Abstract
Soluble guanylate cyclases (sGCs) are gas-binding proteins that control diverse physiological processes such as vasodilation, platelet aggregation, and synaptic plasticity. In the nematode Caenorhabditis elegans, a complex of sGCs, GCY-35 and GCY-36, functions in oxygen (O2) sensing. Previous studies suggested that the neuroglobin GLB-5 genetically interacts with GCY-35, and that the inhibitory effect of GLB-5 on GCY-35 function is necessary for fast recovery from prolonged hypoxia. In this study, we identified mutations in gcy-35 and gcy-36 that impact fast recovery and other phenotypes associated with GLB-5, without undermining sGC activity. These mutations, heb1 and heb3, change conserved amino acid residues in the regulatory H-NOX domains of GCY-35 and GCY-36, respectively, and appear to suppress GLB-5 activity by different mechanisms. Moreover, we observed that short exposure to 35% O2 desensitized the neurons responsible for ambient O2 sensing and that this phenomenon does not occur in heb1 animals. These observations may implicate sGCs in neuronal desensitization mechanisms far beyond the specific case of O2 sensing in nematodes. The conservation of functionally important regions of sGCs is supported by examining site-directed mutants of GCY-35, which suggested that similar regions in the H-NOX domains of O2 and NO-sensing sGCs are important for heme/gas interactions. Overall, our studies provide novel insights into sGC activity and regulation, and implicate similar structural determinants in the control of both O2 and NO sensors.
SIGNIFICANCE STATEMENT Soluble guanylate cyclases (sGCs) control essential and diverse physiological processes, including memory processing. We used Caenorhabditis elegans to explore how a neuroglobin inhibits a complex of oxygen-sensing sGCs, identifying sGC mutants that resist inhibition. Resistance appears to arise by two different mechanisms: increased basal sGC activity or disruption of an interaction with neuroglobin. Our findings demonstrate that the inhibition of sGCs by neuroglobin is essential for rapid adaptation to either low or high oxygen levels, and that similar structural regions are key for regulating both oxygen and nitric oxide sensors. Based on our structural and functional analyses, we present the hypothesis that neuroglobin-sGC interactions may be generally important for adaptation processes, including those in organisms with more complex neurological functions.
Keywords: Caenorhabditis elegans, globin, H-NOX, hypoxia, oxygen-sensing, soluble guanylate cyclase
Introduction
Soluble guanylate cyclases (sGCs) catalyze formation of the second messenger cGMP from GTP. They thereby regulate important processes, such as blood circulation, platelet aggregation, and memory processing (Kleppisch and Feil, 2009; Derbyshire and Marletta, 2012; Shen et al., 2012). The activation of sGCs by nitric oxide (NO) has been extensively investigated (Derbyshire and Marletta, 2012; Follmann et al., 2013), but sGC regulation by proteins is still poorly understood. In this study, we explored how the atypical sGCs GCY-35 and GCY-36 are regulated by the neuroglobin GLB-5 in the Caenorhabditis elegans nematode.
The C. elegans genome encodes seven atypical sGCs (GCY-31 through GCY-37) (Morton et al., 1999). Unlike mammalian sGCs, C. elegans sGCs are thought to bind oxygen (O2) rather than NO (Gray et al., 2004; Derbyshire and Marletta, 2012). Generally, a functional sGC complex includes an α and a β subunit, each of which contains an N-terminal heme NO/O2 binding (H-NOX) domain, a Per/Arnt/Sim-like domain, a coiled-coil domain, and a catalytic cyclase domain (see Fig. 1) (Underbakke et al., 2013). Heterodimerization of α and β creates a functional sGC, as amino acid residues from both catalytic domains form the active site (Derbyshire and Marletta, 2012; Underbakke et al., 2013). GCY-35 and GCY-36 constitute one such functional heterodimer (Cheung et al., 2004).
Figure 1.
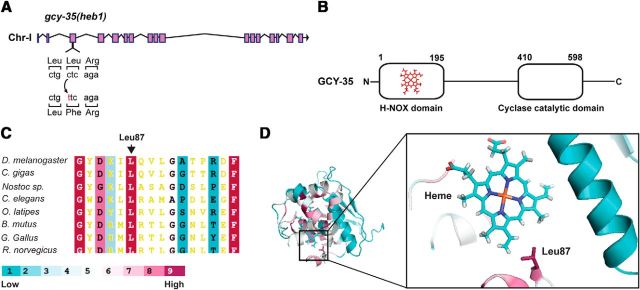
The transition mutation heb1 converts the conserved Leu87 to phenylalanine in the H-NOX domain of GCY-35. A, Schematic diagram of gcy-35 highlighting the transition mutation in the heb1 allele. B, Schematic presentation of GCY-35 domains. C, Amino acid conservation analysis of H-NOX domains from different organisms shows that Leu87 is conserved. Sequence alignment of H-NOX domains from Drosophila melanogaster (ABG02144), Crassostrea gigas (EKC42091), Nostoc sp. (WP_01099643), Caenorhabditis elegans GCY-35 (NP_001252131), Oryzias latipes (NP_001098123), Bos mutus (ELR49741), Gallus gallus (XP_004937100), and Rattus norvegicus (EDL85292). The conservation level is color coded from 1 (low) to 9 (high). D, Leu87 is predicted to be close to the heme moiety. A 3D presentation of the conservation analysis of the GCY-35 H-NOX domain (shown in C). The amino acid residues are colored according to the code shown in C. The heme moiety and Leu87 are labeled.
The GCY-35/36 heterodimer acts as a regulated O2 sensor in the neurons AQR, PQR, and URX (Cheung et al., 2004; Gray et al., 2004). Activation of GCY-35/36 increases cGMP levels, which opens cyclic nucleotide-gated channels, increasing calcium (Ca2+) levels in the cells and triggering neuronal activity (Persson et al., 2009). How neuronal activity upon GCY-35/36 activation translates into behavior is sensitive to genetic background, suggesting that it can be modulated by various factors. One factor that modulates GCY-35/36 is the neuropeptide receptor npr-1 (Gray et al., 2004).
npr-1 is a polymorphic gene. The C. elegans laboratory strain N2 bears the dominant allele of npr-1, npr-1(215 V), and consequently moves slowly on bacteria at 21% O2 and does not form groups on the bacterial lawn border. However, animals bearing the weaker ancestral allele npr-1(215F) or the loss-of-function allele npr-1(ad609) (hereafter referred to as npr-1 worms) move quickly on bacteria at 21% O2 and aggregate on the lawn border (de Bono and Bargmann, 1998). Another phenotype of npr-1 worms is a sharp decrease in speed in response to a shift in O2 concentration from 21% to 7%, but not from 21% to 17%. However, npr-1 animals bearing the dominant Hawaii polymorph of globin 5, glb-5(Haw), show robust deceleration even at 17% O2 (Persson et al., 2009), indicating that GLB-5 is an additional modulator of O2 sensing by GCY-35/36. Most wild C. elegans isolates bear the ancestral glb-5(Haw)/npr-1(215F) allelic combination, whereas N2 worms bear the nonfunctional Bristol allele of glb-5, glb-5(Bri), and npr-1(215V). Therefore, it appears that the glb-5(Bri)/npr-1(215V) combination arose during N2 domestication to laboratory conditions (McGrath et al., 2009; Weber et al., 2010). In this manuscript, we used the bordering behavior and high speed on bacteria at 21% O2 of npr-1 and glb-5(Haw);npr-1 animals as readouts for GCY-35/36 activity.
Like gcy-35/36, glb-5 is expressed in the AQR, PQR, and URX neurons (McGrath et al., 2009). GLB-5(Haw) decreases the activation of these neurons at 17% O2, probably by inhibiting GCY-35/36 activity (Gross et al., 2014). Recently, it was shown that the activity of GLB-5(Haw) is also essential for rapid recovery from prolonged hypoxia (Gross et al., 2014). Hypoxia increases the sensitivity of URX to O2 in both npr-1 and glb-5(Haw);npr-1 worms. However, the rapid readaptation of URX to 21% O2 is GLB-5(Haw)-dependent (Gross et al., 2014). GLB-5(Haw)-dependent O2 responses require the colocalization of GLB-5(Haw) and GCY-35 in the dendritic endings of URX (Gross et al., 2014), suggesting that the proximity of GLB-5(Haw) to GCY-35/36 is functionally important. In principle, GLB-5(Haw) might negatively regulate GCY-35/36 activity by physical interaction or by generating reactive oxygen species.
Materials and Methods
Strains
Strains are as follows: EVG009 glb-5(Haw) V;npr-1(ad609) X; EVG034 npr-1(ad609) X; EVG115 gcy-35(heb1) I;glb-5(Haw) V; npr-1(ad609) X; EVG123 gcy-35(heb1) I;glb-5(Haw) V;npr-1(ad609) X; Ex[Pgcy-35::gcy-35(genomic)::polycismCherry;Pf15e11.1::GFP]; EVG136 gcy-35(heb1) I;glb-5(Haw) V;npr-1(ad609) X;Ex[Pgcy-37::gcy-35(cDNA)::polycismCherry;Pf15e11.1::GFP]; EVG146 gcy-35(heb1) I;glb-5(Haw) V;npr-1(ad609)X; Ex[Pgcy-37::YC2.60+ccRFP]; EVG155 npr-1(ad609) X; Ex[Pgcy-37::YC2.60+ccRFP]; EVG159 glb-5(Haw) V; npr-1(ad609) X; Ex[Pgcy-37::YC2.60+ccRFP]; EVG236 gcy-35(heb1) I; npr-1(ad609) X; EVG301 gcy-35(ok769) I; glb-5(Haw) V; npr-1(ad609) X; Ex[Pgcy-37:: gcy-35 cDNA(L87W)::polycismCherry;Pf15e11.1::GFP]; EVG305 gcy-35(ok769) I; glb-5(Haw) V; npr-1(ad609) X; Ex[Pgcy-37:: gcy-35 cDNA (W67L):: polycismCherry;Pf15e11.1::GFP]; EVG306 gcy-35(ok769) I; glb-5(Haw) V; npr-1(ad609) X; Ex[Pgcy-37:: gcy-35 cDNA(T48W)::polycismCherry;Pf15e11.1::GFP]; EVG312 gcy-35(ok769) I; glb-5(Haw) V; npr-1(ad609) X; Ex[Pgcy-37:: gcy-35 cDNA(W67L+L87F)::polycismCherry;Pf15e11.1::GFP]; EVG332 gcy-35(ok769) I; glb-5(Haw) V; npr-1(ad609) X; Ex[Pgcy-37:: gcy-35 cDNA(T48W+L87F)::polycismCherry;Pf15e11.1::GFP]; EVG333 gcy-35(heb1) I; npr-1(ad609) X; Ex[Pgcy-37::YC2.60+ccRFP]; EVG336 gcy-35(ok769) I; glb-5(Haw) V; npr-1(ad609) X; Ex[Pgcy-37:: gcy-35 cDNA(L87F)::polycismCherry;Pf15e11.1::GFP]; EVG410 gcy-35(heb1) I; glb-5(Haw) V; npr-1(ad609) X; Ex[Pgcy-37::glb-5(cDNA)::polycismCherry;Pf15e11.1::GFP]; EVG556 npr-1(ad609) X; Ex[Pgcy-37::glb-5(cDNA)::polycismCherry;Pf15e11.1::GFP]; EVG629 glb-5(Haw) V; npr-1(ad609) gcy-36(heb3) X; Ex[Pgcy-37::YC2.60+ccRFP]; EVG631 glb-5(Haw) V; npr-1 gcy-36(heb3) X; EVG640 glb-5(Haw) V; npr-1(ad609) gcy-36(heb3) X; Ex[Pgcy-37::gcy-36 (cDNA)::polycismcherry;Pf15e11.1::GFP]; EVG678 gcy-35(heb1) I; glb-5(Haw) V; npr-1(ad609) gcy36(heb3) X; Ex[Pgcy-37::YC2.60+ccRFP]; EVG680 gcy-35(heb1) I; glb-5(Haw) V; npr-1(ad609) gcy36(heb3) X; EVG684 npr-1(ad609) gcy-36(heb3) X; EVG686 gcy-35(ok769) I; glb-5(Haw) V; npr-1(ad609) X.
