Abstract
A proliferative endothelial cell phenotype, inflammation, and pulmonary vascular remodeling are prominent features of pulmonary arterial hypertension (PAH). Bone morphogenetic protein type II receptor (BMPR2) loss-of-function is the most common cause of heritable PAH and has been closely linked to the formation of pathological plexiform lesions. Although some BMPR2 mutations leave ligand-dependent responses intact, the disruption of ligand-independent, noncanonical functions are universal among PAH-associated BMPR2 genotypes, but incompletely understood. This study examined the noncanonical signaling consequences of BMPR2 silencing in human pulmonary artery endothelial cells to identify potential therapeutic targets. BMPR2 siRNA silencing resulted in a proliferative, promigratory pulmonary artery endothelial cell phenotype and disruption of cytoskeletal architecture. Expression profiling closely reflected these phenotypic changes. Gene set enrichment and promoter analyses, as well as the differential expression of pathway components identified Ras/Raf/ERK signaling as an important consequence of BMPR2 silencing. Raf family members and ERK1/2 were constitutively activated after BMPR2 knockdown. Two Raf inhibitors, sorafenib and AZ628, and low-dose nintedanib, a triple receptor tyrosine kinase inhibitor upstream from Ras, reversed the abnormal proliferation and hypermotility of BMPR2 deficiency. Inhibition of dysregulated Ras/Raf/ERK signaling may be useful in reversing vascular remodeling in PAH.
Keywords: pulmonary arterial hypertension, BMPR2, endothelial cell dysfunction, nintedanib
pulmonary arterial hypertension (PAH) is a rare disorder characterized by endothelial cell dysfunction with obliteration of distal pulmonary arteries due to progressive vascular remodeling (42). Despite therapeutic advances, rising pulmonary vascular resistance ultimately culminates in right heart failure and death. Heterozygous germline mutations in bone morphogenetic protein type II receptor (BMPR2) are the most common genetic cause of PAH, accounting for 70% of familial cases (29) and 10–40% of idiopathic PAH (IPAH) (57). BMPR2 expression is markedly reduced in end-stage PAH, even in patients not harboring mutations (2). Importantly, BMPR2 is primarily expressed on endothelial cells (ECs) (2), and dysfunction from its loss appears to drive the formation of plexogenic lesions through abnormal growth and proliferation of underlying vascular smooth muscle (1). A histopathological hallmark of advanced PAH, plexogenic lesions are characterized by dysregulated angiogenesis with EC, vascular smooth muscle, and myofibroblast proliferation, as well as the infiltration of activated inflammatory cells (60).
Model systems of BMPR2 heterozygous mutations (31, 46, 61, 62) and BMPR2 silencing in primary human pulmonary artery ECs (HPAECs) (1, 6, 7, 50, 52, 56) recapitulate important aspects of cellular dysfunction and dysregulated signaling associated with PAH. Ligand-dependent canonical BMPR2 signaling has a central role in maintaining EC homeostasis and regulating proliferation, apoptosis, and cell fate determination. BMP/BMPR2 signaling results in the phosphorylation and activation of SMAD 1/5/8, resulting in the activation of target gene transcription (33). In addition, BMP-dependent, but Smad-independent signal transduction through Wnt/β-catenin (6, 62), PPARγ/apoE (17), and PPARγ/β-catenin (1) regulate apoptosis, cell proliferation, and migration. Loss of BMP-dependent BMPR2 signaling has been shown to increase EC susceptibility to apoptosis, with the subsequent emergence of a proproliferative, apoptosis-resistant endothelial phenotype during the angioproliferative phase of vascular remodeling (32, 43, 55, 56).
Less well understood have been aberrant signaling and functional abnormalities associated with ligand-independent, noncanonical effects of BMPR2 mutations and loss of function. Some mutations in patients with PAH are limited to the cytoplasmic tail of BMPR2 and leave Smad signaling intact (39). The BMPR2 tail regulates stress kinase pathways (46) and interacts with scaffolding proteins and cytoskeletal components (23). Despite variable impact on canonical BMP/Smad signaling, heterogeneous BMPR2 mutants consistently result in the activation of p38 MAPK (46). In cells expressing PAH-associated BMPR2 alleles, altered ERK signaling appeared to increase cell growth, similar to the cellular phenotype seen in PAH vessels (26).
This study examines the ligand-independent, noncanonical consequences of BMPR2 silencing in HPAECs. In the absence of exposure to exogenous BMPs, BMPR2 silencing was assessed for effects on 1) cellular phenotype (proliferation, migration, and cytoskeletal structure), 2) global gene expression, 3) stress kinase activation, and 4) response to therapies targeting these aberrant signal transduction pathways.
MATERIALS AND METHODS
Cell Culture
Primary HPAECs were cultured for a maximum of six passages in endothelial basal medium-2 (EBM-2) supplemented with growth factors [endothelial growth medium (EGM)-2 SingleQuot kit from Lonza (Walkersville, MD)] containing 2% serum on cell culture flasks coated with type I collagen (50 μg/ml in 0.02 N acetic acid) or BD Biocoat flasks (BD Biosciences, Franklin Lakes, NJ). Cells were maintained at 37°C in a humidified incubator with 5% CO2 and 95% air. Primary HPAECs were seeded at a density of 70,000 cells/well in 12-well plates or 150,000 cells/well in 6-well plates for RNA and protein analysis, respectively. Male (n = 8) and female (n = 5) HPAEC donors ranging in age from 19 to 67 yr old were purchased from Lonza and Lifeline (Frederick, MD). Mycoplasmal contamination testing was performed by Lonza and Lifeline.
BMPR2 Silencing
Primary HPAECs were transfected with gene-specific siRNA pools targeting BMPR2 at a final concentration of 10 nM using DharmaFECT-1 (Dharmacon; Lafayette, CO) in Opti-MEM (Invitrogen, Grand Island, NY) for 6 h followed by growth in EGM-2 (minus hydrocortisone) containing 2% charcoal-stripped serum. Nontargeting siRNA pool-1 (siGENOME, Dharmacon) was used as a control. After incubating for an additional 42 h (48 h from the start of transfection), total RNA or protein lysates were collected for analysis.
RNA Isolation, cDNA Synthesis, and Quantitative Real-Time PCR
Total RNA was isolated using the RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions, including DNase I treatment. RNA samples were then subjected to reverse transcription using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Quantitative real-time PCR (qRT-PCR) analysis of target genes was performed using iTaq Universal SYBR Green Supermix with ROX (Bio-Rad) using the Applied Biosystems (Foster City, CA) ViiA7 instrument. Reactions without RT were performed to rule out amplification of any residual genomic DNA. Each experimental condition was performed in triplicate, and each sample was run in duplicate. Relative gene expression was calculated using the 2−ΔΔCT method. Delta cycle thresholds rather than fold changes (FCs) were tested for significance, as the latter are not normally distributed. The geometric means ± SE of quantitative RT-PCR (qRT-PCR) FCs are plotted, whereas other data are presented as arithmetic means ± SE.
The following primer sequences (5′ to 3′) were utilized for qRT-PCR: BMPR2 (forward: CACTGCGGCTGCTTCGCAGA and reverse: AGCAGGTGCTACCTTTCGAGCA); RAF1 (forward: TTT CCTGGATCATGTTCCCCT and reverse: ACTTTGGTGCTACAGTGCTCA); A-RAF (forward: CCTGGCGTTCTGTGACTTCTG and reverse: CGGTTGGTACTCATGTCAACAC); B-RAF (forward: CAAACTTATAGATATTGCACG and reverse: GCTAGACCAAAATCACCTATT); ID1 (forward: GGAATCCGAAGTTGGAACCCCCG and reverse: AGGAACGCATGCCGCCTCG); VEGFA (forward: AGGGCAGAATCATCACGAAGT and reverse: AGGGTCTCGATTGGATGGCA); VEGFB (forward: AGCACCAAGTCCGGATG and reverse: GTCTGGCTTCACAGCACTG); VEGFC (forward: GAGGAGCAGTTACGGTCTGTG and reverse: TCCTTTCCTTAGCTGACACTTGT); ITGB3 (forward: CTCATTGGCCTTGCCGCCCT and reverse: AGTGGGTTGTTGGCTGTGTCCC); THSB (forward: CTGGCCTGGGGACTAGGCGTC and reverse: GGGGCCCTTCACCAGTCGGC); SPRY1 (forward: GAGAGAGATTCAGCCTACTGCT and reverse: GCAGGTCTTTTCACCACCGAA); BCL2L11 (forward: TAAGTTCTGAGTGTGACCGAGA and reverse: GCTCTGTCTGTAGGGAGGTAGG); RB1 (forward: TCACCTTGAATCTGCTTGTCC and reverse: TGGAGATCTTACAGGAGAAAGATACA); CXCR4 (forward: GGGCAATGGATTGGTCATCCT and reverse: TGCAGCCTGTACTTGTCCG); PDE4B (forward: AACGCTGGAGGAATTAGACTGG and reverse: GCTCCCGGTTCAGCATTCT); BTG2 (forward: CCTGTGGGTGGACCCCTAT and reverse: GGCCTCCTCGTACAAGACG); and β-ACTIN (forward: ACCATGGATGATGATATCGC and reverse: TTGCTGATCCACATCTGCTG).
Western Blot
Whole cell protein lysates were isolated from HPAEC transfected with nontargeting siRNA control or BMPR2 siRNA for 48 h with RIPA buffer (Thermo Scientific Pierce, Rockford, IL) supplemented with protease and phosphatase inhibitor cocktail (Thermo Scientific Pierce). Protein lysates (30 μg) were resolved by SDS-PAGE and transferred to a nitrocellulose membrane with iBlot (Invitrogen). Membranes were incubated with 5% blocking solution (Bio-Rad; nonfat milk) in PBS-Tween 20 (0.1%) for 1 h at room temperature and then incubated overnight at 4°C with primary antibody. In some experiments as indicated, Raf inhibitors, sorafenib, or AZ628 (Selleckchem, Houston, TX) or the angiokinase inhibitor, nintedanib (Selleckchem), were added to cells for 1 h or 24 h, respectively, prior to harvesting and assessment of ERK1/2 activation. For Western blots, loading control-normalized densitometry results were analyzed, but FCs from the control condition are plotted for the ease of interpretation. Antibodies against p-ERK (no. 9101, 1:1,000), total ERK (no. 9102, 1:1,000), p-p38 (no. 9211, 1:400), total p38 (no. 9212, 1:800), p-JNK (no. 4671, 1:400), total JNK (no. 9258, 1:1,000), p-RAF-1 (no. 9427, 1:1,000), p-ARAF (no. 4431, 1:500), p-BRAF (no. 2696, 1:500), were purchased from Cell Signaling (Danvers, MA). Total RAF-1 (sc-133, 1:1,000), ARAF (sc408, 1:1,000), and BRAF (sc-166, 1:200) were purchased from Santa Cruz Biotechnology (Dallas, TX). Anti-BMPR2 antibody (no. 612292, 1:500) was obtained from BD Biosciences, anti-BIM antibody (ab7888, 1:1000) was obtained from Abcam (Cambridge, MA) and a horseradish peroxidase-conjugated anti-β-actin antibody (clone AC-15) was obtained from Sigma (St. Louis, MO).
