Abstract
The study aimed to identify proteins regulated by the cardiovascular protective peptide angiotensin-(1-7) and to determine potential intracellular signaling cascades.
Human endothelial cells were stimulated with Ang-(1-7) for 1h, 3h, 6h, and 9h. Peptide effects on intracellular signaling were assessed via antibody microarray, containing antibodies against 725 proteins. Bioinformatics software was used to identify affected intracellular signaling pathways. Microarray data was verified exemplarily by Western blot, Real-Time RT-PCR, and immunohistochemical studies.
The microarray identified 110 regulated proteins after 1h, 119 after 3h, 31 after 6h, and 86 after 9h Ang-(1-7) stimulation. Regulated proteins were associated with high significance to several metabolic pathways like “Molecular Mechanism of Cancer” and “p53 signaling” in a time dependent manner. Exemplarily, Western blots for the E3-type small ubiquitin-like modifier ligase PIAS2 confirmed the microarray data and displayed a decrease by more than 50% after Ang-(1-7) stimulation at 1h and 3h without affecting its mRNA. Immunohistochemical studies with PIAS2 in human endothelial cells showed a decrease in cytoplasmic PIAS2 after Ang-(1-7). The Ang-(1-7) mediated decrease of PIAS2 was reproduced in other endothelial cell types. The results suggest that angiotensin-(1-7) plays a role in metabolic pathways related to cell death and cell survival in human endothelial cells.
Graphical Abstract
Introduction
The renin-angiotensin system (RAS) is one of the best characterized hormonal systems. It is involved in the regulation of several important physiological processes, including blood pressure regulation, sodium and water balance, and electrolyte homeostasis [1]. Initially, the RAS was considered to be an endocrine system whose bioactive metabolite angiotensin II (AngII) was the final product of the system [2, 3] and that the actions of this peptide were mediated by the AngII receptors type 1 (AT1) and type 2 (AT2) [1, 4].
For many years, AngII was considered to be the only biologically active peptide of the RAS, but evidence has accumulated that both shorter and longer angiotensin metabolites, such as angiotensin III (AngIII)[5-7], angiotensin IV (AngIV) [8, 9], angiotensin-(1-9) (Ang-(1-9)) [10] and the heptapeptide angiotensin-(1-7) (Ang-(1-7)) [11, 12] also possess potent biological activity. The main pathway for Ang-(1-7) formation is hydrolysation of AngII by the carboxypeptidase angiotensin converting enzyme 2 (ACE2) [13, 14]. The heptapeptide Ang-(1-7) has been the focus of intense interest due to its ability to counteract the detrimental actions of AngII, including vasoconstriction [11]. Furthermore, Ang-(1-7) has beneficial effects on cardiac function [15, 16], counteracts the growth-stimulating effects of AngII in cardiomyocytes and vascular smooth muscle cells [17, 18], and is involved in the control of water and electrolyte homeostasis [19]. It has also been reported that the heptapeptide participates in wound healing [20], learning and memory [21], hematopoiesis [22], and cancer [23-25].
Despite many publications regarding the physiological actions of Ang-(1-7) and its beneficial effects under pathophysiological circumstances, the intracellular signaling initiated by this peptide has remained elusive. We have previously demonstrated that the receptor Mas is associated with Ang-(1-7)-stimulated intracellular signaling [26]. It has been suggested recently that Ang-(1-7) influences the phosphorylation status of 79 proteins, including several downstream effectors of insulin signaling and the antiproliferative and antitumorigenic FOXO1, which might account for some of the antitumorigenic effects of the heptapeptide [27]. The effects of Ang-(1-7) on intracellular signaling, however, have not be delineated. It was the aim of our study to identify regulated intracellular proteins, verify them, and define possible intracellular signaling pathways activated by the heptapeptide. For this purpose, we used a time resolved antibody microarray approach, which enabled us to screen for the regulation of 725 proteins in endothelial cells. We further evaluated the findings from the microarray by Western blot, Real-Time RT-PCR, and immunohistochemical studies.
Material and methods
Materials and chemicals
Human Umbilical Vein Endothelial Cells (HUVEC), the cell culture medium (EGM-2 Bulletkit), Trypsin/EDTA, and the Trypsin Neutralizing Solution (TNS) were purchased from Lonza (Basel, Switzerland). Human Dermal Microvascular Endothelial Cells (HDMEC) and Mouse Brain Endothelial Cells (bEnd.3) were a kind gift from Dr. Burkhard Wiesner (FMP Berlin, Germany), whereas Dulbecco's Modified Eagle Medium (DMEM-Bulletkit), Dulbecco's Phosphate Buffered Saline (DPBS) and fetal bovine serum (FBS) were from Gibco (Darmstadt, Germany). Ang-(1-7) was purchased from Bachem (Bubendorf, Switzerland). The mono-reactive dyes were from GE Healthcare (Little Chalfont, UK), the Panorama Antibody Microarry-XPRESS Profiler725 Kit from Sigma-Aldrich (St. Louis, USA). For Western blot experiments, the PIAS2 antibody (ab105361) was from abcam (Cambridge, UK), the FBI-1/Pokemon (F9429), GAPDH (G9545), and secondary antibodies (A9169 and A9044) were purchased from Sigma-Aldrich, whereas for the immunofluorescence studies the PIAS2 antibody was purchased from antibodies-online GmbH (Aachen, Germany) and the Cy™3-conjugated AffiniPure goat anti-mouse IgG antibody from DIANOVA (Hamburg, Germany). Real-Time RT-PCR primers for PIAS2 (QT01012172), FBI-1/Pokemon (QT00226688), and GAPDH (QT00079247), and QuantiTect SYBR Green RT-PCR Kit were from Qiagen (Hilden, Germany). All other chemicals were obtained from Sigma-Aldrich.
Cell culture and cell stimulation
Endothelial cells were grown on 100mm dishes according to manufacturer's specifications in EBM-2 or DMEM medium at 37°C in a humidified incubator (Sanyo, Watford, UK) in an atmosphere of 5% CO2. Cells used were passage 6 for HUVEC, passage 10 for HDMEC and passage 11 for bEnd.3. When the cells reached 70% confluence, they were washed two times with DPBS and serum starved for 1h in supplements-free EBM-2 or DMEM. Afterwards, 10−7M Ang-(1-7) was added and cells were incubated for 30min, 1h, 2h, 3h, 6h, and 9h. Cells only treated with the peptide solvent (DPBS) were used as control. The 10−7M concentration used for Ang-(1-7) was derived from data of former cell culture studies (e.g. [28] and unpublished experiments).
Antibody microarray
The antibody microarray was performed as described in the Panorama Antibody Microarray-XPRESS Profiler725 Kit manual. A concentration of 1mg/ml of protein extract from control and Ang-(1-7)-stimulated HUVEC were labeled with Cy™3 or Cy™5 dye. For two approaches, control samples were labeled with Cy™5, whereas stimulated samples were labeled with Cy™3. In an additional set, control samples were labeled with Cy™3, whereas stimulated samples were labeled with Cy™5. Free Cy™3/ Cy™5 was removed, and the Panorama Antibody Microarray slide was incubated with the labeled samples for 45 min. The slide was washed with provided washing buffer and air-dried before images were acquired, using a GenePix 4100A Microarray Scanner (Molecular Devices, Sunnyvale, USA). Scan resolution was 5μm per pixel, and the photomultiplier tube gains were manually set for each dye to ensure a normal ratio of 1 (±0.1). Data was imported into Acuity 4.0 software (Molecular Devices, Sunnyvale, USA) and normalized using the nonlinear Lowess normalization method. Applied criteria for spot quality control were <3% saturated pixels, not flagged as absent, relatively uniform intensity and uniform background, and detectable above background. The log ratios for the relative expression of the spots that passed quality control were converted to fold change. The program IPA (Ingenuity Systems, Redwood City, USA) was used to associate Ang-(1-7) influenced proteins with potential metabolic pathways.
Western Blotting
Endothelial cells were harvested and lysed for Western blot analysis (5 independent samples per blot). The medium was removed, and cells were washed two times with ice-cold DPBS. Cells were lysed and the samples centrifuged for 10s at 10.000 × g in a microcentrifuge to remove cellular debris. Protein content of the lysates was determined by BCA protein assay (Thermo Fisher, Waltham, USA). An amount of 20μg of protein was denatured with Laemmli buffer, incubated at 95°C for 10min, and subjected to SDS-polyacrylamide gel electrophoresis. The proteins were transferred onto Immobilon-P Membrane (Millipore, Billerica, USA), and the membrane was blocked with 5% [w/v] skimmed milk in TBS, pH 7.4 and 0.1% [v/v] Tween 20 for 1h at room temperature. The membrane was then incubated with either PIAS2 (1:1000, 3% [w/v] skimmed milk) or FBI-1/Pokemon (1:1000, 5% [w/v] skimmed milk) antibody overnight at 4°C. After incubation with peroxidase-linked secondary antibody (1:1000, 5% [w/v] skimmed milk) for 1h at room temperature, immunoreactive proteins were visualized by ECL reagent and autoradiography film (GE Healthcare, Little Chalfont, UK). Band density was quantified using ImageJ software (National Institute of Health, http://rsb.info.nih.gov/ij/) and normalized to the amount of GAPDH in each experiment.
