Abstract
Telomerase is a specialized ribonucleoprotein complex that extends the 3′ ends of chromosomes to counteract telomere shortening. However, increased telomerase activity is associated with ∼90% of human cancers. The telomerase enzyme minimally requires an RNA (hTR) and a specialized reverse transcriptase protein (TERT) for activity in vitro. Understanding the structure-function relationships within hTR has important implications for human disease. For the first time, we have tested the physical-connectivity requirements in the 451-nucleotide hTR RNA using circular permutations, which reposition the 5′ and 3′ ends. Our extensive in vitro analysis identified three classes of hTR circular permutants with altered function. First, circularly permuting 3′ of the template causes specific defects in repeat-addition processivity, revealing that the template recognition element found in ciliates is conserved in human telomerase RNA. Second, seven circular permutations residing within the catalytically important core and CR4/5 domains completely abolish telomerase activity, unveiling mechanistically critical portions of these domains. Third, several circular permutations between the core and CR4/5 significantly increase telomerase activity. Our extensive circular permutation results provide insights into the architecture and coordination of human telomerase RNA and highlight where the RNA could be targeted for the development of antiaging and anticancer therapeutics.
INTRODUCTION
Linear eukaryotic chromosomes terminate in repeated DNA sequences, called telomeres, which are bound by specific proteins to protect the ends from degradation and detrimental end joining. However, these termini present an end-replication problem that most eukaryotes overcome by utilizing the ribonucleoprotein (RNP) complex telomerase. Telomerase comprises an RNA (hTR in humans) and a reverse transcriptase (TERT), which catalyzes telomere addition. It has been shown that telomeres shorten with aging, and telomerase upregulation occurs in ∼90% of human cancers (1). Furthermore, mutations in telomerase components have been linked to a variety of short-telomere syndromes, such as dyskeratosis congenita, pulmonary fibrosis, and aplastic anemia (2). Thus, understanding the structure-function relationships of human telomerase RNA is crucial to combat a range of human ailments.
During the telomerase catalytic cycle, a short region of the telomerase RNA, known as the template, pairs with the lagging-strand telomeric 3′ overhang. The template is then used to direct the iterative addition of the telomeric repeats (TTAGGG in humans), catalyzed by an active site within the TERT protein. Once TERT reaches the end of the RNA template, the DNA substrate is realigned so that additional repeats can be added (3). This ability of the RNA-protein enzyme complex to translocate underlies the enzyme's repeat-addition processivity (RAP). In addition to hTR and TERT, additional accessory proteins bind telomerase in vivo (4). Many studies have been done to identify important regions within hTERT and hTR that are required for function (5). Thus far, however, the ways the RNA coordinates its roles have yet to be clearly elucidated.
Despite being critical for cell growth, telomerase RNAs are evolving incredibly rapidly and vary substantially in structure from species to species. Nevertheless, a few structural elements are conserved (6, 7) (Fig. 1A). Telomerase RNAs contain an essential core domain with (i) a single-stranded template that directs species-specific repeat addition (8), (ii) a template boundary element (TBE) that defines the 5′ end of the template (9–12), (iii) a pseudoknot (PK) with catalytically important base triples (13–16), (iv) a core-enclosing helix (CEH) (6, 7), and (v) an area of required connectivity (ARC) that connects the pseudoknot to the template via single-stranded junctions and secondary structures in the CEH and TBE (6). Although these elements appear in >97% of identified telomerase-RNA secondary structures (6), their particular structure and function may vary. For instance, template boundary definition in human telomerase requires the core-enclosing helix P1b, located 10 nucleotides 5′ of the template (10), as well as a particular nucleotide in the template itself (17). In contrast, rodent telomerase RNAs lack a core-enclosing helix (and ARC), with template boundary definition provided by a 5′ trimethylguanosine cap and/or runoff transcription (7, 10), while most species of yeast use a helix adjacent to the template that provides a steric block to further reverse transcription by TERT (10–12, 17). So, even though all telomerases contain template boundary elements to achieve the same function, they have evolved different structures and mechanisms to do so.
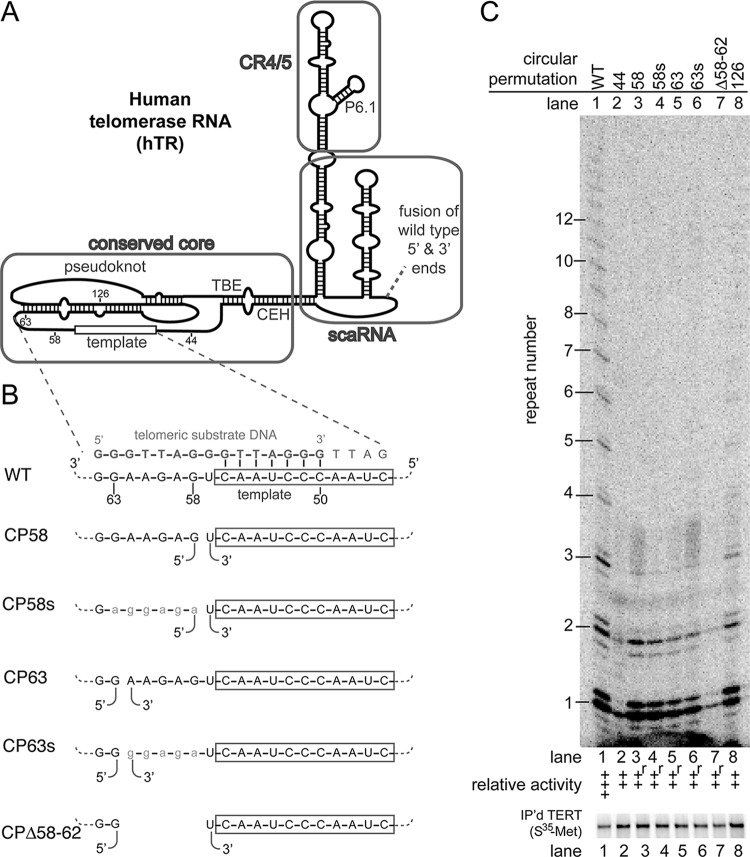
FIG 1.