Cell Proliferation Assays
Primary HPAECs transfected with siControl or siBMPR2 for 48 h were plated in collagen-coated 96-well plates at a density of 7,500 cells/well for 6 h in complete media. Complete medium was replaced with serum-free medium for 24 h before returning HPAECs to complete medium to assess cell proliferation. Cell proliferation over 72 h was quantitated by the presence of ATP using the CellTiter-Glo Luminescent Cell Viability Assay (Promega; Madison, WI) or 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS)/ formazan using CellTiter 96 AQueous One Solution cell proliferation assay (Promega).
Cell Migration Assay
Primary HPAECs transfected with siControl or siBMPR2 for 48 h were plated in Oris collagen-coated 96-well plates (Platypus Technologies; Madison, WI) at a density of 25,000 cells/well for 6 h in complete media. Complete medium was replaced with serum-free media for 24 h, the biocompatible gel insert was removed, and medium was changed to complete medium with or without inhibitors, as indicated. Migration was assessed at 36 h or 48 h with addition of Calcein-AM (Invitrogen; final concentration of 0.5 μg/ml). After incubation for 15 min at 37°C, images were captured, and the area was measured using ImageJ software (http://imagej.nih.gov/ij/), as described previously (53).
Immunocytochemistry
Primary HPAECs were plated at a density of 10,000 cells/well on collagen-coated Lab-Tek II chambered slides (Thermo Scientific Pierce), and on the following day, they were transfected with either control or BMPR2 siRNA. Forty-eight hours following transfection, cells were fixed with 2% paraformaldehyde for 10 min at room temperature, washed with PBS, and costained overnight at 4°C with BMPR2 (no. ab96826, working dilution 0.89 μg/μl; Abcam) and either Alexa Fluor 488 phalloidin (no. A12379, 82.5 nM; Invitrogen) or paxillin (no. 05–417, clone 5H11, 5 μg/ml; Millipore; Billerica, MA). The next day, cells were washed with PBS and incubated with Alexa Fluor 488 or 555 secondary antibody (Invitrogen) for 1 h. Cells were stained with DAPI and mounted using anti-fade, Mowiol mounting medium. Confocal images were acquired on a Zeiss LSM 510 Inverted Meta microscope using a 63× plan-apochromat 1.4 NA oil objective equipped with photomultiplier tubes. Laser Scanning Module 5 software was used for image acquisition, and ImageJ was used for image processing.
Gene Expression Profiling
Total RNA quality was evaluated using RNA 6000 Nano LabChip (Agilent 2100 Bioanalyzer, Santa Clara, CA). All samples had intact 18S and 28S ribosomal RNA bands with RIN values from 9.2 to 10 and 260/280 ratios between 1.98 and 2.03. Total RNA (100 ng) was used as starting material for the synthesis of cDNA and cRNA in vitro transcription following manufacturer's directions (GeneChip 3′ IVT express kit, Affymetrix, Santa Clara, CA). The resulting cRNA was fragmented and labeled using Affymetrix GeneChip WT terminal labeling kit. Fragmented and labeled cRNA was hybridized to GeneChip PrimeView human gene expression arrays (Affymetrix) for 18 h. Arrays were stained and washed in the Affymetrix Fluidics Station 400 and then scanned (Affymetrix 7G). Microarray was performed using HPAECs from a single donor at the same passage number to reduce variability. Four complete experiments were conducted on different days. To determine donor-to-donor variability for a subset of differentially expressed genes, three to five different donors were used for qRT-PCR validation.
Microarray Analysis
Signal intensity values for the oligonucleotide arrays were normalized and transformed using the 3′ Expression Array Robust Multiarray Average (RMA) in Expression Console (Affymetrix). Probe set IDs were annotated using PrimeView Human Gene Expression Array annotation file version 34 (release date 10/24/13). Resulting signal intensity values for each of 49,293 probe sets were processed in JMP statistical package (SAS, Cary, NC) using the Mathematical and Statistical Computing Laboratory Analyst's Toolbox (http://abs.cit.nih.gov/MSCLtoolbox/). The transformed data matrix was subjected to principal components analysis to visualize the relative location of chips in a low-dimensional space allowing for detection of outliers or other relevant patterns. Raw and processed data were submitted to the National Center for Biotechnology Information Gene Expression Omnibus database (NCBI GEO; http://www.ncbi.nlm.nih.gov/projects/geo/index.cgi; accession no. GSE70456), and all data are MIAME compliant.
A one-way ANOVA across four conditions [siControl, siBMRPR2, siControl + phorbol 12-myristate 13-acetate (PMA) and siBMPR2+PMA] was performed, followed by post hoc t-tests for each pairwise contrast using the group mean and mean square error estimates. False discovery rates (FDRs) were calculated on the basis of the corresponding P values. For the main contrast (siControl vs. siBMPR2), selection of differentially expressed transcripts based on a FDR of 1% and a 2-FC cutoff yielded a total of 825 probe set IDs. After removing one unannotated probe set ID and condensing probe set IDs annotated to the same gene (n = 319), 505 probe set IDs remained. Probe set IDs annotated to multiple transcripts (n = 29) were adjudicated using the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/gene) by removing discontinued gene assignments, unknown gene types, noncoding transcripts, and pseudogenes. The final annotated probe set ID list included 488 unique genes and 17 remaining probe set IDs annotated to more than one gene (see Supplemental Table S1). Note that unique gene tallies for these annotated probe sets vary slightly across the various databases used in subsequent analyses.
In a secondary analysis, the transcriptomic effects of BMPR2 gene silencing were investigated in the presence and absence of PMA (Calbiochem, San Diego, CA), a diacylglycerol mimetic that activates PKC isoforms and downstream MAPK pathways (59). For this interaction analysis [(siBMPR2+PMA − siBMPR2) vs. (siControl+PMA − siControl)], selected transcripts demonstrated a significant interaction between BMPR2 silencing and PMA treatment. Applying an FDR <1% for the interaction term yielded a total of 947 probe set IDs.
Bioinformatic Analyses
Ingenuity pathway analysis.
Differentially expressed probe sets from the main contrast (siBMPR2 vs. siControl) were uploaded into Ingenuity Pathway Analysis (IPA) and examined using the Bio Function and Canonical Pathway applications. Significance of enrichment for biological functions and canonical pathways in IPA is calculated using the right-tailed Fisher Exact Test. For canonical pathways, the ratio is calculated by dividing the number of genes within the data set annotated to the pathway by the total number of genes that make up the pathway, as determined by the Ingenuity Knowledge Base. The Upstream Regulator application and the Ingenuity Knowledge Base were used to construct a HPAEC regulatory network based on the dominant pattern of interaction between BMPR2 gene silencing and PMA-induced mitogenesis. Additional connections were manually curated using PubMed (http://www.ncbi.nlm.nih.gov/pubmed/), the Molecular Signatures Database (version 4.0; http://www.broadinstitute.org/gsea/msigdb/index.jsp; Broad Institute, MIT) (28), and STRING v9.1 (http://string-db.org/newstring_cgi/show_input_page.pl?UserId=TkKZPhzFvWK1&sessionId=9nbuyXaGgLPI) (22).
Gene set enrichment analysis.
Gene set enrichment analysis determines whether an a priori defined set of genes shows statistically significant, concordant differences between two biological phenotypes without imparting arbitrary gene selection filters (e.g., FC differences) to the data set (54). Gene sets (Molecular Signatures Database, version 4.0; Broad Institute, MIT) (28) from the oncogenic signatures collection (188/189 gene sets eligible for analysis; one excluded with <15 genes within the set) were tested for significant enrichment between siBMPR2- and siControl-transfected HPAECs (GSEA Software version 2.0.14; Broad Institute, MIT). Using default settings (FDR <25%) (54), genes within our expression data set were rank-ordered on the basis of the difference of mean log2 intensities (siBMPR2-siControl) scaled by the standard deviation. Overall 92/188 gene sets were enriched (see Supplemental Table S2 for complete list). Phenotype permutation was performed to assess the statistical significance of the enrichment score.
Promoter analysis.
Genes upregulated after BMPR2 gene silencing (FDR <1% and FC >2; yes-set; n = 278) were compared with genes with similar expression levels, but without evidence of regulation by either BMPR2 silencing or PMA stimulation (no-set; n = 551) to identify relevant promoter binding site differences. Using Explain 3.1 (BIOBASE Knowledge Library: http//www.biobase-international.com/; Beverly, MA), selected settings were as follows: 1) lung-specific profile of position-weighted matrices; 2) best-supported promoters; 3) maximum promoter window −500 to +100 base pairs; 4) cutoff and window optimization; and 5) P value threshold of 0.01. This F-match (TRANSFAC Pro) promoter analysis was repeated using a predefined set of human housekeeping genes (BIOBASE Knowledge Library) as the no-set to test the validity and stability of the output.
Statistical analysis.
Paired t-tests were used to assess statistical significance between siControl and siBMPR2 using JMP (SAS Institute). Dose-response experimental results across multiple conditions and donors were subjected to linear mixed models with random subject effects to account for the correlation within each donor. Log-transformation was applied when necessary. Two-tailed P < 0.05 was accepted as significant.
RESULTS
Phenotypic Effects of BMPR2 Gene Silencing
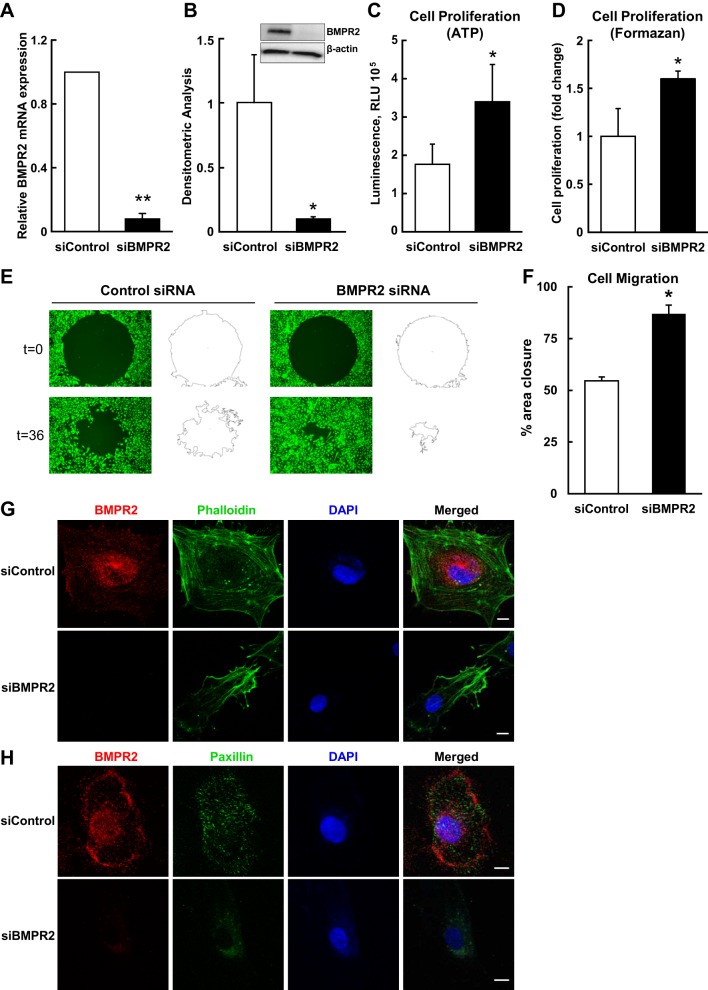
Forty-eight hours after targeted knockdown, BMPR2 mRNA (Fig. 1A) and protein levels (Fig. 1B) were reduced by ≥90% (P ≤ 0.02 for both). Cell proliferation as measured by ATP content (P = 0.01; Fig. 1C) and the MTS/formazan assay (P = 0.04; Fig. 1D) was significantly increased in BMPR2-deficient HPAECs. BMPR2 silencing also enhanced HPAECs migration compared with control cells (Fig. 1E). Nontargeting siRNA control cells migrated to 54% ± 1.9 closure, whereas BMPR2-deficient cells migrated to 87% ± 4.2 closure over 36 h (P = 0.03; Fig. 1F). Importantly, BMPR2 knockdown for 48 h visibly altered cytoskeletal architecture. HPAECs transfected with nontargeting siRNA demonstrated well-defined stress fibers (Fig. 1G) and localization of paxillin at focal adhesions (Fig. 1H), while loss of BMPR2 resulted in poorly organized stress fibers and a more homogenous, less distinct, pattern of paxillin distribution. Taken together, BMPR2 silencing in HPAECs demonstrated a hyperproliferative, promigratory phenotype with disruption of cytoskeletal architecture. The short-term repression of BMPR2 protein in vitro was sufficient for HPAECs to manifest many important functional abnormalities associated with PAH pathogenesis.