Behavioral assays
Bordering.
Bordering experiments were performed as described previously (Gross et al., 2014). In brief, 40 L4 hermaphrodites (grown in 21% O2 at room temperature [RT]) were transferred to a 3.5 cm nematode growth medium (NGM) plate that was seeded 2 d before with 50 μl OP50 bacteria (OD600 ∼ 0.6). The assay plates were put in 1% O2 (Coy hypoxia chamber, COY Lab Products, RT) for 24 h and then brought back to 21% O2 (control plates were maintained in 21% O2 at RT). After 30 min recovery in 21% O2, we calculated the bordering index. Bordering index represents the fraction of worms found on the bacterial lawn border divided by the total number of worms on the plate, multiplied by 100.
Speed.
All speed measurements were done in the presence of food. Low-NGM plates (0.13 g/L bacto-peptone) were seeded with 20 μl OP50 bacteria (OD600 ∼ 0.6) 2 d before the experiment and grown at RT. For each speed measurement assay, we put 7–10 young hermaphrodites on the bacterial lawn. Then, we put a 500-μm-deep rectangular polydimethylsiloxane (PDMS) (12 mm wide, 0.5 mm deep, 17 mm long) on top of the bacterial lawn, thus trapping the worms in the behavioral arena. The PDMS chamber has one inlet and one outlet tubing for gas entry and exit, respectively (Movie 1). Throughout our experiments, we put the outlet tubing inside an Erlenmeyer bottle with water and verified a continuous formation of air bubbles, indicating that the interaction between the PDMS chamber and the agar forms a tight seal. We used a PHD 2000 syringe pump (Harvard Apparatus) to deliver humidified gases to the microfluidic chambers at a flow rate of 0.5 ml/min. In addition, we used Teflon valves, controlled by a ValveBank Controller (Automate Scientific), to rapidly switch between different O2/N2 gas mixtures. We recorded the movies using a Q-Imaging MicroPublisher 5.0 RTV Microscope Camera (QImaging, RHos) mounted onto an Olympus SZ61 stereo microscope (Olympus). The movies were taken at 0.5 frames/s. Video analysis was performed using custom MATLAB software (The MathWorks) (Gross et al., 2014). Notably, in our analysis, we did not track worms that leave the bacterial lawn and thus only calculated the speed of worms on food.
Ethyl methanesulfonate mutagenesis screen
To screen for glb-5(Haw); npr-1 suppressors, we mutagenized glb-5(Haw); npr-1 worms with ethyl methanesulfonate according to standard protocols (Brenner, 1974). In brief, to generate synchronized L4 hermaphrodites, we collected eggs from gravid hermaphrodites using hypochlorite solution (Brenner, 1974) and incubated them in M9 buffer at 21°C for ∼16 h. Hatched L1 larvae were put on NGM plates containing OP50 bacteria, at 21°C. The 360 mutagenized Po hermaphrodites were divided into 35 seeded 9 cm plates. After 4 d, we collected eggs from ∼9000 F1 hermaphrodites and let L1 larvae hatch as described above. Synchronized L1 were grown in 350 NGM plates (3.5 cm, ∼150 worms per plate). After 3 d, we put the L4 hermaphrodites in 1% O2 for 24 h. Then, we transferred the worms back to 21% O2, waited 30 min, and collected worms that remained outside the bacterial lawn border into new plates (350 3.5 cm plates, ∼3 worms per plate). After 4 d, we repeated the assay again and singled nonbordering worms into 3.5 cm plates. The progenies of these worms were reassayed, and lines in which we confirmed phenotypes were kept.
Mapping
We mapped heb1 and heb3 to a 4 Mb and 1.5 Mb genomic interval (on chromosomes I and X, respectively) using single nucleotide polymorphisms (Davis et al., 2005). The genomic regions of gcy-35 and gcy-36 were amplified using PCR, and the heb1 and heb3 mutations were identified by sequencing.
Molecular biology
All molecular manipulations were done according to standard protocols (Sambrook et al., 1989). Genotyping. The duplication in glb-5(Bri) was followed by examining the PCR products amplified using primers that flank the duplication in this allele (Persson et al., 2009). We tracked the npr-1(ad609), gcy-35(heb1), and gcy36(heb3) alleles using a similar strategy. First, we amplified a genomic region that contained these alleles (422, 497, and 374 bp, respectively). Then, we used restriction enzyme digestion to distinguish between mutants from wild-type alleles (using MwoI, DdeI, and Fok-1, respectively).
RNA purification and cDNA synthesis.
In brief, to extract total RNA, ∼120 worms were transferred to Microfuge tubes containing PBS and washed three times with the same buffer. The PBS buffer was replaced with 200 μl TRIzol Reagent (Invitrogen, Thermo Fisher Scientific), and the tubes were put at −80°C for ∼16 h. We then added Bullet Blender pink beads (Next Advance) and lysed the worms using Bullet Blender Homogenizer Cell Disrupter (Next Advance). After centrifugation, we purified the total RNA using the NucleoSpin RNA purification kit (Macherey-Nagel). cDNA was synthesized with the iScript cDNA Synthesis Kit (Bio-Rad). We used this cDNA to amplify the coding regions of gcy-35, gcy-36, and glb-5. In general, all primers were designed based on the Wormbase sequence information (http://www.wormbase.org/).
Transgenes
All rescuing plasmids were generated using a modified polycistronic mCherry pPD95.75 expression vector, as described previously (Gross et al., 2014). For the gcy-35(genomic) rescuing construct, we amplified the gcy-35 promoter (∼4.9 kb) and genomic region (∼8 kb) using N2 genomic DNA. The promoter region was amplified with 5′ GTA CGA ATC CGG ACT ACT GGT TAG AGA AAT ACA GAC GTT TAA AGC 3′ and 5′ GTG GGT TCC TAG GCT ACT CTC CGC AAA AAA GTA ACG TAT ATA ATA TAT ATT T 3′, and inserted using BspEI and AvrII. The genomic sequence of gcy-35 was cloned in two steps. The first genomic part was amplified using 5′ GTA CGA ACC TAG GAA AAT ATA ATG GTG AGT ATT TTT AAG GAA ATA GAT TTT TGT TTA C 3′ and 5′ AGT TAT TGG TAC CTT GAT AAA GTT TCC AAA ATG CT 3′ and inserted using AvrII and KpnI. To clone the second part, we introduced a SalI restriction site to the plasmid backbone (using QuikChange II XL, Agilent Technologies), amplified the second part of the genomic region with 5′ GTA CGA AGG TAC CAA TAA CTA GGC TTC TAA AAT TTT CAA AAT 3′ and 5′ GTG GGT TGT CGA CTT AAG AAA TTG TGC AAG TCG GTG GAT CGG ATG 3′, and inserted it with KpnI and SalI. To create the gcy-35 cDNA (2 kb) rescuing construct, the genomic part of the gcy-35 gene was replaced with gcy-35 cDNA. The coding region of gcy-35 was amplified using 5′ GTA CGA ACC TAG GAA AAT ATA ATG TTC GGC TGG ATT CAC GAA AGC TTC 3′ and 5′ GTG GGT TGT CGA CTT AAG AAA TTG TGC AAG TCG GTG ATC GGA TG 3′ and inserted with AvrII and SalI. To express gcy-35 in the AQR, PQR, and URX neurons, the promoter region of gcy-35 was replaced with the promoter gcy-37(∼1.3 kb). The promoter region was amplified using 5′ GGT GGT TCG ATA TCG GGA CCG ACA TGC TCG GCT TG 3′ and 5′ GTG GGT TCC TAG GCT GAA TTT ACT TTC TTT TTT TTT TGA AAT TTT AGC T 3′ and inserted with EcoRV and AvrII. To express gcy-36 and glb-5 cDNA in the AQR, PQR, URX and neurons, we replaced the coding region of gcy-35 cDNA with the coding region of these genes. The coding region of gcy-36 (∼2.3 kb) was amplified using 5′ GTA CGA ACC TAG GAA AAT ATA ATG TTC GGC TTC ATT CAT GAA TC 3′ and 5′ GTG GGT TGT CGA CTT AGG AAA TCG AGC AAG TTT TGG AAT TCT TTT GAG TAA CCT TGT TAA GGA C 3′, and the coding region of glb-5 (∼1.4 kb) was amplified using 5′ GTA CGA ACC TAG GAA AAT ATA ATG AAC GAG ACG ACA GTC GGA GTT GG 3′ and 5′ GTA CGA ACC TAG GAA AAT ATA ATG AAC GAG ACG ACA GTC GGA GTT GG 3′, and inserted with AvrII and SalI. We used site-directed mutagenesis (QuikChange II XL kit) to introduce the Thr48Trp, Trp67Leu, Leu87Phe, and Leu87Trp replacements. Microinjections were performed as described previously (Mello et al., 1991). Constructs were injected (0.5–2.5 ng/μl) with the PF15E11.1::GFP coinjection marker (47.5 ng/μl). Ca2+ imaging strains, expressing YC2.60 in the AQR, PQR, URX and neurons, were created as described previously (Gross et al., 2014).
Ca2+ imaging
Ca2+ imaging experiments were performed as described previously (Gross et al., 2014). Ca2+ responses were recorded at 2 frames per second using an Olympus IX71S1F-3–5 inverted microscope equipped with UAPON40X Universal apochromatic water-immersion objective (Olympus), DV2 2-channel imager (Photometrics), Rolera EM-C2 (Qimaging), and MetaMorph software (Molecular Devices). To immobilize worms, we glued them to agarose pads (2% agarose in M9) using Dermabond topical skin adhesive (Ethicon). These worms were trapped inside a 500 μm deep rectangular PDMS chamber (12 mm wide, 0.5 mm deep, 17 mm long). For Ca2+ measurements of Tempol-treated animals, we grew glb-5(Haw);npr-1 worms on NGM plates containing 10 mm Tempole (Enzo Life Sciences, catalog #03051432). In all of our imaging experiments, humidified gases were delivered to the microfluidic chamber as described for the speed measurement set-up. Image analysis was performed using custom-written MATLAB software.
Modeling the GCY-35/36 H-NOX structure
I-TASSER (Zhang, 2008; Roy et al., 2010) was used to generate 3D models of the GCY-35 and GCY-36 H-NOX domains. These models were based on H-NOX protein structures from the Nostoc sp. bacterium (H-NOX GCY-35: PDB ID 2O0C, 3TFG, 2O09, H-NOX GCY-36: PDB ID 2O0C, 3TFG, 2O09, and 4IAE). We then used these models, together with protein alignment files that compared H-NOX domains from various organisms (generated by the ClustalW2 server) (Larkin et al., 2007), to calculate an evolutionary conservation score for each amino acid residue using ConSurf (Ashkenazy et al., 2010). The structural-conservation files were visualized using Pymol (The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC).
Paraquat assay
To validate Tempol intake by worms, we did paraquat survival assays. We grew the worms in the presence of Tempol or on regular NGM plates (as control) and measured the survival of worms in paraquat or M9 buffer during a time course of 24 h. To expose worms to Tempol, we prepared NGM plates containing 10 mm Tempol (Enzo Life Sciences, catalog #03051432). Tempol was added to the agar solution after autoclaving and cooling to 55°C. To protect Tempol from light, we dried the plates (as well as control plates) in a dark room, at 21°C for 24 h. Then, we seeded the plates with 150 μl OP50 (OD600 ∼ 0.6) and incubated them, under same conditions for 2 d. Notably, we prepared fresh NGM plates for each experiment. Worms were synchronized by placing 6–8 gravid hermaphrodites (mothers) on Tempol and control NGM plates, and removing them after 5–6 h. The F1 progeny were grown in the dark at 21°C for 4 d. Before the experiment, the adult progeny were washed from the plates with an M9 buffer and further washed for an additional two times with the same buffer. Then, the worms were put in 96-plate wells (Corning, Costar), up to 12 worms per well in 50 μl M9 buffer. To start the experiment, we added 50 μl M9 buffer to control wells and 50 μl M9 plus400 mm paraquat to the experimental (paraquat) wells. In this way, each well contained 100 μl buffer with either no paraquat (control) or 200 mm paraquat (experimental), respectively. The plates were placed on a shaker at 200 rpm, and the survival of worms was measured after 1, 3, 6, and 24 h. We did seven independent assays for both control and experimental groups. The total number of worms in each experiment was at least 180.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 6.02.
