Abstract
DNA methylation is a dynamic epigenetic modification with an important role in cell fate specification and reprogramming. The Ten eleven translocation (Tet) family of enzymes converts 5-methylcytosine to 5-hydroxymethylcytosine, which promotes passive DNA demethylation and functions as an intermediate in an active DNA demethylation process. Tet1/Tet2 double-knockout mice are characterized by developmental defects and epigenetic instability, suggesting a requirement for Tet-mediated DNA demethylation for the proper regulation of gene expression during differentiation. Here, we used whole-genome bisulfite and transcriptome sequencing to characterize the underlying mechanisms. Our results uncover the hypermethylation of DNA methylation canyons as the genomic key feature of Tet1/Tet2 double-knockout mouse embryonic fibroblasts. Canyon hypermethylation coincided with disturbed regulation of associated genes, suggesting a mechanistic explanation for the observed Tet-dependent differentiation defects. Based on these results, we propose an important regulatory role of Tet-dependent DNA demethylation for the maintenance of DNA methylation canyons, which prevents invasive DNA methylation and allows functional regulation of canyon-associated genes.
INTRODUCTION
DNA methylation is an important epigenetic modification that undergoes dynamic changes during cellular differentiation (1, 2). The discovery of the Ten eleven translocation (Tet) family of enzymes and their enzymatic activities identified a novel demethylation pathway triggered by the conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) (3–5). All three mammalian Tet proteins possess catalytic dioxygenase activity to generate 5hmC from existing 5mC and also to further process this modified cytosine to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (4, 5). As the maintenance DNA methyltransferases recognize 5hmC poorly, this modification could promote passive DNA demethylation (4). Moreover, 5hmC and its oxidation derivatives can function as intermediates in a process of active DNA demethylation, where they are enzymatically recognized and removed from the genome by components of the DNA repair machinery (4, 5).
Reduced levels of hydroxymethylation have been described in various human cancers (5). While the mechanistic basis for this loss is not known, it has been associated with reduced levels of Tet expression, in particular of Tet1 and Tet2, and cancer-specific hypermethylation (5). Recently, it has also been shown that 5hmC marks promoters that are resistant to cancer-specific hypermethylation (6). However, even though substantial progress has been made in the characterization of 5hmC as a novel DNA base modification, the epigenetic regulatory function of Tet enzymes remains to be fully understood.
Studies in mouse models with combined null alleles of Tet genes have started to investigate these issues. Embryonic stem cells (ESCs) lacking all three Tet genes are incapable of proper differentiation (7). Tet1/Tet2 double-knockout (DKO) ESCs were characterized by more restricted developmental defects that were associated with DNA hypermethylation (8, 9). DKO mice are characterized by global DNA hypermethylation and developmental plasticity, suggesting an important role of Tet1/Tet2 in the epigenetic regulation of developmental genes (8). In agreement with this notion, recent studies have shown a role of Tet-mediated demethylation in the modulation of enhancer activity (10–12).
Global analyses of mammalian methylomes at single-base resolution have shown that most CpGs are highly methylated (13). Nevertheless, 10% of the genome is comprised of regions mostly smaller than 1.5 kb with low or absent DNA methylation that coincide with promoter-associated CpG islands, gene body, and enhancer regions, as well as transcription factor binding sites (14). Furthermore, whole-genome bisulfite sequencing (WGBS) in human ESCs and mouse hematopoietic stem cells identified around 1,000 larger (>3.5-kb) genomic domains with low average methylation levels, termed DNA methylation valleys or DNA methylation canyons (1, 15). These elements remained unmethylated during differentiation and were often associated with developmental genes.
The roles of Tet enzymes in the establishment and maintenance of the global DNA methylation landscape remain an important topic of current research (16, 17). Here, we have characterized the methylation profiles of Tet1/Tet2 DKO mouse embryonic fibroblasts (MEFs) at single-base resolution. These cells possess impaired demethylation machinery (8) and therefore provide an excellent model to investigate the effects of globally reduced 5hmC. We identified the hypermethylation of DNA methylation canyons that often harbor developmental genes as a key feature of the DKO methylome. DKO MEFs showed pronounced defects in adipogenic differentiation, suggesting functional relevance of the observed methylation changes. We propose that Tet-dependent DNA demethylation plays an important role in the maintenance of hypomethylated canyons and the prevention of invasive hypermethylation.
MATERIALS AND METHODS
Derivation, cell culture, and adipogenic differentiation of MEFs.
MEFs were isolated from Tet-deficient midgestation embryos using a standard protocol (8). In brief, timed pregnancies were set up by intercrossing heterozygous Tet mutant mice. Embryonic day 13.5 (E13.5) embryos were isolated. The extraembryonic layers, internal organs, and heads of the embryos were removed. The heads were used for DNA extraction and genotyping of the embryos/MEFs by quantitative PCR (8). The remaining carcass was minced with a sterile blade and trypsinized for 15 min, followed by pipetting to dissociate the cells. The cell suspension was cultured in 20 ml of MEF medium (Dulbecco's modified Eagle's medium [DMEM] with 10% fetal calf serum [FCS], essential amino acids, and 200 U/ml penicillin and 200 mg/ml streptomycin) in a 15-cm tissue culture plate. Two days later, the confluent cultures (considered passage 0 [P0]) were split 1 to 3 to obtain the P1 culture. The MEFs were then frozen in cryovials in MEF-freezing medium (50% fetal bovine serum [FBS], 40% DMEM, 10% dimethyl sulfoxide [DMSO]) or split again 1 to 3 (to obtain P2) and then grown for an additional 3 days and frozen for long-term storage. For experiments, P2 MEFs were cultured at 37°C and 5% CO2 in DMEM (supplied with 10% FCS and penicillin/streptomycin). For an overview of the different batches of cells employed in the experiments, see Table S1A in the supplemental material.
For adipogenic differentiation, 1 × 105 cells/well were seeded into 12-well plates in duplicate. After 24 h, treatment with adipogenic differentiation medium (ADM) was initiated as described previously (18). After 7, 14, and 21 days, lipid droplets were stained or cells were harvested for further analysis. For staining of lipid droplets, cells were fixed for 1 h in 10% formalin, washed with 60% isopropanol, and stained with a filtered (0.2 μm) Oil-Red-O working solution (a 60% stock solution in water; the stock solution was 0.35% Oil-Red-O [Sigma] in isopropanol) for 10 min. Wells were washed in pure H2O, and the numbers of Oil-Red-O-positive cells were determined in 5 random images per well acquired at ×4 magnification using ImageJ.
RNA extraction, cDNA synthesis, and quantitative RT-PCR.
