Abstract
The majority of biological processes are mediated via protein–protein interactions. Determination of residues participating in such interactions improves our understanding of molecular mechanisms and facilitates the development of therapeutics. Experimental approaches to identifying interacting residues, such as mutagenesis, are costly and time-consuming and thus, computational methods for this purpose could streamline conventional pipelines. Here we review the field of computational protein interface prediction. We make a distinction between methods which address proteins in general and those targeted at antibodies, owing to the radically different binding mechanism of antibodies. We organize the multitude of currently available methods hierarchically based on required input and prediction principles to provide an overview of the field.
Keywords: protein–protein interaction, protein interface prediction, antibody antigen interaction
Protein interfaces
Proteins interact with other proteins, DNA, RNA and small molecules to perform their cellular tasks. Knowledge of protein interfaces and the residues involved is vital to fully understand molecular mechanisms and to identify potential drug targets [1]. The most reliable methods to determine protein complexes and therefore protein interfaces are X-ray crystallography and mutagenesis. Unfortunately these techniques are expensive in time and resources. Therefore, over the past 25 years, there has been a rapid development of computational methods aiming to elucidate protein complexes, such as protein interaction prediction, protein–protein docking and protein interface prediction. These three types of methods all aim at slightly different problems, protein interaction prediction attempts to give a binary answer as to whether two proteins interact, docking aims to recreate the pairwise residue contacts between the two binding partners. The subject of this review is the middle ground between these two problems, protein interface prediction, where one wishes to identify a subset of residues on a protein, which might interact with the presumed binding partner.
Residues involved in these interfaces are normally defined by an intermolecular distance threshold (usually between 4.5 and 8Å [2] with the most common value being 5Å [3]) or a reduction of accessible surface area in a complex compared with the monomer [4] (Supplementary Figure S1 displays an example). Experiments have shown that the choice of interface definition has only a minor impact on a predictors’ performance [5]; the threshold values however are critical for selecting specific features of interfaces [6].
An interface residue predictor receives as input a protein or a pair of proteins. It then predicts a subset of residues on the proteins surface that are involved in intermolecular interactions. When comparing the true interacting residues with the prediction, it is standard to calculate the number of true positives (TP), false positives (FP), true negatives (TN) and false negatives (FN) (Supplementary Figure S2). These four values give rise to a variety of performance metrics (Table 1), which can be used to assess the quality of the predictor.
Table 1.
Commonly used metrics to assess the quality of interface residue predictions
| Metric | Formula |
|---|---|
| Specificity | |
| Sensitivity (also known as recall) | |
| Precision | |
| F1 (harmonic mean of precision and recall) | |
| Accuracy | |
| Matthews correlation coefficient (MCC) |
A single interface prediction consists of a set of residues believed to constitute the binding site and those that do not. Out of those believed to be the binding site, if they are truly binding residues they are called TP, otherwise they are FP. Out of the residues identified as non-binding, if they do not constitute the interface, they are called TN and FN otherwise (see Figure S2). These four numbers are used to calculate a range of performance metrics presented in this table.
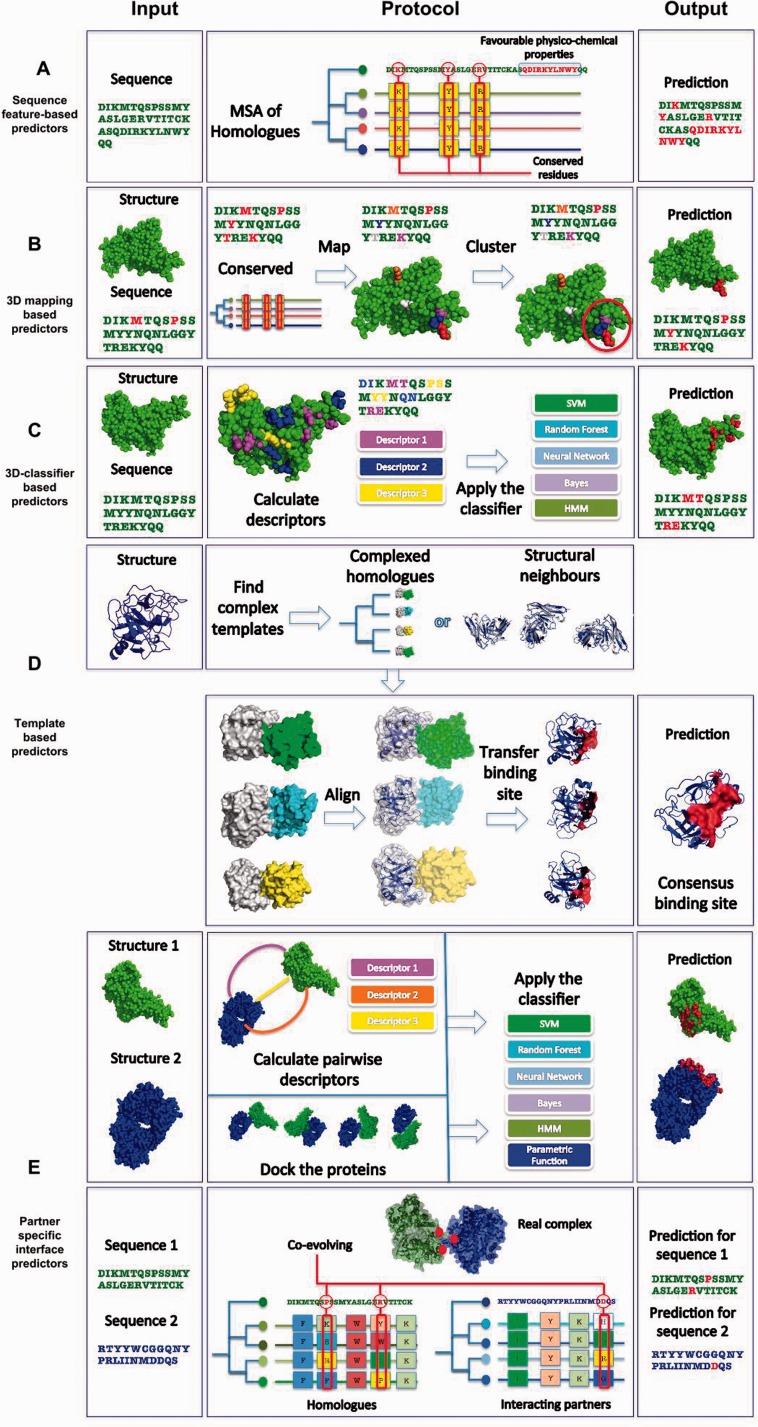
The field of protein–protein interface prediction has diversified into many different approaches (Figure 1) [7]. Methods might use intrinsic features of the sequence or the structure, evolutionary relationships or use an existing complex as a reference template. Predictors make use of many distinct quality measures, different training and testing data sets, thus a fair comparison between them is hard [5]. In this review we attempt to provide a classification for the majority of existing methods in order to get a clear overview of the field. Based on this, we offer suggestions as to how the field could progress, focusing on improved predictions and unified evaluation metrics.
Figure 1.
Classification of existing protein interface prediction methods. In the leftmost column we present the input required by a method. In the middle column, a simplified pipeline for the protocol is presented. In the rightmost, prediction column, the resulting binding site is shown in red. Most methods output a ranked list of possible binding sites. Here for simplicity, we show a single result for each method. (A) Sequence-feature-based predictors: These methods receive a protein sequence. Sequential features of the input are compared with features thought to contribute to a residue being part of an interface, such as conservation scores and physico-chemical properties. (B) 3D mapping-based predictors: These methods receive a protein structure and its sequence as input. Evolutionary conservation is coupled with 3D surface and sequence information. Conserved residues can be grouped according to their surface proximity to form contiguous interface patches. (C) 3D-classifier-based predictors: The input for these methods is a protein structure and its sequence. Distinct sets of attributes (physico-chemical, evolution, 3D structural features, etc.) are used as an input to a learning method such as a SVM or Random Forest. (D) Template-based predictors: These methods receive a protein structure (and thus its sequence) as input. Complex templates are then identified, which can be homologues or structural neighbours (these are shown in white, whereas their binding partners are in green, cyan and yellow). Templates of the input protein are aligned to the query protein. The most commonly aligned contact sites are returned as a prediction. (E) Partner-specific interface predictors: These methods receive the structures/sequences of two proteins that are assumed to interact. The three groups of methods are shown for this category. Partner-specific descriptors can be calculated to predict interfaces. In some cases docking is used to sample possible orientations to identify a consensus binding site. Partner-specific descriptors and docking poses are used as input for parametric functions and classifiers to obtain the final result. In the co-evolution-based strategy, a MSA of interacting homologues is created and sites that appear to mutate in concert (co-evolve) are assumed to constitute the binding site. A colour version of this figure is available at BIB online: http://bib.oxfordjournals.org.
Protein interface predictors
Computational methods for identifying interface residues can be broadly divided into two non-exclusive categories based on their use of protein information: (1) intrinsic-based approaches based on specific features of protein sequences and/or structures and (2) template-based approaches that exploit the conservation found between structurally similar proteins. A simplified overview of all methods is given in Figure 1, and detailed descriptions are provided in the subsequent sections along with a summary in Table 2.
Table 2.
