to calculate the histories of neurons I and J, respectively (Ito et al., 2011). Here, we used k = l = 1, so that only single time bins were considered; we used logarithms with base 2 so that our units would be bits. It is important to note that TE is not merely a time-lagged version of mutual information because TE takes into account the auto-prediction provided by the past of neuron I.
Note also that TE, as defined above, would only measure information transfer from neuron J to neuron I at a delay of one time bin. Given that synaptic delays between cortical neurons could span several milliseconds (Barthó et al., 2004; Sirota et al., 2008), we adopted a version of TE that evaluated multiple delays between neurons I and J. To continue to consider the history of the neuron, we kept a one-bin delay for neuron I, assuming that neuron I depended mostly on its closest previous state (Wibral et al., 2015). To account for synaptic delays between neurons, the time bin of neuron J was delayed by d bins (Fig. 1A). Taking these modifications into account, we used a delayed version of transfer entropy, given by the following:
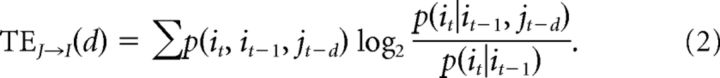 |
Here other terms were as defined in Equation 1. When TE between two neurons was plotted as a function of multiple delays, d, it often showed a distinct peak offset from time 0 (Fig. 1B), because it was likely to be caused by one neuron driving another with axonal delay. We excluded those cases where the peak TE (TEPk) value occurred at time 0, because they were unlikely to be caused by one neuron driving another with axonal delay. We took the nonzero TEPk value to be the single number that represented effective connectivity between two neurons. For calculating TE rapidly over multiple delays, we used the freely available TE toolbox developed by our group (posted at http://code.google.com/p/transfer-entropy-toolbox/; Ito et al., 2011). When this version of TE was applied to a cortical network model for validation, it correctly associated 85% of the total synaptic weights with effective connections. Other toolboxes that calculate transfer entropy are TRENTOOL (Lindner et al., 2011) and MuTE (Montalto et al., 2014).
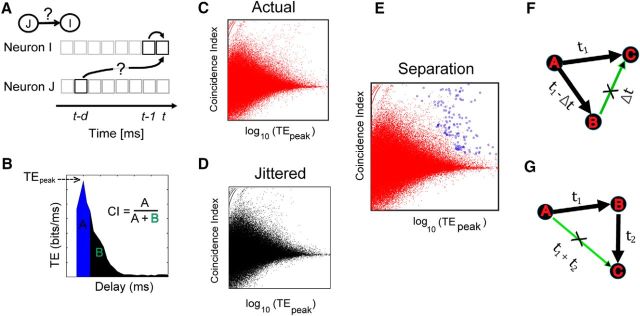
Figure 1.
Overview of transfer entropy analysis. A, To quantify a directed functional link from neuron J to neuron I, we used transfer entropy. It quantifies how much additional information a time bin in the past of neuron J can provide about the current state of neuron I over and above the information provided by the past of neuron I itself. Multiple delays (d) can be used to identify a causal connection. B, Schematic example of transfer entropy plotted as a function of delay. A functional interaction would result in a sharp peak. The sharpness of the peak can be quantified by CI. CI is calculated by evaluating the area formed by a 4 ms region (shaded in blue) centered on the peak and then dividing it by the rest of the area in the plot (shaded in black). Each pair of neurons was characterized by TEpeak and CI. C, Two-dimensional plot of CI and TEpeak for all pairs of neurons in a dataset. D, Two-dimensional plot of CI and TEpeak for all pairs of neurons in the same dataset where spike times of the source neuron have been jittered, as described in the Materials and Methods section. E, Same plot as in C, except the connections that are deemed significant according to the significance testing described in the Materials and Methods section are marked with blue circles. F, Controlling for spurious connections. Neuron A provides common drive to neurons B and C at axonal delays t1 − Δt and t1. As a result, a spurious connection may be detected by the TE analysis from neuron B to C at a delay of Δt. Links that satisfy these delay characteristics were removed from the network. G, Neuron A drives neuron B with a delay t1, and neuron B drives neuron C with a delay of t2. As a result, the TE analysis may detect a spurious link from neuron A to neuron C with delay t1 + t2. All transitive links between triplets of neurons that satisfied these delay characteristics were removed from the network. This last step of spurious link removal generated the final networks on which the network analysis was performed.
Controlling for firing rate.
The information contained in a spike train is provided by the following two components: spike rate and spike timing. In this study, we were especially interested in the component provided by spike timing. To ensure that we primarily measured information from this component, we compared TE measured from spike trains in the actual data to TE measured from jittered spike trains. We measured randomized estimates of TE for each pair of neurons by jittering spikes from the source spike train (J) while keeping the target spike train (I) fixed. This preserved the auto-prediction for the target spike train and only changed the prediction of the target spike train that was provided by the source spike train. The values by which spike times were jittered lay in the range 1–19 ms and were drawn from a normal distribution. As described in our previous study (Shimono and Beggs, 2015), we found that values of TE from cortical data showed a sharp decrease after jittering up to 19 ms, and then remained relatively constant for jitter values of >19 ms. This suggested that TE produced by timing had a resolution of ≤19 ms, and that TE produced by rate had a resolution coarser than 19 ms. We also note that direct synaptic connections in cortex have delays that typically range from 1 to 20 ms (Swadlow, 1994). This jittering method only changed firing times, leaving firing rates and the general form of the interspike interval histogram largely unchanged. With this procedure, we defined IT between a pair of neurons as follows:
where raw and jit indicated unjittered and the average of the jittered data, respectively. Here TEjit was produced by taking the average value of TE among 100 jittered versions of spike train J and the unjittered version of spike train I, at the particular delay where the raw TE peaked. Only positive values of information transfer were used in the analysis. In this manner, we aimed to control for the effects of firing rate on information transfer.
Controlling for network drive.
In addition to the control for firing rate effects outlined above, we also controlled for possible effects produced by network drive, such as might occur during a network burst. During a burst, the firing rates of many neurons can be elevated simultaneously, potentially leading to an increase in coincident spike firings. These increased coincidences could produce an increase in TE. To control for this, we exploited the fact that direct synaptic connections tend to produce relatively narrow peaks (∼5 ms) when TE was plotted against delay, while network bursts tend to produce relatively broad peaks (50–200 ms; Beggs and Plenz, 2003). To measure how narrow a peak was, we used the coincidence index (CI; Ito et al., 2011; Shimono and Beggs, 2015). Intuitively, the CI measured the tendency for TE values to peak sharply at a particular synaptic delay between neurons (Fig. 1B). The CI has been used to identify connections in the context of cross-correlation studies (Juergens and Eckhorn, 1997; Jimbo et al., 1999; Chiappalone et al., 2006). We defined the CI as follows:
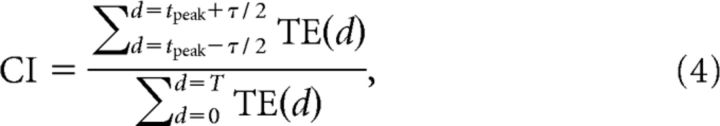 |
where TE(d) was the transfer entropy measured at delay d, tpeak was the delay where the TE value peaks, τ was the coincidence window size, and T was the entire window size of the measure. We used a value of T = 30 ms for the entire window size, as this encompassed the range of monosynaptic delays (1–20 ms) reported in cortex (Swadlow, 1994). We used a value of τ = 4 ms, as this reflected the typical width of peaks found in the data.
Thus, direct connections were expected to have both (1) high values of TE above those produced by jittered data and (2) high values of CI above those produced by common network drive. To select for connections that simultaneously satisfied both of these criteria, we made scatterplots of CI against log10(TEpeak) for all pairs in both unjittered and jittered data, as shown in Figure 1, C and D. When they were plotted together on the same graph as shown in Figure 1F, the jittered data did not extend into the region where both CI and log10(TEpeak) values were simultaneously high, whereas some of the actual data did occupy this region. Intuitively, this upper rightmost region of the plot was populated by connections with tall (high TE) and narrow (high CI) peaks in the TE–delay curve that were unlikely to be the result of merely the firing rate or the network drive. The upper rightmost portion of the jittered data thus formed a decision boundary that allowed us to distinguish significant connections from those produced by chance (Fig. 1E). We found that when this CI versus log10(TEPk) plot was divided at a resolution of 25 × 25 pixels, this decision boundary was smooth and closely followed the contours of the jittered data distribution. The dimensions of this pixel plot were chosen to minimize the squared error between the actual data and the binned histogram. To minimize the error, we followed Terrell and Scott (1985), who noted that the number of pixels needed to be greater than (2n)1/3 (where n is the square root of the number of neurons). Because the maximum value of n in our data was 22.5, we selected 25 as the resolution of the pixels.
Next, we had to decide which pixels in the CI versus log10(TEPk) plot to include. To do this, we defined the maximum fraction of connections in each pixel that were allowed to come from jittered data. We called this measure the rejection threshold (RT), which is given by the following:
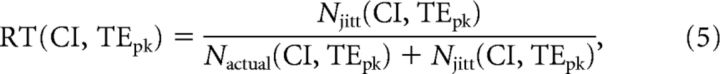 |
where the number of data samples at each pixel was denoted by Nactual for actual data, and by Njitt for jittered data. If a pixel contained connections from jittered data that were below this fraction, all connections from the original data in this pixel were deemed significant. Thus, RT gave the maximum allowable fraction of connections in any pixel that came from randomized data. Note that the majority of pixels that were accepted had a fraction of connections from jittered data that were far below the selected RT. The range of RT values used was determined on the basis of the performance of the TE analysis method in picking out synaptic connections on spiking data generated from a cortical network model where the connectivity was known beforehand. We describe the details of this cortical network model in a dedicated subsection below. The RT value was chosen to maximize the ratio TPR/FPR, where TPR is the true-positive rate and FPR is the false-positive rate. Using the cortical network model, where the connections between neurons were known, we found that this ratio was maximized at an RT of 0.37. To demonstrate robustness, we also examined RT values in the range 0.35–0.50 and found that key network features found in our data were qualitatively similar over this range. Using these methods, we attempted to control for network drive effects.
Controlling for spurious connections caused by common drive and transitivity.
