Summary
Induced pluripotent stem cells (iPSCs) hold great potential not only for human but also for veterinary purposes. The equine industry must often deal with health issues concerning muscle and cartilage, where comprehensive regenerative strategies are still missing. In this regard, a still open question is whether equine iPSCs differentiate toward muscle and cartilage, and whether donor cell type influences their differentiation potential. We addressed these questions through an isogenic system of equine iPSCs obtained from myogenic mesoangioblasts (MAB-iPSCs) and chondrogenic mesenchymal stem cells (MSC-iPSCs). Despite similar levels of pluripotency characteristics, the myogenic differentiation appeared enhanced in MAB-iPSCs. Conversely, the chondrogenic differentiation was augmented in MSC-iPSCs through both teratoma and in vitro differentiation assays. Thus, our data suggest that equine iPSCs can differentiate toward the myogenic and chondrogenic lineages, and can present a skewed differentiation potential in favor of the source cell lineage.
Keywords: equine stem cells, myogenic differentiation, chondrogenic differentiation, equine iPSCs, intrinsic lineage propensity
Highlights
-
•
Equine mesoangioblasts (MABs) are prospectively isolated from syngeneic horses
-
•
Equine iPSCs are obtained from isogenic MABs and MSCs with similar characteristics
-
•
Equine MAB-iPSCs are more prone to the muscle lineage in vivo and in vitro
-
•
Equine MSC-iPSCs display higher propensity for the chondrogenic lineage
The horse industry has a major economic impact worldwide, and needs novel regenerative tools for muscle and cartilage problems. However, a comprehensive strategy for repairing muscle and cartilage is still missing. Equine-induced pluripotent stem cells (iPSCs) have wide potential, but their differentiation toward those tissues remains unaddressed. Furthermore, it is still unknown whether the cell source primes equine iPSCs toward muscle and cartilage differentiation. Sampaolesi, Spaas, and colleagues, by means of isogenic settings and dedicated in vitro assays, report that equine iPSCs retain intrinsic differentiation propensity toward skeletal muscle and cartilage lineages.
Introduction
Horses are invaluable animals for companionship and sport. The equine industry creates an estimated economic impact of US$300 billion worldwide, and novel means for addressing equine health issues are constantly required (Tecirlioglu and Trounson, 2007). Musculoskeletal problems, including pathologies or injuries of muscle and cartilage, constitute a leading health threat among horses (Smith et al., 2014). As an example, equine atypical myopathy has been increasingly reported over recent years (Votion and Serteyn, 2008), and equine osteochondrosis is relatively frequent across different breeds (van Weeren and Jeffcott, 2013). Therefore, the quest for novel tools for muscle and cartilage repair is still compelling. In this regard, stem cells may support the needs of veterinary medicine (Cebrian-Serrano et al., 2013). In equine practice, mesenchymal stem cells (MSCs) are commonly used to treat tendinitis and osteoarthritis (Schnabel et al., 2013). However, a comprehensive regenerative approach tailored to both muscle and cartilage is still missing, especially for large-scale applications. Importantly, considering the obvious difficulties of in vivo trials, in vitro models constitute a useful, first-line trial platform for addressing differentiation and heterogeneity of stem cells (Goodell et al., 2015). To this end, induced pluripotent stem cells (iPSCs) hold great potential, in light of their tremendous expansion capacity and wide differentiation potential (Yamanaka, 2009). Recently, iPSCs have been generated from equine fibroblasts (Breton et al., 2013) and keratinocytes (Sharma et al., 2014); however, their differentiation potential toward myocytes or chondrocytes remains unknown. Furthermore, iPSCs tend to retain, at least partially, the intrinsic fate propensity of the cell source (Sanchez-Freire et al., 2014). In mice, iPSCs derived from resident myogenic pericytes, i.e. mesoangioblasts (MABs), show biased myogenic differentiation in both teratoma and in vitro differentiation assays (Quattrocelli et al., 2011). However, it is still unknown whether it is possible to discriminate the intrinsic equine iPSC propensity toward the myogenic and the chondrogenic lineages. To address this question, isogenic settings need to reduce the variability introduced by genetic background (Kotini et al., 2015). Relevantly for putative veterinary applications, the choice of the cell source should be confined to somatic compartments at facilitated reach, e.g. blood and superficial muscle biopsies. Equine peripheral blood has been recently used to isolate circulating progenitors, exhibiting MSC properties (Spaas et al., 2013) and differentiating in chondrocytes (Broeckx et al., 2014). With regard to the muscle, it is still unknown whether equine MABs can be isolated with similar characteristics to murine, canine, or human MABs (Quattrocelli et al., 2014, Sampaolesi et al., 2006), and to which extent equine MABs display myogenic propensity, once cultured in vitro and after reprogramming.
Thus, the objective of this study is to compare iPSCs obtained from equine MABs and peripheral blood MSCs in isogenic conditions to evaluate the intrinsic iPSC propensity for myogenic and chondrogenic lineages.