Protein interface predictors and their performance
| Input |
Main knowledge source(properties) |
Intrinsic-based |
Template-based |
Output |
Performance |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | Predictor | Sequence | Structure | Both | Sequence | Structure | Both | Additional | Evolution Info. | Intrinsic features | Both | Homologous Structure | Structural Neighbour | Residue-based | Patch-based | Data set* | Recall % | Precision % | Specificity % | Accuracy % | MCC | F1 % | AUC | Numbers taken from* |
| A | [60] | x | x | x | x | [10] | 45.55 | 86.98 | 97.41 | 83.12 | 0.55 | 59.79 | – | |||||||||||
| [181] | x | x | x | x | 57.9 | – | 65 | 62.5 | 0.22 | 52 | – | |||||||||||||
| [35] | x | x | x | x | [45] | 83 | – | 78 | – | 0.76 | – | – | ||||||||||||
| [23] | x | x | + | x | x | 47 | 22.2 | 69 | 66.4 | 0.13 | 25.6 | |||||||||||||
| [10] | x | x | x | x | 42.84 | 81.96 | – | – | – | 56.25 | – | |||||||||||||
| [12] | x | x | x | x | 70 | 37.7 | – | – | – | 49 | – | [10] | ||||||||||||
| [22] | x | x | + | x | x | [23] | 36.6 | 18.9 | 76.1 | 71.9 | 0.09 | 23.2 | – | [23] | ||||||||||
| [15] | x | x | x | x | [64] | 69 | – | 65 | – | 0.28 | 67 | – | [66] | |||||||||||
| [16] | x | x | x | x | 58.8 | 26.3 | – | – | – | 36.3 | [10] | |||||||||||||
| [4] | x | x | x | x | [182] | 39 | – | 58 | 72 | – | – | – | ||||||||||||
| [9] | x | x | x | x | 50 | 62 | – | – | – | 10 | – | [10] | ||||||||||||
| B | [30] | x | x | x | x | [13] | 39.8 | – | 86.9 | 72.6 | – | – | – | |||||||||||
| [13] [183] | x | x | x | x | [13] | 34.2 | – | 85.1 | 68.5 | – | – | – | [30] | |||||||||||
| C | [68] | x | x | x | x | [71] | 63.6 | – | 84.3 | – | 0.37 | – | – | |||||||||||
| [65] | x | x | x | x | [64] | 72.7 | – | 61 | 75.2 | 0.47 | 66.3 | 0.82 | ||||||||||||
| [71] | x | x | x | x | [184] | – | – | – | – | 0.17 | – | 0.69 | ||||||||||||
| [54] | x | x | x | x | 99.08 | 99.91 | – | 80.32 | 1.29 | 99.48 | – | |||||||||||||
| [57] | x | x | x | x | [45] | 45.8 | 69.6 | – | 79.8 | – | – | – | ||||||||||||
| [58] | x | x | x | x | 78.99 | 65.3 | 54.66 | 67.29 | 0.34 | – | – | |||||||||||||
| [66] | x | x | x | x | x | [64] | 68 | – | 73 | 71 | 0.43 | 71 | – | |||||||||||
| [55] | x | x | x | x | [50] | 74.7 | 63.4 | – | – | 0.58 | – | 0.9 | ||||||||||||
| [39] | x | x | x | x | [185] | – | – | – | 70 | – | – | – | ||||||||||||
| [49] | x | x | x | x | [64] | 77 | – | 63 | – | 0.35 | 69 | – | [66] | |||||||||||
| [26] | x | x | x | x | [58] | 78.27 | 63.44 | 51.28 | 65.3 | 0.30 | – | – | [58] | |||||||||||
| [64] | x | x | x | x | 59 | – | 54 | 69 | 0.33 | 56 | – | [66] | ||||||||||||
| [48] | x | x | x | x | 60.7 | – | 41.9 | – | 0.20 | – | – | |||||||||||||
| [63] | x | x | x | x | [45] | – | – | – | – | – | – | – | ||||||||||||
| [38] | x | x | x | x | CAPRI | 41.7 | 40.3 | – | – | – | – | – | ||||||||||||
| [47] | x | x | x | x | [186] | 46.2 | 42.2 | – | 83.2 | 0.30 | 44.1 | – | ||||||||||||
| [67] | x | x | x | x | 37.7 | 57.8 | – | 75.1 | 0.31 | 45.7 | – | |||||||||||||
| [41] | x | x | x | x | CAPRI | 30.1 | 30.4 | – | 76.9 | 0.16 | 30.2 | 0.60 | [101] | |||||||||||
| [70] | x | x | x | x | [64] | 36 | – | 93 | – | 0.33 | 52 | – | [66] | |||||||||||
| [50] | x | x | x | x | 60.3 | 63.7 | – | 74.2 | 0.42 | – | – | |||||||||||||
| [62] | x | x | x | x | – | – | – | – | – | – | – | – | ||||||||||||
| [45] | x | x | x | x | – | – | – | – | – | – | – | – | ||||||||||||
| [46] | x | x | x | x | [187] | 67 | 22 | – | 67 | – | – | – | ||||||||||||
| [188] | x | x | x | x | CAPRI | 34.5 | 37.4 | – | 79.5 | 0.23 | 35.9 | 0.71 | [101] | |||||||||||
| [34] | x | x | x | x | 42.8 | 57.8 | – | 73.3 | – | – | – | |||||||||||||
| [61] | x | x | x | x | CAPRI | 27.3 | 28.7 | – | 76.6 | 0.14 | 28 | 0.62 | [101] | |||||||||||
| [189] | x | x | x | x | [52] | – | – | – | 76 | 0.5 | – | – | ||||||||||||
| [52] | x | x | x | x | – | – | – | 72 | 0.43 | – | – | [189] | ||||||||||||
| [51] | x | x | x | x | [48] | 27.7 | – | 44.2 | – | 0.15 | – | – | [48] | |||||||||||
| D | [72] | [186] | – | 25 | – | 45 | – | – | – | |||||||||||||||
| [74] | [186] | – | 50.5 | – | 49.5 | – | – | – | ||||||||||||||||
| CAPRI | 24 | 38.9 | – | 81.1 | 0.20 | 29.7 | 0.71 | [101] | ||||||||||||||||
| E | [90] | x | x | x | x | [184] | 56.1 | 52.6 | – | 85.4 | 0.45 | 52.5 | – | |||||||||||
| [88] | x | x | x | x | [190] | 43 | 72.7 | – | – | – | – | – | ||||||||||||
| [27] | x | x | x | x | 67.3 | 50 | – | – | – | – | – | |||||||||||||
| F | [101] | x | x | x | x | CAPRI-bound | 46.1 | 45.4 | – | 80.9 | 0.34 | 45.7 | 0.77 | |||||||||||
| CAPRI-unbound | 43.7 | 44 | – | 81.2 | 0.32 | 43.8 | 0.75 | |||||||||||||||||
| [99] | x | x | x | x | [190] | 57.5 | 50.3 | – | 72.6 | 0.34 | 0.53 | 0.73 | ||||||||||||
| CAPRI-bound | 53 | 43 | – | 72.1 | 0.29 | 0.47 | 0.71 | |||||||||||||||||
| CAPRI-unbound | 53.6 | 43.3 | – | 73.2 | 0.30 | 0.48 | 0.72 | |||||||||||||||||
| [100] | x | x | x | x | [190] | 45.7 | 43.60 | – | – | – | – | – | ||||||||||||
| CAPRI-bound | 42.2 | 41.50 | – | – | – | – | – | |||||||||||||||||
| CAPRI-unbound | 44.6 | 39.8 | – | – | – | – | – | |||||||||||||||||
| [97] | x | x | x | x | x | x | [98] | 34 | 32 | – | – | – | 34 | – | ||||||||||
| [98] | x | x | x | x | x | x | 35.3 | 31.5 | – | – | – | 33.3 | – | |||||||||||
| G | [111] | x | x | x | x | [184] | – | – | – | – | – | – | 0.47 | |||||||||||
| [104] | x | x | x | x | [184] | – | – | – | – | – | – | 0.87 | ||||||||||||
| [110] | x | x | x | x | [184] | 62.2 | 40.4 | – | – | – | – | – | ||||||||||||
| [102] | x | x | x | x | [190] | – | – | – | – | – | – | 0.72 | ||||||||||||
| [109] | x | x | x | x | 72.7 | 39.3 | – | – | – | 51 | – | |||||||||||||
| [115] | x | x | + | x | – | – | – | – | – | – | – | |||||||||||||
| [118] | x | x | x | [118] test | 20 | 59 | – | – | – | – | – | [118] | ||||||||||||
| [122] | x | x | x | [118] fitting | 20 | 23 | – | – | – | – | – | [118] | ||||||||||||
| [119] | x | x | x | [118] fitting | 20 | 23 | – | – | – | – | – | [118] | ||||||||||||
| [121] | x | x | x | [118] fitting | 20 | 23 | – | – | – | – | – | [118] | ||||||||||||
| [18] | x | x | x | [118] fitting | 20 | 25 | – | – | – | – | – | [118] | ||||||||||||
| [120] | x | x | x | [118] fitting | 20 | 20 | – | – | – | – | – | [118] | ||||||||||||
The predictors are grouped by their corresponding category from this manuscript, based on the input and methodology used. The numbers in the ‘Method’ column correspond to the heading numbering in the text (except from meta predictors). Performance measures, where available, were collected from the original publications. Where possible, the performance measures were taken from studies benchmarking several studies at once. Empty cells in columns with * correspond to the same study where its reference number is available in the predictor column in the same row. Cells with + refer to ‘predicted structural feature’. In the data set column, CAPRI refers to the targets used in the CAPRI challenge, which can be in the bound or unbound form. The 3D classifier group contains some methods, which are based on scoring function. Columns marked with x correspond to the features the predictor is using. Where data is not available - sign is used. In the Method column for ‘A' see section ‘Sequence Feature-based Predictors', for ‘B' see section ‘3D mapping-based Predictors', for ‘C' see section ‘3D-Classifier Predictors', for ‘D' see meta methods in section ‘Descriptors used by predictors', for ‘E' see section ‘Homologous Template-based Predictors', for ‘F' see section ‘Structural Neighbour-based Predictors' and for ‘G' see section ‘Partner-specific interface predictors'.
Intrinsic-based predictors
Sequence-based interface predictors
Sequence-based interface predictors use only the sequence features of the query proteins to detect interfaces and thus, can be applied to almost any protein. Early work exploited sequence features such as hydrophobicity distribution [8], composition/propensity to be an interface residue [9] and physico-chemical properties [4]. Predictors have also combined such features, using machine learning strategies such as support vector machine (SVM) [4, 10], neural-network [11] or random-forest [12]. Such approaches suffer from low specificity [4] and therefore later predictors proposed integration of evolutionary information to further improve prediction accuracy [4, 9].
Sequence feature-based predictors
The success of evolutionary information in predicting functional sites [13, 14] inspired many interface predictors to combine evolutionary information with other sequence features [15, 16]. Interface residues are more conserved than the rest of the protein surface [17, 18] and these conserved positions are identified from multiple sequence alignments (MSAs) [5, 18, 19] often with phylogenetic trees assisting the procedure [19–21] (Figure 1A).
