Abstract
In humans and in mice, mutations in the Ostm1 gene cause the most severe form of osteopetrosis, a major bone disease, and neuronal degeneration, both of which are associated with early death. To gain insight into Ostm1 function, we first investigated by sequence and biochemical analysis an immature 34-kDa type I transmembrane Ostm1 protein with a unique cytosolic tail. Mature Ostm1 is posttranslationally processed and highly N-glycosylated and has an apparent mass of ∼60 kDa. Analysis the subcellular localization of Ostm1 showed that it is within the endoplasmic reticulum, trans-Golgi network, and endosomes/lysosomes. By a wide protein screen under physiologic conditions, several novel cytosolic Ostm1 partners were identified and validated, for which a direct interaction with the kinesin 5B heavy chains was demonstrated. These results determined that Ostm1 is part of a cytosolic scaffolding multiprotein complex, imparting an adaptor function to Ostm1. Moreover, we uncovered a role for the Ostm1/KIF5B complex in intracellular trafficking and dispersion of cargos from the endoplasmic reticulum to late endosomal/lysosomal subcellular compartments. These Ostm1 molecular and cellular functions could elucidate all of the pathophysiologic mechanisms underlying the wide phenotypic spectrum of Ostm1-deficient mice.
INTRODUCTION
Human autosomal recessive osteopetrosis (ARO) is a severe genetic disease associated with abnormal accumulation of bone tissue, leading to premature death, development of anemia, and increased susceptibility to infections (1–3). This genetic disease is caused by defects in or absence of osteoclasts (OC), which possess the unique physiologic property to resorb bone matrix and ensure constant bone remodeling (4, 5). The OSTM1 gene is responsible for the most severe spontaneous form of ARO in humans and also for neuronal degeneration (6, 7).
Functional conservation was demonstrated by occurrence of the most severe form of ARO in the Ostm1-null gray-lethal gl/gl mouse mutant (8). In gl/gl mice, osteoclast maturation is impaired with defective ruffled border formation, resulting in ineffective bone resorption and mortality at ∼3 weeks of age (9). Functional rescue of Ostm1 skeletal phenotypes in transgenic mice was obtained by targeting multiple hematopoietic lineages that resulted in osteoclast activation (10). The Ostm1 gene was shown to play additional roles in the brain and in hair pigmentation. Recently, characterization of Ostm1 in the brains of gl/gl mice following correction of osteoclast defects revealed impaired autophagy exclusively in neurons, which led to neuronal degeneration and early death (11). These mice also exhibited neuroretinal degeneration. Interestingly, the gl/gl homozygous mice also display a gray coat color resulting from defective melanosome distribution due to yellow pigment “clumping” (12), underlying the major role of Ostm1 in several cell types. The numerous physiologic impacts of null Ostm1 in mice suggest that Ostm1 may have one or several intracellular roles.
Ostm1 expression is prevalent in brain, spleen, kidney, osteoclasts, and melanocytes but at lower level in thymus, liver, testis, heart, and primary osteoblasts (8). The Ostm1/OSTM1 genes are highly conserved, and the human 334-amino-acid (aa) OSTM1 protein is 83% homologous to the 338-aa mouse protein. Our analysis of the Ostm1 protein hydropathy profile and topology structure predicted a primary 34-kDa type I transmembrane secretory protein that would be mainly luminal (8). However, Ostm1 was also reported from in vitro analysis to be a cytosolic protein that belongs to the ring protein family, and it was classified as an E3 ubiquitin ligase (13). The subcellular localization and the ex vivo and in vivo structures and functions of Ostm1 protein remain to be determined.
In vitro studies supported that Ostm1 could be a secretory protein that interacts with the transmembrane ClC-7 exchanger protein (14). The interaction of Ostm1 and ClC-7 appears to be critical for ClC-7 function in ion transport and Cl−/H+ exchange (15, 16). Based on low ClC-7 levels in gl/gl mice, Ostm1 was proposed to protect ClC-7 from degradation (14). In contrast to Ostm1 deficiency, ClC-7 deficiency in humans was shown to cause benign or intermediate osteopetrotic clinical manifestations (3, 17). The more severe Ostm1 osteopetrosis (8, 18, 19) relative to the ClC-7 disease highlights the existence of additional and critical Ostm1-interacting partners that play an important role in Ostm1 function.
To directly address the cellular function of Ostm1, we performed structure-function analysis and investigated posttranslational processing, and we showed that Ostm1 is highly glycosylated and a resident protein of the endoplasmic reticulum (ER) and the trans-Golgi network (TGN). Further, cytosolic Ostm1 protein partners under physiologic conditions were characterized and validated, which indicated that Ostm1 is part of a multiprotein complex. The complex of Ostm1 with motor protein kinesin 5B (KIF5B) conveyed an adaptor role for Ostm1 in intracellular trafficking and in dispersion of cargos to endosomes/lysosomes.
MATERIALS AND METHODS
Plasmid constructs.
The mouse full-length Ostm1 cDNA was cloned into the pHA.CE vector (derived from the pcDNA3), including a Kozak sequence (20) and a hemagglutinin (HA) tag which is fused to COOH of the Ostm1 cDNA (Ostm1-FL-HA). The different fragments were cloned by PCR; in the Ostm1-Δ(CT) pHA.CE construct the C-terminal domain (CT) (FLHSEQKKRKLILPKRLKSSTSFANIQENAT) of Ostm1 is deleted, and in the Ostm1-Δ(TM+CT) pHA.CE construct the transmembrane (TM) and C-terminal domains (VSVVAVSVFILFLPVVFYLSSFLHSEQKKRKLILPKRLKSSTSFANIQENAT) of Ostm1 are deleted. For immunolocalization assays and immunoprecipitation assays, four fragments were cloned in the pEGFP.N1 vector (Clontech) with an enhanced green fluorescent protein (EGFP) tag fused to COOH of the Ostm1 cDNA to generate the constructs Ostm1-FL-EGFP, Ostm-Δ(CT)-EGFP, Ostm1-Δ(TM+CT)-EGFP, and Ostm1-FL(CT: K→L)-EGFP with all lysine residues (K) replaced by leucine residues (L) (FLHSEQLLRLLILPLRLLSSTSFANIQENAT) (Fig. 1B). Mouse full-length ClC-7 cDNA was cloned in the pEGFP.N1 vector and in pIRES-EGFP, producing C-terminally GFP-tagged and V5-tagged proteins. KIF5B-EYFP was generously provided by Chen Gu, Ohio State University, USA.
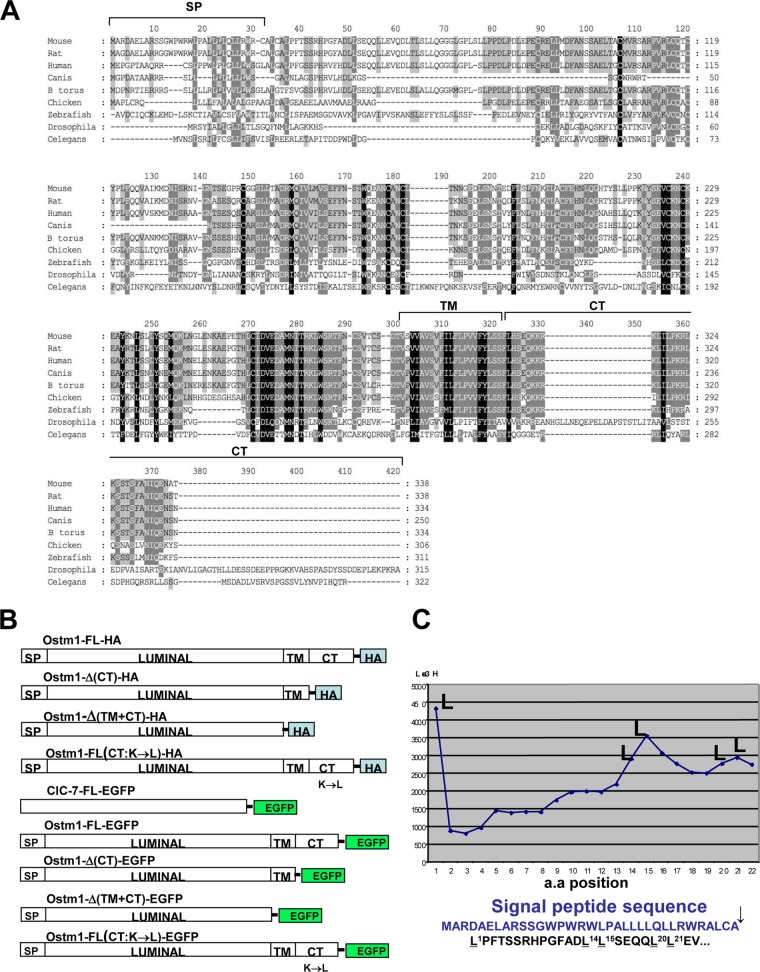
FIG 1.
Ostm1 protein structural characterization. (A) Ostm1 protein alignments demonstrate protein conservation in metazoans. SP, signal peptide; TM, transmembrane domain; CT, C terminus. (B) Schematic representation of the different constructs used in this study for transfection and immunochemistry. (C) Microsequencing of radiolabeled cell lysates of transiently Ostm1-FL-HA-transfected clones immunoprecipitated with HA. The deduced sequences of radiolabeled residues are shown, and Leu peaks/positions confirm the cleavage of the signal peptide compared to the Ostm1 primary sequence. (D) Immunoblot analysis of lysates and culture media of Ostm1-FL-HA and Ostm1-Δ(TM+CT)-HA radiolabeled transient transfectants immunoprecipitated with HA. Compared to the Ostm1-FL control, the truncated form of Ostm1 is secreted into the medium and consists of a single band (←) instead of two (⇐) for the FL form. These results are representative of three independent experiments. (E) Similar subcellular punctate immunolocalization of the different forms of Ostm1-EGFP fusion proteins in transiently transfected COS-7 cells. The low signal detected with Ostm1-Δ(TM+CT)-EGFP is likely due to secretion of the truncated form of the protein. These results are representative of three independent experiments. Bar, 10 μm. (F) Immunolocalization of Ostm1-FL-EGFP with specific subcellular resident control proteins, including calnexin (ER), golgin (TGN), and F-actin (plasma membrane), and with endosomes/lysosomes following LysoTracker staining (merge). Pearson correlation coefficients for colocalization are indicated. Bars, 15 μm.