Real-Time RT-PCR
HUVEC were washed twice with ice-cold DPBS and lysed. Total RNA was isolated using TRIzol Reagent (Invitrogen, Carlsbad, USA) according to manufacturer's instructions. RNA concentration was determined with a NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, USA). The samples were prepared regarding the instructions of the QuantiTect SYBR Green RT-PCR kit with 10ng RNA per reaction. Real-Time RT-PCR was performed in a Roche LightCycler (Roche, Penzberg, Germany) and results were normalized to GAPDH expression.
Immunohistochemical studies
HUVEC were seeded into sterile 8-well μ-Slides (ibidi GmbH, Martinsried/Munich, Germany). At 80% confluence cells were washed once with DPBS and serum starved in supplement-free EBM-2 for 1h followed by stimulation with 10−7M Ang-(1-7) for 1h at 37°C. Cells only treated with the peptide solvent (DPBS) served as control. Stimulation was terminated by cell fixation with 4% paraformaldehyde (Carl Roth GmbH + Co. KG, Karlsruhe, Germany) for 30 min at room temperature. After three times washing with DPBS cells were permeabilized and non-specific binding was blocked with 0.2% Triton-X 100 (Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany) containing 5% goat serum (DIANOVA, Hamburg, Germany) for 30min at room temperature. Cells were incubated with anti-PIAS2 mouse monoclonal antibody (1μg/ml) in DPBS over night at 4°C, washed three times with DPBS and subsequently incubated with Cy™3-conjugated AffiniPure goat anti-mouse IgG (7.5μg/ml) in DPBS for 30min at room temperature. After three times washing with DPBS cells were mounted with Immunoselect Antifading Mounting Medium DAPI (DIANOVA, Hamburg, Germany). Fluorescence images were obtained using an Axio Observer.A1 microscope fitted with an EC Plan-NEOFLUAR 63x/1.25 Oil objective and an AxioCam ICc1 camera (Zeiss, Jena, Germany). The fluorescence was detected using filter set 49 (blue for DAPI DNA stain) and filter set 43 (red for Cy™3). Images were collected at 1392 × 1038 resolution with 10x optical zoom using the AxioVision software (Release 4.8.2) at room temperature. The final composite image was created using Photoshop 10.0 (Adobe Systems GmbH, Munich, Germany).
Statistics
For the antibody microarray data, a significant expression fold change of one value ≥1.8, or of 2 out of 3 values of expression fold change ≥1.5, was applied as cut off criteria. Association of regulated proteins to metabolic pathways was done by IPA software (Ingenuity Systems, Redwood City, USA). The software calculated a p-value using the right tailed Fisher Exact test. The p-value gives the probability that the association between regulated detected proteins and the pathways is due to random chance. The software considers a p-value <0.05 as statistically significant. To compare the differences between Ang-(1-7) stimulation versus solvent stimulation in Western blot and Real-Time RT-PCR experiments, the Student's t-test was used (Graph Pad Prism 5.01; Graph Pad Software Inc., San Diego, USA). So here, a p-value <0.05 was considered to be significant (* P ≤ 0.05, ** P ≤ 0.01, and *** P ≤ 0.001).
Results
Identification of intracellular proteins and metabolic pathways regulated by Ang-(1-7)
To investigate Ang-(1-7) mediated intracellular signaling pathways in human endothelial cells, we used an antibody microarray in a time resolved approach. Stimulation of HUVEC with Ang-(1-7) for 1h changed the protein level of 110, for 3h of 119, for 6h of 31, and for 9h of 86 proteins (data not shown). To rule out possible false-positive results, the datasets were screened for repeatedly identified differentially expressed proteins (RIDEPs) [29]. These are proteins that are detected as differentially expressed in a microarray regardless of the experiment, tissue or species and such regulation can not be confirmed by follow up methods as Western blot [30, 31]. The RIDEPs Zyxin, 14-3-3, BID, etc. were present in the microarray data for all four time points and two of them, 14-3-3 and Zyxin, were even verified by Western blot. As expected, their regulation in the microarray could not be confirmed by Western blots (data not shown).
Subtracting the RIDEPs, the data showed that MAP1b, Pyk2(pTyr579), PIAS2, Cdk3, PP2A, and Mad2 were the six proteins with highest values of expression fold change after 1h of stimulation with the heptapeptide (Table 1). While regulation of Mad2 by Ang-(1-7) was not detected at any other time point, MAP1b was under the highest 25 values after 3h, whereas the four other proteins were still influenced by the heptapetide at a lower level (not among the highest 25 values of expression fold change). However, none of these five was altered at 6h or 9h.
Table 1.
The 25 proteins with the highest detected fold change values after 1h incubation of HUVEC with 10−7M Ang-(1-7). The order of the numbers is oriented on the highest single value. Expression fold change lower than 1.5 is given in hyphen. Data that could not be detected is marked as n.d. Proteins marked in italic show repeatedly identified differentially expressed proteins (RIDEPs). The mentioned dye indicates with which dye the unstimulated sample was labeled with.
| Protein | Antibody id | Cy3 | Cy5 | Cy5 |
|---|---|---|---|---|
| 1. Zyxin | Z0377 | 7.07 | 5.82 | 2.07 |
| 2. MAP1b | M4528 | 5.46 | 3.26 | - |
| 3. Pyk2 (pTyr579) | P7114 | 5.45 | 1.80 | n.d. |
| 4. PIAS2 | P9498 | 5.42 | 2.64 | 1.64 |
| 5. hABH3 | A8353 | 5.20 | 2.96 | - |
| 6. Cdk3 | C9987 | 5.05 | 2.47 | n.d. |
| 7. TRAIL | T9191 | 4.08 | 2.31 | - |
| 8. PP2A | P8109 | 4.02 | 2.31 | - |
| 9. 14-3-3 ϑ/τ | T5942 | - | 3.94 | 2.98 |
| 10. Mad2 | M8694 | 3.92 | - | - |
| 11. F1Aα | F3428 | 3.72 | 1.86 | - |
| 12. GRP75 | G4170 | 3.69 | 1.71 | n.d. |
| 13. BLK | B8928 | - | 3.66 | n.d. |
| 14. BID | B3183 | 3.63 | - | 1.66 |
| 15. PPP2R2C | P5359 | 3.55 | n.d | - |
| 16. FAK (pSer772) | F9051 | 3.34 | - | n.d. |
| 17. MDMX | M0445 | 3.33 | 3.06 | - |
| 18. G9a Methyltransferase | G6919 | 3.32 | - | - |
| 19. NeurabinII | N5162 | 3.20 | - | - |
| 20. OGlcNAc | O6264 | 3.04 | - | - |
| 21. Neurabin I | N4412 | 3.00 | - | - |
| 22. BclxL | B9429 | 2.68 | 2.88 | 1.58 |
| 23. β-Tubulin | T5201 | 2.80 | 2.85 | - |
| 24. RNaseL | R3529 | 1.89 | 2.71 | - |
| 25. TSG101 | T5826 | 2.70 | 1.95 | - |
At 3h of stimulation, FXR2, RAIDD, PINCH-1, Zip Kinase, and DNase II were the proteins with the highest values (Table 2). However, except DNase II, the other four of these proteins were also detected after 1h of stimulation at lower levels of expression fold change. Stimulation of HUVEC with Ang-(1-7) for 6h resulted in the most pronounced regulation of FAK (pTyr577), Heat Shock Factor 1, Protein kinase Cβ1 and Cβ2, DcR1, and Tau (pSer199/202) (Table 3). Except for FAK (pTyr577) and Protein Kinase Cβ1 and Cβ2 none of these proteins were detected in the antibody microarray at any other time point. The 10 proteins with the highest values at 9h (Table 4) are with the exception of H3 (Ac-Lys9) also present at least once among the most effected (25 highest values) proteins for other time points. Interestingly, the regulation of PUMA/bbc3, FBI-1/Pokemon, and Caspase13 could be detected for the first time 6h after the stimulation with Ang-(1-7), where they were between the 10 and 25 highest values. However, the microarray data showed also two proteins, TRAIL and Bmf, which were among the most regulated proteins, but only at three out of four time points.
Table 2.
