Abstract
Centrioles are required to assemble centrosomes for cell division and cilia for motility and signaling. New centrioles assemble perpendicularly to pre-existing ones in G1-S and elongate throughout S and G2. Fully-elongated daughter centrioles are converted into centrosomes during mitosis to be able to duplicate and organize pericentriolar material in the next cell cycle. Here we show that centriole-to-centrosome conversion requires sequential loading of Cep135, Ana1:Cep295 and Asterless:Cep152 onto daughter centrioles during mitotic progression. This generates a molecular network spanning from inner- to outer-most parts of the centriole. Ana1 forms a molecular strut within the network and its essential role can be substituted by an engineered fragment providing an alternative linkage between Asterless and Cep135. This conserved architectural framework is essential for loading Asterless:Cep152, partner of the master regulator of centriole duplication, Plk4. Our study thus uncovers the molecular basis for centriole-to-centrosome conversion that renders daughter centrioles competent for motherhood.
Centrioles are required to assemble both centrosomes and cilia and their dysfunction is associated with multiple inherited diseases and cancer1,2. Centriole duplication and maturation are tightly regulated; the centriole pair of each centrosome disengages as cells exit mitosis allowing new pro-centrioles to assemble in G1-S and elongate during S and G2. Although the newly assembled centriole reaches its full length in early mitosis, it cannot duplicate or organize pericentriolar material (PCM) until it has passed through mitosis3,4. The molecular basis of this conversion from centriole to centrosome has remained mysterious.
Understanding the conversion requires knowledge of which centriolar components are loaded during mitosis, their spatial organization and the timing and dependencies of their recruitment. Recent developments in super resolution microscopy have proved invaluable in analyzing spatial relationships between centrosome components5-10. In Drosophila, Ana2:STIL and Sas-6 first appear at the site of pro-centriole formation9,11 (Zone I, Fig. 2a). Ana2:STIL is phosphorylated by Polo-like kinase 4 (Plk4)11,12, master regulator of centriole duplication13-16, thus recruits Sas-611,12 which can self-assemble into nine dimers to form the nine-fold symmetrical, proximally-located cartwheel17,18. Bld10:Cep135, also in Zone I9, is critical for cartwheel formation in Chlamydomonas and Paramecium19,20 and its human counterpart connects Sas-6 to CPAP and the centriole wall21. Drosophila Sas-4 is part of the Zone II cylinder and Sas-4:CPAP is considered to promote polymerization of centriolar microtubules22-24. The distal cap comprising CP110 and its partners constitutes Zone V that, together with proteins of Zones I and II, form the core of both mother and daughter centrioles. Zone III proteins, Asterless (Asl), Plk4 and Dplp (Drosophila pericentrin-like protein), associate with mother centriole throughout the cell cycle but have not yet assembled onto the daughter in interphase. Thus far Asl is the sole factor known to recruit Plk4 to the centrosome in Drosophila and hence is key for centriole duplication25,26. Zone IV proteins (Polo, Spd-2, Centrosomin and γ-tubulin) also do not associate with daughter centriole, but accumulate robustly around the mother upon mitotic entry.
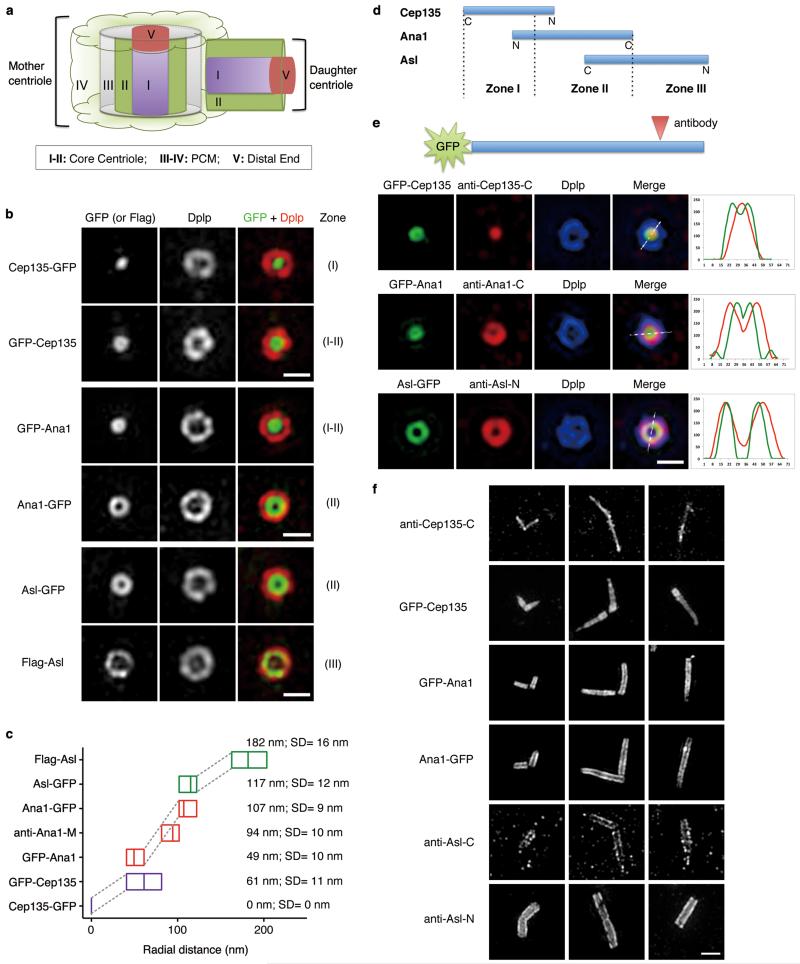
Figure 2. Cep135, Ana1 and Asl are extended molecules that span from inner to outer centriole.
(a) Scheme showing different Zones in Drosophila centrosome (modified from ref. 9).
(b) D.Mel-2 cells constitutively expressing GFP-tagged Cep135, Ana1 or Asl were immunostained to reveal Dplp and DNA (not shown). Cells expressing Flag-Asl were immunostained with additional anti-Flag antibody (bottom panel). The signal of Cep135, Ana1 or Asl appears in different Zones at centriole when the fluorophore-epitope is at different end of the protein. Scale bars, 500 nm.
(c) Average radial distance of different regions of Cep135, Ana1 or Asl. Low-high bar (horizontal) shows the range of radius and vertical line indicates mean. Cep135 in purple; Ana1, red; Asl, green. SD, Standard Deviation. From bottom up, n=35, 22, 14, 22, 15, 32 and 15 centrosomes, respectively.
(d) Map showing the relative positions of Cep135, Ana1 and Asl within single centriole. N and C indicate protein orientation.
(e) D.Mel-2 cells constitutively expressing GFP-tagged Cep135, Ana1 or Asl were immunostained for the respective antibodies recognizing the opposite end of the proteins and Dplp. Line scans reveal the stretched structure of exogenous Cep135, Ana1 and Asl at centriole. Scale bar, 500 nm.
(f) Representative images of centrioles in successive developmental stages of fly spermatocytes. Immunofluorescence signals are seen at positions of successively increasing diameter extending from (inner-most) C-terminus of Cep135; N-terminus of Cep135; N-terminus of Ana1; C-terminus of Ana1; C-terminus of Asl; to (outer-most) N-terminus of Asl. Scale bar, 1 μm.
Here we have determined which proteins known to be required for Drosophila centriole duplication27,28 are loaded onto the daughter late in interphase and during mitosis. This has revealed a conserved architectural network of Cep135-Ana1:Cep295-Asl:Cep152 that is key for centriole-to-centrosome conversion in both Drosophila and human. Our findings thus account for the final stages in the assembly of the daughter centriole that convert it into a mature mother able to duplicate.
Results
Sequential loading of Cep135, Ana1 and Asl in centriole-to-centrosome conversion
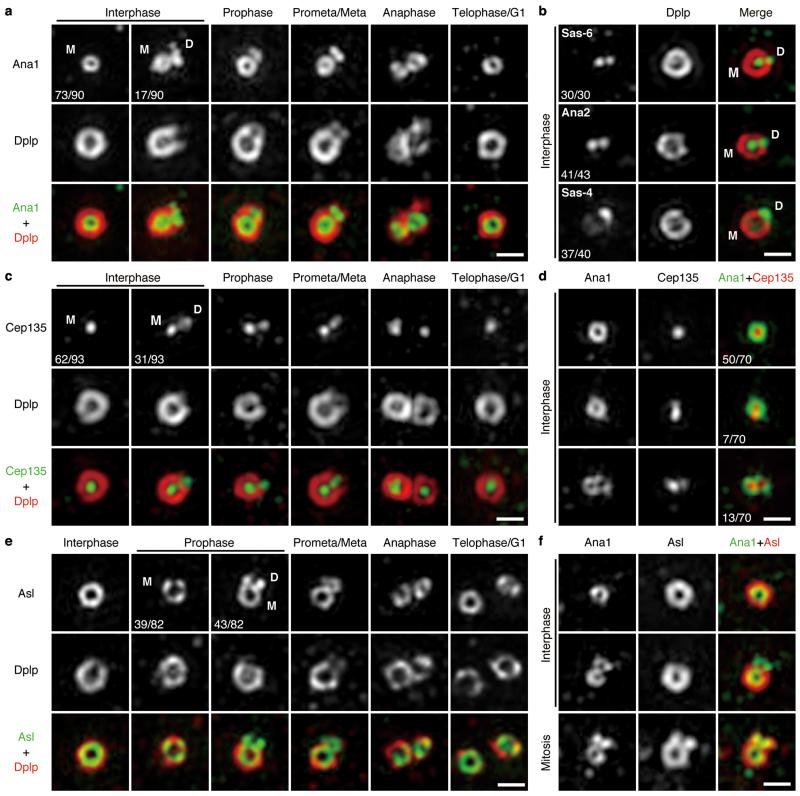
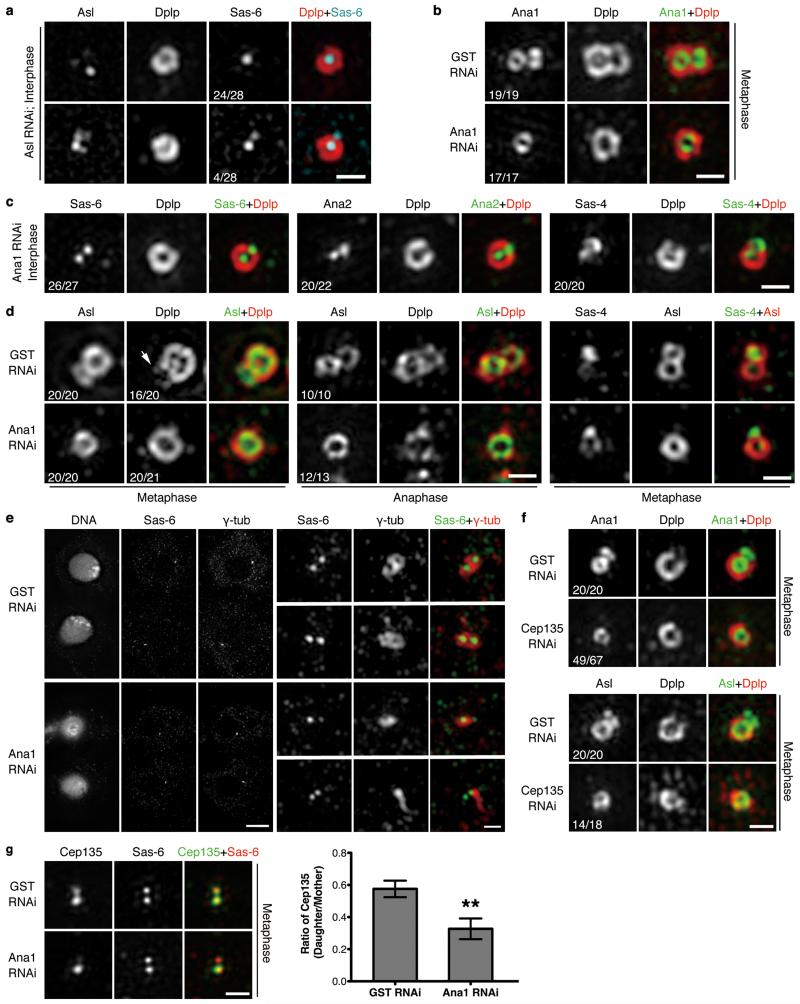
To gain understanding of centriole-to-centrosome conversion, we applied 3D Structured Illumination Microscopy (3D-SIM) to determine the sequential recruitment of centriolar proteins critical for duplication. Dplp is not required for centriole duplication29 and served as a Zone III marker. We found that Cep135, Ana1 and Asl were recruited to the daughter centriole as it matured: Ana1 was on mother centrioles in all interphase cells but only on a small proportion of daughters (19%, n=90, Fig. 1a and Supplementary Fig. 1a), suggesting its recruitment in late interphase. In contrast, Sas-6, Ana2 and Sas-4 were associated with both mother and daughter centrioles in virtually all interphase cells (Fig. 1b), in accord with their roles in early centriole duplication. Ana1 remained associated with daughter centrioles throughout prophase until anaphase, and was on disengaged mother and daughter (now new mother) centrioles in telophase-G1 (Fig. 1a). Cep135 recruitment followed a similar pattern (Fig. 1c and Supplementary Fig. 1b). Of those interphase centrosomes in which Cep135 could be detected on daughters, 65% (n=20) also had Ana1. The remaining 35% had weak Cep135 staining but no detectable Ana1 (Fig. 1d). Thus Cep135 appears to load slightly ahead of Ana1. Asl was first detected on daughter centrioles from prophase onwards (52%, n=82), and formed a complete ring by prometaphase-metaphase, hence ahead of Dplp’s gradual recruitment (Fig. 1e and Supplementary Fig. 1c) and the loading of Ana1 (Fig. 1f). Together Cep135, Ana1 and Asl load sequentially onto daughter centriole from late interphase to early mitosis.