Circular permutations 3′ of the template cause specific defects in repeat-addition processivity. (A) Schematic of hTR structure illustrating the conserved core, CR4/5, and scaRNA domains. The location where the wild-type 5′ and 3′ ends were fused is indicated (dashed line). (B) Sequence of the template (boxed) and adjacent 3′ region binding to the telomeric DNA substrate primer. Below are the locations of the repositioned ends in the circular permutants. Sequence substitutions are lowercase. (C) In vitro telomerase activity assays were performed showing circular permutations 3′ of the template (above). The relative activity was binned into the following categories: –, no detectable telomerase activity, ∼0%; +, low but detectable activity, <∼50%; ++, slightly inhibited activity, >∼50%; +++, wild-type activity, ∼100%; and ++++, greater than wild-type activity, >∼100%. Circular permutants with repeat-addition processivity defects are indicated by an “r.” Immunopurified [35S]methionine-labeled hTERT is shown below.
Although the core alone is sufficient to reconstitute telomerase activity in vitro in budding yeast, many other species, including human, require an additional three-way junction element. In human telomerase RNA, this is the CR4/5 region (Fig. 1A). The core and CR4/5 are sufficient to reconstitute telomerase activity in vitro, even when provided as separate RNA fragments, with each RNA domain binding TERT independently (18, 19). Three-way junction regions, such as CR4/5, have also been identified in the ciliate Tetrahymena thermophila (20) and the yeasts Schizosaccharomyces pombe and Kluyveromyces lactis (21, 22), although the corresponding region is not essential for telomerase activity or basic functionality in vivo in Saccharomyces cerevisiae (23, 24).
Finally, human telomerase requires a scaRNA domain (small Cajal body-specific RNA) containing a box H/ACA motif. In hTR, this domain provides important RNA processing, trafficking, and 3′-end stability functions in vivo (18, 25, 26). In yeasts, similar roles are provided by the Sm7 and/or Lsm complex binding at the 3′ end (11, 27). Thus, as with the template-boundary element, telomerase RNAs have evolved distinct ways to accomplish 3′-end processing and protection.
A multitude of studies have been done using substitution, deletion, and insertion mutations to uncover structure-function relationships in human telomerase RNA, but circular permutations (CPs) have yet to be used. Circular permutations involve relocating the 5′ and 3′ ends of the RNA to novel locations, introducing a break in the phosphate backbone. These unique mutations have proven to be highly informative in the exploration of structure-function relationships in both T. thermophila (20) and S. cerevisiae (6). Specifically, in Tetrahymena, circular permutations identified a template recognition element (TRE) in the single-stranded region 3′ of the template (28). The TRE contributes to repeat addition processivity and is thought to be alternately compressed and extended during the catalytic cycle as part of an accordion model (29). In S. cerevisiae, circular permutations identified essential connections (the area of required connectivity [ARC]) between the pseudoknot and template via the core-enclosing helix and template boundary element. The mechanistic role for the ARC in budding yeast is still under study; it may be required for RNA folding, TERT binding, template positioning, and/or template translocation. Whether human telomerase RNA shares important features, such as those mentioned above, with well-studied model system telomerase RNAs has yet to be determined.
We have examined where physical connections are important in human telomerase RNA. We tested 36 circular permutations of the full-length 451-nucleotide hTR for function in reconstituted telomerase activity assays. We observed phenotypes ranging from complete abolishment to a significant increase in telomerase activity. Notably, we found circular permutations 3′ of the template specifically inhibited repeat-addition processivity, similar to mutations in the TRE region in ciliates. This provides the first evidence that the TRE is a functionally conserved element in telomerase RNA across two distantly related species, increasing the number of highly conserved core elements to six. In contrast, backbone connectivity was dispensable for function at 12 positions tested, four of which significantly increased telomerase activity. Furthermore, circular permutations that abolished telomerase activity cluster in conserved regions near the catalytically important base triples of the pseudoknot and helix P6.1 of CR4/5. These results from an extensive number of hTR circular permutants expand our fundamental understanding of telomerase RNA structure, function, and coordination while also identifying regions that could be targeted to develop treatments for cancer and short-telomere syndromes.
MATERIALS AND METHODS
Cloning of circular permutations.
We ordered a DNA fragment (GenScript) containing two tandem copies of the hTR gene directly linked between positions 451 and 1 and cloned this fragment into vector pUC19. We used this plasmid as a template for PCR to generate circular permutants, using a dedicated set of PCR primers for each (28). For each circular permutant, the corresponding PCR product was cloned into pUC19, and its sequence was confirmed via Sanger sequencing. Each circular permutant was designed to have at least three guanosines present at the 5′ end for efficient T7 transcription and a FokI site at the 3′ end to linearize the plasmid DNA for runoff transcription. One to two untemplated uridine nucleotides may be added to the 3′ end by T7 RNA polymerase. Plasmid DNA was prepared for transcription by FokI digestion, followed by phenol-chloroform extraction and ethanol precipitation.
Reconstitution of human telomerase in a rabbit reticulocyte lysate transcription-translation system.
We provided the rabbit reticulate lysate with two plasmids: one encoding 3×-FLAG-hTERT (30) and the other encoding hTR, each at a final concentration of 20 ng/μl. The transcription-translation reactions were carried out using standard conditions (Promega) at 30°C for 90 min. Telomerase was immunopurified as previously described (31) and resuspended in 50 mM Tris-HCl (pH 8.0), 50 mM KCl, 1 mM MgCl2, 1 mM spermidine, 5 mM β-mercaptoethanol, and 30% glycerol. To verify the hTERT protein was translated and equivalent amounts were purified, we ran 2.5 μl of immunopurified beads on a denaturing 7.5% polyacrylamide gel and imaged on a Typhoon 9410 scanner (GE).
Direct telomerase primer extension assays.
Telomerase activity was measured using a published protocol (32, 33). One-third of the total reaction was run on a 10% acrylamide–7 M urea–1× Tris-borate-EDTA gel at 90 W for 90 min. Gels were imaged using a Typhoon 9410 scanner. Data were quantified with ImageQuant TL software; however, various levels of background radiation prohibited accurate analysis. We therefore chose to have five raters blindly bin the data from two or three independent biological replicates. Binning telomerase activity has been reported previously (20) and is supported by a recent study (33).
RESULTS
To test the effects of circularly permuting human telomerase RNA (hTR) on RNP enzyme activity, we expressed the new hTR alleles, along with affinity-tagged TERT in an in vitro transcription-translation system. One reason we chose an in vitro telomerase RNP reconstitution system as opposed to in vivo expression was to avoid likely complications in RNA biogenesis and stability due to repositioning the 3′-end scaRNA domain inherently caused by circularly permuting the gene (6, 18, 25, 26, 34). We then assayed reconstituted telomerase activity by direct primer extension to obtain detailed information about enzyme functionality. We named each circularly permuted allele based on the hTR nucleotide that follows the position of permutation; for example, CP58 has a 5′ end at position 58 (and its 3′ end terminates with nucleotide 57). No extra nucleotides were inserted between the last nucleotide (i.e., nucleotide 451) and the first (Fig. 1A).