The 25 proteins with the highest detected fold change values after 3h incubation of HUVEC with 10−7M Ang-(1-7). The order of the numbers is oriented on the highest single value. Expression fold change lower than 1.5 is given in hyphen. Data that could not be detected is marked as n.d. Proteins marked in italic show repeatedly identified differentially expressed proteins (RIDEPs). The mentioned dye indicates with which dye the unstimulated sample was labeled with.
| Protein | Antibody id | Cy3 | Cy5 | Cy5 |
|---|---|---|---|---|
| 1. Zyxin | Z0377 | 5.18 | 5.82 | 5.41 |
| 2. Bmf | B1684 | 4.78 | 2.38 | 2.01 |
| 3. FXR2 | F1554 | 1.65 | - | 4.19 |
| 4. MTBP | M3566 | 4.05 | 1.79 | 2.39 |
| 5. 14-3-3 ϑ/τ | T5942 | n.d. | 3.94 | n.d. |
| 6. RAIDD | R9775 | 3.70 | - | 1.52 |
| 7. BLK | B8928 | n.d. | 3.66 | n.d. |
| 8. PINCH-1 | P9371 | 3.57 | 2.61 | 2.39 |
| 9. Zip Kinase | Z0134 | 3.55 | 1.66 | 1.99 |
| 10. DNase II | D1689 | 3.39 | - | 2.47 |
| 11. ADAM17 | T5442 | n.d. | n.d. | 3.34 |
| 12. Rnase L | R3529 | 3.33 | 2.71 | 3.08 |
| 13. HDAC7 | H6663 | 2.47 | 1.55 | 3.28 |
| 14. MAP1b | M4528 | n.d. | 3.26 | n.d. |
| 15. CtBP1 | C8741 | 1.96 | - | 3.23 |
| 16. ERK2 | M7431 | 3.21 | 1.69 | 2.09 |
| 17. C-Raf (pSer621) | R1151 | 3.09 | 2.36 | 1.86 |
| 18. MDMX | M0445 | 2.62 | 3.06 | 2.91 |
| 19. DcR2 | D3188 | 3.00 | - | 2.31 |
| 20. hABH3 | A8353 | n.d. | 2.96 | n.d. |
| 21. BNIP3 | B7931 | n.d. | - | 2.89 |
| 22. APRIL | A1726 | 2.89 | 1.95 | - |
| 23. BclxL | B9429 | 2.02 | 2.88 | - |
| 24. NBS1 | N9287 | - | - | 2.87 |
| 25. EGF | E2520 | 2.00 | - | 2.86 |
Table 3.
The 25 proteins with the highest detected fold change values after 6h incubation of HUVEC with 10−7M Ang-(1-7). The order of the numbers is oriented on the highest single value. Expression fold change lower than 1.5 is given in hyphen. Data that could not be detected is marked as n.d. Proteins marked in italic show repeatedly identified differentially expressed proteins (RIDEPs). The mentioned dye indicates with which dye the unstimulated sample was labeled with.
| Protein | Antibody id | Cy3 | Cy5 | Cy5 |
|---|---|---|---|---|
| 1. Bmf | B1559 | 4.33 | - | - |
| 2. 14-3-3 ϑ/τ | T5942 | n.d. | 3.37 | 2.27 |
| 3. FAK (pTyr577) | F8926 | 3.36 | - | n.d. |
| 4. Heat Shock Factor 1 | H4163 | - | 3.20 | - |
| 5. MTBP | M3566 | 2.90 | 1.60 | - |
| 6. TRAIL | T9191 | 2.86 | n.d. | - |
| 7. Protein Kinase Cβ1 | P6959 | 2.15 | 2.82 | - |
| 8. Protein Kinase Cβ2 | P3203 | 2.58 | - | - |
| 9. DcR1 | D3566 | 2.51 | n.d. | n.d. |
| 10. Tau (pSer199/202) | T6819 | 2.43 | n.d. | - |
| 11. Caspase 13 | C8854 | - | 2.38 | - |
| 12. Sir2 | S5313 | 2.32 | - | - |
| 13. FBI-1/Pokemon | F9429 | 2.31 | n.d. | - |
| 14. c-Raf (pSer621) | R1151 | 1.84 | 2.28 | n.d. |
| 15. FAK (pTyr397) | F7926 | 2.21 | - | 1.65 |
| 16. Zyxin | Z0377 | - | - | 2.18 |
| 17. BclxL | B9429 | - | - | 2.14 |
| 18. Pinin | P0084 | - | 2.07 | - |
| 19. Vinculin | V4505 | 2.03 | - | - |
| 20. Cathepsin D | C0715 | - | 2.03 | - |
| 21. Rad17 | R8029 | 2.02 | n.d. | - |
| 22. Cytokeratin CK5 | C7785 | - | 1.95 | - |
| 23. PUMA/bbc3 | P4743 | 1.91 | - | - |
| 24. p19INK4d | P4354 | - | 1.91 | - |
| 25. MAP1 (Light Chain) | M6783 | 1.89 | n.d. | - |
Table 4.
The 25 proteins with the highest detected fold change values after 9h incubation of HUVEC with 10−7M Ang-(1-7). The order of the numbers is oriented on the highest single value. Expression fold change lower than 1.5 is given in hyphen. Data that could not be detected is marked as n.d. Proteins marked in italic show repeatedly identified differentially expressed proteins (RIDEPs). The mentioned dye indicates with which dye the unstimulated sample was labeled with.
| Protein | Antibody id | Cy3 | Cy5 | Cy5 |
|---|---|---|---|---|
| 1. TRAIL | T9191 | 9.45 | - | - |
| 2. PUMA/bbc3 | P4743 | 6.77 | - | - |
| 3. FBI-1/Pokemon | F9429 | 6.54 | n.d. | - |
| 4. Caspase 13 | C8854 | 6.45 | n.d. | - |
| 5. Bmf | B1559 | 5.45 | 1.99 | - |
| 6. BclxL | B9429 | 2.72 | 2.19 | 5.34 |
| 7. 14-3-3 ϑ/τ | T5942 | n.d. | n.d. | 5.01 |
| 8. GRP75 | G4170 | 4.82 | - | - |
| 9. H3 (Ac-Lys9) | H0913 | 4.52 | - | - |
| 10. MyD88 | M9934 | 4.38 | n.d. | 3.09 |
| 11. p53DINP1/SIP | P4868 | 4.28 | - | 1.61 |
| 12. ILP-2 | I4782 | 4.11 | 1.66 | - |
| 13. Zyxin | Z0377 | 4.06 | 1.91 | 3.40 |
| 14. TRF1 | T1948 | 3.91 | - | - |
| 15. MBNL1 | M3320 | 3.74 | - | - |
| 16. MADD | M5683 | 3.61 | - | - |
| 17. HDAC7 | H2537 | 3.54 | 1.57 | n.d. |
| 18. Protein Kinase C | P5704 | 3.31 | - | - |
| 19. HSP 90 | H1775 | n.d. | 3.16 | - |
| 20. γ Parvin | P5746 | 2.99 | - | n.d. |
| 21. MSK1 | M5437 | 2.90 | - | - |
| 22. hnRNP-A1 | R4528 | 2.84 | - | - |
| 23. Protein Kinase Bα | P1601 | 2.75 | 1.77 | - |
| 24. hBRM/hSNF2α (KR-17) | H9787 | n.d. | 2.73 | - |
| 25. Grb-2 | G2791 | 2.71 | - | - |
Bioinformatics software was used to identify potential intracellular pathways associated to Ang-(1-7) signaling. Three of these pathways, “Molecular Mechanism of Cancer”, “p53 signaling”, and “Cyclins and Cell Cycle regulation”, were not only significantly regulated at all four time points but have been always under the 10 pathways with highest p-values (Table 5). Other metabolic pathways like “Death Receptor signaling”, “Apoptosis signaling”, “Chronic Myeloid Leukemia signaling”, and “Small Cell Lung Cancer signaling” were also associated to the regulated proteins, but with lower significance. Furthermore, the regulation of identified proteins fitted into the signaling cascades in a time dependent manner, as exemplarily comparison of microarray data with the metabolic pathway “Molecular Mechanism of Cancer” showed regulated ERK1/2 and SMAD4 after stimulation for 1 h with Ang-(1-7), whereas after 3 h their downstream targets p19 and p21 were affected (data not shown).
Table 5.
The 10 metabolic pathways with the highest p-values associated by the IPA software to each of the different antibody microarray sets. The 10 highest p-values of each dataset are given in bold, whereas other p-values and ratios are marked in Italic. The ratio states the number of proteins detected in the microarray versus the total number of proteins being part of the pathway. Data that did not reach significance is marked as n.s.