Fig. 1.
Bone morphogenetic protein type II receptor (BMPR2) knockdown in human pulmonary endothelial cells (HPAECs) results in a dysfunctional endothelial cell phenotype. A: BMPR2 mRNA levels were assessed 48 h after BMPR2 knockdown. BMPR2 mRNA was measured in four different donors and is presented as geometric means ± SE. B: BMPR2 protein levels were examined after 48 h to assess the efficiency of gene silencing. For BMPR2 protein, densitometric quantification (means ± SE) relative to β-actin and normalized to the siControl is shown for three different donors; the inset shows a representative Western blot. ATP (C) and MTS (D) assays were used to quantify cell proliferation. Forty-eight hours after BMPR2 knockdown, HPAECs were transferred to 96-well plates for 6 h and serum starved for 24 h, before proliferation was assessed for 72 h in complete media. Data (means ± SE) represent five independent experiments for ATP assay and four independent experiments for formazan assay using different donors. E: representative images of cell migration measured in Oris 96-well plates. F: percent closure (means ± SE) of monolayer defects after 36 h of incubation for three different donors is shown. Cytoskeletal architecture examined by confocal microscopy with costaining for BMPR2 and phalloidin (F-actin) (G) or BMPR2 and paxillin (H). Cells were counterstained with DAPI. Confocal images are representative of results obtained from four different HPAEC donors. Scale bars: 10 μm. *P < 0.05; **P < 0.01.
Genome-Wide Expression Profiling of BMPR2 Gene Silencing in HPAECs
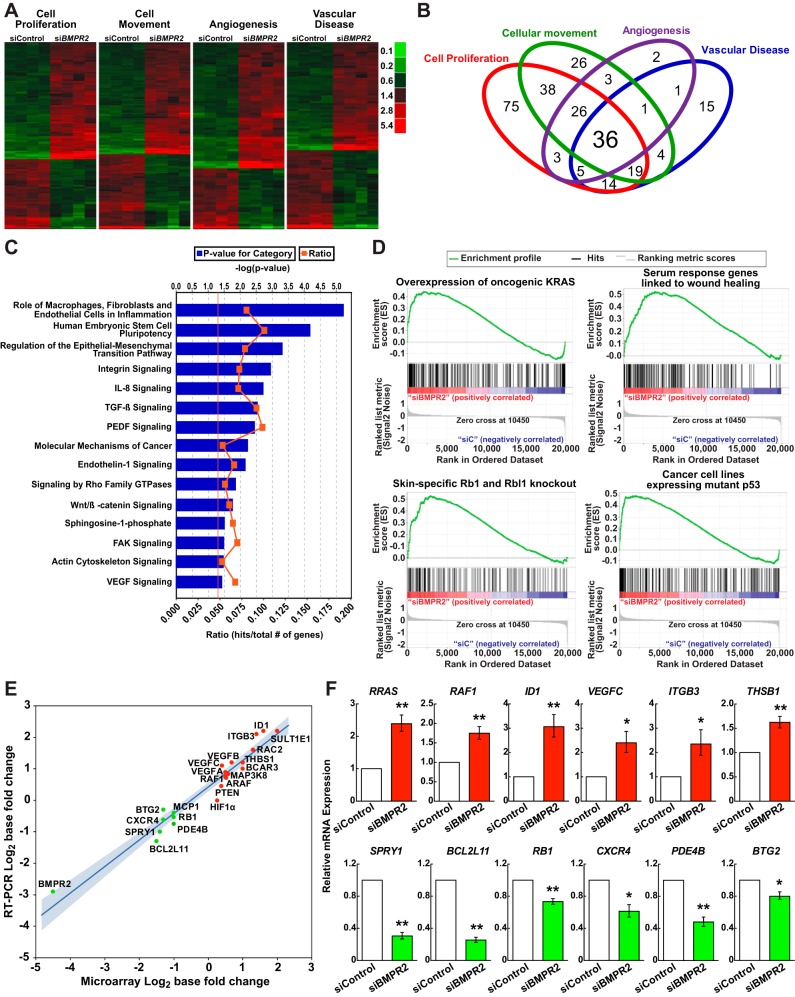
The global impact of BMPR2 silencing on gene expression was investigated in primary HPAECs. Selection of differentially expressed transcripts following BMPR2 siRNA silencing (FDR <1% and FC >2) yielded 505 annotated, nonredundant probe sets (see Supplemental Table S1); 287 were upregulated, and 218 were downregulated. IPA identified cell proliferation (P = 7.76 × 10−20), cellular movement (P = 9.85 × 10−25), angiogenesis (P = 1.92 × 10−19) and vascular disease (P = 6.45 × 10−22), depicted as heat maps (Fig. 2A), among the enriched biological functions. Over half of the 268 unique genes representing these four classifications annotated to more than one of the four categories (Fig. 2B). Top canonical pathways in HPAECs following BMPR2 gene silencing included: macrophage, fibroblast, and endothelial inflammation; epithelial-mesenchymal transition; molecular mechanisms of cancer; and integrin, interleukin 8, transforming growth factor β, endothelin-1, Rho Family GTPase, and Wnt/β-catenin signaling (Fig. 2C).
Fig. 2.
Thematic analyses of differentially expressed genes due to BMPR2 loss-of-function in HPAECs. A: differentially expressed genes within four top Bio Functions are displayed in heat map format: cell proliferation (n = 220 genes; P = 7.77 × 10−20); cellular movement (n = 160 genes; P = 9.85 × 10−25); angiogenesis (n = 77 genes; P = 1.92 × 10−19); and vascular disease (n = 96 genes; P = 6.45 × 10−22). Red signifies expression above and green below the mean value within an individual row. For full annotations, see Supplemental Table S1. B: Venn diagram demonstrating overlap among the functional categories in A. C: canonical pathways significantly associated with BMPR2 loss-of-function. The vertical orange line represents the threshold for significance [−log10(P value) = 1.3 or P = 0.05]. D: gene set enrichment analysis enrichment plots of selected gene sets that are overrepresented following BMPR2 knockdown in HPAECs: 1) epithelial lung cancer cell lines overexpressing KRAS [normalized enrichment score (NES) = 1.48, P < 0.0001, false discovery rate (FDR) q value = 0.08]; 2) serum response genes linked to wound healing and cancer progression (NES = 1.61, P < 0.0001, FDR q value = 0.04]; 3) retinoblastoma 1 (Rb1) and retinoblastoma-like 1 (Rbl1) skin-specific knockout mice (NES = 1.70, P < 0.0001, FDR q value = 0.025); and 4) cancer cell lines expressing a p53 mutant (NES = 1.45, P < 0.0001, FDR q value = 0.086). The running enrichment score (y-axis) and position of gene set members on the rank-ordered list (x-axis) is shown. E: correlation plot, comparing microarray to qRT-PCR FCs expressed as log2 with the 95% confidence interval shaded in blue (P < 0.0001; R2 = 0.94). F: BMPR2 knockdown effect on 12 mRNA transcripts, as assessed by qRT-PCR in multiple HPAEC donors. Data are presented as the geometric means ± SE for 3 to 5 different donors. *P < 0.05; **P < 0.01.
Next, a gene set enrichment analysis was performed to examine the impact of BMPR2 gene silencing without the use of arbitrary gene selection criteria. On the basis of our findings of a hyperproliferative, promigratory phenotype in BMPR2-silenced cells and the association of PAH with cancer-like cellular abnormalities (43, 64), the oncogenic signatures collection in the Molecular Signatures Database (version 4.0; Broad Institute, MIT) was chosen for this analysis. Enriched gene sets (Fig. 2D) included 1) genes upregulated in lung cancer cell lines overexpressing an oncogenic KRAS (3); 2) a serum-induced transcriptional program in fibroblasts previously linked to wound healing and cancer progression (4); 3) genes associated with epidermis-specific deletion of retinoblastoma family members (27); and 4) genes upregulated in cancer cell lines expressing mutant p53 (54). Other gene set associations included RAF1, MAP2K1 (also known as MEK1), and MYC, all of which are downstream effectors of Ras signaling (see Supplemental Table S2).
Promoter Analysis of Transcripts Regulated by siRNA Silencing of BMPR2
The promoter structure of upregulated genes (FDR <1% and FC >2) was compared with expressed genes demonstrating no evidence of regulation. Using only best supported promoters, binding sites for the AP-1 family of transcription factors were prominently represented among the top enriched promoter matrices (enrichment ratio >2 and matched promoter FDR <0.05; Table 1). AP-1 family members c-Jun, c-Fos, and Fra-2 have all been previously implicated in PAH pathogenesis (9, 34). Several other transcription factor binding sites previously associated with PAH were notably enriched in genes upregulated by BMPR2 knockdown, including homeobox A5 (HOXA5) (12), forkhead box O1 (FOXO1) (49), and CCAAT/enhancer binding protein beta (CEBPB) (8).
Table 1.