Results
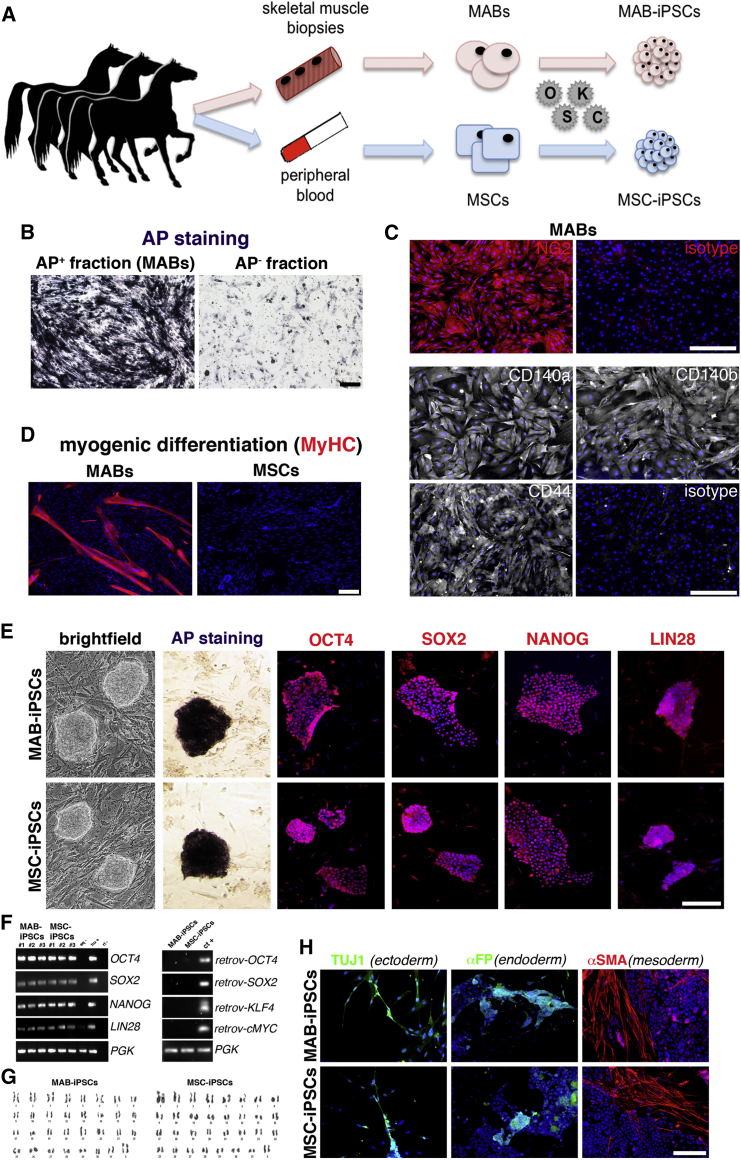
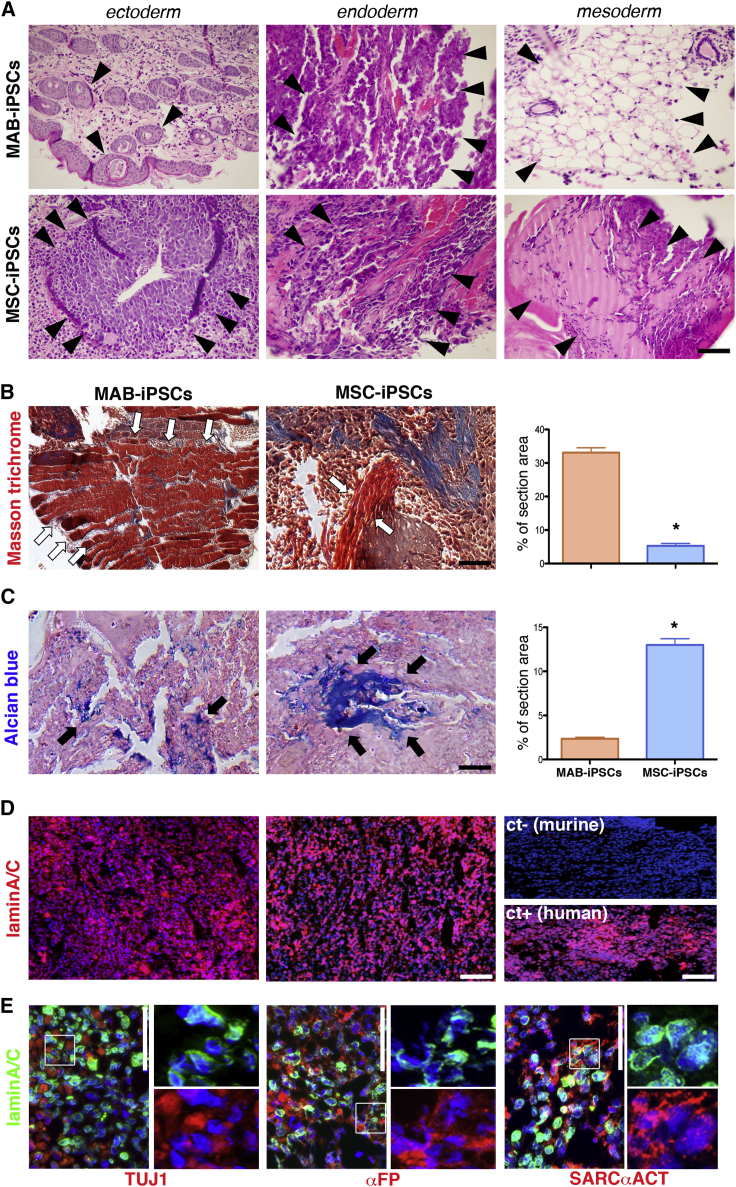
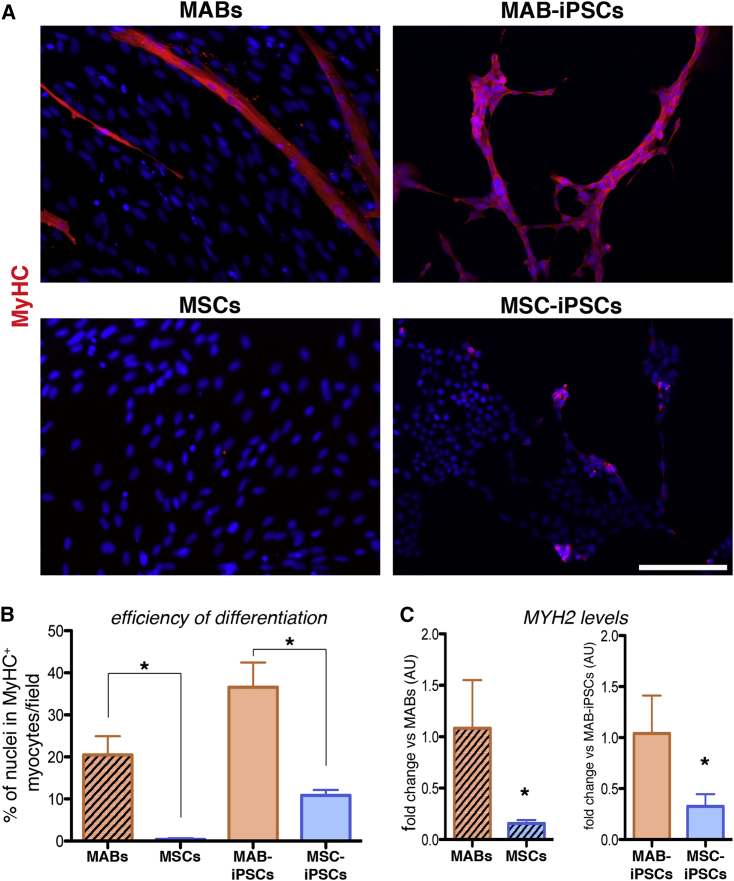
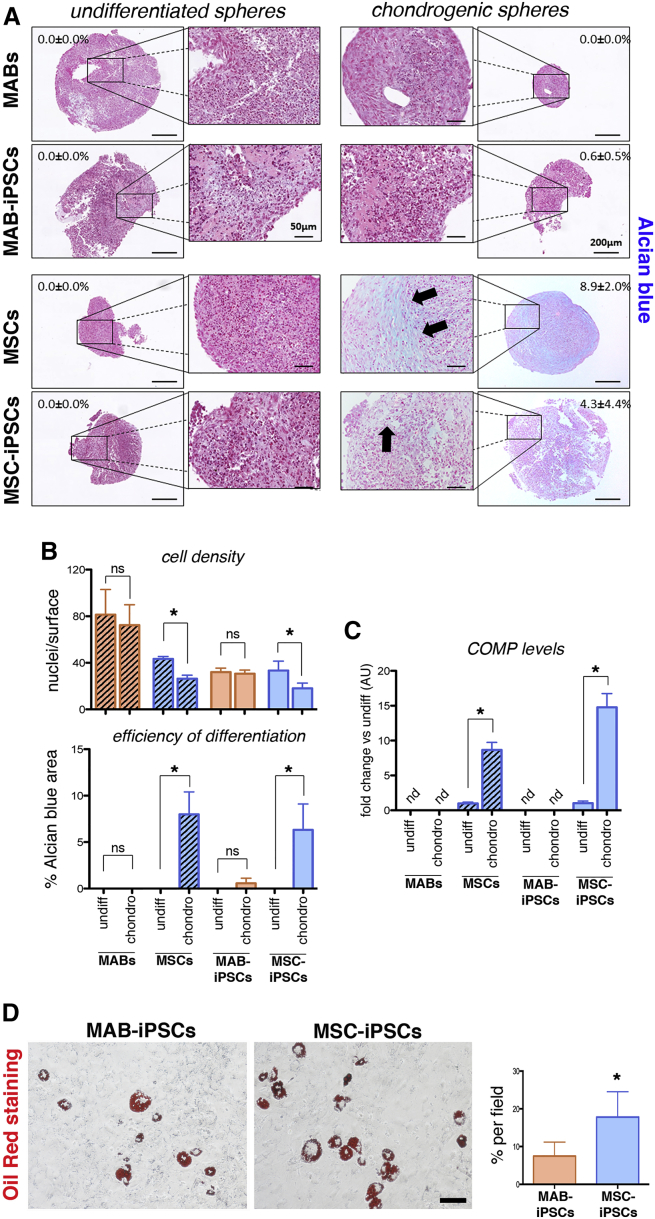
To obtain source cells for reprogramming in isogenic settings (Figure 1A), we isolated equine MABs in isogenic settings with MSC lines, which were already established from peripheral blood (Spaas et al., 2013), and demonstrated chondrogenic commitment (Broeckx et al., 2014). We adapted the procedures that were previously reported for murine and human MABs (Quattrocelli et al., 2012) to biopsies obtained from the splenius muscles of three syngeneic horses, and accordingly sorted the heterogeneous cell population for alkaline phosphatase (AP) (1.5% ± 0.6% [mean ± SD; n = 3]). AP activity was confirmed in the AP+ but not the AP− fraction (Figure 1B). At early passage (<3), AP+ cells proliferated with a doubling time of approximately 27 hr and stained positively for NG2, CD140a, CD140, and CD44, pericytic surface markers reported for murine and human MABs (Quattrocelli et al., 2012) (Figure 1C). Furthermore, after 7 days in serum starvation, the AP+ cells robustly differentiated into multinucleated, MyHC+ myotubes (fusion index, 27.5% ± 7.01% [mean ± SD; n = 3]), unlike the isogenic MSC controls (Figure 1D). We thus identified the AP+ cells as bona fide equine MABs, which show robust myogenic propensity compared with the isogenic MSCs. To obtain iPSCs, we transduced equine isogenic MABs and MSCs with retroviral vectors carrying human reprogramming factors (OCT4, SOX2, KLF4, cMYC), then picked and expanded single-cell clones (n = 3/cell type) on a feeder layer. Reprogramming efficiency was approximately 0.0001% for both cell types. Colonies of both MAB- and MSC-iPSCs appeared round-shaped and with compact borders, and were positive for AP, OCT4, SOX2, NANOG, and LIN28 staining (Figure 1E). The expression of the pluripotency markers OCT4, SOX2, NANOG, and LIN28 was confirmed with specifically cross-reacting primers, using human H9 cells as positive control. Moreover, expression of retroviral transgenes was not detected in both equine iPSC types (Figure 1F). Also, equine iPSCs showed a euploid, donor-matching karyotype (n = 62, XX; Figure 1G). Furthermore, after 20 days of spontaneous in vitro differentiation, both MAB- and MSC-iPSCs differentiated in ectodermal (TUJ1+), endodermal (αFP+), and mesodermal (αSMA+) derivative cells (Figure 1H). Thus, equine isogenic MAB- and MSC-iPSCs shared common markers of pluripotency. To gain insight into iPSC intrinsic propensity, we subcutaneously injected equine iPSCs in immunodeficient mice and analyzed the teratomas at 4–6 weeks after injection. Both iPSC types generated teratomas containing immature derivatives