Figure 1. Sequential loading of Cep135, Ana1 and Asl during centriole-to-centrosome conversion.
(a) Localization of endogenous Ana1 throughout cell cycle. D.Mel-2 cells were immunostained to reveal Ana1, Dplp (Zone III marker) and DNA (to distinguish cell cycle stages, see Supplementary Fig. 1a), and analyzed by 3D-SIM. Interphase, prophase, prometaphase-metaphase, anaphase and telophase-G1 centrioles were shown. Only 19% (n=90) of interphase centrosomes have Ana1 signal on daughter centriole, suggesting Ana1 is recruited to daughter in late interphase. M, mother centriole; D, daughter.
(b) Centrioles immunostained to reveal Dplp (red), DNA (not shown) and Sas-6, Ana2 or Sas-4 (green). Sas-6, Ana2 and Sas-4 appear on both mother and daughter centrioles in majority of interphase centrosomes (100%, 95% and 92%, respectively), indicating their early loading onto the daughters. n=30, 43 and 40 centrosomes.
(c) Localization of endogenous Cep135 throughout cell cycle. 33% (n=93) of interphase centrosomes have Cep135 signal on the daughter. See Supplementary Fig. 1b for DNA staining.
(d) Centrioles immunostained to reveal Ana1 (green), Cep135 (red) and DNA (not shown). Only 10% (n=70) of interphase centrosomes show earlier (less distinct) loading of Cep135 to the daughter than Ana1, indicating a close temporal recruitment of these two.
(e) Localization of endogenous Asl throughout cell cycle. Asl starts to load on the daughter from prophase, when 52% (n=82) of centrosomes show Asl signal on the daughter. See Supplementary Fig. 1c for DNA staining.
(f) Centrioles immunostained to reveal Ana1 (green), Asl (red) and DNA (not shown). Recruitment of Asl to the daughter happens later than Ana1.
All scale bars, 500 nm.
Cep135, Ana1 and Asl extend from inner to outer centriole
To add a spatial dimension to the above temporal framework we tagged Cep135, Ana1 and Asl at their N- or C-termini with GFP for constitutive expression in D.Mel-2 cells. All showed centrosomal localization except for GFP-Asl, which we later replaced by Flag-Asl that did associate with centrosomes. Each tagged protein was able to rescue centriole duplication following depletion of its endogenous counterpart (Supplementary Fig. 2a and 3). Strikingly, the three proteins each extended across different centriolar Zones (Fig. 2a-d). The C-terminal Cep135 tag occupied a dot in Zone I (see also9) whereas its tagged N-terminus lay at the border of Zones I and II as a ring (61±11 nm radius). The N-terminal Ana1 tag occupied a relatively smaller ring (49±10 nm radius) suggesting Ana1 and Cep135 partially overlap. The C-terminal Ana1 tag exhibited a ring of 107±9 nm radius in Zone II, and immunostaining with an antibody against the middle part of Ana1 (622-981aa; Fig. 1a and Supplementary Fig. 2a) revealed an intermediate ring of 94±10 nm radius. Together this suggests Ana1 has an extended linear arrangement in the centriole. The C-terminus of Asl occupied a similar position to the Ana1 C-terminus (117±12 nm radius; see also7), whereas the N-terminal Asl tag occupied a ring of radius 182±16 nm within Zone III (see also7). To visualize the extended structure of each protein within single centriole, we constitutively expressed GFP-tagged Cep135, Ana1 or Asl and immunostained with antibodies against the opposite end of the respective protein (anti-Cep135-C30; anti-Ana1-C, 1400-1729aa; anti-Asl-N, 1-300aa). Line scans of the double end-labelled molecules showed that each had an extended configuration within the same centriole (Fig. 2e). Cep135, Ana1 and Asl also appeared to have their overlapping termini localized to successively increasing radial positions in the elongated centrioles and basal bodies of Drosophila spermatocytes (Fig. 2f; see also9,31). Thus, together Cep135, Ana1 and Asl span from the inner- to the outer-most part of Drosophila centrioles both in cultured cells and spermatocytes.
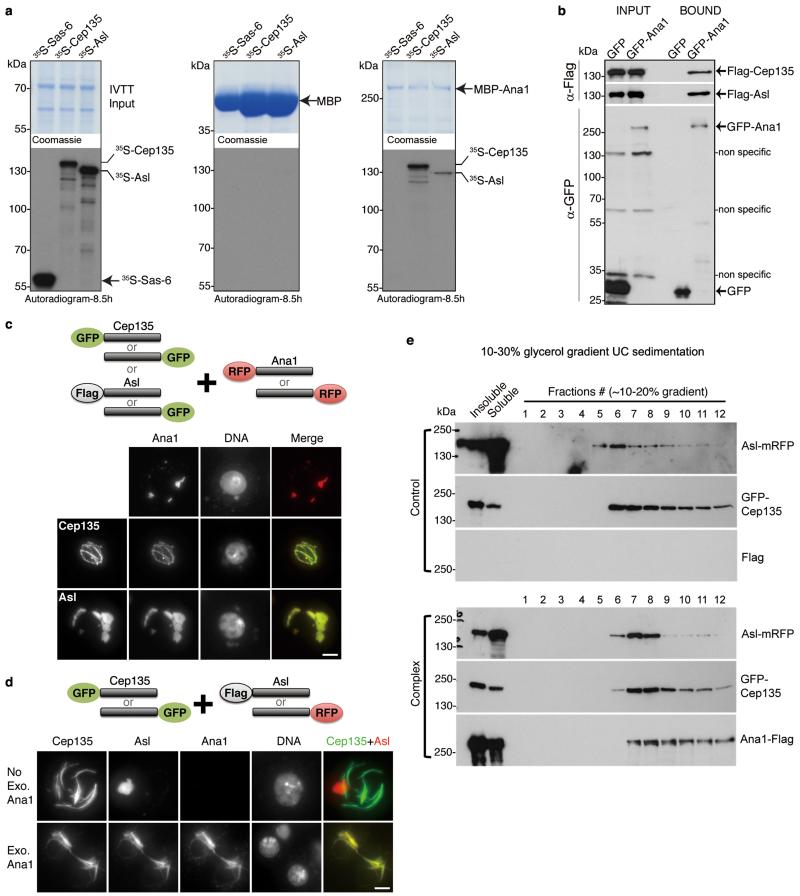
Figure 3. Ana1 provides a molecular linkage between Cep135 and Asl.
(a) Recombinant MBP or MBP-Ana1 was immobilized on amylose resin and incubated with 35S-Met-labelled Cep135, Asl or Sas-6 (negative control) synthesized by coupled in vitro transcription-translation (IVTT). Inputs (9%, left panel) and affinity-purified complexes (middle and right panels) were subjected to SDS-PAGE, stained (Coomassie, upper panels), and dried for autoradiography (lower panels). Note: MBP-Ana1 directly and specifically binds to Cep135 and Asl. Uncropped scans are in Supplementary Fig. 6.
(b) GFP or GFP-Ana1 was transiently co-expressed with Flag-Cep135 or Flag-Asl in D.Mel-2 cells for GFP-Trap pull-downs. Inputs and bound proteins were analyzed by Western blotting. GFP-Ana1 specifically co-purifies with Flag-Cep135 and Flag-Asl. Uncropped scans are in Supplementary Fig. 6.
(c) Cep135 or Asl was transiently co-expressed with Ana1. Proteins were tagged either at the N- or C-terminus as indicated (cartoon, upper part). Representative images show both Cep135 (lower part, middle panel) and Asl (lower part, bottom panel) co-localize with Ana1 in cytoplasm, observed by GFP (green) or mRFP (red) signal. Flag-Asl was detected with anti-Flag antibody (green).