RNA was extracted from frozen cell pellets using TRIzol (Invitrogen), cDNA was synthesized from 1 μg RNA (QuantiTect reverse transcription kit; Qiagen), and quantitative RT-PCRs were performed in triplicate with mouse β-actin and Gapdh as reference genes using ABsolute qPCR SYBR green Mix (Thermo Fisher) and the Lightcycler 480 system (Roche) (for primer sequences, see Table S4 in the supplemental material).
DNA isolation and dot blotting.
For dot blots, 1 μg of sonicated genomic DNA was denatured and neutralized, and serial dilutions were transferred to a nylon membrane (GE Healthcare) using a dot blot apparatus (Bio-Rad). The membrane was washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), air dried, baked at 80°C for 2 h, stained with methylene blue to assess equal loading, destained, blocked, and incubated with primary antibodies (1:1,000) against 5hmC (Active Motif; 39791) for 1 h at room temperature. After 1 h of incubation with a horseradish peroxidase (HRP)-coupled secondary antibody (1:7,500), the membrane was incubated in enhanced chemiluminescence solution (PerkinElmer; Western Lightning Plus-ECL) and exposed to X-ray films.
Whole-genome bisulfite sequencing and analysis of DNA methylation patterns.
Library preparation for paired-end bisulfite sequencing was performed as described previously (19). Reads were trimmed and mapped with BSMAP 2.5 (20) using the mm9 assembly of the mouse genome as a reference sequence. Duplicates were removed using the Picard tool (http://broadinstitute.github.io/picard). Methylation ratios were determined using a Python script (methratio.py) distributed together with the BSMAP package. Inherently metastable partially methylated domains (PMDs) were removed as described previously (19) using a sliding-window approach with a window size of 100 kb. For every single window, the average methylation value was subtracted from the corresponding value of the reference Mus musculus skin methylome. The resulting sequence of average methylation ratio differences was processed using a 2-state first-order hidden Markov model and separated into two kinds of states, representing windows with a strong reduction of methylation (PMDs) and windows with no or only a small reduction. The PMD part was excluded from further analyses, which removed 38% of the genome sequence but retained 83% of the annotated genes. For an unbiased analysis of methylation changes in different samples, ChromHMM (21; http://compbio.mit.edu/ChromHMM/) was applied to published chromatin immunoprecipitation sequencing (ChIP-seq) data sets (22, 23) to partition the MEF genome into 15 chromatin states, and methylation ratios for these genomic segments (E1 to E15) were calculated. The data sets used were as follows (deposited at the Gene Expression Omnibus): GSM307609 (H3K27me3), GSM307610 (H3K36me3), GSM307611 (H3K9me3), GSM723004 (CTCF), GSM723005 (H3K4me1), GSM723006 (H3K4me3), GSM723007 (Pol2), and GSM851277 (H3K27ac).
Analysis of DNA methylation canyons.
DNA methylation canyons were identified as described previously (15, 24). A canyon that overlapped the transcription start site of a gene was assigned to that gene. For every wild-type (WT) canyon, the average level of active (H3K4me3 and H3K27ac) and repressive (H3k27me3 and H3k9me3) histone marks was computed using ChIP-seq data sets available for wild-type MEFs (22, 23). For further analysis, canyons were ordered by increasing amounts of the respective histone mark, and the resulting list was split into 10 equal parts. For every part, the average length of the canyons in wild-type and DKO MEFs was determined.
454 DNA bisulfite sequencing.
Deep DNA bisulfite sequencing was performed with 500 ng of genomic DNA for bisulfite treatment using the EpiTect bisulfite kit (Qiagen) according to the manufacturer's instructions. Treated DNA was amplified with sequence-specific primers (see Table S4 in the supplemental material) containing cell-type-specific barcodes and standard 454 linker sequences. PCR products were gel extracted using the peqGold extraction kit (Peqlab). For sequencing, equimolar amounts of all amplicons were combined in a single tube and processed on a GS Junior sequencer (Roche) according to the manufacturer's instructions. Sequence reads were aligned and displayed as color-coded heat maps.
MeDIP and hydroxymethylated DNA immunoprecipitation (hMeDIP).
Methylated DNA immunoprecipitation (MeDIP) was performed as described previously, using antibodies against 5mC (Active Motif; 39649) and 5hmC (Active Motif; 39791) (25). Immunoprecipitates (IPs) were analyzed by real-time PCR using specific primer pairs (see Table S4 in the supplemental material). The mouse Gapdh and β-actin promoters served as negative controls for unmethylated regions. Final enrichments were calculated relative to these unmethylated controls.
Gene expression analysis.
Transcriptome-sequencing libraries were prepared from total RNA preparations from WT-1 and DKO-1 MEFs (see Table S1A in the supplemental material) using the TruSeq RNA sample preparation kit (Illumina, San Diego, CA). Reads were trimmed to a maximal length of 80 bp, and stretches of bases having a quality score of <30 at the ends of the reads were removed. The reads were mapped using Tophat 2.0.6 (26) against the mm9 assembly of the mouse genome. Differential expression was quantified using DESeq 1.10.1 (27) and Cuffdiff 2.0 (28) and subjected to multiple testing corrections. Genes with a q value smaller than 0.05 were considered differentially expressed.
Statistical analysis.
To compare the frequencies of an occurrence between two groups, a χ2 test was used. Student's t test was performed to obtain P values for quantitative-PCR (qPCR) experiments, MeDIP analyses, and WGBS enrichments. Both tests were performed using implemented functions in R or online tools. P values to indicate significant differential expression for the transcriptome sequencing (RNA-seq) analysis were obtained from DESeq after FDR correction (27). Statistical testing for the overlap between differentially expressed genes and genes associated with hypermethylated canyons was done by a hypergeometric test using R, as described previously (11). Biological pathway analyses were performed using Ingenuity Pathway Analysis software, and bar charts showing the −log (P values) were directly used as figure panels.
WGBS and RNA-seq data accession number.
WGBS and RNA-seq data sets were deposited at the Gene Expression Omnibus database under accession number GSE58611.
RESULTS
Altered DNA methylation patterns in DKO MEFs.
To analyze the roles of Tet1 and Tet2 in somatic cells, we generated MEFs from DKO mouse embryos (8). Expression of Tet1 and Tet2 could not be detected in DKO MEFs, whereas Tet3 was moderately expressed and did not show a significant change compared to the wild type (see Fig. S1A in the supplemental material). Consistent with previous results (8), DKO MEFs retained reduced levels of 5hmC (see Fig. S1B in the supplemental material).