of ectoderm, endoderm, and mesoderm, confirming their pluripotency (Figure 2A). However, MAB-iPSC teratomas showed a significantly higher quantity of immature muscle patches in comparison with MSC-iPSCs (Figure 2B). Conversely, MSC-iPSC teratomas showed significantly larger chondrogenic patches (Figure 2C). To exclude the contribution of host cells to teratoma derivatives, we stained equine iPSC teratoma sections for lamin A/C, using murine iPSC- and human iPSC-derived teratomas as negative and positive controls, respectively. Both MAB- and MSC-iPSC-derived teratomas stained positively to lamin A/C (Figures 2D and 2E), indicating that the teratoma tissues derived from equine iPSCs. Intrigued by the propensities shown in the teratoma assays, we asked whether the source-related propensity was significantly skewing the iPSC fate in dedicated differentiation assays. We tested the myogenic differentiation of iPSCs and related source cells under conditions of bone morphogenetic protein (BMP)/transforming growth factor β (TGF-β) blockade and assayed for MyHC+ myocytes and myotubes. After 30 days, MABs and MAB-iPSCs showed higher differentiation rates compared with isogenic MSCs and MSC-iPSCs (Figures 3A and 3B). Moreover, equine MYH2 expression levels were significantly higher in differentiated MABs and MAB-iPSCs (Figure 3C). We then tested the chondrogenic differentiation in compacted spheres (Spaas et al., 2013) and assayed for Alcian blue-positive structures. Only MSC- and MSC-iPSC-derived spheres showed consistent chondrogenic differentiation, assayed as larger chondrogenic patches and lower cell density when compared with undifferentiated spheres (Figures 4A and 4B). Accordingly, equine COMP expression levels were significantly induced only in differentiated MSC and MSC-iPSC spheres, and appeared non-detectable in MABs and MAB-iPSCs (Figure 4C). Furthermore, in light of the adipogenic potential of equine MSCs (Spaas et al., 2013), we tested the differentiation efficiency of equine iPSCs toward the adipogenic lineage. Intriguingly, albeit generally limited and variable, the differentiation efficiency into Oil Red+ adipocytes appeared higher in MSC-iPSCs than in MAB-iPSCs (Figure 4D), indicating a possible retention of MSC propensity toward other lineages as well. Thus, in both teratoma and in vitro assays, equine isogenic iPSCs showed intrinsic, discriminable propensities toward the lineages of source stem cells, e.g. myogenic in MAB-iPSCs and chondrogenic in MSC-iPSCs.
Figure 1.
Generation of Equine MAB- and MSC-iPSCs in Isogenic Conditions
(A) Schematic experimental plan.
(B) AP activity staining on AP+ (positive signal) and AP− (background signal) cell fractions from the skeletal muscle.
