ABSTRACT
The flagellar basal body is a rotary motor that spans the cytoplasmic and outer membranes. The rod is a drive shaft that transmits torque generated by the motor through the hook to the filament that propels the bacterial cell. The assembly and structure of the rod are poorly understood. In a first attempt to characterize this structure in the alphaproteobacterium Rhodobacter sphaeroides, we overexpressed and purified FliE and the four related rod proteins (FlgB, FlgC, FlgF, and FlgG), and we analyzed their ability to form homo-oligomers. We found that highly purified preparations of these proteins formed high-molecular-mass oligomers that tended to dissociate in the presence of NaCl. As predicted by in silico modeling, the four rod proteins share architectural features. Using affinity blotting, we detected the heteromeric interactions between these proteins. In addition, we observed that deletion of the N- and C-terminal regions of FlgF and FlgG severely affected heteromeric but not homomeric interactions. On the basis of our findings, we propose a model of rod assembly in this bacterium.
IMPORTANCE Despite the considerable amount of research on the structure and assembly of other flagellar axial structures that has been conducted, the rod has been barely studied. An analysis of the biochemical characteristics of the flagellar rod components of the Fla1 system of R. sphaeroides is presented in this work. We also analyze the interactions of these proteins with each other and with their neighbors, and we propose a model for the order in which they are assembled.
INTRODUCTION
The bacterial flagellum is a highly efficient nanomachine that propels the cell in liquid and semisolid media and over surfaces. It is composed of approximately 30 different proteins, whose numbers range from a few to several thousand copies (1). Its structure includes three major components, namely, the basal body, the hook, and the filament.
The basal body contains the flagellum-specific type III secretion system (T3SS) housed in a bell-like structure, named the C ring, which is also the input for chemotactic signals that control the direction of rotation and consequently cell movement (2). The basal body also contains an inner membrane ring (MS ring) and a periplasmic ring (P ring); in addition, Gram-negative bacteria possess an outer membrane ring (L ring). This structure includes the rod that expands from the MS ring through the L and P rings, as well as the rotary motor driven by proton or sodium ions, which is surrounded by stator subunits that are located around the MS ring and harness energy from the electrochemical gradient.
The hook is composed of about 120 copies of FlgE and is connected proximally to the rod and distally to the filament, by means of two hook-associated proteins (HAPs) (HAP1 and HAP3). The filament is composed of thousands of subunits of flagellin (FliC) and is the most abundant component of this organelle. HAP2 acts as a scaffold protein that helps flagellin subunits to polymerize at the distal end of the filament (3).
The structure and polymerization of the filament and the hook in enteric bacteria have been studied extensively (4, 5). The filament is a helical assembly of flagellin subunits, and structural analyses of this protein showed that it consists of four linearly connected domains, namely, D0, D1, D2, and D3 (6, 7), arranged from the inside to the outside of the filament. The N and C termini of the protein are located in the D0 domain and form two coiled-coil α-helices upon polymerization (8); this domain forms the inner core of the filament channel through which the flagellar components are exported. The D1 domain constitutes the next layer of the filament and, like the D0 domain, is highly conserved and is essential for polymerization. The D2 and D3 domains are dispensable for polymerization and are extremely variable among flagellins from different bacterial species (4, 9, 10).
The structure of the hook protein FlgE has also been characterized, and the protein is composed of three domains, i.e., D0, D1, and D2 (11, 12). FlgE shares similarities with flagellin in the D0 domain, which is disordered in solution and acquires a stable conformation upon polymerization (13).
It has been proposed that the rod is also a helical component of the bacterial flagellum, forming a continuous filamentous structure that connects with the hook and the filament, for which helical structures are well characterized (7, 12, 13). In contrast to the filament and the hook, which are composed of single proteins, the rod is composed of the four rod proteins, i.e., FlgB, FlgC, FlgF, and FlgG (14, 15), and an additional protein, FliE (16, 17), which together are referred to as the rod components.
The structure can be divided into a proximal rod and a distal rod. The proximal rod is estimated to have a length of 10 nm and to contain 6 subunits of each of FlgB, FlgC, and FlgF (18, 19) and about 9 FliE subunits per basal body (16). This is consistent with the polymerization of one turn each of the proximal components. In contrast, the distal rod is composed of 26 subunits or four turns of FlgG, with a length of 15 nm (18–20).