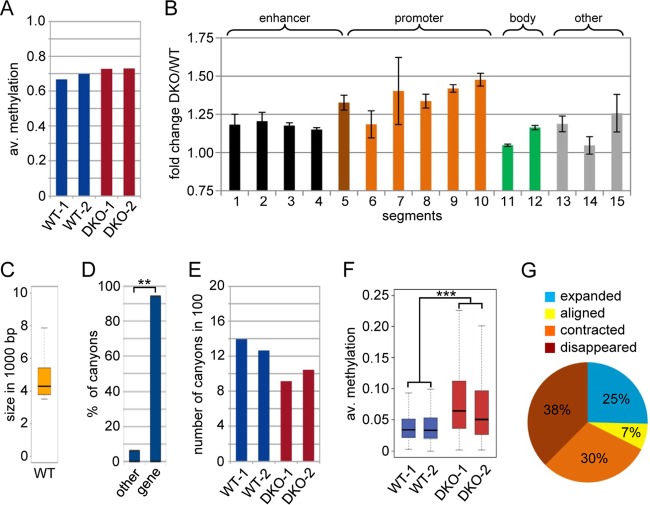
To analyze the methylation patterns of wild-type (WT) and DKO MEFs at the highest possible resolution, we used WGBS of two independent biological replicates for both wild-type and DKO cells (see Table S1A in the supplemental material), with a combined average genome coverage of ∼20× in both WT and DKO MEFs (see Table S1B in the supplemental material). The bisulfite conversion rate was determined by analyzing mitochondrial sequences that we considered unmethylated (29). This revealed conversion rates of >99.9%, suggesting highly effective bisulfite treatment. The average methylation levels of the four samples were similar between replicates but appeared to be increased in DKO cells (Fig. 1A), suggesting specific Tet-dependent methylation changes.
FIG 1.
Methylome of WT and DKO MEFs. (A) Average (av.) methylation ratios of the WT-1, WT-2, DKO-1, and DKO-2 samples. (B) Methylation changes at various subgenomic features shown as fold changes of the average methylation (DKO versus wild type; two replicates). ChromHMM (43) was applied to published ChIP-seq data sets (22, 23) to partition the wild-type MEF genome into 15 chromatin states, and methylation ratios for these genomic segments were calculated. All the segments (except E14) showed variable but significant (P < 0.01; Wilcoxon rank sum test) hypermethylation in DKO MEFs (see Fig. S2 in the supplemental material for a detailed description of segments and values). The error bars represent standard deviations. (C) Box plot showing the size distribution of all canyons. The box shows all canyons between the first and third size quartiles of the data set, whereas the line inside the box depicts the median for the canyon size. The ends of the whiskers mark the minimum and maximum sizes observed. (D) Canyons are strongly and significantly (**, P < 0.01; χ2 test) associated with transcription start sites. (E) Numbers of DNA methylation canyons in WT-1, WT-2, DKO-1, and DKO-2 cells. (F) Box plot showing average methylation ratios of canyons in WT-1, WT-2, DKO-1, and DKO-2 cells. The difference between the wild-type and DKO replicates is highly significant (***, P = 2.2 × 10−16; two-sided paired t test). (G) Pie chart showing canyon size dynamics in DKO MEFs.
We then used multivariate hidden Markov modeling based on published ChIP-seq data sets (22, 23) (see Materials and Methods for the GEO accession numbers) to segment the MEF epigenome into subgenomic features using ChromHMM (21). The identified segments were functionally classified according to their protein or histone modification patterns (30–32), and the average methylation levels of these segments were extracted from the WGBS data sets, revealing widespread hypermethylation in DKO MEFs (Fig. 1B; see Fig. S2 in the supplemental material). Significant (P < 0.01; Wilcoxon rank sum test) hypermethylation was detected for both promoter and enhancer regions, consistent with recent findings in Tet-deficient mouse ESCs (11, 12). As Tet-dependent methylation changes at promoters appeared more prominent (Fig. 1B), we focused our further analysis on promoter regions.
DNA methylation canyons are regions with locally reduced DNA methylation levels that often contain CpG islands and are frequently associated with promoter regions (15). As such, they represent attractive candidate target regions for Tet-dependent epigenetic regulation. Canyons are defined as regions with an average proportion of methylation of ≤10%, requiring at least five CpGs per kilobase (to satisfy the permutation-based false-discovery rate of 5%) and a minimum size of 3.5 kb (15). Applying established hidden Markov models to our WGBS data (15, 24), we identified 1,394 canyons in wild-type MEFs, with an average size of 5.5 kb (Fig. 1C). As expected, the vast majority (95%) of canyons were associated with transcription start sites (Fig. 1D). The number of canyons was distinctly reduced in DKO cells (Fig. 1E), as many canyons failed to reach the size minimum. This effect is directly related to increased canyon methylation in these cells, as average canyon methylation ratios were robustly (2-fold) and significantly (P = 2.2 × 10−16; t test) increased in DKO relative to wild-type MEFs (Fig. 1F).
Collapse of DNA methylation canyons in DKO MEFs.
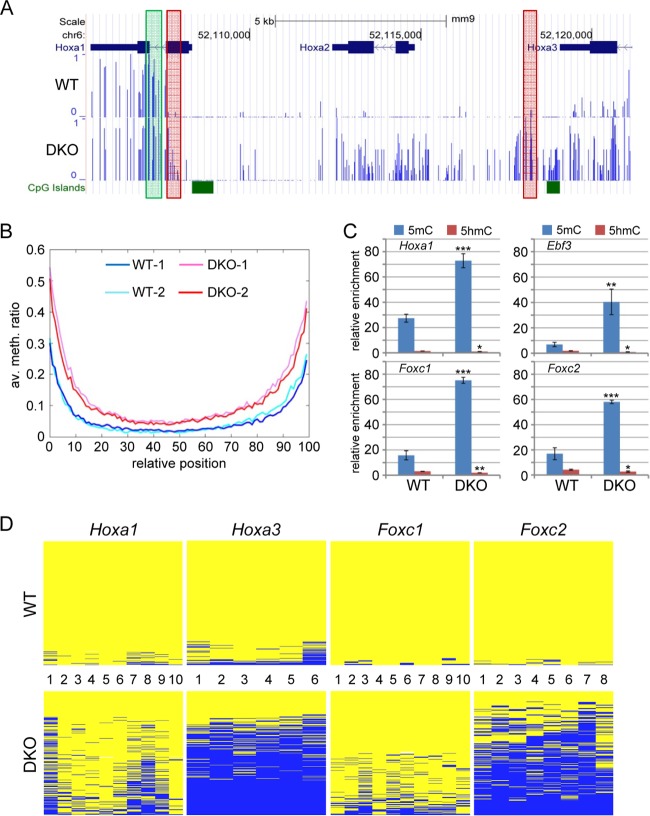
Our findings so far suggested that hypermethylation of canyons is a prominent feature of the DKO methylome. In order to study this further, we took all the canyons detected in wild-type MEFs and analyzed the corresponding canyons in DKO MEFs (Fig. 1G); 38% of the canyons fell below the size limit in DKO MEFs (disappeared), and 30% were contracted. In contrast, 25% of the canyons increased in size in DKO MEFs (expanded), whereas 7% experienced no significant change. These results confirmed that most canyons (68%) showed a pronounced size reduction (collapsed) caused by hypermethylation in DKO cells, which we also observed by visual inspection of differentially methylated canyons (Fig. 2A; see Fig. S3 in the supplemental material). Systematic analysis of biological replicates further confirmed this observation and also demonstrated that hypermethylation in DKO MEFs was particularly evident at canyon borders (Fig. 2B). We therefore used DNA immunoprecipitation (MeDIP) with antibodies against 5mC and 5hmC for an independent analysis of the border regions of four selected canyons in two replicate samples of wild-type and DKO MEFs. The results confirmed the substantially higher levels of 5mC at these regions in DKO MEFs, whereas 5hmC levels were rather low, suggesting fast processing of this mark at canyon borders in MEFs, but were significantly reduced in DKO MEFs (Fig. 2C). In further experiments, we also used deep bisulfite sequencing of 4 selected PCR amplicons to independently confirm the collapse of canyons in replicate samples using 454 technology. The resulting methylation profiles showed pronounced hypermethylation in the DKO sample (Fig. 2D) and thus provided strong confirmation for our WGBS data.
