SUMMARY
To define the C. elegans aging process at the molecular level, we used DNA microarray experiments to identify a set of 1294 age-regulated genes and found that the GATA transcription factors ELT-3, ELT-5, and ELT-6 are responsible for age regulation of a large fraction of these genes. Expression of elt-5 and elt-6 increases during normal aging, and both of these GATA factors repress expression of elt-3, which shows a corresponding decrease in expression in old worms. elt-3 regulates a large number of downstream genes that change expression in old age, including ugt-9, col-144, and sod-3. elt-5(RNAi) and elt-6(RNAi) worms have extended longevity, indicating that elt-3, elt-5, and elt-6 play an important functional role in the aging process. These results identify a transcriptional circuit that guides the rapid aging process in C. elegans and indicate that this circuit is driven by drift of developmental pathways rather than accumulation of damage.
INTRODUCTION
A key approach to understanding how C. elegans age is to characterize differences between young and old animals. Old worms move slowly, become flaccid, and accumulate an age-related pigment called lipofuscin in their intestines (Garigan et al., 2002; Gerstbrein et al., 2005). Electron microscopy has shown that old worms exhibit degeneration of their muscle and intestinal cells but not neural tissue (Herndon et al., 2002). At the molecular level, a GFP reporter for MYO-3 shows disorganization of muscle sarcomeres, and a GFP reporter for yolk protein 170 shows accumulation of yolk protein in the body cavity in old age (Herndon et al., 2002). In addition, analysis of gene expression throughout an aging time course using DNA microarrays has generated a global profile of transcriptional differences during aging (Lund et al., 2002).
By profiling cellular and molecular changes that occur with age, it is possible to identify upstream factors that cause these age-related changes. A widely held view is that aging is driven by cellular and environmental damage that accumulates over time, including damage from cellular stress, free radicals, and pathogenic infection (Garsin et al., 2003). Life span is extended by growing worms on nonpathogenic bacteria or by reducing oxidative damage (Melov, 2000). Furthermore, mutants that have extended life spans, such as daf-2 mutants, also have increased resistance to stress (Garsin et al., 2003). Damage accumulation explains some of the molecular changes observed with age, such as increased levels of protein oxidation and age pigments (Gerstbrein et al., 2005; Ishii et al., 2002). However, damage does not lead to a chronic induction of the stress responses in old worms (Lund et al., 2002).
Besides damage accumulation, another possibility is that aging results from developmental pathways that go awry late in life (antagonistic pleiotropy) (Kirkwood and Rose, 1991; Williams, 1957). Late in life, the force of natural selection is low or absent, permitting deterioration of a broad spectrum of physiological and metabolic pathways (Kirkwood and Rose, 1991). There are few examples of antagonistic pleiotropy in mammals. One example of antagonistic pleiotropy is cell senescence, which limits cancerous growth in young animals but also limits stem cell proliferation late in life (Krtolica et al., 2001). Another example is that Wnt signaling pathway, which promotes the moygenic lineage progression during development (Holowacz et al., 2006), also impairs muscle regeneration in old age (Brack et al., 2007; Liu et al., 2007).
Here, we have repeated DNA microarray experiments to define the aging transcriptome, and we found that much of the age-regulated gene expression arises from changes in a transcriptional circuit characterized by GATA transcription factor binding. We examined elt-3 GATA factor as a potential regulator of these genes since its expression declines in a tissue-specific fashion during normal aging. We found this decline with age results in changes in the expression of the many downstream target genes, including sod-3, ugt-9, and col-144. Decreased expression of elt-3 is not just a marker for old age but is functionally important for shortening life span because mutations in elt-3 suppress the longevity phenotype of daf-2 insulin receptor mutants. To our knowledge, this is the first transcriptional circuit to account for differences in expression in young versus old worms.
Next, we examined age regulation of the elt-3 transcriptional aging circuit and found no evidence that it is caused by cellular damage or environmental stresses. Rather, we found that elt-3 expression in the adult is controlled by increased expression of the repressors elt-5 and elt-6, which also guide elt-3 expression during development. These results suggest that age regulation of elt-3 is caused by age-related drift of an intrinsic developmental program that becomes imbalanced in old age.
RESULTS
A Common Set of Genes Expressed in Old Age, in Dauer Larvae, and in Longevity Mutants
We looked for similarities in gene expression changes during normal aging, in dauer larvae (developmentally arrested worms that can live up to ten times longer than normal worms), and in mutants with either extended or shortened life spans (age-1 or daf-16). To do this, we analyzed expression data from several new DNA microarray experiments as well as those from previously published studies (Lund et al., 2002; McElwee et al., 2003; Murphy et al., 2003; Wang and Kim, 2003). If there were a common transcriptional response across normal aging, dauer, and longevity mutants, we would expect the expression changes in these experiments to be correlated with each other. Furthermore, it may be possible to identify common regulatory motifs found in the upstream regions of genes that change expression in these DNA microarray experiments.
We repeated previous work by performing a genomewide search for genes that change expression during normal aging (Lund et al., 2002). We performed a time course for aging by growing synchronous cultures of hermaphrodites to 4, 7, 10, and 14 days of adulthood in quadruplicate (Experimental Procedures). We used DNA microarrays to measure expression at each aging time point and identified 1254 genes that change expression during aging (ANOVA, p < 0.0001) (Figure 1A and Tables S3 and S4 available online). We found that the list of age-regulated genes was enriched with intestinal and oocyte genes but not neuronal, muscle, or pharyngeal genes (Table 1A). This result suggests that aging affects the intestine and germline tissues more than other tissues such as muscle, pharynx, or neuronal tissues.
Figure 1. An elt-3 Transcriptional Circuit for Aging.
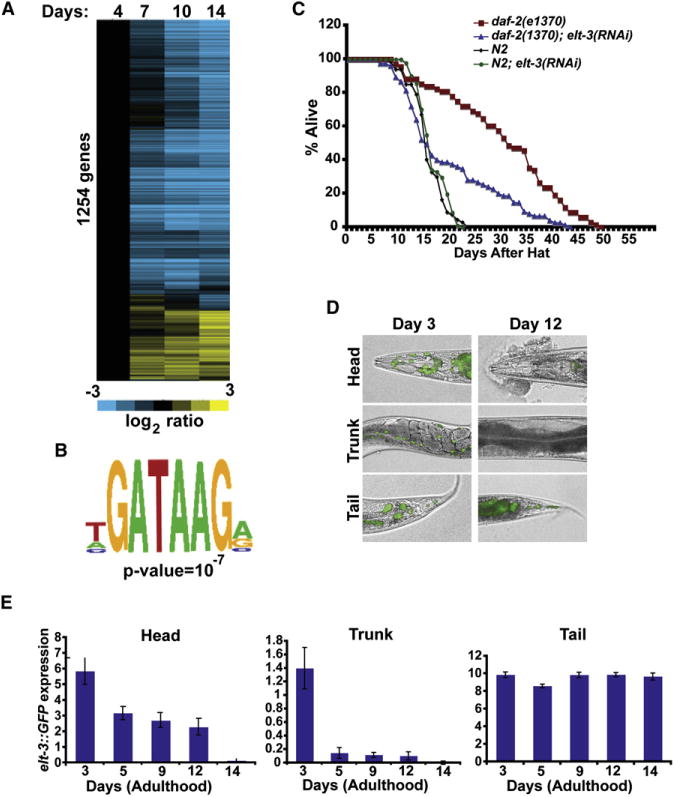
(A) Shown are the log2 average expression levels of 1254 age-regulated genes during aging, normalized to expression on day 4. Rows show age-regulated genes and columns at different aging time points (days of adulthood). Full data showing genes and experimental values for this figure can be found in Table S3. The expression results from this aging time course show a Pearson correlation of 0.429 with results from a similar DNA microarray experiment on aging previously by Lund et al., indicating that the two experiments were generally similar (Table 1A) (Lund et al., 2002). Full DNA microarray data set can be found at http://cmgm.stanford.edu/∼kimlab/elt3/index.html.
(B) A GATA regulatory element that is significantly enriched in the upstream regions of the age-regulated genes was identified using the CompareProspector program (Liu, 2005).