There is reasonable evidence from Salmonella that FliE is the first component of the rod. FlgE (hook) was found in cell extracts of a fliE mutant strain but not in flgB, flgC, flgF, flgG, or flgJ mutant strains (21). This was later shown to be due to a defect of FlgE secretion into the periplasm in the fliE mutant strain but not in the flgB, flgC, flgF, flgG, or flgJ mutant strains (22). It was also shown that secretion of FlgD (hook-cap) was dependent on FliE (23, 24). FliE and FlgB interact physically; therefore, it has been proposed that the two proteins constitute the most proximal end of the rod, with FliE acting as an adaptor protein between the MS ring and the rod (16, 17). It is well established that FlgG is the most distal rod component, given that it was found to be associated at the base of the hook in detached flagella (20). It should be noted that the positions of FlgC and FlgF have not yet been determined.
In this work, we analyzed the biochemical properties and interactions of the rod components of the Fla1 flagellar system of Rhodobacter sphaeroides. Our results showed that the N- and C-terminal regions of these proteins are relevant for heteromeric interactions, while the central domain (also referred to as the core domain) is important for homomeric interactions. We also confirmed the previously reported interaction between FliE and FlgB, and we detected two interactions, between FlgB and FlgF and between FlgC and FlgG, that had never been reported previously. A sequential model for the order of rod assembly is proposed.
MATERIALS AND METHODS
Strains, plasmids, and oligonucleotides.
The bacterial strains, plasmids, and oligonucleotides used in this study are listed in Table 1.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides
| Strain, plasmid, or oligonucleotide | Relevant characteristic(s) or sequence | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| TOP10 | Cloning strain | Invitrogen |
| JM109 | hdsR17 Δ(lac-pro) F′ traD36 proAB lacIqZΔM15 | Novagen |
| M15[pREP4] | thi lac ara gal mtl F′ recA+ uvr+ lon+; pREP4 plasmid; Kanr | Qiagen |
| BL21(DE3)pLysS | F′ ompT hsdSB(rB− mB−) gal dcm(DE3) pLysS | Novagen |
| R. sphaeroides WS8-N | Wild type; spontaneous Nalr | 39 |
| Plasmids | ||
| pQE30 | Expression vector; Ampr; N-terminal His6 tag | Qiagen |
| pET19b | Expression vector; Ampr; N-terminal His10 tag | Novagen |
| pRSFliE | fliE cloned into SacI/HindIII sites of pQE-30 | This study |
| pRSFlgB | flgB cloned into BamHI/HindIII sites of pQE-30 | This study |
| pRSFlgC | flgC cloned into NdeI/BamHI sites of pET-19b | This study |
| pRSFlgF | flgF cloned into KpnI/HindIII sites of pQE-30 | This study |
| pRSF49 | F49 cloned into KpnI/HindIII sites of pQE-30 | This study |
| pRSFlgG | flgG cloned into NdeI/BamHI sites of pET-19b | This study |
| pRSG47 | F47 cloned into NdeI/BamHI sites of pET-19b | This study |
| Oligonucleotides | ||
| fliEfw | CGATGAGCTCATGACCATCCAGTCGATCAGC | This study |
| fliErv | CGATAAGCTTTCAGACCGGCATGTTCATGAT | This study |
| flgBfw | CGATGGATCCATGACGGGATTTCGCGATCAG | This study |
| flgBrv | CGATAAGCTTTCATTCCCCCTTGATCGCCGT | This study |
| flgCfw | GGAATTCCATATGAGCGGGATCGACAGTGTCTTC | This study |
| flgCrv | CGCGGATCCTCACTGCCCCATGCTGGCGGT | This study |
| flgFfw | CGGGGTACCGACCGGCTGATCCACACCGCG | This study |
| flgFrv | CGATAAGCTTTCACTCGGGCGGACGCAGGAG | This study |
| F49fw | CGGGGTACCGCGATGGACGCGGCCTCGGCG | This study |
| F49rv | CGATAAGCTTTCACGTATTGACGTTCGAGCCATA | This study |
| flgGfw | GGAATTCCATATGTCCACCAATGCGATGCATGTC | This study |
| flgGrv | CGCGGATCCTCAGAGCTTGTTGGAAAGATA | This study |
| G47fw | GGAATTCCATATGTACCAGACCTGGAAGCCCGGC | This study |
| G47rv | CGCGGATCCTCACACGTTGACGTTCGAGGCTTC | This study |
Media and growth conditions.