Besides network drive, common drive and transitive effects can confound a pairwise measure (such as the TE we are using here) in the following ways. First, if neurons B and C both receive common drive from neuron A, such that the drive of neuron A to B arrives Δt before the drive of neuron A to C (Fig. 1G), it is possible that the putative connection from neuron B to neuron C, with delay Δt, is spurious. Second if neuron A drives neuron B at a delay (t1) and neuron B drives neuron C at a delay of t2, then there could be a spurious connection from neuron A to C with a delay of t1 + t2, which we called the transitive effect (Fig. 1H). Recent work (Yatsenko et al., 2015; P. Wollstadt, U. Meyer, M. Wibral, unpublished observations) has tried to take into account spurious connections that may be present in pairwise measures. We have used a simpler post hoc scheme to correct for common drive and transitive effects in triplets of neurons. For each neuron (let us call it the source neuron) in the network, we looked at the target neurons that received connections from this neuron. If the target neurons had connections between themselves, we examined whether the delays associated with these connections were equal to the difference of the delays for the links originating from the source neuron. If they were equal, we eliminated those edges from the network as they could possibly be the result of common drive. This was repeated for all other neurons in the network yielding a corrected IT matrix. To correct for transitive edges, we looked at chain connections (A → B → C), and, if there was a connection between A and C, we examined whether the associated delay was the sum of the delays of the connections from A to B and from B to C. In that case, we eliminated the link between A and C as a spurious transitive link. We performed this transitive link removal for all triangular motifs that satisfied the above criteria and generated a corrected IT matrix. The process was repeated 1000 times each time following a different order of the neurons to remove possible order-dependent effects. Finally, the connections that appeared 90% of the time across all of the 1000 corrected networks were used to construct the final common drive corrected or transitive effect corrected matrix of interactions between the neurons. For the in vitro datasets, after eliminating possible spurious connections of either nature, connection density changed by 45 ± 14%, and for the in vivo datasets the connection density changed by 10 ± 5%. The network properties that we report here (e.g., highly concentrated information transfer, rich club) were found to be robust with respect to the elimination of these spurious edges. We note that this procedure was conservative, as many of the connections that it removed might not have been spurious (e.g., a pair of neurons could have both a direct connection and a connection through a third neuron). By this procedure, we ensured that our results were controlled against potential spurious connections.
Validation of effective connectivity.
We used a cortical network model to examine the validity of these methods and to select an appropriate value for the RT. The model used here was composed of 625 Izhikevich model neurons (Izhikevich, 2003). Excitatory neurons took on graded parameterizations between the regular spiking, intrinsically bursting, and chattering regimes (with a strong bias toward regular spiking). Similarly, inhibitory neuronal dynamics were selected using randomized values placing each neuron between the fast-spiking and low-threshold spiking regimes. Inhibitory neurons made up 20% of the total neuronal population. For excitatory (inhibitory) neurons, Inoise was drawn from a normal distribution with a mean of 0 and an SD of 5.0 (2.0). All neurons were placed at random locations within a three-dimensional cube, with the probability of their being a connection between two neurons given as follows (Maass et al., 2002):
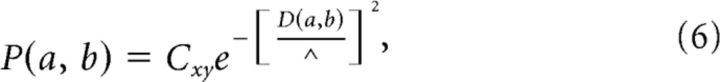 |
where D(a, b) is the Euclidean distance from neuron a to neuron b, where Λ is a parameter that controls both the average number of connections and the average distance between neurons that are synaptically connected, and Cxy is a constant that takes on different values based on the polarity of neurons a and b. This allowed us to control the relative ratios of connections that exist between inhibitory and excitatory neurons. The various C values were set to the following: CEI = 0.4, CEE = 0.3, CIE = 0.2, and CII = 0.1, where CEI represents the value of the constant Cxy for connections between excitatory and inhibitory neurons and CEE, CIE, and CII are defined accordingly. This resulted in an absolute connection density of 4%. Synapses were given temporal delays based on the distance between their presynaptic and postsynaptic cells, with a mean delay of 3.5 ms. Excitatory (inhibitory) synaptic efficacies were randomly drawn from a lognormal distribution with a location parameter of −1.5 (−0.8) and a scale parameter of 1.25 (1.3). The final major difference between this model and the model presented in the study by Izhikevich (2003) was that each postsynaptic response was modeled as an exponential decay from the following equation:
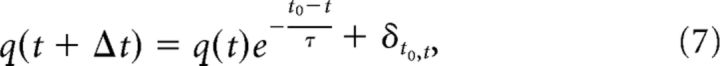 |
where t0 is the time of arrival for the postsynaptic response (the spike time of the presynaptic cell, plus the delay assigned to the synapse in question), ô is a decay time constant set to 3 ms (6 ms) for excitatory (inhibitory) synapses, and δt0,t is the Kronecker δ function (δt0,t = 0 if to ≠ t; δt0,t = 1 if to = t). This model was used to generate spiking activity for 1 h, which was approximately equal to the duration of the in vitro and in vivo recordings. From the model data, we applied the above methods to quantify their performance in detecting existing synaptic connections. This performance was then used to set the level of the RT that was used on the actual data.
Graph theory measures
Graph-theoretic measures were calculated by using algorithms written by the authors, and some of them can be found in the publicly available Brain Connectivity Toolbox (www.brain-connectivity-toolbox.net; Rubinov and Sporns, 2010). To introduce terminology, we begin with the binary directed adjacency matrix A. The element aij of the matrix A represented the presence or absence of a link from neuron i to neuron j and was 1 or 0. W was the corresponding information transfer matrix, where element wij represented the IT value directed from neuron i to neuron j.
Cumulative information transfer.
To examine the distribution of information transfer in the neuronal population, we made cumulative information transfer plots. For these, the neurons were arranged along the x-axis in descending order of their total outgoing (or incoming) IT values. The total outgoing IT for neuron i was found by taking a sum along row i in the weight matrix W. Incoming IT was found by taking a column sum. At every point along the x-axis, the total normalized cumulative value of IT was plotted for the percentage of neurons up to that point. This procedure was used for making Figure 4.
Figure 4.
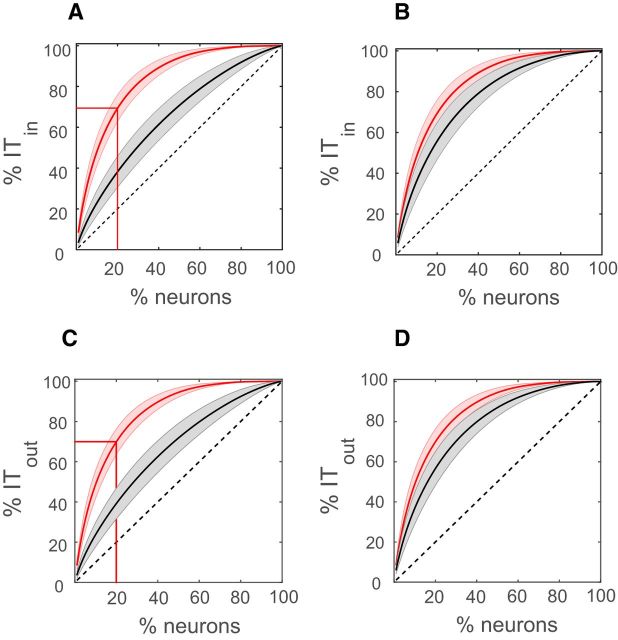
Nonuniform information transfer in cortical networks. A, Cumulative distribution of incoming IT averaged over all datasets (thick curved red line). The shaded (light red) region shows the variation (SD) across datasets. The thick black line represents the average cumulative distribution of incoming IT for a null model where IT values were randomly allocated to neuron pairs from the pairwise IT distribution. The shaded light gray region represents the variation (SD) across datasets. The horizontal and vertical red lines show that 20% of the neurons contributed ∼70% of the total information transfer. B, Same as plot A, except that the thick black line represents the average cumulative distribution of incoming IT for a null model in which weights were allocated randomly, keeping the in and out degree distribution fixed. Here, 20% of the neurons carried approximately the same amount of outgoing IT as in the actual networks. C, D, Same as in A and B except we plot the cumulative outgoing information instead of the cumulative incoming information. The dotted black diagonal line in each of the plots represents what the cumulative information transfer would look like for a completely uniform network where each neuron contributed equally to the total information transfer.
Rich-club coefficient.
Previous work has shown that some neurons have more connections, or stronger connection, than other neurons (Bonifazi et al., 2009; Shimono and Beggs, 2015).The fact that some neurons are “richer” than others naturally leads to the question of how these rich neurons are connected to each other. If rich neurons are more densely/strongly connected to each other than expected by chance, then a (unweighted/weighted) rich club (Zhou and Mondragón, 2004; Colizza et al., 2006; Opsahl et al., 2008) is said to exist (Fig. 2A). Here we describe the methods we used for probing the existence of rich clubs in these data. We closely followed the approach of van den Heuvel and Sporns (2011) to determine the rich-club coefficients. Although a very recent article (Alstott et al., 2014) introduced a new definition of the rich-club measure, here we used the older, and most commonly used, definition of the rich club.
Figure 2.
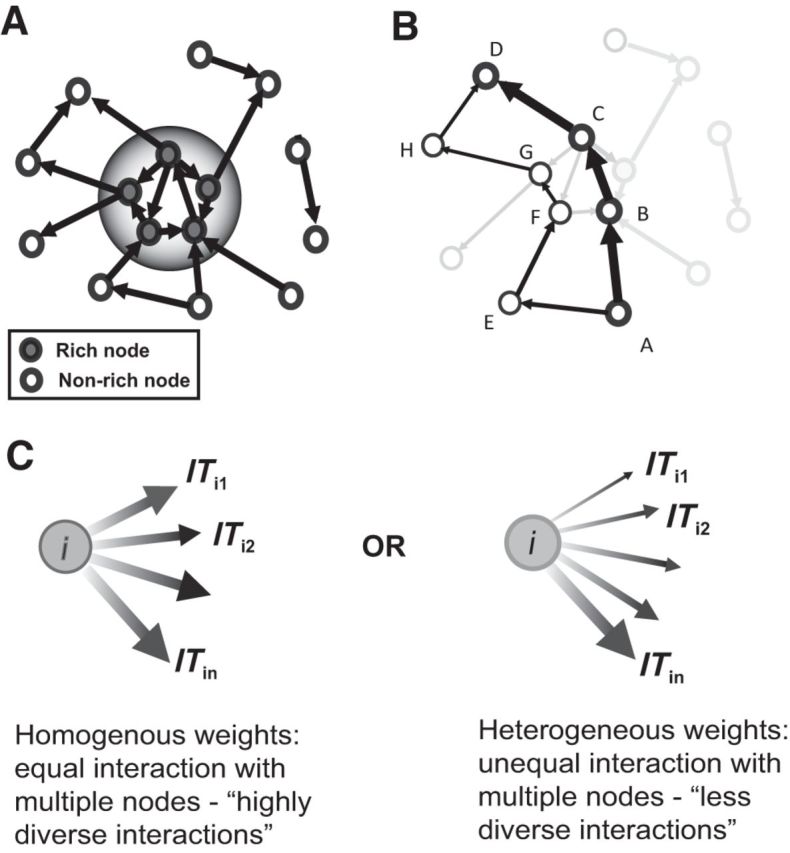
Schematic description of graph-theoretic measures. A, Representation of a rich-club topology in a toy network. The neurons represented by filled gray circles are rich according to a certain chosen parameter, while the neurons represented by filled white circles are nonrich neurons. A rich club is said to exist if the rich neurons preferentially connect to each other more strongly and densely with each other than with the rest of the network. The shaded bigger circle separates this rich-club group from the rest of the network. B, The shortest path in a network is the path involving fewest edges between a source and a target neuron of all possible existing paths. For example, the shortest path from neuron A to D (thick black arrows) goes through neurons B and C. Another alternative path (thin black arrows) exists between neurons A and F, and it goes through neurons E, F, G, and H, which is a longer path in terms of the number of links traveled. A neuron or a link is said to have a high betweenness centrality if a large fraction of shortest paths of the network between all neurons in the network goes through that neuron or link. C, Here we consider the nature of the distribution of outgoing strengths of a neuron. In one case, all of the outgoing connections from neuron i are approximately of the same strength. Hence, this neuron has a homogeneous outgoing weight distribution. It interacts equally with different targets. This makes it more diverse in its interactions. In another case, the same neuron i has outgoing strengths, some of which are very strong, some are medium, and some are very weak in strength. This neuron has a heterogeneous distribution of outgoing weights. Hence, it does not interact with its target nodes equally, making it less diverse. The diversity metric aims to quantify the differences in these two scenarios. A node having a homogenous outgoing weight distribution would have a higher diversity value than a node that has a heterogeneous outgoing weight distribution.