(C) elt-3(RNAi) treatment specifically suppresses the life span extension of daf-2(e1370) mutants (p < 0.001).
(D) Expression of elt-3::GFP declines with age in a tissue-specific manner. Shown are adult animals 3 days and 12 days after adulthood. The GFP images were merged with Nomarski images.
(E) Expression levels of elt-3::GFP during aging were calculated by measuring pixel intensity from GFP images using ImageJ. The y axis denotes GFP expression (arbitrary units), and the x axis denotes days of adulthood. Average expression and SD from 20 animals are shown.
Table 1.
Gene Expression Profiles for Aging
| A. 1254 Age-Regulated Genes Are Enriched for Intestine and Oocyte Genes | ||||
|---|---|---|---|---|
| Data Set | # of Genes | Overlap | Representation Factora | p Valueb |
| Intestine enriched genesc | 609 | 143 | 3.3 | 5.612 × 10−39 |
| Oocyte enriched genesd | 258 | 28 | 2.1 | 2 × 10−4 |
| Muscle enriched genese | 230 | 15 | 0.9 | <0.424 |
| Pharynx enriched genesf | 194 | 0 | 0 | N/A |
| Neuronal enriched genese | 85 | 0 | 0 | N/A |
| B. A GATA Regulatory Motif Is Highly Enriched in the Promoters of Age-Regulated Genes | ||||
|---|---|---|---|---|
| Data Set | # of Genes | Rk (p Value) | GATA Motif | GATA Enrichment (p Value) |
| Aging time courseg | 180 | 0.429 (p < 10−20) |

|
0.009 |
| Dauer/normalh | 478 | 0.301 (p < 10−20) |

|
8 × 10−5 |
| age-1/wt | 758 | 0.048 (p = 0.037) |

|
3 × 10−5 |
| daf-2; daf-16/daf-2i | 214 | −0.369 (p < 10−29) |

|
3 × 10−3 |
| daf-16/wt | 886 | −0.247 (p < 10−29) |

|
1 × 10−06 |
| Data Set | # of Genes | Overlap (p Value) | GATA Motif | GATA Enrichment (p value) |
| daf-16/wt (class1 and 1)j | 467 | 127 (p < 0.0001) |

|
5 × 10−9 |
The representation factor is the number of overlapping genes divided by the expected number expected by chance.
Hypergeometric p value.
Pearson Correlation Coefficient comparing the set of genes in the list to the set of 1254 age-regulated genes.
We compared these aging results to gene expression changes in dauer larvae, using expression data from a previous DNA microarray experiment (Wang and Kim, 2003). We found a significant correlation in gene expression between normal aging and dauer larvae, indicating that there is a common pattern of changes (Table 1B). Specifically, genes that increase expression in old age also tend to increase expression in dauers and vice versa. Such a similarity in the pattern of gene expression changes between old age and the dauer state has been noted previously (Lund et al., 2002).
Next, we compared changes in gene expression associated with normal aging to gene expression changes in two longevity mutants, age-1 and daf-16. An insulin-like signaling pathway specifies longevity in C. elegans, in which a PI3 kinase encoded by age-1 represses the activity of a FOXO transcription factor encoded by daf-16. Loss-of-function mutations in age-1 result in extended longevity, and null mutations in daf-16 suppress the life span extension caused by age-1 (Hekimi et al., 1998).
We used DNA microarray experiments to compare expression in age-1(hx542) mutants and daf-16(m26) mutants to wild-type controls (Experimental Procedures). We prepared ten RNA samples from age-1 mutants, 11 samples from daf-16 mutants, and 12 samples from wild-type worms. Each sample was hybridized to a DNA microarray along with a reference RNA control, and then the ratios of expression between the two mutants and wild-type animals were compared to find differences in gene expression. We identified 758 age-1-regulated and 886 daf-16-regulated genes (Student’s t test, p < 10−4) (Table 1B and Tables S5 and S6). We compared the results for insulin-like signaling mutants to results from the aging time course and found that expression changes during aging are positively correlated with changes in age-1 mutants and negatively correlated with changes in daf-16 mutants (Table 1B). Genes that increase expression during aging also tend to increase expression in age-1 mutants and decrease expression in daf-16 mutants.
Next, we compared our study of age regulation to previous studies of genes that act downstream of the insulin-like signaling pathway (Murphy et al., 2003) and daf-16 (McElwee et al., 2003). McElwee et al. used DNA microarrays to compare expression levels in daf-2 mutants to those in daf-2; daf-16 double mutants and identified 214 genes that act downstream of daf-16. We compared these results of daf-16 mutants to the aging time course and found that there was a strong negative correlation (Table 1B). Murphy et al. performed a series of DNA microarray experiments to find a total of 467 genes that act downstream of three genes in the insulin-like signaling pathway: daf-2, age-1, or daf-16. We compared this list of 467 downstream targets to our list of 1254 age-regulated genes and found 127 genes in common, which is much more than would be expected by chance (p < 10−27) (Table 1B). In summary, the results from each of these DNA microarray experiments show that there is a shared pattern of expression changes between old age and insulin-like signaling mutants.
A second approach to search for a common transcriptional response in these DNA microarray experiments is to identify a common DNA motif in the upstream regulatory regions of genes that change expression in each of these experiments. To do this, we used CompareProspector (Liu, 2005), a program that first selects DNA regions that are conserved between C. elegans and C. briggsae and then uses Gibbs sampling to find DNA sequences that are enriched in the upstream regions of a set of genes (Experimental Procedures). We analyzed six sets of genes that change expression in the aging DNA microar-ray experiments: 1254 age-modulated genes, 478 dauer-enriched genes (Wang and Kim, 2003), 758 age-1-regulated genes, 886 daf-16-regulated genes, 214 daf-16-regulated genes (McElwee et al., 2003, 2004), and 467 genes that are downstream of the insulin-like signaling pathway (Murphy et al., 2003). We found that the upstream regions in each of these gene sets are significantly enriched for a common DNA motif ([T/C/G]GATAA[C/G][A/G]) (Figure 1B and Table 1B). This DNA sequence is a consensus motif recognized by GATA transcription factors. The reverse complement of this GATA transcription binding site was previously identified among the genes responsive to the insulin-like signaling pathway (Murphy et al., 2003). These results indicate that one or more GATA transcription factors may control a common transcriptional network involved in old age, the dauer state, and response to insulin-like signaling.
Control of the Aging Transcriptional Network by the ELT-3 GATA Transcription Factor
There are 14 GATA transcription factor genes in C. elegans (C. elegans Sequencing Consortium, 1998), and none were previously known to have a role in aging. We used RNA interference (RNAi) to investigate whether ten of these GATA transcription factors (egl-18, elt-1, elt-2, elt-3, elt-6, end-1, end-3, egr-1, egl-27, and med-1) play a role in longevity. We found that none of the ten RNAi treatments extended the life span of wild-type worms (Table S1A). Alternatively, if a GATA transcription factor functions specifically to extend life span rather than shorten it, then RNAi treatment may suppress the longevity of a long-lived mutant such as daf-2(e1370) without causing nonspecific early lethality of wild-type worms. Of the ten GATA transcription factor genes, we found that RNAi treatment of three (elt-3, egr-1, and egl-27) behave in this way in multiple independent life span experiments (Table S1A and Figure 1C).
In this paper, we focus on the role of the elt-3 GATA transcription factor gene in aging. elt-3 null mutants (vp1) are viable and exhibit normal growth, development, and behavior (Gilleard et al., 1999). We extended the aging results described above by showing that elt-3(vp1) suppresses the longevity phenotype of both daf-2(e1370) and daf-2(RNAi) animals (Table S1A). In addition, we showed that elt-3(RNAi) suppresses the longevity phenotype of eat-2(ad1116) mutants, which have a defect in pharyngeal pumping that results in dietary restriction (Lakowski and Hekimi, 1998; Table S1B and Figure S1A).