(d) Cep135 and Asl were transiently co-expressed in cells, with or without additional Ana1 construct. Cep135 visualized by GFP fluorescence; Asl and Ana1, by mRFP fluorescence or Flag immunostaining as appropriate (cartoon, upper part). Cep135 and Asl form independent assemblies in cytoplasm in the absence of exogenous Ana1 (lower part, upper panel) but colocalize in the presence of exogenous Ana1 (lower part, lower panel). Scale bars in c and d, 5 μm.
(e) Glycerol gradient sedimentation. D.Mel-2 cells were transiently co-transfected with GFP-Cep135 and Asl-mRFP (Control) or Ana1-Flag, GFP-Cep135 and Asl-mRFP (Complex), lysed and sedimented on 10-30% linear glycerol gradients and collected as 34 fractions. In the presence of Ana1-Flag, GFP-Cep135 and Asl-mRFP perfectly co-sediment (lower panel); when Ana1-Flag is absent, they only partially co-migrate with distinct main peaks (upper panel).
Ana1 provides a molecular linkage between Cep135 and Asl
The overlapping localization and the sequential recruitment of Cep135, Ana1 and Asl led us to ask whether they are physically linked. We approached this in three ways: first, we asked whether Ana1 could interact with Cep135 or Asl directly in vitro. We purified and immobilized recombinant MBP-tagged Ana1 and determined whether it could bind 35S-Methionine-labelled Cep135 or Asl synthesized by in vitro transcription-translation (IVTT). This revealed direct and specific binding of MBP-Ana1 to Cep135 and Asl (Fig. 3a). Second, GFP-Trap pull-downs from D.Mel-2 cells co-expressing GFP-Ana1 and Flag-tagged Cep135 or Asl revealed specific interactions between Ana1 and both Cep135 and Asl (Fig. 3b). Thirdly we co-expressed Ana1 with either Cep135 or Asl at levels higher than required for their centriole incorporation and observed the supramolecular structures that formed in the cytoplasm (Fig. 3c). Ana1 typically formed small globular bodies by itself whereas when co-expressed with Cep135, it co-localized into bundles typical of Cep135 over-expression. When Ana1 was co-expressed with Asl, they co-localized in structures typical of over-expression of Asl alone. Thus Ana1 is able to complex with either Cep135 or Asl in the cytoplasm. We then asked whether Ana1 could bridge Cep135 and Asl to give a trimeric complex (Fig. 3d). When we co-expressed only Cep135 and Asl in cells, they formed characteristic independent assemblies. However, co-expression with Ana1 brought the three molecules into the same complex, indicating that Ana1 can link Cep135 and Asl. We further analyzed the complex formed between GFP-Cep135, Asl-mRFP and Ana1-Flag on glycerol gradient (Fig. 3e). GFP-Cep135 and Asl-mRFP perfectly co-sedimented in the presence of exogenous Ana1, suggesting they are parts of a common assembly. In contrast, when only GFP-Cep135 and Asl-mRFP were co-expressed, they sedimented as distinct peaks providing further proof that Ana1 is needed to link these molecules.
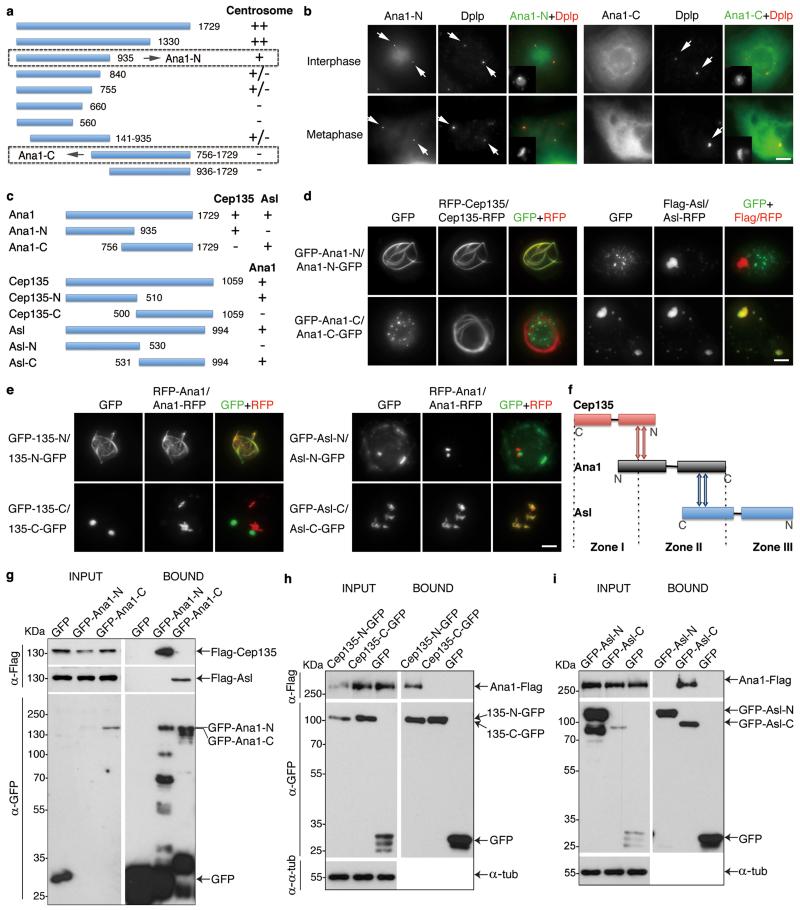
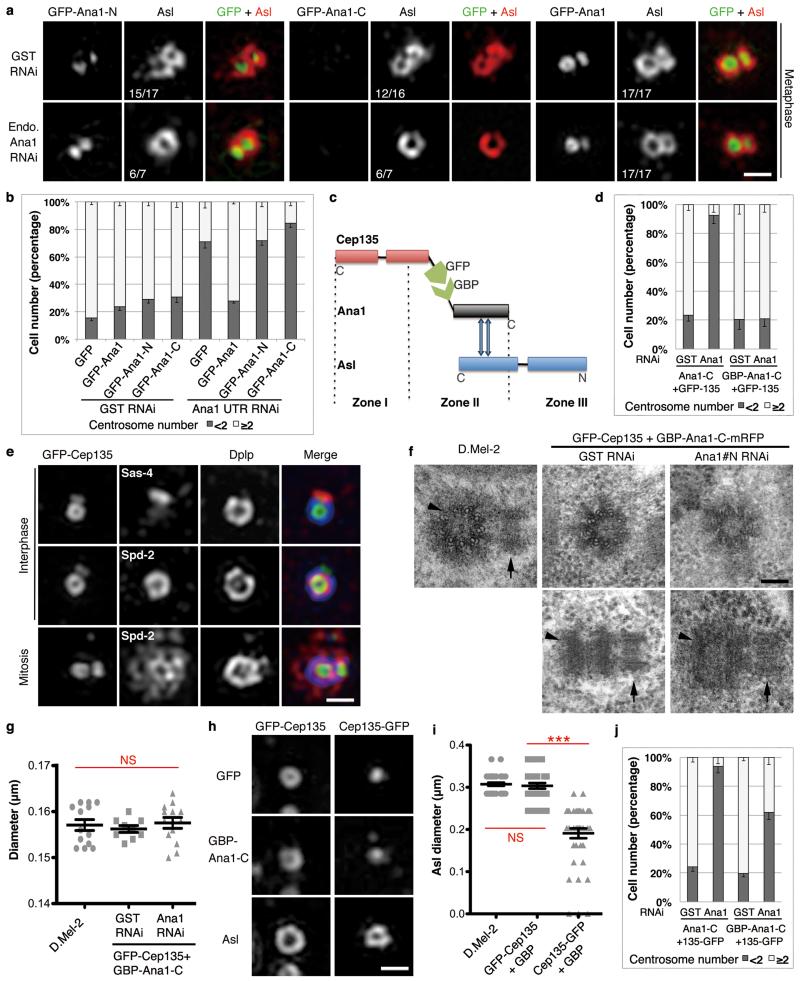
To determine which part of Ana1 was required for centrosomal localization we transiently expressed its GFP-tagged N- or C-terminal truncations (Fig. 4a, b). The N-terminal part (Ana1-N, residues 1-935) showed robust centrosome association whereas the C-terminal part (Ana1-C, residues 756-1729) spread diffusely throughout the cytoplasm. We then asked which part of Ana1 would interact with Cep135 and Asl (Fig. 4c, d). Cep135 co-localized with the N-terminal but not the C-terminal part of Ana1 whereas Asl co-localized only with the C-terminal part of Ana1. Reciprocally, Ana1 co-localized with N-terminal Cep135 (Cep135-N, residues 1-510) and C-terminal Asl (Asl-C, residues 531-994; Fig. 4c, e). Thus the ability of Ana1’s N-terminal part to localize to centrosomes is consistent with the association with Cep135. In parallel, we carried out GFP-Trap pull-downs from cell lysates co-expressing GFP-Ana1-N or -C with either Flag-Cep135 or Flag-Asl (Fig. 4g). This showed that Ana1-N interacted with Cep135 and Ana1-C with Asl. Similarly, we confirmed the interaction between Ana1 and Cep135-N and Asl-C, respectively (Fig. 4h, i). Thus the physical interactions between the terminal regions of Cep135, Ana1 and Asl accord with their overlapping distributions revealed by 3D-SIM (Fig. 4f), indicating the three proteins form an extended network.
Figure 4. Cep135, Ana1 and Asl interact via adjacent regions.
(a) Scheme to identify functional regions of Ana1. Fragments of Ana1 were tagged with GFP, transiently expressed in D.Mel-2 cells and analyzed for their localization using fluorescence microscopy. The N-terminal half (Ana1-N, 1-935aa) is necessary and sufficient for centrosome targeting whereas the C-terminal half (Ana1-C, 756-1729aa) is not.
(b) Representative images of cells transiently expressing N- or C-terminally GFP-tagged Ana1-N or -C and immunostained to reveal Dplp and DNA. Arrows indicate centrosomes.
(c) Summary of the interaction regions between Cep135, Ana1 and Asl.
(d) Cep135 or Asl was transiently co-expressed with N- or C-terminal regions of Ana1 in cells, proteins tagged as indicated. Ana1-N strongly co-localizes with Cep135 (left panel) whereas Ana1-C with Asl (right panel) in cytoplasm, regardless of the positions of the tags.
(e) Fragments of Cep135 or Asl were transiently co-expressed with Ana1 in cells, proteins tagged as indicated. N-terminal half of Cep135 (Cep135-N, 1-510aa) and C-terminal half of Asl (Asl-C, 531-994aa) strongly co-localize with Ana1 in cytoplasm, whereas C-terminal half of Cep135 (Cep135-C, 500-1059aa) and N-terminal half of Asl (Asl-N, 1-530aa) do not.
(f) Summary of interaction surfaces between Cep135, Ana1 and Asl within a single centriole.