The first predictor [16] that combined evolutionary information along with residue composition achieved an accuracy of 64%. This was a 6% increase over the previous sequence-based study [9]. Since then, several methods [12, 15] have experimented with a wide range of sequence-derived features combined with evolutionary information. However, the most recent method in this category [10] showed that using hydrophobicity alone combined with evolutionary information can achieve results similar to methods that use a far larger number of features [12].
In addition to evolutionary information, some sequence-based methods [22, 23] take advantage of predicted structural information (i.e. surface accessibility and secondary structure). Use of predicted structural information in ISIS [22] and PSIVER [23] increased the sensitivity of their predictions, for example, ISIS increased its sensitivity to 20% from a baseline of 0.5% [9]. These results demonstrate that inclusion of predicted structural information can increase the accuracy of interface prediction.
It appears that current sequence-based methods have reached their limit because further combination of available features does not improve accuracy. Therefore, alternative approaches and sources of information should be investigated. In particular, use of structural data has been shown to improve the performance of sequence-based interface predictors.
Structure-based predictors
Structural features are important discriminative attributes for protein interface prediction. These features are associated with the atomic coordinate of the proteins, such as secondary structure [24, 25], solvent-accessible surface area [26, 27], geometric shape of the protein surface [26] and crystallographic B-factor [24]. Historically, methods using structural information were limited by the paucity of available 3D structures. However, in recent years the number of solved structures has been gradually increasing, enabling the development of 3D-based interface predictors. In these predictors, the query 3D structure is either used to identify interface residues in close proximity to each other (see the ‘3D mapping-based predictors’ section) and/or as structural features for detection of interface residues (see the ‘3D-classifier predictors’ section).
3D mapping-based predictors
Conserved residues are an important source of information for interface predictors [28]. If the structure of the query protein is available, one can map the predicted/conserved residues directly onto the structure, identifying clusters of neighbouring residues [13, 28, 29]. This naïve use of structural information improves on sequence-only methods. In addition, including other physico-chemical attributes at the mapping stage can further increase prediction performance [30] (Figure 1B).
3D-classifier predictors
Instead of considering structural information only at the mapping stage, 3D-classifier predictors use 3D structural features (or their combination with sequence features) directly to detect interfaces (Figure 1C). They exploit the fact that the binding interface has different structural properties when compared with the rest of the protein. For instance, Chothia and Janin (1975) [31] discovered that hydrophobicity is a key element to stabilizing protein–protein interactions, which inspired many of the early predictors in this category [24, 32, 33].
To investigate the importance of 3D information for detecting interface residues, predictions based on sequence information alone were compared with predictions including structural data [26, 34]. Results found that using structural information significantly improves prediction accuracy. This is probably mainly owing to the elimination of non-surface residues, greatly reducing the search space [35].
Not one single structural property completely discriminates interface residues from others. Therefore predictors have based their predictions on combining multiple input properties of residues. Methods in this category differ from one another by features employed and the methodology used to combine them. They are broadly divided in two groups [36] (i) score-based and (ii) probabilistic-based predictors. Predictors in both groups are trained using a training set to predict interfaces [36].
Score-based predictors
Score-based predictors calculate an interaction likelihood score for each residue. All residues with a score above a certain cut-off are classified as contacts [36]. Scores can be calculated from a linear [37, 38] or non-linear combination of sequence and structure contributions [36]. Features used include accessible surface area [39], Position Specific Scoring Matrix (PSSM), interface propensity and surface conservation [40], side chain energy scores [41, 42] or desolvation energy [43, 44]. The drawback of constructing such empirical functions is that they rely on specific knowledge of the physical system, which is often error-prone and not suitable for amendments and extensions [36]. This issue is tackled by non-linear combinations of features using machine learning techniques such as SVM [45–48], ensemble methodology [49, 50], Neural Networks [51–54] or Random Forests (RF) [55–59]. As the number of positive samples (interacting residues) is smaller than the negative samples, the training set for machine learning classifiers of interface and non-interface are imbalanced [59]. To deal with this problem, predictors have proposed strategies for splitting the training data into balanced subsets [10] and detecting outliers [60].
Probabilistic-based predictors
An alternative approach to using linear or non-linear combinations is to find the conditional probability of being interface or non-interface, where to are the properties of the residue under study. Conditional probability can be generated from the training sets using Bayesian methods [61–63], Hidden Markov Model [64, 65] or Conditional Random Fields [66–68]. It has been argued that such probabilistic classifiers might offer an increased performance over the machine learning methods described above [62, 67].
Descriptors used by predictors
Machine learning techniques used by score-based and probabilistic-based predictors [59] provide a framework for evaluating the contributions of attributes to the predictive power. Previous studies have investigated which properties play an important role in the discrimination of interface and non-interface residues. The PSSM generated from PSI-BLAST [69] has been argued to be an important factor [47, 70] as well as solvent-accessible surface area, hydrophobicity, conservation and propensity [71]. It was also demonstrated that relative solvent accessibility has more predictive power than other features [50]. Recently it has been demonstrated that only four features, solvent-accessible surface area, hydrophobicity, conservation and propensity of the surface amino acids are sufficient to perform as well as the current state-of-the-art predictors [71]. To the best of our knowledge, the most recent benchmark of the predictive power of attributes was performed by RAD-T [59]. This study named relative solvent-excluded surface area and solvation energy as attributes with the most discriminative power. In the same study, it was established that among the different machine learning methods a random forest-based classifier performed the best. This best combination of attributes and the classifier currently forms the core of RAD-T.
Even though RAD-T performed a rigorous benchmark of the available methods and features to be employed, this predictor relies on one classifier, namely a variant of RF. It was argued that if predictors express a degree of orthogonality, they may be combined in a consensus-based classifier. Therefore, some methods have integrated individual interface predictors into one meta framework [72, 73]. For instance, meta-PPISP [74] combines the prediction scores of PINUP, Cons-PPISP and ProMate using linear regression analysis. One review study [36] confirmed the superiority of meta-PPISP over its constituent PINUP [41], Cons-PPISP [53] and ProMate [61] with accuracies of 50%, 48%, 38% and 36%, respectively.
While meta-predictors are an elegant way to improve the accuracy of individual constituents, significantly better performance is achieved only if the combination of features does not introduce redundancy [59, 75]. It appears that intrinsic-based predictors have reached saturation since further combination of existing features and classifiers has little impact on prediction performance [76]. Therefore, a complementary approach needs to be found in the form of new sources of experimental data or novel classifying methodology. This issue and an increasing number of structures in the Protein Data Bank (PDB) [77] have led to an emergence of an alternative trend in predictors, using existing complexes as templates for interface prediction.
Template-based predictors
The growing number of available structural complexes assists accurate identification of interface templates. Studies have shown that interfaces are conserved among homologous complexes [78–81], inspiring the first category of template-based methods, which relies on homologous complexes. However such homologous structures are not always available. Therefore the second category of template-based predictors uses structurally, but not necessarily evolutionarily, similar complex templates.
Homologous template-based predictors
These methods use known complexes where one of the interacting partners is homologous to the query protein. The interface via which the homologous protein interacts is assumed to be an indicator where the corresponding interface might be found on the query protein. This approach to interface prediction is possible, as it was demonstrated that homologous proteins tend to interact with their partners with a similar orientation [80] and the binding site localization within each family is often conserved regardless of the similarity of binding partner [78, 79, 81]. Physico-chemical properties of the interface residues have higher similarity in homologous proteins than non-homologous ones [82–86]. These observations suggest that integration of homologous structural information into interface predictors should improve performance. The current predictors in this category are HomPPI [35], IBIS [87–89] and T-PIP [90, 91].
HomPPI [35] builds an MSA of the query protein and its homologous complexes. Instead of looking at conservation at a residue level, HomPPI checks if the majority of the homologous residues at that position in the MSA are interface or non-interface. HomPPI implicitly takes advantage of binding site conservation of the homologous complexes. It performs better than 3D classifier methods such as ProMate [61], PIER [38], meta-PPISP [74], cons-PPISP [53] and PSIVER [23].
A combination of sequence and structure conservation scores was introduced in IBIS [87–89]. Initially, homologous complexes with at least 30% sequence similarity to the query protein are extracted. Then, these structures are superposed on to the query protein. Using this alignment, a structure-based-MSA is created, which allows the conserved interface residues to be identified. Comparison with HomPPI (62.8% precision and 50.4% recall) demonstrates the importance of using structure-based MSA (69.7% precision and 72.0% recall).
Recently, T-PIP [90, 91], which outperforms IBIS, was introduced (T-PIP with 52.6% precision and 56.1% recall and IBIS with 42.6% precision and 37.4% recall). Similar to IBIS it builds a structure-based MSA of homologues. The main novelty of T-PIP is that not only is the homology between the query protein and its homologues considered but also the diversity between the interacting partners of the homologues at each specific binding site.
In this category, the main attributes that appear to be contributing to the quality of predictions are the structure-based MSAs and the binding partner information. Although homologous template-based predictors improve the predictions over intrinsic-based methods, they are limited to those proteins where homologous complex structures exist. For instance, HomPPI has lower coverage than the 3D classifier methods and IBIS’s coverage is even lower. Although this issue has been partially addressed in T-PIP by lowering the threshold for selecting homologues, these predictors fail in cases where homologous complexes of the query protein are not available. This issue can be dealt with by using structural neighbours; complexes not necessarily evolutionarily related but with similar folds to the query protein.
Structural neighbour-based predictors
Proteins sharing a similar fold with the query protein, even if not evolutionarily related, can offer similar predictive information to that of homologues. This was established by a study which found that functional relationship can be detected using remote structural neighbours [92]. Furthermore, proteins with similar folds but low sequence identity tend to interact with their partners using the same location [93, 94]. Such structural neighbours are exploited as templates for interface prediction to help overcome the low template coverage that can afflict homology-based methods (Figure 1D) [95–98].
Currently there are two main methods in this category, PredUS [99, 100] and PrISE [101]. PredUS is an earlier method, which identifies structural neighbours by finding structures with a globally similar fold to the query protein. PrISE, on the other hand, uses only the interface structure for template identification, which increases its prediction coverage. PrISE performance is similar to PredUS, as both methods achieve accuracy in the region of 81%. According to [101], PrISE performed better than methods that do not use template information.