Previous work has shown that elt-3 is expressed in hypodermal cells, the intestine, the pharyngeal-intestinal valve cells, and the intestinal-rectal valve cells (Gilleard et al., 1999). Our DNA microarray experiments show that elt-3 decreases expression about 3.3-fold in old age. To more precisely determine how aging affects expression of elt-3, we examined expression of an elt-3 GFP reporter at various ages. In the head, elt-3 GFP expression decreases with age in the hypodermal cells and the pharyngeal-intestinal valve cells, eventually showing little or no expression in old worms (Figures 1D and 1E). In the trunk, elt-3 expression is mostly derived from the hypodermal cells and the intestinal cells, and expression in this region decreases quickly between day 3 and day 5 of adulthood (Figures 1D and 1E). The elt-3 GFP reporter did not change expression in the intestinal-rectal valve cells located in the tail of the worm. In summary, these results show that age-related changes in elt-3 expression are complex, as different tissues show different kinetics of age regulation and some tissues show no age regulation at all.
elt-3 Controls Expression of Age-Regulated Genes
We constructed GFP reporters for 14 genes selected from the set of 602 genes that are age regulated and have GATA sites in their upstream regions. Of these, we found that 12 showed decreased expression in elt-3(RNAi) worms, indicating that they are downstream targets of elt-3 (Table S7).
We examined three GFP reporters (sod-3, ugt-9, and col-144) in more detail. ugt-9 encodes UDP-glucuronosyltransferase and is expressed primarily in pharyngeal cells (Figure 2A). col-144 encodes subunits of nematode cuticle collagen and is expressed specifically in the hypodermis (Figure 2A). sod-3 encodes iron/manganese superoxide dismutase and is expressed in all cells, with highest expression in the pharynx, intestine, and ventral cord motor neurons (Henderson et al., 2006; Figure 2A). ugt-9, sod-3, and col-144 GFP expression decreases in old age, consistent with the DNA microarray data. Specifically, expression of ugt-9 in the pharynx decreases about 30% in day 5 of adulthood and 95% in day 7 of adulthood compared to young adults (Figure 2A). Expression of the col-144 GFP reporter in the hypodermis declines gradually throughout the life span of the worm (Figure 2A). Expression of the sod-3 GFP reporter increases about 30% between day 3 and day 6 of adulthood in all tissues except for the ventral cord motor neurons (Figure 2A, data not shown). After day 6, expression of sod-3::GFP gradually decreases in all tissues except for motor neurons.
Figure 2. Expression of ugt-9::GFP, col-144::GFP, and sod-3::GFP Is Regulated by Age and by elt-3.
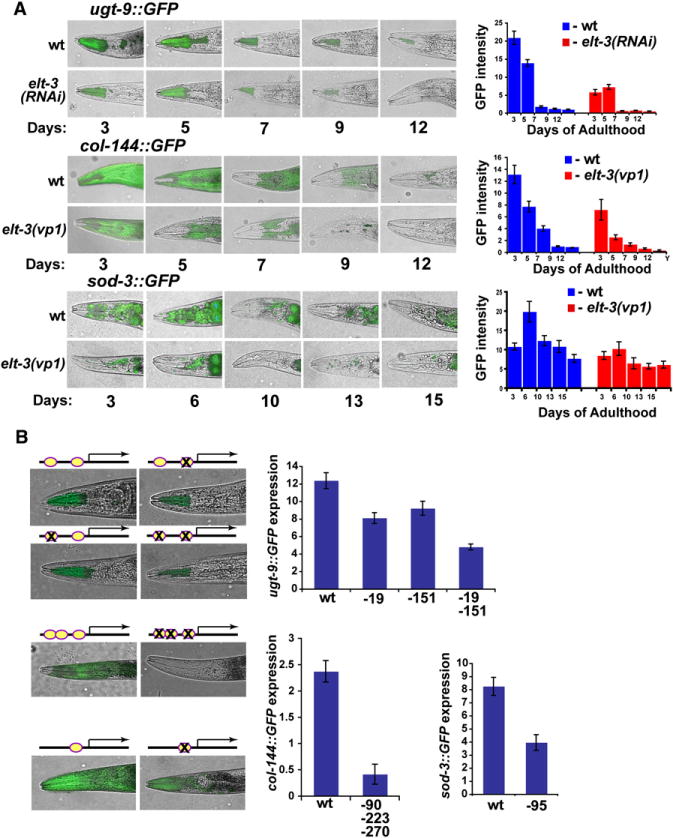
(A) Expression of ugt-9::GFP, col-144::GFP, and sod-3::GFP. (Left) An aging time course of GFP expression merged on Nomarski images for wild-type and elt-3(RNAi) or elt-3(vp1) mutants. (Right) Quantification of GFP expression from 20 worms.
(B) The GATA sequence in the promoters of ugt-9, col-144, and sod-3 was mutated and used to generate transgenic GFP reporter strains. (Left) GFP images/Nomarski of wild-type and mutated promoters. Ovals indicate GATA consensus binding sites, and “X” indicates mutation in the GATA site. (Right) Quantification of GFP expression from 20 animals. Numbers refer to the nucleotide position of the GATA site in the upstream region.
Error bars represent the SEM pixel intensities.
We next determined whether these three downstream aging targets were regulated by elt-3 GATA. First, we examined whether reduction of elt-3 activity affects expression of GFP reporters for each of the downstream targets in an aging time course (Figure 2A). We found that elt-3(RNAi) reduced expression of ugt-9::GFP about 74% at 3 days and 95% at 7 days after adulthood compared to age-matched control worms. elt-3(vp1) reduced expression of col-144::GFP about 45% at day 3 of adulthood and reduced expression of sod-3::GFP about 50% at day 6 of adulthood compared to control worms of similar age. These observations indicate that ugt-9, sod-3, and col-144 are each regulated by elt-3, either directly or indirectly.
For all three age-regulated genes, elt-3(RNAi) or elt-3(vp1) had less effect on expression in old adults than in young adults. This finding indicates that the magnitude of elt-3 regulation changes during aging and that changing levels of elt-3 regulation are partially responsible for age regulation of these downstream genes. However, expression of ugt-9 and col-144 still decreases with age in elt-3(RNAi) or null mutants (although the degree of age regulation in elt-3 mutants is diminished compared to wild-type animals). This observation indicates that elt-3 is not the only factor responsible for age-related changes in these downstream genes.
The elt-3 GATA transcription factor is coexpressed with the downstream aging markers in some tissues but not in others. Specifically, ugt-9 and elt-3 are both expressed in the pharynx, col-144 and elt-3 are both expressed in the hypodermis, and sod-3 is expressed along with elt-3 in the intestinal and pharyngeal cells. In these cases, expression of the downstream aging markers is dependent on elt-3 activity. However, sod-3 is expressed in the ventral cord motor neurons but elt-3 is not, and sod-3::GFP expression is not altered by elt-3(vp1) in these cells. Thus, the downstream aging markers show regulation by elt-3 when they are expressed in the same cells but not when they are expressed in cells that do not express elt-3.
Next, we mutated the GATA DNA sites in the promoters of these three GFP aging reporters to determine whether their GATA motifs are functionally required for expression. For ugt-9, simultaneous elimination of both GATA motifs in the upstream region led to a 60% loss of GFP reporter expression (Figure 2B). Deletion of all three GATA motifs in the col-144 upstream region led to an 80% reduction in GFP expression, and deletion of the single GATA site in the upstream region of the sod-3 gene reduced GFP expression by 50%. Furthermore, GFP expression from the mutated GATA promoters shows constant low-level expression throughout life, similar to expression of the aging GFP reporters in elt-3(RNAi) or elt-3(vp1) mutants (data not shown). These results show that the GATA motifs are required for age regulation of ugt-9, col-144, and sod-3. In summary, elt-3 activity is required in trans and the GATA DNA sites are required in cis for age regulation of these three downstream aging markers.
Regulation of elt-3 GATA by the Insulin-like Signaling Pathway
Our DNA microarray experiments showed that there is a broad overlap between the set of 1254 age-regulated genes and genes that are regulated by the insulin-like signaling pathway. We confirmed this overlap by showing that the insulin-like signaling pathway also regulates the three aging markers. One of the age-regulated genes, sod-3, was previously known to be regulated by daf-16 (Henderson et al., 2006). We showed that mutations in daf-2 insulin-like receptor and age-1 PI3 kinase strongly increase expression of ugt-9::GFP and slightly affect expression of col-144::GFP (Figure S2 and data not shown).