Transcription factor binding sites overrepresented among upregulated genes following BMPR2 gene silencing
| Matrix Name | Associated Transcription Factors | Yes/No | Window Position (Base Pairs From TSS) | Matched Promoters P Value | Matched Promoters FDR |
|---|---|---|---|---|---|
| V$AP1_C | FOS, FOSB, FOSL1/L2, JUN, JUNB/D | 4.8 | −200 to 100 | 1.70E-3 | 8.30E-3 |
| V$AP1_Q4_01 | FOS, FOSB, FOSL1/L2, JUN, JUNB/D | 4.6 | −200 to 100 | 1.61E-5 | 5.79E-4 |
| V$AP1_Q6_01 | FOS, FOSB, FOSL1/L2, JUN, JUNB/D | 4.6 | −200 to 100 | 1.61E-5 | 5.79E-4 |
| V$AP1_01 | FOS, FOSB, FOSL1/L2, JUN, JUNB/D | 4.6 | −200 to 100 | 1.60E-3 | 8.30E-3 |
| V$HOX13_02 | HOXA5, IRF6 | 4.3 | −400 to 100 | 1.69E-4 | 1.70E-3 |
| V$AP1_Q6 | FOS, FOSB, FOSL1/L2, JUN, JUNB/D | 3.6 | −200 to 100 | 1.15E-4 | 1.40E-3 |
| V$AP1_Q4 | FOS, FOSB, FOSL1/L2, JUN, JUNB/D | 3.2 | −200 to 100 | 3.53E-4 | 2.80E-3 |
| V$CEBP_Q2_01 | CEBPA/B/D/E/G | 3.1 | −300 to 100 | 1.30E-3 | 7.10E-3 |
| V$CEBP_Q3 | CEBPA/B/D/E/G | 3.1 | −300 to 100 | 1.10E-3 | 6.50E-3 |
| V$CEBPG_Q6 | CEBPG | 2.8 | −200 to 100 | 7.60E-3 | 2.59E-2 |
| V$FOX_Q2 | FOXC1, FOXD1/3, FOXF1/2, FOXH1, FOXI1, FOXJ1/2, FOXL1, FOXO1/3/4, RRN3 | 2.6 | −300 to 0 | 7.11E-6 | 5.12E-4 |
| V$FRA1_Q5 | FOSL1 | 2.5 | −300 to 0 | 1.12E-4 | 1.60E-3 |
| V$HNF3ALPHA_Q6 | FOXA1/2/3 | 2.3 | −200 to 100 | 2.34E-4 | 2.10E-3 |
| V$AP1_Q2_01 | FOS, FOSB, FOSL1/L2, JUN, JUNB/D | 2.3 | −300 to 0 | 9.70E-3 | 3.16E-2 |
| V$HNF3_Q6 | FOXA1/2/3 | 2.1 | −300 to 0 | 3.93E-4 | 2.80E-3 |
| V$HFH8_01 | FOXC1, FOXD1/3, FOXF1/2, FOXH1, FOXI1, FOXJ1/2 FOXL1, FOXO1/3/4, RRN3 | 2.1 | −300 to 0 | 4.72E-4 | 3.10E-3 |
| V$GLI1_01 | GLI1 | 2.0 | −500 to 0 | 7.20E-3 | 2.72E-2 |
| V$CEBP_Q2 | CEBPA/B | 2.0 | −300 to 100 | 1.03E-2 | 3.22E-2 |
Matrix name, transcription factor binding site matrix from TRANSFAC database; Yes/No score, ratio of transcription factor binding sites of a particular matrix in the set of upregulated genes in BMPR2 KD relative to control siRNA (Yes-set) compared to the background set of genes (No-set). The “no set” was selected based on 1) a P value >0.1 for the interaction term [(siBMPR2+PMA vs. siBMPR2) − (siControl+PMA vs. siControl)] (n = 37526 probe sets); 2) a fold change of <10% for the interaction term (n = 24829 probe sets); 3) a P > 0.1 for each main effect (siBMPR2 and PMA; n = 10427 probe sets); and 4) mean expression intensity across all conditions greater than or equal to the 1st quartile mean expression intensity of probe sets differentially regulated in the siBMPR2 vs. siControl comparison and probe sets that demonstrated a significant interaction between BMPR2 silencing and PMA (n = 686 probe sets). TSS, transcription start site; FDR, false discovery rate.
Validation of Expression Profile Analysis Using Multiple HPAEC Donors
Using the same samples, microarray- and qRT-PCR-determined FC for 22 transcripts were strongly correlated (P < 0.0001; R2 = 0.94), but microarray results tended to overestimate transcript downregulation (Fig. 2E). To further document the validity of the microarray and to determine true donor-to-donor and experimental variability, a subset of differentially expressed genes (FDR < 0.01; no FC cutoff) was quantified across multiple HPAEC donors using qRT-PCR (Fig. 2F). Results were consistent with the microarray data, showing increased expression of RRAS, RAF1, ID1, VEGFC, ITGB3, and THSB1 and decreases in SPRY1, BCL2L11 (also known as BIM), RB1, CXCR4, PDE4B, and BTG2 after BMPR2 knockdown (P < 0.05 for all).
Aberrant Raf/MAPK Signaling in BMPR2 Knockdown Cells
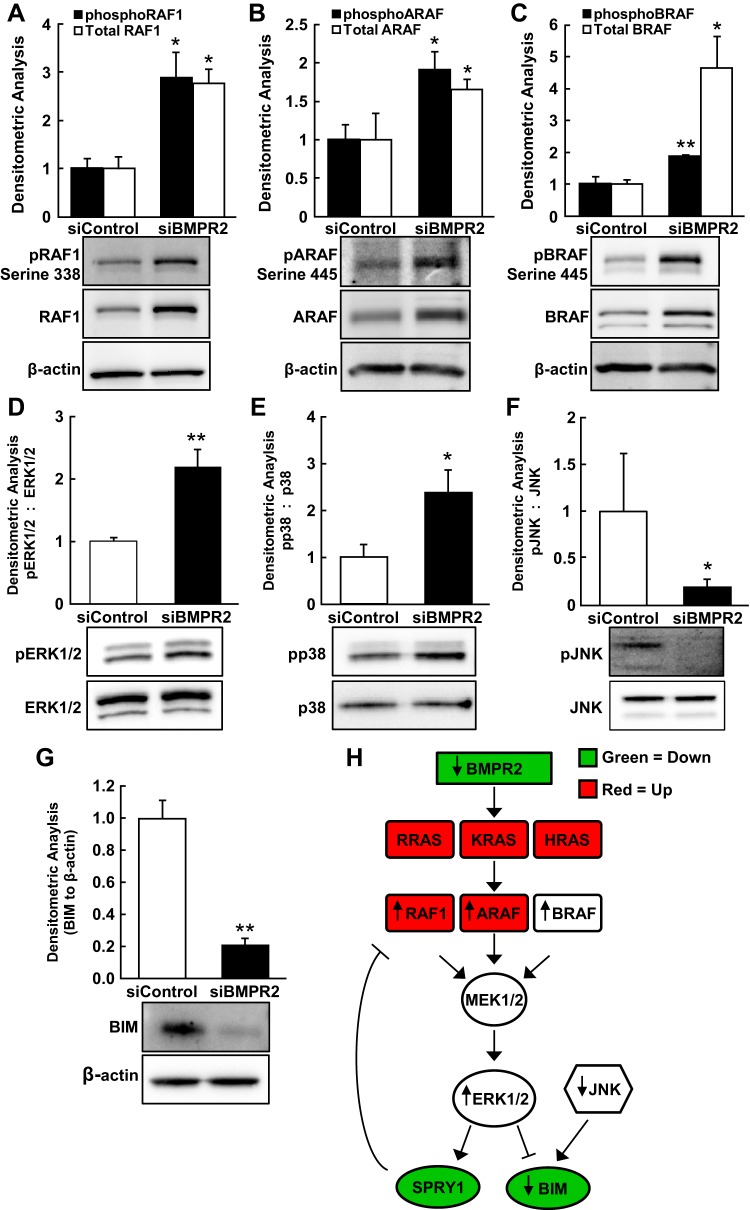
RAF1 mRNA expression was increased by microarray analysis; thus, we evaluated transcript levels, protein expression, and phosphorylation states of RAF1, ARAF, and BRAF in HPAECs deficient in BMPR2. While RAF1 mRNA increased almost twofold after BMPR2 knockdown (P = 0.01; Fig. 2F), ARAF (FC = 1.41, P = 0.03) was only modestly increased, and BRAF (FC = 1.15, P = 0.34) was unaffected, as determined by qRT-PCR (data not shown). BMPR2 silencing in HPAECs significantly increased both total and phosphorylated (active) RAF1 protein (P < 0.03 for both; Fig. 3A), ARAF (P = 0.01 for total and P = 0.02 for phosphorylated protein; Fig. 3B), and BRAF (P = 0.01 for total and P = 0.0005 for phosphorylated protein; Fig. 3C).
Fig. 3.
Aberrant activation of the Raf/ERK signaling pathway in BMPR2-silenced HPAECs. Lysates from control HPAECs and BMPR2-deficient HPAECs were resolved by SDS-PAGE and immunoblotted. pRAF1 (serine 338) and total RAF1 (3 independent experiments in 3 different donors) (A): pARAF (Ser-445) and total ARAF (n = 4) (B), pBRAF (Ser-445) and total BRAF (n = 4) (C), pERK, and total ERK (n = 6) (D), pp38 and total p38 (n = 4) (E), pJNK and total JNK (n = 4) (F), and BIM (BCL2L11; n = 4) (G). Densitometric analyses are normalized to the loading controls and the corresponding siControl condition. Representative blots are shown, and data are presented as means ± SE; *P < 0.05; **P < 0.01. H: schematic diagram of dysregulated Ras/Raf/ERK signaling resulting from BMPR2 loss-of-function. Red (up-regulated) and green (down-regulated) shading indicates the directional changes of significantly regulated transcripts [false discovery rates (FDR) < 0.01; no FC cutoff] by microarray. Black arrows (up or down) indicate experimentally determined directional changes in protein expression and/or activation state.
Because BMPR2 silencing resulted in a proliferative and migratory HPAEC phenotype and Raf family members were induced and activated, downstream activation of ERK1/2, p38 MAPK, and JNK was examined. Silencing BMPR2 in HPAECs increased basal levels of active (phosphorylated) ERK1/2 (P = 0.006; Fig. 3D) and p38 MAPK (P = 0.03; Fig. 3E) greater than twofold. In contrast, phosphorylated JNK decreased by more than twofold with BMPR2 silencing (P < 0.05; Fig. 3F).
BCL2L11 (BIM; FDR < 0.01, FC = −3.12), a proapoptotic protein, and SPRY1 (FDR < 0.01, FC = −2.70), a negative regulator of Raf/MEK/ERK signaling, were both downregulated, as determined by global expression profiling and confirmed by qRT-PCR (Fig. 2F). Accordingly, BMPR2 knockdown in HPAECs was found to substantially decrease BIM protein expression (P = 0.007; Fig. 3G). ERK1/2 activation and JNK inactivation have been shown to suppress BIM protein expression in ECs (16). Conversely, ERK1/2 is a known inducer of SPRY1 transcription (41), so its decreased expression here cannot be explained by the activation of this pathway. Nonetheless, down-regulation of SPRY1 by BMPR2 silencing (Fig. 2F) likely reinforced the activation of ERK1/2 through loss of its suppressive effect on Raf signaling (48). The impact of BMPR2 silencing on the Ras/Raf/ERK signaling pathway is depicted in Fig. 3H, where red (upregulated) and green (downregulated) shading indicates the directional changes of significantly regulated transcripts (FDR < 0.01, no FC cutoff) by microarray and black arrows (up or down) indicate experimentally determined directional changes in protein expression and/or activation state.