Strains of Escherichia coli were grown in LB broth or agar at 37°C. When required, the following antibiotics were added at the following final concentrations: ampicillin, 200 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 34 μg/ml.
Engineering of rod component proteins.
The fliE, flgB, flgC, flgF, and flgG genes were amplified from chromosomal DNA of R. sphaeroides WS8N by PCR, using primers containing the corresponding restriction sites (Table 1) to facilitate cloning, with incorporation of a His6 tag from pQE-30 and a His10 tag from pET-19b at the N terminus. The internal regions of flgF (nucleotides 147 to 636) and flgG (nucleotides 141 to 681) were amplified from pRSFlgF and pRSFlgG, respectively, by PCR using the primers listed in Table 1, which contained the same restriction sites as the wild-type genes.
Overproduction and purification of rod component proteins.
E. coli strain M15[pREP4] carrying the pQE-30 derivative vector and strain BL21(DE3) pLysS carrying the pET-19b derivative vector were inoculated into LB medium supplemented with ampicillin (200 μg/ml) and kanamycin (50 μg/ml) or chloramphenicol (34 μg/ml), respectively, and were incubated at 37°C until they reached the early exponential phase (optical density at 600 nm [OD600] of ∼0.6). Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM or 1.0 mM, and expression was induced at 37°C for 3 h or 2 h, respectively. For E. coli strain BL21(DE3) pLysS carrying pRSFlgC, cells were incubated at 37°C until an OD600 of 0.8 was reached and then expression was induced with 1 mM IPTG for 1 h at 37°C.
FlgF and FlgG, as well as mutant versions expressing only the central regions of these proteins (FlgF49–212 and FlgG47–227, respectively), were purified under native conditions. Cells were harvested by centrifugation (10,000 × g for 10 min at 4°C) and frozen at −20°C. Thawed cell pellets were resuspended in lysis buffer (20 mM Tris-HCl [pH 8.5]) containing complete EDTA-free protease inhibitor mixture tablets (Roche) and were incubated on ice for 15 min prior to disruption by sonication. Cell debris was removed by centrifugation (5,000 × g for 10 min at 4°C), and the collected soluble cell fractions were mixed with Ni2+-nitrilotriacetic acid (NTA) agarose (500 μl per liter of starting culture) and incubated at 4°C for 2 h. The resin slurry was poured into a disposable column, and the flowthrough fractions were collected. Contaminating proteins were removed from the resin by washing with 20 ml of lysis buffer followed by 10 ml of wash buffer (lysis buffer containing 20 mM imidazole). Purified FlgF and FlgG were eluted in 5 to 10 ml of elution buffer (lysis buffer containing 250 mM imidazole) and then were dialyzed against 20 mM Tris-HCl (pH 8.5) at 4°C.
FliE, FlgB, and FlgC were purified under denaturing conditions. In the cases of FliE and FlgB, cells were harvested and lysed as mentioned above. FlgC is highly susceptible to cleavage by endogenous proteases; therefore, disruption of the cells was carried out under denaturing conditions (6 M guanidinium chloride [Gu-HCl], 20 mM Tris-HCl [pH 8.5]) (25). After 1 h of incubation at 4°C, the cell debris was removed by centrifugation (10,000 × g for 10 min at 4°C). Inclusion bodies containing FliE, FlgB, and soluble FlgC were dissolved in denaturing buffer, mixed with Ni2+-NTA agarose (500 μl per liter of starting culture), and incubated for 2 h at 4°C. The resin slurry was poured into a disposable column, and the flowthrough fractions were collected. Contaminating proteins were removed from the resin by washing with 10 ml of denaturing buffer followed by 10 ml of wash buffer (denaturing buffer containing 30 mM imidazole). Purified FliE, FlgB, and FlgC were eluted in 5 to 10 ml of elution buffer (denaturing buffer containing 250 mM imidazole). Proteins were refolded by dialysis against 20 mM Tris-HCl (pH 8.5) at 4°C.