In general, template-based methods show better recall scores, while intrinsic-based methods have better precision [90, 100, 101]. This suggests that intrinsic-based methods predict a smaller set of correct interface residues with higher confidence, which is especially important for mutagenesis studies. Also, T-PIP, a homology-based template method, has been shown to perform better (precision 52.6% and recall 56.1%) than the structural neighbour methods of PredUs (precision 47.3% and recall 58.2%) and PrISE (precision 38.5% and recall 48.9%). This improvement may be the positive impact of the consideration of interacting partners of the structural neighbours.
Partner-specific interface predictors
The methods described above predict interfaces for one query protein, but proteins may display different interface patterns depending on their binding partner (e.g. antibodies [102]). Therefore, partner-specific predictors identify interacting residue pairs between two query proteins that are assumed to interact. One of the main challenges for these predictors is when unbound query protein structures are used. Therefore, performance of these methods decreases with the increase of conformational changes of the protein pairs on binding [102].
Partner-specific methods can be broadly divided into three groups, intrinsic-based methods, docking-based methods and co-evolution-based predictors. Intrinsic-based methods are similar in nature to the 3D classifier methods. The core difference is that the set of features that is being computed for training and testing is complemented by partner-specific features such as propensities and electrostatic complementarity [35, 102, 103]. The most recent method in this category is PAIRpred [104]. Application of these methods is seen in re-ranking docked decoys based on similarity to the predicted interface [90, 102, 105, 106].
Another type of approach uses protein–protein docking (Figure 1E) to generate potential interfaces (for a review on docking see [107, 108]). These methods generate docked poses of the two query proteins and detect interfaces based on contact energy and frequency scores [109]. The two main methods in this category are DoBi and RCF [110, 111]. DoBi (F-scores ∼ 0.55) outperformed the 3D classifiers such as MetaPPI, meta-PPISP, PPI-Pred, PINUP and ProMate (F-scores of 0.35, 0.43, 0.32, 0.43 and 0.21, respectively) [109]. While direct comparison between RCF and DoBi is not available, these results demonstrate the advantage of including partner information into the interface prediction. The main drawback is the requirement of the two protein structures. In addition, docking-based methods are slower, as generating docked poses is computationally expensive.
Co-evolution strategies have also been used to detect interfaces [18, 112]. The co-evolution principle suggests that mutations on one protein in a complex are often compensated for by correlated mutations within the same chain or on a binding partner. Such correlated mutations are assumed to maintain the stability of the protein or protein–protein complex [112]. By creating MSAs of the input proteins, one identifies the columns that appear to change in concert indicating spatial proximity. This paradigm has been used in protein structure prediction [113–116], scoring of docking decoys [117] as well as in protein–protein interface prediction [115, 118] (Figure 1E).
Early applications of co-evolution to protein interface prediction include OMES [119], MI [120] SCA [121], McBASC [18], ELSC [122] and the more recent i-Patch [118] and EVComplex [115]. The earlier methods generally suffer from low precision (20–25% precision at 20% recall) [118]. The more recent method, i-Patch, achieves higher precision (59%) for the same recall values, owing to the incorporation of structural information. The most recent method, EVComplex is capable of providing predictions from sequence alone, as it uses a structural model of the input. Its applicability was demonstrated by delivering interface predictions in accord with experimental data from a de novo model of ATP synthase complex. Co-evolution methods have over the past few years improved dramatically and this new approach has only just been tested on protein interface prediction. Since protein interaction data and sequence information is increasing exponentially, it is likely that this will further improve the quality and the applicability of co-evolution predictors in the future.
Predictors taking the binding partner into consideration [90] have shown promising avenues to better detection of binding sites. Therefore, predictors specialized to a specific type of protein such as antibodies may well yield better predictive power.
Antibody–antigen complex modelling
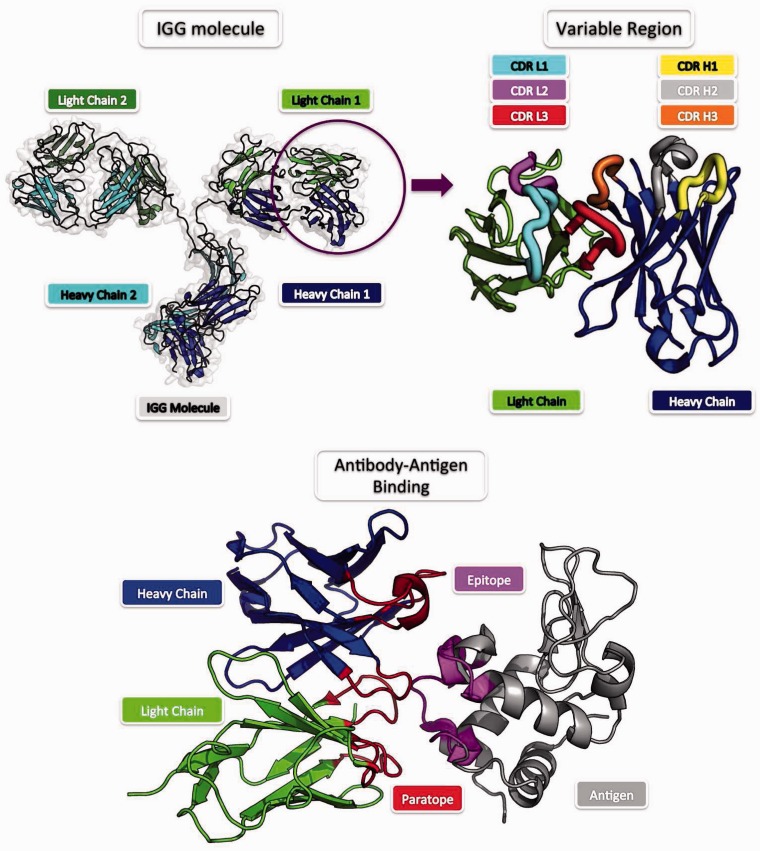
Antibodies are currently the most important class of biopharmaceuticals [123]. The success of antibodies as therapeutics depends on their intrinsic binding mechanism, which allows them to be adjusted toward almost any antigen target by mutations in a well-defined binding region (see Figure 2). The antibody–antigen binding mechanism is radically different to that of general proteins [124] and thus methods attempting antibody–antigen interaction prediction have developed into a separate domain [124–127]. Antibody–antigen interface predictors can be broadly classified into methods that predict the binding residues on either the antibody (paratope prediction) [128] or the antigen side (epitope prediction) [129].
Figure 2.
Antibody structure and binding. The most common form of an antibody is the IgG (upper left). IgG is composed of two pairs of heavy and light chains. The tip of an antibody that carries the binding site (symmetrical in an IgG) is the variable region (upper right). The variable region harbours the six CDR loops, which form the majority of the antigen recognition site, the paratope (lower). The CDR regions are distinct between different antibodies whereas the rest of the antibody remains largely unchanged. The paratope recognizes a specific epitope, the corresponding binding site on the antigen (lower). A colour version of this figure is available at BIB online: http://bib.oxfordjournals.org.
Paratope prediction
The antibody binding site is chiefly composed of six loops known as complementarity determining regions (CDRs). These CDRs have been described using a variety of definitions [127, 130–133], which suggest they contain between 40 and 50 residues. Examinations of antibody complexes show that there are on average 10–15 paratope residues, the majority of which are within the CDRs.
It was recently demonstrated that the residues contained within the boundaries of these CDRs contain only about 80% of the paratope [127]. On the basis of this finding a more robust definition of the antibody binding region was introduced and implemented—PARATOME [127]. Given a sequence or structure of an antibody, PARATOME aligns sequentially similar antibodies with solved complexes. The contacts from the aligned sequences are used in a consensus score to define the binding region for the query. This methodology maximizes the recall (∼94%) at the cost of precision (∼30%) because, just as the CDR definitions, it generates an annotation for the entire binding region neighbourhood rather than singling out possible contact residues.
In contrast to region-wide annotations given by CDR definitions and PARATOME, over the past 2 years there has been an increasing interest in developing methodologies that predict specific paratope residues. There are currently three methods which address this problem: proABC [128], Antibody i-Patch [124] and ISMBLab-PPI [134]. ProABC is a RF-based machine learning protocol, which requires only the sequence of the antibody on input. Antibody i-Patch is a statistical method, which relies on the structure of the antibody; however, it was demonstrated that it is robust to the use of homology models. The most recent method, ISMBLab-PPI, is a neural-network protocol. In contrast to proABC and Antibody i-Patch, its training set is not restrained to antibody–antigen complexes only. This might explain why it underperforms against proABC (comparison with Antibody i-Patch was not performed).
The field of paratope residue contact annotation appears to be greatly underdeveloped, mostly as a result of the assumption that knowing the CDRs is sufficient for antibody engineering through mutagenesis. The antibody binding region however contains on average 40–50 residues and thus complete mutagenesis of this entire region is currently not tractable while only around 18–19 residues are in contact with antigen [135]. For this reason, knowledge of particular paratope residues that might be important for binding would greatly reduce the search.
Epitope prediction
Identifying regions on the antigen that are capable of binding an antibody is an important problem from the point of view of vaccine development and immunogenicity [136–138]. This is particularly difficult because epitope patches appear to be barely distinguishable from general protein surfaces [126, 134, 139]. There exist several experimental methods to identify epitope residues but all of them are costly in time and resources. For this reason, the field of computational B-cell epitope prediction has been developed intending to provide information on potentially immunogenic structures and sequences.
Computational epitope predictors can be divided into linear and conformational predictors. Linear epitope predictors aim to identify contiguous stretches in the antigen sequence, which constitute the epitope, while conformational ones focus on identifying patches of sequence on the antigen, which, when folded, constitute the linearly discontinuous epitope. Around 90% of all known epitopes are conformational [139]. Nevertheless, most of the methods developed over the past 20 years addressed the easier problem of linear epitope identification [129, 140]. Here we focus exclusively on conformational epitopes.
Classes of conformational epitope prediction
Conformational B-cell epitope predictions can be classified into two types, those using antibody information and those that do not. The vast majority of them do not use any antibody information (e.g. CEP [141], DiscoTope [142, 143], ElliPro [144, 145], PEPITO [146], PEPOP [147], SEPPA [148, 149], EPITOPIA [150] and others [151, 152]). Consensus-based methods such as EPCES [153] or the meta-server EPSVR/EpMETA [154] are currently among the best-performing algorithms in this area [152].