Less Than Additive Effects of BMPR2 Silencing and PMA Stimulation
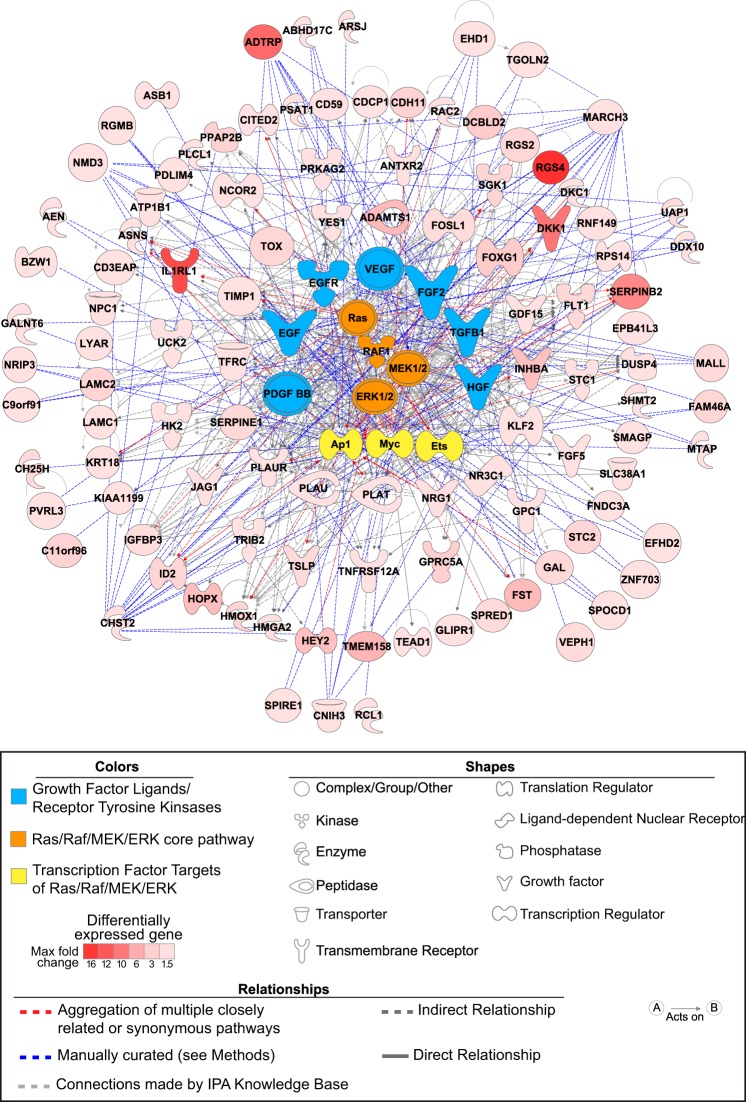
PMA is a potent mitogen and strong inducer of stress kinase pathways (59) that have been implicated in PAH pathogenesis (21, 26, 46, 50). Importantly, as shown above, basal activation of Raf family members ERK1/2 and p38 MAPK in BMPR2-silenced HPAECs regulates downstream genes associated with PAH pathogenesis and may, at least in part, drive the proliferative and migratory phenotype of these altered cells. To further explore signaling pathways relevant to these phenotypic traits, evidence was sought for potential interactions between mitogenic cell activation by PMA and BMPR2 gene silencing. A total of 947 probe sets had an FDR < 1% for an interaction between BMPR2 knockdown and PMA-induced cell activation. The largest subgroup of interacting probe sets (357/947; Table 2 for the distribution of interaction patterns) were upregulated by both BMPR2 knockdown and PMA exposure (FDR < 5% for each contrast), but their combined effects were less than additive (i.e., negatively synergistic). This dominant pattern suggested nonadditive, shared mechanisms of gene induction and is consistent with our findings demonstrating the impact of BMPR2 loss of function on mitogen-activated stress signaling. After applying a FC > 1.5 stringency filter for the BMPR2 silencing effect, a regulatory network of potential therapeutic targets (Fig. 4) was constructed from 107 unique genes demonstrating concordant, but less-than-additive, effects with PMA (Supplemental Table S3).
Table 2.
Distribution of interaction patterns between BMPR2 silencing and PMA stimulation
| siBMPR2 Versus siC | PMA_siC Versus siC | Interaction | Breakdown of 947 Interacting Probe Sets |
|---|---|---|---|
| 1 | −1 | −1 | 15 |
| 1 | 0 | −1 | 43 |
| 1 | 1 | −1 | 357 |
| 0 | −1 | −1 | 5 |
| 0 | 0 | −1 | 5 |
| 0 | 1 | −1 | 125 |
| −1 | −1 | −1 | 6 |
| −1 | 0 | −1 | 1 |
| −1 | 1 | −1 | 25 |
| 1 | −1 | 1 | 42 |
| 1 | 0 | 1 | 4 |
| 1 | 1 | 1 | 6 |
| 0 | −1 | 1 | 85 |
| 0 | 0 | 1 | 8 |
| 0 | 1 | 1 | 10 |
| −1 | −1 | 1 | 121 |
| −1 | 0 | 1 | 25 |
| −1 | 1 | 1 | 64 |
In 2nd and 3rd columns, 1 indicates FDR <0.05 and a positive difference, −1 indicates FDR <0.05 and a negative difference, and 0 indicates FDR >0.05. In column 4, 1 and −1 represent the directional fold change for the interaction term. PMA, phorbol 12-myristate 13-acetate.
Fig. 4.
Regulatory network derived from the less-than-additive interaction between BMPR2 silencing and mitogenic stimulation. HPAECs expression profiles in the presence or absence of BMPR2 knockdown and/or a mitogen (phorbol 12-myristate 13-acetate, PMA) were examined for significant interactions. Upregulation by both BMPR2 silencing and PMA (FDR < 5%, for both) with less-than-additive combined effects was the predominant pattern identified. Regulatory nodes were chosen based on the following algorithm: 1) IPA-predicted upstream regulators present among the 107 interacting genes; 2) components of the Ras/Raf/MEK/ERK shown to be activated after BMPR2 knockdown and predicated by IPA as upstream regulators (P < 0.001); 3) tyrosine kinase receptors/ligands upstream of Ras/Raf with IPA activation prediction z-scores ≥ 2 and associated with PAH pathogenesis (EGF, EGFR, FGF2, TGFB1, VEGF, HGF, and PDGF BB); and 4) transcription factors downstream of Raf/ERK associated with PAH pathogenesis (AP1, ETS, and MYC family members). The Ingenuity Knowledge Base connected 77 of 107 interacting genes and connections for the remaining 30 genes were manually curated using PubMed, the Molecular Signatures Database (MSigDB) version 4.0, and STRING version 9.1.
Evaluation of Two Raf Inhibitors and Nintedanib, an Angiogenic Receptor Tyrosine Kinase Inhibitor, in BMPR2-Silenced HPAECs
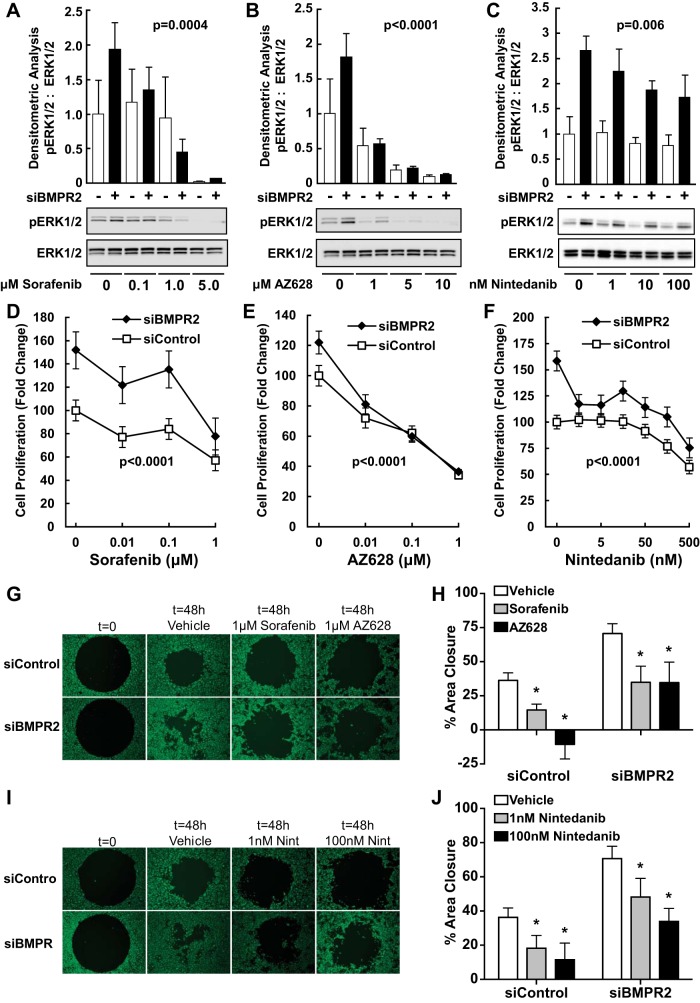
Sorafenib (P = 0.0004; Fig. 5A) and AZ628 (P < 0.0001; Fig. 5B), two different Raf kinase inhibitors, dose-dependently decreased phosphorylated (active) ERK1/2 similarly in both control and BMPR2-silenced HPAECs over 1 h. Low clinically achievable concentrations of nintedanib (40) over 1 h (data not shown) or 24 h only moderately inhibited ERK1/2 phosphorylation in BMPR2-silenced HPAECs (P = 0.006 for a dose-response in both control and BMPR2-silenced cells; Fig. 5C).
Fig. 5.
Inhibition of Raf/ERK signaling and the dysfunctional endothelial cell phenotype associated with BMPR2 knockdown. Effect of sorafenib (A), AZ628 (B), or nintedanib (C) on ERK1/2 activation. HPAECs were transfected with control or BMPR2 siRNAs for 48 h and then exposed to sorafenib or AZ628 for 1 h or to nintedanib for 24 h at the indicated concentrations. Densitometric quantification is expressed as the ratio (means ± SE) of active pERK1/2 to total ERK normalized to the siControl. Sorafenib (P = 0.0004), AZ628 (P < 0.0001), and nintedanib (P = 0.006) decreased ERK1/2 phosphorylation (all P values are main effects for both siControl and siBMPR2 transfected cells). A representative blot from three independent experiments each performed with a different donor is shown. Effects of sorafenib (D), AZ628 (E), or nintedanib (F) on cell proliferation. Sorafenib (P < 0.0001), AZ628 (P < 0.0001), and nintedanib (P < 0.0001) suppressed cell proliferation (all P values are main effects for both siControl and siBMPR2-transfected cells). Cell migration for sorafenib and AZ628 (G and H) or nintedanib (I and J) was measured in Oris 96-well plates. Sorafenib (*P < 0.001), AZ628 (*P < 0.001), and nintedanib (*P < 0.0001), suppressed cell migration (all P values are main effects for both siControl and siBMPR2 transfected cells). D, E, F, H, and J: data are presented as the means ± SE for three different donors.