One possible explanation for the overlap in downstream targets is that elt-3 might be regulated by the insulin-like signaling pathway. In our DNA microarray experiments, elt-3 expression increased 1.9-fold in daf-2 mutants and 2.3-fold in age-1 mutants. To extend this result, we examined age-1 regulation of an elt-3 GFP reporter throughout the life span of the worm. We used RNAi to partially reduce age-1 activity starting in young adults and then examined elt-3::GFP expression as the worms age. We found that age-1(RNAi) slightly increased expression of the elt-3 GFP reporter in the head and trunk hypodermis at all ages (Figure 3). elt-3 expression decreases during aging in the head and trunk in age-1 mutants, but the absolute level of expression is slightly higher at every age than in the control. elt-3 expression in the tail is not regulated by age-1. These results indicate that the insulin-like signaling pathway exerts a constant level of regulation on elt-3 expression throughout life and that decreased expression of elt-3 late in life is not caused by increased repression from the insulin-like signaling pathway. Although the insulin-like signaling pathway regulates genes via the FOXO transcription factor DAF-16, elt-3 does not have any DAF-16 consensus binding sites (T[G/A]TTTAC) in its upstream region. This result suggests that regulation of elt-3 by age-1 is either independent of daf-16 or regulated via daf-16 but indirectly.
Figure 3. Regulation of elt-3 GATA by age-1.
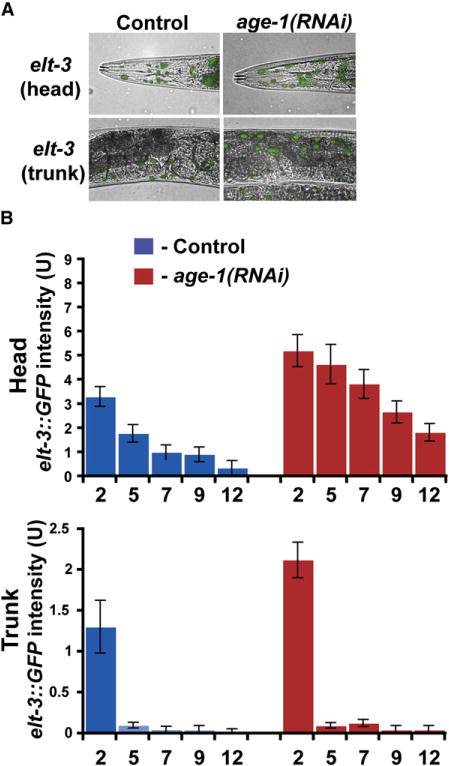
(A) GFP expression of elt-3 in wild-type and age-1(RNAi) animals at day 3 of adulthood. Images show expression in the head and trunk regions. age-1(RNAi) results in increased elt-3 GATA expression in the head and trunk hypodermal cells but not in intestinal-rectal valve cells and the tail hypodermis. (B) Quantification of levels of elt-3::GFP expression from 20 worms in wild-type and age-1(RNAi) mutants at five times during aging. Expression levels were determined in the head area of the worm by measuring pixel intensity from GFP images.
Error bars represent the SEM pixel intensities.
We also tested whether elt-3::GFP expression was affected in two other mutants with extended longevity. eat-2 mutants are calorically restricted and clk-1 mutants are defective in biosynthesis of ubiquinone and mitochondrial respiration (Lakowski and Hekimi, 1998; Wong et al., 1995). We found that neither expression of elt-3 nor the aging markers (sod-3, col-144, ugt-9) was significantly affected in eat-2(RNAi) or clk-1 (RNAi) animals compared to wild-type controls (Figure S1B and data not shown).
Environmental Stress Does Not Regulate the elt-3 Transcriptional Circuit
One possibility is that age-related changes in elt-3 expression could be caused by the accumulation of cellular damage, environmental stress, or pathogenic infection in old age (damage accumulation). Another possibility is that changes in the elt-3 transcriptional circuit may be caused by changes in regulatory pathways used to control elt-3 during development (developmental drift).
To test the first possibility, we determined whether environmental stress (heat shock), oxidative stress (paraquat treatment), DNA damage (γ-irradiation), or pathogenic infection cause a decrease in expression of either elt-3 or the downstream aging markers, mimicking the effects of old age. We found that heat shock or γ-irradiation had no effect on expression of GFP reporters for elt-3, ugt-9, col-144, or sod-3. We induced oxidative stress by subjecting worms to paraquat treatment and saw increased levels of expression of GFP reporters for elt-3 and the three downstream aging markers. However, it seems unlikely that changes in the elt-3 transcriptional circuit in old age are due to changes in oxidative damage because oxidative damage increases with age and would, thus, be expected to increase expression of elt-3 and its target genes rather than decrease expression as is observed in old age.
Finally, pathogen infection can limit C. elegans life span (Garsin et al., 2003). To test the possibility that changes in the elt-3 transcription circuit may be due to increased levels of pathogen infection in old age, we analyzed DNA microarray data showing changes in expression following infection with Pseudomonas aeruginosa (Shapira et al., 2006). We found that expression of elt-3 or its targets (ugt-9, col-144, or sod-3) did not change in P. aeruginosa compared to E. coli. In summary, heat shock, oxidative damage, DNA damage, or pathogen infection do not appear to be responsible for driving transcriptional change of the elt-3 circuit during aging.
Regulation of the elt-3 Transcriptional Cascade by a Developmental Regulatory Pathway
Rather than extrinsic damage, another possibility is that developmental pathways that are beneficial to the young worm may become unbalanced and cause changes in gene expression in old worms (antagonistic pleiotropy) (Kirkwood and Rose, 1991; Williams, 1957). elt-3 is part of a GATA factor transcriptional hierarchy that specifies hypodermal development in the embryo. In this hierarchy, elt-3 expression is activated by elt-1(+) but repressed by elt-5(+) and elt-6(+) (elt-1, elt-5, and elt-6 each encode GATA transcription factors) (Gilleard and McGhee, 2001; Koh and Rothman, 2001). Furthermore, elt-3 has nine GATA sequence motifs in its promoter, suggesting that regulation by ELT-1, ELT-5, or ELT-6 might be direct (Liu, 2005).
First, we examined expression of elt-1, elt-5, and elt-6 at different ages. We found that expression of elt-5::Cherry and elt-6::Cherry reporters increases with age but that expression of elt-1::GFP is steady between young and old worms (Figure 4 and data not shown). Second, we used RNAi to reduce elt-1, elt-5, and elt-6 activity during aging and then observed effects on expression of an elt-3::GFP reporter. elt-5(RNAi) and elt-6(RNAi) both resulted in an increase in expression of elt-3::GFP in the trunk hypodermis (Figure 5A). In elt-5(RNAi) or elt-6 (RNAi) animals, elt-3 shows little or no age regulation, as elt-3::GFP expression remains consistently high from middle-aged worms to old worms. elt-1(RNAi) caused a decrease in elt-3::GFP expression in young worms (data not shown).
Figure 4. Age Regulation of elt-5 and elt-6.
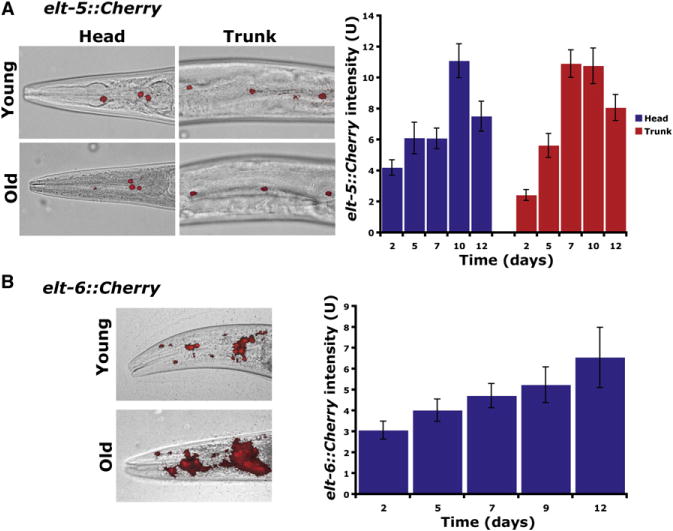
(A) Expression of elt-5::Cherry. (Left) Expression in young (2 days of adulthood) and old (12 days of adulthood) worms. (Right) Quantification of expression levels by measuring pixel intensity from Cherry images using ImageJ. The y axis denotes Cherry expression (arbitrary units), and the x axis denotes days of adulthood. Average expression and SE from 20 animals are shown.