Cell culture, transfection, and biochemical assays.
HEK293 cells grown in alpha minimal essential medium (αMEM) with 10% fetal bovine serum (FBS) were transfected with individual plasmid DNAs (0.5 μg/P3.5 and 1.0 μg/P6) of pHA.CE constructs using Effectene (Qiagen). For microsequencing, cells were incubated in a medium without leucine for 1 h before a pulse of 3 h with 500 μCi/ml of [3H]Leu (Amersham). Cells were lysed, and the supernatant was immunoprecipitated with HA-agarose antibody (Santa Cruz). The beads were washed, boiled, and loaded on an 8% Tricine-SDS-polyacrylamide denaturing gel. After transfer to an Immobilon membrane (Amersham), the radiolabeled protein was excised and sequenced as described previously (21).
In pulse-chase experiments, cell medium was replaced 48 h after transfection with a medium without methionine and cysteine and left for 1 h before a pulse of 15 min with 125 μCi/ml methionine and 125 μCi/ml cysteine (35S-Met or EasyTag and 35S-Cys; Amersham). Before the pulse period, the medium was changed to an amino acid-enriched (50×) medium. Cell lysates in radioimmunoprecipitation assay (RIPA) buffer were immunoprecipitated with HA-agarose beads, loaded on a denaturing gel, and analyzed following autoradiography. For the glycosylation assays, cells were incubated in a medium without methionine and cysteine and pulsed for 3 h with 125 μCi/ml methionine and 125 μCi/ml cysteine (35S-Met or EasyTag and 35S-Cys; Amersham). Cells were then treated with 5 μg/ml tunicamycin, 2.5 μg/ml brefeldin A (BFA), 10 μg/ml kifunensine (KIFU), 10 μg/ml castanospermin (CST), 10 μg/ml 1-deoxymannojirimycin (DMJ), or 10 μg/ml N-methyl-deoxynojirimycin (MDNJ) (Sigma). Endo-β-N-acetylglucosaminidase H (endo H) and peptide:N-glycosidase F (PNGase F) treatments were carried out after precipitation with HA-agarose beads. All samples were resolved on 8% Tricine-SDS-polyacrylamide denaturing gels.
RAW 264.7 cells were cultured in αMEM supplemented with RANKL (receptor activator of nuclear factor κB ligand) (80 ng/ml) for 4 to 6 days to generate mature multinucleated osteoclasts (OC). For immunoprecipitation experiments, RAW cell-derived OC were transfected with Ostm1-EGFP, ClC-7-EGFP, and KIF5B-EYFP expression vectors using the calcium phosphate method.
Stable clones and siRNA.
COS-7 cells were transfected with Ostm1-FL-pEGFP using Effectene (Qiagen) and grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FBS. Individual Ostm1-FL-EGFP clones were isolated after selection with G418 (400 μg/ml). For small interfering RNA (siRNA) transfections, COS-7 cells and COS-7 Ostm1-FL-EGFP stable clones were transfected with KIF5-B or OSTM1 siRNA pools or a scramble pool (SMARTpool; Thermo Fisher Scientific) to a final concentration of 55 or 50 nM, using Dharmafect2 buffer (Thermo Fisher Scientific). After 48 h, cells were stained with LysoTracker Red DND-99 or MitoTracker (Molecular Probes) and subjected to live imaging. To evaluate siRNA efficiency, cells were lysed at 48 h posttransfection on ice in 1× RIPA buffer. Total protein (15 μg) was loaded on a 10% SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride (PVDF) membrane. The membrane was blocked for 1 h in 5% milk, 16 h with primary antibodies at 4°C with shaking, and 1 h with secondary antibodies in 5% milk before Western blotting and detection by the enhanced chemiluminescence procedure (GE Healthcare). The primary antibodies were diluted as follows: GFP (Roche), 1/2,500; KIF5B (Cell Signaling), 1/1,000; V5 (Invitrogen), 1/200; and β-actin (Sigma), 1/1,000. Corresponding secondary antibodies (Bio-Rad) were used at a 1/10,000 dilution.
Immunofluorescence.
COS-7 or EcR293 cells (1 × 105) were plated on glass coverslips and grown in DMEM supplemented with 10% FBS. After 24 h, cells were transfected with individual plasmid DNAs (0.5 μg) of pEGFP.N1 constructs using Effectene (Qiagen). After 48 h, cells were treated for 120 min with LysoTracker Red DND-99 (Molecular Probes), were fixed for 15 min in 4% paraformaldehyde (PFA), permeabilized for 15 min with 0.1% Triton X-100 in phosphate-buffered saline (PBS), washed in PBS, and incubated for 30 min with 3% bovine serum albumin (BSA) in PBS. Cells were then labeled with 1:100 and 1:300 dilutions of primary anti-golgin 97 monoclonal antibodies (Molecular Probes), calnexin rabbit polyclonal antibody (Sigma), KIF5B antibody (Abcam), HA rabbit polyclonal antibody (Zymed), or rhodamine-conjugated phalloidin (Molecular Probes) for 60 min. After washing with PBS containing 0.01% Tween 20, cells were stained with an Alexa Fluor 633-conjugated goat anti-mouse secondary antibody (Molecular Probes). Cells were visualized by confocal microscopy (LSM710; Carl Zeiss) at 488 nm (EGFP), 514 nm (yellow fluorescent protein [YFP]), 543 nm (LysoTracker, MitoTracker, rhodamine-phalloidin, and HA) with Alexa Fluor 546 and 633 (calnexin and golgin), and KIF5B was visualized with Alexa Fluor 633. Nuclei were visualized (405 nm) following Hoechst dye (Sigma) staining. Image analysis was performed and Pearson correlation coefficients for colocalization were calculated using ImageJ software (22).
TAP of Ostm1 and protein identification.
The 30-aa CT cytosolic tail of mouse Ostm1 cDNA (93.3% identity with the human sequence) was cloned into the mammalian expression vectors pMZI and AB-0411 (23, 24) in such a way that the C-terminal domain of Ostm1 carries a tandem affinity purification (TAP) tag at its C terminus and N terminus, respectively (25). Individual EcR293 stable cell lines were produced following transfection (Effectene; Qiagen), and expression of TAP-tagged Ostm1 cells was induced for 24 h with 1 μM Ponasterone A (Invitrogen) (26). The TAP-tagged complexes were then purified and concentrated as described previously (25). Each TAP eluate was run in a 10% SDS-polyacrylamide gel and silver stained. The protein bands were individually excised, reduced, alkylated, and subjected to tryptic digestion as described previously (27). The resulting tryptic peptides were purified and identified by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) (28).
GST pulldown.
The PCR fragment encoding the Ostm1 CT cytosolic domain fused with glutathione S-transferase (GST) was cloned into the pGEX-2T plasmid (GE Healthcare). The expression vector was transformed into Escherichia coli BL21 (Novagen), and bacterial cultures grown in LB at 37°C to an optical density of 0.5 to 0.8 (595 nm) were induced for 3 h by the addition of 1.0 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The GST recombinant fusion protein was purified using a GST module (Amersham), and the protein concentration was determined by the Bradford method (Bio-Rad) using BSA as a standard. For protein affinity chromatography, extracts of GST and GL-CT-GST were immobilized on glutathione-Sepharose beads at a concentration of 5 mg/ml. Beads (∼150 μl) were equilibrated in 450 μl of ACB buffer (10 mM Tris [pH 8.0], 0.1 mM EDTA, 0.1 mM dithiothreitol [DTT], 10% glycerol) containing 0.1 M NaCl and then loaded with 100 μg of EcR293 whole-cell extract that had been previously depleted on glutathione-Sepharose. Cell extracts (6 to 8 mg/ml) were dialyzed against 10 mM HEPES (pH 7.9)–0.1 M NaCl–0.1 M potassium acetate (KAc)–0.1 mM EDTA–0.1 mM DTT–10% glycerol buffer. After 16 h of agitation at 4°C, beads and extracts were loaded on a 1-ml Bio-Spin column (Bio-Rad) and washed 4 times with 500 μl of ACB buffer containing 0.1 M NaCl. The bound proteins were eluted with 450 μl of ACB buffer containing 0.1 M NaCl and 20 mM reduced glutathione, followed by elution with 450 μl of 0.1% SDS (29).
Coimmunoprecipitation and Western blotting.
EcR293 cells were grown in DMEM supplemented with 10% FBS. After 24 h, cells were transfected with individual plasmid DNAs (0.5 μg/P3.5) of pIRES2-EGFP constructs using Effectene (Qiagen) or the calcium phosphate precipitation method. The cells were lysed 30 min on ice with 1× coimmunoprecipitation buffer (120 mM NaCl, 1 mM EDTA, 50 mM Tris [pH 8.0], 0.5% NP-40) and centrifuged, and the supernatant was immunoprecipitated with GFP antibody (Roche) with rotation for 1 h at 4°C before addition of protein A/G-agarose beads or HA-agarose (Santa Cruz) and treatment for 16 h with rotation at 4°C. The beads were washed four times with 1× coimmunoprecipitation buffer before boiling and loading on 8 to 10% SDS-polyacrylamide denaturing gels. After transfer to nitrocellulose membranes, incubations were carried out for 1 h in 5% milk, 16 h with primary antibodies at 4°C with agitation, and 1 h with secondary antibodies in 5% milk before Western blotting and detection by the enhanced chemiluminescence procedure (GE Healthcare). The primary antibodies were diluted as follows: GFP (Roche), 1/2,500; KPNB1 and EDD1 (Bethyl Laboratories), 1/5,000; HA (Zymed), 4/1,000; KLC2 (Abgent), 16/1,000; KIF5B (Cell Signaling), 1/1,000; and ClC-7 (Abcam), 1/500. The secondary antibodies (Bio-Rad) were used at a 1/10,000 dilution.