(g-i) D.Mel-2 cells were transiently co-transfected with (g) GFP-tagged Ana1 fragments and Flag-tagged Cep135 or Asl, (h) GFP-tagged Cep135 fragments and Ana1-Flag, or (i) GFP-tagged Asl fragments and Ana1-Flag. Extracts were then subjected to GFP-Trap purification, and input and purified proteins (bound) were analyzed by Western blotting. α-tubulin (α-tub) served as loading control in (h) and (i). Note: Ana1 specifically binds Cep135-N via its N-terminal part, and Asl-C via its C-terminal part. Uncropped scans are in Supplementary Fig. 6.
All scale bars, 5 μm.
Ana1 loads Asl in centriole-to-centrosome conversion
The outer-most component of the above network is Asl, which in Drosophila is solely responsible for recruiting Plk425, the master regulator of centriole duplication. Accordingly, when Asl was depleted from cultured cells, most centrioles (86%, n=28) did not harbor Sas-6 at a site for pro-centriole formation (Fig. 5a; 100% control cells have Sas-6 on both mothers and daughters, Fig. 1b, n=30). Thus loading of Asl onto the daughter appears to be required for subsequent duplication. This accords with the ability of anti-Asl antibody to block Asl’s loading onto daughters in Drosophila embryos so that when daughters disengage from mothers, they fail to incorporate new Sas-426. To next address whether Ana1 was required to recruit Asl to daughter centriole, we depleted Ana1 from cells and confirmed it led to reduction of centrosome numbers (Supplementary Fig. 4a, b) as reported27,28. Electron microscopy (EM) showed no obvious changes to the diameter, length or symmetry of residual centrioles in Ana1-depleted cells; we also observed successful disengagement of mother and daughter centrioles in an anaphase cell (Supplementary Fig. 4c). Interestingly, serial EM sections revealed that Ana1-depleted centrosomes were devoid of daughter centrioles (Supplementary Fig. 4d, upper panel). Random EM sections from Ana1-depleted cells revealed significantly more single centrioles compared to wild type cells (Supplementary Fig. 4d, lower panel). Thus Ana1 depletion appears not to affect assembly of the walls of daughters but prevents their maturation to become mothers. We then selected cells with a single centrosome (marked with Asl or Dplp) indicating compromised duplication, and found that they all had Ana1 on the mother but not daughter during metaphase when it should have been recruited (Fig. 5b). The great majority of these centrosomes had Sas-6, Ana2 and Sas-4 associated with both mother and daughter centrioles (Fig. 5c), suggesting the initial steps of duplication were not affected. Asl was associated only with mother and not daughter centrioles in all metaphase and anaphase centrosomes (Fig. 5d, left and central part). In the absence of Ana1, Sas-4 was present on the daughter centrioles whereas Asl was not (Fig. 5d, right part). This suggests that Ana1, and not Sas-4, recruits Asl in centriole-to-centrosome conversion. We found Dplp failed to localize to metaphase daughter centrioles after depletion of Ana1 (Fig. 5d, left part), and another PCM marker, γ-tubulin, failed to load on disengaging daughters (Fig. 5e). This suggests that PCM recruitment is also affected by Ana1 depletion.
Figure 5. Ana1 loads Asl to daughter centriole for centriole-to-centrosome conversion in cultured Drosophila cells.
(a) D.Mel-2 cells were depleted of endogenous Asl and immunostained to reveal Asl, Dplp, Sas-6 and DNA. Note: Asl loses its ring structure whereas Dplp remains intact. 86% (n=28) of interphase centrosomes fail to recruit Sas-6 to a site for pro-centriole formation.
(b) D.Mel-2 cells were depleted of GST (control) or Ana1 and immunostained to reveal Ana1, Dplp and DNA. Cells with single centrosome were selected indicating compromised duplication. Depletion of Ana1 does not affect existing centrioles but prevents Ana1 recruitment onto newly formed daughters.
(c) Ana1-depleted cells were immunostained to reveal Dplp, DNA and Sas-6, Ana2 or Sas-4. Recruitment of Sas-6, Ana2 and Sas-4 to interphase daughter centrioles is not affected.
(d) Control or Ana1-depleted cells were immunostained to reveal Asl, Dplp and DNA or Sas-4, Asl and DNA. Depletion of Ana1 leads to failure of Asl recruitment to daughters in metaphase and anaphase. Recruitment of Dplp is similarly impaired in metaphase. Sas-4 is seen on mitotic mother and daughter whereas Asl only on mother in the absence of Ana1. Arrow indicates Dplp on daughter centriole.
(e) D.Mel-2 cells were depleted of GST or Ana1 and immunostained to reveal Sas-6, γ-tubulin and DNA. Note: in control telophase cells, mother and daughter (new mother-to-be) centrioles are both surrounded by γ-tubulin (upper panel, 30 cases out of 30); daughter centrioles in Ana1-depleted cells fail to recruit γ-tubulin by this stage (lower panel, 8 cases out of 8).
(f) D.Mel-2 cells were depleted of GST or Cep135 and immunostained to reveal Dplp, DNA and Ana1 or Asl. Note: 73% (n=67) and 78% (n=18) of metaphase centrosomes fail to load Ana1 and Asl to daughter centrioles, respectively.
(g) D.Mel-2 cells were depleted of GST or Ana1 and immunostained to reveal Cep135, Sas-6 and DNA. Loading of Cep135 to the daughter is impaired by Ana1 depletion. Error bars, SEM (Standard Error of the Mean); **, p<0.005 (two-tailed student’s t-test); n=20 and 21 centrosomes pooled across 3 independent experiments for GST and Ana1 RNAi, respectively.
Scale bars, 5 μm for complete images of cells and 500 nm for magnified centrioles.
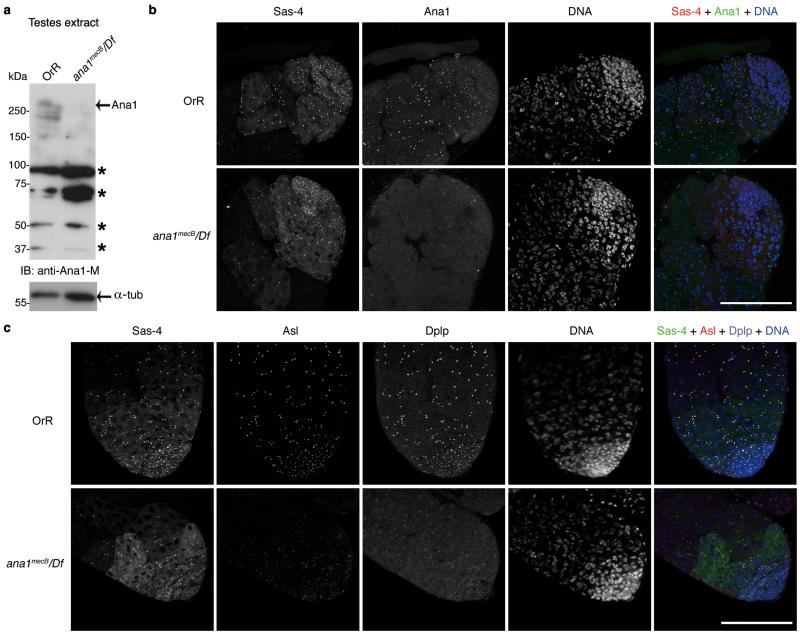
To determine if this loading pathway operates in the fly, we examined the ana1mecB mutant that has a premature stop codon at residue 1120. Such flies are uncoordinated, sterile and lack the giant spermatocyte centrioles32. Western blotting and immunostaining revealed depletion of Ana1 in the mutant testes (Fig. 6a, b) and recruitment of Asl and Dplp was greatly reduced (Fig. 6c), confirming that Ana1 is required for the correct recruitment and (or) maintenance of Asl and Dplp at the centrosome. As in cultured cells, Sas-4 was not obviously affected and could be used as a marker of spermatogonial centrosomes.
Figure 6. Ana1 in centriole-to-centrosome conversion in Drosophila testes.
(a) Western blot analysis of testes extract from ana1mecB/Df(3R)Exel7357 males reveals loss of full-length Ana1. Control, wild type OregonR (OrR) testes. * nonspecific band.
(b) Immunofluorescence showing absence of Ana1 at testes tips in ana1mecB/Df(3R)Exel7357 mutant flies. Sas-4 marks centrosomes in spermatogonia in either presence or absence of Ana1.
(c) Immunofluorescence showing that recruitment of Asl and Dplp at the centrosomes in spermatogonia is greatly reduced in the absence of Ana1.
Scale bars, 100 μm.
Co-dependency of Cep135 and Ana1
We then depleted Cep135 from D.Mel-2 cells to assess its role in the hierarchy and found centrosome loss as previously28,33. This also impaired recruitment of Ana1 and Asl to the daughter centriole (73% of centrosomes lacked Ana1 at the daughter and 78% lacked Asl, n=67 and 18; Fig. 5f). Depletion of Ana1 in turn affected Cep135 recruitment (Fig. 5g) pointing to the importance of Cep135-Ana1 interaction in the first stage of conversion. The requirement of Cep135 for centriole duplication in Drosophila cultured cells (both D.Mel-2 and S2) seems to go against the finding that Cep135 mutant flies show few centriole defects30-34. However, we noted that the mutant used in these studies (bld10c04199) is predicted to retain the N-terminal 369 amino acids30. Indeed using an antibody generated against Cep135’s N-terminal part we observed depletion of full-length protein and generation of a fragment that accorded with the predicted truncation in testes extract from bld10c04199 mutant flies (Supplementary Fig. 2b). This suggests that it would be prudent to generate a null allele at this locus in order to re-examine the role of Cep135 at the centriole in the whole organism.
Physical interactions of Cep135, Ana1 and Asl enable centriole-to-centrosome conversion
We then showed that full-length transgenic GFP-Ana1, but neither of its N-terminal or C-terminal parts, could support Asl’s loading onto the daughter centriole following endogenous Ana1 depletion (Fig. 7a). Consistently, only full-length GFP-Ana1 was able to rescue centriole duplication following the depletion of endogenous Ana1 (Fig. 7b). Together this accords with a requirement for one end of Ana1 to bind Asl and the other to provide a link to internal centriole components.
Figure 7. Cep135-Ana1-Asl interactions enable centriole-to-centrosome conversion.
(a) D.Mel-2 cells constitutively expressing GFP-tagged Ana1-N, -C or full length were depleted of endogenous Ana1 and stained to reveal Asl and DNA. Only full-length Ana1 can rescue Asl recruitment. Scale bar, 500 nm.
(b) Cells stably expressing GFP, GFP-tagged Ana1-N, -C or full length were depleted of endogenous Ana1 and immunostained to reveal Dplp. Only GFP-Ana1 can support centriole duplication following endogenous Ana1 depletion. n=3 independent experiments each scoring 500 cells; error bars, mean +/− SD.
(c) Schematic of GBP-Ana1-C chimera binding GFP-tagged Cep135 and endogenous Asl.