(C) Immunofluorescence staining for pericytic markers and related isotypes of equine MABs (AP+ cells).
(D) MyHC immunofluorescence staining of equine MABs and MSCs after serum starvation. Myogenic differentiation is apparent as multinucleated myotubes.
(E) Panel of pluripotency characterization for equine iPSCs.
(F) (Left) RT-PCR with specifically cross-reacting (equine-human) primers (eq-, negative equine control, parental cells; hu+, human positive control, H9 ESCs; rt-, negative control of reverse transcription). (Right) RT-PCR for expression of retroviral (retrov-) reprogramming factors (ct+, positive control, fibroblasts freshly transduced with the reprogramming retroviruses).
(G) Euploid karyograms of equine iPSCs at passage 3.
(H) Immunofluorescence analysis for markers of ectodermal (TUJ1+), endodermal (αFP+), and mesodermal (αSMA+) derivative cells after spontaneous iPSC differentiation. All results shown were obtained from cell clones from all three donors (n = 3 independent experiments/cell type).
Scale bars, 100 μm.
Figure 2.
Equine iPSC Intrinsic Propensities to Muscle and Cartilage Derivatives in Teratomas
(A) Representative pictures of derivatives (arrowheads) of ectoderm (upper, ectodermal cysts; lower, neural tube), endoderm (unstructured, gland-like acinar tissues), and mesoderm (upper, adipose tissue; lower, immature bone structures).
(B) Quantitation of striated muscle patches (white arrows) by means of Masson trichrome staining (∗p < 0.05, Mann-Whitney U test, n = 3 independent experiments per cell type).
(C) Quantitation of chondrogenic patches (black arrows) by means of Alcian blue staining (∗p < 0.05, Mann-Whitney U test, n = 3 independent experiments per cell type).
(D) Immunofluorescence staining for lamin A/C on equine iPSC teratomas (n = 4/cell type; ct-, negative control, murine iPSC teratoma; ct+, positive control, human iPSC teratoma).
(E) Staining for lamin A/C and markers of ectodermal (TUJ1), endodermal (αFP), and mesodermal (SARCαACT) derivatives in equine MAB-iPSC-derived teratomas (n = 4 technical replicates per cell type). Magnifications of highlighted fields are shown to the right of original panels to evidence co-localization of lamin A/C (nuclear membrane) and the cytoplasmic markers. Comparable results were obtained with MSC-iPSCs (data not shown).
Histograms represent average values; error bars indicate SD. Scale bars, 100 μm.
Figure 3.
Assessment of Equine iPSC Myogenic Differentiation
(A) Immunofluorescence staining for MyHC of parental cells and iPSCs under BMP/TGF-β blockade.
(B) Quantitation of differentiation efficiency as fraction of cells participating to nascent MyHC+ myotubes (fusion index; ∗p < 0.05, one-way ANOVA with Bonferroni comparison, n = 3 independent experiments per cell type).