FIG 2.
Collapse of DNA methylation canyons in DKO MEFs. (A) Example of a DNA methylation canyon in the Hoxa cluster. Methylation profiles in WT-1 (top) and DKO-1 (bottom) are shown as methylation ratios. The green box indicates the region analyzed by (h)MeDIP (C). The red boxes indicate the genomic regions analyzed in panel D. (B) Average methylation levels of all canyons, comparing WT-1, WT-2, DKO-1, and DKO-2 MEFs. The canyons were size normalized before superposition. (C) (h)MeDIP analysis of borders of canyons overlapping Hoxa1, Erbf3, Foxc1, and Foxc2. The results are shown as normalized enrichments calculated relative to the unmethylated Gapdh and β-actin promoters (see Fig. S4 in the supplemental material). The regions analyzed are shown as green boxes in panel A and in Fig. S3 in the supplemental material. All differences comparing WT with DKO MEFs are significant (t test; ***, P <0.001; **, P <0.01; *, P <0.05). The error bars represent standard deviations (n = 3). (C) Validation of DNA methylation by targeted 454 bisulfite sequencing at canyon borders near Hoxa1, Hoxa3, Foxc1, and Foxc2. The results are shown as heat maps in which each row represents one sequence read. The individual blue boxes indicate methylated CpGs, and the yellow boxes indicate unmethylated CpGs. The CpGs covered by the individual amplicons are numbered. The regions analyzed are shown as red boxes in panel A and in Fig. S3 in the supplemental material.
Recent publications have suggested distinct roles for Tet1 and Tet2 in mouse embryonic stem cells, particularly with respect to the subgenomic localization of Tet1- or Tet2-dependent 5hmC and the degree of hypermethylation in Tet1- or Tet2-deficient cell lines (12, 33). We therefore analyzed the same canyons in Tet1 and Tet2 single-knockout MEFs. The results showed more moderate canyon border hypermethylation in Tet2-deficient and, to a lesser extent, also in Tet1-deficient MEFs (see Fig. S5 in the supplemental material). This suggests that the combined activities of Tet1 and Tet2 protect canyon borders against methylation invasion from surrounding methylated sequences.
Altered gene expression patterns and differentiation defects in DKO MEFs.
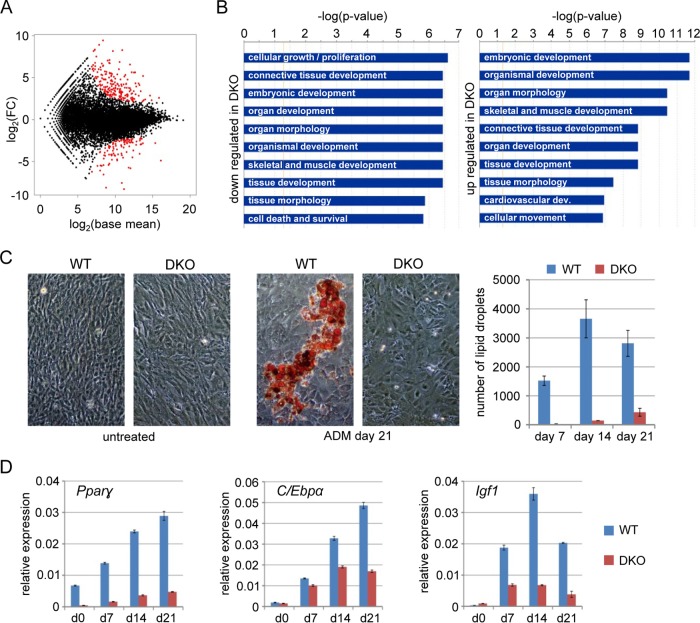
In order to correlate phenotypic differences between wild-type and DKO MEFs with gene expression changes, we analyzed the transcriptomes of both cell lines using RNA-seq. A total of 301 differentially expressed genes were identified using DESeq (Fig. 3A; see Table S2 in the supplemental material). Differential gene expression was validated by qRT-PCR analysis of 14 selected genes in independent samples (see Fig. S6 in the supplemental material). Biological pathway analysis identified cellular growth and various developmental categories as the most significantly deregulated functions (Fig. 3B), suggesting an altered developmental state of DKO cells.
FIG 3.
DKO transcriptome and defective adipogenic differentiation in DKO MEFs. (A) MA plot comparing the WT and DKO transcriptomes. The average base mean (the average normalized read pair counts) of WT and DKO genes [log2(base mean)] is plotted against the expression difference [log2(FC)]. FC, fold change. Differentially expressed genes are shown in red. (B) Biological pathway analysis of differentially expressed genes in DKO MEFs. (C) Bright-field microscopy images of WT and DKO MEFs stained with Oil-Red-O before treatment and after 21 days of ADM treatment. Quantifications of lipid droplets after 7, 14, and 21 days of treatment are shown in the diagram on the right. The data represent mean numbers of particles per five view fields ± standard deviations. (D) Relative expression of adipogenic marker genes Pparγ, C/ebpα, and Igf1 in untreated (day 0) and ADM-treated WT and DKO MEFs normalized to β-actin and Gapdh. The error bars represent standard deviations (n = 3).