Data resources for epitope prediction
The main aim of methods that use no antibody information is to identify epitope-like sites on proteins as a means to improve vaccine design. Their mode of operation is similar in nature to that of general protein–protein interface prediction introduced in the earlier sections. In contrast to general protein predictors, epitope predictors use antibody-antigen-specific data from the PDB, AntigenDB [155], the Conformational Epitope Database [156], DIGIT [157], Immune Epitope Database [158–160], IMGT [161], Structural Antibody Database [162] and others [163]. The main issue is that virtually any part of a protein can be an epitope for some kind of a monoclonal antibody; thus including antibody information may be crucial [125, 164].
Antibody-specific epitope prediction
The field of antibody-specific conformational B-cell epitope predictors is relatively underdeveloped—only six methods exist to address this problem [125, 164–168]. The earliest used only 26 antibody–antigen complexes (those available in 2007) to produce its predictions [165]. They combined the program FADE [169] for paratope–epitope complementarity with FastContact [170] for physicochemical descriptor calculations. On their small test set they achieved 18% sensitivity and 87% specificity.
Another method that attempted to obtain antibody-specific predictions relied on the coupling of ASEP and DiscoTope [166]. The ASEP potential was computed by counting residue–residue interface preferences from a non-redundant set of antibody–antigen complexes from the PDB. This potential was then used to constrain general epitope predictions made by DiscoTope, with respect to a single antibody.
Following their study of antibody–antigen complexes [167, 171], Zhang et al. developed a method that treats antibody–antigen interactions as a Hidden Markov Model. They used 80 antibody–antigen complexes to train their method, achieving 43% sensitivity and 71% specificity. The testing procedure was performed using leave-one-out validation, which, as the authors admit, given the redundancy of their data set might have led to over-fitting [167].
Recently a mixed computational-experimental method was proposed to predict antibody-specific epitopes [164]. An RF-based computational method assesses the propensity of possible antibody–antigen residue matches to be in contact. Their first protocol, ‘per-residue’, requiring sequence of the antibody and structure of an antigen outperforms EPSVR, which relies on the antigen structure. Their second protocol, ‘patch-per Ab’, requiring the structure of an antigen, performed even better. They demonstrated its application in combination with blocking experiments in making good predictions for the antibody D8 for VACV. Such combination of computational and experimental techniques holds a particular promise in being able to identify epitopes with a much higher throughput than crystallization.
The most recent general antibody-specific epitope predictor is EpiPred [125]. Its protocol requires the structure of an antibody (which can be a homology model) and the structure of the antigen. Antigenic epitopes are identified by performing simplified surface matching complemented by antibody-antigen-specific statistical scoring. This method (44% recall at 14% precision) outperforms the antibody-ignoring Discotope (23% recall at 14% precision), demonstrating the value of introducing antibody information into predictions.
There has not yet been a comprehensive study benchmarking the antibody-specific methods. Because antibody information improves the quality of predictions, we expect the field to investigate further antibody-specific predictions. One of the main challenges remains the lack of understanding of antibody specificity. A comprehensive study contrasting different epitopes on a single antigen (e.g. lysozyme) with respect to their binding antibodies could improve our understanding of the specificity of antibodies, providing ground for better epitope predictions.
Conclusion
In this review we have discussed the myriad features and techniques used by protein interface predictors (summarized in Table 2). Although considerable effort has been expended to develop the field thus far, no method yet yields excellent results and objective comparison between approaches is difficult.
However, usage of 3D structural and evolutionary properties tends to improve results over predictions based on sequence alone. It appears that feature-based methods have reached saturation, and the inclusion of more properties does not improve predictive performance. A possible solution to this problem would be to diversify the predictions into specific protein types, such as antibodies, kinases and GPCRs. Such predictions would exploit the intrinsic features of these particular protein complexes, a property that is lost if all the proteins are considered together [172].
With the increasing availability of structural templates [173, 174], a new trend in protein interface prediction methodology uses structural homologues or structural neighbours for template-based predictions. Although, in many cases, the binding partner of the template is disregarded, taking it into account could contribute to better predictive power in a similar way as knowledge of the antibody contributes to epitope prediction.
Furthermore the increasing amount of complex structural data available has made it possible to perform large-scale protein–protein interaction predictions [175–178]. As such proteome-scale approaches are one novel way to address the protein interface prediction problem.
Benchmarking of protein interface prediction methods has so far not been systematic. Because predictors are assessed on different data sets by distinct metrics, it is currently difficult to fairly evaluate the multitude of methods and identify clear areas for improvement. This would be facilitated if protein interface predictors consistently formed a subcategory in the Critical Assessment of Prediction of Interactions (CAPRI) challenge [3, 179, 180, 191] or developed their own assessment scheme. Thus, introducing unified training and test data sets as well as blind benchmarking is essential for the further development of the field.
Key Points.
There is a plethora of available protein interface predictors and the field in its current state appears to be saturated. This calls for new methodologies or sources of information to be exploited. Recent methods use existing complexes as templates or use co-evolution to inform predictions.
One avenue of recent interest is the specialization of methods with respect to a single protein type, e.g. antibodies, which could improve predictions and make benchmarking more transparent.
There is an urgent need to benchmark the available methods in a consistent manner. Available protocols rarely perform comprehensive comparisons. Therefore it is impossible to precisely identify areas where improvement is necessary. Consistent participation of available predictors in the CAPRI challenge or development of a protein interface predictor-specific assessment scheme would address this issue.
Supplementary data
Supplementary data are available online at http://bib.oxfordjournals.org/.
Funding
2020 Science Programme (UK Engineering and Physical Sciences Research Council (EPSRC) Cross-Discipline Interface Programme, EP/I017909/1).
Supplementary Material
Biographies
Reyhaneh Esmaielbeiki is a postdoctoral researcher in Computational Structural Biology at University of Oxford. She has been working on protein interface prediction and modelling of membrane proteins.
Konrad Krawczyk is a research fellow in Structural Biology at the Department of Statistics and the Department of Computer Science, Oxford University.
Bernhard Knapp is a postdoctoral research fellow in Computational Structural Biology at the University of Oxford. His research interest is in the modelling of immune system-related protein structures and their dynamics.
Jean-Christophe Nebel is an associate professor in Computing Science and Bioinformatics in the Faculty of Science, Engineering and Computing at Kingston University, London. His research interests include protein interaction and structure prediction.
Charlotte M. Deane is a professor in the Department of Statistics, University of Oxford. Her research interests include the areas of protein structure prediction, evolution and interaction.
References
- 1.Sudha G, Nussinov R, Srinivasan N. An overview of recent advances in structural bioinformatics of protein-protein interactions and a guide to their principles. Prog Biophys Mol Biol 2014;116:141–50. [DOI] [PubMed] [Google Scholar]
- 2.Cazals F. Revisiting the Voronoi description of protein-protein interfaces: Algorithms. Pattern Recognit Bioinform 2010;6282:419–30. [Google Scholar]
- 3.Janin J, Henrick K, Moult J, et al. CAPRI: a critical assessment of predicted interactions. Proteins 2003;52:2–9. [DOI] [PubMed] [Google Scholar]
- 4.Yan C, Dobbs D, Honavar V. A two-stage classifier for identification of protein-protein interface residues. Bioinformatics 2004;20:i371–8. [DOI] [PubMed] [Google Scholar]
- 5.Ezkurdia I, Bartoli L, Fariselli P, et al. Progress and challenges in predicting protein-protein interaction sites. Brief Bioinform 2009;10:233–46. [DOI] [PubMed] [Google Scholar]
- 6.De Vries SJ, Bonvin AM. How proteins get in touch: interface prediction in the study of biomolecular complexes. Curr Protein Pept Sci 2008;9:394–406 [DOI] [PubMed] [Google Scholar]
- 7.Tuncbag N, Kar G, Keskin O, et al. A survey of available tools and web servers for analysis of protein–protein interactions and interfaces. Brief Bioinform 2009;10:217–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallet X, Charloteaux B, Thomas A, et al. A fast method to predict protein interaction sites from sequences. J Mol Biol 2000;302:917–26 [DOI] [PubMed] [Google Scholar]
- 9.Ofran Y, Rost B. Predicted protein-protein interaction sites from local sequence information. FEBS Lett 2003;544:236–9. [DOI] [PubMed] [Google Scholar]
- 10.Chen P, Li J. Sequence-based identification of interface residues by an integrative profile combining hydrophobic and evolutionary information. BMC Bioinformatics 2010;11:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ofran Y, Rost B. Analysing six types of protein-protein interfaces. J Mol Biol 2003;325:377–87. [DOI] [PubMed] [Google Scholar]
- 12.Chen XW, Jeong JC. Sequence-based prediction of protein interaction sites with an integrative method. Bioinformatics 2009;25:585–91. [DOI] [PubMed] [Google Scholar]
- 13.Lichtarge O, Bourne HR, Cohen FE. An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol 1996;257:342–58. [DOI] [PubMed] [Google Scholar]
- 14.Madabushi S, Gross AK, Philippi A, et al. Evolutionary trace of G protein-coupled receptors reveals clusters of residues that determine global and class-specific functions. J Biol Chem 2004;279:8126–32. [DOI] [PubMed] [Google Scholar]