(C) qRT-PCR with equine-specific MYH2 primers; data are depicted as fold change versus MABs or MAB-iPSCs (AU, arbitrary units; ∗p < 0.05, unpaired t test, n = 3 independent experiments per cell type). Both analyses revealed higher myogenic propensity in MAB-iPSCs than in MSC-iPSCs (n = 3 independent experiments per cell type).
Histograms represent average values; error bars indicate SD. Scale bars, 100 μm.
Figure 4.
Assessment of Equine iPSC Chondrogenic Differentiation
(A) Alcian blue staining for undifferentiated and chondrogenic microspheres. Indicated percentages refer to the Alcian blue-positive areas (arrows) across the depicted spheres.
(B) Quantitation of cell density and differentiation efficiency in parental cells and iPSCs, comparing undifferentiated (undiff) and differentiated (chondro) spheres (∗p < 0.05, one-way ANOVA with Bonferroni comparison; ns, not significant).
(C) qRT-PCR with equine-specific COMP primers; data are depicted as fold change versus undiff (AU, arbitrary units; nd, not detectable; ∗p < 0.05, one-way ANOVA with Bonferroni comparison; n = 3 independent experiments [3 technical replicates per experiment/cell type]).
(D) Oil Red staining on MAB- and MSC-iPSCs after adipogenic differentiation (red lipid vacuoles denote adipocyte-like cells). Differentiation rate is quantitated as percentage of adipocytes per field (∗p < 0.05, Mann-Whitney U test, n = 3/cell type, n = 3 independent experiments per cell type).
Histograms represent average values; error bars indicate SD. Scale bars, 100 μm.
Discussion
Inconsistencies among cell source isolation and reprogramming techniques can result in iPSCs that may appear similar, but have very different capacities to modulate or contribute to tissue regeneration (Bar-Nur et al., 2011, Hiler et al., 2015). In addition, several studies on the epigenetic variations in iPSCs have raised concerns that these dissimilarities may compromise the clinical applications of iPSCs (Shtrichman et al., 2013). Our data indicate that source-related intrinsic propensity is retained in iPSCs, and quantitatively primes their differentiation toward skeletal muscle and cartilage lineages. The iPSCs used in our study were obtained with human reprogramming factors, and appeared genuinely reprogrammed as equine endogenous genes were reactivated (NANOG, LIN28) and retroviral transgenes were silenced. Albeit far from comprehensive, the pluripotency characterization of our cells is consistent with that of other equine iPSC lines previously reported by independent groups (Breton et al., 2013, Sharma et al., 2014). However, a more translationally relevant insight is expected when non-integrative vectors carrying the equine ortholog factors will be available. Xeno-free conditions for reprogramming and expansion will be necessary also for properly addressing the durability of such propensity (our studies are limited to passage 8 after clone isolation). Such a strategy will indeed allow more refined studies, including epigenetic analyses. In this regard, our in vitro data support the notion that not all epigenetic signatures of parental stem cells are equally erased during reprogramming, thus altering the differentiation efficiency of iPSCs. The effects appeared relevant for myogenic and chondrogenic lineages. Our results showing enhanced differentiation with equine iPSCs seem consistent with the findings previously reported with murine MAB-iPSCs (Quattrocelli et al., 2011) and human reprogrammed chondrocytes (Borestrom et al., 2014). In this regard, an important issue to be considered is that we reprogrammed MABs and MSCs, and not differentiated cells, i.e. myocytes and chondrocytes. This was based on clinical and experimental reasons. From a translational perspective for iPSC applications, stem cells are likely superior to terminally differentiated progenies, due to greater expansion ability and higher susceptibility to reprogramming protocols (Cai et al., 2010). Moreover, MABs and MSCs are currently under clinical trial scrutiny in human patients for the treatment of muscle (EudraCT#2011-000176-33) and cartilage conditions (US-CT#NCT01879046). Also, our attempts to reprogram the differentiated progenies of equine MABs and MSCs failed (data not shown), likely due to very low transduction efficiency and halted proliferation. Importantly, as shown by the results of parallel differentiation of parental cells and iPSCs, MAB- and MSC-iPSCs efficiently retained the myogenic and chondrogenic differentiation biases, respectively. However, it remains unknown whether MSCs from other body regions (e.g. bone marrow, adipose tissue) will exhibit different levels of chondrogenic bias and reprogramming potency. Furthermore, our results for the adipogenic lineage, although limited to the iPSC pools, suggest that other differentiation propensities might be retained too, albeit likely with different efficiencies. Dedicated studies are needed to address this point, as the purpose of our study was centered on muscle and cartilage. In conclusion, the observations reported here hint at further studies of in vivo/in vitro differentiation, possibly in combination with known genetic defects and tailored corrections, in order to better harness the potential of equine iPSCs for veterinary disease modeling and regenerative medicine.