Circular permutations upstream of the template cause repeat-addition processivity defects.
The T. thermophila telomerase RNA requires connectivity between the 3′ end of the template and the pseudoknot to promote efficient repeat-addition processivity via the TRE (28). To test whether a similar element exists in human telomerase RNA, we made circular permutations 3′ of the template. We found that repositioning the 5′ end to position 58 or 63 caused profound repeat-addition processivity defects (Fig. 1B and C, lanes 3 and 5). Specifically, in these replicate samples, telomeric DNA products corresponding to repeats 1 and 2 were equivalently abundant to wild type, whereas repeat 3 was reduced, and there were no detectable four-repeat or longer products (Fig. 1C). Biological replicates of these circular permutants similarly showed a RAP defect, though in this case weak repeats up to 5 were detectable. The defect caused by the circular permutations 3′ of the template is RAP specific, since it is distinct from the uniform reduction in products of many other mutants with decreased activity (e.g., Fig. 1C, lane 8). We conclude that connectivity between the template and the pseudoknot, 3′ of the template, is important for repeat-addition processivity.
To investigate what aspect of the putative TRE region in human telomerase is important for repeat-addition processivity, we first substituted nucleotides 58 to 63 with the corresponding conservative purines (Fig. 1B, CP58s and CP63s). These sequence-substituted circular permutants exhibited RAP defects similar to their unsubstituted counterparts (Fig. 1C, compare lanes 3 and 4 to lanes 5 and 6). These data suggest that the precise sequence in the junction between the template and pseudoknot is not essential for residual repeat-addition activity in these circular permutants. To test whether deleting the nucleotides between the template and pseudoknot further disrupts telomerase activity, we made a circular permutation with the 5′ end at nucleotide 63 and the 3′ end at nucleotide 57, deleting nucleotides 58 to 62 (Fig. 1B, CP58-62Δ). This mutant exhibited a significant decrease in adding the first and second repeats, suggesting that deleting nucleotides 58 to 62 further impaired telomerase activity (Fig. 1C, lane 7), although a biological replicate showed a repeat-addition defect similar to CP58 and CP63. In summary, our data demonstrate that a connection between the 3′ end of the template and pseudoknot is important for repeat-addition processivity.
Circular permutations near the pseudoknot base triples abolish telomerase activity.
The pseudoknot of the conserved catalytic center of hTR comprises over half of the core domain, and contains base triples at the junction between the paired stems (Fig. 2A and B) (14, 35, 36). In budding yeast, analogous base triples have been shown to contribute directly to catalysis through an unknown mechanism and are located within cross-linking distance from the template (13). Solved nuclear magnetic resonance structures of pieces of the human pseudoknot reveal that the 5-nucleotide bulge, J2a/b, introduces a significant bend in the pseudoknot, serving as a hinge within this larger structure (37). Previous data indicate that not all connections within the pseudoknot are essential, since a three-piece telomerase RNA that disrupts connections in loop II of the human pseudoknot has been shown to reconstitute telomerase activity (38).
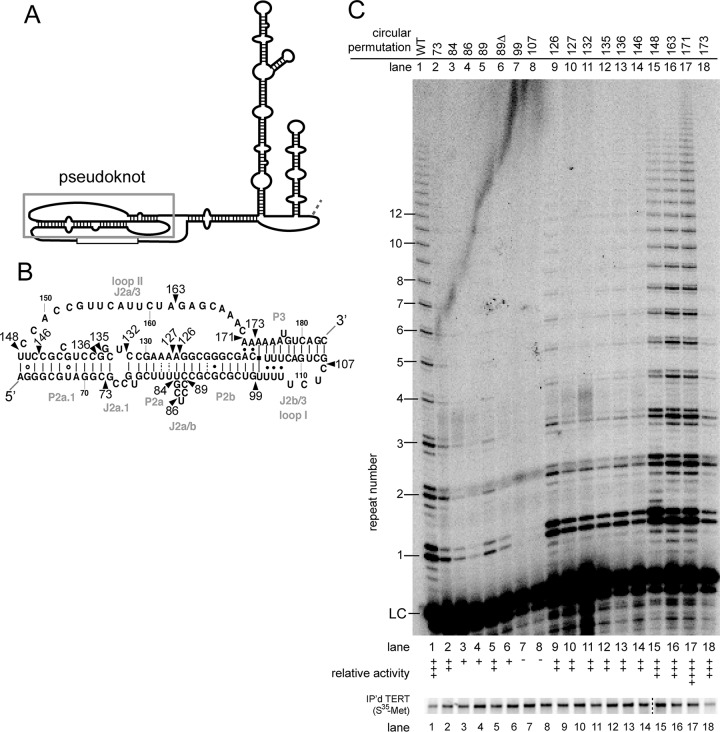
FIG 2.
Circular permutations throughout the pseudoknot cause a wide range of phenotypes. (A) Secondary structure of hTR with the pseudoknot region indicated. (B) Detailed structure of the pseudoknot, based a previously published study (35). Wild-type nucleotide numbers are indicated. Circular permutations are indicated by arrowheads and are named by the corresponding nucleotide at the new 5′ end. (C) In vitro telomerase assays exhibit various levels of activity. LC, internal radiolabeled recovery and loading control. The level of activity was binned as in Fig. 1. Immunopurified [35S]methionine-labeled FLAG-hTERT shown below. Protein samples in lanes 1 to 14 and lanes 15 to 18 were run on two separate gels in parallel, as indicated by the dashed line.
To determine what connections within the pseudoknot of hTR are functionally important, we made 16 circular permutations in this region. Circular permutations in specific subregions of this structure were well tolerated, while others completely abolished telomerase activity. The range of these phenotypes highlight regions that are particularly sensitive to backbone disruption, as we describe below.
First, circular permutations near the base triples in loop I (J2b/3) show the most severe phenotypes. Specifically, circular permutations at positions 99 and 107 completely abolish any detectable telomerase activity (Fig. 2C, lanes 7 and 8). These mutations are located near a well-studied hTR mutation linked to the disease dyskeratosis congenita (PKDKC) (39).
Second, circular permutations on either side of stem I (P2a.1, J2a.1, P2a, J2a/b, and P2b) showed decreased, but detectable, telomerase activity (Fig. 2C, lanes 2 to 6 and lanes 9 to 14). Of particular note, circular permutations surrounding the J2a/b bulge (37) decreased activity (Fig. 2C, lanes 3 to 6 and lane 9), suggesting that backbone connectivity may be required to adopt a particular bend at this region of the pseudoknot in the RNP and increasing the potential flexibility by disrupting backbone interactions within this region is deleterious.