(B) Expression of elt-6::Cherry.
Error bars represent the SEM pixel intensities.
Figure 5. Effect of elt-5(RNAi) and elt-6(RNAi) on elt-3 Expression and Longevity.
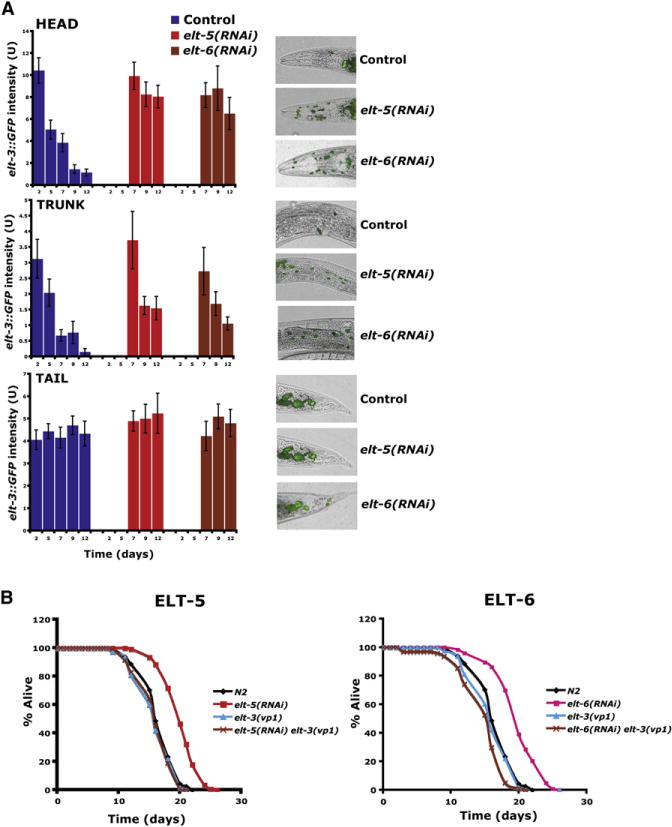
(A) elt-3::GFP expression is increased in elt-5(RNAi) or elt-6(RNAi) animals. RNAi was induced starting at day 5 of adulthood by feeding worms bacteria expressing dsRNA. elt-3::GFP expression was measured starting at day 7. The y axis denotes GFP expression (arbitrary units). Average expression and SE from 20 animals are shown.
(B) elt-5(RNAi) or elt-6(RNAi) extends life span compared to N2, and this longevity effect is suppressed by elt-3(vp1) (p < 0.0001).
Error bars represent the SEM pixel intensities.
Third, we determined the effects of elt-5(RNAi) and elt-6(RNAi) on life span. We used RNAi to reduce elt-5 or elt-6 activity starting at day 5 of adulthood (earlier RNAi treatment causes worms to become sick) and then measured their life span. We performed the experiment multiple times and found that elt-5 (RNAi) and elt-6(RNAi) extended life span compared to wild-type controls each time (Figure 5B).
Fourth, we showed that the effects of elt-5(RNAi) and elt-6 (RNAi) on life span are elt-3 dependent. We used RNAi to reduce elt-5 or elt-6 activity in elt-3 null mutants and found that elt-5(RNAi) and elt-6(RNAi) fail to extend life span of elt-3 null mutants compared to wild-type controls (Figure 5B).
Fifth, we determined whether changes in life span caused by an elt-3 null mutation or elt-5(RNAi) were associated with changes in the relative resistance of worms to heat or oxidative stress. To test for sensitivity to heat shock, we determined how long worms could survive after they were moved from 20°C to 35°C. To test for sensitivity to oxidative stress, we determined the length of survival of worms after they were exposed to 100 mM paraquat, which is a powerful oxidant. We found that elt-3(vp1) mutants were more sensitive to heat shock and oxidative stress than wild-type controls (Figure 6A). Conversely, elt-5(RNAi) animals are slightly resistant to heat shock and paraquat treatment (Figure 6B).
Figure 6. Contrasting Functions of elt-3 and elt-5 in Thermotolerance and Resistance to Oxidative Stress.
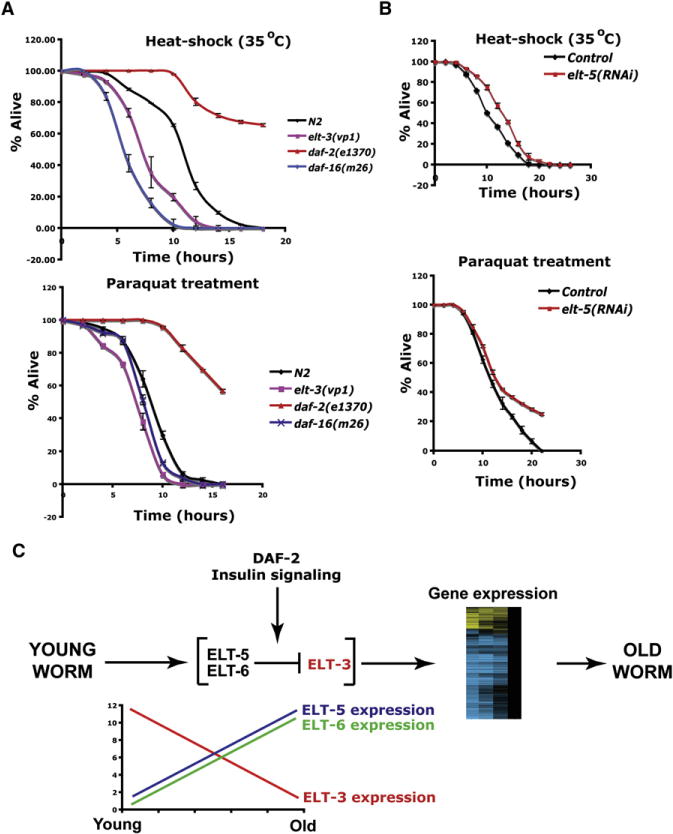
(A) elt-3(vp1) animals are more sensitive than wild-type to heat shock or oxidative stress. Shown are survival comparisons of elt-3(vp1), wild-type, daf-2(e1370) (a control known to show resistance), and daf-16(m26) (a control known to be sensitive to stress) worms under acute thermal stress at 35°C and under oxidative stress (paraquat, 100 mM).
(B) elt-5(RNAi) animals are more resistant to heat shock and paraquat treatment than wild-type animals.
(C) Model for transcriptional changes during aging. Expression of elt-5 GATA and elt-6 GATA increases as worms age, leading to increased repression of elt-3 in old worms. Changes in the elt-3 GATA transcription factor activate a cascade of downstream changes in expression of 1254 age-regulated genes in old age. Expression of elt-3 is also controlled by the insulin-like signaling pathway.
DISCUSSION
An elt-3/elt-5/elt-6 GATA Transcriptional Circuit for Aging
We used DNA microarray experiments to profile expression changes associated with old age and found that elt-3 GATA plays a key role in directing changes in gene expression during aging (Figure 6C). To our knowledge, this is the first transcriptional circuit accounting for global changes in expression during aging in any organism. Expression of the elt-3 GATA transcription factor gene shows a tissue-specific decline in expression with age. elt-3 expression decreases rapidly in the intestinal and trunk hypodermal cells, more gradually in the hypodermal and pharyngeo-intestinal cells in the head, and is not age regulated in the intestinal-rectal valve cells in the tail. Age regulation of elt-3 results in changes in expression of a large battery of downstream genes, including ugt-9, col-144, and sod-3. In addition to these three genes, elt-3 may also regulate as many as 602 other age-regulated genes that have GATA motifs in their upstream regions.