A weighted rich-club analysis (Opsahl et al., 2008) was performed, with the total outgoing and incoming IT from and into each neuron as the richness parameter. The analysis was performed on the largest weakly connected component of the network. The largest weakly connected component of a network was composed of neurons that could be reached from any other neuron following an existing link without taking directionality into account. The weighted rich-club coefficients were then calculated in the following way.
First, a list (ITrank) of the pairwise IT values in the network that were found to be significant was created, ranked from largest to smallest (ITmax, IT2…ITn−1, ITmin). We defined the richness parameter r to be the normalized total outgoing IT from each neuron. Then a list of the unique values of this richness parameter r, was constructed from smallest to largest (rmax, r2…rn−1, rmin). Second, we looked at the subnetwork where each of the neurons had a richness parameter, >rk. We counted the number of edges in that subnetwork and called it E>rk. Then we summed all the pairwise IT values between this subset of neurons and called it W>rk. The weighted rich-club coefficient Φactw(rk) was defined as the ratio between W>rk and the sum of the E>rk strongest pairwise IT values in the network obtained from the list (ITrank).
 |
The above ratio represents what fraction of the strongest weights in the whole network is present in the subnetwork. Third, to observe how much the observed coefficient was above chance, we generated 1000 randomized versions of the actual networks such that the richness parameters of the neurons were unchanged and the largest connected component of the network also remained the same. The weighted rich-club coefficients were also calculated for these randomized networks Φrandw(rk), and the normalized rich-club coefficient for a particular richness parameter rk was given by the following:
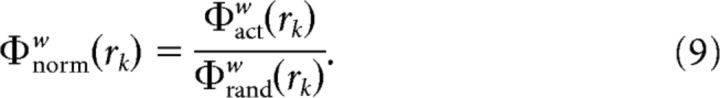 |
The normalized rich-club coefficients were determined for each value of the ranked richness parameter (rmin, r2…rk−1, rmax). If Φnormw was >1 for a range of the richness parameter, then a rich club existed in that regime of the richness parameter. To determine the statistical significance of the rich-club coefficients, the coefficients obtained from the actual network were compared with the null distribution of rich-club coefficients generated from the 1000 randomized networks, and a one sided p value was generated. Fourth, to correct for multiple comparisons over the range of values of the richness parameter, false discovery rate correction (Benjamini and Yekutieli, 2001) was performed, limiting the false discovery rate to 0.05.
Betweenness centrality.
Measures of node and link centrality attempt to quantify the global importance of individual neurons and links between pairs of neurons (Freeman, 1979; Brandes, 2001). We used node and edge betweenness centrality to characterize the importance of the neurons and links in the network. Betweenness centrality is closely tied to the concept of shortest path in a network. The shortest path is the minimum number of edges that one has to travel to go from one neuron to another. A neuron is said to have a high betweenness centrality if many of the shortest paths of the network go through that neuron. Similarly, an edge has a high betweenness centrality if many of the shortest paths of the network go through that particular edge (Fig. 2B). Since we had weighted directed connectivity matrices, we used the weighted versions of betweenness centrality (Rubinov and Sporns, 2010). Weighted betweenness centrality was computed by inverting the normalized weights of the connections to create a distance matrix, where stronger connections implied smaller distance.
Dynamic importance.
To quantify the importance of neurons in a network, we used dynamic importance (Restrepo et al., 2006). Given the adjacency matrix of the network, we found the largest eigenvalue of the matrix, λ, which, according to Perron's theorem, is real and positive. The largest eigenvalue of the adjacency matrix has been shown to determine important aspects of the dynamics of complex networks, like the critical coupling strength at which transition occurs from incoherence to coherence in a network of coupled oscillators (Restrepo et al., 2005), the emergence of a giant connected component in the case of weighted percolation in complex networks (Restrepo et al., 2008), and whether a network will have chaotic, critical, or damped responses to perturbation (Larremore et al., 2011). The dynamic importance of a neuron (Ik) was defined as fractional change in the largest eigenvalue of the adjacency matrix upon removal of the neuron from the network. Similarly, the dynamic importance of an edge (Iij) connecting two neurons (i and j) was the fractional change in the maximum eigenvalue upon removal of the edge from the network, as follows:
 |
 |
Neuron diversity.
To quantify how heterogeneous or homogenous the outgoing/incoming links were in terms of their strengths (Fig. 2C), we used the diversity metric (Eagle et al., 2010). It was defined as follows: let neuron i have k outgoing connections, the strength of connections being ITi1, ITi2…ITik. We then normalized these IT values by their sum, as follows:
 |
These normalized values were then used to define the diversity of neuron i, as follows:
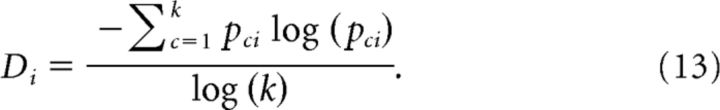 |
If all the outgoing IT values were approximately of the same value (i.e., they were homogenous), then Di was close to its maximum value of 1. This implied that if a neuron had the same amount of interaction with a large number of other neurons, then it was maximally diverse and minimally heterogeneous. If the values of the outgoing IT values were different, then Di would assume a value <1. In this case, the neuron was less diverse because it had different levels of interaction with different neurons. Hence, higher diversity values indicated a homogenous set of incoming/outgoing IT values, and lower values indicated a more heterogeneous set.
Results
The Results are composed of three main sections. First, we describe the effective connectivity obtained from TE and the pattern of highly concentrated information transfer. Second, we calculate various graph-theoretic measures for weighted networks to show that there is “central/rich” core in the network. Third, we describe the properties of the “rich” neurons, the connections between them, and how these differ from what is found in the rest of the network.
Effective connectivity from transfer entropy analysis
The average number of neurons detected by spike-sorting analysis was 347 ± 119, with the maximum number being 500. Figure 3A shows the positions of identified neurons (filled purple circles) overlaid on the rectangular micro-electrode array (white rectangle) in a representative experiment. The duration of these recordings was ∼1 h, and the average firing rate of the neurons across all cultures was 2.61 ± 1.27 Hz, which remained fairly constant over the duration of the recording. Similar to what has been reported in vivo, the distribution of firing rates was found to be approximately lognormal (Fig. 3B). Transfer entropy analysis was performed on the spike trains of the detected neurons to generate effective connectivity maps of the networks. We obtained both binary and weighted effective connectivity matrices for these networks. The resulting networks were found to be sparse, having an average connection density of 6.1 ± 4.7%. We found that the distribution of pairwise IT values was heavy tailed, with a few strong links and a large number of weak links. The logarithms of the pairwise IT values were found to approximately follow a normal distribution (Fig. 3C), which implied that the IT values had an approximately lognormal distribution. This was similar to the distribution of synaptic strengths reported from acute slice data (Song et al., 2005). Thus, in terms of firing rate distribution, connection strength distribution, and sparsity, the data from these slice cultures were similar to what has been reported in acute slices and in vivo preparations.
Figure 3.
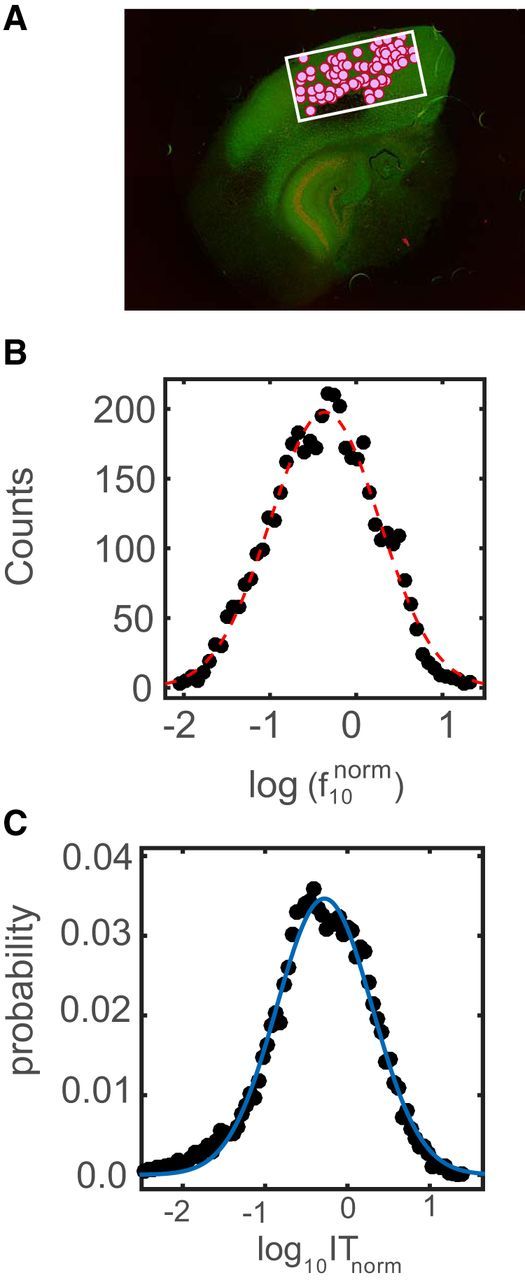
Lognormal distributions of firing rates and pairwise IT values. A, Stained organotypic culture tissue with the array (white rectangle) and neurons identified after spike sorting (filled purple circles). B, Distribution of the logarithm of the pooled normalized firing rates (N = 15 slices). The firing rates of all neurons in a particular dataset were divided by the mean firing rate of the dataset to obtain normalized firing rates for each neuron. The black dots represent the actual distribution, while the dotted red line represents the Gaussian fit generated by the curve fitting toolbox in MATLAB. C, Distribution of the logarithm of the pooled pairwise IT values (N = 15) normalized by the mean IT of each dataset. The black dots represent the actual distribution, while the solid blue line represents the Gaussian fit generated by the curve fitting toolbox in MATLAB.