Third, mutations within loop II (J2a/3) were well tolerated with essentially wild-type phenotypes (Fig. 2C, lanes 15 to 17). This is consistent with phenotypes observed in the three-piece system (38), as well as in yeast telomerase RNA (6), suggesting backbone connectivity within loop II is largely dispensable. Interestingly, the circular permutation at nucleotide 173 leads to robust activity (Fig. 2C, lane 18), even though it is located within newly proposed base triples (35).
Previous data showed deletion of the J2a/b bulge decreased telomerase activity (37), presumably by preventing hinging. To test whether breaking the phosphate backbone with a circular permutation would be able to overcome stiffening caused by J2a/b deletion, we tested a circular permutation missing nucleotides 84 to 88 (CP89Δ). This circular permutant had very weak but detectable telomerase activity (Fig. 2C, lane 6), suggesting that disrupting the backbone does not overcome the defect observed in the J2a/b deletion mutant. Furthermore, CP89Δ has a more severe defect than CP89 (Fig. 2C, compare lanes 5 and 6), indicating that the bulge nucleotides may contribute to enzyme function beyond permitting the large bend in the pseudoknot.
Circular permutations in CR4/5 abolish telomerase activity.
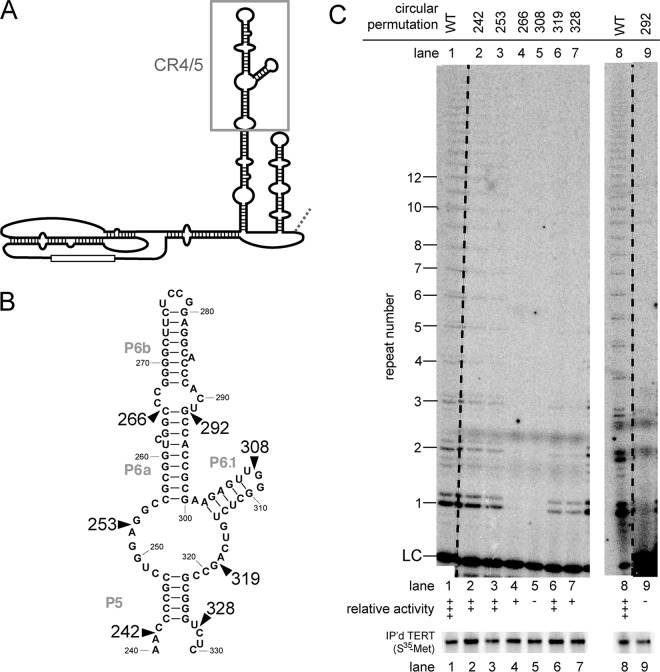
It has been shown that human telomerase activity requires not only the conserved core domain but also the CR4/5 domain (Fig. 3A and B) (18, 19). Within the CR4/5 domain, the loop and base of helix P6.1 have been shown to be especially important (40–42). Using our circular-permutation analysis, we found that disrupting the backbone between CR4/5 and the core is tolerated to different degrees. Relocating the ends to within the central portion of CR4/5 completely abolished telomerase function (Fig. 3C, lanes 4, 5, and 9). Other circular permutations in CR4/5 had intermediate effects: those in the 5′ portion were better tolerated than in the 3′ portion (Fig. 3C, compare lanes 3 and 4 to lanes 6 and 7). These results confirm that the CR4/5 domain is fundamentally important for catalytic function.
FIG 3.
Multiple circular permutations in the CR4/5 domain completely abolish telomerase activity. (A) Secondary structure of hTR highlighting the CR4/5 region (gray box). (B) Sequence of the CR4/5 region with the wild-type nucleotide numbering labeled. (C) In vitro telomerase activity assays were performed, with relative activity indicated as in Fig. 1. While in this experiment CP266 shows essentially no telomerase activity, a biological replicate indicated very low levels of detectable activity. The data in lanes 1 to 7 were from the same gel, with the dashed line indicating some intervening lanes that were cropped out. Lanes 8 and 9 were run on another gel, with the dashed line similarly indicating lanes cropped out. [35S]methionine-labeled immunopurified hTERT from each assayed telomerase preparation is shown below.
Several circular permutations in the native 5′ region and 3′ scaRNA domain increase telomerase activity.
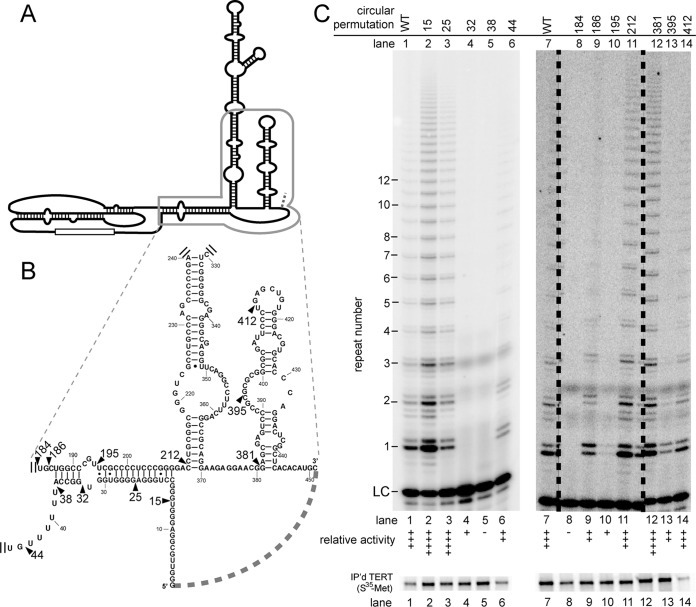
The wild-type 5′ end of hTR contains several runs of at least three guanosines and has been proposed to form G-quadruplexes that may compete with formation of P1 (43). The formation of helix P1b has been shown to aid in proper template-boundary definition (10). We tested the effect of circular permutations in the 5′ region of hTR (Fig. 4A and B). Notably, circular permutations at positions 15 and 25 caused increased telomerase activity (Fig. 4C, lanes 2 and 3), whereas circular permutations closer to the core, at positions 32 and 38, completely abolished telomerase activity (Fig. 4C, lanes 4 and 5). Circular permutant 44 (CP44) showed decreased but detectable telomerase activity, with no apparent template boundary definition defects (Fig. 4C, lane 6). Together, these data indicate that disrupting RNA connectivity near the 5′ end of hTR is beneficial, while circular permutations nearer the core are detrimental.