(d) Cells co-expressing GFP-Cep135 (constitutively) with Ana1-C-mRFP or GBP-Ana1-C-mRFP (inducible) were depleted of endogenous Ana1 (Ana1#N dsRNA, 4 days) and immunostained to reveal Dplp. GBP-Ana1-C-mRFP complements loss of endogenous Ana1 in presence of GFP-Cep135 to permit centriole duplication whereas Ana1-C-mRFP cannot. n=3 independent experiments each scoring 200 cells; error bars, mean +/− SD.
(e) Cells co-expressing GFP-Cep135 (green) and GBP-Ana1-C-mRFP were depleted of endogenous Ana1 (Ana1#N, 5 days) and immunostained to reveal Dplp (blue), DNA and Sas-4 or Spd-2 (red). Rescued centrioles are positive for Sas-4, Spd-2, Dplp and capable of duplicating and recruiting PCM. Scale bar, 500 nm.
(f, g) EM of cells co-expressing GFP-Cep135 and GBP-Ana1-C-mRFP with or without endogenous Ana1 (Ana1#N, 5 days). (f) Engineered centrioles maintain nine-fold symmetry and duplicate. Arrowheads, mothers; arrows, daughters. Scale bar, 100 nm. (g) Engineered centrioles are similar in diameter to controls. Error bars, SEM; NS, not significant (two-tailed student’s t-test p>0.1); from left to right, n=13, 9 and 13 centrosomes.
(h, i) Cells co-expressing GFP-Cep135 or Cep135-GFP with GBP-Ana1-C-mRFP were depleted of endogenous Ana1 (Ana1#N, 5 days) and immunostained for Asl. GFP-Cep135 and GBP-Ana1-C-mRFP recruits Asl to similar radial position as in control cells; Cep135-GFP and GBP-Ana1-C-mRFP recruit Asl more interiorly. Scale bar, 500 nm. Error bars, SEM; NS, not significant (two-tailed student’s t-test p>0.1); ***, p<0.001; from left to right, n=40, 33 and 42 centrosomes.
(j) Cells co-expressing Cep135-GFP with Ana1-C-mRFP or GBP-Ana1-C-mRFP were depleted of endogenous Ana1 (Ana1#N, 4 days) and immunostained to reveal Dplp. Note limited rescue of centriole duplication by GBP-Ana1-C-mRFP and no rescue by Ana1-C-mRFP. n=3 independent experiments each scoring 200 cells. Error bars, mean +/− SD.
To test the hypothesis that Ana1 provides a molecular link, we asked whether we might re-engineer this molecular network in alternative ways. We therefore replaced the N-terminal segment of Ana1 with a GFP-binding protein (GBP)35 and co-expressed it with N-terminally GFP-tagged Cep135 (GFP-Cep135, Fig. 7c). This enabled Ana1’s C-terminal part to localize to the centrosome rather than throughout the cytoplasm (Supplementary Fig. 5a). We then established cell lines constitutively expressing GFP-Cep135 and depleted endogenous Ana1 while inducing GBP-Ana1-C-mRFP or Ana1-C-mRFP (Supplementary Fig. 5b). Ana1-C-mRFP failed to support centriole duplication after endogenous Ana1 depletion whereas GBP-Ana1-C-mRFP restored centrosome numbers (Fig. 7d and Supplementary Fig. 5c). Sas-4, Dplp and Spd-2 were normally distributed on these centrioles, which actively undertook duplication (indicated by Sas-4 on the daughter) and PCM recruitment in mitosis (indicated by Spd-2; Fig. 7e). EM revealed centrioles in cells expressing GFP-Cep135 and GBP-Ana1-C-mRFP with or without endogenous Ana1 had the same symmetry and diameter of those in control cells (Fig. 7f, g) and were able to undergo duplication (Fig. 7f). Asl was recruited to Zone III of the rescued centrioles and occupied a ring similar in diameter to that in normal D.Mel-2 centrioles (Fig. 7h, i). In parallel, we depleted endogenous Ana1 from cells expressing a combination of C-terminally GFP-tagged Cep135 (Cep135-GFP) and GBP-Ana1-C-mRFP. This gave some rescue of centriole duplication but at a remarkably lower efficiency (Fig. 7j). The diameter of Asl ring in these cells was significantly decreased as Cep135-GFP brought Asl to a more inner part of the centriole (Fig. 7h, i). Together this indicates that the N-terminal part of Ana1 provides a physical linkage to internal centriole components that can be replaced through the molecular transplantation of interacting domains. The C-terminal part of Ana1, on the other hand, is required to recruit Asl. Thus Ana1 provides a molecular link between the inner and outer parts of the centriole that allows Asl to be recruited to the daughter at an appropriate radial position giving it the ability to duplicate.
Conserved mechanism of centriole-to-centrosome conversion in human cells
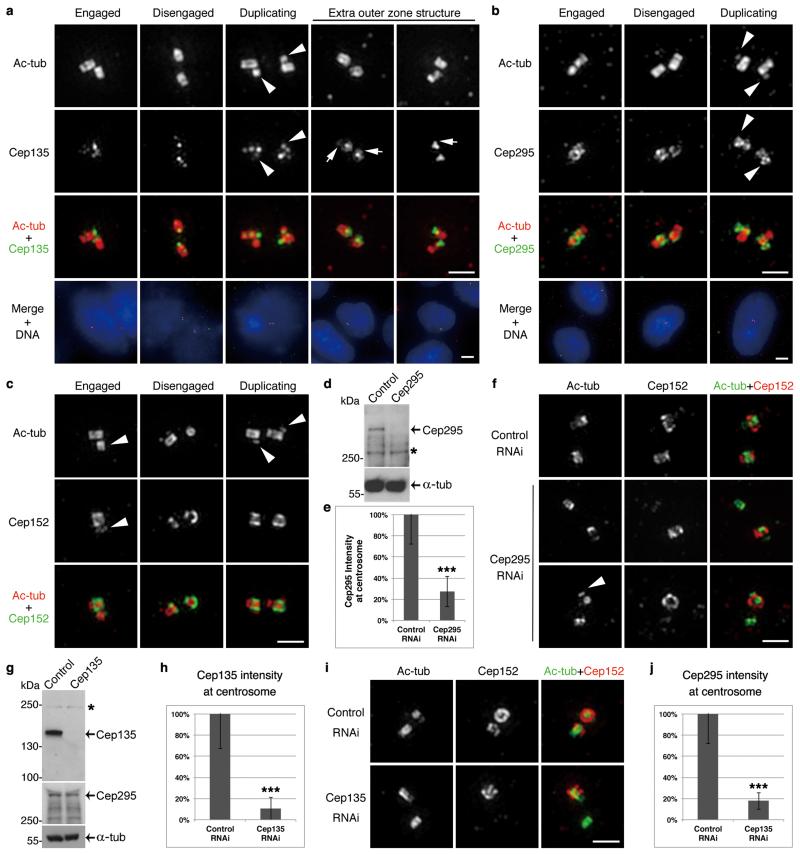
To investigate if the mechanism for centriole-to-centrosome conversion was used beyond Drosophila, we first studied the localization of the human counterparts of Cep135-Ana1-Asl known as Cep135, Cep295 and Cep152, respectively (Fig. 8a-c). Cep135 was concentrated at the proximal end of the centriole within the microtubule wall and occasionally, in an additional ring at the base of microtubule wall as reported6. Cep295 closely surrounded the proximal part of the microtubule wall. In agreement with earlier studies6, Cep152 extended from the inner PCM just outside the microtubule wall (300 nm diameter; see also Fig. 8c) to the intermediate PCM (400 nm diameter; see also Fig. 8f, upper panel). Cep135 and Cep295 were loaded onto daughter centrioles at similarly early stages as their Drosophila counterparts. Moreover, Cep152 localized predominantly on the mother and was not fully recruited onto the daughter until it started to disengage36,37 as in Drosophila.
Figure 8. Mechanism for centriole-to-centrosome conversion is conserved in human cells.
(a-c) U2OS cells immunostained to reveal acetylated tubulin (ac-tub), DNA (by DAPI), and Cep135 (a), Cep295 (b) or Cep152 (c, antibody against C-terminus). Centrioles from different stages of centrosome cycle (engaged, disengaged and undergoing duplication) are shown. Cep135 and Cep295 are recruited onto daughter centrioles (arrowheads) early in centriole assembly; Cep152 is only fully loaded when daughter disengages from mother. Cep135 strongly localizes to the proximal end of centriole within microtubule wall, and in some cases at the base of the wall (arrows). Cep295 closely surrounds proximal part of microtubule wall.
(d, e) U2OS cells were transfected with control or Cep295 siRNA, synchronized to G2 and subjected to Western blotting (d) or immunostaining for Cep295 and γ-tubulin (e; γ-tub as centrosome marker). Overall Cep295 is reduced by 96% (d, * nonspecific band), and at centrosome by 73% (e, n=30 centrosomes each; error bars, mean +/− SD; ***, p<0.001 (two-tailed student’s t-test)).
(f) U2OS cells were depleted of Cep295, synchronized to G2 and immunostained to reveal ac-tub, Cep152 (antibody to N-terminus) and DNA. Note: newly disengaged daughter fails to load Cep152 when Cep295 is depleted (middle panel), correlating with failure of duplication (lower panel; arrowhead, daughter centriole.)
(g, h) U2OS cells were transfected with control or Cep135 siRNA, synchronized to G2 and subjected to Western blotting (g) or immunostaining to reveal Cep135 and γ-tubulin (h). Overall Cep135 level is reduced by 99% (g, * nonspecific band), and at centrosome by 89% (h, n=30 centrosomes each; error bars, mean +/− SD; ***, p<0.001 (two-tailed student’s t-test)). Overall Cep295 level is not affected by Cep135 depletion (g).
(i) U2OS cells were depleted of Cep135, synchronized to G2 and immunostained to reveal ac-tub, Cep152 (antibody to N-terminus) and DNA. Newly disengaged daughter fails to load Cep152 when Cep135 is depleted.
(j) U2OS cells treated as in (i) were immunostained to reveal Cep295 and γ-tubulin. Note: intensity of Cep295 at centrosome is reduced by 82% following Cep135 depletion (n=30 centrosomes each; error bars, mean +/− SD; ***, p<0.001 (two-tailed student’s t-test)).
Scale bars, 5 μm for complete images and 1 μm for magnified centrioles.
Depletion of Cep295 from U2OS cells (overall protein level reduced by 96% on Western blot and at the centrosome 73% by immunofluorescence; Fig. 8d, e) resulted in 30% of G2 cells having one or no centrosomes in accord with earlier reports4,38. Although some cells still had two centrosomes, the new mother failed to load Cep152 (Fig. 8f, middle panel). Consequently, we observed G2 cells with one doublet indicating successful duplication and one singlet that failed to duplicate; this correlated with the absence of Cep152 (Fig. 8f, lower panel). Thus Cep295 has a conserved function in loading Cep152 and converting the daughter centriole into a centrosome.