Highly concentrated information transfer
We next sought to quantify how these few strong links were distributed among different neurons. This question was important because a previous model (Koulakov et al., 2009) predicted that for a lognormal firing rate distribution and a lognormal weight distribution to coexist, some neurons would have to receive much stronger net inputs than others. Also, they showed that a Hebbian learning rule would not only generate strong inputs to some neurons but would also cause some neurons to send out much stronger net outputs than others. Accordingly, we calculated the cumulative distribution of total incoming IT (Fig. 4A, solid red lines) and total outgoing IT (Fig. 4C, solid red lines) from each neuron averaged over all 15 datasets. We found that ∼20% of the neurons contributed to ∼70% of the total incoming/outgoing IT, suggesting that some neurons were richer than others in terms of how much information they received and how much they sent out. This is shown in (Fig. 4A,C), where neurons are sorted in descending order of total incoming/outgoing IT contribution. To check whether this nonuniformity was different from what could be expected by chance, for each dataset we assigned the calculated pairwise IT values to randomly chosen pairs of neurons. This preserved the approximately lognormal pairwise IT distribution but randomized any possible correlations between weights. This procedure also randomized the degree of distribution. The resulting average cumulative distribution of outgoing/incoming IT (N = 15) from the random assignment of weights was found to be significantly different [Kolmogorov–Smirnov (KS) test; p < 10−6] from the cumulative distribution found in the actual datasets. In the null model, 20% of the neurons accounted for ∼40% of the total incoming/outgoing IT (Fig. 4A,C, solid black lines) in the network, which was significantly less than the nonuniformity observed in the actual networks. This result showed that a lognormal distribution of information transfer values coupled with a random network topology could not account for the nonuniformity observed in the actual datasets. To explore a possible mechanism underlying this nonuniformity, we next looked at the contribution of the particular network topology. To do this, we randomized the weights in the networks, but preserved both the pairwise IT and degree (out and in) distributions. In this case, there was no significant difference between the cumulative curves of the actual data and the null model (KS test, p = 0.09; Fig. 4B,D) for both incoming and outgoing IT. From this, we conclude that the network topology observed in the datasets played an important role in producing the nonuniform pattern of both outgoing and incoming information transfer.
Edge effects and subsampling effects in observed nonuniformity
It could be argued that because of the limited size of the array, potential biases could produce artifacts in the structure of the estimated network. These could be quantified in terms of (1) edge effects, in which rich neurons could potentially be concentrated toward the center of the array (rich neurons on the edges would have invisible connections and would not be seen as rich); and (2) subsampling effects, in which the observed nonuniformity could be a result of observing only a small part of the network. To check whether this was the case, we calculated the correlation coefficient between the distance of a neuron from the center of the array and its total degree. We did not find a significant correlation, implying that high-degree neurons were not concentrated at the center of the array. To check for bias created by subsampling, we created a random network of 50,000 neurons with the same connection density as observed in the actual networks. We also constructed the networks so that each neuron sent out approximately the same amount of information, which made the networks close to being uniform. In these uniformly constructed networks, we observed that 20% of the neurons accounted for ∼30% of the total information transfer. Then we randomly subsampled from these networks the same number of neurons as observed in our datasets 1000 times. We calculated the average cumulative outgoing IT curves for these subsampled networks. We found that subsampling introduced some amount of nonuniformity in the cumulative curves. Here, 20% of the neurons carried ∼45% of the total information (15% increase in nonuniformity) but could not account for all of the nonuniformity observed in the actual datasets, where 20% of the neurons accounted for ∼70% of the information transfer. From this, we conclude that the effect of nonuniform IT is unlikely to be a result of subsampling from a larger, more evenly distributed network.
Graph-theoretic analysis
The above results confirmed that the information transfer weights were unevenly distributed, but they did not tell us how neurons with high total weights connected to each other. To probe this issue, we turned to graph-theoretic measures.
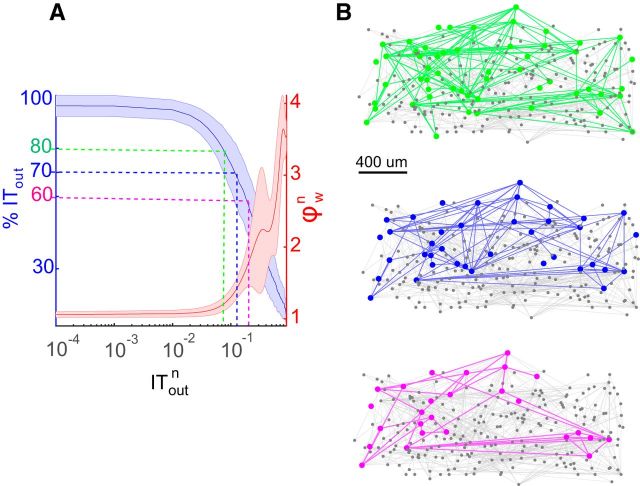
To look at how the rich neurons in the network interacted with each other, we calculated the weighted rich-club coefficient for all datasets (Fig. 5A). The richness parameter was chosen to be the total outgoing IT from each neuron. From the plots, we see that there is a regime of the richness parameter where the normalized coefficient was >1, indicating that the actual rich-club coefficients were significantly greater than those found in the randomized networks, where the significance testing was performed as described in the Materials and Methods section. Hence, in this regime the rich neurons were connected with each other more strongly than would be expected by chance. Figure 5A also shows that the subnetwork formed by the rich neurons, as defined in the previous section (70% cumulative outgoing information transfer), has a rich-club coefficient of >1. We also observed similar rich-club topology if the richness of the neurons was classified according to the total IT flowing in instead of out. Hence, neurons that received a large amount of net incoming information were strongly connected to neurons that also received a large amount of incoming information. Figure 5B shows the rich-club network topology in one representative effective connectivity network from one of the cultures for three different thresholds picked for defining rich neurons. We saw similar rich-club properties in networks of in vivo recordings in the mouse orbitofrontal cortex (Fig. 6A). The mean (±SD) number of neurons in the datasets was 141 ± 35. In these networks, subnetworks of rich neurons were more strongly connected to each other than expected by chance. This was evident from the rich-club coefficient being >1 over a range of values of the richness parameter (Fig. 6B).
Figure 5.
Rich neurons interact strongly with each other, forming a rich club. A, Weighted rich-club coefficient (y-axis) for different values of the normalized richness parameter (x-axis), which is the total outgoing IT from each neuron. The red line represents the average rich-club coefficient across all datasets, and the shaded pink region represents the SD. We also plotted the cumulative outgoing information (blue curve) for the various subnetworks generated at different values of the richness parameter. The shaded region represents the SD of the values for all the datasets. The dashed lines (green, blue, and magenta) are drawn to show that subnetworks that cumulatively account for 80%, 70%, and 60% of the total outgoing information all have rich-club coefficients of >1. B, Effective connectivity maps showing rich-club topology for one representative culture. The three maps correspond to three definitions of rich neurons. For the top right network, neurons that cumulatively contribute 80% of the total outgoing information are classified as rich. Those neurons are represented by filled green circles, and connections between them are represented by green lines. All other connections are labeled with a gray line, and the nonrich neurons are represented by filled gray circles. Similarly for the same network, in the middle right panel, neurons that cumulatively contribute 70% of the total outgoing information are defined to be rich and are represented by filled blue circles, and connections between them are represented by blue lines. Finally, in the third network we define neurons to be rich if their cumulative contribution is 60% of the total outgoing information. The networks are represented as undirected for the sake of clarity.
Figure 6.
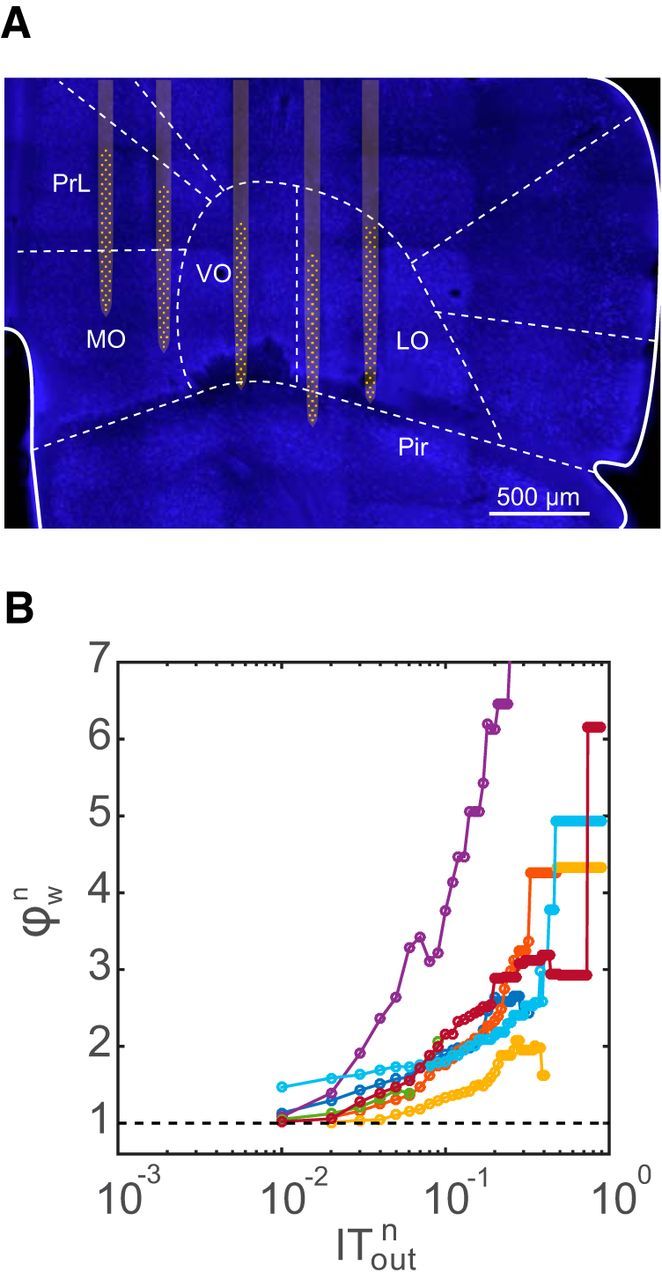
Rich-club structure in vivo. A, Placement of a 256-electrode recording array in the mouse orbitofrontal cortex and surrounding regions. Image shows a brain section with neuronal nuclei fluorescently labeled with a NeuN antibody (blue). The electrode positions were determined from tracks made by the silicon prongs visible in the sectioned tissue. For clarity, the five silicon prongs of the electrode array are superimposed on the image together with the recording sites (yellow). PrL, Prelimbic cortex; MO, medial orbitofrontal cortex; VO, ventral orbitofrontal cortex; LO, lateral orbitofrontal cortex; Pir, piriform cortex. B, Rich-club plot for each in vivo dataset (N = 7) for a richness parameter equal to the normalized total outgoing IT. Note the similarity with the rich-club plot for in vitro data in Figure 5A.