FIG 4.
Several circular mutations in the 5′ end and scaRNA domain cause increased telomerase activity. (A) Structure of hTR with the region of interest boxed. (B) Sequence of the 5′ end and scaRNA regions. Wild-type nucleotide numbers are indicated. (C) In vitro telomerase activity assay. The relative telomerase activity is indicated as in Fig. 1. The data in lanes 7 to 14 were run on the same gel; the dashed lines indicate where intervening lanes were cropped from the gel. Below is immunopurified [35S]methionine-labeled hTERT.
The observed defects of 5′-region circular permutations could result from disrupting P1. If so, we would expect circular permutations on the opposite of the helix to have similar phenotypes. However, circular permutations at positions 186 and 195 have detectable telomerase activity (Fig. 4C, lanes 9 and 10), unlike the circular permutants 32 and 38 (Fig. 4C, lanes 4 and 5). This suggests that the loss of telomerase activity is not likely caused by disrupting connectivity of P1b to adjacent regions but is specific to the 5′ end. Interestingly, a circular permutation just two nucleotides 5′ of 186, CP184, has no detectable telomerase activity (Fig. 4C, lane 8), indicating that circular permutations between the pseudoknot and helix P1b have strikingly different effects on telomerase activity.
In vivo, the scaRNA domain is required for RNA stability, processing, and trafficking (18, 25, 26, 34). However, this region is dispensable for function in vitro (18). We found that several circular permutations in this region were well tolerated. Strikingly, circularly permuting at 381 increased activity (Fig. 4C, lane 12). Circular permutation at position 412 had essentially wild-type activity (Fig. 4C, lane 14), while a circular permutation at position 395 had slightly decreased activity (Fig. 4C, lane 13). It is interesting that two of these circular permutants have phenotypes in vitro, even though this domain is dispensable.
DISCUSSION
In order to gain insights into the architecture of human telomerase RNA, we undertook a large-scale circular permutation analysis to determine regions where physical connectivity is, and is not, required for core enzyme activity. By repositioning the ends of the RNA, we interrupted the covalent linkage of the backbone and potentially increase local flexibility. Using reconstituted RNP enzyme and direct telomerase activity assays, we tested 36 circular permutants and found distinct classes of phenotypes that highlight several functionally important regions in the RNA (Fig. 5). This approach revealed 12 positions where backbone connectivity is dispensable for robust telomerase activity, highlighting substantial tolerance of hTR to backbone disruption. These positions reside in rapidly evolving portions of vertebrate telomerase RNAs (44). In contrast, circular permutations that greatly decrease or abolish telomerase function tended to cluster in the most conserved parts of the core and CR4/5 domains (Fig. 5). In addition to identifying mutations that alter overall activity, we found mutants 3′ of the template that alter the pattern of telomerase activity product formation, indicating decreased repeat-addition processivity (Fig. 5, CP58 and CP63). Strikingly, this shows that human telomerase RNA shares a functionally conserved template recognition element (TRE) with the distantly related ciliated protozoan, Tetrahymena thermophila. Together, our results provide an informative map of functional connectivity requirements between and within structural elements of this disease-relevant RNA.
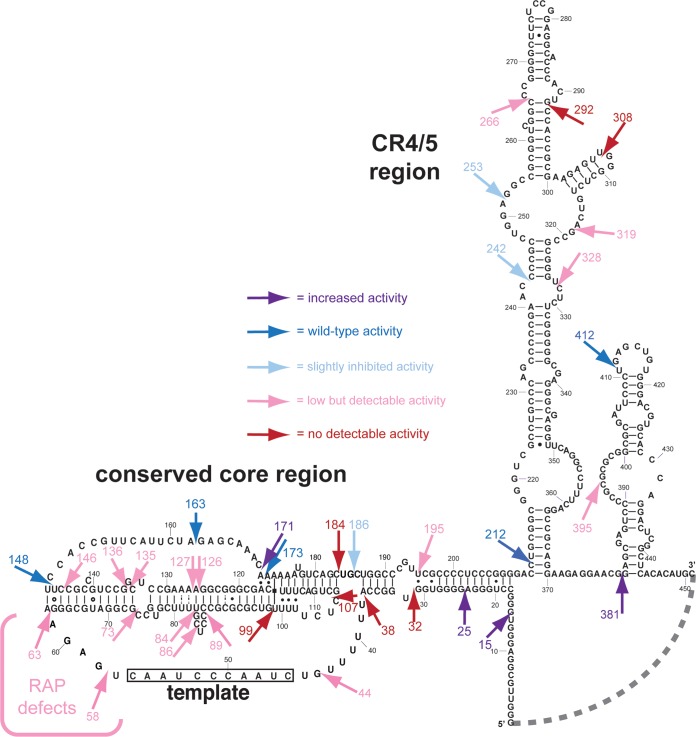
FIG 5.
Human telomerase RNA circular permutations reveal structure-function correlations. The sequence of the 451-nucleotide hTR is depicted. The dashed line between positions 1 and 451 indicates the joined ends in the circular permutants. Each circular permutation allele phenotype is indicated by an arrow pointing to the location of the repositioned 5′ and 3′ ends. The color of the arrow represents the relative activity bin.
This inaugural circular-permutation analysis of hTR allows us to compare these phenotypes with telomerase RNA circular permutants of the well-studied T. thermophila (20) and S. cerevisiae species (6). Although circular permutations may cause defects beyond breaking the phosphate backbone, such as cotranscriptional RNA misfolding or TERT binding, they have proven useful in discovering functionally important RNA regions. Circular permutations in T. thermophila revealed a TRE that is required for repeat-addition processivity (28) and the accordion model of template translocation (29), while circular permutations in S. cerevisiae reveal an area of required connectivity (ARC) that may coordinate the pseudoknot base triples and TERT-binding site(s) with the template in the active site (6). Our data suggest that human telomerase RNA shares both functional connectivity similarities and differences with this protozoan and fungus, as we highlight below.
RNA 3′ of the template contributes to repeat-addition processivity.
Circularly permuting hTR 3′ of the template, at positions 58 and 63, allows telomerase to add three to five detectable repeats, but beyond this point these circular permutants did not catalyze more detectable rounds of synthesis (Fig. 1). The ability of telomerase to synthesize the first three to five telomeric repeats is not strictly dependent on the sequence of the nucleotides between positions 58 and 63 (Fig. 1). The lack of a sequence requirement for nucleotides 3′ of the template is similar to previous results using a multipartite reconstitution system that adds a single repeat (45). These phenotypes are reminiscent of ciliate TRE circular permutants, which also exhibit a decrease in repeat-addition processivity in vitro (28). Thus, we propose that human telomerase RNA has a functionally analogous, conserved TRE required for repeat-addition processivity. This is the first evidence that a TRE 3′ of the template may be a sixth functionally conserved element of some telomerase RNA cores.