A key question is what causes the downward trend in elt-3 expression with age, as this provides insight into molecular mechanisms that specify the rate of normal aging. During embryonic development, elt-3 expression is activated by the GATA transcription factor gene elt-1 and repressed by the GATA transcription factor genes elt-5 and elt-6 (Gilleard and McGhee, 2001; Koh and Rothman, 2001). Our results show that elt-5 and elt-6 are responsible for changes in elt-3 expression during aging. Expression of elt-5 and elt-6 increases in old age, leading to increased repression of elt-3. In elt-5(RNAi) or elt-6(RNAi) mutants, elt-3 expression remains high throughout life and life span is increased. These results indicate that elt-3 is functionally important for aging and that low levels of elt-3 expression in old age are likely to be detrimental for life span.
Many genes can affect aging in C. elegans, such as genes in the insulin-like signaling pathway (daf-2 insulin-like receptor, age-1 PI3 kinase, or daf-16 FOXO transcription factor), sir-2.1 protein deacetylase, or clk-1 demethoxyubiquinone (DMQ) hydroxylase (Ewbank et al., 1997; Friedman and Johnson, 1988; Guarente and Kenyon, 2000; Kenyon et al., 1993; Lakowski and Hekimi, 1996; Morris et al., 1996; Tissenbaum and Guarente, 2001; Tissenbaum and Ruvkun, 1998). Although these genes can be genetically altered to change life span, it is not clear whether their activity changes during normal aging, and, thus, it is unclear whether they play a role in normal aging. For instance, it is not known whether the activity of the insulin-like signaling pathway changes during aging; the FOXO transcription factor DAF-16, which regulates most or all of the outputs from this pathway, is not observed in the nucleus during normal aging (Lin et al., 2001). The SIR-2.1 protein deacetylase gene extends life span by inducing ER stress, but it is unclear whether ER stress is part of normal aging (Viswanathan et al., 2005). Mutations in genes in the mitochondrial electron transport chain, such as clk-1, may extend life span either by inducing stress or by reducing generation of oxygen radicals to an extent that does not occur in wild-type worms (Anson and Hansford, 2004; Rodriguez-Aguilera et al., 2005). These and other longevity mutants may perturb C. elegans signaling, transcription, or metabolism in a manner that does not reflect normal aging. A key difference between the elt-3/elt-5/elt-6 transcriptional network and these other aging genes is that the elt-3/elt-5/elt-6 network changes during the normal aging process.
Molecular Markers for Aging
The age-regulated GFP reporters can be used as molecular markers for aging. sod-3::GFP was previously known to decrease expression with age (Essers et al., 2005), and our results show that elt-3::GFP, ugt-9::GFP, and col-144::GFP are similarly age regulated. We found that the kinetics of expression of these GFP reporters scales with life span for worms grown at different temperatures and in age-1 mutants. Expression of the four aging GFP markers declines slowly at 15°C and rapidly at 25°C (A. Sanchez-Blanco, personal communication and unpublished data). Furthermore, expression of the aging GFP markers is increased in long-lived age-1 mutants. These results show that expression of these aging markers indicates age of the worm and not chronological time, such that high GFP expression identifies young worms and low GFP expression indicates old worms. In addition to these aging GFP markers, previous work has shown that accumulation of the aging pigment lipofuscin, disorganized appearance of myo-3::GFP, or presence of YP170::GFP in the body cavity can be used as molecular markers for aging (Herndon et al., 2002). Compared to a population-based life span assay, GFP markers are an attractive metric because they are easy to score and can be calculated for individual worms.
Regulation by the Insulin-like Signaling Pathway
In addition to regulation by aging, we found that elt-3 is also regulated by the insulin-like signaling pathway. In age-1 mutants, elt-3 expression increases slightly, which in turn alters expression of downstream GATA targets that comprise a large fraction of the aging transcriptional profile. Regulation of elt-3 by the insulin-like signaling pathway accounts for the overlap in transcriptional profiles from the aging and daf-2/age-1/daf-16 DNA microarray experiments (Figure 6C).
Although the insulin-like signaling pathway can regulate elt-3 expression, this regulation is not responsible for age-related changes in elt-3 expression. If the insulin-like signaling pathway was itself age regulated (e.g., low AGE-1 activity in young animals and high activity in old animals), one would expect that age-1 mutations would not affect elt-3 expression in young animals and would increase elt-3 expression substantially in old animals. Further, if age-related changes in the insulin-like signaling pathway were mainly responsible for age-related changes in elt-3 gene expression, one would expect that elt-3 expression would show little age regulation in age-1 mutants. This is not the case, as age-1 mutants show a similar downward trend in expression of elt-3 as wild-type, except that expression is increased proportionally at each age.
Drift of the elt-3 Transcriptional Hierarchy during Aging
A widely held view is that aging is caused by accumulation of damage (Golden et al., 2002; Harman, 1992; Martin and Grotewiel, 2006; Rattan, 2006; Sohal and Weindruch, 1996), and, thus, one might expect that age-related changes in the elt-3 transcriptional network would be caused by a lifelong accumulation of damage or stress. In mammals, there is abundant evidence that aging is the result of damage accumulation, such as oxidative damage, somatic DNA mutation, telomere shortening, protein glycation, and inflammation (Aviv, 2004; Ayub and Hallett, 2004; Blasco, 2007; Kadenbach et al., 1995; Lu et al., 2004; McGeer and McGeer, 2004; Ulrich and Cerami, 2001). However, worms age very rapidly compared to mammals, and it is unclear whether the rate of damage accumulation is high enough to account for the short worm life span. We found no evidence that age regulation of the elt-3 transcriptional network is caused by accumulation of damage, stress, or inflammation.
Besides damage accumulation, another possibility is that aging might result from developmental pathways that go awry late in life (antagonistic pleiotropy) (Kirkwood and Rose, 1991). In worms, we found that decreased expression of elt-3 GATA in old age is caused by increased expression of elt-5 GATA or elt-6 GATA, which act as repressors. The activities of elt-5 or elt-6 are not known to be affected by cellular damage or environmental stressors, and, thus, drift in the GATA transcriptional hierarchy might be due to intrinsic processes. However, we cannot completely rule out that age-related changes in the elt-3/elt-5/elt-6 GATA transcriptional circuit are caused by extrinsic factors, and further work on the nature of age-related changes will help resolve this issue.
How could the elt-3/elt-5/elt-6 transcriptional network for aging evolve? It seems unlikely that any changes in old age could provide a selective advantage and be under natural selection. In the wild, worms usually die of predation rather than old age, and traits that are only evident in old worms would have little effect on fitness. Rather than evolving under the force of natural selection, another possibility is that age-related changes in the elt-3/elt-5/elt-6 transcriptional network have a neutral effect on fitness in the wild and have become fixed in the C. elegans genome (the mutation accumulation theory) (Kirkwood, 1989). Regulation of elt-3 by elt-5 and elt-6 would be under evolutionary selection because of its important early role during development. In old worms, there is little or no advantage to maintaining proper elt-3 expression, and decreased expression of elt-3 might occur as a secondary consequence. This could shorten the life of old worms but would have a neutral effect on population fitness, as old worms are extremely rare in the wild. Thus, the elt-3/elt-5/elt-6 hierarchy is a developmental program that may change during aging simply because proper homeostatic maintenance in late life is not under the force of natural selection.
Gene expression profiles for aging have been defined in many other animals, including flies, mice, and humans. It will be interesting to determine whether transcriptional changes during aging in other animals are also caused by imbalances in developmental regulatory hierarchies.
EXPERIMENTAL PROCEDURES
Strains
All C. elegans strains (Table S2) were maintained and handled as described previously (Brenner, 1974).