Structural analysis by circular dichroism spectrometry.
Circular dichroism (CD) spectra were recorded with an AVIV spectrophotometer (model 202-01; Aviv Biomedical, Lakewood, NJ, USA), in the far-UV range (195 to 260 nm), for proteins at a concentration of 20 μM in 20 mM Tris-HCl (pH 8.5). Spectra were recorded at 25°C in 1-mm quartz cells, with a scan speed of 20 nm/min and a data interval of 1 nm.
Size exclusion liquid chromatography.
Size exclusion liquid chromatography was performed with an ÄKTA fast protein liquid chromatography (FPLC) system (GE Healthcare), using a Superdex 200 HR 10/30 column equilibrated with 20 mM Tris-HCl (pH 8.5) buffer (250 mM NaCl was added when indicated), at a flow rate of 0.2 ml/min; 500 μg of each purified protein was loaded in a final volume of 500 μl.
Antibody production and immunoblotting.
Polyclonal antibodies against each flagellar rod component protein were raised in male rabbits. Antiserum was obtained from whole blood, and gamma globulins were precipitated twice with saturated ammonium sulfate (26).
Purified proteins were subjected to SDS-PAGE on 17.5% polyacrylamide gels (27) and were transferred to nitrocellulose membranes (Bio-Rad). Membranes were blocked for 1 h at room temperature with Tris-buffered saline (20 mM Tris-HCl [pH 7.5], 500 mM NaCl) containing 0.1% (vol/vol) Tween 20 (TTBS) plus 5% nonfat milk powder. Immunoblotting was carried out with polyclonal anti-FliE, anti-FlgB, anti-FlgF, and anti-FlgG gamma globulins in TTBS, at the dilutions indicated below; monoclonal anti-FLAG (Sigma-Aldrich) was used for FlgC. Detection was performed by using SuperSignal West Pico (Thermo Scientific).
Prediction of three-dimensional structures of flagellar rod component proteins.
The models of the 5 rod protein components were prepared by using two different servers, i.e., the Robetta server (http://robetta.bakerlab.org) of the University of Washington (28) and the Quark program (iTasser) at the Zhang Lab server (http://zhanglab.ccmb.med.umich.edu/QUARK) of the University of Michigan (29). A set of 5 possible models of each protein was obtained. The models chosen were as follows: from the Robetta server, FlgB (PDB accession no. 3A69), FlgC (PDB accession no. 4PFP), and FlgF (PDB accession no. 4UT1), and from the iTasser (Zhang Lab) server, FliE (PDB accession no. 4AKV) and FlgG (PDB accession no. 3A69).
Protein-protein interactions assayed by affinity blotting.
Affinity blotting analysis was carried out according to the method described by Hall (30). Purified proteins (0.05 nmol of each) were subjected to SDS-PAGE on 17.5% polyacrylamide gels and then were transferred to nitrocellulose membranes. Membranes containing immobilized rod component proteins were incubated for 1 h at room temperature in TTBS (20 mM Tris-HCl [pH 7.5], 500 mM NaCl, with 0.1% Tween 20), in the presence of externally added purified probe proteins at the following concentrations: FliE, 1.2 μg/ml; FlgB, 1.5 μg/ml; FlgC, 8.0 μg/ml; FlgF, 1.2 μg/ml; FlgG, 2.0 μg/ml. Membranes incubated with the probe were washed three times with the corresponding buffer. Then the antibody against the probe was added, i.e., anti-FliE, anti-FlgB, and anti-FlgF at 1:20,000 dilutions and anti-FlgG at a 1:100,000 dilution, followed by the secondary antibody at a 1:10,000 dilution. For FlgC, monoclonal anti-FLAG (Sigma-Aldrich) at a 1:5,000 dilution was used. Detection was performed by immunoblotting, as described above.
RESULTS
Overexpression and purification of rod component proteins.