Properties of the rich subnetwork
Now that graph-theoretic measures identified a subnetwork of rich neurons, we turned to further investigate the properties of rich neurons to see whether they differed from those of other neurons. As shown in Figure 4A, we defined the rich subpopulation of the network to be made up of those neurons whose cumulative contribution to the total outgoing IT in the network was ∼70%. In each dataset, these subnetworks had a rich-club coefficient of >1 and hence formed a rich club. In the following sections, we look at the properties of these rich neurons. The reported properties were robust with respect to the percentage of the cumulative contribution that was chosen to define rich neurons. We looked at two other values: 60% and 80%. The properties reported below were the same in this range and were not limited to merely the cumulative contribution value of 70%.
Rich neurons are the most active in the network
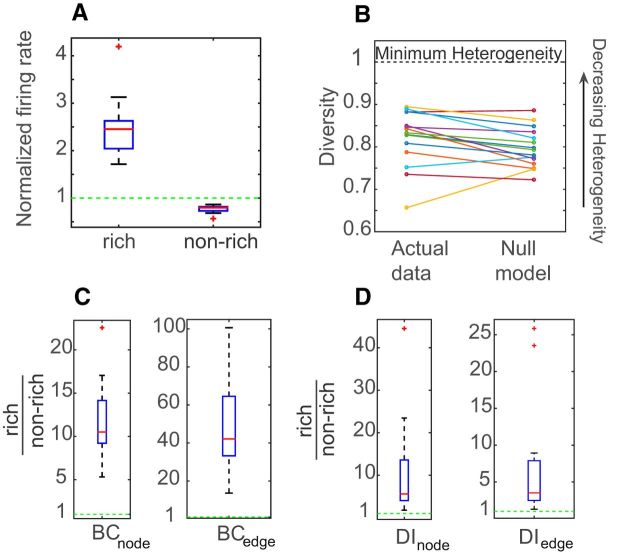
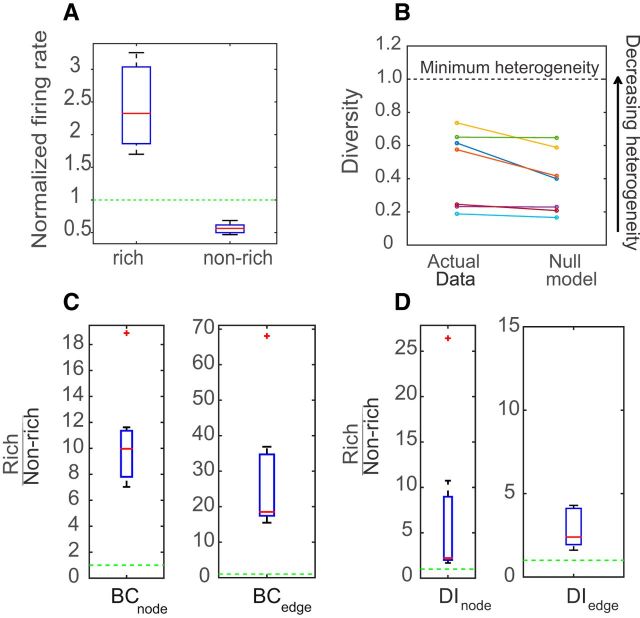
The firing rates of the rich neurons and nonrich neurons were calculated and were normalized by the mean firing rate of the dataset to allow comparison. A two-sample KS test showed that the distributions of firing rates between these two populations were significantly different (p < 10−4), with the rich neurons having a higher firing rate than the nonrich neurons. To summarize the results for all datasets, we pooled together the mean normalized firing rate of rich and nonrich neurons from each dataset and plotted the values in a box plot (Fig. 7A). The rich neurons on average were found to have more than two times (2.4 ± 0.6) the mean firing rate of the networks, while the nonrich neurons had ∼0.8 times (0.77 ± 0.07) the mean firing rate of the networks. We observed very similar results in the in vivo networks as well (Fig. 8A).
Figure 7.
Distinctive properties of rich-club neurons in in vitro networks. A, Box plot of mean normalized firing rates for rich neurons and nonrich neurons for N = 15 in vitro datasets The firing rate of each neuron in each dataset was normalized by the mean firing rate of the whole dataset. B, Mean diversity of rich neurons is plotted for each dataset along with the mean diversity of rich neurons in the null model, as defined previously in the Materials and Methods section. Higher values of diversity indicate less heterogeneity in the distribution of outgoing weights. C, Boxplot (from left to right) of ratios of neuron betweenness centrality (left) and edge betweenness centrality (right) for rich neurons to that of nonrich neurons. D, Boxplots of pooled ratios of the mean dynamic importance of rich neurons/edges to nonrich neurons/edges.
Figure 8.
Distinctive properties of rich-club neurons in in vivo networks. A, Box plot of mean normalized firing rates for rich neurons and nonrich neurons for N = 7 in vivo datasets. The firing rate of each neuron in each dataset was normalized by the mean firing rate of the whole dataset. B, Mean diversity of rich neurons is plotted for each dataset along with the mean diversity of rich neurons in the null model as defined previously in the Materials and Methods section. Higher values of diversity indicate less heterogeneity in the distribution of outgoing weights. C, Boxplot (from left to right) of ratios of neuron betweenness centrality (left) and edge betweenness centrality (right) for rich neurons to that of nonrich neurons. D, Boxplots of pooled ratios of the mean dynamic importance of rich neurons/edges to nonrich neurons/edges.
Spike-timing contributions to rich club
The above results might suggest that the strong connections between rich-club neurons are simply the result of the high firing rates of these neurons. This could be expected, to some extent, as high-firing rate neurons have higher entropy rates than low-firing rate neurons. We observed that there was indeed a positive correlation (Pearson's correlation coefficient, r = 0.28; p < 10−4) between the significant pairwise IT values and the average firing rates of the source and target neuron pairs. Recall, however, that we defined IT as the difference between the actual and the jittered TE values (Eq. 3) in an effort to control for firing rate effects. Thus, all of the connection strengths reported here were the result of spike timing at a scale of ≤19 ms. Although rich-club neurons indeed tend to have high firing rates, the rich-club structure identified here is not caused by them, but is caused by information contained in spike timing around the 20 ms scale.
Rich neuron connections are not more diverse
Diversity analysis on the outgoing connections of the rich neurons showed that their diversity coefficients were less than unity, implying a heterogeneous outgoing IT distribution (Fig. 7B). To check whether the strength of the outgoing connections of the rich neurons were being sampled from the entire range of the IT distribution, we created a null model where each neuron had the same fixed out degree as in the actual data but was allowed to sample freely from the available weights. The average mean diversity of rich neurons was not found to be significantly different than that in the null model (Wilcoxon rank sum test, p = 0.9).We observed the same results in the in vivo networks as well (Fig. 8B). This shows that the rich neurons do not become rich by just grabbing on to the strongest links in the network. Rather, they sample from a wide range of weights, as explained in Figure 2C.
Rich neurons are highly central to the network
To assess the importance of the rich-club neurons and the connections between them, we calculated the weighted neuron and edge betweenness centrality of the rich and the nonrich neurons (Fig. 7C). A significantly higher number of shortest paths went through the rich neurons and through the edges connecting the rich neurons to one another, than through the nonrich neurons or edges connecting nonrich neurons to other nonrich neurons. We observed similar results in the in vivo networks as well (Fig. 8C). These results indicate that rich neurons and their connections are positioned so that they could facilitate efficient communication in cortical networks.
Rich neurons are dynamically important
In all 15 in vitro datasets, the mean dynamic importance of rich neurons and edges was significantly higher than that of nonrich neurons and edges (KS test, p < 10−4). We also calculated the ratio of the mean dynamic importance of the rich neurons and edges connecting rich neurons to each other to that of nonrich neurons and edges connecting nonrich neurons to each other for all datasets (Fig. 7D), and observed that the ratios were greater than unity. We observed the same results in the in vivo networks as well (Fig. 8D). This shows that rich neurons and their connections are very influential to the dynamics of the network.
Discussion
The main result of this work is that information transfer in local cortical networks is highly concentrated. Some neurons transfer much more information than others, and these neurons in turn connect with each other more than chance, forming a rich club. This causes 70% of the total transfer entropy to pass through only 20% of the neurons in local cortical networks.
Validity
The in vitro networks self-organized to a state where they spontaneously produced an approximately lognormal distribution of firing rates, as observed in vivo (Hromádka et al., 2008). In addition, they had an approximately lognormal distribution of information transfer strengths, as seen for synaptic strengths in paired patch-clamp studies (Song et al., 2005). Our previous work in slice cultures (Shimono and Beggs, 2015) has shown that the patterns of information transfer between clusters of six to eight neurons are qualitatively similar to the patterns of synaptic connectivity reported in acute slice experiments (Perin et al., 2011). Furthermore, we also analyzed in vivo data from rodent cortex. Although these data generally had fewer total neurons, and sometimes had larger interelectrode spacing, we obtained results that were qualitatively similar to those found in vitro. In addition, we checked that our results were not confounded by edge effects, subsampling, firing rate effects, or transitive or common drive, and were valid over a wide range of values of the rejection threshold (see Materials and Methods, Eq. 4). Together, this suggests that highly nonuniform information transfer is a robust, general feature of mammalian cortical networks.
Information transfer versus synaptic connections
The drive to understand the connectome of the brain (Sporns et al., 2005) has included impressive work that has identified detailed synaptic connections in organisms ranging from the worm Caenorhabditis elegans (White et al., 1986; Jarrell et al., 2012) to the fly (Takemura et al., 2013), to mammalian patches of retina (Helmstaedter et al., 2013) and cortex (Bock et al., 2011). This work will form an essential component in developing activity maps of brains (Alivisatos et al., 2013). Even complete knowledge of synaptic connections, though, is not sufficient to reveal how neural activity is routed. For example, decades after the complete wiring diagram for C. elegans became known (White et al., 1986), a vibrant body of work still aims to record (Larsch et al., 2013) and model (Izquierdo and Beer, 2013) its neural activity patterns. For similar reasons, it is also essential to study directed relationships of neural activity in mammalian cortex, using, for example, information transfer or Granger causality (Brovelli et al., 2004; Nakhnikian et al., 2014). Such work is complementary to studies of synaptic connectivity in the same way that knowledge of traffic patterns complements a roadmap.