DNA Microarray Experiments
All microarray experiments were performed as previously described (Jiang et al., 2001; Lund et al., 2002) and also can be found at http://cmgm.stanford.edu/%7Ekimlab/elt3/index.html. Briefly, we used temperature-sensitive fer-15(b26) worms for the aging time course experiments. These worms are sterile at 25°C but show normal rates of aging (Fabian and Johnson, 1994). RNA was isolated from age-synchronous cultures of hermaphrodite worms grown at 25°C at 2, 5, 8, or 11 days of adulthood. We observed no deaths in the worm population at days 2 and 5 of adulthood, 30% death at day 8, and 93% death at day 11. The aging time course was repeated four times. To identify changes in gene expression in age-1 mutants, we compared expression in fer-15(b26) animals to expression in fer-15(b26); age-1(hx542) mutants. We prepared RNA from young adult fer-15(b26) mutants grown at 25°C (five samples prepared at Stanford University and seven samples prepared at the University of Colorado) and young adult fer-15(b26); age-1(hx542) mutants (four samples at Stanford University and six samples at the University of Colorado). To identify changes in gene expression in daf-16 mutants, we compared expression in fer-15 young adults to expression in fer-15(b26); daf-16(m26) young adults grown at 25°C. At Stanford University, five samples of fer-15 and five samples of fer-15; daf-16 animals were prepared. At the University of Colorado, seven samples of fer-15 and six samples of fer-15; daf-16 were prepared.
cy5-labeled cDNA samples from RNA at each time point were compared to a standard cy3-labeled reference cDNA (prepared from mixed stage hermaphrodite mRNA). We calculated log2(cy5/cy3 expression) at each repeat and then calculated the average log2 expression ratio. The expression data from the aging time course, age-1(hx542) mutants, and daf-16(m26) mutants are shown in Tables S3, S5, and S6, respectively.
Analysis of Life Span
Life span analyses were conducted at 20°C as previously described (Apfeld and Kenyon, 1999; Kenyon et al., 1993). At least 200 worms were used for each experiment. Age refers to days following adulthood, and p values were calculated using the log-rank (Mantel-Cox) method (Lawless, 1982).
RNAi Experiments
HT115 bacteria transformed with RNAi vectors expressing dsRNA of the genes of interest were grown at 37°C in LB with 100 μg/ml ampicillin and 10 μg/ml tetracycline and then seeded onto NG-ampicillin plates supplemented with 100 μl of 0.1 M IPTG. Worms at the L4 larvae stage were added to the plates and transferred to new plates every 4 days.
Construction of GFP and Cherry Reporters
The promoter::GFP constructs for ugt-9, col-144, and sod-3 were obtained from D. Dupuy (Dupuy et al., 2004). elt-3pro::GFP::H2B and sod-3pro::GFP::H2B were constructed using the Gateway recombinatorial cloning system (Cheo et al., 2004). Transgenic strains expressing GFP from the promoter of each gene were made by microinjecting pha-1(ts) animals with promoter::GFP (50 ng/mg) and pha-1(+) (pC1, 100 ng/ml, a gift from A. Fire), generating an extrachromasomal array. The promoter::wCherry constructs presented in Table S7 were constructed using the Gateway recombinatorial cloning system. Transgenic lines were made by microparticle bombardment (Praitis et al., 2001) of unc-119(ed3) animals with promoter::wCherry: unc-199(+) (10 μg), and the transformants were screened for stable integration. The resulting strains for each gene are listed in Table S2.
Site-Directed Mutagenesis
Changes in the GATA transcription binding site in the upstream promoter regions of ugt-9, col-144, and sod-3 were introduced by PCR-based, site-directed mutagenesis (Quickchange mutagenesis kit, Stratagene) using primers described in Table S8. A KpnI restriction site was used as a diagnostic for mutagenesis. All mutants were verified by DNA sequencing. The mutated promoter::GFP constructs were used to generate transgenic strains as described above.
Imaging and Quantification of GFP and Cherry Expression
To examine changes in expression of GFP and Cherry reporters with respect to age, we picked 20 worms at five different ages (3, 5, 7, 9, and 12 days of adulthood) and then measured the level of GFP and Cherry expression using quantitative fluorescence microscopy. Specifically, we used pixel intensity to quantify the level of GFP expression for both wild-type and mutant constructs for each gene. Twenty hermaphrodites from each strain were analyzed for GFP and Cherry expression using a Zeiss Axioplan microscope. Comparison of all images was carried out on the same day with the same microscope settings. Images were analyzed using ImageJ, a public domain Java image-processing program (Rasband, 2004).
Oxidative Stress Assay
Transgenic lines expressing ugt-9pro::GFP, col-144pro::GFP, sod-3pro::GFP::H2B, and elt-3pro::GFP::H2B were synchronized by hypochloride treatment and hatching on unseeded NGM agar plates and then grown to the L4 stage. Fifty L4 animals from each transgenic line were picked onto NGM plates (Essers et al., 2005) containing 0.25 mM paraquat (methyl viologen, Sigma). GFP expression was measured 48 hr later. Oxidative stress resistance assays were performed in 24-well plates as previously described (Fisher and Lithgow, 2006). Briefly, 4-day-old adult hermaphrodites were immersed in S-basal media containing 100 mM of paraquat. Worms were scored every hour until all worms were scored as dead by touch-provoked movement. Three independent trials were pooled for analysis.
Heat Shock Treatment
We tested whether heat shock would affect expression of ugt-9pro::GFP, col-144pro::GFP, sod-3pro::GFP::H2B, and elt-3pro::GFP::H2B. Fifty animals at day 2 of adulthood were picked onto fresh NGM agar plates and incubated for 30 min at 33°C. Worms were then allowed to recover for an hour at 20°C before imaging. Levels of GFP expression were measured as described above. Heat shock survival assay was performed by placing worms at 35°C and recording their rate of death as previously described (Lithgow et al., 1995). Three independent trials were pooled for analysis.
γ-Irradiation Treatment
To examine the effect of DNA damage on expression of ugt-9pro::GFP, col-144pro::GFP, sod-3pro::GFP::H2B, and elt-3pro::GFP::H2B, 50 L4 animals were picked onto fresh NGM plates and γ irradiated with a 137Cs source (Cesium Irradiator) at 3000, 3500, and 4000 Ray doses. Levels of GFP expression were measured in all three groups of irradiated animals 48 hr later as described above.
Supplementary Material
Acknowledgments
We thank Andrew Fire, Denis Dupuy, and Mark Vidal for providing plasmids and nematode strains; James McGhee and John Gilleard for elt-3 strains; and all members of Kim lab for discussions and comments on the manuscript. This work was supported by NIH grants number R01AG025941 (S.K.K.) and RO1AG16219 (T.E.J.), The Ellison Medical Foundation (S.K.K. and T.E.J.), and The Larry L. Hillblom Foundation.
Footnotes
ACCESSION NUMBERS
The microarray data can be found in the Gene Expression Omnibus (GEO) of NCBI through accession numbers GSE12094.
SUPPLEMENTAL DATA
Supplemental Data for this article include figures and tables and can be found with this article online at http://www.cell.com/cgi/content/full/134/2/291/DC1/.