Experimental Procedures
Cell Isolation and Expansion
All procedures involving live horses were performed under the specifications of the Ethical Approval Forms EC_2012_001 for blood withdrawal and EC_2014_003 for muscle tissue sampling (Ethics Committee, Global Stem Cell Technology). Equine MABs were isolated from three syngeneic female siblings of the French trotter breed aged between 5 and 7 years. Splenius muscle biopsies were taken in sterile conditions under local anesthesia, and equine MABs were isolated using previously reported procedures for murine and human MABs (Quattrocelli et al., 2012). In brief, muscle biopsy pieces (approximately 2 × 2 × 2 mm) were cultured on collagen (Sigma-Aldrich)-coated dishes (NUNC) and MABs were isolated as AP+-sorted population from the outgrowths. Peripheral blood MSCs from the same horses were already isolated and characterized (Spaas et al., 2013). Equine MABs were then passaged and expanded on collagen-coated culture vessels (NUNC) in IMDM10% medium (IMDM supplemented with 10% FBS, 1% penicillin-streptomycin [pen/strep], 1% L-glutamine, 1% sodium pyruvate, 1% non-essential amino acids [NEAA], 1% insulin-transferrin-selenium supplement [Thermo Fisher], 0.2% 2-mercaptoethanol, and 5 ng/ml basic fibroblast growth factor [bFGF; Peprotech]) at 5% O2/5% CO2 and 37°C. Equine MSCs were passaged and expanded on tissue culture vessels (NUNC) in DMEM10% medium (DMEM supplemented with 10% FBS, 1% pen/strep, 1% L-glutamine, and 1% sodium pyruvate) at 5% O2/5% CO2 and 37°C. Equine cells were reprogrammed analogously to procedures described for murine iPSCs (Quattrocelli et al., 2011). In brief, cells were transduced with retroviral particles carrying human OCT4, SOX2, KLF4, and CMYC cDNA sequences (pMX vectors, Addgene) and seeded at low confluence on a feeder layer of mitomycin-treated primary murine embryonic fibroblasts (iMEFs). Reprogramming efficiency was calculated as percentage of ESC-like colonies versus starting cell number prior to transduction. Equine iPSC colonies were picked at 4–5 weeks after transduction, clonally expanded, and characterized for pluripotency marker expression. Equine MAB- and MSC-iPSCs were cultured on a feeder layer of iMEFs in equine iPSC medium (DMEM-F12 with Nutrient mix and HEPES, supplemented with 20% KnockOut serum, 1% pen/strep, 1% NEAA, 0.2% 2-mercaptoethanol, and 5 ng/ml bFGF) at 5%O2/5%CO2 and 37°C.
Differentiation Assays
The myogenic propensity of somatic MABs and MSCs was tested in serum starvation conditions, i.e. in DMEM2% medium (high-glucose DMEM supplemented with 2% horse serum [Thermo Fisher], 1% pen/strep, and 1% L-glutamine) for 7 days. Fusion index was calculated as the percentage of nuclei enclosed in myotubes per field. For myogenic differentiation, iMEF-deprived iPSCs were seeded at 4,000 cells/cm2 on gelatin (Millipore)-coated vessels, and kept in F12-20% medium (DMEM-F12 with Nutrient mix and HEPES, supplemented with 20% FBS, 1% pen/strep, 1% NEAA, and 0.2% 2-mercaptoethanol) for 20 days. Cells were then reseeded at 4,000 cells/cm2 on gelatin-coated vessels and kept in F12-20% medium, supplemented with 1% insulin-transferrin-selenium (ITS) supplement (Thermo Fisher), for 15 days. Terminal myogenic maturation was achieved in DMEM2% medium, supplemented with 1% ITS, 50 ng/ml recombinant human NOGGIN (BMP inhibitor; Thermo Fisher), and 1 μM SB431542 hydrate (TGF-β inhibitor; Sigma-Aldrich) for 15 days. All aforementioned differentiation assays were performed at 5% O2/5% CO2 and 37°C. To initiate chondrogenic differentiation, 3 × 105 cells were resuspended in 5 ml of medium and centrifuged for 5 min at 150 × g. Subsequently, 0.5 ml of chondrogenic differentiation medium (basal differentiation medium [Lonza] supplemented with 10 ng/ml TGF-β3 [Sigma]) or expansion medium were added per pellet and refreshed twice a week. Four chondrospheres and four undifferentiated control spheres were established per cell type per horse, and evaluated after 10 days with Alcian blue staining and PCR. Adipogenic differentiation was induced with the Mesenchymal Adipogenesis Assay (Millipore) on plated embryoid bodies after cell sprouting (>95% confluence). The manufacturer's instructions were followed for differentiation cocktails, timing, and staining.
Teratoma Assay
All procedures involving live mice were performed under the specifications of the Ethical Approval #P150/2014 (Ethics Committee, KU Leuven). Immunodeficient 2-month-old Rag2-null/Il2rg-null mice were injected with a 100-μl suspension of 350,000 equine iPSCs in 1:2 diluted Matrigel (Thermo Fisher) subcutaneously in the nape region. Teratomas (n = 3/iPSC type) were retrieved between 4 and 6 weeks after injection, when visible and not yet ulcerated.
Immunofluorescence, Histological, and Colorimetric Staining
Immunofluorescence staining was performed following the commonly used steps of Triton-based (Sigma-Aldrich) permeabilization (avoided for MAB surface marker staining), background blocking with donkey serum (Sigma-Aldrich), overnight incubation with primary antibody at 4°C, 1 hr of incubation with 1:500 AlexaFluor-conjugated donkey secondary antibodies (Thermo Fisher), and final counterstain with Hoechst. Here follows the list of primary antibodies and relative dilutions: mouse anti-NG2 PE-conjugated (R&D Systems #MAB6689), 1:100; mouse anti-CD140a and anti-CD140b APC-conjugated (Antibodies Online #ABIN910520/ABIN457302), 1:50; mouse anti-CD44 APC-conjugated (eBioscience #17-0441-81), 1:200; goat anti-OCT4 (Abcam #ab19857), 1:500; goat anti-SOX2 (Santa Cruz #sc-17320), 1:100; rabbit anti-Nanog (Abcam #ab80892), 1:500; goat anti-Lin28 (Santa Cruz #sc-54032), 1:100; mouse anti-MyHC (DSHB, #MF20), 1:8; rabbit anti-TUJ1 (Millipore #04-1049), 1:300; rabbit anti-αFP (Abcam #ab87635), 1:500; mouse anti-αSMA (Sigma-Aldrich #A2547), 1:300; mouse anti-lamin A/C (Millipore #05-714), 1:100. Histological staining (H&E and Alcian blue, reagents from Roche-Ventana; Masson trichrome, reagents from Sigma-Aldrich) was performed on 5-μm-thick slices from paraffin-embedded tissue pieces following the manufacturer's instructions. AP staining was performed exposing 2% PFA-fixed cells to the BCT/NBT reagent (Sigma-Aldrich; resuspended in dH2O) for 1 hr. Quantitation of myogenic differentiation was performed on at least ten independent fields per cell type. Quantitation of teratoma derivatives and chondrogenic differentiation of the microspheres was performed on at least ten serial sections (approximately 100-μm interval between consecutive sections).