Lognormal firing rates
Any detailed model of local cortical networks must simultaneously satisfy the constraints of a lognormal firing rate distribution (Hromádka et al., 2008) and a lognormal connection weight distribution (Song et al., 2005), as observed in the data. One type of model (Roxin et al., 2011) has shown that a lognormal firing rate distribution can be produced when neuronal firing rates, f, are an exponential function of I, the synaptic input currents (f = eI). In this type of model, the lognormal firing rate is primarily a consequence of the transfer function, and not the network connectivity. Another type of model, in contrast, emphasizes that particular patterns of network connectivity can produce a lognormal firing rate distribution (Koulakov et al., 2009). In a random network, if synaptic input strengths are drawn randomly from a lognormal distribution, they tend to produce a relatively narrow distribution of total input currents, due to the central limit theorem. This leads to a Gaussian distribution of firing rates, contradicting the data. There are two ways to overcome this. First, if connection strengths are drawn in a correlated manner, a small population of neurons receives much stronger connections, driving them to firing rates several orders of magnitude larger than the mean. Second, if some neurons receive many more connections than others, they could also be driven to fire at much higher rates. Either or both mechanisms would lead to a long-tailed, and approximately lognormal, firing rate distribution. Our results are consistent with those of Koulakov et al. (2009), who predicted that some neurons would receive much more drive than others. By randomizing both the correlations in connection strengths and the topology, we found that the topology contributed most to this effect (Fig. 4). However, this does not exclude possible contributions of an exponential f–I relationship to the firing rate distribution.
Rich-club structure
The fact that some neurons receive much more input than others does not, by itself, necessarily imply that neurons with high total information transfer will preferentially transfer information to each other. Even though we previously found that highly connected neurons (hubs) exist in local cortical networks (Shimono and Beggs, 2015), until now we had not investigated whether these hubs strongly connected to each other more than chance. To assess group structure and move beyond neuron-wise measures, we measured the rich-club coefficient. Rich-club analysis (unweighted and weighted) has been performed on a variety of real-world and biological networks.(Zhou and Mondragón, 2004; Colizza et al., 2006; Opsahl et al., 2008).
In the brain at the macroscopic scale, rich-club structure has been shown to exist in the structural connectome of cat (de Reus and van den Heuvel, 2013), macaque (Harriger et al., 2012), adult human (van den Heuvel and Sporns, 2011), and newborn infants (Ball et al., 2014). In these studies, though, the analysis has been performed at the macroscopic scale of structural connections between brain regions and not at the scale of individual neurons. There is only one study (Towlson et al., 2013) to have reported a rich club at the level of individual neurons, and that was in the structural connectome of the worm C. elegans. Very recent work by Schroeter et al. (2015) reported a rich-club structure at the level of electrodes, not neurons, in dissociated cultures. Our findings are consistent with these previous reports, but go beyond them by identifying individual neurons as well as the information transfer strengths between them. Thus, we are the first to show the existence of a rich club for incoming and outgoing information transfer among hundreds of neurons. This finding is consistent with the significant clustering values obtained by Perin et al. (2011) for synaptic connections among 6–12 cortical neurons. It may also harmonize with the findings of a recent study (Cossell et al., 2015) in the visual cortex that found neurons with similar receptive fields were functionally more correlated and structurally more strongly coupled than neurons with dissimilar receptive fields.
Role of rich-club neurons
Several clues to the possible roles of rich-club neurons may be gleaned from previous work. Studies at the macroscopic scale in humans performing diverse cognitive tasks have shown the existence of a rich club in functional coactivation networks (Crossley et al., 2013; Baggio et al., 2015). Rich-club structures also have been suggested to facilitate control of synchronization (Gómez-Gardeñes et al., 2010; Batista et al., 2012; Watanabe, 2013), to reduce network path length (Zhou and Mondragón, 2004), and to elevate the network clustering coefficient (van den Heuvel and Sporns, 2011). The potential benefits of these features could justify the costly long-distance synaptic connections that are necessary to establish a rich-club structure in the human brain (Collin et al., 2014). Interestingly, a recent study (van den Heuvel et al., 2013) showed that patients with schizophrenia had significantly smaller rich-club density than healthy control subjects. This suggests the importance of the rich-club organization for healthy brain function.
At the neuronal scale, modeling studies (Klinshov et al., 2014) have shown that networks with clustered subnetworks have bistable low-firing and high-firing network states. Random networks with no clustering just show one stable, low-firing state. Bistability has been shown to play an important role in stored-trace reactivation (Johnson et al., 2010) and working memory (Goldman-Rakic, 1995). Recent work by Senden et al. (2014) has shown that in a spin glass model, the number of attractors was found to be larger for scale-free networks with rich-club characteristics than for non-rich-club networks that had scale-free, small-world regular or random patterns of connectivity. This suggests that networks with rich clubs can sustain a larger repertoire of distinct network activity states, enhancing versatility.
Teramae et al. (2012) have even suggested a role for the nonrich neurons. Using a computational model, they noted that a strong cortical synapse, by itself, is unable to drive a postsynaptic neuron to fire. A noisy background produced by many weak connections, though, could enhance the ability of single strong inputs to drive their postsynaptic target neurons in a process called stochastic resonance.
Future questions
This division of labor between information-rich and information-poor neurons naturally raises questions about how these two populations would interact in vivo as an animal is encoding, learning, and remembering. We view the identification of these neuron populations and their information flow structure as a first step toward future studies on these topics.
Footnotes
This research was supported by National Science Foundation Grants 1513779 (J.M.B.), 1429500 (J.M.B.), IIS-0904413 (A.M.L.), CBET-1263785 (S.C.M.), CNS-0521433 (Indiana University computing resources), and CNS-0723054 (Indiana University computing resources). The research was also supported through a Japan Society for the Promotion of Science Research Abroad fellowship to M.S. from MEXT (The Ministry of Education, Culture, Sports, Science, and Technology of Japan) and by National Institutes of Health Grant 5R01-DA-034178-03 (S.C.M.). Additionally, via the use of computing resources at Indiana University, this research was supported in part by Lilly Endowment, Inc., through its support for the Indiana University Pervasive Technology Institute, and in part by the Indiana METACyte Initiative.
The authors declare no competing financial interests.
The funders had no role in study design, data collection, and analysis, the decision to publish, or preparation of the manuscript.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License Creative Commons Attribution 4.0 International, which permits unrestricted use, distribution and reproduction in any medium provided that the original work is properly attributed.
References
- Alivisatos AP, Chun M, Church GM, Deisseroth K, Donoghue JP, Greenspan RJ, McEuen PL, Roukes ML, Sejnowski TJ, Weiss PS, Yuste R. The brain activity map. Science. 2013;339:1284–1285. doi: 10.1126/science.1236939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alstott J, Panzarasa P, Rubinov M, Bullmore ET, Vértes PE. A unifying framework for measuring weighted rich clubs. Sci Rep. 2014;4:7258. doi: 10.1038/srep07258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggio HC, Segura B, Junque C, de Reus MA, Sala-Llonch R, Van den Heuvel MP. Rich club organization and cognitive performance in healthy older participants. J Cogn Neurosci. 2015;27:1801–1810. doi: 10.1162/jocn_a_00821. [DOI] [PubMed] [Google Scholar]
- Baker RE, Van Pelt J. Cocultured, but not isolated, cortical explants display normal dendritic development: a long-term quantitative study. Brain Res Dev Brain Res. 1997;98:21–29. doi: 10.1016/S0165-3806(96)00163-0. [DOI] [PubMed] [Google Scholar]
- Baker RE, Corner MA, van Pelt J. Spontaneous neuronal discharge patterns in developing organotypic mega-co-cultures of neonatal rat cerebral cortex. Brain Res. 2006;1101:29–35. doi: 10.1016/j.brainres.2006.05.028. [DOI] [PubMed] [Google Scholar]
- Ball G, Aljabar P, Zebari S, Tusor N, Arichi T, Merchant N, Robinson EC, Ogundipe E, Rueckert D, Edwards AD, Counsell SJ. Rich-club organization of the newborn human brain. Proc Natl Acad Sci U S A. 2014;111:7456–7461. doi: 10.1073/pnas.1324118111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrat A, Barthelemy M, Vespignani A. Dynamical processes on complex networks. Cambridge, UK: Cambridge UP; 2008. [Google Scholar]
- Barthó P, Hirase H, Monconduit L, Zugaro M, Harris KD, Buzsáki G. Characterization of neocortical principal cells and interneurons by network interactions and extracellular features. J Neurophysiol. 2004;92:600–608. doi: 10.1152/jn.01170.2003. [DOI] [PubMed] [Google Scholar]
- Batista CA, Lameu EL, Batista AM, Lopes SR, Pereira T, Zamora-López G, Kurths J, Viana RL. Phase synchronization of bursting neurons in clustered small-world networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2012;86 doi: 10.1103/PhysRevE.86.016211. 016211. [DOI] [PubMed] [Google Scholar]
- Beggs JM, Plenz D. Neuronal avalanches in neocortical circuits. J Neurosci. 2003;23:11167–11177. doi: 10.1523/JNEUROSCI.23-35-11167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs JM, Plenz D. Neuronal avalanches are diverse and precise activity patterns that are stable for many hours in cortical slice cultures. J Neurosci. 2004;24:5216–5229. doi: 10.1523/JNEUROSCI.0540-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001:1165–1188. [Google Scholar]