In T. thermophila, the TRE is proposed to function together with the region 5′ of the template to promote repeat-addition processivity via the accordion model (29). Whereas circular permutations 5′ of the template in ciliates exhibit repeat-addition processivity defects (29), we found that circularly permuting 5′ of the template significantly decreased overall telomerase activity, without an obvious repeat-addition defect. Thus, there appear to be some functional differences between ciliate and human telomerase RNA. However, in both the ciliate and the human circular permutants, disrupting connectivity 3′ of the template causes more severe defects than disrupting connectivity 5′ of the template.
Interestingly, S. cerevisiae telomerase lacks detectable repeat-addition processivity and exhibits weak nucleotide addition processivity under standard in vitro assay conditions (23, 46, 47). Additionally, circular permutations in the analogous region 3′ of the template cause no detectable defects in vitro (6). Furthermore, S. cerevisiae and most other yeast telomerase RNAs contain at least one helical structure between the template and the pseudoknot, while a phylogenetic analysis of ciliate, vertebrate, and all other telomerase RNAs has shown that there are no naturally occurring helices in the corresponding region (6). Thus, perhaps the lack of robust repeat-addition observed in most yeast species is related to the presence of a helix within the TRE region.
Circular permutations in the pseudoknots of different species show similar phenotypes.
Circular permutations have now been tested in human (the present study), ciliate (20), and budding yeast (6) telomerase RNAs, and the analogous circular permutants show strikingly similar phenotypes, suggesting structural and functional homology. The pseudoknot is one of five phylogenetically conserved elements of the telomerase core and contains conserved base triples that contribute to catalysis (6, 7, 13–16). In yeast, stem I of the pseudoknot is largely dispensable for function (7), and we find that circular permutations in human telomerase RNA stem I cause substantially decreased, yet detectable, enzyme activity (Fig. 2). In contrast, mutations in stem II of yeast telomerase show more severe phenotypes. These phenotypes are probably due to disrupted base-triple formation and/or TERT binding (6, 7, 13). The base triples in yeast and human telomerase RNA have been shown to contribute to catalysis (13, 16, 36). Consistent with disruption of base triples causing significant defects, circular permutations in loop I of the pseudoknot (J2b/3), which contains the third strand of the base triples, completely abolish human telomerase activity (Fig. 2) (6). On the other hand, circular permutations in loop II (J2a/3) of the pseudoknot are well tolerated, causing no defects in human or, as we showed in vitro and in vivo, in S. cerevisiae (Fig. 2) (6).
There is a bulge spanning nucleotides 84 to 88 in hTR that has been reported to introduce an important bend in the human pseudoknot (37). Although circular permutations in this region should provide nearly unrestricted flexibility, we found that all circular permutations tested in this region significantly decrease telomerase activity (Fig. 2). Specifically, circular permutations at the beginning (CP84), middle (CP86), end (CP89), or opposite of the bulge (CP126) all decrease telomerase activity. Furthermore, a circular permutant with a deletion of the bulge nucleotides also exhibits decreased activity (Fig. 2, CP89Δ). Thus, it seems likely that RNA backbone connectivity in this bulge is important for adopting a specific bend observed in this RNA; circular permutations could introduce too great a range of motion if this is important for pseudoknot function.
Completely inactivating hTR circular permutations cluster near the 3′ end of the pseudoknot and in the CR4/5 domain.
We identified seven circular permutations that completely abolish telomerase activity. These positions cluster in specific elements within the core domain and CR4/5 regions essential for telomerase activity. The first group of circular permutations are at 32, 38, 99, 107, and 184 (Fig. 2 and 4; Fig. 5). In three-dimensional space, given their locations in the secondary structure and what is known from pseudoknot structures, these positions may not be far apart. One possibility is that these circular permutations disrupt or destabilize base-triple formation. Alternatively, these circular permutations could cause significant pseudoknot misfolding or disruption of TERT binding. Interestingly, the circular permutation at position 173 has wild-type activity, even though there is a base-triple interaction with nucleotide 99, where circular permutation is completely inactivating (Fig. 2) (14, 35). Furthermore, other circular permutations in this vicinity do not completely abolish telomerase activity, including CP171, CP186, CP195, and CP44. Together, these data provide insights into the overall coordination within this region, which generally corresponds with the essential ARC delineated in S. cerevisiae (6). While the activity of human circular permutants CP186 and CP144 indicates that the hTR ARC region analogous to that in yeast is not strictly required, the decreased activity of these mutants suggests that this region is indeed important, as was observed in T. thermophila (20, 29). Perhaps the more active human and ciliate telomerases compared to yeast is evidence that these enzymes are more robust and are more protected from mutational disruption of conserved ARC function(s).
The second group of inactivating circular permutations (CP292, CP308 and, to a lesser extent, CP266), are in CR4/5, and cluster near the catalytically important P6.1 helix (Fig. 3). These mutations could affect the essential CR4/5 TERT-binding function (18) or RNA conformational changes. For instance, the medaka fish CR4/5 binds to the RNA-binding domain of TERT and undergoes a dramatic rearrangement of the three-way junction compared to unbound RNA (40). In contrast, circular permutations at the 5′ and 3′ borders of CR4/5 (CP242 and CP328) show strong or detectable activity (Fig. 3), consistent with the previously described two-piece RNP-enzyme reconstitution systems (18, 19). Furthermore, circular permutations at 253 and 319 also exhibit detectable activity (Fig. 3), consistent with the minimally mapped essential CR4/5 region (14). Overall, our circular permutation results demonstrate that RNA backbone disruptions within the flanks of the CR4/5 domain are less disruptive than those in or just before P6.1 helix.
Four circular permutations increase telomerase activity.
We found that circular permutations at positions 15, 25, 171, and 381 reproducibly increase telomerase activity. These circular permutations are located in the native 5′ domain (Fig. 4, CP15 and CP25), loop II (J2a3) of the pseudoknot (Fig. 2, CP171), and the scaRNA domain near the natural 3′ end of hTR (Fig. 4, CP381). General explanations for increased activity in these circular permutations include improved RNA folding, including altering cotranscriptional RNA folding pathways, destabilizing inactive final folds, and/or altering RNA conformational dynamics that occur throughout the catalytic cycle of nucleotide- and repeat-addition processivity. Additionally, circular permutations could increase the binding affinity of the RNA to TERT, either by themselves or via RNA folding changes.