Live-cell imaging.
Live-cell imaging (by time-lapse video microscopy) was performed with a Zeiss LSM710 laser scanning confocal microscope (Carl Zeiss). Briefly, EcR293 and COS-7 cells were cultured on glass-bottom petri dishes (MatTek) in DMEM and transiently transfected with 0.1 μg of Ostm1-FL-EGFP constructs using Effectene (Qiagen). Twenty-four hours later, cells were transfected with 0.1 μg of KIF5B-EYFP and incubated for 6 to 8 h before the localization of Ostm1 (green channel) and KIF5B (yellow channel; pseudocolored red for convenience) was recorded at 1 image/5 to 10 s for 60 cycles with a 63× objective lens. Ostm1-FL-EGFP clones transfected with siRNA against KIF5-B or OSTM1 or a scramble siRNA (SMARTpool; Thermo Fisher Scientific) were treated with LysoTracker Red DND-99 (Molecular Probes) and subjected to live imaging at 1 image/4 s for 50 cycles with a 63× objective lens. All images were scanned at a 512 × 512 resolution with a scan speed of 8 and an average of 4 scans for fine resolution. During image acquisition, cells were buffered in Hanks balanced salt solution (HBSS) and maintained in a temperature-controlled incubator. Images and video were analyzed using ImageJ and presented at a frame rate of 7 per second. For OSTM1-FL-EGFP and endosome/lysosome dispersion, the area of organelles on the overall cell area was quantified (30).
Statistical analysis.
Values are expressed as means ± standard errors of the means (SEM). An unpaired two-sample Student t test was used for statistical analysis, with a P value of <0.05 considered significant.
RESULTS
Structural conservation of Ostm1 from fly to human.
To interrogate Ostm1 protein conservation across species, protein databases were searched for orthologous proteins based on the mouse full-length Ostm1 sequence. Full-length orthologous Ostm1 was identified in Danio rerio (zebrafish) but also in invertebrates such as Drosophila melanogaster and Caenorhabditis elegans. High homologies of specific motifs and residues within the vertebrate and invertebrate Ostm1 proteins were delineated with a BLAST engine program (Fig. 1A), suggesting conserved cellular function. Interestingly, no yeast homolog was detected, indicating that Ostm1 is evolutionarily restricted to metazoans.
Ostm1 protein processing and structural properties.
To gain insight into Ostm1 intracellular processing, we first analyzed whether the full-length Ostm1 undergoes maturation of the protein by metabolic [3H]Leu labeling of HEK 293 cells expressing the Ostm1-HA protein (Ostm1-FL-HA) (Fig. 1B). The Ostm1-FL-HA protein from cell lysates was immunoprecipitated with HA antibodies and microsequenced. We detected a unique N-terminal sequence in the first 35 residues of the protein, with leucine residues occupying positions 1, 14, 15, 20, and 21 (Fig. 1C). Based on the primary protein sequence (Fig. 1A), this clearly demonstrates that pre-Ostm1 is cleaved at ALCA35↓L, consistent with a consensus cleavage site of a signal peptide. This was the first evidence that in HEK293 cells, Ostm1 is a secretory membrane-bound protein that is not cytosolic. Furthermore, our sequencing analysis did not detect other cleavage sites or additional Ostm1 fragments, in contrast to a previously suggested hypothesis (31).
We next set out to determine Ostm1 protein orientation. Analysis of the mouse Ostm1 protein topology and hydropathy predicted a C-terminal (CT) cytosolic domain of 30 aa (aa 309 to 338) and a single hydrophobic transmembrane (TM) stretch of 23 aa (aa 286 to 308) (8). Based on these predictions, we generated two different truncated constructs, expressing the Ostm1-Δ(CT)-HA protein and a potentially soluble Ostm1-Δ(TM+CT)-HA protein, that were transiently transfected in HEK293 cells in comparison to the full-length Ostm1 protein in the presence of [35S]Cys. While the full-length protein, Ostm1-Δ(TM+CT), and even Ostm1-Δ(CT)-HA are clearly detected in cell extracts, only the Ostm1-Δ(TM+CT) form is secreted into the medium, indicating that the N-terminal region of the protein is likely luminal (Fig. 1D and data not shown). These data characterized Ostm1 as a type I transmembrane protein and pointed to the importance of the transmembrane and cytosolic domains for cellular localization.
Ostm1 is an atypical type I transmembrane protein.
To define the role of the Ostm1 subdomains in controlling cellular localization, Ostm1 fused to EGFP full-length (Ostm1-FL-EGFP) and truncated [Ostm1-Δ(CT)-EGFP and Ostm1-Δ(TM+CT)-EGFP] forms were transfected in COS-7 cells (Fig. 1B). The native Ostm1 as well as the truncated Δ(CT) form localized to the ER and displayed a punctate cellular distribution throughout the cytosol (Fig. 1E). In contrast, the weakness of the EGFP cellular signal detected with Ostm1-Δ(TM+CT)-EGFP, likely due to extracellular secretion, precluded precise analysis (Fig. 1D). These data suggested that the transmembrane domain is critical for Ostm1 localization in the ER and in punctate, vesicular-like cytosolic structures.
Since retention of type I proteins in the ER typically depends on the presence at their C termini of a dilysine (KK) retention signal at positions −3 and −4 (KKXX) or −3 and −5 (KXKXX) (32, 33), the role of the lysine residues in the CT of Ostm1 was assessed. Notably, the Ostm1 CT domain contains five K residues and one KK signal, although not at the C terminus (Fig. 1A). All were replaced by Leu in Ostm1-FL(CT:K→L)-EGFP. This mutant in fact displayed a punctate cellular distribution similar to that of the full-length Ostm1 control (Fig. 1E), suggesting that Ostm1 subcellular localization and ER retention are not dependent on lysine residues and that Ostm1 is not a classical ER-retained type I transmembrane protein (Fig. 1D). To monitor Ostm1 subcellular localization, costaining of full-length Ostm1-EGFP cells was carried out with calnexin for the ER, golgin for the TGN, LysoTracker for late endosomes/lysosomes, and F-actin for the plasma membrane. Costaining analysis using a correlation coefficient revealed that Ostm1 localized to the ER and TGN and also to the endosomes/lysosomes (Fig. 1F). However, Ostm1 did not appear to localize at the plasma membrane. Interestingly, this subcellular localization of full-length Ostm1 indicates that it may play a role in trafficking.
Ostm1 posttranslational modification.
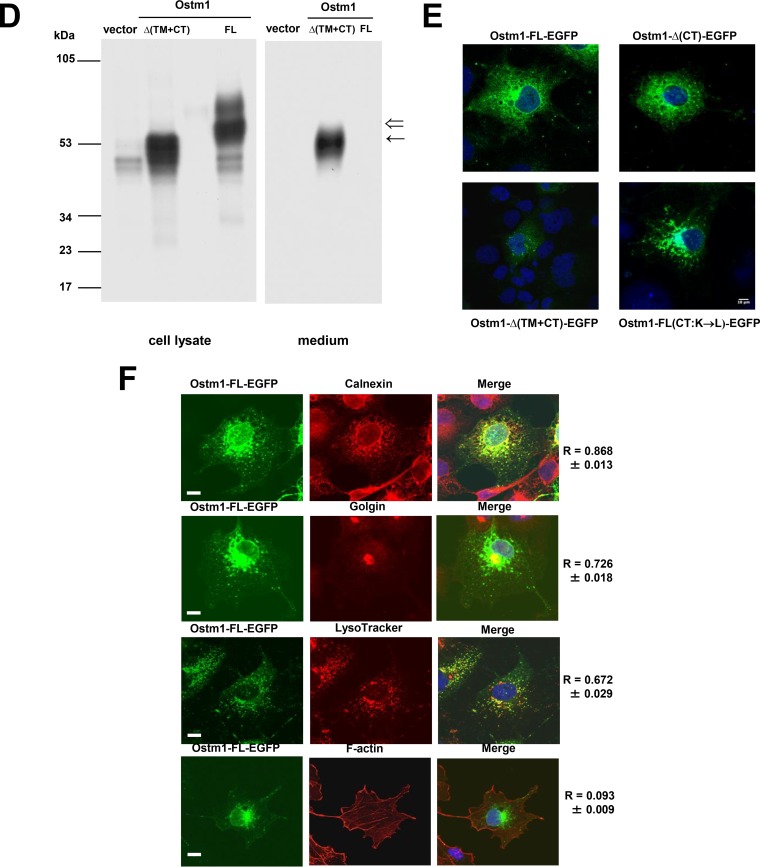
To determine whether Ostm1 is posttranslationally modified, Ostm1 protein was analyzed for glycosylation. The protein sequence predicted N-linked glycosylation at 10 luminal consensus sites (Fig. 2A). Inhibition of Ostm1 N-glycosylation by tunicamycin showed that the two ∼60-kDa and ∼80-kDa protein forms of Ostm1 shifted to an ∼34-kDa protein band corresponding to the predicted unmodified Ostm1 size (Fig. 2B). These data point to N-linked glycosylation as a major Ostm1 modification. Analysis of Ostm1 protein in the presence of brefeldin A, an inhibitor of protein trafficking from the ER to the Golgi compartment, showed a major band of ∼60 kDa for Ostm1 protein and the loss of the ∼80-kDa form relative to that in the untreated sample, indicating that the ∼80-kDa protein likely contains complex N-glycosylation achieved in the medial/trans-Golgi compartments, whereas the ∼60-kDa form is an intermediate, not yet fully mature N-glycosylated Ostm1 (Fig. 2B). N-glycan modification of Ostm1 was also analyzed with the N-deglycosylases peptide:N-glycosidase F (PNGase F) and endoglycosidase H (endo H), which indirectly monitor protein trafficking along the secretory pathway. Because PNGase F cleaves all types of N-glycans from glycoproteins, the Ostm1 forms were shifted to an ∼34-kDa band as obtained with tunicamycin (Fig. 2C). Treatment with the endo H does not process complex N-glycans found in core glycosylated Ostm1 forms in the ER and early cis-Golgi compartments. This processing results in the appearance of two distinct forms, a weak endo H-resistant form at ∼60 kDa and a strong ∼34-kDa endo H-sensitive form, suggesting that the major fraction of Ostm1 is resident in the ER/cis-Golgi compartments, whereas a significant minority has egressed from the ER and transited through late Golgi compartments (Fig. 2C).