- Bock DD, Lee WC, Kerlin AM, Andermann ML, Hood G, Wetzel AW, Yurgenson S, Soucy ER, Kim HS, Reid RC. Network anatomy and in vivo physiology of visual cortical neurons. Nature. 2011;471:177–182. doi: 10.1038/nature09802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolz J, Novak N, Götz M, Bonhoeffer T. Formation of target-specific neuronal projections in organotypic slice cultures from rat visual cortex. Nature. 1990;346:359–362. doi: 10.1038/346359a0. [DOI] [PubMed] [Google Scholar]
- Bonifazi P, Goldin M, Picardo MA, Jorquera I, Cattani A, Bianconi G, Represa A, Ben-Ari Y, Cossart R. GABAergic hub neurons orchestrate synchrony in developing hippocampal networks. Science. 2009;326:1419–1424. doi: 10.1126/science.1175509. [DOI] [PubMed] [Google Scholar]
- Brandes U. A faster algorithm for betweenness centrality. J Math Sociol. 2001;25:163–177. doi: 10.1080/0022250X.2001.9990249. [DOI] [Google Scholar]
- Brovelli A, Ding M, Ledberg A, Chen Y, Nakamura R, Bressler SL. Beta oscillations in a large-scale sensorimotor cortical network: directional influences revealed by Granger causality. Proc Natl Acad Sci U S A. 2004;101:9849–9854. doi: 10.1073/pnas.0308538101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomano DV. Timing of neural responses in cortical organotypic slices. Proc Natl Acad Sci U S A. 2003;100:4897–4902. doi: 10.1073/pnas.0736909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsáki G, Mizuseki K. The log-dynamic brain: how skewed distributions affect network operations. Nat Rev Neurosci. 2014;15:264–278. doi: 10.1038/nrn3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caeser M, Bonhoeffer T, Bolz J. Cellular organization and development of slice cultures from rat visual cortex. Exp Brain Res. 1989;77:234–244. doi: 10.1007/BF00274981. [DOI] [PubMed] [Google Scholar]
- Chiappalone M, Bove M, Vato A, Tedesco M, Martinoia S. Dissociated cortical networks show spontaneously correlated activity patterns during in vitro development. Brain Res. 2006;1093:41–53. doi: 10.1016/j.brainres.2006.03.049. [DOI] [PubMed] [Google Scholar]
- Colizza V, Flammini A, Serrano MA, Vespignani A. Detecting rich-club ordering in complex networks. Nat Phys. 2006;2:110–115. doi: 10.1038/nphys209. [DOI] [Google Scholar]
- Collin G, Sporns O, Mandl RC, van den Heuvel MP. Structural and functional aspects relating to cost and benefit of rich club organization in the human cerebral cortex. Cereb Cortex. 2014;24:2258–2267. doi: 10.1093/cercor/bht064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossell L, Iacaruso MF, Muir DR, Houlton R, Sader EN, Ko H, Hofer SB, Mrsic-Flogel TD. Functional organization of excitatory synaptic strength in primary visual cortex. Nature. 2015;518:399–403. doi: 10.1038/nature14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley NA, Mechelli A, Vértes PE, Winton-Brown TT, Patel AX, Ginestet CE, McGuire P, Bullmore ET. Cognitive relevance of the community structure of the human brain functional coactivation network. Proc Natl Acad Sci U S A. 2013;110:11583–11588. doi: 10.1073/pnas.1220826110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Reus MA, van den Heuvel MP. Rich club organization and intermodule communication in the cat connectome. J Neurosci. 2013;33:12929–12939. doi: 10.1523/JNEUROSCI.1448-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagle N, Macy M, Claxton R. Network diversity and economic development. Science. 2010;328:1029–1031. doi: 10.1126/science.1186605. [DOI] [PubMed] [Google Scholar]
- Field GD, Gauthier JL, Sher A, Greschner M, Machado TA, Jepson LH, Shlens J, Gunning DE, Mathieson K, Dabrowski W, Paninski L, Litke AM, Chichilnisky EJ. Functional connectivity in the retina at the resolution of photoreceptors. Nature. 2010;467:673–677. doi: 10.1038/nature09424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman LC. Centrality in social networks conceptual clarification. Soc Networks. 1979;1:215–239. [Google Scholar]
- Friedman N, Ito S, Brinkman BA, Shimono M, DeVille RE, Dahmen KA, Beggs JM, Butler TC. Universal critical dynamics in high resolution neuronal avalanche data. Phys Rev Lett. 2012;108:208102. doi: 10.1103/PhysRevLett.108.208102. [DOI] [PubMed] [Google Scholar]
- Garofalo M, Nieus T, Massobrio P, Martinoia S. Evaluation of the performance of information theory-based methods and cross-correlation to estimate the functional connectivity in cortical networks. PLoS One. 2009;4:e6482. doi: 10.1371/journal.pone.0006482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard F, Pipa G, Lima B, Neuenschwander S, Gerstner W. Extraction of network topology from multi-electrode recordings: is there a small-world effect? Front Comput Neurosci. 2011;5:4. doi: 10.3389/fncom.2011.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gireesh ED, Plenz D. Neuronal avalanches organize as nested theta-and beta/gamma-oscillations during development of cortical layer 2/3. Proc Natl Acad Sci U S A. 2008;105:7576–7581. doi: 10.1073/pnas.0800537105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman-Rakic PS. Cellular basis of working memory. Neuron. 1995;14:477–485. doi: 10.1016/0896-6273(95)90304-6. [DOI] [PubMed] [Google Scholar]
- Gómez-Gardeñes J, Zamora-López G, Moreno Y, Arenas A. From modular to centralized organization of synchronization in functional areas of the cat cerebral cortex. PLoS One. 2010;5:e12313. doi: 10.1371/journal.pone.0012313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz M, Bolz J. Formation and preservation of cortical layers in slice cultures. J Neurobiol. 1992;23:783–802. doi: 10.1002/neu.480230702. [DOI] [PubMed] [Google Scholar]
- Gourévitch B, Eggermont JJ. Evaluating information transfer between auditory cortical neurons. J Neurophysiol. 2007;97:2533–2543. doi: 10.1152/jn.01106.2006. [DOI] [PubMed] [Google Scholar]
- Harriger L, van den Heuvel MP, Sporns O. Rich club organization of macaque cerebral cortex and its role in network communication. PLoS One. 2012;7:e46497. doi: 10.1371/journal.pone.0046497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-White ME, Zanotti SA, Frautschy SA, Charles AC. Spiral intercellular calcium waves in hippocampal slice cultures. J Neurophysiol. 1998;79:1045–1052. doi: 10.1152/jn.1998.79.2.1045. [DOI] [PubMed] [Google Scholar]
- Helmstaedter M, Briggman KL, Turaga SC, Jain V, Seung HS, Denk W. Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature. 2013;500:168–174. doi: 10.1038/nature12346. [DOI] [PubMed] [Google Scholar]
- Hlaváčková-Schindler K, Paluš M, Vejmelka M, Bhattacharya J. Causality detection based on information-theoretic approaches in time series analysis. Phys Rep. 2007;441:1–46. doi: 10.1016/j.physrep.2006.12.004. [DOI] [Google Scholar]
- Hromádka T, Deweese MR, Zador AM. Sparse representation of sounds in the unanesthetized auditory cortex. PLoS Biol. 2008;6:e16. doi: 10.1371/journal.pbio.0060016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegaya Y, Aaron G, Cossart R, Aronov D, Lampl I, Ferster D, Yuste R. Synfire chains and cortical songs: temporal modules of cortical activity. Science. 2004;304:559–564. doi: 10.1126/science.1093173. [DOI] [PubMed] [Google Scholar]
- Ito S, Hansen ME, Heiland R, Lumsdaine A, Litke AM, Beggs JM. Extending transfer entropy improves identification of effective connectivity in a spiking cortical network model. PLoS One. 2011;6:e27431. doi: 10.1371/journal.pone.0027431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Yeh FC, Hiolski E, Rydygier P, Gunning DE, Hottowy P, Timme N, Litke AM, Beggs JM. Large-scale, high-resolution multielectrode-array recording depicts functional network differences of cortical and hippocampal cultures. PLoS One. 2014;9:e105324. doi: 10.1371/journal.pone.0105324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izhikevich EM. Simple model of spiking neurons. IEEE Trans Neural Netw. 2003;14:1569–1572. doi: 10.1109/TNN.2003.820440. [DOI] [PubMed] [Google Scholar]
- Izquierdo EJ, Beer RD. Connecting a connectome to behavior: an ensemble of neuroanatomical models of C. elegans klinotaxis. PLoS Comput Biol. 2013;9:e1002890. doi: 10.1371/journal.pcbi.1002890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell TA, Wang Y, Bloniarz AE, Brittin CA, Xu M, Thomson JN, Albertson DG, Hall DH, Emmons SW. The connectome of a decision-making neural network. Science. 2012;337:437–444. doi: 10.1126/science.1221762. [DOI] [PubMed] [Google Scholar]
- Jimbo Y, Tateno T, Robinson HP. Simultaneous induction of pathway-specific potentiation and depression in networks of cortical neurons. Biophys J. 1999;76:670–678. doi: 10.1016/S0006-3495(99)77234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson HA, Buonomano DV. Development and plasticity of spontaneous activity and Up states in cortical organotypic slices. J Neurosci. 2007;27:5915–5925. doi: 10.1523/JNEUROSCI.0447-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LA, Euston DR, Tatsuno M, McNaughton BL. Stored-trace reactivation in rat prefrontal cortex is correlated with down-to-up state fluctuation density. J Neurosci. 2010;30:2650–2661. doi: 10.1523/JNEUROSCI.1617-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juergens E, Eckhorn R. Parallel processing by a homogeneous group of coupled model neurons can enhance, reduce and generate signal correlations. Biol Cybern. 1997;76:217–227. doi: 10.1007/s004220050334. [DOI] [PubMed] [Google Scholar]
- Klinshov VV, Teramae JN, Nekorkin VI, Fukai T. Dense neuron clustering explains connectivity statistics in cortical microcircuits. PLoS One. 2014;9:e94292. doi: 10.1371/journal.pone.0094292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klostermann O, Wahle P. Patterns of spontaneous activity and morphology of interneuron types in organotypic cortex and thalamus—cortex cultures. Neuroscience. 1999;92:1243–1259. doi: 10.1016/S0306-4522(99)00009-3. [DOI] [PubMed] [Google Scholar]
- Koulakov AA, Hromádka T, Zador AM. Correlated connectivity and the distribution of firing rates in the neocortex. J Neurosci. 2009;29:3685–3694. doi: 10.1523/JNEUROSCI.4500-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larremore DB, Shew WL, Restrepo JG. Predicting criticality and dynamic range in complex networks: effects of topology. Phys Rev Lett. 2011;106:58101. doi: 10.1103/PhysRevLett.106.058101. [DOI] [PubMed] [Google Scholar]
- Larsch J, Ventimiglia D, Bargmann CI, Albrecht DR. High-throughput imaging of neuronal activity in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2013;110:E4266–E4273. doi: 10.1073/pnas.1318325110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman AL, Seil FJ. Influence of subcortical neurons on the functional development of cerebral neocortex in tissue culture. Brain Res. 1986;365:205–210. doi: 10.1016/0006-8993(86)91631-8. [DOI] [PubMed] [Google Scholar]
- Lindner M, Vicente R, Priesemann V, Wibral M. TRENTOOL: a Matlab open source toolbox to analyse information flow in time series data with transfer entropy. BMC Neurosci. 2011;12:119. doi: 10.1186/1471-2202-12-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litke A, Bezayiff N, Chichilnisky E, Cunningham W, Dabrowski W, Grillo A, Grivich M, Grybos P, Hottowy P, Kachiguine S. What does the eye tell the brain? Development of a system for the large-scale recording of retinal output activity. IEEE Trans Nucl Sci. 2004;51:1434–1440. doi: 10.1109/TNS.2004.832706. [DOI] [Google Scholar]