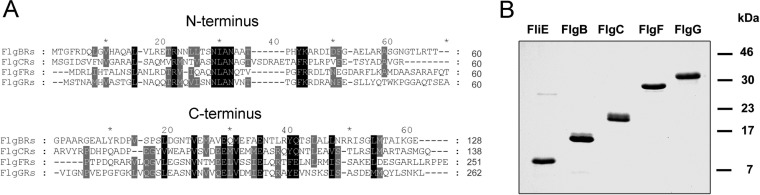
The rod proteins show a considerable degree of conservation at the N and C termini. A characteristic signature (NLAN) is conserved in the N termini, whereas heptad repeats of hydrophobic residues are observed in both terminal regions; these are important for the formation of α-helical coiled coils (14) (Fig. 1A). FliE was not included in the protein alignment because it is not homologous to the rest of the rod components. We overexpressed fliE, flgB, and flgF from R. sphaeroides cloned into the overexpression vector pQE-30, which adds 6 histidine residues at the N terminus of the induced protein (see Materials and Methods). The flgC and flgG genes were cloned into pET19b in order to obtain higher induction levels, and this vector adds 10 histidine residues at the N terminus of the protein. Two of the five rod components (FliE and FlgB) form inclusion bodies in cells; therefore, we carried out their solubilization in 6.0 M Gu-HCl. FlgC did not form inclusion bodies but was rapidly degraded by endogenous proteases (data not shown), as reported previously for FlgF of Salmonella (31). Therefore, we disrupted the cells in a buffer containing 6.0 M Gu-HCl, to inhibit protein degradation through protease activity (25). The purification of the three proteins was carried out under denaturing conditions, and the proteins were refolded by dialysis. FlgF and FlgG retained their soluble state after overexpression, and their purification was carried out under native conditions. All of the rod components were purified to homogeneity by means of Ni-NTA agarose affinity chromatography (Fig. 1B), and we did not pursue an additional purification step. By using circular dichroism (CD), we evaluated the secondary structures of the three rod proteins that were renatured from Gu-HCl. CD spectra showed that FliE, FlgB, and FlgC acquired secondary structure elements that suggested that the proteins were refolded correctly (data not shown).
FIG 1.
Sequence analysis and purification of the flagellar rod components from R. sphaeroides. (A) Amino acid sequence alignment of the N and C termini of the four rod proteins. Sixty residues of each terminal region were aligned using the program MUSCLE. Conserved residues are highlighted, and the length of each protein is listed at the end of the alignment. (B) SDS-PAGE analysis of purified N-terminal His-tagged rod component proteins, with Coomassie blue staining.
In vitro homo-oligomerization analyzed by size exclusion chromatography.
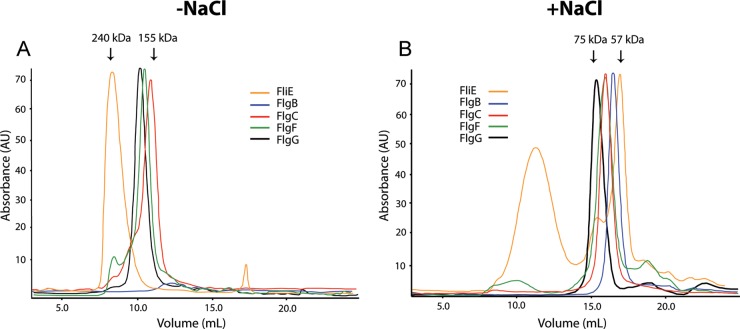
The rod components are filamentous proteins that should self-associate during polymerization. We analyzed the oligomerization state of the five rod components by using size exclusion liquid chromatography (see Materials and Methods). Figure 2A and B show chromatograms of the rod components in the absence and presence of NaCl, respectively. We observed that, in the absence of NaCl, these proteins eluted in fractions that corresponded to molecular masses ranging from 240 kDa to 155 kDa. These values were higher than would be expected for these proteins in their monomeric state (Table 2). Furthermore, FlgB did not enter the column, given that it precipitated under these conditions (Fig. 2A). We tested the effects of various buffer modifications (glycerol, pH, and ionic strength) on FlgB solubility and, in contrast to the situation observed for the rod proteins of Salmonella, NaCl was effective in reducing the aggregation of this protein; NaCl reached its maximum effect on FlgB solubility at 250 mM (data not shown). Given that, under these conditions, FlgB eluted in a volume corresponding to a lower molecular mass, we analyzed whether NaCl also had effects on the oligomerization of the rest of the rod components. We subjected these proteins to size exclusion chromatography in the presence of 250 mM NaCl, and all of the proteins eluted as complexes with lower molecular masses, as can be observed in Fig. 2B. It should be noted that FliE eluted in three different fractions, which is in accordance with a previous report indicating that FliE showed the strongest tendency to self-associate (31). Table 2 shows the estimated molecular masses of the rod components under the two conditions we tested. In the absence of NaCl, FliE formed aggregates based on a molecular mass of 240 kDa, which is equivalent to an oligomer of 18 subunits; in the presence of NaCl, the fraction corresponding to the smallest complex was about 57 kDa, equivalent to an aggregate of only 4 subunits. The rest of the proteins behaved in a similar way; in the presence of NaCl, the size of the homo-oligomers decreased.