Similarly, depletion of Cep135 (overall protein level reduced by 99% on Western blot and at the centrosome 89% by immunofluorescence; Fig. 8g, h) resulted in 38% of G2 cells having only one or zero centrosomes consistent to an earlier report21. Cep135 depletion was associated with failure to load Cep152 onto newly disengaged daughters as with depletion of Cep295 (Fig. 8i), indicating that Cep135 and Cep295 work in a similar pathway to their Drosophila counterparts. Indeed fluorescence intensity of Cep295 at the centrosome was reduced by 83% after Cep135 depletion (Fig. 8j) whereas its total amount in cells was not affected (Fig. 8g). Thus loading of Cep295 onto centrosomes depends on Cep135 in human cells.
Discussion
Here we account for the final stages in the assembly of daughter centriole that convert it into a mature mother that is able to duplicate. The newly formed pro-centriole has Sas-6, Ana2:STIL and Sas-4 at its core, yet only incorporates Cep135, Ana1:Cep295 and Asl:Cep152 at later stages. Their sequential recruitment correlates with radial position suggesting that these final stages of assembly occur in layers, from inside out. Cep135 is the most interior component and in human cells has also been reported to bind Sas-621, the major cartwheel component. In Drosophila, the most external component, Asl, is the sole factor known to recruit Plk425 whereas its human counterpart, Cep152, competes with another Plk4 binding partner Cep192 at the inner centriole to relocate Plk4 to the outer ring36. Despite the differences, human Cep152 is key for Plk4 function since loss of Cep152-Plk4 interaction affects Sas-6 recruitment to the pro-centriole assembly site36,37,39-41. In both species, Asl:Cep152 binds Plk4 through its N-terminal region that extends outward into Zone III as does Plk46,7,9,25,36,37,39,40. Without loading of Asl:Cep152 onto the daughter, Plk4 could not direct centriole duplication in the next cell cycle.
Our findings allocate a role to Drosophila Ana1, previously known from RNAi and mutant studies to be required for centriole duplication27,28,32. Its human counterpart Cep295-KIAA1731, with 15% similarity in primary sequence32,38,42, was previously suggested to function in centriole-to-centrosome conversion4. We now provide a mechanistic basis for its role in loading Cep152 to the newly disengaged daughter in human just as Ana1 does in Drosophila. It remains possible that once loaded, Asl:Cep152 could interact with other molecules such as Cep63 and Cep192 in human or Sas-4 in Drosophila to be maintained at the centriole25,36,37,43-45.
Together our findings suggest centriole-to-centrosome conversion involves a cascade of interactions in which Cep135 could be first recruited to Sas-6, permitting the sequential loading of Ana1:Cep295 and Asl:Cep152 and thereby Plk4. We also demonstrate the high similarity of this process between insect and mammalian cells. Centriole-to-centrosome conversion takes place following a strict timetable during mitosis in which the mitotic kinase, Plk1, has been suggested to play an important role3. It seems likely that the protein-protein interactions we describe can be further regulated by phosphorylation both in time and space to control the sequence of events. Unraveling the potential roles of Polo:Plk1 in regulating these interactions will be a fascinating topic for future study.
Methods
DNA Constructs
cDNA clones for cep135, ana1, sas-6 and asl were described in11, sas-4 in15, and gbp was a gift from Iain Hagan (University of Manchester, UK) 35. Entry clones with CDS encoding full-length or fragments of these genes were generated using Gateway System (Life Technologies). Expression constructs were made by recombination between entry clones and the following destination vectors: pDEST15 (for N-terminal GST fusion in E. coli, Life Technologies), pDEST17 (for N-terminal 6×His fusion in E. coli, Life Technologies), pET-T7-DEST42 (for IVTT expression under the regulation of T7 promoter; Life Technologies; due to the stop codon between the CDS and 6×His, the tag is not translated), pKM596 (for N-terminal MBP fusion in E. coli, Addgene), pAFW or pAWF (for actin5C promoter-driven N- or C-terminal 3×Flag fusion in D.Mel-2 cells, Drosophila Genomics Resource Center (DGRC, NIH grant 2P40OD010949-10A1)), pAGW or pAWG (for actin5C promoter-driven N- or C-terminal GFP fusion in cells, DGRC), pMT-N-mRFP or -C-mRFP (for inducible metallothionein promoter-driven N- or C-terminal mRFP fusion in cells), pAct5c-C-mRFP (for actin5C promoter-driven C-terminal mRFP fusion in cells), pUGW or pUWG (for ubiquitin promoter-driven N- or C-terminal GFP fusion in flies, DGRC). For cloning GBP and Ana1-C into a Gateway destination vector, MultiSite Gateway Pro 2.0 (Life Technologies) was used.
Recombinant Protein Expression and Purification
Maltose binding protein (MBP) or MBP-tagged Ana1 were expressed in E. coli strain Rosetta™2(DE3)pLysS Singles™ (Novagen) induced with 0.3 mM isopropyl 1-thio-β-D-galactopyranoside for 6 h at 24°C. Cells were lysed by sonication in ice cold LBM buffer (20 mM Tris pH 7.4, 200 mM NaCl, 1 mM EDTA, 1 mM DTT, 2 mM MgCl2, 5% glycerol) supplemented with EDTA-free complete protease inhibitor cocktail (PIC, Roche) and 0.2 mg/ml lysozyme (Sigma). Recombinant proteins were immobilized on Amylose resin (NEB) following the manufacturer’s instruction. Beads were resuspended in LBM + 50 % glycerol and stored at −20°C before used in in vitro binding assay.
Glutathione S-transferase (GST)-tagged Sas-4-301-600aa, Sas-4-601-901aa or Ana1-622-981aa (Ana1-M-antigen) were expressed in E. coli strain BL21Star(DE3)pLysS (Life Technologies) as detailed above. Cells were lysed by sonication in PBS supplemented with 1 mM PMSF, PIC and 2% N-Lauroylsarcosine (Sigma, 61747). 4% Triton X-100 was then added and the lysate was incubated for 20 min at 4°C followed by centrifugation at 34,000g, at 4°C for 20 min. Recombinant proteins were purified on Glutathione Sepharose 4B beads (GE Healthcare). GST-tagged Sas-4-301-600aa and Sas-4-601-901aa were eluted from the resin using 100 mM Glycine-HCl (pH 2.1), while Ana1-622-981aa eluted with reduced glutathione solution (50 mM Tris pH 8.0, 20 mM glutathione). His-tagged Ana1-1400-1729aa (Ana1-C-antigen) or Cep135-1-225 (Cep135-N-antigen) were expressed in E. coli strain BL21Star(DE3)pLysS or Rosetta™2(DE3)pLysS Singles™ (Novagen). Ana1 antigen was purified on Ni-NTA agarose (Qiagen) under native conditions and eluted following the manufacturer’s guide. Cep135 antigen was purified on Ni-NTA resin under denaturing condition and refolded following standard protocol. Purified proteins were concentrated and dialyzed against PBS or PBS + 1 M Urea overnight before used for immunization.
Antibodies
The following primary antibodies were used for immunofluorescence in Drosophila cells or tissues: rat anti-Sas-611 (1:100, against GST-Sas-6-236-472aa); rabbit anti-Ana211 (1:500, against GST-Ana2-1-280aa); rat anti-Sas-4 (1:500, against the mixture of GST-Sas-4-301-600aa and GST-Sas4-601-901aa, serum produced by Animal Facility at IBMC, Porto, Portugal and purified as below); mouse anti-Sas-445 (1:500, monoclonal, courtesy of Tomer Avidor-Reiss (University of Toledo, USA)); rabbit anti-Asl25 (1:500, against full length); rabbit anti-Spd-29 (1:500); chicken anti-Dplp15 (1:500); mouse anti-γ-tubulin (GTU-88, Sigma T6557); mouse anti-Flag (1:1000, M2, Sigma, F3165); rat anti-RFP (1:500, 5F8, ChromoTek). Antibody against C terminus of Cep135 was kindly provided by Timothy Megraw (Florida State University, USA) 30 (rabbit anti-Cep135-C, 1:500); Serum against N terminus of Cep135 was produced by Moravian Biotechnology, Czech Republic, and purified as below (Rabbit anti-Cep135-N, 1:500, against His-Cep135-1-225aa). Serum against middle region of Ana1 was produced by Animal Facility at IBMC (rat anti-Ana1-M, 1:500, against GST-Ana1-622-981aa); Serum against C terminus of Ana1 was produced by Scottish National Blood Transfusion Service and purified as below (rabbit anti-Ana1-C, 1:5000, against His-Ana1-1400-1729aa). Antibody against N terminus of Asl was purified as below (rabbit anti-Asl-N, 1:500, against 1-300aa), and antibody that recognizes 16-mer peptide within the C-terminal of Asl was kindly provided by Cayetano Gonzalez (IRB Barcelona, Spain) 46.
Rat anti-Sas-4 and anti-Ana1-M immunoglobulins (IgGs) were purified using a mixture of Protein-A and Protein-G Sepharose (GE Healthcare), and rabbit anti-Ana1-C IgG was purified on Protein-A Sepharose according to manufacturer’s guidelines. Rabbit anti-Asl-N or anti-Cep135-N IgGs were affinity purified on Asl-1-300aa or Cep135-1-225aa antigens immobilized on nitrocellulose membranes following standard procedures. Specificity of these antibodies was verified by Western blotting (Supplementary Fig. 2a, b).
The following primary antibodies were used for immunofluorescence in human cells: rabbit anti-Cep13521 (1:500, against Cep135-650-1140aa-His, courtesy of Tang Tang (Institute of Biomedical Sciences, Taiwan)); rabbit anti-Cep152-N40 (1:500, against Cep152-26-39aa, courtesy of Ingrid Hoffmann (German Cancer Research Center, Germany)); rabbit anti-Cep152-C40 (1:500, against Cep152-1140-1308aa, courtesy of Ingrid Hoffmann); rabbit anti-Cep295 (KIAA1731, 1:500, Abcam 122490); mouse anti-acetylated tubulin (1:1000, Sigma T7451).
The following primary antibodies were used for Western blotting: mouse anti-Flag (1:10000, M2, Sigma); rabbit anti-GFP (1:10000, ab280, Abcam); mouse anti-α-tubulin (1:6000, DM1A, Sigma); rabbit anti-Asl (1:8000); rabbit anti-Asl-N IgG (1:500); rat anti-Ana1-M IgG (1:5000); rabbit anti-Cep135-N IgG (1:3000); rabbit anti-Cep135-C IgG (1:5000); rat anti-Sas-4 (1:5000); rabbit anti-human Cep135 IgG (1:500); rabbit anti-Cep295 (1:2000).
Secondary antibodies conjugated with Alexa Fluor 488, 594 or 405 were diluted 1:500 for immunofluorescence (Life Technologies); secondary antibodies conjugated with HRP were diluted 1:10000 for Western blotting (Jackson ImmunoResearch).