| 1h | 3h | 6h | 9h | |||||
|---|---|---|---|---|---|---|---|---|
| Pathway | p-value | ratio | p-value | ratio | p-value | ratio | p-value | ratio |
| 1. Molecular Mechanism of Cancer | 3.67 | 17/379 | 1.20 | 32/379 1.33 | 9/379 1.14 | 19/379 | ||
| E-11 | (4.5%) | E-22 | (8.4%) | E-09 | (2.4%) | E-15 | (5.0%) | |
| 6.57 | 9/96 | 3.00 | 19/96 | 2.99 | 5/96 | 1.73 | 15/96 | |
| 2. p53 signaling | E-09 | (9.4%) | E-20 | (19.7%) | E-07 | (5.2%) | E-19 | (15.6%) |
| 3. Glucocorticoid Receptor signaling | 9.70 | 13/295 | 5.01 | 15/295 | n.s. | n.s. | 1.26 | 12/295 |
| E-09 | (4.4%) | E-08 | (5.1%) | 6.31 | 3/73 | E-08 | (4.1%) | |
| 4. Chemokine signaling | 2.44 | 7/73 | 5.74 | 8/73 | E-04 | (4.1%) | n.s. | n.s. |
| 5. Cyclins and Cell Cycle regulation | E-07 | (9.6%) | E-08 | (11.0%) | 1.31 | 5/89 | 2.51 | 10/89 |
| 6.24 | 7/89 | 2.70 | 14/89 | E-07 | (5.6%) | E-12 | (11.2%) | |
| 6. PI3K/AKT signaling | E-07 | (7.9%) | E-14 | (15.7%) | 3.31 | 4/140 | 1.58 | 9/140 |
| 7. ATM signaling | 7.24 | 8/140 | 1.58 | 13/140 | E-05 | (2.9%) | E-08 | (6.4%) |
| 8. VEGF signaling | E-07 | (5.7%) | E-10 | (9.3%) | n.s. | n.s. | 3.94 | 10/54 |
| 9. Apoptosis signaling | 7.59 | 6/54 | 5.01 | 9/54 | 2.14 | 5/99 | E-14 | (18.5%) |
| 10. Death Receptor signaling | E-07 | (11.1%) | E-09 | (16.7%) | E-07 | (5.1%) | 3.47 | 7/99 |
| 8.91 | 7/99 | 5.01 | 12/99 | 6.66 | 3/96 | E-07 | (7.1%) | |
| 11. Chronic Myeloid Leukemia signaling | E-07 | (7.1%) | E-11 | (12.1%) | E-04 | (3.1%) | 4.07 | 7/96 |
| 1.26 | 7/96 | 2.51 | 11/96 | n.s. | n.s. | E-07 | (7.3%) | |
| 12. Pancreatic Adenocarcinoma signaling | E-06 | (7.3%) | E-09 | (11.5%) | 3.16 | 5/105 | n.s. | n.s. |
| 1.58 | 6/95 | 3.51 | 12/65 | E-07 | (4.8%) | 1.71 | 12/105 | |
| E-06 | (9.9%) | E-13 | (18.5%) | 2.29 | 4/119 | E-14 | (11.4%) | |
| 13. PTEN signaling | 2.00 | 7/105 | 1.59 | 15/105 | E-05 | (3.4%) | 7.94 | 10/119 |
| 14. Huntington's Disease signaling | E-06 | (6.7%) | E-14 | (14.3%) | 2.69 | 4/124 | E-11 | (8.4%) |
| 5.01 | 7/119 | 2.00 | 11/119 | E-05 | (3.2%) | n.s. | n.s. | |
| 15. Small Cell Lung Cancer signaling | E-06 | (5.9%) | E-08 | (9.2%) | 2.19 | 5/238 | 1.48 | 14/238 |
| 6.31 | 7/124 | 3.16 | 13/124 | E-05 | (2.1%) | E-12 | (5.9%) | |
| 16. Cell Cycle: G1/S Checkpoint regulation | E-06 | (5.6%) | E-11 | (10.5%) | 7.65 | 5/89 | n.s. | n.s. |
| 1.26 | 8/238 | 7.94 | 16/238 | E-08 | (5.6%) | 1.91 | 7/61 | |
| E-04 | (3.4%) | E-10 | (6.7%) | 8.91 | 3/61 | E-08 | (11.5%) | |
| 17. Glioma signaling | 1.58 | 5/89 | 3.80 | 18/89 | E-05 | (4.9%) | 3.16 | 10/112 |
| 18. GM-CSF signaling | E-04 | (5.6%) | E-09 | (11.2%) | 3.98 | 5/112 | E-11 | (8.9%) |
| 19. IL-8 signaling | n.s. | n.s. | 3.98 | 11/61 | E-07 | (4.5%) | n.s. | n.s. |
| 20. HGF signaling | n.s. | n.s. | E-12 | (18.0%) | 3.16 | 4/67 | n.s. | n.s. |
| 21. Role of NFAT in Cardiac Hypertrophy | n.s. | n.s. | 6.31 | 11/112 | E-06 | (6.0%) | 2.75 | 8/105 |
| n.s. | n.s. | E-09 | (9.8%) | 6.31 | 5/193 | E-08 | (7.6%) | |
| 22. Hereditary Breast Cancer signaling | n.s. | n.s. | n.s. | n.s. | E-06 | (2.6%) | 6.31 | 12/208 |
| n.s. | n.s. | 3.98 | 14/193 | 2.00 | 4/105 | E-11 | (5.8%) | |
| n.s. | n.s. | E-09 | (7.3%) | E-05 | (3.8%) | 2.00 | 10/129 | |
| n.s. | n.s. | n.s. | n.s. | E-10 | (7.8%) | |||
| 7.59 | 10/208 | 1.02 | 3/129 | |||||
| E-06 | (4.8%) | E-03 | (2.3%) | |||||
| 5.25 | 10/129 | |||||||
| E-08 | (7.8%) | |||||||
Verification of antibody microarray data of Ang-(1-7)-regulated proteins PIAS2 and FBI-1/Pokemon
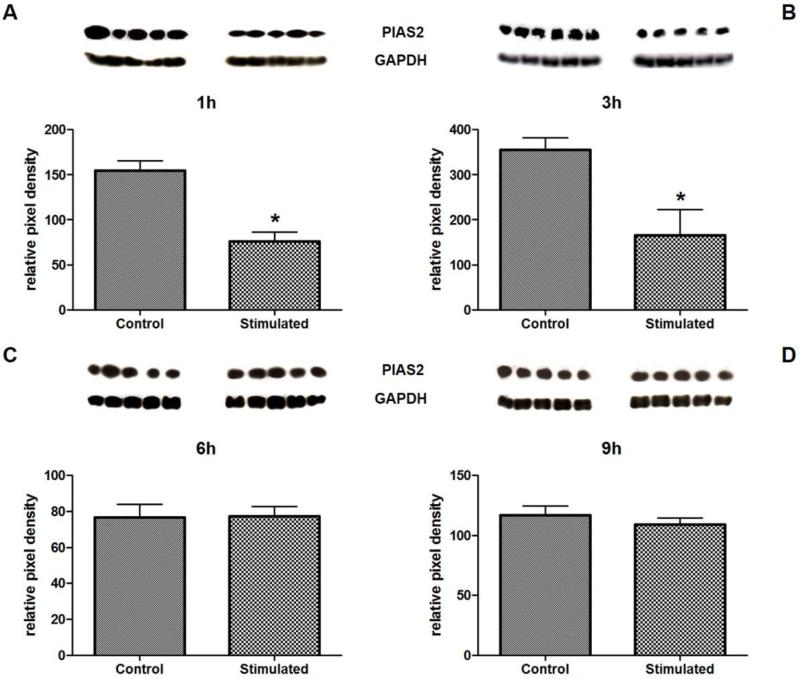
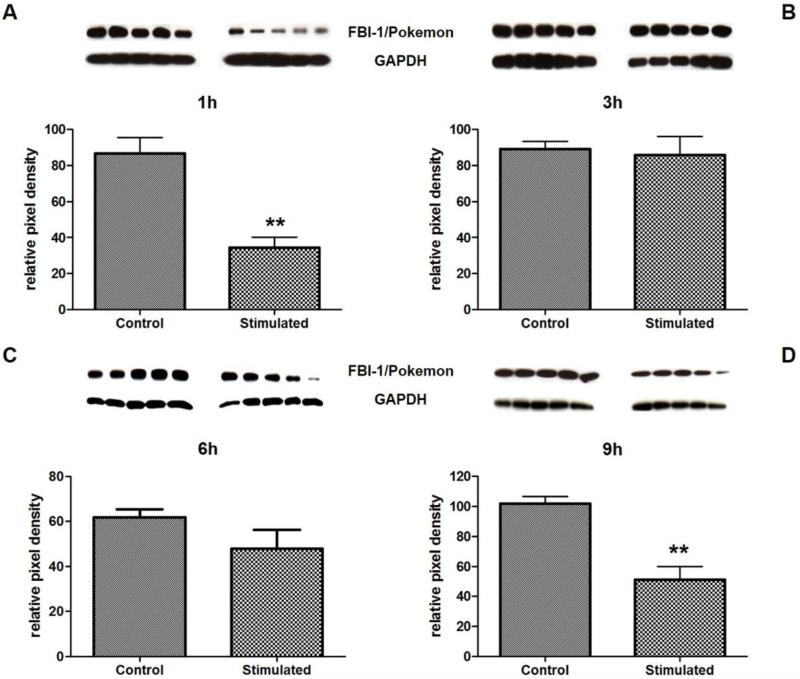
Because of the large number of proteins regulated by Ang-(1-7), we selected for further validation one protein that has been regulated by the heptapeptide at an early stage and one protein regulated at the late stage. The antibody microarray data displayed a significant Ang-(1-7)-mediated effect on MAP1b, Pyk2(pTyr579), PIAS2, Cdk3, and PP2A at an early stage (1h) of stimulation, whereas it detected a distinct regulation of PUMA/bbc3, FBI-1/Pokemon, and Caspase13 after 6h of stimulation with the heptapeptide that increased in significance at the late stage (9h) of stimulation. Therefore, we chose one of the proteins that showed regulation by Ang-(1-7) at an early stage, PIAS2, and another one, FBI-1/Pokemon that was affected at a late stage of stimulation with the heptapeptide and used Western blots to verify the antibody microarray results for these two proteins. For PIAS2, stimulation of HUVEC with Ang-(1-7) for 1h and 3h showed a significant decrease by 48% and 46% respectively, but no change in protein level for 6h and 9h stimulation (Figure 1 A-D), being congruent with the array data. In case of FBI-1/Pokemon, Western blot results showed a decrease in protein level for 1h (by 40%) and 9h (by 50%) (Figure 2 A-D), whereas stimulation for 3h and 6h with the heptapeptide did not change the FBI-1/Pokemon protein concentration. These results were only partially in accordance with the antibody microarray data that showed Ang-(1-7)-induced change in protein level for 6h and 9h stimulation, but none for 1h and 3h.