FIG 2.
The Ostm1 protein is N-glycosylated in the endoplasmic reticulum. (A) Prediction of potential N-glycosylation sites in the Ostm1 protein sequence using NetNGlyc 1.0 Server. (www.cbs.dtu.dk). (B) Immunoblot analysis of lysates immunoprecipitated with HA-agarose beads from radiolabeled transient OSTM1-FL-HA and control vector transfections, treated with tunicamycin and brefeldin. Tunicamycin treatment shows that Ostm1is highly glycosylated, shifting from an ∼60-kDa molecular mass to ∼34 kDa (←). BFA treatment impairs formation of a post-ER remodeling of the protein (⇐). These results are representative of three independent experiments. (C) Immunoblot analysis of lysates immunoprecipitated with HA-agarose beads from radiolabeled transient OSTM1-FL-HA and control vector transfections, treated with endo H and PNGase F. The Ostm1 protein is deglycosylated by both enzymes (←). These results are representative of three independent experiments. (D) Transient transfections of Ostm1-FL-HA with pulse metabolic labeling and chase reveal a rapid glycosylation and sugar remodeling of Ostm1. At chase time zero, the sugar remodeling is partial, revealing a protein that migrates to a higher molecular weight (*). By 45 min, the sugar remodeling is over (←), and a second band appears and becomes more intense at 90 min (⇐). These results are representative of three independent experiments. (E) Immunoblot analysis of lysates immunoprecipitated with HA-agarose beads from radiolabeled transient OSTM1-FL-HA and control vector transfections, treated or not with different inhibitors of enzymes implicated in sugar remodeling in the ER (CST and MDNJ) or Golgi apparatus (KIFU and DMJ). Inhibition of the ER enzymes impairs sugar remodeling and leads to degradation of the protein (*). Inhibiting the Golgi-specific enzymes stabilizes the protein (←) by preventing its traffic to TGN and formation/degradation of the higher form (⇐). These results are representative of four independent experiments.
The kinetics of Ostm1 N-glycosylation was monitored following transient transfection, metabolic labeling, and pulse-chase analysis. Very early on, Ostm1 is modified to result in an ∼60-kDa band, providing evidence of rapid N-glycosylation and carbohydrate remodeling (Fig. 2D). In addition, the ∼80-kDa Ostm1 band was detectable by 45 min and readily apparent at 90 min postchase, indicating a swift Ostm1 processing in late Golgi compartments. To further characterize these modifications, Ostm1 was treated with specific glucosidase inhibitors in the ER, i.e., castanospermine (CST) and N-methyl-deoxynojirimycin (MDNJ), which prevent glucose trimming, and with the mannosidase inhibitors kifunensine (KIFU) and 1-deoxymannojirimycin (DMJ) (Fig. 2E). Inhibition of glucose trimming in the ER produced an ∼70-kDa band, whereas inhibition of mannosidase remodeling enzymes resulted in higher levels of immature ∼60-kDa Ostm1 protein (Fig. 2E). These data indicate that the different N-glycosylation modifications likely modulate Ostm1 intracellular trafficking.
Ostm1 cytosolic interactions with potentially multiple roles.
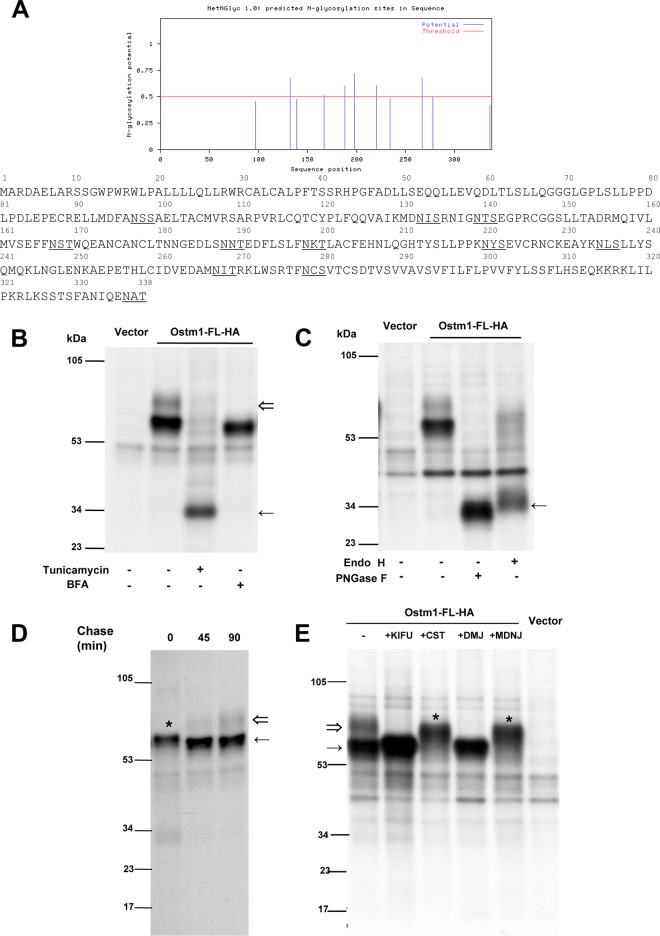
To characterize key cytosolic interacting Ostm1 partners, a TAP tag screen was designed with the Ostm1 short 30-amino-acid cytosolic CT domain (Ostm1-CT) acting as a bait in EcR293 cells (26, 27). Of the stable Ostm1 clones, only clones with near physiologic expression levels upon induction were selected (25) in comparison to noninduced controls. Affinity-purified protein complexes from several experiments were eluted, resolved on SDS-PAGE (Fig. 3A), and processed for MS analysis. A total of 16 unique polypeptides were identified, which were classified into four subgroups of proteins that potentially interact with Ostm1 (Fig. 3B). Among these, the kinesin motor KIF5B yielded by far the highest score. To validate these interactions in cells, GST pulldown assays were carried out on potential partners with different tandem MS (MS/MS) scores in the trafficking, nuclear pore, and protein modification subgroups. As shown in Fig. 3C, interactions of Ostm1-CT-GST were detected with the protein remodeler EDD1/UBR5, the nuclear pore importin KPNB1, and the trafficking motor kinesin KIF5B. These results suggest that Ostm1 can have multiple interactions with cytosolic proteins and could participate in a multifunctional protein platform, implicated in nucleocytoplasmic transport processes in addition to intracellular trafficking.
FIG 3.
Ostm1-interacting partners detected using TAP tag. (A) Representative pattern of affinity-purified Ostm1 TAP eluates from noninduced and induced EcR293 clones resolved on SDS-PAGE. These results are representative of three independent experiments. (B) Representative list of proteins form TAP eluates subjected to liquid chromatography (LC)-MS analysis and identified based on their unique peptide signature. Classification in four classes is related to cellular function. (C) Immunoblot analysis of GST-Ostm1-CT pulldown assays. Cell extracts were incubated with GST or GST-Ostm1-CT. Bound proteins were eluted with glutathione and revealed with EDD1/UBR5, KPNB1, KIF5B, and GST antibodies. (D) Cell extracts from EcR293 cells transiently transfected with Ostm1-EGFP expression vector were subjected to immunoprecipitation (IP) with anti-GFP antibody. The resulting precipitates, as well as a portion (10% of the input of IP) of the cell extracts as controls, were subjected to immunoblotting (IB) with KIF5B and KLC antibodies. Ostm1 coprecipitates with KIF5B heavy chains but not with KLC light chains, suggesting a direct interaction. (E) Representative immunostaining of EcR293 cells expressing Ostm1-FL-EGFP with KIF5B antibody demonstrate colocalization by confocal microscopy (63× objective). (F) Representative imaging of EcR293 cells coexpressing Ostm1-FL-EGFP/KIF5B-YFP, acquired by time-lapse confocal microscopy. Composite images are presented from a stack (63× objective). Enlarged image areas are not to scale.
Dynamic interaction of Ostm1 with KIF5B heavy chains.