Cell culture, Transfection, and RNAi
D.Mel-2 cells (Life Technologies) were grown at 25°C in Express Five SFM (Life Technologies) supplemented with L-glutamine (2 mM; Gibco) and penicillin-streptomycin (50 units/ml-50 μg/ml; Gibco). Transfection of plasmids was performed using X-tremeGENE HP DNA Transfection Reagent (Roche). For transient expression, cells were collected and subjected to immunostaining, GFP-trap pull-down or glycerol gradient sedimentation after 36 h. To establish stable cell lines, 20 μg/ml blasticidin (Life Technologies) was added to the medium 48 h after transfection. Established cell lines were authenticated by both western blotting and immunostaining. D.Mel-2 cells were transfected with dsRNA using TransFast™ Transfection Reagent (Promega). For repeated rounds of depletion, cells were harvested every 4 days and re-submitted to the same transfection protocol. RNAi efficiency was tested by Western blotting (Supplementary Fig. 2a, all experiments were independently performed three times).
Double-stranded RNAs (dsRNA) directed against the coding sequence were synthesized from templates of Drosophila cDNA clones using T7 RiboMAX Express RNAi System (Promega), primers listed in Supplementary Table 1. To selectively knock down endogenous proteins, 5’-UTR-3’-UTR hybrid was generated by Overlap Extension PCR and used as templates for dsRNA. Full sequences are listed in Supplementary Table 1.
U2OS cells (ATCC) were cultured at 37°C with 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 10% heat inactivated Fetal Bovine Serum (FBS, Gibco) and penicillin-streptomycin (50 units/ml-50 μg/ml; Gibco). RNAi was performed using RNAiMAX (Life Technologies). siRNAs targeting specific genes (siCep295: 5’-GUGAUACACUAACAAUUGA-3’4; siCep135-1: 5’-CAAGCAGAUUGAGCUAAGA-3’47; siCep135-2: 5’-GACUGAGUGAUGAACUCCUUGUAAA-3’21; siCep135-3: 5’-GCGAAGAUCUUGCUCUACAAGUUAU-3’21) and a MISSION® siRNA Universal Negative Control #1 were obtained from Sigma. Because Cep135 is difficult to be stripped from centrioles21, two consecutive RNAi were performed for a total of 6 days using combination of three different siRNAs (100 nM each). Cep295 depletion was performed for 3 days (100 nM of single siRNA).
To enrich G2 population, U2OS cells were synchronized by double thymidine (2.5 mM, Sigma) block and released into fresh medium for 9 h before subjecting to immunostaining or Western-blotting. All experiments in Figure 8 (including RNAi and the following analysis) were independently performed three times.
Structured Illumination Microscopy and Data Processing
These procedures were described in9. Briefly, D.Mel-2 cells were plated on Concanavalin A (Sigma)-coated coverslips (#1.5, 0.17mm thickness, Zeiss) 3 h prior to fixation. Cells were washed with PBS, fixed with pre-cooled methanol for 6 min at −20°C, and washed three times in 0.1% Triton X-100-PBS. Cells were then blocked with 3% BSA-0.1% Triton X-100-PBS for 30 min and incubated with the primary antibody overnight at 4°C. Cells were washed again for three times in 0.1% Triton X-100-PBS and incubated with the secondary antibody for 45 min at room temperature. Coverslips were mounted onto slides using VECTASHIELD mounting medium with or without DAPI (VECTOR laboratories). U2OS cells grown on coverslips were fixed and immunostained similarly to above, but using 0.1% Tween-PBS instead of Triton X-100 for the whole procedure. To visualize the centrioles by acetylated tubulin staining, cells were first kept on ice for 2 h to depolymerize the cytoplasmic microtubules, pre-extracted with 0.5% Triton X-100-PBS for 2 min and fixed with pre-cooled methanol for 10 min at −20°C.
Super-resolution images were acquired using a Deltavision OMX 3D-SIM System V3 BLAZE from Applied Precision (GE Healthcare) equipped with 3 sCMOS cameras, 405, 488, 592.5nm diode laser illumination, an Olympus Plan Apo N 60× 1.42 NA oil objective, and standard excitation and emission filter sets. Imaging of each channel was done sequentially using three angles and five phase shifts of the illumination pattern. The refractive index of the immersion oil (Cargille) was adjusted to minimize spherical aberrations. Sections were acquired at 0.125 μm z steps.
Raw OMX data was reconstructed and channel registered in SoftWoRx software version 5.5 (Applied Precision, GE Healthcare). Reconstructions were carried out using channel specific Optical Transfer Functions (OTFs), a Wiener filter of 0.002, and channel specific K0 angles. OTFs were generated within the SoftWoRx software by imaging 100 nm beads (Life Technologies) using appropriate immersion oils to match the data. Channel registration was carried out using the Image Registration parameters generated within the SoftWoRx software and checked for accuracy by imaging Tetraspeck beads (Life Technologies) and control samples where one primary antibody was labeled with two secondary antibodies. Channel registration was accurate to within one pixel.
To measure the radial distance of different proteins or regions given in Fig. 2c and 7i, the line scan and plot profiles of Fiji were applied to centrioles that are perpendicular to the coverslips. The distance between two farthest peak intensities of a ring was defined as diameter, and radius=diameter/2. Total centriole numbers were stated in the figure legends and SD (Standard Deviation), SEM (Standard Error of the Mean) and p value determined using Prism 5. To analyze distribution of signals detected in different channels in Fig. 2e, RGB merged files were subjected to line scan and RGB profiler. To measure the ratio of Cep135 on daughter and mother centrioles in Fig. 5g, the mean intensity was subtracted by the background before multiplied by area using Fiji and Excel.
All experiments were independently performed three times.
Fluorescence Microscopy
D.Mel-2 cells were fixed and immunostained as above. Microscopic analysis was performed on a Carl Zeiss Axiovert 200M microscope with 40×/1 or 100×/1.4 Plan Apochromat objectives. Images were acquired with a Photometrics Cool SNAP HQ2 camera and the image analysis software Metamorph (v7.7). All experiments were independently performed three times.
For co-localization analysis in Fig. 3 and 4, D.Mel-2 cells were co-transfected with indicated constructs, plated on Concanavalin A (Sigma)-coated coverslips 3 h prior to fixation. Coverslips were then mounted onto slides using VECTASHIELD mounting medium with DAPI (VECTOR laboratories). Protein localization was observed by native GFP or mRFP signal. In the case of Flag-tagged constructs, cells were immunostained with anti-Flag antibody as described above.
Electron Microscopy and Data Processing
The procedure was described in48. Briefly, cultured cells on coverslips were fixed in 4% paraformaldehyde in PBS for 15 min and immersed in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) for 2 h at 4°C. They were then washed three times for 30 min in phosphate buffer, postfixed with 1% OsO4 for 1 h at 4°C, washed once in phosphate buffer, and then in distilled water. Samples were stained for 1 h in uranyl acetate. They were washed again and then dehydrated in a graded series of ethanol and flat-embedded in a mixture of Epon and Araldite. After polymerization for 2 days at 60°C, the coverslips were removed from the resin after a short immersion in liquid nitrogen. Ultrathin serial sections were obtained with a LKB ultratome, stained with uranyl acetate and lead citrate, and observed and photographed with a Philips CM10 electron microscope at 80 kV.
To measure centriole diameters in Fig. 7g and Supplementary Fig. 4c, cross section images were used and a line was drawn between two opposite B-tubules of the microtubule wall.
Fly Stocks
Fly stocks were maintained at 25 °C on standard Drosophila food. OregonR (OrR) flies were used as wild type control. The ana1 mutant line (w; CyO GFP/Sco; ana1mecB/TM6B GFP) was kindly provided by Tomer Avidor-Reiss (University of Toledo, USA) and the line carrying the deficiency uncovering ana1 (w; Df(3R)Exel7357/TM6B) was obtained from the Bloomington Drosophila Stock Center. The cep135 mutant line (bld10c04199/TM6B) and the line carrying the deficiency uncovering cep135 (w; If /CyO act GFP; Df(3L)Brd15/TM6B) were kindly provided by Clemens Cabernard (University of Basel, Switzerland). Trans-heterozygous males for ana1 or cep135, obtained by crossing the aforementioned stocks, were used to prepare testes extracts for Western blotting or immunofluorescence analysis. polyubiquitin GFP-Ana1/FM7 and polyubiquitin Ana1-GFP/FM7 transgenic flies were generated by University of Cambridge Department of Genetics Fly Facility, and polyubiquitin GFP-Cep135/CyO was kindly provided by Mónica Bettencourt-Dias (Instituto Gulbenkian de Ciência, Portugal).
Preparation of Testes Extracts
For Western blot analysis, testes from pharate adults were dissected in PBS (20 pairs) and pestle homogenized in 40 μl 2× Laemmli sample buffer (Sigma), incubated on ice for 5 min and boiled. Experiments were independently performed twice.
For immunofluorescence analysis, testes from pharate adults were dissected in PBS, transferred to 5% glycerol-PBS and squashed between microscope slide and coverslip. After snap freezing in liquid nitrogen, testes on slides were fixed in methanol, rehydrated in 0.5% Triton-X100-PBS for 30 seconds, rinsed in PBS for 10 min and incubated with primary antibodies (diluted in PBS) at 4°C overnight. Slides were then rinsed for 30 seconds in PBS, incubated again in PBS for 10 min and with secondary antibodies (1:200 in PBS) for 4 h at room temperature. Finally, slides were rinsed in PBS for 30 seconds followed by a 10 min wash and mounting in Vectashield containing DAPI (Vector Laboratories).
Images were either acquired using Structured Illumination Microscopy as described above, or a Zeiss LSM 510 Meta confocal microscope equipped with a 63× oil objective, NA 1.4, using the 488 nm laser line of an Argon Ion laser (green), the 561 nm laser line of an DPSS laser (red), the 633 nm laser line of a HeNe laser (far red) and a diode laser with a 405 nm laser line (blue). Stack of images were acquired at a 0.5 μm z step. Figures shown are the maximum-intensity projections of optical sections prepared with Fiji. All experiments were independently performed twice.
In Vitro Transcription-Translation (IVTT)
35S-methionine-labelled Cep135, Asl and Sas-6 were expressed in vitro using the TNT T7 Quick Coupled Transcription-Translation System (IVTT; Promega) following the procedure described earlier11. Briefly, pET-DEST42-T7-Cep135 (90 ng), -Asl (90 ng) or -Sas-6 (90 ng) plasmids were added into 11 μl IVTT reaction mixture (9.9 μl TNT Quick Master Mix (Promega, L1170), 0.9 μl RNasin Plus (Promega), 0.9 μl 50× PIC (Roche) and 0.48 MBq Methionine-L [35S] (Perkin Elmer)), and incubated at 30°C for 1.5 h followed by centrifugation at room temperature at 21,000g for 5 min. Supernatant (Input) was used in in vitro binding assay.