Figure 1.
Verification of antibody microarray data for PIAS2 by Western blot. Western blots were performed with lysates from HUVEC stimulated for 1h (A), 3h (B), 6h (C), and 9h (D) with 10−7M Ang-(1-7). Protein quantities were measured by densitometry and were normalized to GAPDH levels. Cells stimulated with the solvent were used as control.
Figure 2.
Verification of antibody microarray data for FBI-1/Pokemon by Western blot. Western blots were performed with lysates from HUVEC stimulated for 1h (A), 3h (B), 6h (C), and 9h (D) with 10−7M Ang-(1-7). Protein quantities were measured by densitometry and were normalized to GAPDH levels. Cells stimulated with the solvent were used as control.
Notably, a few more regulated proteins were verified by Western blot as e.g. AKT and p21 (data not shown). In both cases the Western blot results showed an increase in protein level, for AKT by 20% and 30% for 3h and 9h, and for p21 by 39% for 3h stimulation with Ang-(1-7), confirming the microarray data.
Ang-(1-7)-mediated effect on PIAS2 and FBI-1/Pokemon is dynamic over time
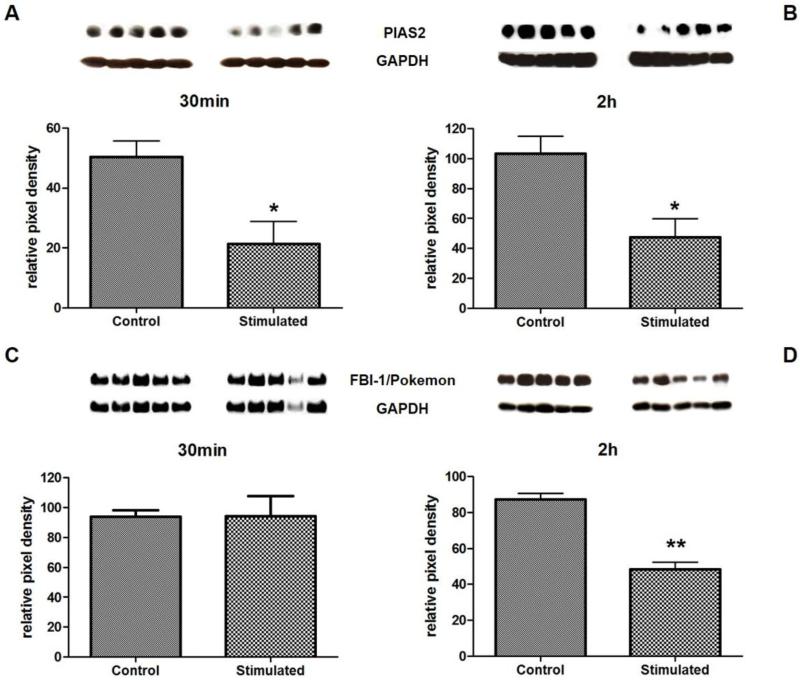
To investigate whether Ang-(1-7) influences the levels of both proteins already on an earlier stage than 1h, we investigated the heptapeptide's effect on both proteins at additional time points. In case of PIAS2, protein concentration was already influenced by Ang-(1-7) after 30min (Figure 3A) and remained low after 2h of stimulation (Figure 3B), indicating that Ang-(1-7) is not just effective at a certain time point, but down-regulates PIAS2 over a longer time period between at least 30 min and 3h after stimulation. In contrast, stimulation with Ang-(1-7) for 30 min did not lead to any effect on FBI-1/Pokemon, determining the start point of the heptapeptide's earliest influence on this protein somewhere between 30 min and 1h (Figure 3C). Since the Ang-(1-7) mediated reduction of FBI-1/Pokemon protein level after 1h (Figure 2A) was also seen after 2h of stimulation (Figure 3D), the early phase of the significant effect of Ang-(1-7) on FBI-1/Pokemon ends consequently somewhere between 2h and 3h.
Figure 3.
Identification of Ang-(1-7)-mediated effect on PIAS2 (A and B) and FBI-1/Pokemon (C and D) after 30min and 2h. Western blots were performed with lysates from HUVEC stimulated for 30min and 2h with 10−7M Ang-(1-7). Protein quantities were measured by densitometry and were normalized to GAPDH levels. Cells stimulated with the solvent were used as control.
Ang-(1-7) does not regulate PIAS2 and FBI-1/Pokemon on mRNA level
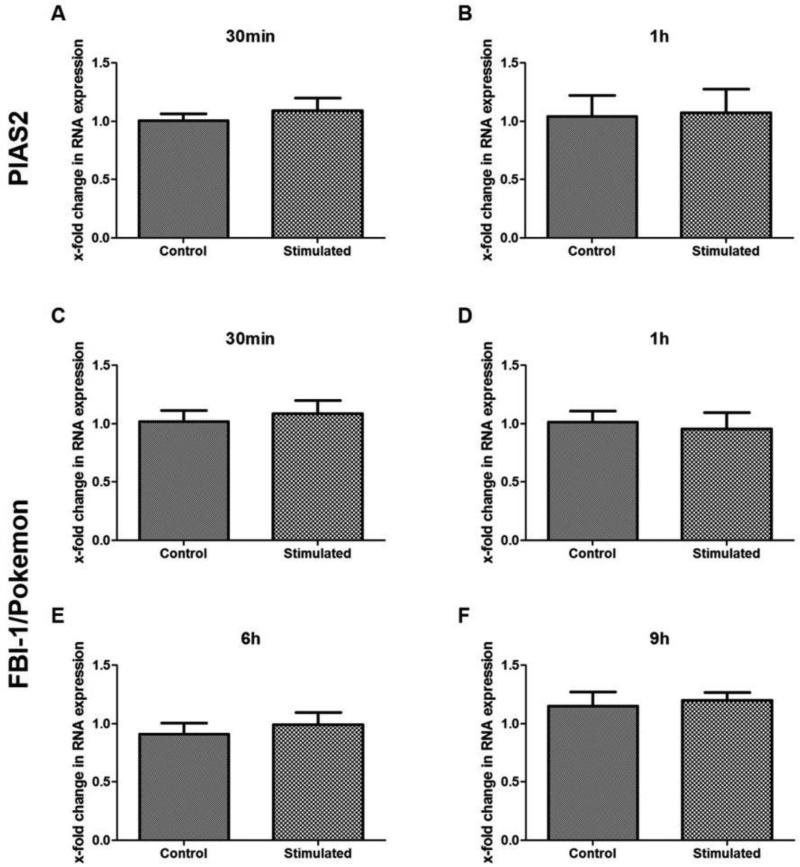
To determine, whether the Ang-(1-7)-mediated early effect on PIAS2 and FBI-1/Pokemon takes place on mRNA or protein level, we quantified their RNA expression in a Real-Time RT-PCR approach. As shown in Figure 4A-D, no change in mRNA level was detected for either proteins after 30 min and 1h of stimulation with the heptapeptide. Furthermore, the late effect of the heptapeptide on FBI-1/Pokemon (Figure 2D) was not mediated by alteration in mRNA, since 9h of stimulation with Ang-(1-7) did not affect the mRNA expression (Figure 4E-F). These results suggest that the Ang-(1-7) mediated effect on PIAS2 and FBI-1/Pokemon takes place on protein, but not on mRNA level.
Figure 4.
Effect of Ang-(1-7) on PIAS2 and FBI-1/Pokemon on mRNA level. For PIAS2, mRNA was quantified in HUVEC stimulated for 30min (A) and 1h (B) with 10−7M Ang-(1-7), whereas mRNA concentration of FBI-1/Pokemon was determined for 30 min (C), 1h (D), 6h (E), and 9h (F) of stimulation. Changes in mRNA concentration were normalized to GAPDH expression. Cells stimulated with the solvent were used as control.