FIG 2.
Analysis of purified rod component proteins by size exclusion liquid chromatography. Elution profiles of the rod components were determined using a Superdex 200 HR 10/30 column. (A) The column was equilibrated with a buffer containing 20 mM Tris-HCl (pH 8.5) (−NaCl). (B) The column was equilibrated with a buffer containing 20 mM Tris-HCl and 250 mM NaCl (pH 8.5) (+NaCl). A total amount of 500 μg of each protein was loaded in a volume of 500 μl.
TABLE 2.
Molecular masses of purified rod components, as deduced by size exclusion liquid chromatography
| Protein | Predicted molecular mass (kDa) | Without NaCl |
With NaCl |
||
|---|---|---|---|---|---|
| Molecular mass (kDa) | No. of subunits | Molecular mass(es) (kDa) | No. of subunits | ||
| FliE | 12.9 | 240 | 18 | 147, 74, and 57 | 11, 5, and 4 |
| FlgB | 15.3 | NDa | ND | 62 | 4 |
| FlgC | 17.8 | 155 | 8 | 68 | 3 |
| FlgF | 29.3 | 167 | 5 | 68 | 2 |
| FlgG | 30.5 | 176 | 5 | 75 | 2 |
ND, not determined.
Three-dimensional structure predictions.
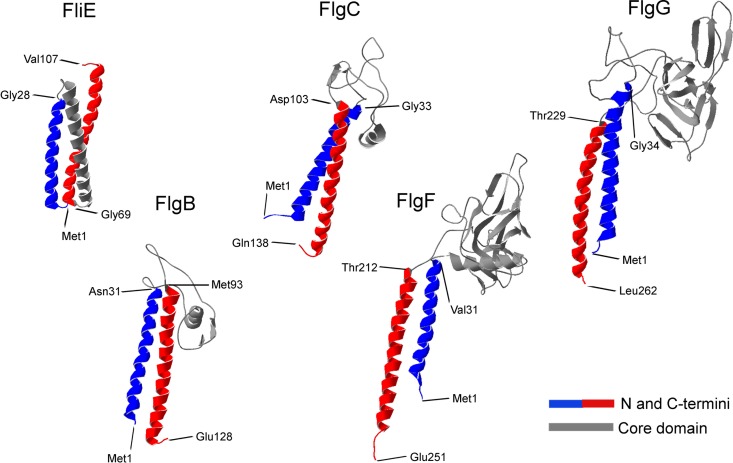
There have been several studies on the structures of flagellin and the hook protein that included atomic models of the filament and the hook (7, 11–13). In contrast, there has been only one study reporting the successful crystallization of a fragment that corresponds to the core domain of FlgG, and the protein structure has yet to be solved (32). In order to gain information on the tertiary structures of the rod components, we generated computational models. Figure 3 shows the predicted three-dimensional structures of the five proteins, with four of them resembling the previously reported crystallographic structures of FlgE and FliC (7, 11, 12, 33). We identified two clearly distinct structural domains, corresponding to the N and C termini, which are composed of α-helices, and the central region, which is formed of β-sheets and random coils. In accordance with the structures of the filament and the hook, these regions would correspond to the D0 and D1 domains, respectively (6, 7, 13). In contrast, FliE was predicted to fold into three packed α-helices.
FIG 3.