- 15.Wang B, Chen P, Huang D-S, et al. Predicting protein interaction sites from residue spatial sequence profile and evolution rate. FEBS Lett 2006;580:380–4. [DOI] [PubMed] [Google Scholar]
- 16.Reš I, Mihalek I, Lichtarge O. An evolution based classifier for prediction of protein interfaces without using protein structures. Bioinformatics 2005;21:2496–501. [DOI] [PubMed] [Google Scholar]
- 17.Lovell SC, Robertson DL. An integrated view of molecular coevolution in protein-protein interactions. Mol Biol Evol 2010;27:2567–75. [DOI] [PubMed] [Google Scholar]
- 18.Pazos F, Helmer-Citterich M, Ausiello G, et al. Correlated mutations contain information about protein-protein interaction. J Mol Biol 1997;271:511–23. [DOI] [PubMed] [Google Scholar]
- 19.Valencia A, Pazos F. Prediction of protein-protein interactions from evolutionary information. Methods Biochem Anal 2003;44:411–26. [PubMed] [Google Scholar]
- 20.Del Sol Mesa A, Pazos F, Valencia A. Automatic methods for predicting functionally important residues. J Mol Biol 2003;326:1289–302. [DOI] [PubMed] [Google Scholar]
- 21.Rausell A, Juan D, Pazos F, et al. Protein interactions and ligand binding: from protein subfamilies to functional specificity. Proc Natl Acad Sci 2010;107:1995–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ofran Y, Rost B. ISIS: interaction sites identified from sequence. Bioinformatics 2007;23:e13–16. [DOI] [PubMed] [Google Scholar]
- 23.Murakami Y, Mizuguchi K. Applying the Naive Bayes classifier with kernel density estimation to the prediction of protein-protein interaction sites. Bioinformatics 2010;26:1841–8. [DOI] [PubMed] [Google Scholar]
- 24.Jones S, Thornton JM. Analysis of protein-protein interaction sites using surface patches. J Mol Biol 1997;272:121–32. [DOI] [PubMed] [Google Scholar]
- 25.Talavera D, Robertson DL, Lovell SC. Characterization of protein-protein interaction interfaces from a single species. PLoS One 2011;6:e21053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Šikić M, Tomić S, Vlahoviček K. Prediction of protein-protein interaction sites in sequences and 3D structures by random forests. PLoS Comput Biol 2009;5:e1000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung JL, Wang W, Bourne PE. Exploiting sequence and structure homologs to identify protein-protein binding sites. Proteins 2005;62:630–40. [DOI] [PubMed] [Google Scholar]
- 28.Ashkenazy H, Erez E, Martz E, et al. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 2010;38:W529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pupko T, Bell RE, Mayrose I, et al. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics 2002;18:S71–7. [DOI] [PubMed] [Google Scholar]
- 30.Engelen S, Trojan LA, Sacquin-Mora S, et al. Joint evolutionary trees: a large-scale method to predict protein interfaces based on sequence sampling. PLoS Comput Biol 2009;5:e1000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chothia C, Janin J. Principles of protein-protein recognition. Nature 1975;256:705–8. [DOI] [PubMed] [Google Scholar]
- 32.Jones S, Thornton JM. Prediction of protein-protein interaction sites using patch analysis. J Mol Biol 1997;272:133–43. [DOI] [PubMed] [Google Scholar]
- 33.Murakami Y, Jones S. SHARP2: protein-protein interaction predictions using patch analysis. Bioinformatics 2006;22:1794–5. [DOI] [PubMed] [Google Scholar]
- 34.Koike A, Takagi T. Prediction of protein-protein interaction sites using support vector machines. Protein Eng Des Sel 2004;17:165–73. [DOI] [PubMed] [Google Scholar]
- 35.Xue LC, Dobbs D, Honavar V. HomPPI: a class of sequence homology based protein-protein interface prediction methods. BMC Bioinformatics 2011;12:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou HX, Qin S. Interaction-site prediction for protein complexes: a critical assessment. Bioinformatics 2007;23:2203–9. [DOI] [PubMed] [Google Scholar]
- 37.Li J-J, Huang D-S, Wang B, et al. Identifying protein–protein interfacial residues in heterocomplexes using residue conservation scores. Int J Biol Macromol 2006;38:241–7. [DOI] [PubMed] [Google Scholar]
- 38.Kufareva I, Budagyan L, Raush E, et al. PIER: protein interface recognition for structural proteomics. Proteins 2007;67:400–17. [DOI] [PubMed] [Google Scholar]
- 39.Negi SS, Schein CH, Oezguen N, et al. InterProSurf: a web server for predicting interacting sites on protein surfaces. Bioinformatics 2007;23:3397–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Vries SJ, Van Dijk AD, Bonvin AM. WHISCY: what information does surface conservation yield? Application to data-driven docking. Proteins 2006;63:479–89. [DOI] [PubMed] [Google Scholar]
- 41.Liang S, Zhang C, Liu S, et al. Protein binding site prediction using an empirical scoring function. Nucleic Acids Res 2006;34:3698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cole C, Warwicker J. Side-chain conformational entropy at protein-protein interfaces. Protein Sci 2002;11:2860–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandez-Recio J, Totrov M, Skorodumov C, et al. Optimal docking area: a new method for predicting protein-protein interaction sites. Proteins 2005;58:134–43. [DOI] [PubMed] [Google Scholar]
- 44.Fernández-Recio J. Prediction of protein binding sites and hot spots. Wiley Interdiscip. Rev Comput Mol Sci 2011;1:680–98. [Google Scholar]
- 45.Bradford JR, Westhead DR. Improved prediction of protein-protein binding sites using a support vector machines approach. Bioinformatics 2005;21:1487–94. [DOI] [PubMed] [Google Scholar]
- 46.Bordner AJ, Abagyan R. Statistical analysis and prediction of protein-protein interfaces. Proteins 2005;60:353–66. [DOI] [PubMed] [Google Scholar]
- 47.Dong Q, Wang X, Lin L, et al. Exploiting residue-level and profile-level interface propensities for usage in binding sites prediction of proteins. BMC Bioinformatics 2007;8:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li N, Sun Z, Jiang F. Prediction of protein-protein binding site by using core interface residue and support vector machine. BMC Bioinformatics 2008;9:553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng L, Guan J, Dong Q, et al. Prediction of protein-protein interaction sites using an ensemble method. BMC Bioinformatics 2009;10:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Porollo A, Meller J. Prediction-based fingerprints of protein-protein interactions. Proteins 2006;66:630–45. [DOI] [PubMed] [Google Scholar]
- 51.Zhou HX, Shan Y. Prediction of protein interaction sites from sequence profile and residue neighbor list. Proteins 2001;44:336–43. [DOI] [PubMed] [Google Scholar]
- 52.Fariselli P, Pazos F, Valencia A, et al. Prediction of protein-protein interaction sites in heterocomplexes with neural networks. Eur J Biochem 2002;269:1356–61. [DOI] [PubMed] [Google Scholar]
- 53.Chen H, Zhou HX. Prediction of interface residues in protein-protein complexes by a consensus neural network method: test against NMR data. Proteins 2005;61:21–35. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y, Xu J, Yang B, et al. A novel method for prediction of protein interaction sites based on integrated RBF neural networks. Comput Biol Med 2012;42:402–7. [DOI] [PubMed] [Google Scholar]
- 55.Segura J, Jones PF, Fernandez-Fuentes N. Improving the prediction of protein binding sites by combining heterogeneous data and Voronoi Diagrams. BMC Bioinformatics 2011;12:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Segura J, Jones PF, Fernandez-Fuentes N. A holistic in silico approach to predict functional sites in protein structures. Bioinformatics 2012;28:1845–50. [DOI] [PubMed] [Google Scholar]
- 57.Qiu Z, Wang X. Prediction of protein-protein interaction sites using patch-based residue characterization. J Theor Biol 2012;293:143–50. [DOI] [PubMed] [Google Scholar]
- 58.Li B-Q, Feng K-Y, Chen L, et al. Prediction of protein-Protein interaction sites by random forest algorithm with mRMR and IFS. PLoS One 2012;7:e43927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bendell CJ, Liu S, Aumentado-Armstrong T, et al. Transient protein-protein interface prediction: datasets, features, zalgorithms, and the RAD-T predictor. BMC Bioinformatics 2014;15:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen P, Wong L, Li J. Detection of outlier residues for improving interface prediction in protein heterocomplexes. IEEE/ACM Trans Comput Biol Bioinforma 2012;9:1155–65. [DOI] [PubMed] [Google Scholar]
- 61.Neuvirth H, Raz R, Schreiber G, et al. ProMate: a structure based prediction program to identify the location of protein-protein binding sites. J Mol Biol 2004;338:181. [DOI] [PubMed] [Google Scholar]
- 62.Bradford JR, Needham CJ, Bulpitt AJ, et al. Insights into protein-protein interfaces using a Bayesian network prediction method. J Mol Biol 2006;362:365–86. [DOI] [PubMed] [Google Scholar]
- 63.Higa RH, Tozzi CL. A simple and efficient method for predicting protein-protein interaction sites. Genet Mol Res 2008;7:898–909. [DOI] [PubMed] [Google Scholar]
- 64.Liu B, Wang X, Lin L, et al. Prediction of protein binding sites in protein structures using hidden Markov support vector machine. BMC Bioinformatics 2009;10:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu B, Liu B, Liu F, et al. Protein binding site prediction by combining hidden markov support vector machine and profile-based propensities. Sci World J 2014;2014:464093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Savojardo C, Fariselli P, Piovesan D, et al. Machine-learning methods to predict protein interaction sites in folded proteins. In: Biganzoli E, et al. (eds). Computational Intelligence Methods for Bioinformatics and Biostatistics, Vol. 7548, Springer Berlin Heidelberg, 2012, pp. 127–35. [Google Scholar]
- 67.Li M-H, Lin L, Wang X-L, et al. Protein-protein interaction site prediction based on conditional random fields. Bioinformatics 2007;23:597–604. [DOI] [PubMed] [Google Scholar]
- 68.Dong Z, Wang K, Dang TKL, et al. CRF-based models of protein surfaces improve protein-protein interaction site predictions. BMC Bioinformatics 2014;15:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997;25:3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nguyen MN, Rajapakse JC. Protein-protein interface residue prediction with SVM using evolutionary profiles and accessible surface areas. In: CIBCB ’06. 2006 IEEE Symposium on Computational Intelligence and Bioinformatics and Computational Biology IEEE, 2006; pp. 1–5. Toronto, Ontario. [Google Scholar]
- 71.Zellner H, Staudigel M, Trenner T, et al. Prescont: predicting protein-protein interfaces utilizing four residue properties. Proteins 2012;80:154–68. [DOI] [PubMed] [Google Scholar]
- 72.Huang B, Schroeder M. Using protein binding site prediction to improve protein docking. Gene 2008;422:14–21. [DOI] [PubMed] [Google Scholar]
- 73.Huang J, Deng R, Wang J, et al. MetaPIS: a sequence-based meta-server for protein interaction site prediction. Protein Pept Lett 2013;20:218–30. [DOI] [PubMed] [Google Scholar]
- 74.Qin S, Zhou HX. meta-PPISP: a meta web server for protein-protein interaction site prediction. Bioinformatics 2007;23:3386–7. [DOI] [PubMed] [Google Scholar]