Ang-(1-7) regulates PIAS2 and FBI-1/Pokemon in other endothelial cell lines
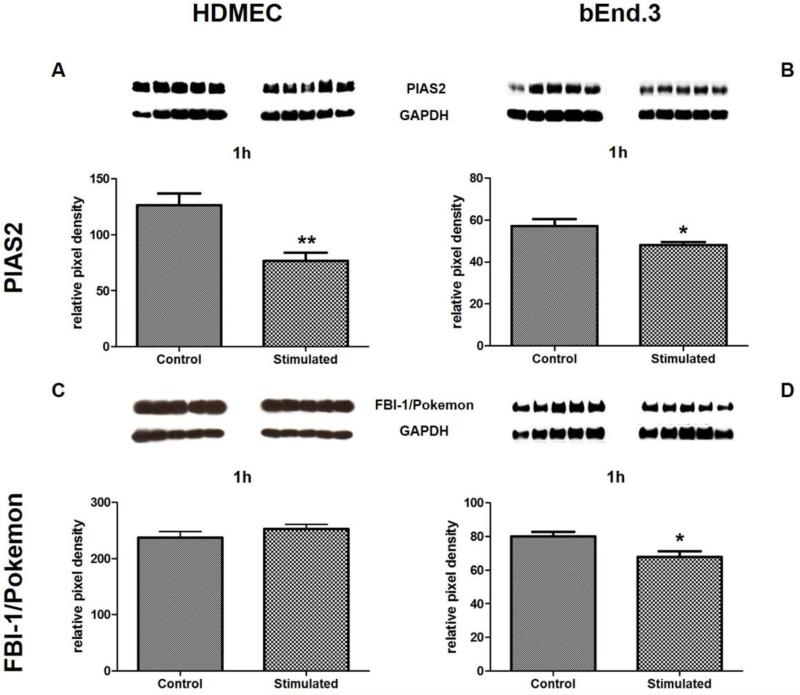
Next, we investigated if our findings in HUVEC could be reproduced in other human endothelial cell lines. For PIAS2, stimulation of HDMEC with Ang-(1-7) resulted in decreased protein level (Figure 5A), as observed earlier in HUVEC, while Ang-(1-7) had no effect on FBI-1/Pokemon in these cells (Figure 5C). Furthermore, we tested the effect of the heptapeptide in endothelial cells of another species. We stimulated mouse brain endothelial cells with the heptapeptide for 1h. As seen for both human endothelial cell lines, a reduction in PIAS2 protein was also found in bEnd.3 cells (Figure 5B). In such cells, Ang-(1-7) could also decrease the protein level of FBI-1/Pokemon (Figure 5D).
Figure 5.
Identification of Ang-(1-7)-mediated effect on PIAS2 (A and B) and FBI-1/Pokemon (C and D) in human dermal microvascular endothelial cells (HDMEC) and mouse brain endothelial cells (bEnd.3). The endothelial cell lines were stimulated for 1h with 10−7M Ang-(1-7). Protein quantities were measured by densitometry and were normalized to GAPDH levels. Cells stimulated with the solvent were used as control.
Ang-(1-7) decreases cytoplasmic PIAS2 concentration
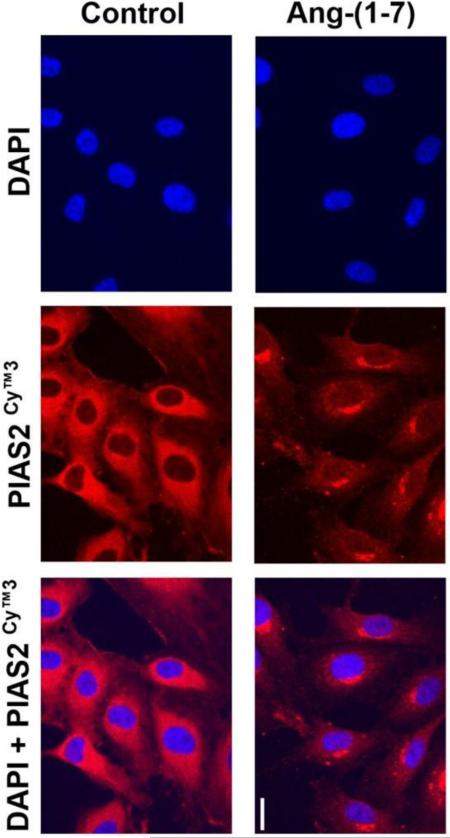
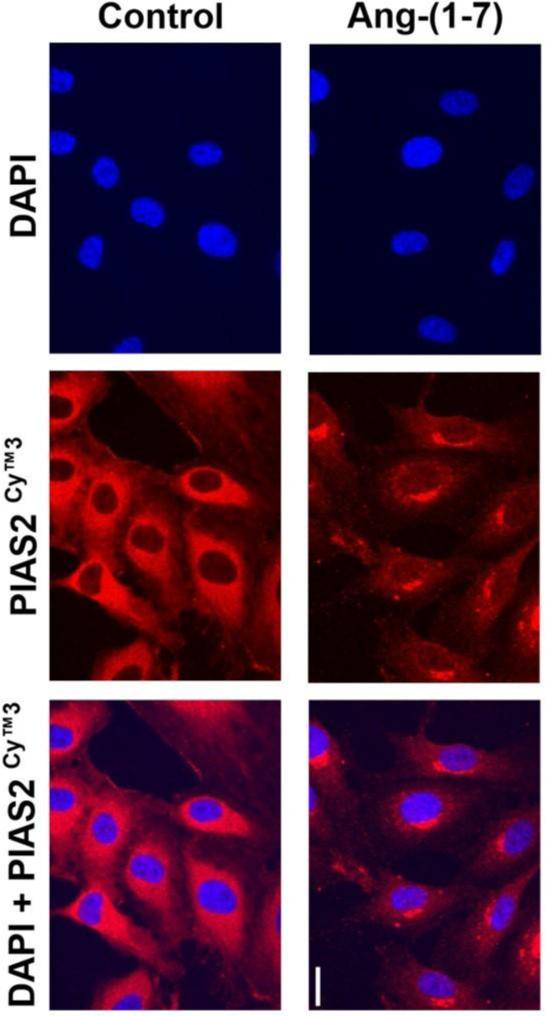
Finally we investigated whether the Ang-(1-7)-induced decrease in PIAS2 protein concentration could be also visualized in immunofluorescence studies in HUVEC. Unstimulated cells showed an intense and diffuse signal for PIAS2 in the cytoplasm (Figure 6, left panel). Stimulation of HUVEC with Ang-(1-7) for 1h dramatically decreased cytoplasmic PIAS2 (Figure 6, right panel). Furthermore, the remaining PIAS2 appeared to be confined to a distinct compartment at the edge of the nucleus. Notably, there was no evidence of translocation of PIAS2 from the cytoplasm into the nucleus after stimulation with Ang-(1-7).
Figure 6.
Effect of Ang-(1-7) on intracellular location of PIAS2. The localization of PIAS2 (red) was determined using immunofluorescence in untreated HUVEC (control) or cells treated with 10−7M Ang-(1-7) for 60 min (bar: 20 μm). The nuclei were visualized with DAPI (blue).
Discussion
Although both the number of publications related to Ang-(1-7) and recognition of its effects on physiological and pathophysiological processes have dramatically increased over the last few years, there is almost nothing known about the intracellular signaling stimulated by this heptapeptide. Whereas some single proteins like DUSP1 were described as targets of Ang-(1-7) [32], there is to our knowledge only a single publication that used an array approach to examine its effects [27]. Since this approach aimed to discover Ang-(1-7)-mediated changes in phosphorylation, and therefore focused on short-term changes, it was the aim of our study to investigate the long-term effects of the heptapeptide on the regulation of a widespread number of proteins.
In the pool of 725 proteins tested, the number of regulated proteins detected by the antibody microarray was unexpectedly high. Stimulation of HUVEC with the hetapeptide affected an average of 11% of the total amount of proteins that can be examined with the array. Only after stimulation for 3h less than a tenth of the proteins were influenced by the heptapeptide. However, the number of identified regulated proteins depends on the defined cut-off value and the higher the cut-off, the less protein appears to be regulated. In our case the relatively high number of identified proteins could be due to the second cut-off of ≥1.5, which was chosen deliberately low to ensure that no interesting potential candidates were overlooked. This cut point was selected with the knowledge that sometimes small changes in the concentration of a critical protein might have intracellular effects that result in a substantial impact on physiological/pathophysiological processes. Although the microarray results provided total values of expression fold change to identify Ang-(1-7)-regulated proteins, it has to be stated that a prediction for the direction of regulation was not possible, a clear disadvantage of the microarray, as it necessarily requires further evaluation before concluding on up- or downregulation.
The authors have deliberately omitted the investigation of later time points like 24 h or 48 h, as their experience gained from cell culture work shows that at later time points it cannot be distinguished between primary, secondary and tertiary effects. In addition, at later time points (24-48 h) morpho-histological changes as well as alterations in the number of cells can result in changes in intracellular signalling that are not directly triggered or influenced by intracellular signalling stimulated by Ang-(1-7).
False positive detection of proteins might also influence the results of the antibody microarray. Hodgkinson et al. identified 13 RIDEPs specifically for the Panorama Antibody Microarray-XPRESS Profiler725 Kit from Sigma-Aldrich [29], the majority of which were also found in our microarray data. However, it is possible that there were even more false positive results among the proteins assayed. Given the fact that TRAIL and Bmf were always among the most regulated proteins at three out of four time points, these two might be also false positives. However, this possibility requires further investigation when testing for other proteins than PIAS2 and FBI-1/Pokemon we investigated here in detail.
Due to such RIDEP issues and also to quantify the regulation of specific proteins, it is essential to validate the antibody microarray data with an additional method. Because of the very large number of proteins regulated by Ang-(1-7), we selected candidates of interest for further validation. Thus we decided to validate the microarray data for PIAS2, a protein that showed regulation by Ang-(1-7) at an early stage and for FBI-1/Pokemon, a protein that was affected at a late stage of stimulation with the heptapeptide.