- Lungarella M, Pitti A, Kuniyoshi Y. Information transfer at multiple scales. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;76 doi: 10.1103/PhysRevE.76.056117. 056117. [DOI] [PubMed] [Google Scholar]
- Maass W, Natschläger T, Markram H. Real-time computing without stable states: a new framework for neural computation based on perturbations. Neural Comput. 2002;14:2531–2560. doi: 10.1162/089976602760407955. [DOI] [PubMed] [Google Scholar]
- Mason A, Nicoll A, Stratford K. Synaptic transmission between individual pyramidal neurons of the rat visual cortex in vitro. J Neurosci. 1991;11:72–84. doi: 10.1523/JNEUROSCI.11-01-00072.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalto A, Faes L, Marinazzo D. MuTE: a MATLAB toolbox to compare established and novel estimators of the multivariate transfer entropy. PLoS One. 2014;9:e109462. doi: 10.1371/journal.pone.0109462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhnikian A, Rebec GV, Grasse LM, Dwiel LL, Shimono M, Beggs JM. Behavior modulates effective connectivity between cortex and striatum. PLoS One. 2014;9:e89443. doi: 10.1371/journal.pone.0089443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatan M, Wilson MA, Brown EN. Analyzing functional connectivity using a network likelihood model of ensemble neural spiking activity. Neural Comput. 2005;17:1927–1961. doi: 10.1162/0899766054322973. [DOI] [PubMed] [Google Scholar]
- Opsahl T, Colizza V, Panzarasa P, Ramasco JJ. Prominence and control: the weighted rich-club effect. Phys Rev Lett. 2008;101:168702. doi: 10.1103/PhysRevLett.101.168702. [DOI] [PubMed] [Google Scholar]
- Orlandi JG, Stetter O, Soriano J, Geisel T, Battaglia D. Transfer Entropy reconstruction and labeling of neuronal connections from simulated calcium imaging. PLoS One. 2014;9:e98842. doi: 10.1371/journal.pone.0098842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzeri S, Petersen RS, Schultz SR, Lebedev M, Diamond ME. The role of spike timing in the coding of stimulus location in rat somatosensory cortex. Neuron. 2001;29:769–777. doi: 10.1016/S0896-6273(01)00251-3. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The rat brain in stereotaxic coordinates. San Diego: Academic; 1986. [DOI] [PubMed] [Google Scholar]
- Perin R, Berger TK, Markram H. A synaptic organizing principle for cortical neuronal groups. Proc Natl Acad Sci U S A. 2011;108:5419–5424. doi: 10.1073/pnas.1016051108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrusca D, Grivich MI, Sher A, Field GD, Gauthier JL, Greschner M, Shlens J, Chichilnisky EJ, Litke AM. Identification and characterization of a Y-like primate retinal ganglion cell type. J Neurosci. 2007;27:11019–11027. doi: 10.1523/JNEUROSCI.2836-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillow JW, Shlens J, Paninski L, Sher A, Litke AM, Chichilnisky EJ, Simoncelli EP. Spatio-temporal correlations and visual signalling in a complete neuronal population. Nature. 2008;454:995–999. doi: 10.1038/nature07140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenz D, Aertsen A. Neural dynamics in cortex-striatum co-cultures—II. Spatiotemporal characteristics of neuronal activity. Neuroscience. 1996;70:893–924. doi: 10.1016/0306-4522(95)00405-X. [DOI] [PubMed] [Google Scholar]
- Restrepo JG, Ott E, Hunt BR. Onset of synchronization in large networks of coupled oscillators. Phys Rev E Stat Nonlin Soft Matter Phys. 2005;71 doi: 10.1103/PhysRevE.71.036151. 036151. [DOI] [PubMed] [Google Scholar]
- Restrepo JG, Ott E, Hunt BR. Characterizing the dynamical importance of network nodes and links. Phys Rev Lett. 2006;97 doi: 10.1103/PhysRevLett.97.094102. 094102. [DOI] [PubMed] [Google Scholar]
- Restrepo JG, Ott E, Hunt BR. Weighted percolation on directed networks. Phys Rev Lett. 2008;100:58701. doi: 10.1103/PhysRevLett.100.058701. [DOI] [PubMed] [Google Scholar]
- Roxin A, Brunel N, Hansel D, Mongillo G, van Vreeswijk C. On the distribution of firing rates in networks of cortical neurons. J Neurosci. 2011;31:16217–16226. doi: 10.1523/JNEUROSCI.1677-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Schreiber T. Measuring information transfer. Phys Rev Lett. 2000;85:461–464. doi: 10.1103/PhysRevLett.85.461. [DOI] [PubMed] [Google Scholar]
- Schroeter MS, Charlesworth P, Kitzbichler MG, Paulsen O, Bullmore ET. Emergence of rich-club topology and coordinated dynamics in development of hippocampal functional networks in vitro. J Neurosci. 2015;35:5459–5470. doi: 10.1523/JNEUROSCI.4259-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senden M, Deco G, de Reus MA, Goebel R, van den Heuvel MP. Rich club organization supports a diverse set of functional network configurations. Neuroimage. 2014;96:174–182. doi: 10.1016/j.neuroimage.2014.03.066. [DOI] [PubMed] [Google Scholar]
- Shimono M, Beggs JM. Functional clusters, hubs, and communities in the cortical microconnectome. Cereb Cortex. 2015;25:3743–3757. doi: 10.1093/cercor/bhu252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlens J, Field GD, Gauthier JL, Grivich MI, Petrusca D, Sher A, Litke AM, Chichilnisky EJ. The structure of multi-neuron firing patterns in primate retina. J Neurosci. 2006;26:8254–8266. doi: 10.1523/JNEUROSCI.1282-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shobe JL, Claar LD, Parhami S, Bakhurin KI, Masmanidis SC. Brain activity mapping at multiple scales with silicon microprobes containing 1024 electrodes. J Neurophysiol. 2015;114:2043–2052. doi: 10.1152/jn.00464.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirota A, Montgomery S, Fujisawa S, Isomura Y, Zugaro M, Buzsáki G. Entrainment of neocortical neurons and gamma oscillations by the hippocampal theta rhythm. Neuron. 2008;60:683–697. doi: 10.1016/j.neuron.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Sjöström PJ, Reigl M, Nelson S, Chklovskii DB. Highly nonrandom features of synaptic connectivity in local cortical circuits. PLoS Biol. 2005;3:e68. doi: 10.1371/journal.pbio.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kötter R. The human connectome: a structural description of the human brain. PLoS Comput Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal JA, Alexander SR, Liu Y, Dickson TD, Vickers JC. Characterization of cortical neuronal and glial alterations during culture of organotypic whole brain slices from neonatal and mature mice. PLoS One. 2011;6:e22040. doi: 10.1371/journal.pone.0022040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetter O, Battaglia D, Soriano J, Geisel T. Model-free reconstruction of excitatory neuronal connectivity from calcium imaging signals. PLoS Comput Biol. 2012;8:e1002653. doi: 10.1371/journal.pcbi.1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swadlow HA. Efferent neurons and suspected interneurons in motor cortex of the awake rabbit: axonal properties, sensory receptive fields, and subthreshold synaptic inputs. J Neurophysiol. 1994;71:437–453. doi: 10.1152/jn.1994.71.2.437. [DOI] [PubMed] [Google Scholar]
- Takemura SY, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, Katz WT, Olbris DJ, Plaza SM, Winston P, Zhao T, Horne JA, Fetter RD, Takemura S, Blazek K, Chang LA, Ogundeyi O, Saunders MA, Shapiro V, Sigmund C, et al. A visual motion detection circuit suggested by Drosophila connectomics. Nature. 2013;500:175–181. doi: 10.1038/nature12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A, Jackson D, Hobbs J, Chen W, Smith JL, Patel H, Prieto A, Petrusca D, Grivich MI, Sher A, Hottowy P, Dabrowski W, Litke AM, Beggs JM. A maximum entropy model applied to spatial and temporal correlations from cortical networks in vitro. J Neurosci. 2008;28:505–518. doi: 10.1523/JNEUROSCI.3359-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrell GR, Scott DW. Oversmoothed nonparametric density estimates. J Am Stat Assoc. 1985;80:209–214. [Google Scholar]
- Teramae JN, Tsubo Y, Fukai T. Optimal spike-based communication in excitable networks with strong-sparse and weak-dense links. Sci Rep. 2012;2:485. doi: 10.1038/srep00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme N, Ito S, Myroshnychenko M, Yeh FC, Hiolski E, Hottowy P, Beggs JM. Multiplex networks of cortical and hippocampal neurons revealed at different timescales. PLoS One. 2014;9:e115764. doi: 10.1371/journal.pone.0115764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towlson EK, Vértes PE, Ahnert SE, Schafer WR, Bullmore ET. The rich club of the C. elegans neuronal connectome. J Neurosci. 2013;33:6380–6387. doi: 10.1523/JNEUROSCI.3784-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O. Rich-club organization of the human connectome. J Neurosci. 2011;31:15775–15786. doi: 10.1523/JNEUROSCI.3539-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O, Collin G, Scheewe T, Mandl RC, Cahn W, Goñi J, Hulshoff Pol HE, Kahn RS. Abnormal rich club organization and functional brain dynamics in schizophrenia. JAMA Psychiatry. 2013;70:783–792. doi: 10.1001/jamapsychiatry.2013.1328. [DOI] [PubMed] [Google Scholar]
- Vicente R, Wibral M, Lindner M, Pipa G. Transfer entropy—a model-free measure of effective connectivity for the neurosciences. J Comput Neurosci. 2011;30:45–67. doi: 10.1007/s10827-010-0262-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T. Rich-club network topology to minimize synchronization cost due to phase difference among frequency-synchronized oscillators. Physica A Stat Mech Appl. 2013;392:1246–1255. doi: 10.1016/j.physa.2012.11.041. [DOI] [Google Scholar]
- Wehr M, Zador AM. Balanced inhibition underlies tuning and sharpens spike timing in auditory cortex. Nature. 2003;426:442–446. doi: 10.1038/nature02116. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wibral M, Lizier JT, Priesemann V. Bits from brains for biologically-inspired computing. Front Robot AI. 2015;2:5. doi: 10.3389/frobt.2015.00005. [DOI] [Google Scholar]
- Wollstadt P, Martínez-Zarzuela M, Vicente R, Díaz-Pernas FJ, Wibral M. Efficient transfer entropy analysis of non-stationary neural time series. PLoS One. 2014;9:e102833. doi: 10.1371/journal.pone.0102833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassin L, Benedetti BL, Jouhanneau JS, Wen JA, Poulet JF, Barth AL. An embedded subnetwork of highly active neurons in the neocortex. Neuron. 2010;68:1043–1050. doi: 10.1016/j.neuron.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko D, Josić K, Ecker AS, Froudarakis E, Cotton RJ, Tolias AS, Pillow JW. Improved estimation and interpretation of correlations in neural circuits. PLoS Comput Biol. 2015;11:e1004083–e1004083. doi: 10.1371/journal.pcbi.1004083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Mondragón RJ. The rich-club phenomenon in the Internet topology. Commun Lett IEEE. 2004;8:180–182. doi: 10.1109/LCOMM.2004.823426. [DOI] [Google Scholar]