Results
A novel mutation in gcy-35 inhibits the rapid GLB-5(Haw)-dependent recovery from prolonged hypoxia
To explore how GLB-5(Haw) regulates GCY-35/36 function, we took a forward genetics approach. We mutagenized glb-5(Haw);npr-1 worms and looked for suppressors that showed attenuated bordering behavior recovery after 24 h exposure to 1% O2. To avoid isolating mutants with nonfunctional gcy-35 or gcy-36, we considered only mutants that regained strong bordering and high speed on bacteria after 4–6 h recovery at 21% O2. Our mutagenesis screen identified 14 suppressors. To focus on gcy-35 and gcy-36, we initially screened for mutants that mapped to either chromosome I or chromosome X, in which gcy-35 and gcy-36, respectively, are located. We selected two recessive mutations, heb1 and heb3, for further chromosomal interval mapping. We mapped heb1 to an ∼4 Mb genomic interval of chromosome I containing gcy-35. Sequencing this genomic region identified a transition mutation in exon 3 that changes cytosine 1122 to thymine and thus converts leucine 87 in GCY-35 to phenylalanine (Fig. 1A). Leucine 87 (Leu87) is a conserved amino acid within the H-NOX domain (Fig. 1B,C).
To understand how the Leu87Phe mutation may affect GCY-35 function, we generated a homology model of the GCY-35 H-NOX domain. This structure model suggested that Leu87 is close to the heme moiety (Fig. 1D). A previous study suggested that a phenylalanine residue in position 78 in a H-NOX protein from Thermoanaerobacter tengcongensis (Tt) bacterium could interact with heme via π-stacking and that this interaction affects the affinity of the heme toward O2 (Weinert et al., 2013). Indeed, the Phe78Leu mutant showed increased affinity toward O2 compared with the wild-type protein (5.4 nm vs 48 nm). Because Leu87 is replaced with phenylalanine in the H-NOX domain of GCY-35(heb1), it is intriguing to hypothesize that π-stacking interaction between Phe87 and the heme could affect the affinity of GCY-35 for O2.
GCY-35(heb1) inhibits glb-5(Haw) function in the AQR, PQR, and URX neurons
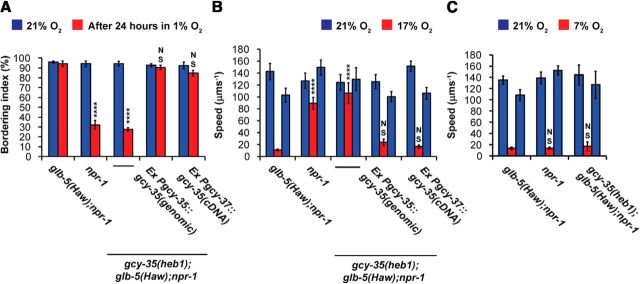
We further explored how Leu87Phe affects GLB-5(Haw) activity using behavioral assays. As expected from previous experiments (Gross et al., 2014), glb-5(Haw);npr-1 animals recovered quickly from prolonged hypoxia and showed bordering behavior after 30 min recovery at 21% O2 similar to naive animals that were kept in 21% O2 (Fig. 2A). However, npr-1 (bearing the nonactive allele of glb-5; glb-5(Bri)) and gcy-35(heb1);glb-5(Haw);npr-1 animals displayed poor bordering behavior recovery after prolonged hypoxia. Importantly, animals from these two strains that were not subjected to hypoxia showed robust bordering behavior, similarly to glb-5(Haw);npr-1 worms, indicating that GLB-5-independent activity of GCY-35/36 was not compromised (Fig. 2A). Expression of either the genomic sequence of gcy-35 under its own promoter or the coding sequence of gcy-35 just in the AQR, PQR, and URX neurons (using the gcy-37 promoter region) in gcy-35(heb1);glb-5(Haw);npr-1 animals restored the fast bordering behavior recovery seen in glb-5(Haw);npr-1 animals (Fig. 2A), demonstrating that the mutation in gcy-35 was indeed responsible for the failure of the heb1 animals to rapidly recover bordering behavior.
Figure 2.
gcy-35(heb1) suppresses glb-5(Haw)-dependent O2 responses in the AQR, PQR, and URX neurons. A, gcy-35(heb1) inhibits the rapid recovery of bordering in glb-5(Haw); npr-1 animals returned to 21% O2 after 24 h exposure to 1% O2. Bordering was measured 30 min after return to 21% O2. Asterisks indicate significance for comparisons with glb-5(Haw); npr-1 animals (after 24 h in 1% O2). B, gcy-35(heb1) suppresses the switch from rapid to slow movement when O2 levels drop from 21% to 17%. Asterisks indicate significance for comparisons with glb-5(Haw); npr-1 at 17% O2. C, gcy-35(heb1);glb-5(Haw);npr-1 animals are able to switch from rapid to slow movement when O2 levels drop from 21% to 7%. n = 6 assays performed over at least 3 d. ****p < 0.0001 (two-way ANOVA with Bonferroni post-test). NS, Not significant. Error bars indicate SEM.
Because GLB-5(Haw) is also important for the robust slowing response at 17% O2 (Persson et al., 2009), we asked whether gcy-35(heb1) affects GLB-5(Haw) function in this behavioral paradigm. To answer this question, we compared the speed of worms at 21% and 17% O2 (Fig. 2B). At 21% O2, npr-1, glb-5(Haw);npr-1 and gcy-35(heb1);glb-5(Haw);npr-1 animals moved quickly on the bacterial lawn. However, at 17% O2, the speed of glb-5(Haw);npr-1 animals significantly decreased, whereas npr-1 and gcy-35(heb1);glb-5(Haw);npr-1 worms continued to move quickly. Similarly to the bordering behavior recovery assay, the expression of either the genomic region of gcy-35 (under its own promoter) or gcy-35 coding sequence in AQR, PQR, and URX was sufficient to rescue the slowing behavior of gcy-35(heb1);glb-5(Haw);npr-1 worms to glb-5(Haw);npr-1 levels (Fig. 2B). Importantly, gcy-35(heb1);glb-5(Haw);npr-1 worms showed a robust slowing response when shifted from 21% to 7% O2 (Fig. 2C), indicating that GLB-5(Haw)-independent activity in these animals is intact. Together, our results show that gcy-35(heb1) blocks the effect of GLB-5(Haw) in the O2-sensing neurons AQR, PQR, and URX, while having little effect on glb-5(Haw)-independent behaviors.
GCY-35(heb1) blocks the rapid desensitization of URX after prolonged hypoxia
The activation of GCY-35/36 by O2 is thought to trigger a cGMP-dependent signaling cascade that maintains high tonic calcium (Ca2+) levels in AQR, PQR, and URX at 21% O2 (Busch et al., 2012; Couto et al., 2013). Indeed, ratiometric Ca2+ measurements using the genetically encoded yellow chameleon Ca2+ sensors YC2.60 and YC3.60 (Nagai et al., 2004) detected a high Ca2+ level in the URX neurons at 21% O2, which dropped upon transition to lower O2 concentrations (Persson et al., 2009; Busch et al., 2012; Gross et al., 2014). Therefore, Ca2+ imaging provides an efficient tool to follow the activity of GCY-35/36 in the URX neurons.
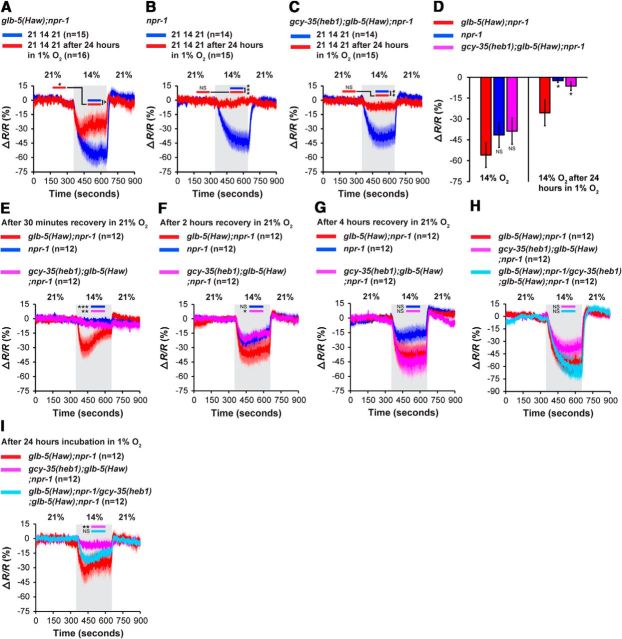
Previous studies demonstrated that glb-5(Haw);npr-1 animals show rapid recovery of Ca2+ responses in the URX neurons after prolonged exposure to 1% O2 (Gross et al., 2014). To determine whether the failure of gcy-35(heb1);glb-5(Haw);npr-1 animals to resume robust bordering behavior after hypoxia is due to attenuated recovery of Ca2+ responses, we created npr-1, glb-5(Haw);npr-1, and gcy-35(heb1);glb-5(Haw);npr-1 transgenic worms that express YC2.60 in the URX neurons. We exposed these strains to 1% O2 for 24 h (or left them at 21% O2 as controls) and measured O2-evoked Ca2+ responses using a 21%–14%-21% O2 stimulus (Fig. 3). We chose the 21%–14% O2 protocol to mimic the O2-concentration shift that the worms experience when they are returned to 21% O2 after exposure to hypoxia and then move to the bacterial lawn border (Gray et al., 2004). The shift from 21% to 14% O2 inhibited the activity of URX in control animals from the three strains (Fig. 3A–D), indicating that 14% O2 is below the activation threshold of URX in worms that did not experience hypoxia. However, after 24 h exposure to hypoxia, the activity of the URX neurons of npr-1, and gcy-35(heb1);glb-5(Haw);npr-1 worms was not significantly inhibited at 14% O2, whereas the activity of URX in glb-5(Haw);npr-1 animals was substantially reduced (Fig. 3A–D).
Figure 3.
O2-evoked Ca2+ responses in the URX neurons. A–C, O2-evoked Ca2+ responses were measured in animals expressing the chameleon YC2.60 in the URX neurons. The responses were measured in naive worms (grown in 21% O2, blue Ca2+ traces) or in worms exposed to 1% O2 (red Ca2+ traces). Plots represent relative ratio change (ΔR/R) compared with the baseline in the first 21% O2 period (the mean ratio during 2 min, just before the shift to 14% O2). Asterisks indicate significant differences at times specified by bars between the Ca2+ levels (unpaired t test with Welch's correction). Gray shading represents 14% O2. n = the number of worms imaged in each strain. Error bars (lighter shading) indicate SEM. D, Bar graph comparing tonic URX Ca2+ levels at 14% O2 (2 min before the switch to 21% O2) in animals that were exposed to 1% O2 for 24 h or grown at 21% O2 (data from A–C). Unpaired t test with Welch's correction. Error bars indicate SEM. E–G, The 21%–14%-21% O2-evoked Ca2+ responses in URX neurons at three recovery time-points after prolonged hypoxia; after 30 min (E), 2 h (F), and 4 h (G) in 21% O2. The 21%–14%-21% O2-evoked Ca2+ responses in URX neurons of glb-5(Haw);npr-1/gcy-35(heb1);glb-5(Haw);npr-1 heterozygous worms. H, Naive animals that were not exposed to hypoxia. I, After 24 h exposure to hypoxia. The data for glb-5(Haw);npr-1 and gcy-35(heb1);glb-5(Haw);npr-1 worms were taken from A, C. Asterisks indicate significant compared with glb-5(Haw);npr-1 Ca2+ levels at times specified by bars (two-way ANOVA with Bonferroni post-test). Gray shading represents 14% O2. n = the number of worms imaged in each strain. Error bars (lighter shading) indicate SEM. *p < 0.05. **p < 0.01. ***p < 0.001. NS, Not significant.