To investigate whether Ostm1 and KIF5B interact, immunoprecipitation experiments were first performed with the full-length Ostm1-FL-EGFP protein and KIF5B. Since conventional kinesin proteins consist of heterotetramers with two heavy chains (KHC) and two light-chain isoforms (KLC) (34), Ostm1-FL-EGFP protein was analyzed for interaction with KHC and KLC. Immunoblots revealed that Ostm1 coprecipitated with the KHC but not with the KLC of KIF5B relative to an IgG control (Fig. 3D), revealing interaction of Ostm1 specifically with the KIF5B heavy chains that move along microtubules. To provide compelling evidence that Ostm1 interacts with KIF5B, both static and dynamic cellular colocalization experiments were carried out. Immunostaining of Ostm1-FL-EGFP and endogenous KIF5B displayed widespread punctate colocalization of the two proteins (Fig. 3E). Analysis of the dynamic interaction was monitored by live time lapse imaging, since KIF5B participates in trafficking of multiple cellular cargos and organelles (35). Ostm1-FL-EGFP and KIF5B-YFP colocalization was clearly observed as fluorescent puncta over background, and the vast majority of complexes were in constant dynamic movement (Fig. 3F). These data support that Ostm1 plays a role in cargo trafficking through interaction with KIF5B along the microtubules.
Characterization of Ostm1 interaction regions.
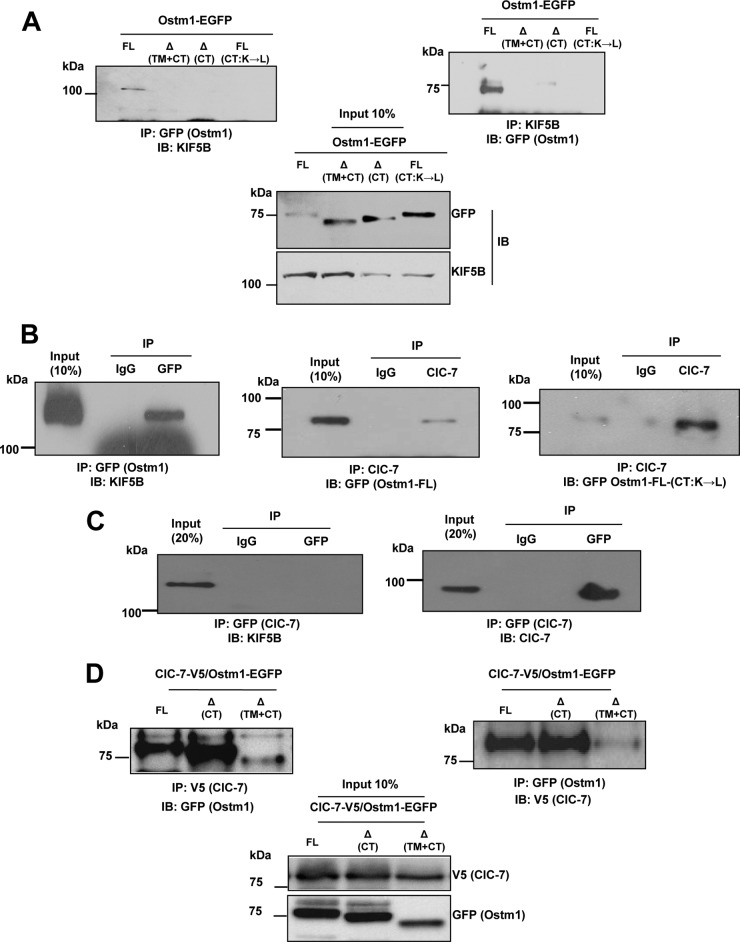
To delineate the region responsible for Ostm1-KIF5B interactions, a series of EGFP-tagged mutants of Ostm1 were tested by reciprocal coimmunoprecipitation. Different Ostm1 forms with deletions of the native CT domain, of both the TM and cytosolic CT domains (mimicking ARO mutations in humans [7, 8]), or with modified Ostm1-FL(CT: K→L)-EGFP were generated. The Ostm1 truncated and modified forms failed to coimmunoprecipitate KIF5B, whereas full-length Ostm1 was able to interact with KIF5B (Fig. 4A). Reciprocally, KIF5B can also coimmunoprecipitate full-length Ostm1 but not the truncated or the modified forms. These data indicate that the native C-terminal region of Ostm1 is critical for the interactions with KIF5B and that one or more Lys residues in the CT are implicated in Ostm1-KIF5B interaction.
FIG 4.
Ostm1 interacts with KIF5B independently of ClC-7. (A) Immunoblot analysis of ECR293 cell lysates from four different OSTM1-EGFP forms of transiently transfected clones, subjected reciprocally to IP with GFP and KIF5B antibodies. Western blot analysis with KIF5B and GFP antibodies shows that only Ostm1-FL interacts with KIF5B. (B) Immunoblot analysis of RAW cell-derived OC cell extracts transiently transfected with Ostm1-FL-EGFP, KIF5B-YFP, or Ostm1 FL(CT:K→L)-EGFP subjected to IP with GFP (Ostm1) and ClC-7 antibodies. Western blot analysis with GFP antibody shows that Ostm1 coprecipitates with ClC-7 and that the lysine residues of the Ostm1 CT are not important for this interaction. (C) Immunoblot analysis of RAW cell-derived OC cell extracts transiently transfected with ClC-7-EGFP and KIF5B-YFP and subjected to IP with ClC-7 antibody. Western blot analysis with KIF5B and ClC-7 antibodies shows that ClC-7 does not coprecipitate with KIF5B. (D) Immunoblot analysis of ECR293 cell lysates from ClC-7-V5/Ostm1-FL-EGFP cotransfectants reciprocally subjected to IP with V5 and GFP antibodies. Western blot analysis shows that ClC-7 interacts with all forms of Ostm1. These results are representative of three independent experiments.
To determine whether the interaction of Ostm1 and KIF5B that is ubiquitously expressed occurs in other cell types, cellular interaction of Ostm1 with KIF5B was monitored in osteoclasts. Analysis showed that Ostm1-EGFP can readily immunoprecipitate KIF5B protein compared to the IgG control (Fig. 4B), suggesting that Ostm1-KIF5B could play a role in functional activities such as intracellular vesicular trafficking.
Since Ostm1 was reported to associate with the ClC-7 exchanger in fibroblasts, we first monitored whether full-length Ostm1 can interact with ClC-7 in osteoclast-like RAW-derived cells. As shown in Fig. 4B, ClC-7 and full-length Ostm1 can mutually coprecipitate each other (Fig. 4B). Interestingly, ClC-7 could also immunoprecipitate modified Ostm1-FL(CT:K→L)-EGFP protein (Fig. 4B), indicating that the Lys residues in the Ostm1 CT domain are not important for this interaction. We then inquired whether ClC-7 could interact with KIF5B. No direct interaction was detected from coprecipitation of KIF5B-YFP by a ClC-7 antibody despite substantial ClC-7 levels being immunoprecipitated (Fig. 4C). In addition, we demonstrated that ClC-7 interacts with the truncated forms of Ostm1 (Fig. 4D). These data indicate that Ostm1 protein, via different interaction sites, is associated with distinct functions.
KIF5B is essential for Ostm1 intracellular dispersion.
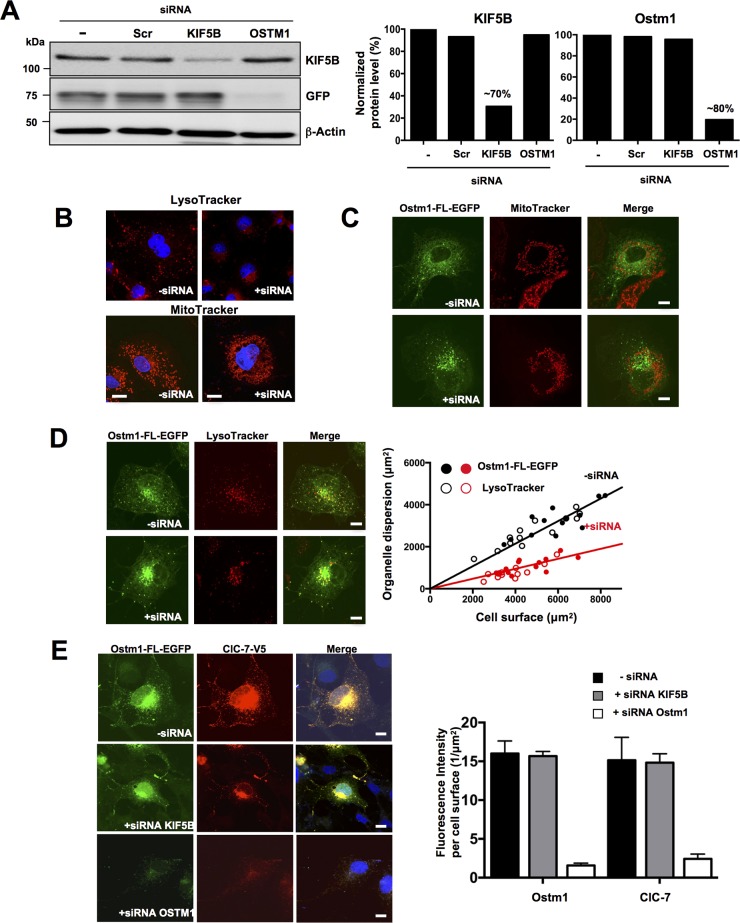
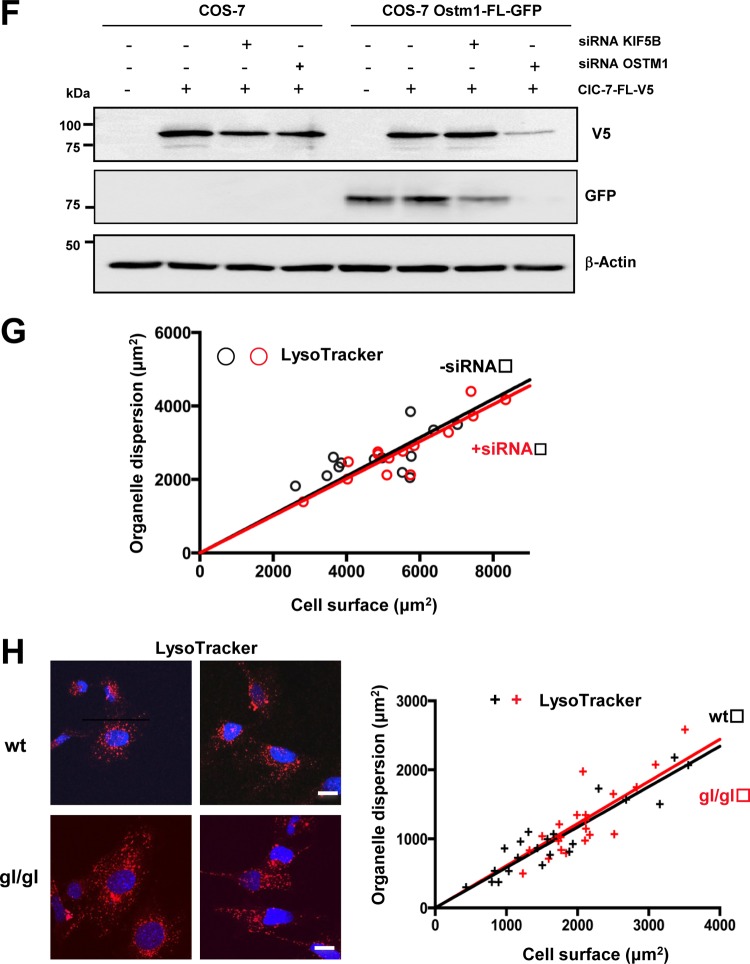
To interrogate the role of KIF5B in Ostm1 subcellular localization, KIF5B depletion was carried out using siRNA, as the null allele Kif5B in the mouse is embryonically lethal (30). KIF5B expression was reduced to ∼70% of the intracellular level with and without Ostm1-EGFP, as evaluated by immunoblotting (Fig. 5A). Under these defined conditions, the intracellular organelle distribution was monitored following LysoTracker and MitoTracker staining. Depletion of KIF5B in COS-7 cells resulted in clustering of perinuclear lysosomes and mitochondria compared to the wide cellular dispersion observed with KIF5B endogenous levels (Fig. 5B). Subsequently, KIF5B siRNA treatment of Ostm1-EGFP-expressing clones also showed mitochondrial clustering (Fig. 5C) but no colocalization of Ostm1 in mitochondria, suggesting that Ostm1 does not participate in the KIF5B-Milton complex or in mitochondrial transport (Fig. 5C). In contrast, upon depletion of KIF5B, Ostm1 showed impaired dynamic dispersion and consistently clustered to the perinuclear region with lysosomes (Fig. 5D; see Movies S1 and S2 in the supplemental material). Semiquantitative sublocalization analysis of Ostm1 and lysosomes was performed in individual cells in comparison to the overall cell surface area from confocal images and showed that dispersion upon KIF5B depletion was markedly abrogated (Fig. 5D, right panel). To further investigate ClC-7 status in the Ostm1-KIF5B complex and dispersion, ClC-7 was introduced in Ostm1-FL-EGFP COS-7 cells. Both ClC-7 and Ostm1 colocalized in cells, consistent with Ostm1 interaction with ClC-7 and even appear compatible with subcellular dispersion, supporting that ClC-7 is a cargo of Ostm1 (Fig. 5E). Upon KIF5B depletion, ClC-7 dispersion is impaired, as shown by perinuclear clustering localization (Fig. 5E). Since Ostm1, ClC-7, and lysosome dispersion is significantly impaired upon reduced expression of KIF5B, this suggests that Ostm1/KIF5B interaction is essential for Ostm1 dispersion within endosome/lysosome organelles.