Previous proposals about the function of hTR suggest more specific hypotheses. For instance, it has been suggested that the guanosine-rich 5′ end of hTR may form inhibitory G-quadruplex structures that compete with formation of a noninhibitory helix P1 (43). Thus, it is possible that the circular permutations at positions 15 and 25 (which break up the cotranscription of consecutive runs of guanosines) disrupt the ability of inhibitory RNA G-quadruplexes to form.
Interestingly, we found that circular permutations near the 5′-end cause nearly opposite phenotypes compared to previously tested deletion mutants in this region (10). The first 14 or 24 nucleotides of hTR are dispensable for activity in vitro, but the equivalent circular permutations that put these nucleotides at the 3′ end increase activity (Fig. 4). Reciprocally, deletion of the first 31 or 43 nucleotides increases activity (10), while moving them to the 3′ end by circular permutation inhibits or abolishes activity (Fig. 4). Further highlighting differences between deletions and circular permutations, the 43-nucleotide deletion exhibited template boundary defects (10), while no boundary defects are exhibited by the circular permutant at position 44 (Fig. 2 and 4).
Alternatively, it has been proposed that the 5′ G-quadruplexes and/or the scaRNA domain may promote human telomerase dimerization (48). It is possible that activity-increasing circular permutations in these regions promote a conformation more amenable to dimerization. However, the functional importance of human telomerase dimerization in vivo remains a topic of debate.
The regions described above where circularly permuting hTR increased activity are noteworthy, since they show that increasing physical flexibility may ameliorate intrinsic negative constraints on the RNA. Also, our circular permutation analysis shows that most regions (85%) within human telomerase RNA accommodate repositioning the ends while retaining detectable activity. This mutational tolerance illustrates perturbability of hTR at these locations, given that circularly permuting alters cotranscriptional folding and the break in the phosphodiester backbone can change structure and dynamics even if the RNA does still fold appropriately. The permutable portions of hTR may already be flexible (and thus breaking backbone connectivity does not increase local motion), be structured but dispensable for activity, or else could have a structure that is not abolished by permutation within it. The fact that the majority of circular permutants we tested retain detectable activity shows that hTR function is sufficiently robust to not be eradicated by the perturbation of circularly permuting and highlights the unique regions where circular permutation completely abolishes activity.
In conclusion, our extensive circular permutation analysis of human telomerase RNA has identified many areas of interest that merit further mechanistic investigation in the future. The results yield insights into the conserved structure-function relationships within the telomerase RNP in humans and permit us to discern similarities and differences between the best-studied telomerase RNAs of model organisms. We have identified seven specific locations where RNA connectivity is essential within the human telomerase RNA. This knowledge could potentially be used as the basis for designing antisense DNA oligonucleotides as telomerase-targeting therapeutics for treating cancers (49), based on endogenous RNase H cleavage at these sites, to inhibit the telomerase aberrantly expressed in the vast majority of human cancer cells.
ACKNOWLEDGMENTS
We thank the Cech, Stone, and Prasad labs for sharing valuable reagents and protocols.
This study was supported by U.S. National Institutes of Health (National Institute of General Medical Sciences) funding (R00 GM80400 to D.C.Z.) and startup funds from The Johns Hopkins University.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Shay JW, Bacchetti S. 1997. A survey of telomerase activity in human cancer. Eur J Cancer 33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 2.Armanios M, Blackburn EH. 2012. The telomere syndromes. Nat Rev Genet 13:693–704. doi: 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greider CW. 1991. Telomerase is processive. Mol Cell Biol 11:4572–4580. doi: 10.1128/MCB.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egan ED, Collins K. 2010. Specificity and stoichiometry of subunit interactions in the human telomerase holoenzyme assembled in vivo. Mol Cell Biol 30:2775–2786. doi: 10.1128/MCB.00151-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q, Kim NK, Feigon J. 2011. Architecture of human telomerase RNA. Proc Natl Acad Sci U S A 108:20325–20332. doi: 10.1073/pnas.1100279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mefford MA, Rafiq Q, Zappulla DC. 2013. RNA connectivity requirements between conserved elements in the core of the yeast telomerase RNP. EMBO J 32:2980–2993. doi: 10.1038/emboj.2013.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin J, Ly H, Hussain A, Abraham M, Pearl S, Tzfati Y, Parslow TG, Blackburn EH. 2004. A universal telomerase RNA core structure includes structured motifs required for binding the telomerase reverse transcriptase protein. Proc Natl Acad Sci U S A 101:14713–14718. doi: 10.1073/pnas.0405879101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greider CW, Blackburn EH. 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 9.Lai CK, Miller MC, Collins K. 2002. Template boundary definition in Tetrahymena telomerase. Genes Dev 16:415–420. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen JL, Greider CW. 2003. Template boundary definition in mammalian telomerase. Genes Dev 17:2747–2752. doi: 10.1101/gad.1140303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Box JA, Bunch JT, Zappulla DC, Glynn EF, Baumann P. 2008. A flexible template boundary element in the RNA subunit of fission yeast telomerase. J Biol Chem 283:24224–24233. doi: 10.1074/jbc.M802043200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seto AG, Umansky K, Tzfati Y, Zaug AJ, Blackburn EH, Cech TR. 2003. A template-proximal RNA paired element contributes to Saccharomyces cerevisiae telomerase activity. RNA 9:1323–1332. doi: 10.1261/rna.5570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiao F, Cech TR. 2008. Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol 15:634–640. doi: 10.1038/nsmb.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim NK, Zhang Q, Zhou J, Theimer CA, Peterson RD, Feigon J. 2008. Solution structure and dynamics of the wild-type pseudoknot of human telomerase RNA. J Mol Biol 384:1249–1261. doi: 10.1016/j.jmb.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cash DD, Cohen-Zontag O, Kim NK, Shefer K, Brown Y, Ulyanov NB, Tzfati Y, Feigon J. 2013. Pyrimidine motif triple helix in the Kluyveromyces lactis telomerase RNA pseudoknot is essential for function in vivo. Proc Natl Acad Sci U S A 110:10970–10975. doi: 10.1073/pnas.1309590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shefer K, Brown Y, Gorkovoy V, Nussbaum T, Ulyanov NB, Tzfati Y. 2007. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol Cell Biol 27:2130–2143. doi: 10.1128/MCB.01826-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown AF, Podlevsky JD, Qi X, Chen Y, Xie M, Chen JJ. 2014. A self-regulating template in human telomerase. Proc Natl Acad Sci U S A 111:11311–11316. doi: 10.1073/pnas.1402531111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell JR, Collins K. 2000. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol Cell 6:361–371. doi: 10.1016/S1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 19.Tesmer VM, Ford LP, Holt SE, Frank BC, Yi X, Aisner DL, Ouellette M, Shay JW, Wright WE. 1999. Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hTERT) in vitro. Mol Cell Biol 19:6207–6216. doi: 10.1128/MCB.19.9.6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mason DX, Goneska E, Greider CW. 2003. Stem-loop IV of tetrahymena telomerase RNA stimulates processivity in trans. Mol Cell Biol 23:5606–5613. doi: 10.1128/MCB.23.16.5606-5613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi X, Li Y, Honda S, Hoffmann S, Marz M, Mosig A, Podlevsky JD, Stadler PF, Selker EU, Chen JJ. 2013. The common ancestral core of vertebrate and fungal telomerase RNAs. Nucleic Acids Res 41:450–462. doi: 10.1093/nar/gks980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown Y, Abraham M, Pearl S, Kabaha MM, Elboher E, Tzfati Y. 2007. A critical three-way junction is conserved in budding yeast and vertebrate telomerase RNAs. Nucleic Acids Res 35:6280–6289. doi: 10.1093/nar/gkm713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zappulla DC, Goodrich K, Cech TR. 2005. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat Struct Mol Biol 12:1072–1077. doi: 10.1038/nsmb1019. [DOI] [PubMed] [Google Scholar]
- 24.Livengood AJ, Zaug AJ, Cech TR. 2002. Essential regions of Saccharomyces cerevisiae telomerase RNA: separate elements for Est1p and Est2p interaction. Mol Cell Biol 22:2366–2374. doi: 10.1128/MCB.22.7.2366-2374.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell JR, Cheng J, Collins K. 1999. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol 19:567–576. doi: 10.1128/MCB.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu D, Collins K. 2003. Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol Cell 11:1361–1372. doi: 10.1016/S1097-2765(03)00196-5. [DOI] [PubMed] [Google Scholar]
- 27.Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 28.Miller MC, Collins K. 2002. Telomerase recognizes its template by using an adjacent RNA motif. Proc Natl Acad Sci U S A 99:6585–6590. doi: 10.1073/pnas.102024699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berman AJ, Akiyama BM, Stone MD, Cech TR. 2011. The RNA accordion model for template positioning by telomerase RNA during telomeric DNA synthesis. Nat Struct Mol Biol 18:1371–1375. doi: 10.1038/nsmb.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drosopoulos WC, Direnzo R, Prasad VR. 2005. Human telomerase RNA template sequence is a determinant of telomere repeat extension rate. J Biol Chem 280:32801–32810. doi: 10.1074/jbc.M506319200. [DOI] [PubMed] [Google Scholar]
- 31.Parks JW, Stone MD. 2014. Coordinated DNA dynamics during the human telomerase catalytic cycle. Nat Commun 5:4146. doi: 10.1038/ncomms5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen JL, Greider CW. 2003. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J 22:304–314. doi: 10.1093/emboj/cdg024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaug AJ, Crary SM, Jesse Fioravanti M, Campbell K, Cech TR. 2013. Many disease-associated variants of hTERT retain high telomerase enzymatic activity. Nucleic Acids Res 41:8969–8978. doi: 10.1093/nar/gkt653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Egan ED, Collins K. 2012. An enhanced H/ACA RNP assembly mechanism for human telomerase RNA. Mol Cell Biol 32:2428–2439. doi: 10.1128/MCB.00286-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niederer RO, Zappulla DC. 2015. Refined secondary-structure models of the core of yeast and human telomerase RNAs directed by SHAPE. RNA 21:1053. [PMC free article] [PubMed] [Google Scholar]
- 36.Theimer CA, Blois CA, Feigon J. 2005. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell 17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Q, Kim NK, Peterson RD, Wang Z, Feigon J. 2010. Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA. Proc Natl Acad Sci U S A 107:18761–18768. doi: 10.1073/pnas.1013269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen JL, Greider CW. 2005. Functional analysis of the pseudoknot structure in human telomerase RNA. Proc Natl Acad Sci U S A 102:8080–8089. doi: 10.1073/pnas.0502259102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I. 2001. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature 413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 40.Huang J, Brown AF, Wu J, Xue J, Bley CJ, Rand DP, Wu L, Zhang R, Chen JJ, Lei M. 2014. Structural basis for protein-RNA recognition in telomerase. Nat Struct Mol Biol 21:507–512. doi: 10.1038/nsmb.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen JL, Opperman KK, Greider CW. 2002. A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucleic Acids Res 30:592–597. doi: 10.1093/nar/30.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robart AR, Collins K. 2010. Investigation of human telomerase holoenzyme assembly, activity, and processivity using disease-linked subunit variants. J Biol Chem 285:4375–4386. doi: 10.1074/jbc.M109.088575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Booy EP, Meier M, Okun N, Novakowski SK, Xiong S, Stetefeld J, McKenna SA. 2012. The RNA helicase RHAU (DHX36) unwinds a G4-quadruplex in human telomerase RNA and promotes the formation of the P1 helix template boundary. Nucleic Acids Res 40:4110–4124. doi: 10.1093/nar/gkr1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen JL, Blasco MA, Greider CW. 2000. Secondary structure of vertebrate telomerase RNA. Cell 100:503–514. doi: 10.1016/S0092-8674(00)80687-X. [DOI] [PubMed] [Google Scholar]
- 45.Wu RA, Collins K. 2014. Human telomerase specialization for repeat synthesis by unique handling of primer-template duplex. EMBO J 33:921–935. doi: 10.1002/embj.201387205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bosoy D, Lue NF. 2004. Yeast telomerase is capable of limited repeat addition processivity. Nucleic Acids Res 32:93–101. doi: 10.1093/nar/gkg943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohn M, Blackburn EH. 1995. Telomerase in yeast. Science 269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 48.Martadinata H, Phan AT. 2014. Formation of a stacked dimeric G-quadruplex containing bulges by the 5′-terminal region of human telomerase RNA (hTERC). Biochemistry 53:1595–1600. doi: 10.1021/bi4015727. [DOI] [PubMed] [Google Scholar]
- 49.Zvereva MI, Zatsepin TS, Azhibek DM, Shubernetskaya OS, Shpanchenko OV, Dontsova OA. 2015. Oligonucleotide inhibitors of telomerase: prospects for anticancer therapy and diagnostics. Biochemistry 80:251–259. doi: 10.1134/S0006297915030013. [DOI] [PubMed] [Google Scholar]