Predictions of the tertiary structures of the flagellar rod components from R. sphaeroides. In the three-dimensional structure models of the five rod component proteins, the structural domains are labeled as follows: N-terminal domain, blue; C-terminal domain, red; central domain, gray. The amino acids that limit the two domains are indicated.
Role of the central domain in homo-oligomerization.
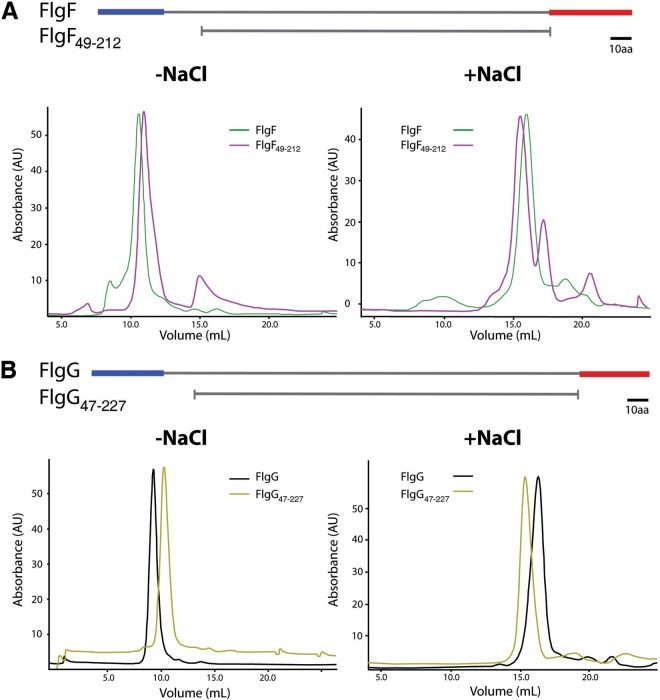
Based on the models obtained for the rod components, we designed mutant versions of FlgF and FlgG lacking the N- and C-terminal regions, in order to analyze the role of the central domain in homo-oligomerization. We decided to work only with these two proteins because they both possess a large central domain, in comparison with the rest of the rod components (Fig. 3). The mutants were named FlgF49–212 (polypeptide chain from Ala49 to Thr212) and FlgG47–227 (from Tyr47 to Val227); this mutant version of FlgG is equivalent to the mutant version of the Salmonella FlgG that was crystallized recently (32). The mutant proteins were purified under the same conditions as the wild-type proteins (see Materials and Methods). Figure 4 shows that FlgF49–212 and FlgG47–227 formed large aggregates in the absence of NaCl, whereas the size of the aggregates decreased in the presence of NaCl, in a manner similar to the behavior shown for the wild-type proteins (Fig. 4A and B). These results suggest that the N and C termini do not participate in the oligomerization process. It could be possible that the central domain is responsible for homo-oligomerization.
FIG 4.
Effects of the terminal regions of FlgF and FlgG on protein oligomerization, analyzed by size exclusion liquid chromatography. (A) Elution profiles of FlgF and FlgF49–212. (B) Elution profiles of FlgG and FlgG47–227. Schematic representations of the wild-type and mutant proteins are shown above each panel. We used a Superdex HR 10/30 column equilibrated with 20 mM Tris-HCl (pH 8.5) (−NaCl) or 20 mM Tris-HCl, 250 mM NaCl (pH 8.5) (+NaCl). A total amount of 500 μg of each protein was loaded in a volume of 500 μl.
Interactions between rod components detected by affinity blotting experiments.
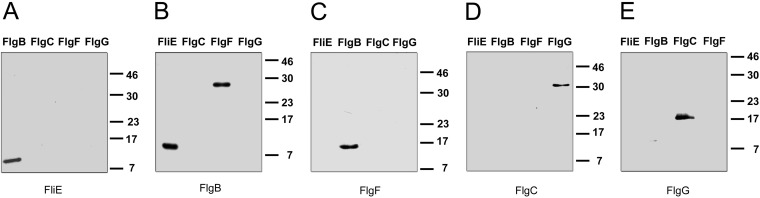
The hook and the filament are composed of multiple subunits of a single protein; this suggests that each subunit interacts only with an identical partner. The rod is a helical assembly of 5 different proteins and, based on the estimated copy number, it is thought that each protein polymerizes to complete at least one turn. The interactions that take place in this structure suggest that each protein subunit interacts with an identical partner and, at the same time, with a different neighboring rod component. In a previous report, the interaction between FliE and FlgB of Salmonella enterica was detected by affinity blotting and was confirmed genetically (17). Furthermore, affinity blotting has been used as direct proof of interactions between different flagellar proteins (17, 34–36). The assay relies on the fact that proteins that have been denatured by SDS are partially renatured when transferred to the blotting membrane.