The recovery of npr-1 worms from prolonged hypoxia is a gradual process that is completed within 4 h incubation in 21% O2 (Gross et al., 2014). In the above Ca2+ experiments, we compared the adaptation of the URX neurons of npr-1, glb-5(Haw);npr-1, and gcy-35(heb1);glb-5(Haw);npr-1 worms after 5–10 min recovery in 21% O2 (Fig. 3A–C). To further explore the recovery time course of URX in these strains, we measured the Ca2+ activity in URX after 30 min, 2 h, and 4 h recovery at 21% O2. We chose the 30 min and 4 h time-points because previous experiments showed that at these time-points the bordering indexes of npr-1 worms after prolonged hypoxia were ∼30% and ∼90%, respectively (Gross et al., 2014). Therefore, these time-points represent poor and full bordering behavior recovery in npr-1 animals, respectively. The 2 h time-point was chosen to represent an intermediate bordering recovery (∼50%). After 30 min recovery in 21% O2, the transition from 21% to 14% O2 evoked a substantial decrease in Ca2+ levels in glb-5(Haw);npr-1 worms that was significantly stronger compared with the [Ca2+] drop in npr-1 and gcy-35(heb1);glb-5(Haw);npr-1 worms (Fig. 3E). These results mirror the differences in bordering recovery of the three strains after 30 min exposure to 21% O2 (Fig. 2A). Moreover, we show that the Ca2+ responses after 5–10 and 30 min recovery in 21% O2 are similar (Fig. 3A–C,E). After 2 h recovery, the 21%–14% shift triggered a robust and sustainable [Ca2+] drop in the three strains (Fig. 3F). However, the Ca2+ levels of gcy-35(heb1);glb-5(Haw);npr-1 worms were slightly (but significantly) higher compared with glb-5(Haw);npr-1 animals. Finally, after 4 h recovery, the 21%–14% shift elicited similar Ca2+ responses in the three strains (Fig. 3G), as expected from the behavioral recovery time course of npr-1 worms.
Our Ca2+ imaging experiments show that gcy-35(heb1) increases the sensitivity of URX to 14% O2 even after 30 min recovery in 21% O2 (Fig. 3E), supporting the hypothesis that heb1 ameliorates the activity of GCY-35. To explore how gcy-35(heb1) activity is affected by gcy-35(wt), we generated glb-5(Haw);npr-1/gcy-35(heb1);glb-5(Haw);npr-1 heterozygous animals that expressed YC2.60 in URX and measured Ca2+ responses evoked by a 21%–14%-21% O2 stimulus. The Ca2+ responses of either naive glb-5(Haw);npr-1/gcy-35(heb1);glb-5(Haw);npr-1 animals that were not exposed to hypoxia or animals that were exposed to hypoxia were similar to glb-5(Haw);npr-1 controls (Fig. 3H,I, respectively). By contrast, gcy-35(heb1);glb-5(Haw);npr-1 animals showed elevated Ca2+ level in 14% O2 (after exposure to hypoxia) compared with glb-5(Haw);npr-1 (Fig. 3I). Together, our results demonstrate that gcy-35(heb1) is recessive to gcy-35(wt).
In summary, our Ca2+ experiments mirror the behavioral results and further support data from a previous study (Gross et al., 2014) suggesting that long exposure to hypoxia increases the sensitivity of URX to O2 and that GLB-5(Haw) rapidly desensitizes URX upon returning to 21% O2. Moreover, we show that GCY-35(heb1) is resistant to this inhibitory effect of GLB-5(Haw) and thus facilitates high neuronal activity at 14% O2 shortly after returning to 21% O2. Because Ca2+ imaging and behavioral assays proved to be efficient ways to study GCY-35(heb1) function, we next combined these techniques to explore how GCY-35(heb1) affects the dynamic range and sensitivity of O2 responses.
GCY-35(heb1) affects the dynamic range of O2 responses
Does heb1 block the inhibitory effect of GLB-5(Haw) by increasing the sensitivity of GCY-35 to O2? If so, one can predict that gcy-35(heb1) should enhance O2 responses independently of glb-5(Haw) function. To test this hypothesis, we generated a gcy-35(heb1);npr-1 strain and measured its speed at 21% and 7% O2 (Fig. 4A). We chose this O2 stimulus because it evokes a similar slowing response in both npr-1 and glb-5(Haw);npr-1 animals (Persson et al., 2009) and thus represents a glb-5(Haw)-independent O2 behavior. The speed of gcy-35(heb1);npr-1 animals at 7% O2 was significantly higher compared with the other strains (Fig. 4A), indicating that GCY-35(heb1) affects GLB-5(Haw)-independent O2 responses. Next, we asked whether GCY-35(heb1) could increase the sensitivity of worms to 17% O2. The speed of gcy-35(heb1);npr-1 worms at 17% O2 was higher compared with npr-1 animals (Fig. 4B), but the difference did not reach significance. Moreover, the speed of gcy-35(heb1);npr-1 worms at 17% O2 was similar to gcy-35(heb1);glb-5(Haw);npr-1 worms, indicating that heb1 efficiently blocks GLB-5(Haw) activity.
Figure 4.
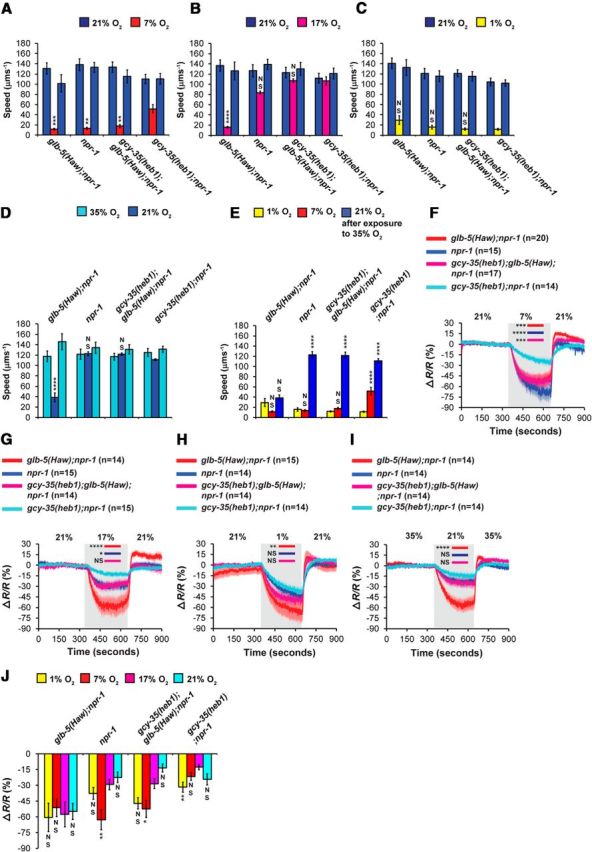
GCY-35(heb1) regulates both glb-5(Haw)-dependent and -independent O2 responses. A–D, O2-evoked speed responses were measured in npr-1, glb-5(Haw);npr-1, gcy-35(heb1);glb-5(Haw);npr-1, and gcy-35(heb1);npr-1 animals. Four O2 stimuli were used: (A) 21%–7%-21%, (B) 21%–17%-21%, (C) 21%–1%-21%, and (D) 35%–21%-35%. Asterisks indicate significance for comparisons with gcy-35(heb1); npr-1 animals at 7% O2, 17% O2, 1% O2, and 21% O2, respectively. Two-way ANOVA with Bonferroni post-test. n = 6 assays performed over at least 3 d. E, Bar graph comparing speed at 1%, 7%, and 21% O2 (2 min before the switch to 21% or 35% O2; data taken from A, C, D). Asterisks indicate significance for comparisons between speeds at 1%, 7%, and 21% O2, within each strain. One-way ANOVA with Dunn's post-test. F–I, Four O2-stimuli (described in A–D) were used to evoke Ca2+ responses in animals expressing the chameleon YC2.60 in the URX neurons. Asterisks indicate significant differences at times indicated by bars between the Ca2+ levels. Two-way ANOVA with Bonferroni post-test. Gray shading represents periods at 7%, 17%, 1%, and 21% O2, respectively. Error bars (lighter shading) indicate SEM. n = the number of worms imaged in each strain. J, Bar graph comparing tonic URX Ca2+ levels at 1%, 7%, 17%, and 21% O2 (2 min before the switch to 21% O2 or 35% O2; data taken from F–I). Asterisks indicate significance for comparisons with Ca2+ levels at 17% O2, within each strain. One-way ANOVA with Dunn's post-test. *p < 0.05. **p < 0.01. ***p < 0.001. ****p < 0.0001. NS, Not significant. Error bars indicate SEM.
To further explore the dynamic range of GCY-35(heb1), we measured the speed of worms at two extreme O2 shifts: 21%–1%-21% and 35%–21%-35% (Fig. 4C,D). The transition from 21% to 1% O2 evoked strong slowing responses in all strains (Fig. 4C), suggesting that the sensitivity of GCY-35(heb1) and GCY-35(wt) to 1% O2 is similar. Next, we measured the speed of worms while stimulated by a 35%–21%-35% O2 shift (Fig. 4D). Strikingly, the exposure of glb-5(Haw);npr-1 worms to 35% O2 for 5 min caused a robust slowing response upon shifting to 21% O2 (Fig. 4D). The speed of glb-5(Haw);npr-1 worms at 21% O2 resembled their speed at 1% O2 (Fig. 4E). Importantly, glb-5(Haw);npr-1 worms did not lose their ability to respond to O2 and displayed a sharp increase in speed upon returning to 35% O2, indicating that GCY-35 did not lose its heme group due to oxidative damage (Rogers et al., 2014). The transition from 35% to 21% O2 did not trigger a significant slowing response in the other strains (Fig. 4D), indicating that the inhibitory activity of GLB-5(Haw) is essential for this adaptation and that GCY-35(heb1) is resistant to this inhibition. To explore how GCY-35(heb1) affects the O2-dynamic range at the neuronal level, we next measured the activity of URX using Ca2+ imaging.
O2-evoked Ca2+ responses in gcy-35(heb1) mutants
To explore whether GCY-35(heb1) changes the dynamic range of O2 responses by directly modulating the sensitivity of URX to O2, we recorded Ca2+ responses in these neurons using the same O2 stimuli as described in Figure 4A–D. As expected from the behavioral assays, the Ca2+ levels in the URX neurons of gcy-35(heb1);npr-1 worms at 7% O2 were significantly higher compared with the other strains (Fig. 4F), indicating that GCY-35(heb1) increases the sensitivity of URX to O2. The transition from 21% to 17% O2 evoked a small drop in Ca2+ levels in the URX neurons of npr-1, gcy-35(heb1);npr-1, and gcy-35(heb1);glb-5(Haw);npr-1 worms (Fig. 4G). However, the Ca2+ levels of glb-5(Haw);npr-1 worms at 17% O2 were significantly lower compared with the other strains (p < 0.001 compared with npr-1 and gcy-35(heb1);glb-5(Haw);npr-1 worms, and p < 0.0001 compared with gcy-35(heb1);npr-1 animals, two-way ANOVA with Bonferroni post-test), indicating that GCY-35(heb1) enhances the activity of URX even in the presence of GLB-5(Haw). The shift from 21% to 1% O2 elicited a robust decrease in Ca2+ levels in all strains (Fig. 4H). However, counter to expectation from the behavioral assay results (Fig. 3C), Ca2+ levels in the URX neurons of glb-5(Haw);npr-1 worms in 1% O2 were significantly lower compared with gcy-35(heb1);npr-1 worms. Finally, the transition from 35% to 21% O2 evoked a significantly stronger decrease in Ca2+ level in glb-5(Haw);npr-1 worms (Fig. 4I) compared with the other strains (p < 0.0001, two-way ANOVA with Bonferroni post-test).