While the Western blot data for PIAS2 completely confirmed the microarray data, it was only partially consistent for FBI-1/Pokemon as this protein was regulated in the array at 6h and 9h, but in Western blot experiments only at 9h. However, the results show a high reproducibility of the antibody microarray data, which has been also shown by other groups [33-35]. Nonetheless, it seems vital to validate changes in proteins seen in the microarray by other methods in the future. However, it is important to note that there may be deviations in the results due to different sensitivities of the validation methods.
The validity of the antibody microarray data that we have reported is also supported by the fact that most of the identified proteins are part of specific pathways, with most of them clustered in a few networks (Table 5). The most significant pathways were the “Molecular Mechanism of Cancer”, “p53 signaling”, and “Cyclins and Cell Cycle regulation”, a finding that is consistent with published data showing that Ang-(1-7) negatively influences the progression of cancer by reducing cell proliferation [23, 24, 32, 36-38]. In addition, another one of the pathways identified is the “Chronic Myeloid Leukemia” signaling, which is in keeping with our recent findings that Ang-(1-7) stimulates the production of hematopoetic progenitor cells [22]. Furthermore, the fact that bioinformatic software associated the regulation of identified proteins to the signaling cascades in a time dependent manner as e.g. regulation of the early protein SMAD4 in the case of “Molecular Mechanism of Cancer” after 1h lead to changes in downstream proteins p19 and p21 at 3h, strongly supports the quality of our array data.
Unfortunately, both of the proteins that we investigated in detail cannot easily be linked into biological and physiological/pathophysiological consequences caused by their down-regulation by Ang-(1-7), e.g. PIAS2 was originally identified as a protein inhibitor of activated STAT2, but also interacts with STAT1 and STAT4 [39, 40]. Moreover, PIAS2 has been reported to interact with RACK1, PARK7, DNMT3A, and the androgen receptor [41-45]. Our findings, however, should stimulate further studies to determine the upstream/downstream targets of PIAS2 and to identify the receptor by which Ang-(1-7) affects the protein.
One of the main findings is that PIAS2 protein is regulated without any change in mRNA quantity at all time points RNA was investigated. This identifies the level of regulation, because no change in mRNA but a significant alteration in protein implicates posttranslational regulation. The reduction of PIAS2 protein levels could result from dissolving the PIAS2 complex during SUMOylation of STATs by stimulation with Ang-(1-7), making PIAS2 more vulnerable to proteolytic activity. Furthermore, our results suggest that the Ang-(1-7)-mediated effect on PIAS2 is not restricted to either the source of the vessel or species, suggesting that the down-regulation of PIAS2 by Ang-(1-7) is a generalized effect of the intracellular signaling that is stimulated by the heptapeptide in endothelial cells.
In summary, our experiments identified a significant number of previously unknown proteins associated with Ang-(1-7). Our data is the needed add to the short-term regulated (phosphorylated) proteins described by Verano-Braga et al. [27] to understand the Ang-(1-7)-mediated signaling and illustrates the complexity of intracellular network been altered by the heptapeptide.
Conclusion
We are the first, to our knowledge, to use a broad approach to identify proteins quantitatively regulated by Ang-(1-7). We detected a significant amount of proteins that are affected by the heptapeptide in endothelial cells, many of which have not been associated with Ang-(1-7) previously. These findings open potentially new insights into the role of Ang-(1-7) not only in endothelial cells and in vascular reactivity, but also in other physiologic and pathophysiologic processes throughout the body.
Significance.
Despite many publications regarding the physiological actions of Ang-(1-7) and its beneficial effects under pathophysiological circumstances, the intracellular signalling initiated by Ang-(1-7) remains still elusive. Since no studies are available related to longer-term effects of Ang-(1-7) on intracellular signalling, we are the first using a broad approach to identify proteins quantitatively regulated by Ang-(1-7). We aimed to identify such proteins, to verify them, and based on this to define possible intracellular signalling pathways activated by the heptapeptide. We detected a significant amount of affected proteins that have not been associated with Ang-(1-7) before. They have been mainly part of metabolic pathways related to cell death and cell survival.
Taken together, our results identified a variety of new proteins being associated with the intracellular signalling of Ang-(1-7). This is all the more important as they might be associated with the beneficial cardiovascular effects of the heptapeptide and thus could represent new targets for the treatment of cardiovascular diseases. Finally, our data will significantly stimulate further research on the Ang-(1-7) related intracellular signalling.
Highlights.
Ang-(1-7) regulates proteins in a time-dependent manner in endothelial cells.
Such proteins associate to metabolic pathways related to cell death and cell survival.
Microarrays and Western blots identified downregulation of PIAS2 FBI-1/Pokemon.
Changes in both proteins but not their RNA implicate posttranscriptional regulation.
Regulation pattern of both proteins was similar in different endothelial cell types.
Acknowledgments
We want to thank Victoria Hodgkinson for introducing us into the analysis of the antibody microarray data. The technical assistance of Stephanie Müller within the immunohistochemical studies is kindly acknowledged.
Source of funding
The project was supported by the NIH (R01HL091191-01A2).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None.
References
- 1.Fyhrquist F, Saijonmaa O. Renin-angiotensin system revisited. J Intern Med. 2008;264:224–36. doi: 10.1111/j.1365-2796.2008.01981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carey RM, Siragy HM. Newly recognized components of the renin-angiotensin system: potential roles in cardiovascular and renal regulation. Endocr Rev. 2003;24:261–71. doi: 10.1210/er.2003-0001. [DOI] [PubMed] [Google Scholar]
- 3.Leung PS, Carlsson PO. Tissue renin-angiotensin system: its expression, localization, regulation and potential role in the pancreas. J Mol Endocrinol. 2001;26:155–64. doi: 10.1677/jme.0.0260155. [DOI] [PubMed] [Google Scholar]
- 4.Burson JM, Aguilera G, Gross KW, Sigmund CD. Differential expression of angiotensin receptor 1A and 1B in mouse. Am J Physiol. 1994;267:E260–7. doi: 10.1152/ajpendo.1994.267.2.E260. [DOI] [PubMed] [Google Scholar]
- 5.Ardaillou R. Active fragments of angiotensin II: enzymatic pathways of synthesis and biological effects. Curr Opin Nephrol Hypertens. 1997;6:28–34. doi: 10.1097/00041552-199701000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Vauquelin G, Michotte Y, Smolders I, Sarre S, Ebinger G, Dupont A, et al. Cellular targets for angiotensin II fragments: pharmacological and molecular evidence. J Renin Angiotensin Aldosterone Syst. 2002;3:195–204. doi: 10.3317/jraas.2002.041. [DOI] [PubMed] [Google Scholar]
- 7.Wright JW, Harding JW. Important role for angiotensin III and IV in the brain renin-angiotensin system. Brain Res Brain Res Rev. 1997;25:96–124. doi: 10.1016/s0165-0173(97)00019-2. [DOI] [PubMed] [Google Scholar]
- 8.Kerins DM, Hao Q, Vaughan DE. Angiotensin induction of PAI-1 expression in endothelial cells is mediated by the hexapeptide angiotensin IV. J Clin Invest. 1995;96:2515–20. doi: 10.1172/JCI118312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chai SY, Fernando R, Peck G, Ye SY, Mendelsohn FA, Jenkins TA, et al. The angiotensin IV/AT4 receptor. Cell Mol Life Sci. 2004;61:2728–37. doi: 10.1007/s00018-004-4246-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kramkowski K, Mogielnicki A, Leszczynska A, Buczko W. Angiotensin-(1-9), the product of angiotensin I conversion in platelets, enhances arterial thrombosis in rats. J Physiol Pharmacol. 2010;61:317–24. [PubMed] [Google Scholar]
- 11.Ferrario CM, Chappell MC, Dean RH, Iyer SN. Novel angiotensin peptides regulate blood pressure, endothelial function, and natriuresis. J Am Soc Nephrol. 1998;9:1716–22. doi: 10.1681/ASN.V991716. [DOI] [PubMed] [Google Scholar]