FIG 5.
Ostm1/KIF5B interaction and organelle dispersion. (A) Immunoblot analysis of lysates from stable COS-7 clones expressing Ostm1-FL-EGFP treated with siRNA. Protein levels were decreased by ∼70% for KIF5B and ∼80% for OSTM1 compared to untreated and scramble (scr) controls (right panels). β-Actin was used as internal loading control. (B) Representative confocal images of COS-7 cells immunostained with LysoTracker (top panels) and MitoTracker (bottom panels) display lysosomes and mitochondrial clustering around the nucleus (blue, DAPI [4′,6′-diamidino-2-phenylindole]) with KIF5B siRNA (+siRNA) compared to untreated control cells (−siRNA) that show normal organelle dispersion. Bars, 20 μm. (C) COS-7 stable Ostm1-FL-EGFP clones (green) and MitoTracker (red) staining show no colocalization but cytosolic dispersion of Ostm1 and mitochondria (upper panels, merge). KIF5B siRNA leads to Ostm1-FL-EGFP and mitochondrial perinuclear clustering without colocalization. Bars, 20 μm. (D) COS-7 stable Ostm1-FL-EGFP clones (green) and LysoTracker staining (red) display cytosolic dispersion and colocalization of Ostm1 and lysosomes (upper panels, merge) that is impaired with KIF5B siRNA (lower panels, merge). Bars, 20 μm. Dispersion (right panel) in control (−siRNA; black circles) and KIF5B siRNA (+siRNA; red circles) cells from four independent experiments was measured, and the regression slopes of 0.55 (−siRNA) and 0.25 (+siRNA) indicate reduced organelle dispersion with KIF5B depletion. (E) COS-7 stable Ostm1-FL-EGFP clones (green) and ClC-7-FL-V5 fusion protein (red) exhibit colocalization in both cytosolic dispersion and perinuclear clustering when untreated or treated with KIF5B siRNA. Depletion of Ostm1 (lower panel) resulted in loss of the ClC-7 fluorescence signal (right panel). Bars, 20 μm. (F) Immunoblot of control COS-7 cells and stable Ostm1-FL-EGFP clones treated with Ostm1 siRNA, showing a major reduction of the ClC-7 protein level. (G) Lysosome dispersion in controls (black circles) or cells with OSTM1 siRNA (red circles) was measured, and the regression slopes of 0.52 (−siRNA) and 0.51 (+siRNA) indicate no effect on organelle dispersion. (H) Immunostaining of wild-type (wt) and gl/gl MEF cells with LysoTracker and DAPI (nucleus) is similar, and lysosome dispersion (right panel) in control (wt; black crosses) and in gl/gl (Ostm1 null; red crosses) cells was not reduced, with regression slopes of 0.59 and 0.61, respectively. Bars, 20 μm.
To analyze the role of Ostm1 in KIF5B organelle dispersion, we performed a reciprocal experiment consisting of depletion of Ostm1 expression induced upon siRNA treatment of Ostm1-EGFP-expressing clones. Efficiency of Ostm1 depletion (∼80%) was confirmed by a barely detectable GFP signal (Fig. 5A). The virtual lack of ClC-7 signal precluded analysis of protein localization (Fig. 5E, left lower panels). Consequently, Ostm1 siRNA cells showed a major decrease in ClC-7 signals, as determined by the significant 7-fold reduction of fluorescence intensity per cell surface area (Fig. 5E, right panel). The absence of ClC-7 signal was corroborated by the major decrease of ClC-7 protein levels seen by immunoblotting (Fig. 5F), indicating that Ostm1 is critical for maintenance of ClC-7 steady-state levels. Nonetheless, the role of Ostm1 in organelle dispersion was assessed in Ostm1-depleted cells. Reduced Ostm1 did not affect lysosomal dispersion in COS-7 cells (Fig. 5G), consistent with the Ostm1 nonmotor protein structure. Further, mouse embryo fibroblasts (MEFs) derived from gl/gl Ostm1-null mutant and wild-type mice were monitored for lysosomal dispersion. As illustrated in Fig. 5H, genetic loss of Ostm1 led to unaffected lysosomal dispersion comparable to that in wild-type controls, providing physiologic evidence that Ostm1 is most likely a KIF5B adaptor.
DISCUSSION
Insights into the molecular basis of protein biogenesis, posttranslational modification, and trafficking are central to understanding intracellular networks and function and the subsequent global impact on physiologic responses. This study defined Ostm1 as a secretory transmembrane protein that resides primarily in the ER and progresses through the secretory pathway. The wide protein screen for Ostm1 cytosolic partners revealed for the first time that Ostm1 likely participates in multiprotein complexes regulating cellular relocalization and trafficking of protein partners. Most significantly, the role of Ostm1 in endosomal/lysosomal intracellular dispersion can elucidate all of the pathophysiologic mechanisms underlying the large gl/gl phenotypic spectrum.
OSTM1 protein showed high conservation in metazoans implying that it plays a critical role in pluricellular organisms. Our characterization of a unique posttranslational Ostm1 signal peptide cleavage supported that Ostm1 is destined for the secretory pathway. Evidence of a single transmembrane domain for Ostm1 was provided upon deletion of this region, which led to protein secretion. Our data showed not only the importance of the transmembrane domain for subcellular localization of Ostm1 but also that the N-terminal region of Ostm1 is luminal. Significantly, the crucial role of the Ostm1 transmembrane domain was demonstrated in vivo from osteopetrotic patients expressing human OSTM1 lacking this domain (6, 7). Our analysis revealed that the luminal N-terminal orientation defined Ostm1 as a type I transmembrane protein, as we previously proposed (8). Interestingly, Ostm1 appears to be an atypical type I transmembrane protein that is partially retained in the ER by a mechanism independent of a lysine retention signal.
Similar to type I membrane proteins that exit the ER toward the TGN, Ostm1 is substantially and rapidly N-glycosylated in the ER. Further, a considerable amount of Ostm1 resides primarily in the ER, whereas a small fraction of Ostm1 progress through the secretory pathway. Interestingly, the trafficking and relative distribution of the modified Ostm1 appear to be mostly dependent on the rate of glucose trimming, which is likely crucial for proper folding and stability of Ostm1 to transit through the Golgi compartments. Consistent with the biochemical characterization, Ostm1 was localized in the ER by colocalization with calnexin, in the TGN with golgin, and in the late endosomes/lysosomes with LysoTracker staining. Notably, Ostm1 was detected in the perinuclear region but not on the plasma membrane. Collectively, the protein structure and cellular localization demonstrate that Ostm1 is a membrane protein that is not only resident in the ER but also implicated in protein sorting and transport within the endomembrane system and on secretory vesicles.