In order to investigate the functional relevance of the observed epigenetic defects, we utilized an established paradigm for MEF differentiation and induced adipogenesis using insulin-containing medium (ADM) (34). A substantial fraction of ADM-treated wild-type MEFs started to form lipid droplets visible under a microscope, whereas DKO MEFs showed strongly reduced lipid droplet formation, indicating incomplete adipogenesis (Fig. 3C). Consistent with this, wild-type MEFs showed ADM-dependent induction of the established adipogenic marker genes Pparγ, C/ebpα, and Igf1 (34), whereas the response of DKO MEFs was markedly reduced (Fig. 3D). Also, all three Tet genes were upregulated in wild-type MEFs upon ADM treatment (see Fig. S1C in the supplemental material), consistent with a functional requirement for these enzymes. Our results thus uncovered a pronounced differentiation phenotype in DKO MEFs that may be related to altered epigenetic reprogramming.
Defective regulation of canyon-associated developmental genes in DKO MEFs.
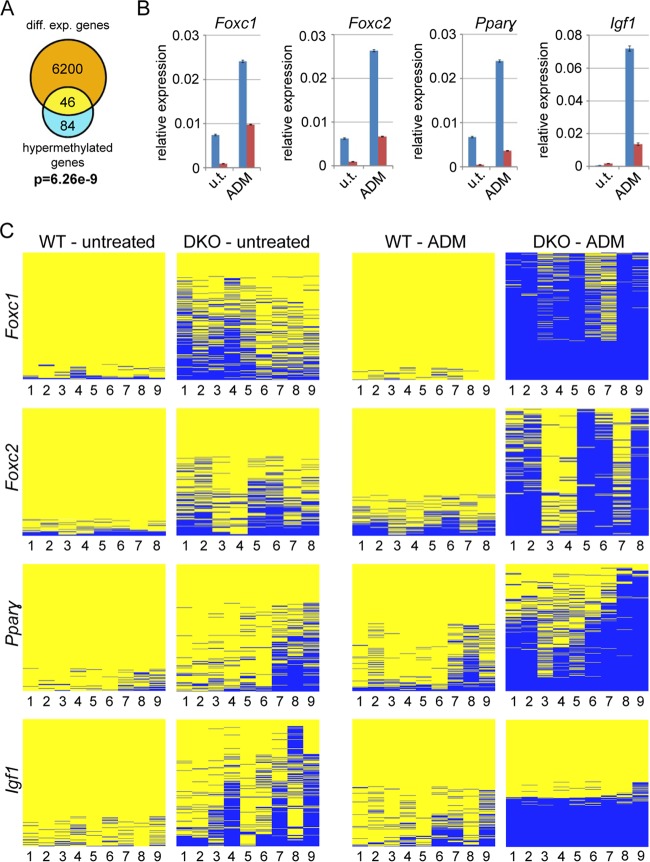
In order to establish a functional connection between Tet-dependent hypermethylation and the differentiation defects observed, we analyzed canyon association and canyon hypermethylation for all deregulated genes in DKO cells. To do this, we took all the genes with a log fold change of greater than or equal to ±0.5 (WT versus DKO) from our RNA-seq data (6,200 genes) and then selected those genes residing in a hypermethylated canyon (average methylation difference > 0.1) from the WGBS data set (84 genes) and calculated the overlap. This identified 46 deregulated genes that were associated with hypermethylated canyons in DKO cells (see Table S3 in the supplemental material). Statistical testing revealed that the overlap between gene deregulation and the hypermethylation of gene-associated canyons was highly significant (P = 6.26e−9; hypergeometric test) (Fig. 4A). This association remained highly significant when less stringent parameters were used for the definition of hypermethylated canyons, which increased the number of genes included in the statistical analysis (see Fig. S7A in the supplemental material). Next, we asked if there is a similar overlap when all genes residing in canyons were considered. This association was also significant (see Fig. S7B in the supplemental material), albeit to a lesser degree. Half of the genes associated with a hypermethylated canyon were also deregulated in DKO MEFs (see Fig. S7A in the supplemental material), whereas only one-third of all canyon genes were deregulated (see Fig. S7B in the supplemental material), indicating that genes with a transcription start site in a hypermethylated canyon are very likely misregulated. Of note, the expression of the majority of deregulated genes in DKO MEFs does not seem to depend on Tet-regulated canyons. However, due to the minimum size for canyons (3.5 kb), many deregulated genes are not considered to be associated with a canyon. Their expression might therefore depend on the presence of smaller Tet-dependent hypomethylated regions (see Fig. S7A in the supplemental material, which shows that the reduction of the size minimum for canyons leads to a strong increase of the number of overlapping deregulated genes), or their misregulation was caused by indirect effects.
FIG 4.
Differentiation defects in DKO MEFs are accompanied by canyon hypermethylation. (A) Venn diagram showing overlap between deregulated genes [log(FC) ≥ ±0.5] and genes associated with a hypermethylated canyon (P = 7.55 × 10−7; hypergeometric test). diff. exp., differentially expressed. (B) Relative expression values for the developmental canyon genes Foxc1, Foxc2, Pparγ, and Igf1 in the four cell types normalized to β-actin and Gapdh (WT, blue; DKO, red). The error bars indicate standard deviations (n = 3). u.t., untreated. (C) Validation of DNA methylation by targeted 454 bisulfite sequencing at promoter regions near sites of the developmental canyon genes Foxc1, Foxc2, Pparγ, and Igf1. The results are shown as heat maps as in Fig. 2D. DNA from untreated and induced MEFs (14 days of ADM) was analyzed. The CpGs covered by the individual amplicons are numbered. The regions analyzed are shown as yellow boxes in Fig. S3 in the supplemental material.
Biological pathway analysis again identified mostly developmental categories as enriched in the 46 deregulated genes associated with hypermethylated canyons (see Fig. S8 in the supplemental material), consistent with the previous analysis (Fig. 3B) and the altered expression patterns of developmental genes in DKO MEFs. Prominent examples of genes that overlap a hypermethylated canyon and that were downregulated in DKO MEFs are Hoxa1 and Hoxa2 (Fig. 2A; see Table S3 in the supplemental material), whereas several genes of the other three Hox clusters associated with hypermethylated canyons in DKO MEFs showed increased expression (see Table S3 in the supplemental material).