References
- Anson RM, Hansford RG. Mitochondrial influence on aging rate in Caenorhabditis elegans. Aging Cell. 2004;3:29–34. doi: 10.1111/j.1474-9728.2003.00077.x. [DOI] [PubMed] [Google Scholar]
- Apfeld J, Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402:804–809. doi: 10.1038/45544. [DOI] [PubMed] [Google Scholar]
- Aviv A. Telomeres and human aging: facts and fibs. Sci Aging Knowledge Environ. 2004;2004:pe43. doi: 10.1126/sageke.2004.51.pe43. [DOI] [PubMed] [Google Scholar]
- Ayub K, Hallett MB. Signalling shutdown strategies in aging immune cells. Aging Cell. 2004;3:145–149. doi: 10.1111/j.1474-9728.2004.00100.x. [DOI] [PubMed] [Google Scholar]
- Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, Rando TA. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Cheo DL, Titus SA, Byrd DR, Hartley JL, Temple GF, Brasch MA. Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination: functional analysis of multi-segment expression clones. Genome Res. 2004;14:2111–2120. doi: 10.1101/gr.2512204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy D, Li QR, Deplancke B, Boxem M, Hao T, Lamesch P, Sequerra R, Bosak S, Doucette-Stamm L, Hope IA, et al. A first version of the Caenorhabditis elegans Promoterome. Genome Res. 2004;14:2169–2175. doi: 10.1101/gr.2497604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers MA, de Vries-Smits LM, Barker N, Polderman PE, Burgering BM, Korswagen HC. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science. 2005;308:1181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- Ewbank JJ, Barnes TM, Lakowski B, Lussier M, Bussey H, Hekimi S. Structural and functional conservation of the Caenorhabditis elegans timing gene clk-1. Science. 1997;275:980–983. doi: 10.1126/science.275.5302.980. [DOI] [PubMed] [Google Scholar]
- Fabian TJ, Johnson TE. Production of age-synchronous mass cultures of Caenorhabditis elegans. J Gerontol. 1994;49:B145–B156. doi: 10.1093/geronj/49.4.b145. [DOI] [PubMed] [Google Scholar]
- Fisher AL, Lithgow GJ. The nuclear hormone receptor DAF-12 has opposing effects on Caenorhabditis elegans lifespan and regulates genes repressed in multiple long-lived worms. Aging Cell. 2006;5:127–138. doi: 10.1111/j.1474-9726.2006.00203.x. [DOI] [PubMed] [Google Scholar]
- Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, Ruvkun G, Ausubel FM. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- Gerstbrein B, Stamatas G, Kollias N, Driscoll M. In vivo spectrofluorimetry reveals endogenous biomarkers that report healthspan and dietary restriction in Caenorhabditis elegans. Aging Cell. 2005;4:127–137. doi: 10.1111/j.1474-9726.2005.00153.x. [DOI] [PubMed] [Google Scholar]
- Gilleard JS, McGhee JD. Activation of hypodermal differentiation in the Caenorhabditis elegans embryo by GATA transcription factors ELT-1 and ELT-3. Mol Cell Biol. 2001;21:2533–2544. doi: 10.1128/MCB.21.7.2533-2544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard JS, Shafi Y, Barry JD, McGhee JD. ELT-3: A Caenorhabditis elegans GATA factor expressed in the embryonic epidermis during morphogenesis. Dev Biol. 1999;208:265–280. doi: 10.1006/dbio.1999.9202. [DOI] [PubMed] [Google Scholar]
- Golden TR, Hinerfeld DA, Melov S. Oxidative stress and aging: beyond correlation. Aging Cell. 2002;1:117–123. doi: 10.1046/j.1474-9728.2002.00015.x. [DOI] [PubMed] [Google Scholar]
- Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- Harman D. Free radical theory of aging. Mutat Res. 1992;275:257–266. doi: 10.1016/0921-8734(92)90030-s. [DOI] [PubMed] [Google Scholar]
- Hekimi S, Lakowski B, Barnes TM, Ewbank JJ. Molecular genetics of life span in C. elegans: how much does it teach us? Trends Genet. 1998;14:14–20. doi: 10.1016/S0168-9525(97)01299-7. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Bonafe M, Johnson TE. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J Gerontol A Biol Sci Med Sci. 2006;61:444–460. doi: 10.1093/gerona/61.5.444. [DOI] [PubMed] [Google Scholar]
- Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, Sakano Y, Paupard MC, Hall DH, Driscoll M. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- Holowacz T, Zeng L, Lassar AB. Asymmetric localization of numb in the chick somite and the influence of myogenic signals. Dev Dyn. 2006;235:633–645. doi: 10.1002/dvdy.20672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Goto S, Hartman PS. Protein oxidation during aging of the nematode Caenorhabditis elegans. Free Radic Biol Med. 2002;33:1021–1025. doi: 10.1016/s0891-5849(02)00857-2. [DOI] [PubMed] [Google Scholar]
- Jiang M, Ryu J, Kiraly M, Duke K, Reinke V, Kim SK. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2001;98:218–223. doi: 10.1073/pnas.011520898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadenbach B, Munscher C, Frank V, Muller-Hocker J, Napiwotzki J. Human aging is associated with stochastic somatic mutations of mitochondrial DNA. Mutat Res. 1995;338:161–172. doi: 10.1016/0921-8734(95)00021-w. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kirkwood TB. DNA, mutations and aging. Mutat Res. 1989;219:1–7. doi: 10.1016/0921-8734(89)90035-0. [DOI] [PubMed] [Google Scholar]
- Kirkwood TB, Rose MR. Evolution of senescence: late survival sacrificed for reproduction. Philosophical Transactions: Biological Sciences. 1991;332:15–24. doi: 10.1098/rstb.1991.0028. [DOI] [PubMed] [Google Scholar]
- Koh K, Rothman JH. ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development. 2001;128:2867–2880. doi: 10.1242/dev.128.15.2867. [DOI] [PubMed] [Google Scholar]
- Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci USA. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawless J. Models and Methods for Lifetime Data. New York: Wiley; 1982. [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci USA. 1995;92:7540–7544. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Fergusson MM, Castilho RM, Liu J, Cao L, Chen J, Malide D, Rovira II, Schimel D, Kuo CJ, et al. Augmented Wnt signaling in a mammalian model of accelerated aging. Science. 2007;317:803–806. doi: 10.1126/science.1143578. [DOI] [PubMed] [Google Scholar]
- Liu Y. Genome-wide Discovery of DNA Regulatory Motifs in C elegans. Stanford: Computer Science, Stanford University; 2005. [Google Scholar]
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- Lund J, Tedesco P, Duke K, Wang J, Kim SK, Johnson TE. Transcriptional profile of aging in C. elegans. Curr Biol. 2002;12:1566–1573. doi: 10.1016/s0960-9822(02)01146-6. [DOI] [PubMed] [Google Scholar]
- Martin I, Grotewiel MS. Oxidative damage and age-related functional declines. Mech Ageing Dev. 2006;127:411–423. doi: 10.1016/j.mad.2006.01.008. [DOI] [PubMed] [Google Scholar]
- McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- McGeer PL, McGeer EG. Inflammation and the degenerative diseases of aging. Ann N Y Acad Sci. 2004;1035:104–116. doi: 10.1196/annals.1332.007. [DOI] [PubMed] [Google Scholar]
- Melov S. Mitochondrial oxidative stress. Physiologic consequences and potential for a role in aging. Ann N Y Acad Sci. 2000;908:219–225. doi: 10.1111/j.1749-6632.2000.tb06649.x. [DOI] [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Pauli F, Liu Y, Kim YA, Chen PJ, Kim SK. Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans. Development. 2006;133:287–295. doi: 10.1242/dev.02185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband W. ImageJ. Bethesda, MD: National Institute of Health; 2004. [Google Scholar]
- Rattan SI. Theories of biological aging: genes, proteins, and free radicals. Free Radic Res. 2006;40:1230–1238. doi: 10.1080/10715760600911303. [DOI] [PubMed] [Google Scholar]
- Reinke V, Smith HE, Nance J, Wang J, Van Doren C, Begley R, Jones SJ, Davis EB, Scherer S, Ward S, et al. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Aguilera JC, Gavilan A, Asencio C, Navas P. The role of ubiquinone in Caenorhabditis elegans longevity. Ageing Res Rev. 2005;4:41–53. doi: 10.1016/j.arr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Roy PJ, Stuart JM, Lund J, Kim SK. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature. 2002;418:975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- Shapira M, Hamlin BJ, Rong J, Chen K, Ronen M, Tan MW. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc Natl Acad Sci USA. 2006;103:14086–14091. doi: 10.1073/pnas.0603424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA, Ruvkun G. An insulin-like signaling pathway affects both longevity and reproduction in Caenorhabditis elegans. Genetics. 1998;148:703–717. doi: 10.1093/genetics/148.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Ulrich P, Cerami A. Protein glycation, diabetes, and aging. Recent Prog Horm Res. 2001;56:1–21. doi: 10.1210/rp.56.1.1. [DOI] [PubMed] [Google Scholar]
- Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell. 2005;9:605–615. doi: 10.1016/j.devcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Wang J, Kim SK. Global analysis of dauer gene expression in Caenorhabditis elegans. Development. 2003;130:1621–1634. doi: 10.1242/dev.00363. [DOI] [PubMed] [Google Scholar]
- Williams GC. Pleiotropy, natural selection, and the evolution of senescence. Evolution Int J Org Evolution. 1957;11:398–411. [Google Scholar]
- Wong A, Boutis P, Hekimi S. Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics. 1995;139:1247–1259. doi: 10.1093/genetics/139.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