- 75.Neuvirth H, Heinemann U, Birnbaum D, et al. ProMateus—an open research approach to protein-binding sites analysis. Nucleic Acids Res 2007;35:W543–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hamp T, Rost B. More challenges for machine learning protein interactions. Bioinformatics 2015:pii: btu857v1. [DOI] [PubMed] [Google Scholar]
- 77.Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res 2000;28:235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma B, Elkayam T, Wolfson H, et al. Protein-protein interactions: structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc Natl Acad Sci 2003;100:5772–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu Z, Ma B, Wolfson H, et al. Conservation of polar residues as hot spots at protein interfaces. Proteins 2000;39:331–42. [PubMed] [Google Scholar]
- 80.Aloy P, Ceulemans H, Stark A, et al. The relationship between sequence and interaction divergence in proteins. J Mol Biol 2003;332:989–98. [DOI] [PubMed] [Google Scholar]
- 81.Korkin D, Davis FP, Sali A. Localization of protein-binding sites within families of proteins. Protein Sci 2005;14:2350–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martin J. Beauty is in the eye of the beholder: proteins can recognize binding sites of homologous proteins in more than one way. PLoS Comput Biol 2010;6:e1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shoemaker BA, Panchenko AR, Bryant SH. Finding biologically relevant protein domain interactions: conserved binding mode analysis. Protein Sci 2006;15:352–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Han J-H, Kerrison N, Chothia C, et al. Divergence of interdomain geometry in two-domain proteins. Structure 2006;14:935–45. [DOI] [PubMed] [Google Scholar]
- 85.Kim WK, Ison JC. Survey of the geometric association of domain–domain interfaces. Proteins 2005;61:1075–88. [DOI] [PubMed] [Google Scholar]
- 86.Littler SJ, Hubbard SJ. Conservation of orientation and sequence in protein domain-domain interactions. J Mol Biol 2005;345:1265–79. [DOI] [PubMed] [Google Scholar]
- 87.Shoemaker BA, Zhang D, Thangudu RR, et al. Inferred Biomolecular Interaction Server—a web server to analyze and predict protein interacting partners and binding sites. Nucleic Acids Res 2010;38:D518–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tyagi M, Thangudu RR, Zhang D, et al. Homology inference of protein-protein interactions via conserved binding sites. PLoS One 2012;7:e28896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shoemaker BA, Zhang D, Tyagi M, et al. IBIS (Inferred Biomolecular Interaction Server) reports, predicts and integrates multiple types of conserved interactions for proteins. Nucleic Acids Res 2012;40:D834–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Esmaielbeiki R, Nebel J-C. Scoring docking conformations using predicted protein interfaces. BMC Bioinformatics 2014;15:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Esmaielbeiki R, Nebel J-C. Unbiased protein interface prediction based on ligand diversity quantification. Ger Conf Bioinforma 2012;2012:119–30. [Google Scholar]
- 92.Petrey D, Fischer M, Honig B. Structural relationships among proteins with different global topologies and their implications for function annotation strategies. Proc Natl Acad Sci USA 2009;106:17377–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Russell RB, Sasieni PD, Sternberg MJE. Supersites within superfolds. Binding site similarity in the absence of homology. J Mol Biol 1998;282:903–18. [DOI] [PubMed] [Google Scholar]
- 94.Brylinski M, Skolnick J. A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. Proc Natl Acad Sci USA 2008;105:129–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Konc J, Janežič D. ProBiS algorithm for detection of structurally similar protein binding sites by local structural alignment. Bioinformatics 2010;26:1160–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Konc J, Janežič D. ProBiS: a web server for detection of structurally similar protein binding sites. Nucleic Acids Res 2010;38:W436–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Carl N, Konc J, Vehar B, et al. Protein-protein binding site prediction by local structural alignment. J Chem Inf Model 2010;50:1906–13. [DOI] [PubMed] [Google Scholar]
- 98.Carl N, Konc J, Janezic D. Protein surface conservation in binding sites. J Chem Inf Model 2008;48:1279–86. [DOI] [PubMed] [Google Scholar]
- 99.Zhang QC, Deng L, Fisher M, et al. PredUs: a web server for predicting protein interfaces using structural neighbors. Nucleic Acids Res 2011;39:W283–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang QC, Petrey D, Norel R, et al. Protein interface conservation across structure space. Proc Natl Acad Sci 2010;107:10896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jordan RA, Yasser ELM, Dobbs D, et al. Predicting protein-protein interface residues using local surface structural similarity. BMC Bioinformatics 2012;13:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ahmad S, Mizuguchi K. Partner-aware prediction of interacting residues in protein-protein complexes from sequence data. PLoS One 2011;6:e29104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Amos-Binks A, Patulea C, Pitre S, et al. Binding site prediction for protein-protein interactions and novel motif discovery using re-occurring polypeptide sequences. BMC Bioinformatics 2011;12:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Minhas A, ul Amir F, Geiss BJ, et al. PAIRpred: partner-specific prediction of interacting residues from sequence and structure. Proteins 2014;82:1142–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xue LC, Jordan RA, EL-Manzalawy Y, et al. DockRank: ranking docked conformations using partner-specific sequence homology based protein interface prediction. Proteins 2014;82:250–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.De Vries SJ, Bonvin AM. CPORT: a consensus interface predictor and its performance in prediction-driven docking with HADDOCK. PLoS One 2011;6:e17695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rodrigues JP, Bonvin AM. Integrative computational modeling of protein interactions. FEBS J 2014;281:1988–2003. [DOI] [PubMed] [Google Scholar]
- 108.Vreven T, Hwang H, Pierce BG, et al. Evaluating template-based and template-free protein-protein complex structure prediction. Brief Bioinform 2014;15:169–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fernández-Recio J, Totrov M, Abagyan R. Identification of protein–protein interaction sites from docking energy landscapes. J Mol Biol 2004;335:843–65. [DOI] [PubMed] [Google Scholar]
- 110.Guo F, Li S, Wang L, et al. Protein-protein binding site identification by enumerating the configurations. BMC Bioinformatics 2012;13:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hwang H, Vreven T, Weng Z. Binding interface prediction by combining protein–protein docking results. Proteins 2014;82:57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.De Juan D, Pazos F, Valencia A. Emerging methods in protein co-evolution. Nat Rev Genet 2013;14:249–61. [DOI] [PubMed] [Google Scholar]
- 113.Jones DT, Buchan DW, Cozzetto D, et al. PSICOV: Precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics 2012;28:184–90. [DOI] [PubMed] [Google Scholar]
- 114.Kaján L, Hopf TA, Kalaš M, et al. FreeContact: fast and free software for protein contact prediction from residue co-evolution. BMC Bioinformatics 2014;15:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hopf TA, Schärfe CPI, Rodrigues JP, et al. Sequence co-evolution gives 3D contacts and structures of protein complexes. arXiv Prepr 2014:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Morcos F, Pagnani A, Lunt B, et al. PNAS Plus: direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc Natl Acad Sci USA 2011;108:E1293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Andreani J, Faure G, Guerois R. InterEvScore: a novel coarse-grained interface scoring function using a multi-body statistical potential coupled to evolution. Bioinformatics 2013;29:1742–9. [DOI] [PubMed] [Google Scholar]
- 118.Hamer R, Luo Q, Armitage JP, et al. i-Patch: interprotein contact prediction using local network information. Proteins 2010;78:2781–97. [DOI] [PubMed] [Google Scholar]
- 119.Kass I, Horovitz A. Mapping pathways of allosteric communication in GroEL by analysis of correlated mutations. Proteins 2002;48:611–17. [DOI] [PubMed] [Google Scholar]
- 120.Korber BT, Farber RM, Wolpert DH, et al. Covariation of mutations in the V3 loop of human immunodeficiency virus type 1 envelope protein: an information theoretic analysis. Proc Natl Acad Sci USA 1993;90:7176–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lockless SW, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science 1999;286:295–9. [DOI] [PubMed] [Google Scholar]
- 122.Dekker JP, Fodor A, Aldrich RW, et al. A perturbation-based method for calculating explicit likelihood of evolutionary co-variance in multiple sequence alignments. Bioinformatics 2004;20:1565–72. [DOI] [PubMed] [Google Scholar]
- 123.Reichert JM. Antibodies to watch in 2014. MAbs 2013;6:5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Krawczyk K, Baker T, Shi J, et al. Antibody i-Patch prediction of the antibody binding site improves rigid local antibody-antigen docking. Protein Eng Des Sel 2013;26:621–9. [DOI] [PubMed] [Google Scholar]
- 125.Krawczyk K, Liu X, Baker T, et al. Improving B-cell epitope prediction and its application to global antibody-antigen docking. Bioinformatics 2014;30:2288–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kunik V, Ofran Y. The indistinguishability of epitopes from protein surface is explained by the distinct binding preferences of each of the six antigen-binding loops. Protein Eng Des Sel 2013;26:599–609. [DOI] [PubMed] [Google Scholar]
- 127.Kunik V, Peters B, Ofran Y. Structural consensus among antibodies defines the antigen binding site. PLoS Comput Biol 2012;8:e1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Olimpieri PP, Chailyan A, Tramontano A, et al. Prediction of site-specific interactions in antibody-antigen complexes: the proABC method and server. Bioinformatics 2013;29:2285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kringelum JV, Lundegaard C, Lund O, et al. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput Biol 2012;8:e1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chothia C, Lesk AM. Canonical structures for the hypervariable regions of immunoglobulins. J Mol Biol 1987;196:901–17. [DOI] [PubMed] [Google Scholar]
- 131.Al-Lazikani B, Lesk AM, Chothia C. Standard conformations for the canonical structures of immunoglobulins. J Mol Biol 1997;4:927–48. [DOI] [PubMed] [Google Scholar]
- 132.Wu TT, Kabat EA. An analysis of the sequences of the variable regions of Bence Jones proteins and myeloma light chains and their implications for antibody complementarity. J Exp Med 1970;132:211–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.MacCallum RM, Martin ACR, Thornton JM. Antibody-antigen interactions: contact analysis and binding site topography. J Mol Biol 1996;262:732–45. [DOI] [PubMed] [Google Scholar]