Because canyon hypermethylation might also prevent efficient gene regulation and transcriptional maintenance during adipogenic differentiation, we used 454 bisulfite sequencing to analyze the methylation status of the promoter regions of the canyon genes Foxc1, Foxc2, Pparγ, and Igf1 during differentiation. Igf1 and Pparγ expression is a key feature of adipogenesis (34), whereas both Foxc1 and Foxc2 have broader roles in regulating adipogenic processes (35, 36) but have also been reported to function during mesenchymal-epithelial transition (MET) (37–39). This is of particular interest, as a previous study suggested that Tet-deficient MEFs show a reduced capacity to undergo reprogramming due to a block in MET (40). With the exception of Igf1, all the genes were expressed in wild-type MEFs and significantly downregulated in DKO MEFs (Fig. 4B). Upon adipogenesis, expression of all four genes was strongly increased in wild-type MEFs, whereas induction was greatly reduced in DKO MEFs (Fig. 4B). These findings could be correlated with the methylation state of the corresponding canyons and also the promoters of the four genes. In wild-type cells, the promoter regions were unmethylated (see Fig. S3 in the supplemental material) and showed only a moderate increase upon differentiation (Fig. 4C). In DKO cells, canyons collapsed and promoters showed pronounced hypermethylation, which was further exacerbated during the 2 weeks of ADM treatment (Fig. 4C). Together, these findings suggest that the activation of the assayed genes during adipogenesis requires Tet-mediated maintenance of low methylation levels in the corresponding canyons.
Epigenetic features of collapsing canyons.
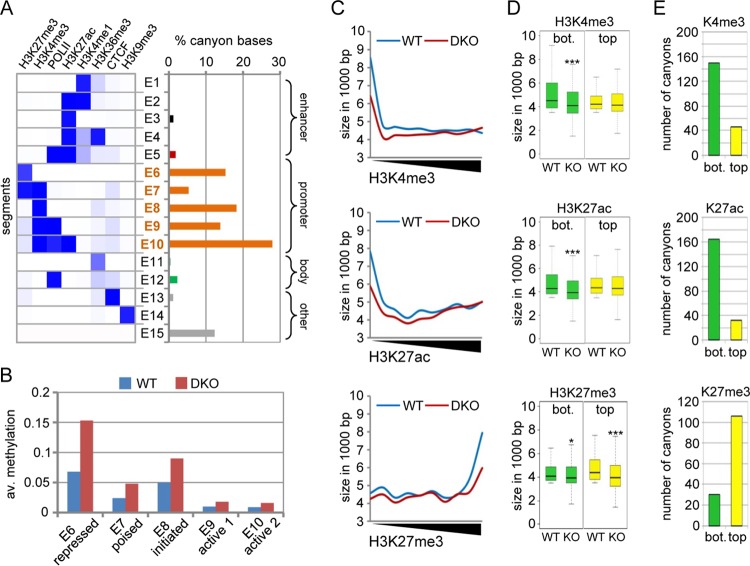
To further characterize hypermethylated canyons in DKO MEFs, we analyzed the genomic features that were identified in wild-type canyons by ChromHMM (see Fig. S2 in the supplemental material). As shown in Fig. 5A, 80% of canyon sequences showed promoter features (segments E6 to E10), whereas 12% were not marked (E15) and less than 4% showed enhancer features (E1 to E5). The related promoter features were mainly characterized by chromatin marks that correspond to repressed promoters (E6; H3K27me3), poised promoters (E7; H3K27me3 and H3K4me3), initiated promoters (E8; H3K4me3), and active promoters (E9 and E10; H3K4me3, H3K27ac, and PolII). When extracting the average methylation levels of the segments E6 to E10 from our WGBS data sets, regions corresponding to repressed promoters (segment E6) showed the strongest increase in DNA methylation in DKO MEFs (Fig. 5B).
FIG 5.
Tet1/Tet2 are required for the epigenetic regulation of poorly expressed genes. (A) Chromatin segments identified previously (left) (see Fig. S2 in the supplemental material) and relative percentages of bases for each segment belonging to a mapped canyon in WT MEFs (right). Features were grouped for enhancers (black), promoters (orange), gene body (green), and others (gray). (B) Average methylation levels in the ChromHMM segments E6 to E10 (promoters in panel A) in WT and DKO MEFs. (C) All the canyons in wild-type MEFs that retained a length of >1.5 kb in DKO cells were identified, and the average H3K4me3, H3K27ac, and H3K27me3 levels were calculated using published ChIP-seq data sets. The graphs show the lengths of canyons in wild-type and DKO MEFs in relation to histone methylation levels. (D) Box plots showing the lengths of canyons that are comparably lowly (bottom 50% [bot.]; green) or highly (top 50%; yellow) enriched for the indicated histone H3-specific methylation marks. The asterisks indicate statistically significant differences between wild-type and DKO cells (*, P < 0.05; ***, P < 0.001; two-sided paired t test). (E) Bar plots showing the numbers of canyons that disappear in DKO cells (as defined by falling below the minimum size required for a canyon). Numbers are shown for canyons that are comparably lowly (bottom 50%; green) or highly (top 50%; yellow) enriched for the indicated histone H3-specific methylation marks.
Next, we again used published ChIP-seq data sets for wild-type MEFs (22, 23) to analyze the correlation between canyon size and enrichment for the three histone marks we found associated with canyons. Canyons with low levels of active marks (H3K4me3 and H3K27ac) in wild-type MEFs showed size reduction in DKO cells (Fig. 5C). Similarly, canyons marked with high levels of H3K27me3 in the wild type also showed a pronounced length reduction (Fig. 5C). This effect was not observed for canyons that were enriched for these active marks (Fig. 5C). Rather, we could observe a trend toward enlargement for canyons highly tagged with H3K4me3 (Fig. 5C, top). Next, we compared enrichments for a specific histone modification in wild-type canyons with DNA hypermethylation and expression changes of associated genes in the same region in DKO MEFs. We found that low tagging of canyons with active marks or high tagging with H3K27me3 coincided with canyon hypermethylation and transcriptional deregulation (see Fig. S9 in the supplemental material). Further, when we analyzed the average canyon size in wild-type and DKO MEFs for lowly tagged (bottom 50%) and highly tagged (top 50%) canyons, a significant (P < 0.001; t test) size reduction was again observed for canyons with low levels of H3K4me3 and H3K27ac and high levels of H3K27me3 (Fig. 5D). Finally, when we counted the canyons that disappeared in DKO cells (as defined by failure to reach the minimum size for a canyon), we observed substantially higher numbers for the low-H3K4me3, low-H3K27ac, or high-H3K27me3 fractions (Fig. 5E). These data suggest that repressed canyons are particularly prone to canyon collapse.