Our studies revealed several direct Ostm1-interacting proteins, supporting a model whereby Ostm1 possibly acts within multiprotein complexes and/or fulfills various cellular functions. While we and others showed that Ostm1 is a partner of the 12-transmembrane protein ClC-7 (36), no cytosolic interaction of ClC-7 and Ostm1 was detected either from our Ostm1 screen or from direct interaction with the Ostm1 CT domain. Our data provide evidence that ClC-7 interaction with Ostm1 is likely luminal within one or more compartments of the ER/Golgi/endosome/lysosome system. Moreover, our results show that Ostm1 is essential for ClC-7 steady-state levels and subcellular dispersion. Accordingly, the most severe phenotype triggered by the Ostm1 null allele relative to that of ClC-7 implicates not only ClC-7 function but also additional regulatory partners and targets. Interestingly, the Ostm1 direct cytosolic partners from our protein screen, KPNB1/importin β1/karyopherin, EDD1/UBR5 ubiquitin ligase, and KIF5B, may all participate in protein transport or trafficking with the Ostm1 cargo. Ostm1 interaction with KPNB1/importin β1/karyopherin, a nucleocytoplasmic trafficking protein (37), is consistent with Ostm1 perinuclear localization toward potential delivery of KPNB1/importin β1/karyopherin at the nuclear pore. The EDD1/UBR5 ubiquitin ligase together with Ostm1 may have a role in the proteasome, but this complex is highly reminiscent of the Nedd4 ubiquitin ligase (38, 39) interaction with the lysosomal transmembrane LAPTM5 protein. The Nedd4/LAPTM5 complex was shown to traffic from the Golgi apparatus to lysosomes (40), suggesting a similar sorting system for the EDD1/UBR5 ubiquitin ligase and Ostm1 complex. The most prevalent Ostm1 cytosolic partner, kinesin KIF5B, associates directly via its heavy chains with lysine residues in the Ostm1 CT domain. Interestingly, KIF5B interacts with importin β/karyopherin like Ostm1 and is a member of a multiprotein complex in human neuroretina (41). These shared interactions suggest that this protein triad could play a role in gl/gl retinal neurodegeneration (11).
KIF5B, a member of the kinesin-1 family (35), was shown to move along the microtubule cytoskeleton and promote, through adaptors, anterograde trafficking of multiple cellular cargos and organelles such as mitochondria and lysosomes (30, 42–44). Based on intracellular colocalization, Ostm1 cargos linked to KIF5B traffic specifically to lysosomes, whereas cargos destined for mitochondria likely involve other adaptors. Consistently, Ostm1 in a motor complex with KIF5B would serve to transport EDD1/UBR5 ubiquitin ligase to lysosomes. Moreover, our data argue that the Ostm1/KIF5B complex could also relocate KPNB1/importin β1/karyopherin at the nuclear pore, correlating with the perinuclear localization of importin β1/karyopherin in neuroretina (41). Such functional roles for Ostm1 are supported by our dynamic studies that showed intracellular organelle dispersion and protein corelocalization. Since both Ostm1 and KIF5B are ubiquitously expressed in a wide range of cells, this interaction could underlie a general cellular mechanism where Ostm1 in a complex with KIF5B would transport and deliver key molecules by trafficking to organelles or to specific sites within cells (45–47).
Our cellular and biochemical studies uncovered a key intracellular role for Ostm1 in trafficking and endosome/lysosome dispersion. Indeed, the phenotype of the Ostm1-null gray-lethal gl/gl mouse in osteoclasts, immunodeficiency, neuronal cells, and melanosomes can be elucidated by a defective adaptor function endosome/lysosome dispersion mechanism. Normally, the secretory lysosomes produced in osteoclasts (48–50) participate in the formation of a specific membrane structure, the ruffled border, and regulate secretion of specific proton pumps and enzymes essential for bone matrix degradation (51, 52). In Ostm1-null osteoclasts, the lack of Ostm1 vesicles linked to KIF5B cargos would elucidate the virtual absence of ruffled border, resulting in nonfunctional secretory cells, inefficient bone resorption, and osteopetrosis (9). Similarly, defective secretory lysosome and/or organelle dispersion due to loss of Ostm1 is likely the cellular mechanism causing the altered gl/gl immune cell response and hair pigmentation by melanosome clumping (8, 10, 11). Moreover, the role of Ostm1 in trafficking can also underlie the severe neuronal degeneration with impaired autophagosome targeting to lysosomes in gl/gl mice (11). Together, the gl/gl mouse phenotypes can result from the critical cellular role of Ostm1 in the relocalization of cargos and organelle-associated cargos to various specific compartments and destinations.
Our studies established that Ostm1 is an atypical type I transmembrane protein and part of a multiprotein scaffolding cytosolic complex with KIF5B. Furthermore, we demonstrated a key role of this complex in dynamic intracellular endosome/lysosome dispersion, a cellular mechanism known to be critical in osteoclast physiology and bone remodeling, in melanosome trafficking and hair pigmentation, and in autophagy and neuronal homeostasis. Of major physiologic and clinical relevance, the regulation of the newly identified Ostm1 interactors in trafficking function will lead to development of innovative therapeutic strategies.
Supplementary Material
ACKNOWLEDGMENTS
We thank B. Coulombe, C. Géronimo, J. Archambault, W. Y. Tsang, and C. Gu for invaluable advice and reagents. We thank D. Faubert for mass spectrometry analysis and D. Fillon for confocal microscopy.
This work was supported by Canadian Institutes of Health Research grant MOP 86655 to J.V. and by Canadian Institutes of Health Research grant 44363 and Canada Chair no. 216684 to N.G.S. J.B. was supported by an award from the CIHR Training Program in Skeletal Health Research.
We have no conflicting financial interests.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00656-15.
REFERENCES
- 1.Tolar J, Teitelbaum SL, andOrchard PJ. 2004. Osteopetrosis. N Engl J Med 351:2839–2849. doi: 10.1056/NEJMra040952. [DOI] [PubMed] [Google Scholar]
- 2.Balemans W, Van Wesenbeeck L, Van Hul W. 2005. A clinical and molecular overview of the human ostopetroses. Calcif Tissue Int 77:263–274. doi: 10.1007/s00223-005-0027-6. [DOI] [PubMed] [Google Scholar]
- 3.Sobacchi C, Schulz A, Coxon FP, Villa A, Helfrich MH. 2013. Osteopetrosis: genetics, treatment and new insights into osteoclast function. Nat Rev Endocrinol 9:522–536. doi: 10.1038/nrendo.2013.137. [DOI] [PubMed] [Google Scholar]
- 4.Teitelbaum SL, Ross P. 2003. Genetic regulation of osteoclast development and function. Nat Rev Genet 4:638–649. doi: 10.1038/nrg1122. [DOI] [PubMed] [Google Scholar]
- 5.Zaidi M. 2007. Skeletal remodeling in health and disease. Nat Med 13:791–801. doi: 10.1038/nm1593. [DOI] [PubMed] [Google Scholar]
- 6.Quarello P, Forni M, Barbereis L, Defilippi C, Campagnoli MF, Frattini A, Chalhoub N, Vacher J, Ramenghi U. 2004. Severe malignant osteopetrosis due to a Gl gene mutation. J Bone Miner Res 19:1194–1199. doi: 10.1359/JBMR.040407. [DOI] [PubMed] [Google Scholar]
- 7.Maranda B, Chabot G, Décarie J-C, Pata M, Azeddine B, Moreau A, Vacher J. 2008. Clinical and cellular manifestations of OSTM1 related infantile osteopetrosis. J Bone Miner Res 23:296–300. [DOI] [PubMed] [Google Scholar]
- 8.Chalhoub N, Benachenhou N, Rajapurohitam V, Pata M, Ferron M, Frattini A, Villa A, Vacher J. 2003. Grey-lethal mutation induces severe malignant autosomal recessive osteopetrosis in mouse and human. Nat Med 9:399–406. doi: 10.1038/nm842. [DOI] [PubMed] [Google Scholar]
- 9.Rajapurohitam V, Chalhoub N, Benachenhou N, Neff L, Baron R, Vacher J. 2001. The mouse osteopetrotic grey-lethal mutation induces a defect in osteoclast maturation/function. Bone 28:513–523. doi: 10.1016/S8756-3282(01)00416-1. [DOI] [PubMed] [Google Scholar]
- 10.Pata M, Héraud C, Vacher J. 2008. OSTM1 bone defect reveals an intercellular hematopoietic crosstalk. J Biol Chem 283:30522–30530. doi: 10.1074/jbc.M805242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Héraud C, Griffiths A, Pandruvada SNM, Kilimann MW, Pata M, Vacher J. 2014. Severe neurodegeneration with impaired autophagy mechanism triggered by Ostm1 deficiency. J Biol Chem 289:13912–13925. doi: 10.1074/jbc.M113.537233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orlow S. 1995. Melanosomes are specialized members of the lysosomal lineage of organelles. J Investig Dermatol 105:3–7. doi: 10.1111/1523-1747.ep12312291. [DOI] [PubMed] [Google Scholar]
- 13.Fischer T, De Vries L, Meerloo T, Farquhar MG. 2003. Promotion of Gαi3 subunit down-regulation by GIPN, a putative E3 ubiquitin ligase that interacts with RGS-GAIP. Proc Natl Acad Sci U S A 100:8270–8275. doi: 10.1073/pnas.1432965100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lange P, Wartosch L, Jentsch T, Fuhrmann J. 2006. ClC-7 requires Ostm1 as a beta-subunit to support bone resorption and lysosomal function. Nature 440:220–223. doi: 10.1038/nature04535. [DOI] [PubMed] [Google Scholar]