- 12.Ferrario CM, Chappell MC. Novel angiotensin peptides. Cell Mol Life Sci. 2004;61:2720–7. doi: 10.1007/s00018-004-4243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vickers C, Hales P, Kaushik V, Dick L, Gavin J, Tang J, et al. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J Biol Chem. 2002;277:14838–43. doi: 10.1074/jbc.M200581200. [DOI] [PubMed] [Google Scholar]
- 14.Rice GI, Thomas DA, Grant PJ, Turner AJ, Hooper NM. Evaluation of angiotensin-converting enzyme (ACE), its homologue ACE2 and neprilysin in angiotensin peptide metabolism. Biochem J. 2004;383:45–51. doi: 10.1042/BJ20040634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loot AE, Roks AJ, Henning RH, Tio RA, Suurmeijer AJ, Boomsma F, et al. Angiotensin-(1-7) attenuates the development of heart failure after myocardial infarction in rats. Circulation. 2002;105:1548–50. doi: 10.1161/01.cir.0000013847.07035.b9. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Qian C, Roks AJ, Westermann D, Schumacher SM, Escher F, et al. Circulating rather than cardiac angiotensin-(1-7) stimulates cardioprotection after myocardial infarction. Circ Heart Fail. 2010;3:286–93. doi: 10.1161/CIRCHEARTFAILURE.109.905968. [DOI] [PubMed] [Google Scholar]
- 17.Tallant EA, Ferrario CM, Gallagher PE. Angiotensin-(1-7) inhibits growth of cardiac myocytes through activation of the mas receptor. Am J Physiol Heart Circ Physiol. 2005;289:H1560–6. doi: 10.1152/ajpheart.00941.2004. [DOI] [PubMed] [Google Scholar]
- 18.Strawn WB, Ferrario CM, Tallant EA. Angiotensin-(1-7) reduces smooth muscle growth after vascular injury. Hypertension. 1999;33:207–11. doi: 10.1161/01.hyp.33.1.207. [DOI] [PubMed] [Google Scholar]
- 19.Santos RA, Simoes e Silva AC, Magaldi AJ, Khosla MC, Cesar KR, Passaglio KT, et al. Evidence for a physiological role of angiotensin-(1-7) in the control of hydroelectrolyte balance. Hypertension. 1996;27:875–84. doi: 10.1161/01.hyp.27.4.875. [DOI] [PubMed] [Google Scholar]
- 20.Rodgers KE, Espinoza T, Felix J, Roda N, Maldonado S, diZerega G. Acceleration of healing, reduction of fibrotic scar, and normalization of tissue architecture by an angiotensin analogue, NorLeu3-A(1-7). Plast Reconstr Surg. 2003;111:1195–206. doi: 10.1097/01.PRS.0000047403.23105.66. [DOI] [PubMed] [Google Scholar]
- 21.Hellner K, Walther T, Schubert M, Albrecht D. Angiotensin-(1-7) enhances LTP in the hippocampus through the G-protein-coupled receptor Mas. Mol Cell Neurosci. 2005;29:427–35. doi: 10.1016/j.mcn.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 22.Heringer-Walther S, Eckert K, Schumacher SM, Uharek L, Wulf-Goldenberg A, Gembardt F, et al. Angiotensin-(1-7) stimulates hematopoietic progenitor cells in vitro and in vivo. Haematologica. 2009;94:857–60. doi: 10.3324/haematol.2008.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallagher PE, Tallant EA. Inhibition of human lung cancer cell growth by angiotensin-(1-7). Carcinogenesis. 2004;25:2045–52. doi: 10.1093/carcin/bgh236. [DOI] [PubMed] [Google Scholar]
- 24.Menon J, Soto-Pantoja DR, Callahan MF, Cline JM, Ferrario CM, Tallant EA, et al. Angiotensin-(1-7) inhibits growth of human lung adenocarcinoma xenografts in nude mice through a reduction in cyclooxygenase-2. Cancer Res. 2007;67:2809–15. doi: 10.1158/0008-5472.CAN-06-3614. [DOI] [PubMed] [Google Scholar]
- 25.Rodgers KE, Oliver J, diZerega GS. Phase I/II dose escalation study of angiotensin 1-7 [A(1-7)] administered before and after chemotherapy in patients with newly diagnosed breast cancer. Cancer Chemother Pharmacol. 2006;57:559–68. doi: 10.1007/s00280-005-0078-4. [DOI] [PubMed] [Google Scholar]
- 26.Santos RA, Simoes e Silva AC, Maric C, Silva DM, Machado RP, de Buhr I, et al. Angiotensin-(1-7) is an endogenous ligand for the G protein-coupled receptor Mas. Proc Natl Acad Sci U S A. 2003;100:8258–63. doi: 10.1073/pnas.1432869100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verano-Braga T, Schwammle V, Sylvester M, Passos-Silva DG, Peluso AA, Etelvino GM, et al. Time-Resolved Quantitative Phosphoproteomics: New Insights into Angiotensin-(1-7) Signaling Networks in Human Endothelial Cells. J Proteome Res. 2012;11:3370–81. doi: 10.1021/pr3001755. [DOI] [PubMed] [Google Scholar]
- 28.Gembardt F, Grajewski S, Vahl M, Schultheiss HP, Walther T. Angiotensin metabolites can stimulate receptors of the Mas-related genes family. Mol Cell Biochem. 2008;319:115–23. doi: 10.1007/s11010-008-9884-4. [DOI] [PubMed] [Google Scholar]
- 29.Hodgkinson VC, ElFadl D, Drew PJ, Lind MJ, Cawkwell L. Repeatedly identified differentially expressed proteins (RIDEPs) from antibody microarray proteomic analysis. J Proteomics. 2011;74:698–703. doi: 10.1016/j.jprot.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 30.Petrak J, Ivanek R, Toman O, Cmejla R, Cmejlova J, Vyoral D, et al. Deja vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics. 2008;8:1744–9. doi: 10.1002/pmic.200700919. [DOI] [PubMed] [Google Scholar]
- 31.Mariman EC. 2DE-proteomics meta-data indicate the existence of distinct cellular stress-responsive mechanisms. Expert Rev Proteomics. 2009;6:337–9. doi: 10.1586/epr.09.50. [DOI] [PubMed] [Google Scholar]
- 32.Cook KL, Metheny-Barlow LJ, Tallant EA, Gallagher PE. Angiotensin-(1-7) reduces fibrosis in orthotopic breast tumors. Cancer Res. 2010;70:8319–28. doi: 10.1158/0008-5472.CAN-10-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agarwal V, Hodgkinson VC, Eagle GL, Scaife L, Lind MJ, Cawkwell L. Proteomic (antibody microarray) exploration of the molecular mechanism of action of the specific COX-2 inhibitor DuP 697. Int J Oncol. 2013;42:1088–92. doi: 10.3892/ijo.2013.1784. [DOI] [PubMed] [Google Scholar]
- 34.Simara P, Koutna I, Stejskal S, Krontorad P, Rucka Z, Peterkova M, et al. Combination of mRNA and protein microarray analysis in complex cell profiling. Neoplasma. 2009;56:141–9. doi: 10.4149/neo_2009_02_141. [DOI] [PubMed] [Google Scholar]
- 35.Smith L, Watson MB, O'Kane SL, Drew PJ, Lind MJ, Cawkwell L. The analysis of doxorubicin resistance in human breast cancer cells using antibody microarrays. Mol Cancer Ther. 2006;5:2115–20. doi: 10.1158/1535-7163.MCT-06-0190. [DOI] [PubMed] [Google Scholar]
- 36.Krishnan B, Smith TL, Dubey P, Zapadka ME, Torti FM, Willingham MC, et al. Angiotensin-(1-7) attenuates metastatic prostate cancer and reduces osteoclastogenesis. Prostate. 2013;73:71–82. doi: 10.1002/pros.22542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krishnan B, Torti FM, Gallagher PE, Tallant EA. Angiotensin-(1-7) reduces proliferation and angiogenesis of human prostate cancer xenografts with a decrease in angiogenic factors and an increase in sFlt-1. Prostate. 2013;73:60–70. doi: 10.1002/pros.22540. [DOI] [PubMed] [Google Scholar]
- 38.Soto-Pantoja DR, Menon J, Gallagher PE, Tallant EA. Angiotensin-(1-7) inhibits tumor angiogenesis in human lung cancer xenografts with a reduction in vascular endothelial growth factor. Mol Cancer Ther. 2009;8:1676–83. doi: 10.1158/1535-7163.MCT-09-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arora T, Liu B, He H, Kim J, Murphy TL, Murphy KM, et al. PIASx is a transcriptional co-repressor of signal transducer and activator of transcription 4. J Biol Chem. 2003;278:21327–30. doi: 10.1074/jbc.C300119200. [DOI] [PubMed] [Google Scholar]
- 40.Rogers RS, Horvath CM, Matunis MJ. SUMO modification of STAT1 and its role in PIAS-mediated inhibition of gene activation. J Biol Chem. 2003;278:30091–7. doi: 10.1074/jbc.M301344200. [DOI] [PubMed] [Google Scholar]
- 41.Kotaja N, Aittomaki S, Silvennoinen O, Palvimo JJ, Janne OA. ARIP3 (androgen receptor-interacting protein 3) and other PIAS (protein inhibitor of activated STAT) proteins differ in their ability to modulate steroid receptor-dependent transcriptional activation. Mol Endocrinol. 2000;14:1986–2000. doi: 10.1210/mend.14.12.0569. [DOI] [PubMed] [Google Scholar]
- 42.Ling Y, Sankpal UT, Robertson AK, McNally JG, Karpova T, Robertson KD. Modification of de novo DNA methyltransferase 3a (Dnmt3a) by SUMO-1 modulates its interaction with histone deacetylases (HDACs) and its capacity to repress transcription. Nucleic Acids Res. 2004;32:598–610. doi: 10.1093/nar/gkh195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shuai K, Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat Rev Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi K, Taira T, Niki T, Seino C, Iguchi-Ariga SM, Ariga H. DJ-1 positively regulates the androgen receptor by impairing the binding of PIASx alpha to the receptor. J Biol Chem. 2001;276:37556–63. doi: 10.1074/jbc.M101730200. [DOI] [PubMed] [Google Scholar]
- 45.Zheng Y, Zhang L, Jia X, Wang H, Hu Y. Interaction of protein inhibitor of activated STAT 2 (PIAS2) with receptor of activated C kinase 1, RACK1. FEBS Lett. 2012;586:122–6. doi: 10.1016/j.febslet.2011.12.013. [DOI] [PubMed] [Google Scholar]