In Vitro Binding Assay
These procedures were described in49. Specifically, MBP or MBP-Ana1 immobilized on beads were mixed with 500 μl Binding Buffer (BB: 50 mM HEPES pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.1% Triton X-100, PIC (Roche), 1 mg/ml BSA) and 5 μl IVTT reaction mixture (containing 35S-Met-Sas-6, Cep135 or Asl) and incubated for 3 h at 4°C on an end-over-end rotator. Beads were then settled by mild centrifugation and washed 3 times with Washing Buffer-1 (WB-1: BB without BSA) by gently pipetting followed by incubation for 5 min on the rotator. Beads were then washed twice in WB-2 (WB-1 supplemented with 100 mM NaCl and 0.1% Triton X-100), resuspended in 20 μl 2× Laemmli sample buffer, boiled and subjected to SDS-PAGE. Gels were stained with Bio-Safe Coomassie (Bio-Rad), scanned, dried and used for autoradiography. All experiments were independently performed twice.
GFP-Trap Affinity Purification
D.Mel-2 cells were washed in PBS, homogenized in ice cold Extraction Buffer-1 (EB-1: 50 mM HEPES pH 7.5, 100 mM K-acetate, 100 mM NaCl, 50 mM KCl, 2 mM MgCl2, 2 mM EGTA, 0.1% NP-40, 5% glycerol, PIC (Roche), 25 μM MG132) supplemented with 10 U/ml Bensonaze (Novagen) by passing through a pre-chilled G25 needle (10 times) followed by incubation on ice for 20 min. Lysates were then centrifuged (3,000g, 4°C, 20 min) and supernatant (Input) were subjected to GFP-Trap purification (magnetic agarose beads, ChromoTek) as described before50. After final wash, beads were mixed with 2× Laemmli sample buffer, boiled and subjected to Western blotting. All experiments were independently performed three times.
Glycerol gradient sedimentation
D.Mel-2 cells were washed in PBS and lysed in EB-2 buffer (EB-1 supplemented with 1 mM DTT, 25 μM MG132 and 20U/ml Benzonase) as above. After centrifugation (2000 g, 4 °C, 10 min) pellets were resuspended in EB-2 and loaded onto a 11 ml 10-30% linear glycerol gradient made in EB-2 buffer (without MG132 and NP-40). Sedimentation was carried out by centrifugation in a SW40 Ti rotor at 35,000 rpm (RCFmax=217,874) for 20 h at 4°C. Fractions (350 μl each) were collected from the top of the gradient and precipitated with 25% Trichloroacetic-acid followed by ice cold acetone wash. Precipitates were resuspended in 2× Laemmli sample buffer, boiled and subjected to Western-blotting analysis. Experiments were independently performed twice.
Supplementary Material
Acknowledgments
J.F., Z.L., S.S. and N.S.D. are supported from Programme Grant to D.M.G. from Cancer Research UK. H.R. is supported from MRC Programme Grant to D.M.G. J.F. thank the British Academy and the Royal Society for Newton International Fellowship and Z.L. thanks the Federation of European Biochemical Societies for the Long-Term postdoctoral Fellowship. The authors thank Nicola Lawrence and Alex Sossick for assistance with 3D-SIM.
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Bornens M, Gonczy P. Centrosomes back in the limelight. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu J, Hagan IM, Glover DM. The Centrosome and Its Duplication Cycle. Cold Spring Harb. Perspect. Biol. 2015;7 doi: 10.1101/cshperspect.a015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang WJ, Soni RK, Uryu K, Tsou MF. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J. Cell Biol. 2011;193:727–739. doi: 10.1083/jcb.201101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Izquierdo D, Wang WJ, Uryu K, Tsou MF. Stabilization of cartwheel-less centrioles for duplication requires CEP295-mediated centriole-to-centrosome conversion. Cell Rep. 2014;8:957–965. doi: 10.1016/j.celrep.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sillibourne JE, et al. Assessing the localization of centrosomal proteins by PALM/STORM nanoscopy. Cytoskeleton. 2011;68:619–627. doi: 10.1002/cm.20536. [DOI] [PubMed] [Google Scholar]
- 6.Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open. 2012;1:965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mennella V, et al. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 2012;14:1159–1168. doi: 10.1038/ncb2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawo S, Hasegan M, Gupta GD, Pelletier L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 2012;14:1148–1158. doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- 9.Fu J, Glover DM. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2012;2:120104. doi: 10.1098/rsob.120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau L, Lee YL, Sahl SJ, Stearns T, Moerner WE. STED microscopy with optimized labeling density reveals 9-fold arrangement of a centriole protein. Biophys. J. 2012;102:2926–2935. doi: 10.1016/j.bpj.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dzhindzhev NS, et al. Plk4 phosphorylates Ana2 to trigger Sas6 recruitment and procentriole formation. Curr. Biol. 2014;24:2526–2532. doi: 10.1016/j.cub.2014.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohta M, et al. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat. Commun. 2014;5:5267. doi: 10.1038/ncomms6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bettencourt-Dias M, et al. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 14.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 15.Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007;316:1046–1050. doi: 10.1126/science.1142950. [DOI] [PubMed] [Google Scholar]
- 16.Kleylein-Sohn J, et al. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 17.van Breugel M, et al. Structures of SAS-6 suggest its organization in centrioles. Science. 2011;331:1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- 18.Kitagawa D, et al. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011;144:364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiraki M, Nakazawa Y, Kamiya R, Hirono M. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr. Biol. 2007;17:1778–1783. doi: 10.1016/j.cub.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 20.Jerka-Dziadosz M, et al. Basal body duplication in Paramecium: the key role of Bld10 in assembly and stability of the cartwheel. Cytoskeleton. 2010;67:161–171. doi: 10.1002/cm.20433. [DOI] [PubMed] [Google Scholar]
- 21.Lin YC, et al. Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J. 2013;32:1141–1154. doi: 10.1038/emboj.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang CJ, Fu RH, Wu KS, Hsu WB, Tang TK. CPAP is a cell-cycle regulated protein that controls centriole length. Nat. Cell Biol. 2009;11:825–831. doi: 10.1038/ncb1889. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt TI, et al. Control of centriole length by CPAP and CP110. Curr. Biol. 2009;19:1005–1011. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 24.Kohlmaier G, et al. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr. Biol. 2009;19:1012–1018. doi: 10.1016/j.cub.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dzhindzhev NS, et al. Asterless is a scaffold for the onset of centriole assembly. Nature. 2010;467:714–718. doi: 10.1038/nature09445. [DOI] [PubMed] [Google Scholar]
- 26.Novak ZA, Conduit PT, Wainman A, Raff JW. Asterless licenses daughter centrioles to duplicate for the first time in Drosophila embryos. Curr. Biol. 2014;24:1276–1282. doi: 10.1016/j.cub.2014.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goshima G, et al. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobbelaere J, et al. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 2008;6:e224. doi: 10.1371/journal.pbio.0060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Campos M, Basto R, Baker J, Kernan M, Raff JW. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 2004;165:673–683. doi: 10.1083/jcb.200402130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mottier-Pavie V, Megraw TL. Drosophila bld10 is a centriolar protein that regulates centriole, basal body, and motile cilium assembly. Mol. Biol. Cell. 2009;20:2605–2614. doi: 10.1091/mbc.E08-11-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roque H, et al. Drosophila Cep135/Bld10 maintains proper centriole structure but is dispensable for cartwheel formation. J. Cell Sci. 2012;125:5881–5886. doi: 10.1242/jcs.113506. [DOI] [PubMed] [Google Scholar]
- 32.Blachon S, et al. A proximal centriole-like structure is present in Drosophila spermatids and can serve as a model to study centriole duplication. Genetics. 2009;182:133–144. doi: 10.1534/genetics.109.101709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carvalho-Santos Z, et al. Stepwise evolution of the centriole-assembly pathway. J. Cell Sci. 2010;123:1414–1426. doi: 10.1242/jcs.064931. [DOI] [PubMed] [Google Scholar]
- 34.Carvalho-Santos Z, et al. BLD10/CEP135 is a microtubule-associated protein that controls the formation of the flagellum central microtubule pair. Dev. Cell. 2012;23:412–424. doi: 10.1016/j.devcel.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 35.Grallert A, et al. Centrosomal MPF triggers the mitotic and morphogenetic switches of fission yeast. Nat. Cell Biol. 2013;15:88–95. doi: 10.1038/ncb2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park SY, et al. Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis. Nat. Struct. Mol. Biol. 2014;21:696–703. doi: 10.1038/nsmb.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonnen KF, Gabryjonczyk AM, Anselm E, Stierhof YD, Nigg EA. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J. Cell Sci. 2013;126:3223–3233. doi: 10.1242/jcs.129502. [DOI] [PubMed] [Google Scholar]
- 38.Knorz VJ, et al. Centriolar association of ALMS1 and likely centrosomal functions of the ALMS motif-containing proteins C10orf90 and KIAA1731. Mol. Biol. Cell. 2010;21:3617–3629. doi: 10.1091/mbc.E10-03-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 2010;191:721–729. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cizmecioglu O, et al. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 2010;191:731–739. doi: 10.1083/jcb.201007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim TS, et al. Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc. Natl. Acad. Sci. USA. 2013;110:E4849–4857. doi: 10.1073/pnas.1319656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avidor-Reiss T, Khire A, Fishman EL, Jo KH. Atypical centrioles during sexual reproduction. Front. Cell Dev. Biol. 2015;3:21. doi: 10.3389/fcell.2015.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sir JH, et al. A primary microcephaly protein complex forms a ring around parental centrioles. Nat. Genet. 2011;43:1147–1153. doi: 10.1038/ng.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lukinavicius G, et al. Selective chemical crosslinking reveals a Cep57-Cep63-Cep152 centrosomal complex. Curr. Biol. 2013;23:265–270. doi: 10.1016/j.cub.2012.12.030. [DOI] [PubMed] [Google Scholar]
- 45.Gopalakrishnan J, et al. Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat. Commun. 2011;2:359. doi: 10.1038/ncomms1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varmark H, et al. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr. Biol. 2007;17:1735–1745. doi: 10.1016/j.cub.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 47.Kim K, Lee S, Chang J, Rhee K. A novel function of CEP135 as a platform protein of C-NAP1 for its centriolar localization. Exp. Cell Res. 2008;314:3692–3700. doi: 10.1016/j.yexcr.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 48.Delgehyr N, et al. Klp10A, a microtubule-depolymerizing kinesin-13, cooperates with CP110 to control Drosophila centriole length. Curr. Biol. 2012;22:502–509. doi: 10.1016/j.cub.2012.01.046. [DOI] [PubMed] [Google Scholar]
- 49.Lipinszki Z, et al. Centromeric binding and activity of Protein Phosphatase 4. Nat. Commun. 2015;6:5894. doi: 10.1038/ncomms6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lipinszki Z, et al. Affinity purification of protein complexes from Drosophila embryos in cell cycle studies. Methods Mol. Biol. 2014;1170:571–588. doi: 10.1007/978-1-4939-0888-2_33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.