DISCUSSION
Low levels of hydroxymethylation have been described in various human cancers (5), but the significance of this effect for epigenetic gene regulation has remained unclear. While the mechanistic basis for the loss of 5hmC in cancer is not known, it has been associated with reduced levels of Tet expression, in particular of Tet1 and Tet2, and cancer-specific hypermethylation (5). MEFs lacking Tet1 and Tet2 possess an impaired demethylation machinery and therefore provide an excellent model to investigate the effects of globally reduced 5hmC levels on the epigenome of differentiated cells. Our unbiased analysis of genome-wide methylation patterns showed widespread hypermethylation in DKO MEFs, which included promoter and enhancer regions. These findings are consistent with recent studies of Tet-deficient mouse ESCs (11, 12) and provide a mechanistic explanation for the epigenetic regulatory defects of Tet-deficient cells.
Our data strongly suggest that a subset of hypomethylated canyons is maintained by the cooperative activity of Tet proteins, in particular Tet1 and Tet2. In cells lacking both enzymes, many canyons decreased in size or collapsed, leading to the transcriptional deregulation of associated genes. This is particularly the case for canyons that are not enriched for active histone modifications and that can be considered poorly active or epigenetically silent. Prominent examples identified in this study include Hox genes and a subset of genes related to adipogenesis and mesenchymal-to-epithelial transition. The latter pathway represents a key step for the reprogramming of MEFs and the generation of MEF-derived induced pluripotent stem cells. As such, the observed inability of Tet-deficient MEFs to undergo reprogramming (40) is very likely also influenced by inefficient transcriptional regulation of canyon-associated epithelial genes. A recent study has also shown that 5-hydroxymethylcytosine marks promoters that resist DNA hypermethylation in colon cancer (6). This suggests that Tet1/Tet2-dependent prevention of promoter hypermethylation is also relevant for a key feature of the cancer epigenome.
Canyon collapse and invasive hypermethylation affect not only promoters but also gene bodies, regulatory sequences outside genes, and enhancers associated with the canyon. Hypermethylation can therefore have context-dependent effects on gene expression, leading to up- or downregulation of canyon-associated genes (see Table S3 in the supplemental material). In addition, especially for Tet1, gene-regulatory functions (repressive and activating) independent of its enzymatic activity have been described (41, 42). This might in part account for Tet-dependent effects on gene expression that are apparently not correlated with hypermethylation.
On the mechanistic level, our results also expand on previously published data that suggested a role for Tet1 as a demethylase to counteract aberrant DNA methylation and the spread of DNA methylation into CpG islands (41, 43). We observed strong demethylation defects at selected canyon borders already in Tet2 single-knockout cells. Because the deficiency of Tet1 also led to significant, albeit weaker, hypermethylation in the same regions, we suggest an additive and collaborative role for both enzymes, at least in the regions we analyzed. Lineage-specific genes residing in hypomethylated domains appear to be mostly regulated by chromatin modifications and not directly by DNA methylation (1, 15, 44). Canyon collapse in Tet-deficient cells then leads to the disruption of this chromatin-based transcription-regulatory network. Indeed, canyons marked by H3K27me3 and most likely overlapping poorly active or inactive genes were particularly affected by canyon collapse, which supports earlier findings that 5hmC and Tet1 coincide to a significant degree with Polycomb group target regions (41, 45). Thus, repressed or poised canyons appear to be more susceptible to collapse in Tet-deficient cells than active or moderately expressed canyons. When Dnmt3a was deleted in the hematopoietic system of the mouse, borders of canyons marked by active chromatin modifications became eroded and hypomethylated canyons expanded (15).
Our study suggests that the maintenance of hypomethylated canyons associated with developmental genes represents a major activity of Tet proteins in differentiating cells. We envisage a scenario where Tet1/Tet2 and Dnmt3a play cooperative but antagonistic roles in the maintenance of DNA methylation canyons to allow undisturbed access for chromatin-based transcriptional regulators (see Fig. S10 in the supplemental material). Tet deficiency leads to an imbalance in these methylating and demethylating activities, inducing canyon hypermethylation and thus destabilizing canyons and related gene-regulatory mechanisms. Our results, therefore, identify the protection of hypomethylated canyons as an important epigenetic regulatory role of Tet-dependent DNA demethylation, which provides a conceptual framework for understanding Tet-dependent epigenetic gene regulation.
Supplementary Material
ACKNOWLEDGMENTS
We thank the DKFZ Genomics and Proteomics Core Facility for sequencing services and Tobias Reber for highly competent information technology support.
A.B., R.J., and F.L. conceived the study. A.B., L.W., and F.L. designed the experiments and interpreted the results. M.M.D. and R.J. provided cell lines. L.W., T.M., and A.B. performed the experiments. G.R. performed the bioinformatics analyses. A.B. and F.L. wrote the manuscript, with contributions from all.
We declare that we have no conflict of interest.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00587-15.