In conclusion, the Ca2+ imaging data mirror the behavioral results and suggest that GCY-35(heb1) increases the sensitivity of URX to O2. Nevertheless, the activation of URX by 7% O2 is higher in gcy-35(heb1);npr-1 animals compared with gcy-35(heb1);glb-5(Haw);npr-1 worms (Fig. 4F,J), indicating that that GCY-35(heb1) is not completely resistant to the inhibitory effect of GLB-5(Haw). Finally, we show, for the first time, that short exposure of worms to 35% O2 desensitizes URX to a subsequent activation by 21% O2. This adaptation requires the inhibitory activity of GLB-5(Haw) and is suppressed by GCY-35(heb1) activity. We next continued to explore how GLB-5(Haw) inhibits GCY-35(heb1) activity.
GLB-5(Haw) regulates GCY-35 function in a dose-dependent manner
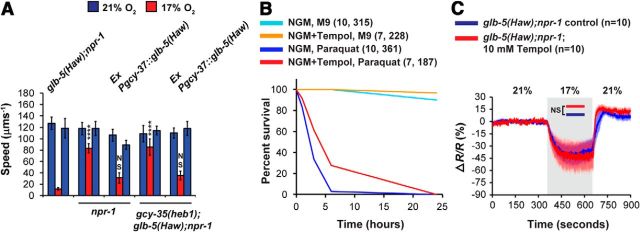
So far, our results support the hypothesis that GLB-5(Haw) inhibits GCY-35 activity, and that GCY-35(heb1) is partially resistant to this inhibition. Therefore, we asked whether overexpression of glb-5(Haw) in the AQR, PQR, and URX neurons could overcome this resistance and thus restore the glb-5(Haw)-dependent slowing response at 17% O2. To test this possibility, we created a gcy-35(heb1);glb-5(Haw);npr-1 strain that overexpresses the coding sequence of glb-5(Haw) under the gcy-37 promoter and measured the speed of these worms at 21% and 17% O2 (Fig. 5A). The overexpression of glb-5(Haw) in AQR, PQR, and URX restored the strong slowing response to gcy-35(heb1);glb-5(Haw);npr-1 animals (Fig. 5A), indicating that the inhibition of GCY-35 by GLB-5(Haw) is dose-dependent. Of note, the same chromosomal array induced a robust slowing response in npr-1 worms, verifying that the rescuing construct restores the function of GLB-5(Haw) in the AQR, PQR, and URX neurons (Persson et al., 2009).
Figure 5.
GLB-5(Haw) regulates GCY-35 function in a dose-dependent manner, independently of superoxide signaling. A, Overexpression of glb-5(Haw) in the AQR, PQR, and URX neurons of gcy-35(heb1);glb-5(Haw);npr-1 animals restores the robust slowing response at 17% O2. Asterisks indicate significance for comparisons with glb-5(Haw); npr-1 animals at 17% O2. Two-way ANOVA with Bonferroni post-test. ****p < 0.0001. n = 6 assays performed over at least 3 d. Error bars indicate SEM. B, Tempol intake significantly increases the survival of worms in paraquat. Survival curves of glb-5(Haw);npr-1 worms in M9 buffer or 200 mm paraquat. Worms were grown on NGM plates containing 10 mm Tempol or on regular NGM plates as control. The Tempol treatment significantly improved the survival in paraquat (p < 0.0001, log-rank test). Numbers in parentheses indicate the number of independent experiments and total number of adult worms, respectively. C, O2-evoked Ca2+ responses in the URX neurons of glb-5(Haw);npr-1 animals that were grown on NGM plates containing 10 mm Tempol or regular NGM plates as control (similarly to worms in B). Gray shading represents periods at 17% O2. n = the number of worms imaged in each strain. Error bars (lighter shading) indicate SEM. Significance for comparisons between strains at 17% was calculated using unpaired t test with Welch's correction. NS, Not significant.
Previous studies suggested that the C. elegans globins GLB-6 and GLB-26 may generate superoxide radicals (Yoon et al., 2010; Tiso et al., 2011). Because these molecules could potentially inhibit GCY-35 activity, we asked whether GLB-5(Haw) inhibits GCY-35 activity via superoxide radicals. To explore this question, we grew glb-5(Haw);npr-1 worms on NGM plates containing the superoxide scavenger Tempol (10 mm) or on regular NGM plates as control. We synchronized the worms by letting 6–8 gravid hermaphrodites (mothers) lay eggs on the Tempol and control NGM plates, and subsequently removed the mothers after 5–6 h. In this way, the synchronized worms were exposed to the Tempol throughout their life cycle (from the egg to the adult stage). Before examining the effect of Tempol on the URX Ca2+ activity of glb-5(Haw);npr-1 worms, we set out to demonstrate that Tempol indeed enters and is effective in the worms. For this aim, we measured the survival of worms in 200 mm paraquat in a time course of 24 h. Paraquat triggers mitochondrial superoxide generation (Drechsel and Patel, 2009; Yang and Hekimi, 2010). Therefore, we hypothesized that Tempol intake by the worms should provide protection against paraquat compared with control worms that grew on regular NGM plates. In addition, we did a control experiment in which we measured the survival of worms, grown on Tempol and regular NGM, in M9 buffer during a time course of 24 h. The Tempol treatment significantly increased the survival of worms in paraquat (Fig. 5B; p < 0.0001, log-rank, Mantel-Cox), indicating that Tempol functioned in the worms. Interestingly, the Tempol treatment improved the survival of worms in M9 (Fig. 5B; p < 0.0034, log-rank, Mantel-Cox), suggesting that 24 h incubation in M9 may induce a mild oxidative stress. Next, we recorded 21%–17%-21% O2 evoked Ca2+ responses in URX in Tempol-treated and control worms. We used this O2 stimulus because the 21%–17% O2 downshift evokes a significantly bigger decrease in Ca2+ level in glb-5(Haw);npr-1 worms compared with npr-1 animals. Therefore, it can be used as a measure for GLB-5(Haw) activity. The Ca2+ responses of glb-5(Haw);npr-1 animals in the presence of Tempol were similar to glb-5(Haw);npr-1 controls (Fig. 5C), indicating that, at least under these experimental conditions, superoxide radicals are not important for GCY-35 inhibition. To further understand how GCY-35 activity is regulated, we explored the function of other conserved amino acid residues in this protein.
Threonine 48 and tryptophan 67 are essential for GCY-35 activity
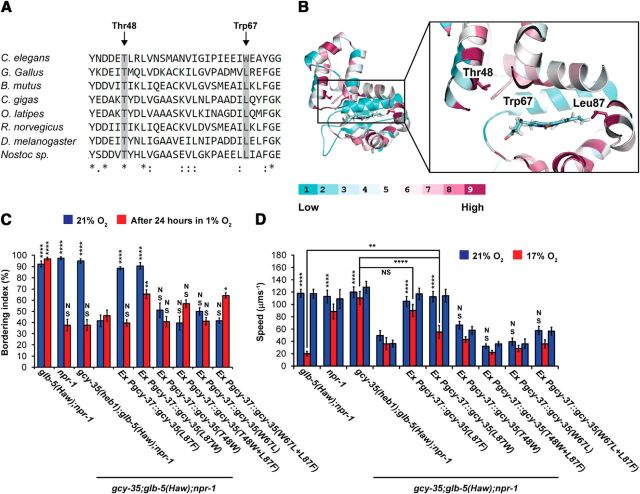
The discovery of heb1 prompted us to search for other regulatory amino acid residues in the H-NOX domain of GCY-35. The crystal structure of the NO-binding H-NOX protein from Nostoc bacterium revealed a “Y-shaped” channel system inside this protein (composed of tunnels 1 and 2) that appears to facilitate gas migration to and from the heme cavity (Winter et al., 2011). Indeed, blocking tunnel 2 by replacing leucine 67 with the larger amino acid tryptophan (Leu67Trp) increased the binding affinity for carbon monoxide (CO), probably by extending its duration at the heme cavity (Winter et al., 2011). Interestingly, this conserved leucine is naturally replaced with tryptophan in the H-NOX domain of GCY-35 (Fig. 6A,B), as well as in the other putative O2 sensors GCY-32, GCY-34, GCY-36, and GCY-37. Therefore, we asked whether tryptophan 67 (Trp67) is important for GCY-35 O2-sensing activity. We used site-directed mutagenesis to replace Trp67 with leucine in the gcy-35 (cDNA) rescuing construct that we previously used (Fig. 2). The gcy-35 (Trp67Leu) construct was then injected into worms bearing the gcy-35(ok769) deletion allele of gcy-35. Because the deletion allele of gcy-35 (hereafter referred to as gcy-35) suppresses the bordering behavior and high speed on food of gcy-35;glb-5(Haw);npr-1 worms at 21% O2 (Persson et al., 2009), we used these behaviors to examine the functionality of the gcy-35 (Trp67Leu) rescuing construct. The bordering and speed behavior of gcy-35 (Trp67Leu) transgenic animals were similar to gcy-35;glb-5(Haw);npr-1 worms (Fig. 6C,D), indicating that GCY-35(Trp67Leu) was not functional. As a control, we replaced leucine 87 in the gcy-35(cDNA) rescuing construct with either phenylalanine or tryptophan. Transgenic gcy-35(Leu87Phe) and gcy-35(Leu87Trp) animals showed high bordering and speed behavior at 21% O2 (similarly to glb-5(Haw);npr-1 worms; Fig. 6C,D), indicating that our rescuing protocol is effective. Moreover, transgenic gcy-35(Leu87Phe) and gcy-35(heb1);glb-5(Haw);npr-1 animals showed similar attenuated bordering recovery after hypoxia and high speed at 17% O2 (Fig. 6C,D), demonstrating that the gcy-35(Leu87Phe) rescuing construct recapitulated the activity of GCY-35(Heb1). Interestingly, the magnitudes of recovery after hypoxia and slowing response at 17% O2 of gcy-35(Leu87Trp) animals were intermediates between glb-5(Haw);npr-1 and gcy-35(heb1);glb-5(Haw);npr-1 worms (Fig. 6C,D), indicating that GCY-35(Leu87Trp) is less resistant to the inhibitory activity of GLB-5(Haw) compared with GCY-35(Leu87Phe).
Figure 6.
Thr48 and Trp67 are essential for GCY-35 activity. A, Protein sequence alignment (generated by the ClustalW2 server) of H-NOX domains from Caenorhabditis elegans GCY-35 (NP_001252131), Gallus gallus (XP_004937100), Bos mutus (ELR49741), Crassostrea gigas (EKC42091), Oryzias latipes (NP_001098123), Rattus norvegicus (EDL85292), Drosophila melanogaster (ABG02144), and Nostoc sp.(WP_01099643). B, A 3D model of the GCY-35 H-NOX domain was generated. The side-chains of Thr48, Trp67, and Leu87 are shown. The degree of conservation is color coded, from 1 (low) to 9 (high). C, D, Thr48 and Trp67 are essential for normal bordering behavior and high-foraging speed at 21% O2. C, Bordering was measured 30 min after return to 21% O2. Asterisks indicate significance for comparisons with gcy-35;glb-5(Haw);npr-1 animals that were either kept at 21% O2 or exposed to 1% O2 for 24 h. Two-way ANOVA with Bonferroni post-test. D, The 21%–17%-21% O2-evoked speed responses. In case of comparison between strains at 21% O2 (first period), asterisks indicate significance for comparisons with gcy-35;glb-5(Haw); npr-1 animals. Two-way ANOVA with Bonferroni post-test. *p < 0.05. **p < 0.01. ****p < 0.0001. NS, Not significant. Error bars indicate SEM. n = 6 assays performed over at least 3 d.