- 15.Leisle L, Ludwig CF, Wagner FA, Jentsch TJ, Stauber T. 2011. ClC-7 is a slowly voltage-gated 2Cl(−)/1H(+)-exchanger and requires Ostm1 for transport activity. EMBO J 30:2140–2152. doi: 10.1038/emboj.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinert S, Jabs S, Hohensee S, Chan WL, Kornak U, Jentsch TJ. 2014. Transport activity and presence of ClC-7/Ostm1 complex account for different cellular functions. EMBO Rep 15:784–791. doi: 10.15252/embr.201438553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frattini A, Pangrazio A, Susani L, Sobacchi C, Mirolo M, Abinun M, Andolina M, Flanagan A, Horwitz EM, Mihci E, Notarangelo LD, Ramenghi U, Teti A, Van Hove J, Vujic D, Young T, Albertini A, Orchard P, Vezzoni JP, Villa A. 2003. Chloride channel ClCN7 mutations are responsible for severe recessive, dominant, and Intermediate osteopetrosis. J Bone Miner Res 18:1740–1747. doi: 10.1359/jbmr.2003.18.10.1740. [DOI] [PubMed] [Google Scholar]
- 18.Mazzolari E, Forino C, Razza A, Porta F, Villa A, Notarangelo LD. 2009. A single-center experience in 20 patients with infantile malignant osteopetrosis. Am J Hematol 84:473–479. doi: 10.1002/ajh.21447. [DOI] [PubMed] [Google Scholar]
- 19.Kornak U, Kasper D, Bösl MR, Kaiser E, Schweizer M, Schulz A, Friedrich W, Delling G, Jentsch TJ. 2001. Loss of the CIC-7 chloride channel leads to osteopetrosis in mice and man. Cell 104:205–215. doi: 10.1016/S0092-8674(01)00206-9. [DOI] [PubMed] [Google Scholar]
- 20.Kozak M. 1984. Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature 308:241–246. doi: 10.1038/308241a0. [DOI] [PubMed] [Google Scholar]
- 21.Benjannet S, Elagoz A, Wickham L, Mamarbachi M, Munzer JS, Basak A, Lazure C, Cromlish JA, Sisodia S, Checler F, Chrétien M, Seidah NG. 2001. Post-translational processing of γ-secretase (β-amyloid-converting enzyme) and its ectodomain shedding. J Biol Chem 276:10879–10887. doi: 10.1074/jbc.M009899200. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt MR, Maritzena T, Kukhtinaa V, Higmanb VA, Doglioc L, Baraka NN, Straussb H, Oschkinatb H, Dottic CG, Hauckea V. 2009. Regulation of endosomal membrane traffic by a Gadkin/AP-1/kinesin KIF5 complex. Proc Acad Sci U S A 106:15344–15349. doi: 10.1073/pnas.0904268106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeghouf M, Li J, Butland G, Borkowska A, Canadien V, Richards DP, Beattie BK, Emili A, Greenblatt JF. 2004. Sequence peptide affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J Proteome Res 3:463–468. doi: 10.1021/pr034084x. [DOI] [PubMed] [Google Scholar]
- 24.No D, Yao TP, Evans RM. 1996. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc Natl Acad Sci U S A 93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 26.Jeronimo C, Langelier MF, Zeghouf M, Cojocaru M, Bergeron D, Baali D, Forget D, Mnaimneh S, Davierwala AP, Pootoolal J, Chandy M, Canadien V, Beattle BK, Richards DP, Workman JL, Hughes TR, Greenblatt J, Coulombe B. 2004. RPAP1, a novel human RNA polymerase II-associated protein affinity purified with recombinant wild-type and mutated polymerase subunits. Mol Cell Biol 24:7043–7058. doi: 10.1128/MCB.24.16.7043-7058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Thérien C, Bergeron D, Bourassa S, Greenblatt J, Chabot B, Poirier GG, Hughes TR, Blanchette M, Price DH, Coulombe B. 2007. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell 27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, Beatlle BK, Lalev A, Zhang W, Davierwala AP, Mnaimneh S, Starostine A, Tikuisis AP, Grigull J, Datta N, Bray JE, Hughes TR, Emili A, Greenblatt J. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell 13:225–239. doi: 10.1016/S1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 29.Sénéchal H, Poirier GG, Coulombe B, Laimins LA, Archambault J. 2007. Amino acid substitutions that specifically impair the transcriptional activity of papillomavirus E2 affect binding to the long isoform of Brd4. Virology 358:10–17. doi: 10.1016/j.virol.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka Y, Kanai Y, Okada Y, Nonaka S, Takeda S, Harada A, Hirokawa N. 1998. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell 93:1147–1158. doi: 10.1016/S0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- 31.Lange A, Yutzey KE. 2006. NFATc1 expression in the developing heart valves is responsive to the RANKL pathway and is required for endocardial expression of cathepsin K. Dev Biol 292:407–417. doi: 10.1016/j.ydbio.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 32.Teasdale RD, Jackson MR. 1996. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the golgi apparatus. Annu Rev Cell Dev Biol 12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- 33.Jackson MR, Nilsson T, Peterson PA. 1990. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J 9:3153–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirokawa N, Takemura R. 2005. Molecular motors and mechanisms of diretional transport in neurons. Nat Rev Neurosc 6:201–214. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- 35.Hirokawa N, Noda Y, Tanaka Y, Niwa S. 2009. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol 10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 36.Jentsch TJ, Friedrich T, Schriever A, Yamada H. 1999. The CLC chloride channel family. Pflügers Arch 437:783–795. [DOI] [PubMed] [Google Scholar]
- 37.Jäekel S, Görlich D. 1998. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J 17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cojocaru M, Bouchard A, Cloutier P, Cooper JJ, Varzavand K, Price DH, Coulombe B. 2011. Transcription factor IIS cooperates with the E3 ligase UBR5 to ubiquitinate the CDK9 subunit of the positive transcription elongation factor B. J Biol Chem 286:5012–5022. doi: 10.1074/jbc.M110.176628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, Xiong Y, Guan KL, Zhao S. 2011. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell 43:33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pak Y, Glowacka WK, Bruce MC, Pham N, Rotin D. 2006. Transport of LAPTM5 to lysosomes requires association with ubiquitin ligase Nedd4, but not LAPTM5 ubiquitination. J Cell Biol 175:631–645. doi: 10.1083/jcb.200603001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mavlyutov TA, Cai Y, Ferreira PA. 2002. Identification of RanBP2- and kinesin-mediated transport pathways with restricted neuronal and subcellular localization. Traffic 3:630–640. doi: 10.1034/j.1600-0854.2002.30905.x. [DOI] [PubMed] [Google Scholar]
- 42.Semiz S, Park JG, Nicoloro SM, Furcinitti P, Zhang C, Chawla A, Leszyk J, Czech MP. 2003. Conventional kinesin KIF5B mediates insulin-stimulated GLUT4 movements on microtubules. EMBO J 22:2387–2399. doi: 10.1093/emboj/cdg237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trejo HE, Lecuona E, Grillo D, Szleifer I, Nekrasova OE, Gelfand VI, Sznajder JI. 2010. Role of kinesin light chain-2 of kinesin-1 in the traffic of Na,K-ATPase-containing vesicles in alveolar epithelial cells. FASEB J 24:374–382. doi: 10.1096/fj.09-137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cardoso CMP, Groth-Pedersen L, Høyer-Hansen M, Kirkegaard T, Corcelle E, Andersen JS, Jäättelä M, Nylandsted J. 2009. Depletion of kinesin 5B affects lysosomal distribution and stability and induces peri-nuclear accumulation of autophagosomes in cancer cells. PLoS One 4:e4424. doi: 10.1371/journal.pone.0004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho K-I, Cai Y, Yi H, Yeh A, Aslankov A, andFerreira PA. 2007. Association of the kinesin-binding domain of RanBP2 to KIF5B and KIF5C determines mitochondria localization and function. Traffic 8:1722–1735. doi: 10.1111/j.1600-0854.2007.00647.x. [DOI] [PubMed] [Google Scholar]
- 46.Wang X, Schwarz TL. 2009. The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell 136:163–174. doi: 10.1016/j.cell.2008.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nabavi N, Urukova Y, Cardelli M, Aubin JE, Harrison RE. 2008. Lysosome dispersion in osteoblasts accommodates enhanced collagen production during differentiation. J Biol Chem 283:19678–19690. doi: 10.1074/jbc.M802517200. [DOI] [PubMed] [Google Scholar]
- 48.Blott EJ, Griffiths GM. 2002. Secretory lysosomes. Nat Rev Mol Cell Biol 3:122–131. doi: 10.1038/nrm732. [DOI] [PubMed] [Google Scholar]
- 49.Luzio JP, Pryor PR, Bright NA. 2007. Lysosomes: fusion and function. Nat Rev Mol Cell Biol 8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 50.Holt OJ, Gallo F, Griffiths GM. 2006. Regulating secretory lysosomes. J Biochem 140:7–12. doi: 10.1093/jb/mvj126. [DOI] [PubMed] [Google Scholar]
- 51.Coxon FP, Taylor A. 2008. Vesicular trafficking in osteoclasts. Sem Cell Dev Biol 19:424–433. doi: 10.1016/j.semcdb.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 52.Lacombe J, Karsenty G, Ferron M. 2013. Regulation of lysosome biogenesis and functions in osteoclasts. Cell Cycle 12:2744–2752. doi: 10.4161/cc.25825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.