As seen before, BMPR2 silencing in HPAECs increased cell proliferation (P ≤ 0.01 for all). Treatment with either sorafenib (P < 0.0001; Fig. 5D) or AZ628 (P < 0.0001; Fig. 5E) for 72 h dose-dependently decreased cell proliferation in both control and BMPR2-silenced cells. Likewise, low-dose nintedanib reversed the hyperproliferative phenotype associated with BMPR2 silencing (P < 0.0001 for an overall dose-dependent effect; Fig. 5F). An effect on control HPAECs was only seen at nintedanib concentrations >10 nM.
As previously shown, BMPR2 silencing increased cell motility compared with nontargeting siRNA control cells (P < 0.0001; Fig. 5, G–J). Sorafenib and AZ628 (1 μM each) for 48 h decreased the migration of both BMPR2-deficient and control HPAECs alike (P < 0.001; Fig. 5, G and H). Low dose nintedanib similarly blocked both control and BMPR2-deficient cells (P < 0.001; Fig. 5, I and J) with the highest dose tested (100 nM) normalizing migration. Collectively, these results support a role for receptor tyrosine kinase (RTK) signaling and Raf/ERK activation in mediating both the proliferative and promigratory phenotype of BMPR2 knockdown.
DISCUSSION
BMPR2 silencing in HPAECs produced phenotypic, transcriptomic, and functionally significant signaling changes that closely recapitulated many of the abnormalities and pathogenic mechanisms associated with advanced PAH. This conclusion is based on the following evidence: 1) BMPR2 knockdown resulted in a proliferative, promigratory cellular phenotype with disruption of normal cytoskeletal architecture; 2) global expression profiling highly mirrored these phenotypic changes; and 3) top molecules and networks affected by BMPR2 silencing have all been previously associated with PAH pathogenesis in vivo. Analyses for oncogenic gene enrichment and shared promoter sites, as well as the differential expression and constitutive activation of specific pathway components, identified Ras/Raf/MEK/ERK/AP1 signaling as an important consequence of BMPR2 silencing. Notably, Raf family members and ERK1/2 were constitutively activated after BMPR2 knockdown. In addition, BMPR2 silencing produced a signature that extensively overlapped with the expression profile induced by PMA, a mitogenic activator of stress kinase pathways. The network map derived from this overlap was highly consistent with prevailing views on the molecular drivers of PAH pathophysiology. Finally, Raf inhibitors and nintedanib, a triple RTK inhibitor, chosen to target upstream components of this signaling network, reversed some of the phenotypic abnormalities of BMPR2 deficiency.
Pulmonary artery ECs from patients with IPAH grow faster in culture due to both increased proliferation and resistance to apoptosis (32). In addition, these late-stage IPAH cells demonstrated increased migration, but less organized angiogenesis, as measured by tube formation. Late-outgrowth endothelial progenitor cells from PAH patients with BMPR2 mutations are also hyperproliferative with an impaired capacity to form vascular networks (58). Here, we demonstrated that in vitro BMPR2 silencing in HPAECs produced a similar proliferative and motogenic cellular phenotype. Importantly, changes in gene expression closely reflected these alterations in cell phenotype. Previously, BMPR2 knockdown was shown to increase HPAEC susceptibility to apoptosis (7, 56), whereas cell migration was only modestly increased (6), and cell proliferation was not affected (6, 7). In an angioproliferative rat model of PAH, caspase inhibition not only blocked apoptosis, but also prevented disease, suggesting that early cell death led to the selection of apoptosis-resistant ECs and vascular remodeling (55). Supporting this paradigm, VEGF receptor 2 blockade and shear stress in human pulmonary microvascular ECs caused early elevations in caspase 3 activity (47). A proliferative phenotype subsequently emerged 7 days after the experimental challenge. Likewise, cell proliferation and migration were both significantly increased in our in vitro model more than 3 days after highly efficient BMPR2 knockdown (reductions in mRNA and protein were 92% and 90%, respectively). Here, ATP production and formazan generation, measures of cellular metabolism, were used as surrogate markers of cell proliferation. While ATP and formazan correlate strongly with cell number, factors independent of cell proliferation (5, 38) could have also affected these metabolic products. Likewise, enhanced wound closure might reflect changes in both cell motility and proliferation (45).
Cytoskeletal reorganization is another important EC phenotype associated with PAH pathobiology across multiple different BMPR2 mutations (23). Murine pulmonary microvascular ECs with the Bmpr2R899X mutation, which does not disrupt Smad signaling (61), have a defective cytoskeletal architecture characterized by lack of stress fibers, truncated microtubules, and disorganized VE-cadherin (23). Noncanonical and ligand-independent interactions of the BMPR2 cytoplasmic tail with proteins, such as LIMK (10), TCTEX (30), and SRC (63), key regulators of cytoskeletal organization, are thought to play a role in PAH pathogenesis (23). We demonstrated that BMPR2 silencing in HPAECs also caused stress fiber loss (a promigratory phenotype) and redistribution of paxillin away from focal adhesions. Paxillin is a multidomain docking protein important for motility and barrier integrity (51). The orderly formation and disassembly of focal adhesions controls endothelial remodeling during angiogenesis, inflammation, and vascular repair.
Global gene expression profiling after BMPR2 siRNA silencing identified proliferation, motility, and angiogenesis as among the most highly enriched biological functions (Fig. 2A). In addition, focal adhesion kinase and actin cytoskeletal signaling were significantly affected by BMPR2 knockdown, reflecting the disruption of normal cytoskeletal architecture. Most notably, many of the altered networks and pathways (Fig. 2C), including endothelial inflammation (15), epithelial-mesenchymal transition (14), molecular mechanisms of cancer (64), growth factors, Rho family GTPases (15) and Wnt/β-catenin (1, 6, 62) have been previously recognized as important contributors to PAH pathogenesis.
An enrichment analysis, using oncogenic gene sets from curated experimental data and our microarray results without prior gene selection, identified Ras and Raf among the most significantly enriched pathways. Additionally, a promoter level analysis of genes upregulated by BMPR2 knockdown returned a strong signal for AP1 DNA binding sites (Table 1), a downstream target of Ras/Raf signaling. Direct investigation of this signal transduction pathway revealed that Raf family members were induced and activated (i.e., phosphorylated) in BMPR2 silenced cells and ERK1/2, a downstream intermediary that culminates in AP1 transcriptional responses, was constitutively activated. ERK1/2 is known to induce SPRY1, a negative feedback inhibitor of RTK/Ras/Raf signaling (41). However, SPRY1 was suppressed by BMPR2 deficiency (Fig. 2F), an effect that likely acted to reinforce and prolong RTK/Ras/Raf/MEK/ERK signaling.
The cytoplasmic tail of BMPR2 has been shown to physically interact with PKC and stress kinase signaling components (18). More importantly, PAH-associated BMPR2 mutations, regardless of their impact on Smad signaling, uniformly activate ERK (26) and p38 MAPK (46), an effect mirrored by BMPR2 silencing (Fig. 3F). Furthermore, aberrant ERK activation has been described in the pulmonary vasculature of patients with advanced PAH (26). More recently, a gain-in-function RAF1 mutation has been associated with the development of rapidly fatal PAH in two infants (21). Exploring the importance of Raf/ERK signaling in PAH, Raf-1 kinase inhibitor protein knockout mice were found to exhibit exaggerated hypoxia-induced pulmonary hypertension (36). Collectively, sustained Ras/Raf/MEK/ERK signaling, downstream from growth factors and their associated RTKs, may represent a broadly applicable paradigm for new therapeutic approaches to pathological vascular remodeling in PAH.
Two Raf kinase inhibitors, sorafenib and AZ628, and a triple RTK inhibitor, nintedanib, were tested in our in vitro model system of BMPR2 deficiency. In addition to inhibiting RAF1 and BRAF, sorafenib also blocks VEGF and PDGF RTKs. Sorafenib has been shown to improve right ventricular (RV) function and reverse pressure overload-induced RV remodeling (25) and to prevent pulmonary vascular remodeling (24) in the monocrotaline rat model of PAH. Using the VEGF receptor blockade/hypoxia rat model of PAH, sorafenib abolished activation of the MAPK cascade (37). However, in a phase 1b clinical trial, sorafenib was associated with decreases in cardiac output and poor tolerability (13). In contrast, the triple angiokinase inhibitor nintedanib was beneficial in patients with idiopathic pulmonary fibrosis and very well tolerated for up to 52 wk (44). By inhibiting VEGF, basic FGF and PDGF BB signaling and the downstream activation of ERK1/2 and AKT (19), nintedanib targets many of the dysregulated pathways seen in HPAECs after BMPR2 silencing. Sorafenib, AZ628, and nintedanib all decreased ERK1/2 activation, proliferation, and migration in BMPR2-deficient HPAECs. Sorafenib and AZ628 required relatively high concentrations and similarly affected control and BMPR2-silenced HPAECs alike. Nintedanib had only modest effect on ERK1/2 activation, but very low doses nearly normalized proliferation (Fig. 5F). Importantly, concentrations with somewhat selective effects on BMPR2 knockdown cells are achievable in vivo at nintedanib doses lower (40) than those used in the idiopathic pulmonary fibrosis trial (44).
Phase II (11) and III trials (20) of imatinib, a RTK inhibitor used to treat chronic myelogenous leukemia (CML), have yielded some evidence of efficacy in PAH, but the latter also raised concern due to a substantial incidence of subdural hematoma. In stark contrast, precapillary pulmonary hypertension has developed in patients taking dasatinib for imatinib-resistant CML (35). Both dasatinib and imatinib inhibit the PDGF receptor, but dasatanib also targets a broader range of RTKs, including LIMK1/2, CSK, and FAK (35), that regulate focal adhesions and the cytoskeleton. Although blocking a select subset of RTKs may stop or reverse pulmonary vascular remodeling, RTKs are functionally diverse with complex downstream networks, and some inhibitors apparently have the potential to trigger or worsen PAH. Importantly, nintedanib has not been associated with the development of either systemic or pulmonary hypertension (44). Preclinical studies of nintedanib in animal models of PAH are warranted as a first step toward randomized human trials.
BMPR2 silencing in HPAECs reproduced many of the cellular abnormalities that have been associated with PAH in vivo and can serve as a useful in vitro model for exploring therapeutic targets. The in vitro profiling of endothelial cells with heterogeneous PAH-associated molecular defects, such as those involving CAV1, RAF1, and BMPR2, might be helpful in developing a comprehensive picture of pathogenic mechanisms and therapeutic targets. The comparative biology of seemingly unrelated molecular defects may lead to both individualized and possibly universal approaches for arresting or even reversing pathologic vascular remodeling.
GRANTS
This study was supported through intramural National Institutes of Health (NIH) funds, including a 2011–2013 Bench to Bedside Award from the NIH Office of Women's Health.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: K.S.A., J.M.E., M.A.S., and R.L.D. conception and design of research; K.S.A. and G.A.F. performed experiments; K.S.A., J.M.E., S.W., S.G., and R.L.D. interpreted results of experiments; K.S.A. and J.M.E. prepared figures; K.S.A., J.M.E., and R.L.D. drafted manuscript; K.S.A., J.M.E., S.W., S.G., G.A.F., R.C., J.S., M.A.S., and R.L.D. edited and revised manuscript; K.S.A., J.M.E., S.W., S.G., G.A.F., R.C., J.S., M.A.S., and R.L.D. approved final version of manuscript; S.W., R.C., and J.S. analyzed data.