The crystal structure of the H-NOX(Leu67Trp) protein reveals a close interaction between Trp67 and threonine 48 (Winter et al., 2011). Indeed, mutating threonine 48 (Thr48) to tryptophan (Thr48Trp) blocks tunnels 1 and 2 and increases the binding affinity of the heme for CO (Winter et al., 2011). Thr48 is conserved in C. elegans GCY-35 (Fig. 6A,B) and is predicted to be in close proximity to Trp67 H-NOX domain (Fig. 6B). To determine whether Thr48 is important for GCY-35 activity, we replaced Thr48 with tryptophan in the gcy-35 rescuing construct, generated transgenic worms, and tested their O2 responses as described above (Fig. 6C,D). The bordering and speed behavior of gcy-35 (Thr48Trp) transgenic animals were similar to gcy-35;glb-5(Haw);npr-1 controls (Fig. 6C,D), indicating that GCY-35 (Thr48Trp) is not functional.
In conclusion, we show that Thr48 and Trp67 are essential for GCY-35 activity. Because equivalent amino acid positions regulate the interaction of heme with gaseous ligands in the Nostoc H-NOX protein, it is tempting to speculate that similar structural regions regulate gas/heme interaction in both NO- and O2-binding H-NOX proteins. To expand our knowledge about sGC activity regulation, we next cloned the heb3 mutation.
The heb3 mutation changes a conserved valine to methionine in the H-NOX domain of GCY-36
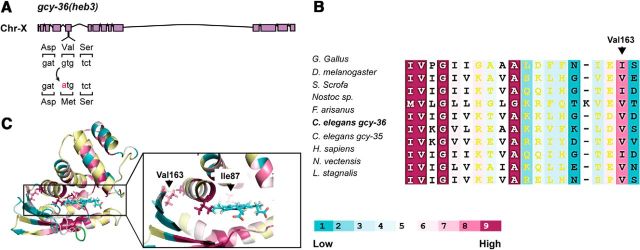
The second mutant from our screen that met our interval mapping criteria was heb3. Using snip single nucleotide polymorphism mapping, we mapped heb3 to ∼1.5 Mb genomic interval of chromosome X containing gcy-36. Sequencing this genomic region identified a transition mutation in exon 5 that changes guanine 805 to adenine, and thus converts valine 163 (Val163) to methionine (Fig. 7A). Like Leu67, Val163 is a conserved amino acid within the H-NOX domain (Fig. 7A,B). To explore how the Val163Met mutation may affect GCY-36 function, we generated a homology model of the GCY-36 H-NOX domain (Fig. 7C). This structure model suggested that Val163 is not located near the heme group (Fig. 7C). Moreover, a previous study proposed that equivalent valine residues in H-NOX proteins from Nostoc and Tt bacteria are not involved in gas migration (Zhang et al., 2010). Therefore, based on this information, we hypothesized that heb3 affects GCY-36 activity indirectly, by regulating its interaction with GLB-5(Haw). To explore this hypothesis, we first asked whether gcy-36(heb3) regulates the function of GLB-5(Haw) in the AQR, PQR, and URX neurons.
Figure 7.
heb3 is a transition mutation that converts Val163 to methionine in the H-NOX domain of GCY-36. A, Schematic diagram of gcy-36, highlighting the transition mutation in the heb3 allele. B, Amino acid conservation analysis of the H-NOX domain from different organisms. Aligned sequences are from Homo sapiens (BAG37638), Sus scrofa (XP_003356977), Gallus gallus (XP_426684), Nostoc sp. (WP_010996435), Nematostella vectensis (XP_001639149), Drosophila melanogaster (ABG02144), Caenorhabditis elegans GCY-36 (NP_510557), Caenorhabditis elegans GCY-35 (NP_001252131), Fopius arisanus (XP_011299736), and Lymnaea stagnalis (ACC95432). The degree of conservation is color coded, from 1 (low) to 9 (high). Arrow indicates the conserved Val163 in GCY-36. C, 3D presentation of the protein sequence alignment data displayed on a homology model of the GCY-36 H-NOX domain. The side-chains of isoleucine 87 and valine 163 are shown.
gcy-36(heb-3) inhibits glb-5(Haw)-dependent O2 responses in AQR, PQR, and URX
Our mapping results suggest that GCY-36(heb3) is resistant to the inhibitory effect of GLB-5(Haw) in the AQR, PQR, and URX neurons. To validate this, we measured the bordering recovery and slowing response at 17% O2 of npr-1, glb-5(Haw);npr-1, and glb-5(Haw);npr-1 gcy-36(heb3) animals (Fig. 8A,B). The bordering recovery and slowing response of glb-5(Haw);npr-1 gcy-36(heb3) animals were significantly reduced compared with glb-5(Haw);npr-1 controls (Fig. 8A,B), indicating that gcy-36(heb3) is generally important for GLB-5(Haw) function. Of note, in addition to exhibiting decreased bordering recovery after hypoxia, glb-5(Haw);npr-1 gcy-36(heb3) animals that were not exposed to 1% O2 also showed decreased bordering (Fig. 8A). To demonstrate that gcy-36(heb3) regulates GLB-5(Haw) function in the AQR, PQR, and URX neurons, we expressed the coding region of gcy-36(wt) under the gcy-37 promoter in glb-5(Haw);npr-1 gcy-36(heb3) animals. The resulting transgenic strain displayed both rapid recovery from prolonged hypoxia and a robust slowing response at 17% O2 (Fig. 8A,B). Moreover, these transgenic worms showed high bordering index at 21% O2 (Fig. 8A). Therefore, our results demonstrate that gcy-36(heb3) is important for GLB-5(Haw)-dependent O2 responses in the AQR, PQR, and URX neurons.
Figure 8.
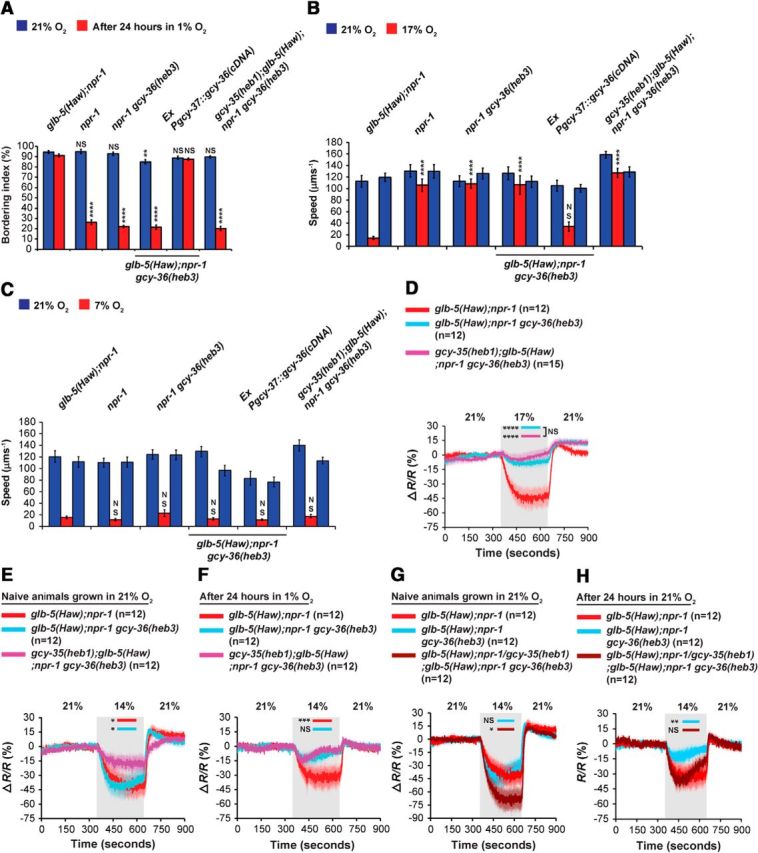
gcy-36(heb3) is a recessive mutation that regulates glb-5(Haw)-dependent O2 responses. A, The Val163Met replacement (heb3) inhibits the fast bordering recovery from prolonged hypoxia. Statistical significance was calculated against the bordering index of glb-5(Haw); npr-1 animals (naive and after 24 h in 1% O2). Two-way ANOVA with Bonferroni post-test. B, C, Speed measurements at 17% and 7% O2, respectively. The Val163Met replacement suppresses the switch from rapid to slow movement when O2 levels drop from 21% to 17%. Asterisks indicate significance for comparisons with glb-5(Haw); npr-1 at 17% O2 (B). The slowing response, induced by the 21%–7% O2 shift, of glb-5(Haw); npr-1 gcy-36(heb3) animals is similar to glb-5(Haw);npr-1 controls (C). Two-way ANOVA with Bonferroni post-test. n = 6 assays performed over at least 3 d. Error bars indicate SEM. D–F, Ca2+ imaging in animals expressing the chameleon YC2.60 in the URX neurons. Two O2 stimulus train profiles were used to evoke Ca2+ responses: 21%–17%-21% (D), 21%–14%-21% (in naive worms (E), and worms after 24 incubation in 1% O2 (F). The 21%–14%-21% O2-evoked Ca2+ responses in URX neurons of glb-5(Haw);npr-1/glb-5(Haw);npr-1 gcy-36(heb3) heterozygous worms. G, Naive animals that were not exposed to hypoxia. H, After 24 h exposure to hypoxia. The data for glb-5(Haw);npr-1 and glb-5(Haw);npr-1 gcy-36(heb3) worms were taken from E, F. Asterisks indicate significance compared with glb-5(Haw);npr-1 Ca2+ levels at times specified by bars. Two-way ANOVA with Bonferroni post-test. Gray shading represents periods at 17%, and 14% O2. Error bars (lighter shading) indicate SEM. n = the number of worms imaged in each strain. *p < 0.05. **p < 0.01. ***p < 0.001. ****p < 0.0001. NS, Not significant.
The function of gcy-36(heb3) in glb-5(Haw)-independent O2 responses
To explore whether GCY-36(heb3) affects glb-5(Haw)-independent O2-responses, we generated an npr-1 gcy-36(heb3) strain and compared its speed with npr-1, glb-5(Haw);npr-1, and glb-5(Haw);npr-1 gcy-36(heb3) animals at 7% O2 (Fig. 8C). The npr-1 gcy-36(heb3) animals were not significantly faster than glb-5(Haw);npr-1 controls (Fig. 8C), indicating that gcy-36(heb3) does not regulate glb-5(Haw)-independent O2 responses. To test whether GCY-36(heb3) is completely resistant to the inhibitory effect of GLB-5(Haw), we compared the recovery after hypoxia and slowing response at 17% O2 of npr-1 gcy-36(heb3) and glb-5(Haw);npr-1 gcy-36(heb3) worms (Fig. 8A,B). The recovery after hypoxia and speed at 17% O2 were similar in these strains, indicating that GCY-36(heb3) is fully resistant to GLB-5(Haw) inhibition.
Together, our results show that GCY-36(heb3) escapes the inhibitory effect of GLB-5(Haw) without increasing the sensitivity of URX to O2. Because GCY-35(heb1) increases the sensitivity of URX to O2, we conclude that GCY-35(heb1) and GCY-36(heb3) resist the inhibitory effect of GLB-5(Haw) by different mechanisms. GCY-35(heb1) appears to act by increasing the affinity toward O2 and therefore the activity of the cyclase at lower O2 levels. GCY-36(heb3), in turn, may interfere with the binding of GLB-5(Haw) to the GCY-35/GCY-36(heb3) complex. Next, we explored the combined effect of GCY-35(heb1) and GCY-36(heb3) on O2 responses.