Molecular Assays
For pluripotency marker expression, 1:5 diluted cDNA was obtained from 1 μg of total RNA (SSIII cDNA production kit and RNA extraction kit from Thermo Fisher). RT-PCR was performed using Taq polymerase-associated reagents (Thermo Fisher) and conditions (95°C 30 s, 60°C 30 s, 72°C 60 s; 40× for endogenous, 25× for retroviral genes). Primers: OCT4 Fw GAGGCTCTGCAGCTCAGTTT, Rev CTCCAGGTTGCCTCTCACTC; SOX2 Fw AACCAGCGCATGGACAGTTA, Rev GACTTGACCACCGAACCCAT; NANOG Fw ATACCTCAGCCTCCAGCAGA, Rev AGCCCCGGGTAGTCTGTAGT; LIN28 Fw TGTAAGTGGTTCAACGTGCG, Rev CAGCTTACTCTGGTGCACAA; PGK Fw CTGGGCAAGGATGTTTTGTT, Rev TATTTTGGCTGGCTCAGCTT; retrov-OCT4 Fw CCCCAGGGCCCCATTTTGGTACC; retrov-SOX2 Fw GGCACCCCTGGCATGGCTCTTGGCTC; retrov-KLF4 Fw ACGATCGTGGCCCCGGAAAAGGACC; retrov-cMYC Fw CAACAACCGAAAATGCACCAGCCCCAG; pMX Rev (for all retrov-primers) TTATCGTCGACCACTGTGCTGCTG. MYH2 qRT-PCR was performed by means of SybrGreen mix (Thermo Fisher) using the Viia7 384-plate reader (Thermo Fisher; final primer concentration, 100 nM; final volume, 10 μl; PGK, internal reference; thermal profile, 95°C 15 s, 60°C 60 s, 40×). Primers: MYH2 Fw TGAGTCCCAGGTCAACAAGC, Rev TTCCATAGCGTGAAGGCATGA. For the COMP qRT-PCR assay, total RNA (totRNA) was isolated with the High Pure Isolation Kit (Roche Diagnostics). TotRNA integrity was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies; RIN values, 8.8–10). TotRNA was reverse transcribed with the iScript cDNA Kit (Bio-Rad). qRT-PCR assay for COMP and reference genes SDHA and ACTB (selected with geNorm) was performed with FastStart essential DNA Green Master (Roche Diagnostics) on the LightCycler 480 (Roche) with following cycling conditions: 20 s at 95°C, 40 s at annealing temperature (sdha, 59°C; actb, comp, 60°C), 1 min at 72°C, 45×. SDHA, ACTB primers: see De Schauwer et al., 2014. COMP primers: Fw GACTACGCGGGCTTCATCTT, rev CTGCTCCATCTGCTTCCACA. Specificity was proved through melting curve analysis. Expression levels were analyzed and normalized using qBase+ (version 3.0). Assays were performed according to the MIQE guidelines (Bustin et al., 2009).
Statistical Analysis
Power analysis was performed using Sample Size Calculator (power, 0.80; alpha, 0.05). Statistical tests were chosen based on data distribution. For teratoma derivatives, the Mann-Whitney U test was applied (p < 0.05). For MYH2 levels, the unpaired t test was applied (p < 0.05). For myogenic efficiency, chondrogenic efficiency, COMP levels, and cell density, one-way ANOVA with Bonferroni multi-comparison was applied (p < 0.05). All tests were performed by means of Prism v5.0 (GraphPad).
Acknowledgments
This work has been funded by “Opening The Future” Campaign (EJJ-OPTFUT-02010), CARE-MI FP7, AFM, CARIPLO, FWO (#G060612N, #G0A8813N and #G088715N), GOA, IUAP, and OT grants. M.Q. is supported by FWO (Postdoctoral Fellowship #1263314N) and AFM (Trampoline Grant #18373). We are grateful to Michael David for technical help with the COMP qRT-PCR assay. We thank Christina Vochten and Vicky Raets for professional administrative assistance. We would also like to thank Paolo Luban and Rondoufonds voor Duchenne Onderzoek for kind donations. J.H.S. declares competing financial interests as shareholder in Global Stem Cell Technology (GST). S.Y.B. and J.H.S. are both employed by GST and are inventors of several pending patents owned by GST (BE2012/0656; WO2014053418A9; WO2014053420A1; PCT/EP2013/075782).
Published: January 12, 2016
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Contributor Information
Jan H. Spaas, Email: jan.spaas@anacura.com.
Maurilio Sampaolesi, Email: maurilio.sampaolesi@med.kuleuven.be.