- 134.Peng H-P, Lee KH, Jian J-W, et al. Origins of specificity and affinity in antibody-protein interactions. Proc Natl Acad Sci USA 2014;111:E2656–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Stave JW, Lindpaintner K. Antibody and antigen contact residues define epitope and paratope size and structure. J Immunol 2013;191:1428–35. [DOI] [PubMed] [Google Scholar]
- 136.Idrees S, Ashfaq UA. Structural analysis and epitope prediction of HCV E1 protein isolated in Pakistan: an in-silico approach. Virol J 2013;10:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gershoni JM, Roitburd-Berman A, Siman-Tov DD, et al. Epitope mapping. BioDrugs 2007;21:145–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Irving MB, Pan O, Scott JK. Random-peptide libraries and antigen-fragment libraries for epitope mapping and the development of vaccines and diagnostics. Curr Opin Chem Biol 2001;5:314–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sun J, Xu T, Wang S, et al. Does difference exist between epitope and non-epitope residues? Analysis of the physicochemical and structural properties on conformational epitopes from B-cell protein antigens. Immunome Res 2011;7:1–11. [Google Scholar]
- 140.Reimer U. Prediction of linear B-cell epitopes. Methods Mol Biol 2009;524:335–44. [DOI] [PubMed] [Google Scholar]
- 141.Kulkarni-Kale U, Bhosle S, Kolaskar AS. CEP: a conformational epitope prediction server. Nucleic Acids Res 2005;33:W168–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kringelum JV, Lundegaard C, Lund O, et al. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput Biol 2012;8:e1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Haste Andersen P, Nielsen M, Lund O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci 2006;15:2558–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ponomarenko J, Bui H-H, Li W, et al. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics 2008;9:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ravindranath MH, Pham T, El-Awar N, et al. Anti-HLA-E mAb 3D12 mimics MEM-E/02 in binding to HLA-B and HLA-C alleles: web-tools validate the immunogenic epitopes of HLA-E recognized by the antibodies. Mol Immunol 2011;48:423–30. [DOI] [PubMed] [Google Scholar]
- 146.Sweredoski MJ, Baldi P. PEPITO: improved discontinuous B-cell epitope prediction using multiple distance thresholds and half sphere exposure. Bioinformatics 2008;24:1459–60. [DOI] [PubMed] [Google Scholar]
- 147.Moreau V, Fleury C, Piquer D, et al. PEPOP: computational design of immunogenic peptides. BMC Bioinformatics 2008;9:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sun J, Wu D, Xu T, et al. SEPPA: a computational server for spatial epitope prediction of protein antigens. Nucleic Acids Res 2009;37:W612–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Qi T, Qiu T, Zhang Q, et al. SEPPA 2.0 - more refined server to predict spatial epitope considering species of immune host and subcellular localization of protein antigen. Nucleic Acids Res 2014;42:W59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rubinstein ND, Mayrose I, Martz E, et al. Epitopia: a web-server for predicting B-cell epitopes. BMC Bioinformatics 2009;10:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wu WK, Chung WC, Chang HT, et al. B-cell conformational epitope prediction based on structural relationship and antigenic characteristics. Proceeding of the International Conference on Complex, Intelligent and Software Intensive Systems 2009; pp. 830–5. Fukuoka. [Google Scholar]
- 152.Sun P, Ju H, Liu Z, et al. Bioinformatics resources and tools for conformational B-cell epitope prediction. Comput Math Methods Med 2013;2013:943636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liang S, Zheng D, Zhang C, et al. Prediction of antigenic epitopes on protein surfaces by consensus scoring. BMC Bioinformatics 2009;10:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liang S, Zheng D, Standley DM, et al. EPSVR and EPMeta: prediction of antigenic epitopes using support vector regression and multiple server results. BMC Bioinformatics 2010;11:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ansari HR, Flower DR, Raghava GPS. AntigenDB: an immunoinformatics database of pathogen antigens. Nucleic Acids Res 2010;38:D847–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Huang J, Honda W. CED: a conformational epitope database. BMC Immunol 2006;7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chailyan A, Tramontano A, Marcatili P. A database of immunoglobulins with integrated tools: DIGIT. Nucleic Acids Res 2012;40:D1230–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ponomarenko J, Papangelopoulos N, Zajonc DM, et al. IEDB-3D: structural data within the immune epitope database. Nucleic Acids Res 2011;39:D1164–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kim Y, Ponomarenko J, Zhu Z, et al. Immune epitope database analysis resource. Nucleic Acids Res 2012;40:W525–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Vita R, Zarebski L, Greenbaum JA, et al. The immune epitope database 2.0. Nucleic Acids Res 2010;38:D854–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ehrenmann F, Kaas Q, Lefranc M. IMGT/3Dstructure-DB and IMGT/DomainGapAlign: a database and a tool for immunoglobulins or antibodies, T cell receptors, MHC, IgSF and MhcSF. Nucleic Acids Res 2010;38:D301–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Dunbar J, Krawczyk K, Leem J, et al. SAbDab: the structural antibody database. Nucleic Acids Res 2013;42:1140–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shirai H, Prades C, Vita R, et al. Antibody informatics for drug discovery. Biochim Biophys Acta 2014;1844:2002–15. [DOI] [PubMed] [Google Scholar]
- 164.Sela-Culang I, Benhnia MR, Matho MH, et al. Using a combined computational-experimental approach to predict antibody-specific B cell epitopes. Structure 2014;22:646–57. [DOI] [PubMed] [Google Scholar]
- 165.Rapberger R, Lukas A, Mayer B. Identification of discontinuous antigenic determinants on proteins based on shape complementarities. J Mol Recognit 2007;20:113–21. [DOI] [PubMed] [Google Scholar]
- 166.Soga S, Kuroda D, Shirai H, et al. Use of amino acid composition to predict epitope residues of individual antibodies. Protein Eng Des Sel 2010;23:441–8. [DOI] [PubMed] [Google Scholar]
- 167.Zhao L, Wong L, Li J. Antibody-specified B-cell epitope prediction in line with the principle of context-awareness. IEEE/ACM Trans Comput Biol Bioinformatics 2011;8:1483–94. [DOI] [PubMed] [Google Scholar]
- 168.Chuang G-Y, Acharya P, Schmidt SD, et al. Residue-level prediction of HIV-1 antibody epitopes based on neutralization of diverse viral strains. J Virol 2013;87:10047–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mitchell JC, Kerr R, Ten Eyck LF. Rapid atomic density methods for molecular shape characterization. J Mol Graph Model 2001;19:325–30. [DOI] [PubMed] [Google Scholar]
- 170.Camacho CJ, Zhang C. FastContact: rapid estimate of contact and binding free energies. Bioinformatics 2005;21:2534–36. [DOI] [PubMed] [Google Scholar]
- 171.Zhao L, Li J. Mining for the antibody-antigen interacting associations that predict the B cell epitopes. BMC Struct Biol 2010;10:S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ross GA, Morris GM, Biggin PC. One size does not fit all: the limits of structure-based models in drug discovery. J Chem Theory Comput 2013;9:4266–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Vakser IA. Low-resolution structural modeling of protein interactome. Curr Opin Struct Biol 2013;23:198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kundrotas PJ, Vakser IA. Accuracy of protein-protein binding sites in high-throughput template-based modeling. PLoS Comput Biol 2010;6:e1000727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang QC, Petrey D, Garzón JI, et al. PrePPI: a structure-informed database of protein–protein interactions. Nucleic Acids Res 2013;41:D828–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhang QC, Petrey D, Deng L, et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature 2012;490:556–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wass MN, Fuentes G, Pons C, et al. Towards the prediction of protein interaction partners using physical docking. Mol Syst Biol 2011;7:469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Schoenrock A, Samanfar B, Pitre S, et al. Efficient prediction of human protein-protein interactions at a global scale. BMC Bioinformatics 2014;15:383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Janin J. Docking predictions of protein-protein interactions and their assessment: the CAPRI experiment. In: Identification of Ligand Binding Site and Protein-Protein Interaction Area. 2013, Vol. 8, Springer Netherlands, pp. 87–104. [Google Scholar]
- 180.Lensink MF, Wodak SJ. Docking, scoring, and affinity prediction in CAPRI. Proteins 2013;81:2082–95. [DOI] [PubMed] [Google Scholar]
- 181.Wang B, Chen P, Zhang J. Protein interface residues prediction based on amino acid properties only. Bio-Inspired Comput Appl 2012;448–52. [Google Scholar]
- 182.Chakrabarti P, Janin J. Dissecting protein-protein recognition sites. Proteins Struct Funct Genet 2002;47:334–43. [DOI] [PubMed] [Google Scholar]
- 183.Lichtarge O, Sowa ME. Evolutionary predictions of binding surfaces and interactions. Curr Opin Struct Biol 2002;12:21–7. [DOI] [PubMed] [Google Scholar]
- 184.Hwang H, Vreven T, Janin J, et al. Protein–protein docking benchmark version 4.0. Proteins Struct Funct Bioinformatics 2010;78:3111–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Negi SS, Braun W. Statistical analysis of physical–chemical properties and prediction of protein–protein interfaces. J Mol Model 2007;13:1157–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Mintseris J, Wiehe K, Pierce B, et al. Protein–protein docking benchmark 2.0: an update. Proteins Struct Funct Bioinformatics 2005;60:214–6. [DOI] [PubMed] [Google Scholar]
- 187.Nooren I, Thornton JM. Structural characterisation and functional significance of transient protein–protein interactions. J. Mol. Biol 2003;325:991–1018. [DOI] [PubMed] [Google Scholar]
- 188.Chen H, Zhou H-X. Prediction of solvent accessibility and sites of deleterious mutations from protein sequence. Nucleic Acids Res 2005;33:3193–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Fariselli P, Zauli A, Rossi I, et al. A neural network method to improve prediction of protein-protein interaction sites in heterocomplexes. In: Neural Networks Signal Process. 2003, NNSP’03. 2003 IEEE 13th Work 2003; pp. 33–41. [Google Scholar]
- 190.Hwang H, Pierce B, Mintseris J, et al. Protein–protein docking benchmark version 3.0. Proteins Struct. Funct. Bioinformatics 2008;73:705–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lensink MF, Wodak SJ. Blind predictions of protein interfaces by docking calculations in CAPRI. Proteins 2010;78:3085–95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.