The combined effect of gcy-35(heb1) and gcy-36(heb3) activity on O2 responses
GCY-35 and GCY-36 appear to form a functional heterodimer in the AQR, PQR, and URX neurons (Cheung et al., 2004; Gray et al., 2004). To explore the function of the putative GCY-35(heb1)/GCY-36(heb3) heterodimer, we created a gcy-35(heb1);glb-5(Haw);npr-1 gcy-36(heb3) double mutant strain and used bordering and speed assays to test its activity. The bordering behavior and slowing responses of the double mutant strain at 7% and 17% O2 were similar to the glb-5(Haw);npr-1 gcy-36(heb3) single mutant strain (Fig. 8A–C), indicating that the interaction between these alleles is not synergistic. Next, we explored the function of GCY-35(heb1)/GCY-36(heb3) directly in the URX neurons. For this aim, we introduced the Pgcy-37::YC2.60 array into glb-5(Haw);npr-1 gcy-36(heb3) and gcy-35(heb1);glb-5(Haw);npr-1 gcy-36(heb3) animals and recorded O2-evoked Ca2+ responses (Fig. 8D–F). First, we recorded Ca2+ responses evoked by a 21%–17%-21% O2 stimulus (Fig. 8D). The Ca2+ levels at 17% O2 of glb-5(Haw);npr-1 gcy-36(heb3) and gcy-35(heb1);glb-5(Haw);npr-1 gcy-36(heb3) animals were similar, and both were significantly higher compared with glb-5(Haw);npr-1 controls. Therefore, our results suggest that GCY-35(heb1) does not further activate the GCY-35(heb1)/GCY-36(heb3) heterodimer at 17% O2 compared with GCY-35(wt)/GCY-36(heb3). Next, we measured Ca2+ responses evoked by a 21%–14%-21% O2 stimulus in naive worms and worms that experienced 1% O2 for 24 h (Fig. 8E,F, respectively). The Ca2+ levels at 14% O2 of naive gcy-35(heb1);glb-5(Haw);npr-1 gcy-36(heb3) animals were significantly higher compared with glb-5(Haw);npr-1 gcy-36(heb3) and glb-5(Haw);npr-1 worms (Fig. 8E), suggesting that gcy-35(heb1) activity is additive to that of gcy-36(heb3) at this O2 concentration. The exposure to hypoxia significantly suppressed the Ca2+ responses of glb-5(Haw);npr-1 gcy-36(heb3) and gcy-35(heb1);glb-5(Haw);npr-1 gcy-36(heb3) worms compared with glb-5(Haw);npr-1 controls at 14% O2 (Fig. 8F; p < 0.01 and p < 0.001, respectively; two-way ANOVA with Bonferroni post-test). Notably, the Ca2+ responses of glb-5(Haw);npr-1 gcy-36(heb3) and gcy-35(heb1);glb-5(Haw);npr-1 gcy-36(heb3) worms at 14% O2 were similar, suggesting that gcy-35(heb1) activity is not additive to that of gcy-36(heb3) at this behavioral paradigm. Together, our behavioral and Ca2+ imaging experiments show that the interaction between GCY-35(heb1) and GCY-36(heb3) does not provide additional protection against the inhibitory effect of GLB-5(Haw) after exposure to hypoxia or at 17% O2 but may be additive at 14% O2 in naive worms that were not exposed to hypoxia.
The activity of gcy-36(heb3) ameliorates the sensitivity of URX to O2 (Fig. 8D,F). However, unlike gcy-35(heb1), gcy-36(heb3) does not affect glb-5(Haw)-independent O2 responses (Fig. 8C), suggesting that gcy-36(heb3) increases the activity of URX by protecting against the inhibitory activity of GLB-5(Haw). To explore how gcy-36(heb3) affects glb-5(Haw) activity in the presence of gcy-36(wt), we created heterozygous glb-5(Haw);npr-1/glb-5(Haw);npr-1 gcy-36(heb3) worms and measured Ca2+ responses evoked by a 21%–14%-21% O2 stimulus in URX. Naive heterozygous worms that were not exposed to hypoxia displayed a robust decrease in Ca2+ level upon transition from 21% to 14% O2 (Fig. 8G). Interestingly, the drop in Ca2+ level was slightly stronger than in glb-5(Haw);npr-1 controls. The Ca2+ responses of the heterozygous worms after hypoxia were similar to glb-5(Haw);npr-1 controls (Fig. 8H), indicating that gcy-36(heb3) is recessive to gcy-36(wt). Together, our results show that, although gcy-35(heb1) and gcy-36(heb3) increase the sensitivity of URX to O2, both the mutations are recessive.
Discussion
cGMP homeostasis is important for animal physiology and therefore should be tightly regulated. Indeed, abnormal cGMP levels are associated with the pathophysiology of diseases, including atherosclerosis, stroke, myocardial ischemia, moyamoya, and achalasia (Stasch et al., 2011; Hervé et al., 2014). The production of cGMP is catalyzed by sGCs. Therefore, understanding sGC regulation is crucial for understanding cGMP homeostasis. In this study, we explore how the atypical sGCs GCY-35 and GCY-36 are regulated by the neuroglobin GLB-5.
The glb-5(Haw)-dependent bordering recovery assay as a tool to identify regulatory amino acid residues in GCY-35 and GCY-36
Previous studies showed that glb-5(Haw);npr-1 worms recover quickly after prolonged hypoxia, whereas npr-1 worms recover slowly (Gross et al., 2014). Here we used this “recovery lag-time” to identity glb-5(Haw) suppressors. Because GLB-5(Haw) function depends on GCY-35/36 activity (Persson et al., 2009), we predicted that our screen would identify regulatory amino acid residues in these proteins. Indeed, two of three mutations we cloned are found in these genes. Moreover, we were able to use the recovery lag-time phenotype to explore by direct mutagenesis the function of other amino acid residues in GCY-35. In conclusion, we would like to suggest that the glb-5(Haw)-dependent bordering recovery assay could be used as an efficient tool to identify and study regulatory amino acid residues in sGC. We anticipate that many of these amino acid residues would be conserved, such that findings regarding sGC regulation in nematodes might be relevant to humans.
How does heb1 regulate GCY-35 activity?
The heb1 mutation converts leucine 87 to phenylalanine (Leu87Phe) in the H-NOX domain of GCY-35. Based on the structure model of the GCY-35 H-NOX domain (Fig. 1D), Phe87 is located near the heme group (Fig. 1). As aromatic side chains can make π-stacking interactions with heme (Weinert et al., 2013), it is tempting to hypothesize that Phe87 interacts with the heme in this manner, thereby increasing the affinity of the heme for O2. The hypothesis that the Leu87Phe mutation increases the O2 affinity of GCY-35 leads to the following predictions: (1) GCY-35(heb1) would be activated by lower O2 levels compared with GCY-35(wt); and (2) GCY-35(heb1) would be more resistant to the inhibitory effect of GLB-5(Haw). Our results are consistent with these predictions. For example, both speed and URX activity of gcy-35(heb1);npr-1 animals at 7% O2 are higher compared with npr-1 worms (Fig. 4B,G). Moreover, both speed and URX activity of gcy-35(heb1);glb-5(Haw);npr-1 animals at 17% O2 are higher compared with glb-5(Haw);npr-1 worms (Figs. 2B, 4F, respectively). Although these data are consistent with increased O2 affinity in GCY-35(heb1), it is important to consider other models that could account for the enhanced activity of GCY-35(heb1). For example, previous studies on sGC from rat suggested an auto-inhibitory model for sGC activity. In this model, the H-NOX domain inhibits the cyclase activity by direct interaction, and the binding of NO to the H-NOX domain relieves this interaction and thus activates the enzyme (Winger and Marletta, 2005). It could be that the Leu87Phe mutation decreases the interaction between the H-NOX and the catalytic domain, and therefore facilitates GCY-35 activation. Notably, this model is consistent with the fact that heb1 is a recessive gain-of-function mutation. Recessive gain-of-function mutations are relatively rare, but they are found in genes encoding transcription factors (e.g., LIN-1, HMBX-1) (Lesch and Bargmann, 2010) and ion channels (e.g., DEG-1 and NAPts) (Kernan et al., 1991; García-Añoveros et al., 1995). The unique situation in which the gain-of-function allele recessive to the wild-type allele but dominant over the null allele could be explained whether these alleles are part of a multimeric protein or whether the recessive gain-of-function allele attenuates an inhibitory protein interaction (Lesch and Bargmann, 2010; Luhur et al., 2014). In the case of heb1, it could be that the GCY-35/36 complex is multimeric and one copy of GCY-35(wt) can suppress the activity of GCY-35(heb1). Indeed, this scenario is consistent with a combined model in which GCY-35(heb1) has increased affinity for O2 but decreased affinity to the catalytic domain. However, the conformation of these hypotheses awaits future structural and biophysical studies.
Similar structural determinants regulate NO- and O2-sensor activity
Threonine 48 (Thr48) and leucine 67 (Leu67) are amino acid residues in a tunnel system (composed of tunnels 1 and 2) that facilitates gas migration in the NO-binding H-NOX protein from Nostoc cyanobacterium (Winter et al., 2011). Either replacing Thr48 with tryptophan to block tunnels 1 and 2 or replacing Leu67 with tryptophan to block just tunnel 2 increases the binding affinity of the heme to CO, suggesting that these amino acid residues regulate the sensitivity to NO. Thr48 is conserved in the O2 sensor GCY-35, whereas Leu67 is naturally replaced with tryptophan. Interestingly, Trp67 is conserved in the H-NOX domains of GCY-32, GCY-34, GCY-35, GCY-36, and GCY-37, which are all thought to be activated by high O2 concentrations. However, Trp67 is not conserved in GCY-31 and GCY-33, which are inhibited by high O2 levels (Zimmer et al., 2009).
The Thr48Trp and Trp67Leu mutations inactivate GCY-35 (Fig. 6C,D), suggesting that Thr48 and Trp67 function in O2 sensing. These results are surprising because, unlike the crystal structure of the H-NOX protein from Nostoc sp., the structure of the O2-binding H-NOX protein from Thermoanaerobacter tengcongensis bacterium did not reveal a continuous tunnel system for gas migration (Winter et al., 2011), suggesting that O2 does not diffuse through discrete tunnels inside the H-NOX domain. A possible explanation for the loss of function of the GCY-35 Thr48Trp and Trp67Leu mutants is that these mutations interfere with the correct folding of the H-NOX domain. However, we do not favor this explanation because replacing Thr48 and Leu67 with tryptophan did not interfere with the correct folding of the Nostoc H-NOX mutants and even increased their affinity to CO (Winter et al., 2011). We anticipate that structural analysis of the H-NOX domain of GCY-35 would help to understand the function of Thr48 and Trp67 in O2 sensing.
GLB-5(Haw) function in GCY-35/36-dependent O2 responses
In the introduction, we put forward two hypotheses that aim to explain how GLB-5(Haw) inhibits GCY-35/36 activity. The first suggested that GLB-5(Haw) inhibits GCY-35/36 activity through physical interaction, and the second considers the possibility that the inhibition is mediated by reactive oxygen/nitrogen species. In this work, we provide evidence that supports the first hypothesis. First, we show that GLB-5(Haw) inhibits GCY-35/36 activity in a dose-dependent manner (Fig. 5A) and that this inhibition is not mediated by reactive oxygen species signaling (Fig. 5C). Second, we show that the heb3 mutation in GCY-36 specifically interferes with glb-5(Haw)-dependent O2 responses and not glb-5(Haw)-independent responses (Fig. 8), indicating that heb3 does not suppress GLB-5(Haw) by changing the intrinsic activity of GCY-36. Notably, similarly to gcy-35(heb1), gcy-36(heb3) is a recessive gain-of-function allele. Following the hypothesis mentioned above for GCY-35(heb1) function, we suggest that the gain-of-function activity of GCY-36(heb3) is due to decreased affinity for GLB-5(Haw). Therefore, the presence of GCY-36(wt), with high affinity to GLB-5(Haw), suppresses the activity of GCY-36(heb3) in the GCY-35/36 multimeric complex. Together, these results support a model in which GLB-5(Haw) attenuates the activity of GCY-35/36 by physical interaction. However, validation of this model awaits further biochemical studies.