References
- Bar-Nur O., Russ H.A., Efrat S., Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Borestrom C., Simonsson S., Enochson L., Bigdeli N., Brantsing C., Ellerstrom C., Hyllner J., Lindahl A. Footprint-free human induced pluripotent stem cells from articular cartilage with redifferentiation capacity: a first step toward a clinical-grade cell source. Stem Cells Transl. Med. 2014;3:433–447. doi: 10.5966/sctm.2013-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton A., Sharma R., Diaz A.C., Parham A.G., Graham A., Neil C., Whitelaw C.B., Milne E., Donadeu F.X. Derivation and characterization of induced pluripotent stem cells from equine fibroblasts. Stem Cell Dev. 2013;22:611–621. doi: 10.1089/scd.2012.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeckx S., Zimmerman M., Crocetti S., Suls M., Marien T., Ferguson S.J., Chiers K., Duchateau L., Franco-Obregon A., Wuertz K. Regenerative therapies for equine degenerative joint disease: a preliminary study. PLoS One. 2014;9:e85917. doi: 10.1371/journal.pone.0085917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Cai J., Li W., Su H., Qin D., Yang J., Zhu F., Xu J., He W., Guo X., Labuda K. Generation of human induced pluripotent stem cells from umbilical cord matrix and amniotic membrane mesenchymal cells. J. Biol. Chem. 2010;285:11227–11234. doi: 10.1074/jbc.M109.086389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrian-Serrano A., Stout T., Dinnyes A. Veterinary applications of induced pluripotent stem cells: regenerative medicine and models for disease? Vet. J. 2013;198:34–42. doi: 10.1016/j.tvjl.2013.03.028. [DOI] [PubMed] [Google Scholar]
- De Schauwer C., Goossens K., Piepers S., Hoogewijs M.K., Govaere J.L., Smits K., Meyer E., Van Soom A., Van de Walle G.R. Characterization and profiling of immunomodulatory genes of equine mesenchymal stromal cells from non-invasive sources. Stem Cell Res. Ther. 2014;5:6. doi: 10.1186/scrt395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell M.A., Nguyen H., Shroyer N. Somatic stem cell heterogeneity: diversity in the blood, skin and intestinal stem cell compartments. Nat. Rev. Mol. Cell Biol. 2015;16:299–309. doi: 10.1038/nrm3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiler D., Chen X., Hazen J., Kupriyanov S., Carroll P.A., Qu C., Xu B., Johnson D., Griffiths L., Frase S. Quantification of retinogenesis in 3D cultures reveals epigenetic memory and higher efficiency in iPSCs derived from rod photoreceptors. Cell Stem Cell. 2015;17:101–115. doi: 10.1016/j.stem.2015.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotini A.G., Chang C.J., Boussaad I., Delrow J.J., Dolezal E.K., Nagulapally A.B., Perna F., Fishbein G.A., Klimek V.M., Hawkins R.D. Functional analysis of a chromosomal deletion associated with myelodysplastic syndromes using isogenic human induced pluripotent stem cells. Nat. Biotechnol. 2015;33:646–655. doi: 10.1038/nbt.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocelli M., Palazzolo G., Floris G., Schoffski P., Anastasia L., Orlacchio A., Vandendriessche T., Chuah M.K., Cossu G., Verfaillie C. Intrinsic cell memory reinforces myogenic commitment of pericyte-derived iPSCs. J. Pathol. 2011;223:593–603. doi: 10.1002/path.2845. [DOI] [PubMed] [Google Scholar]
- Quattrocelli M., Palazzolo G., Perini I., Crippa S., Cassano M., Sampaolesi M. Mouse and human mesoangioblasts: isolation and characterization from adult skeletal muscles. Methods Mol. Biol. 2012;798:65–76. doi: 10.1007/978-1-61779-343-1_4. [DOI] [PubMed] [Google Scholar]
- Quattrocelli M., Costamagna D., Giacomazzi G., Camps J., Sampaolesi M. Notch signaling regulates myogenic regenerative capacity of murine and human mesoangioblasts. Cell Death Dis. 2014;5:e1448. doi: 10.1038/cddis.2014.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaolesi M., Blot S., D'Antona G., Granger N., Tonlorenzi R., Innocenzi A., Mognol P., Thibaud J.L., Galvez B.G., Barthelemy I. Mesoangioblast stem cells ameliorate muscle function in dystrophic dogs. Nature. 2006;444:574–579. doi: 10.1038/nature05282. [DOI] [PubMed] [Google Scholar]
- Sanchez-Freire V., Lee A.S., Hu S., Abilez O.J., Liang P., Lan F., Huber B.C., Ong S.G., Hong W.X., Huang M. Effect of human donor cell source on differentiation and function of cardiac induced pluripotent stem cells. J. Am. Coll. Cardiol. 2014;64:436–448. doi: 10.1016/j.jacc.2014.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel L.V., Fortier L.A., McIlwraith C.W., Nobert K.M. Therapeutic use of stem cells in horses: which type, how, and when? Vet. J. 2013;197:570–577. doi: 10.1016/j.tvjl.2013.04.018. [DOI] [PubMed] [Google Scholar]
- Sharma R., Livesey M.R., Wyllie D.J., Proudfoot C., Whitelaw C.B., Hay D.C., Donadeu F.X. Generation of functional neurons from feeder-free, keratinocyte-derived equine induced pluripotent stem cells. Stem Cell Dev. 2014;23:1524–1534. doi: 10.1089/scd.2013.0565. [DOI] [PubMed] [Google Scholar]
- Shtrichman R., Germanguz I., Itskovitz-Eldor J. Induced pluripotent stem cells (iPSCs) derived from different cell sources and their potential for regenerative and personalized medicine. Curr. Mol. Med. 2013;13:792–805. doi: 10.2174/1566524011313050010. [DOI] [PubMed] [Google Scholar]
- Smith R.K., Garvican E.R., Fortier L.A. The current ‘state of play’ of regenerative medicine in horses: what the horse can tell the human. Regen. Med. 2014;9:673–685. doi: 10.2217/rme.14.42. [DOI] [PubMed] [Google Scholar]
- Spaas J.H., De Schauwer C., Cornillie P., Meyer E., Van Soom A., Van de Walle G.R. Culture and characterisation of equine peripheral blood mesenchymal stromal cells. Vet. J. 2013;195:107–113. doi: 10.1016/j.tvjl.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Tecirlioglu R.T., Trounson A.O. Embryonic stem cells in companion animals (horses, dogs and cats): present status and future prospects. Reprod. Fertil. Dev. 2007;19:740–747. doi: 10.1071/rd07039. [DOI] [PubMed] [Google Scholar]
- van Weeren P.R., Jeffcott L.B. Problems and pointers in osteochondrosis: twenty years on. Vet. J. 2013;197:96–102. doi: 10.1016/j.tvjl.2013.03.048. [DOI] [PubMed] [Google Scholar]
- Votion D.M., Serteyn D. Equine atypical myopathy: a review. Vet. J. 2008;178:185–190. doi: 10.1016/j.tvjl.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Yamanaka S. A fresh look at iPS cells. Cell. 2009;137:13–17. doi: 10.1016/j.cell.2009.03.034. [DOI] [PubMed] [Google Scholar]