REFERENCES
- 1.Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, Yang H, Wang T, Lee AY, Swanson SA, Zhang J, Zhu Y, Kim A, Nery JR, Urich MA, Kuan S, Yen CA, Klugman S, Yu P, Suknuntha K, Propson NE, Chen H, Edsall LE, Wagner U, Li Y, Ye Z, Kulkarni A, Xuan Z, Chung WY, Chi NC, Antosiewicz-Bourget JE, Slukvin I, Stewart R, Zhang MQ, Wang W, Thomson JA, Ecker JR, Ren B. 2013. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, Shalek AK, Kelley DR, Shishkin AA, Issner R, Zhang X, Coyne M, Fostel JL, Holmes L, Meldrim J, Guttman M, Epstein C, Park H, Kohlbacher O, Rinn J, Gnirke A, Lander ES, Bernstein BE, Meissner A. 2013. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 153:1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu H, Zhang Y. 2014. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang Y, Rao A. 2014. Connections between TET proteins and aberrant DNA modification in cancer. Trends Genet 30:464–474. doi: 10.1016/j.tig.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uribe-Lewis S, Stark R, Carroll T, Dunning MJ, Bachman M, Ito Y, Stojic L, Halim S, Vowler SL, Lynch AG, Delatte B, de Bony EJ, Colin L, Defrance M, Krueger F, Silva AL, Ten Hoopen R, Ibrahim AE, Fuks F, Murrell A. 2015. 5-Hydroxymethylcytosine marks promoters in colon that resist DNA hypermethylation in cancer. Genome Biol 16:69. doi: 10.1186/s13059-015-0605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawlaty MM, Breiling A, Le T, Barrasa MI, Raddatz G, Gao Q, Powell BE, Cheng AW, Faull KF, Lyko F, Jaenisch R. 2014. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell 29:102–111. doi: 10.1016/j.devcel.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, Faull KF, Lyko F, Jaenisch R. 2013. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell 24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hackett JA, Dietmann S, Murakami K, Down TA, Leitch HG, Surani MA. 2013. Synergistic mechanisms of DNA demethylation during transition to ground-state pluripotency. Stem Cell Reports 1:518–531. doi: 10.1016/j.stemcr.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sérandour AA, Avner S, Oger F, Bizot M, Percevault F, Lucchetti-Miganeh C, Palierne G, Gheeraert C, Barloy-Hubler F, Péron CL, Madigou T, Durand E, Froguel P, Staels B, Lefebvre P, Métivier R, Eeckhoute J, Salbert G. 2012. Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. Nucleic Acids Res 40:8255–8265. doi: 10.1093/nar/gks595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu F, Liu Y, Jiang L, Yamaguchi S, Zhang Y. 2014. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev 28:2103–2119. doi: 10.1101/gad.248005.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hon GC, Song CX, Du T, Jin F, Selvaraj S, Lee AY, Yen CA, Ye Z, Mao SQ, Wang BA, Kuan S, Edsall LE, Zhao BS, Xu GL, He C, Ren B. 2014. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell 56:286–297. doi: 10.1016/j.molcel.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schübeler D. 2015. Function and information content of DNA methylation. Nature 517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 14.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, Schübeler D. 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 15.Jeong M, Sun D, Luo M, Huang Y, Challen GA, Rodriguez B, Zhang X, Chavez L, Wang H, Hannah R, Kim SB, Yang L, Ko M, Chen R, Göttgens B, Lee JS, Gunaratne P, Godley LA, Darlington GJ, Rao A, Li W, Goodell MA. 2014. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat Genet 46:17–23. doi: 10.1038/ng.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delatte B, Deplus R, Fuks F. 2014. Playing TETris with DNA modifications. EMBO J 33:1198–1211. doi: 10.15252/embj.201488290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrlich M, Ehrlich KC. 2014. DNA cytosine methylation and hydroxymethylation at the borders. Epigenomics 6:563–566. doi: 10.2217/epi.14.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. 2012. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 19:900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 19.Raddatz G, Gao Q, Bender S, Jaenisch R, Lyko F. 2012. Dnmt3a protects active chromosome domains against cancer-associated hypomethylation. PLoS Genet 8:e1003146. doi: 10.1371/journal.pgen.1003146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xi Y, Li W. 2009. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ernst J, Kellis M. 2012. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, Ren B. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun D, Xi Y, Rodriguez B, Park HJ, Tong P, Meong M, Goodell MA, Li W. 2014. MOABS: model based analysis of bisulfite sequencing data. Genome Biol 15:R38. doi: 10.1186/gb-2014-15-2-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bocker MT, Tuorto F, Raddatz G, Musch T, Yang FC, Xu M, Lyko F, Breiling A. 2012. Hydroxylation of 5-methylcytosine by TET2 maintains the active state of the mammalian HOXA cluster. Nat Commun 3:818. doi: 10.1038/ncomms1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong EE, Okitsu CY, Smith AD, Hsieh CL. 2013. Regionally specific and genome-wide analyses conclusively demonstrate the absence of CpG methylation in human mitochondrial DNA. Mol Cell Biol 33:2683–2690. doi: 10.1128/MCB.00220-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zentner GE, Tesar PJ, Scacheri PC. 2011. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res 21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Jørgensen M, Kolde R, Zhao X, Parker B, Valen E, Wen J, Sandelin A. 2011. Prediction of RNA polymerase II recruitment, elongation and stalling from histone modification data. BMC Genomics 12:544. doi: 10.1186/1471-2164-12-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y, Chavez L, Chang X, Wang X, Pastor WA, Kang J, Zepeda-Martínez JA, Pape UJ, Jacobsen SE, Peters B, Rao A. 2014. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc Natl Acad Sci U S A 111:1361–1366. doi: 10.1073/pnas.1322921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosen ED, MacDougald OA. 2006. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 35.Omatsu Y, Seike M, Sugiyama T, Kume T, Nagasawa T. 2014. Foxc1 is a critical regulator of haematopoietic stem/progenitor cell niche formation. Nature 508:536–540. doi: 10.1038/nature13071. [DOI] [PubMed] [Google Scholar]
- 36.Davis KE, Moldes M, Farmer SR. 2004. The forkhead transcription factor FoxC2 inhibits white adipocyte differentiation. J Biol Chem 279:42453–42461. doi: 10.1074/jbc.M402197200. [DOI] [PubMed] [Google Scholar]
- 37.Thiery JP, Acloque H, Huang RY, Nieto MA. 2009. Epithelial-mesenchymal transitions in development and disease. Cell 139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 38.Hader C, Marlier A, Cantley L. 2010. Mesenchymal-epithelial transition in epithelial response to injury: the role of Foxc2. Oncogene 29:1031–1040. doi: 10.1038/onc.2009.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bard JB, Lam MS, Aitken S. 2008. A bioinformatics approach for identifying candidate transcriptional regulators of mesenchyme-to-epithelium transitions in mouse embryos. Dev Dyn 237:2748–2754. doi: 10.1002/dvdy.21652. [DOI] [PubMed] [Google Scholar]
- 40.Hu X, Zhang L, Mao SQ, Li Z, Chen J, Zhang RR, Wu HP, Gao J, Guo F, Liu W, Xu GF, Dai HQ, Shi YG, Li X, Hu B, Tang F, Pei D, Xu GL. 2014. Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell 14:512–522. doi: 10.1016/j.stem.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 41.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. 2011. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai YP, Chen HF, Chen SY, Cheng WC, Wang HW, Shen ZJ, Song C, Teng SC, He C, Wu KJ. 2014. TET1 regulates hypoxia-induced epithelial-mesenchymal transition by acting as a co-activator. Genome Biol 15:513. doi: 10.1186/s13059-014-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin C, Lu Y, Jelinek J, Liang S, Estecio MR, Barton MC, Issa JP. 2014. TET1 is a maintenance DNA demethylase that prevents methylation spreading in differentiated cells. Nucleic Acids Res 42:6956–6971. doi: 10.1093/nar/gku372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamura R, Tsukahara T, Qu W, Ichikawa K, Otsuka T, Ogoshi K, Saito TL, Matsushima K, Sugano S, Hashimoto S, Suzuki Y, Morishita S, Takeda H. 2014. Large hypomethylated domains serve as strong repressive machinery for key developmental genes in vertebrates. Development 141:2568–2580. doi: 10.1242/dev.108548. [DOI] [PubMed] [Google Scholar]
- 45.Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. 2011. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev 25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.