Supplementary Material
ACKNOWLEDGMENTS
Lynne Holtzclaw at the Microscopy & Imaging Core Facility (National Institute of Child Health and Human Development, NIH) provided invaluable assistance with immunohistochemistry and confocal imaging. We also thank Kelly Byrne for editing and formatting the manuscript and figures.
REFERENCES
- 1.Alastalo TP, Li M, Perez Vde J, Pham D, Sawada H, Wang JK, Koskenvuo M, Wang L, Freeman BA, Chang HY, Rabinovitch M. Disruption of PPARgamma/beta-catenin-mediated regulation of apelin impairs BMP-induced mouse and human pulmonary arterial EC survival. J Clin Invest 121: 3735–3746, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson C, Stewart S, Upton PD, Machado R, Thomson JR, Trembath RC, Morrell NW. Primary pulmonary hypertension is associated with reduced pulmonary vascular expression of type II bone morphogenetic protein receptor. Circulation 105: 1672–1678, 2002. [DOI] [PubMed] [Google Scholar]
- 3.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, Frohling S, Chan EM, Sos ML, Michel K, Mermel C, Silver SJ, Weir BA, Reiling JH, Sheng Q, Gupta PB, Wadlow RC, Le H, Hoersch S, Wittner BS, Ramaswamy S, Livingston DM, Sabatini DM, Meyerson M, Thomas RK, Lander ES, Mesirov JP, Root DE, Gilliland DG, Jacks T, Hahn WC. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462: 108–112, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, Chi JT, van de Rijn M, Botstein D, Brown PO. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol 2: E7, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crouch SP, Kozlowski R, Slater KJ, Fletcher J. The use of ATP bioluminescence as a measure of cell proliferation and cytotoxicity. J Immunol Methods 160: 81–88, 1993. [DOI] [PubMed] [Google Scholar]
- 6.de Jesus Perez VA, Alastalo TP, Wu JC, Axelrod JD, Cooke JP, Amieva M, Rabinovitch M. Bone morphogenetic protein 2 induces pulmonary angiogenesis via Wnt-beta-catenin and Wnt-RhoA-Rac1 pathways. J Cell Biol 184: 83–99, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diebold I, Hennigs JK, Miyagawa K, Li CG, Nickel NP, Kaschwich M, Cao A, Wang L, Reddy S, Chen PI, Nakahira K, Alcazar MA, Hopper RK, Ji L, Feldman BJ, Rabinovitch M. BMPR2 preserves mitochondrial function and DNA during reoxygenation to promote endothelial cell survival and reverse pulmonary hypertension. Cell Metab 21: 596–608, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Kasmi KC, Pugliese SC, Riddle SR, Poth JM, Anderson AL, Frid MG, Li M, Pullamsetti SS, Savai R, Nagel MA, Fini MA, Graham BB, Tuder RM, Friedman JE, Eltzschig HK, Sokol RJ, Stenmark KR. Adventitial fibroblasts induce a distinct proinflammatory/profibrotic macrophage phenotype in pulmonary hypertension. J Immunol 193: 597–609, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fantozzi I, Zhang S, Platoshyn O, Remillard CV, Cowling RT, Yuan JX. Hypoxia increases AP-1 binding activity by enhancing capacitative Ca2+ entry in human pulmonary artery endothelial cells. Am J Physiol Lung Cell Mol Physiol 285: L1233–L1245, 2003. [DOI] [PubMed] [Google Scholar]
- 10.Foletta VC, Lim MA, Soosairajah J, Kelly AP, Stanley EG, Shannon M, He W, Das S, Massague J, Bernard O. Direct signaling by the BMP type II receptor via the cytoskeletal regulator LIMK1. J Cell Biol 162: 1089–1098, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghofrani HA, Morrell NW, Hoeper MM, Olschewski H, Peacock AJ, Barst RJ, Shapiro S, Golpon H, Toshner M, Grimminger F, Pascoe S. Imatinib in pulmonary arterial hypertension patients with inadequate response to established therapy. Am J Respir Crit Care Med 182: 1171–1177, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golpon HA, Geraci MW, Moore MD, Miller HL, Miller GJ, Tuder RM, Voelkel NF. HOX genes in human lung: altered expression in primary pulmonary hypertension and emphysema. Am J Pathol 158: 955–966, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomberg-Maitland M, Maitland ML, Barst RJ, Sugeng L, Coslet S, Perrino TJ, Bond L, Lacouture ME, Archer SL, Ratain MJ. A dosing/cross-development study of the multikinase inhibitor sorafenib in patients with pulmonary arterial hypertension. Clin Pharmacol Ther 87: 303–310, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Good RB, Gilbane AJ, Trinder SL, Denton CP, Coghlan G, Abraham DJ, Holmes AM. Endothelial to mesenchymal transition contributes to endothelial dysfunction in pulmonary artery hypertension. Am J Pathol 185: 1850–1858, 2015. [DOI] [PubMed] [Google Scholar]
- 15.Guignabert C, Tu L, Girerd B, Ricard N, Huertas A, Montani D, Humbert M. New molecular targets of pulmonary vascular remodeling in pulmonary arterial hypertension: importance of endothelial communication. Chest 147: 529–537, 2015. [DOI] [PubMed] [Google Scholar]
- 16.Gupta K, Kshirsagar S, Li W, Gui L, Ramakrishnan S, Gupta P, Law PY, Hebbel RP. VEGF prevents apoptosis of human microvascular endothelial cells via opposing effects on MAPK/ERK and SAPK/JNK signaling. Exp Cell Res 247: 495–504, 1999. [DOI] [PubMed] [Google Scholar]
- 17.Hansmann G, de Jesus Perez VA, Alastalo TP, Alvira CM, Guignabert C, Bekker JM, Schellong S, Urashima T, Wang L, Morrell NW, Rabinovitch M. An antiproliferative BMP-2/PPARγ/apoE axis in human and murine SMCs and its role in pulmonary hypertension. J Clin Invest 118: 1846–1857, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassel S, Eichner A, Yakymovych M, Hellman U, Knaus P, Souchelnytskyi S. Proteins associated with type II bone morphogenetic protein receptor (BMPR-II) and identified by two-dimensional gel electrophoresis and mass spectrometry. Proteomics 4: 1346–1358, 2004. [DOI] [PubMed] [Google Scholar]
- 19.Hilberg F, Roth GJ, Krssak M, Kautschitsch S, Sommergruber W, Tontsch-Grunt U, Garin-Chesa P, Bader G, Zoephel A, Quant J, Heckel A, Rettig WJ. BIBF 1120: triple angiokinase inhibitor with sustained receptor blockade and good antitumor efficacy. Cancer Res 68: 4774–4782, 2008. [DOI] [PubMed] [Google Scholar]
- 20.Hoeper MM, Barst RJ, Bourge RC, Feldman J, Frost AE, Galie N, Gomez-Sanchez MA, Grimminger F, Grunig E, Hassoun PM, Morrell NW, Peacock AJ, Satoh T, Simonneau G, Tapson VF, Torres F, Lawrence D, Quinn DA, Ghofrani HA. Imatinib mesylate as add-on therapy for pulmonary arterial hypertension: results of the randomized IMPRES study. Circulation 127: 1128–1138, 2013. [DOI] [PubMed] [Google Scholar]
- 21.Hopper RK, Feinstein JA, Manning MA, Benitz W, Hudgins L. Neonatal pulmonary arterial hypertension and Noonan syndrome: Two fatal cases with a specific RAF1 mutation. Am J Med Genet A 167: 882–885, 2015. [DOI] [PubMed] [Google Scholar]
- 22.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37: D412–D416, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson JA, Hemnes AR, Perrien DS, Schuster M, Robinson LJ, Gladson S, Loibner H, Bai S, Blackwell TR, Tada Y, Harral JW, Talati M, Lane KB, Fagan KA, West J. Cytoskeletal defects in Bmpr2-associated pulmonary arterial hypertension. Am J Physiol Lung Cell Mol Physiol 302: L474–L484, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein M, Schermuly RT, Ellinghaus P, Milting H, Riedl B, Nikolova S, Pullamsetti SS, Weissmann N, Dony E, Savai R, Ghofrani HA, Grimminger F, Busch AE, Schafer S. Combined tyrosine and serine/threonine kinase inhibition by sorafenib prevents progression of experimental pulmonary hypertension and myocardial remodeling. Circulation 118: 2081–2090, 2008. [DOI] [PubMed] [Google Scholar]
- 25.Kojonazarov B, Sydykov A, Pullamsetti SS, Luitel H, Dahal BK, Kosanovic D, Tian X, Majewski M, Baumann C, Evans S, Phillips P, Fairman D, Davie N, Wayman C, Kilty I, Weissmann N, Grimminger F, Seeger W, Ghofrani HA, Schermuly RT. Effects of multikinase inhibitors on pressure overload-induced right ventricular remodeling. Int J Cardiol 167: 2630–2637, 2013. [DOI] [PubMed] [Google Scholar]
- 26.Lane KB, Blackwell TR, Runo J, Wheeler L, Phillips JA 3rd, Loyd JE. Aberrant signal transduction in pulmonary hypertension. Chest 128: 564S–565S, 2005. [DOI] [PubMed] [Google Scholar]
- 27.Lara MF, Garcia-Escudero R, Ruiz S, Santos M, Moral M, Martinez-Cruz AB, Segrelles C, Lorz C, Paramio JM. Gene profiling approaches help to define the specific functions of retinoblastoma family in epidermis. Mol Carcinog 47: 209–221, 2008. [DOI] [PubMed] [Google Scholar]
- 28.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27: 1739–1740, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Machado RD, Aldred MA, James V, Harrison RE, Patel B, Schwalbe EC, Gruenig E, Janssen B, Koehler R, Seeger W, Eickelberg O, Olschewski H, Elliott CG, Glissmeyer E, Carlquist J, Kim M, Torbicki A, Fijalkowska A, Szewczyk G, Parma J, Abramowicz MJ, Galie N, Morisaki H, Kyotani S, Nakanishi N, Morisaki T, Humbert M, Simonneau G, Sitbon O, Soubrier F, Coulet F, Morrell NW, Trembath RC. Mutations of the TGF-β type II receptor BMPR2 in pulmonary arterial hypertension. Hum Mutat 27: 121–132, 2006. [DOI] [PubMed] [Google Scholar]
- 30.Machado RD, Rudarakanchana N, Atkinson C, Flanagan JA, Harrison R, Morrell NW, Trembath RC. Functional interaction between BMPR-II and Tctex-1, a light chain of Dynein, is isoform-specific and disrupted by mutations underlying primary pulmonary hypertension. Hum Mol Genet 12: 3277–3286, 2003. [DOI] [PubMed] [Google Scholar]