The function of GLB-5(Haw) is required for rapid adaptation to high O2 levels. The exposure of glb-5(Haw);npr-1 worms to 35% O2 for just 5 min decreases the sensitivity of URX to O2 and thus inhibits the activation of these neurons at 21% O2 (Fig. 4D,I). The desensitization of URX by GLB-5(Haw) is inhibited by GCY-35(heb1), indicating that fast adaptation to high O2 levels is regulated by cGMP signaling. Intriguingly, cGMP signaling plays a major role in the adaptation of cone and rod photoreceptor cells to strong illumination (Govardovskii et al., 2000; Calvert et al., 2002; Hurley, 2002). This adaptation prevents the saturation of these cells and thus maintains sensitivity to a wide range of light conditions. Because neuroglobin and soluble guanylate cyclases are expressed in the eye (Koch et al., 1994; Schmidt et al., 2003; Vielma et al., 2012; Di Giulio et al., 2013), it is intriguing to speculate that neuroglobin regulates cGMP signaling in this organ and thus modulates vision physiology.
In conclusion, our studies provide strong evidence that glb-5(Haw)-dependent O2 responses are generally mediated by URX desensitization. This conclusion is supported by previous studies that showed that URX desensitization is essential for rapid recovery from prolonged hypoxia (Gross et al., 2014) and for fine-tuned O2 responses at ambient O2 levels (Persson et al., 2009). Moreover, our studies suggest that GLB-5(Haw) exerts its inhibitory activity by interacting with the H-NOX domain of GCY-36. This possibility is intriguing and raises the question whether H-NOX domains are generally important for the regulation of sGCs by other proteins.
Footnotes
The research leading to these results was supported by the European Research Council under the European Union's Seventh Framework Programme FP/2007–2013/ERC Grant Agreement 281844 to Z.A., A.K.C., B.Z., and E.G. Some strains were provided by the CGC, which is funded by National Institutes of Health Office of Research Infrastructure Programs P40 OD010440, the C. elegans Knockout Consortium, and the National BioResource Project (Japan). We thank Deborah Fass for helpful discussions and critical reading of the manuscript; and Norman Grover and members of the E.G. laboratory members for comments and advice.
The authors declare no competing financial interests.
References
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch KE, Laurent P, Soltesz Z, Murphy RJ, Faivre O, Hedwig B, Thomas M, Smith HL, de Bono M. Tonic signaling from O(2) sensors sets neural circuit activity and behavioral state. Nat Neurosci. 2012;15:581–591. doi: 10.1038/nn.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert PD, Govardovskii VI, Arshavsky VY, Makino CL. Two temporal phases of light adaptation in retinal rods. J Gen Physiol. 2002;119:129–145. doi: 10.1085/jgp.119.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung BH, Arellano-Carbajal F, Rybicki I, de Bono M. Soluble guanylate cyclases act in neurons exposed to the body fluid to promote C. elegans aggregation behavior. Curr Biol. 2004;14:1105–1111. doi: 10.1016/j.cub.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Couto A, Oda S, Nikolaev VO, Soltesz Z, de Bono M. In vivo genetic dissection of O2-evoked cGMP dynamics in a Caenorhabditis elegans gas sensor. Proc Natl Acad Sci U S A. 2013;110:E3301–E3310. doi: 10.1073/pnas.1217428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MW, Hammarlund M, Harrach T, Hullett P, Olsen S, Jorgensen EM. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics. 2005;6:118. doi: 10.1186/1471-2164-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bono M, Bargmann CI. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell. 1998;94:679–689. doi: 10.1016/S0092-8674(00)81609-8. [DOI] [PubMed] [Google Scholar]
- Derbyshire ER, Marletta MA. Structure and regulation of soluble guanylate cyclase. Annu Rev Biochem. 2012;81:533–559. doi: 10.1146/annurev-biochem-050410-100030. [DOI] [PubMed] [Google Scholar]
- Di Giulio C, Zara S, De Colli M, Ruffini R, Porzionato A, Macchi V, De Caro R, Cataldi A. Cytoglobin and neuroglobin in the human brainstem and carotid body. Adv Exp Med Biol. 2013;788:59–64. doi: 10.1007/978-94-007-6627-3_9. [DOI] [PubMed] [Google Scholar]
- Drechsel DA, Patel M. Differential contribution of the mitochondrial respiratory chain complexes to reactive oxygen species production by redox cycling agents implicated in parkinsonism. Toxicol Sci. 2009;112:427–434. doi: 10.1093/toxsci/kfp223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follmann M, Griebenow N, Hahn MG, Hartung I, Mais FJ, Mittendorf J, Schäfer M, Schirok H, Stasch JP, Stoll F, Straub A. The chemistry and biology of soluble guanylate cyclase stimulators and activators. Angewandte Chemie. 2013;52:9442–9462. doi: 10.1002/anie.201302588. [DOI] [PubMed] [Google Scholar]
- García-Añoveros J, Ma C, Chalfie M. Regulation of Caenorhabditis elegans degenerin proteins by a putative extracellular domain. Curr Biol. 1995;5:441–448. doi: 10.1016/S0960-9822(95)00085-6. [DOI] [PubMed] [Google Scholar]
- Govardovskii VI, Calvert PD, Arshavsky VY. Photoreceptor light adaptation: untangling desensitization and sensitization. J Gen Physiol. 2000;116:791–794. doi: 10.1085/jgp.116.6.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, Marletta MA, Bargmann CI. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature. 2004;430:317–322. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- Gross E, Soltesz Z, Oda S, Zelmanovich V, Abergel Z, de Bono M. GLOBIN-5-dependent O2 responses are regulated by PDL-1/PrBP that targets prenylated soluble guanylate cyclases to dendritic endings. J Neurosci. 2014;34:16726–16738. doi: 10.1523/JNEUROSCI.5368-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé D, Philippi A, Belbouab R, Zerah M, Chabrier S, Collardeau-Frachon S, Bergametti F, Essongue A, Berrou E, Krivosic V, Sainte-Rose C, Houdart E, Adam F, Billiemaz K, Lebret M, Roman S, Passemard S, Boulday G, Delaforge A, Guey S, et al. Loss of alpha 1 beta 1 soluble guanylate cyclase, the major nitric oxide receptor, leads to moyamoya and achalasia. Am J Hum Genet. 2014;94:385–394. doi: 10.1016/j.ajhg.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JB. Shedding light on adaptation. J Gen Physiol. 2002;119:125–128. doi: 10.1085/jgp.119.2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernan MJ, Kuroda MI, Kreber R, Baker BS, Ganetzky B. napts, a mutation affecting sodium channel activity in Drosophila, is an allele of mle, a regulator of X chromosome transcription. Cell. 1991;66:949–959. doi: 10.1016/0092-8674(91)90440-A. [DOI] [PubMed] [Google Scholar]
- Kleppisch T, Feil R. cGMP signalling in the mammalian brain: role in synaptic plasticity and behaviour. Handb Exp Pharmacol. 2009;2009:549–579. doi: 10.1007/978-3-540-68964-5_24. [DOI] [PubMed] [Google Scholar]
- Koch KW, Lambrecht HG, Haberecht M, Redburn D, Schmidt HH. Functional coupling of a Ca2+/calmodulin-dependent nitric oxide synthase and a soluble guanylyl cyclase in vertebrate photoreceptor cells. EMBO J. 1994;13:3312–3320. doi: 10.1002/j.1460-2075.1994.tb06633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lesch BJ, Bargmann CI. The homeodomain protein hmbx-1 maintains asymmetric gene expression in adult C. elegans olfactory neurons. Genes Dev. 2010;24:1802–1815. doi: 10.1101/gad.1932610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhur A, Chawla G, Wu YC, Li J, Sokol NS. Drosha-independent DGCR8/Pasha pathway regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2014;111:1421–1426. doi: 10.1073/pnas.1318445111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Rockman MV, Zimmer M, Jang H, Macosko EZ, Kruglyak L, Bargmann CI. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron. 2009;61:692–699. doi: 10.1016/j.neuron.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton DB, Hudson ML, Waters E, O'Shea M. Soluble guanylyl cyclases in Caenorhabditis elegans: NO is not the answer. Curr Biol. 1999;9:R546–R547. doi: 10.1016/S0960-9822(99)80349-2. [DOI] [PubMed] [Google Scholar]
- Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci U S A. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson A, Gross E, Laurent P, Busch KE, Bretes H, de Bono M. Natural variation in a neural globin tunes oxygen sensing in wild Caenorhabditis elegans. Nature. 2009;458:1030–1033. doi: 10.1038/nature07820. [DOI] [PubMed] [Google Scholar]
- Rogers NM, Seeger F, Garcin ED, Roberts DD, Isenberg JS. Regulation of soluble guanylate cyclase by matricellular thrombospondins: implications for blood flow. Front Physiol. 2014;5:134. doi: 10.3389/fphys.2014.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Ed 2. New York: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schmidt M, Giessl A, Laufs T, Hankeln T, Wolfrum U, Burmester T. How does the eye breathe? Evidence for neuroglobin-mediated oxygen supply in the mammalian retina. J Biol Chem. 2003;278:1932–1935. doi: 10.1074/jbc.M209909200. [DOI] [PubMed] [Google Scholar]
- Shen F, Li YJ, Shou XJ, Cui CL. Role of the NO/sGC/PKG signaling pathway of hippocampal CA1 in morphine-induced reward memory. Neurobiol Learn Mem. 2012;98:130–138. doi: 10.1016/j.nlm.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Stasch JP, Pacher P, Evgenov OV. Soluble guanylate cyclase as an emerging therapeutic target in cardiopulmonary disease. Circulation. 2011;123:2263–2273. doi: 10.1161/CIRCULATIONAHA.110.981738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiso M, Tejero J, Basu S, Azarov I, Wang X, Simplaceanu V, Frizzell S, Jayaraman T, Geary L, Shapiro C, Ho C, Shiva S, Kim-Shapiro DB, Gladwin MT. Human neuroglobin functions as a redox-regulated nitrite reductase. J Biol Chem. 2011;286:18277–18289. doi: 10.1074/jbc.M110.159541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underbakke ES, Iavarone AT, Marletta MA. Higher-order interactions bridge the nitric oxide receptor and catalytic domains of soluble guanylate cyclase. Proc Natl Acad Sci U S A. 2013;110:6777–6782. doi: 10.1073/pnas.1301934110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielma AH, Retamal MA, Schmachtenberg O. Nitric oxide signaling in the retina: what have we learned in two decades? Brain Res. 2012;1430:112–125. doi: 10.1016/j.brainres.2011.10.045. [DOI] [PubMed] [Google Scholar]
- Weber KP, De S, Kozarewa I, Turner DJ, Babu MM, de Bono M. Whole genome sequencing highlights genetic changes associated with laboratory domestication of C. elegans. PLoS One. 2010;5:e13922. doi: 10.1371/journal.pone.0013922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert EE, Phillips-Piro CM, Marletta MA. Porphyrin π-stacking in a heme protein scaffold tunes gas ligand affinity. J Inorg Biochem. 2013;127:7–12. doi: 10.1016/j.jinorgbio.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winger JA, Marletta MA. Expression and characterization of the catalytic domains of soluble guanylate cyclase: interaction with the heme domain. Biochemistry. 2005;44:4083–4090. doi: 10.1021/bi047601d. [DOI] [PubMed] [Google Scholar]
- Winter MB, Herzik MA, Jr, Kuriyan J, Marletta MA. Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain. Proc Natl Acad Sci U S A. 2011;108:E881–E889. doi: 10.1073/pnas.1114038108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Hekimi S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol. 2010;8:e1000556. doi: 10.1371/journal.pbio.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J, Herzik MA, Jr, Winter MB, Tran R, Olea C, Jr, Marletta MA. Structure and properties of a bis-histidyl ligated globin from Caenorhabditis elegans. Biochemistry. 2010;49:5662–5670. doi: 10.1021/bi100710a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu M, Cheng Y, Li Z. H-NOX domains display different tunnel systems for ligand migration. J Mol Graph Model. 2010;28:814–819. doi: 10.1016/j.jmgm.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Zimmer M, Gray JM, Pokala N, Chang AJ, Karow DS, Marletta MA, Hudson ML, Morton DB, Chronis N, Bargmann CI. Neurons detect increases and decreases in oxygen levels using distinct guanylate cyclases. Neuron. 2009;61:865–879. doi: 10.1016/j.neuron.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.