- 31.Majka S, Hagen M, Blackwell T, Harral J, Johnson JA, Gendron R, Paradis H, Crona D, Loyd JE, Nozik-Grayck E, Stenmark KR, West J. Physiologic and molecular consequences of endothelial Bmpr2 mutation. Respir Res 12: 84, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masri FA, Xu W, Comhair SA, Asosingh K, Koo M, Vasanji A, Drazba J, Anand-Apte B, Erzurum SC. Hyperproliferative apoptosis-resistant endothelial cells in idiopathic pulmonary arterial hypertension. Am J Physiol Lung Cell Mol Physiol 293: L548–L554, 2007. [DOI] [PubMed] [Google Scholar]
- 33.Massague J. Integration of Smad and MAPK pathways: a link and a linker revisited. Genes Dev 17: 2993–2997, 2003. [DOI] [PubMed] [Google Scholar]
- 34.Maurer B, Reich N, Juengel A, Kriegsmann J, Gay RE, Schett G, Michel BA, Gay S, Distler JH, Distler O. Fra-2 transgenic mice as a novel model of pulmonary hypertension associated with systemic sclerosis. Ann Rheum Dis 71: 1382–1387, 2012. [DOI] [PubMed] [Google Scholar]
- 35.Montani D, Bergot E, Gunther S, Savale L, Bergeron A, Bourdin A, Bouvaist H, Canuet M, Pison C, Macro M, Poubeau P, Girerd B, Natali D, Guignabert C, Perros F, O'Callaghan DS, Jais X, Tubert-Bitter P, Zalcman G, Sitbon O, Simonneau G, Humbert M. Pulmonary arterial hypertension in patients treated by dasatinib. Circulation 125: 2128–2137, 2012. [DOI] [PubMed] [Google Scholar]
- 36.Morecroft I, Doyle B, Nilsen M, Kolch W, Mair K, Maclean MR. Mice lacking the Raf-1 kinase inhibitor protein exhibit exaggerated hypoxia-induced pulmonary hypertension. Br J Pharmacol 163: 948–963, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Moreno-Vinasco L, Gomberg-Maitland M, Maitland ML, Desai AA, Singleton PA, Sammani S, Sam L, Liu Y, Husain AN, Lang RM, Ratain MJ, Lussier YA, Garcia JG. Genomic assessment of a multikinase inhibitor, sorafenib, in a rodent model of pulmonary hypertension. Physiol Genomics 33: 278–291, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65: 55–63, 1983. [DOI] [PubMed] [Google Scholar]
- 39.Nishihara A, Watabe T, Imamura T, Miyazono K. Functional heterogeneity of bone morphogenetic protein receptor-II mutants found in patients with primary pulmonary hypertension. Mol Biol Cell 13: 3055–3063, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogura T, Taniguchi H, Azuma A, Inoue Y, Kondoh Y, Hasegawa Y, Bando M, Abe S, Mochizuki Y, Chida K, Kluglich M, Fujimoto T, Okazaki K, Tadayasu Y, Sakamoto W, Sugiyama Y. Safety and pharmacokinetics of nintedanib and pirfenidone in idiopathic pulmonary fibrosis. Eur Respir J 45: 1382–1392, 2015. [DOI] [PubMed] [Google Scholar]
- 41.Ozaki K, Kadomoto R, Asato K, Tanimura S, Itoh N, Kohno M. ERK pathway positively regulates the expression of Sprouty genes. Biochem Biophys Res Commun 285: 1084–1088, 2001. [DOI] [PubMed] [Google Scholar]
- 42.Rabinovitch M. Molecular pathogenesis of pulmonary arterial hypertension. J Clin Invest 122: 4306–4313, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rai PR, Cool CD, King JA, Stevens T, Burns N, Winn RA, Kasper M, Voelkel NF. The cancer paradigm of severe pulmonary arterial hypertension. Am J Respir Crit Care Med 178: 558–564, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richeldi L, du Bois RM, Raghu G, Azuma A, Brown KK, Costabel U, Cottin V, Flaherty KR, Hansell DM, Inoue Y, Kim DS, Kolb M, Nicholson AG, Noble PW, Selman M, Taniguchi H, Brun M, Le Maulf F, Girard M, Stowasser S, Schlenker-Herceg R, Disse B, Collard HR, Investigators IT. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N Engl J Med 370: 2071–2082, 2014. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez LG, Wu X, Guan JL. Wound-healing assay. Methods Mol Biol 294: 23–29, 2005. [DOI] [PubMed] [Google Scholar]
- 46.Rudarakanchana N, Flanagan JA, Chen H, Upton PD, Machado R, Patel D, Trembath RC, Morrell NW. Functional analysis of bone morphogenetic protein type II receptor mutations underlying primary pulmonary hypertension. Hum Mol Genet 11: 1517–1525, 2002. [DOI] [PubMed] [Google Scholar]
- 47.Sakao S, Taraseviciene-Stewart L, Lee JD, Wood K, Cool CD, Voelkel NF. Initial apoptosis is followed by increased proliferation of apoptosis-resistant endothelial cells. FASEB J 19: 1178–1180, 2005. [DOI] [PubMed] [Google Scholar]
- 48.Sasaki A, Taketomi T, Kato R, Saeki K, Nonami A, Sasaki M, Kuriyama M, Saito N, Shibuya M, Yoshimura A. Mammalian Sprouty4 suppresses Ras-independent ERK activation by binding to Raf1. Nat Cell Biol 5: 427–432, 2003. [DOI] [PubMed] [Google Scholar]
- 49.Savai R, Al-Tamari HM, Sedding D, Kojonazarov B, Muecke C, Teske R, Capecchi MR, Weissmann N, Grimminger F, Seeger W, Schermuly RT, Pullamsetti SS. Pro-proliferative and inflammatory signaling converge on FoxO1 transcription factor in pulmonary hypertension. Nat Med 20: 1289–1300, 2014. [DOI] [PubMed] [Google Scholar]
- 50.Sawada H, Saito T, Nickel NP, Alastalo TP, Glotzbach JP, Chan R, Haghighat L, Fuchs G, Januszyk M, Cao A, Lai YJ, Perez Vde J, Kim YM, Wang L, Chen PI, Spiekerkoetter E, Mitani Y, Gurtner GC, Sarnow P, Rabinovitch M. Reduced BMPR2 expression induces GM-CSF translation and macrophage recruitment in humans and mice to exacerbate pulmonary hypertension. J Exp Med 211: 263–280, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schaller MD. Paxillin: a focal adhesion-associated adaptor protein. Oncogene 20: 6459–6472, 2001. [DOI] [PubMed] [Google Scholar]
- 52.Spiekerkoetter E, Tian X, Cai J, Hopper RK, Sudheendra D, Li CG, El-Bizri N, Sawada H, Haghighat R, Chan R, Haghighat L, de Jesus Perez V, Wang L, Reddy S, Zhao M, Bernstein D, Solow-Cordero DE, Beachy PA, Wandless TJ, Ten Dijke P, Rabinovitch M. FK506 activates BMPR2, rescues endothelial dysfunction, and reverses pulmonary hypertension. J Clin Invest 123: 3600–3613, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Straatman K. Wound healing assay. http://www.le.ac.uk/biochem/microscopy/pdf/Wound%20healing%20assay.pdf. [Google Scholar]
- 54.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15,545–15,550, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taraseviciene-Stewart L, Kasahara Y, Alger L, Hirth P, Mc Mahon G, Waltenberger J, Voelkel NF, Tuder RM. Inhibition of the VEGF receptor 2 combined with chronic hypoxia causes cell death-dependent pulmonary endothelial cell proliferation and severe pulmonary hypertension. FASEB J 15: 427–438, 2001. [DOI] [PubMed] [Google Scholar]
- 56.Teichert-Kuliszewska K, Kutryk MJ, Kuliszewski MA, Karoubi G, Courtman DW, Zucco L, Granton J, Stewart DJ. Bone morphogenetic protein receptor-2 signaling promotes pulmonary arterial endothelial cell survival: implications for loss-of-function mutations in the pathogenesis of pulmonary hypertension. Circ Res 98: 209–217, 2006. [DOI] [PubMed] [Google Scholar]
- 57.Thomson JR, Machado RD, Pauciulo MW, Morgan NV, Humbert M, Elliott GC, Ward K, Yacoub M, Mikhail G, Rogers P, Newman J, Wheeler L, Higenbottam T, Gibbs JS, Egan J, Crozier A, Peacock A, Allcock R, Corris P, Loyd JE, Trembath RC, Nichols WC. Sporadic primary pulmonary hypertension is associated with germline mutations of the gene encoding BMPR-II, a receptor member of the TGF-β family. J Med Genet 37: 741–745, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toshner M, Voswinckel R, Southwood M, Al-Lamki R, Howard LS, Marchesan D, Yang J, Suntharalingam J, Soon E, Exley A, Stewart S, Hecker M, Zhu Z, Gehling U, Seeger W, Pepke-Zaba J, Morrell NW. Evidence of dysfunction of endothelial progenitors in pulmonary arterial hypertension. Am J Respir Crit Care Med 180: 780–787, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Troppmair J, Bruder JT, Munoz H, Lloyd PA, Kyriakis J, Banerjee P, Avruch J, Rapp UR. Mitogen-activated protein kinase/extracellular signal-regulated protein kinase activation by oncogenes, serum, and 12-O-tetradecanoylphorbol-13-acetate requires Raf and is necessary for transformation. J Biol Chem 269: 7030–7035, 1994. [PubMed] [Google Scholar]
- 60.Tuder RM, Chacon M, Alger L, Wang J, Taraseviciene-Stewart L, Kasahara Y, Cool CD, Bishop AE, Geraci M, Semenza GL, Yacoub M, Polak JM, Voelkel NF. Expression of angiogenesis-related molecules in plexiform lesions in severe pulmonary hypertension: evidence for a process of disordered angiogenesis. J Pathol 195: 367–374, 2001. [DOI] [PubMed] [Google Scholar]
- 61.West J, Harral J, Lane K, Deng Y, Ickes B, Crona D, Albu S, Stewart D, Fagan K. Mice expressing BMPR2R899X transgene in smooth muscle develop pulmonary vascular lesions. Am J Physiol Lung Cell Mol Physiol 295: L744–L755, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.West JD, Austin ED, Gaskill C, Marriott S, Baskir R, Bilousova G, Jean JC, Hemnes AR, Menon S, Bloodworth NC, Fessel JP, Kropski JA, Irwin D, Ware LB, Wheeler L, Hong CC, Meyrick B, Loyd JE, Bowman AB, Ess KC, Klemm DJ, Young PP, Merryman WD, Kotton D, Majka SM. Identification of a common Wnt-associated genetic signature across multiple cell types in pulmonary arterial hypertension. Am J Physiol Cell Physiol 307: C415–C430, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong WK, Knowles JA, Morse JH. Bone morphogenetic protein receptor type II C-terminus interacts with c-Src: implication for a role in pulmonary arterial hypertension. Am J Respir Cell Mol Biol 33: 438–446, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yeager ME, Halley GR, Golpon HA, Voelkel NF, Tuder RM. Microsatellite instability of endothelial cell growth and apoptosis genes within plexiform lesions in primary pulmonary hypertension. Circ Res 88: E2–E11, 2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.