Therefore, we explored the physical interactions between the rod components by using affinity blotting. Figure 5 shows the results obtained with the five rod components. We confirmed the previously reported interaction between FliE and FlgB (17) (Fig. 5A and B), and we also detected an interaction between FlgB and FlgF (Fig. 5B and C) and an interaction between FlgC and FlgG (Fig. 5D and E). It should be noted that these interactions were evident regardless of which rod component was in solution or immobilized on the membrane.
FIG 5.
Heteromeric interactions of flagellar rod components detected by affinity blotting. The protein loaded in each lane is indicated; 0.05 nmol of each protein was loaded. The proteins used as probes were FliE (A), FlgB (B), FlgF (C), FlgC (D), and FlgG (E). Detection was performed by using specific antibodies against the probes. Molecular mass markers are shown on the right (in kilodaltons).
Roles of N- and C-terminal regions in heterologous interactions.
We showed that the N- and C-terminal domains do not contribute to the oligomerization of FlgF and FlgG. Therefore, we decided to explore whether these regions play a role in the interactions of these proteins with their neighboring partners. Figure 6A shows that FlgB did not interact with FlgF49–212, as was observed in the case of the wild-type protein. Figure 6B shows that FlgC barely recognized FlgG47–227, in contrast to the full-length protein. These results strongly suggest that the N and C termini are essential for heteromeric interactions among the rod components.
FIG 6.
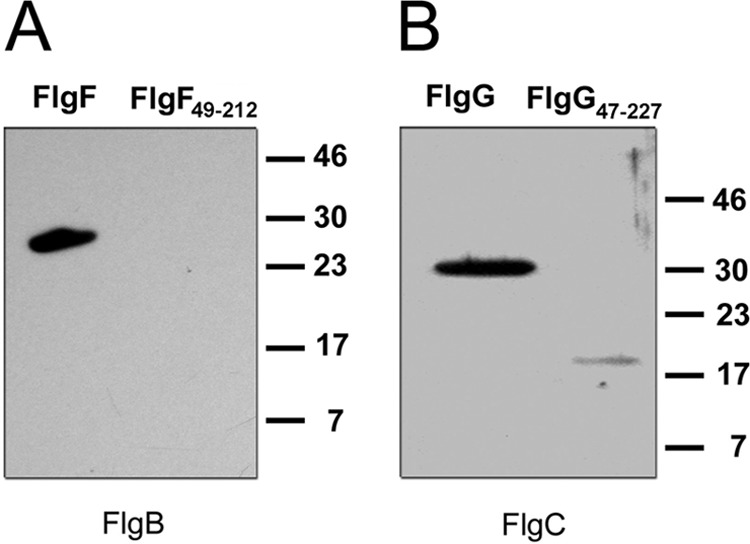
Effects of the N- and C-terminal regions of FlgF and FlgG on heteromeric interactions, tested by affinity blotting. (A) FlgF and FlgF49–212 (0.05 nmol) were transferred to a nitrocellulose membrane, and FlgB was used as the probe. (B) FlgG and FlgG47–227 (0.05 nmol) were transferred to a nitrocellulose membrane, and FlgC was used as the probe. Detection was performed by using specific antibodies against the probes. Molecular mass markers are shown on the right (in kilodaltons).
DISCUSSION
The flagellar rod can be regarded as the most proximal portion of the axial structure that extends to the hook, and it plays an important role in transmitting motor torque to the helical propeller for bacterial locomotion. The rod is composed of five different proteins, i.e., FliE, FlgB, FlgC, FlgF, and FlgG, which have been studied only in S. enterica (31). In the present study, we characterized biochemically the rod components of R. sphaeroides. We purified N-terminal His-tagged FlgF and FlgG under native conditions and N-terminal His-tagged FliE, FlgB, and FlgC under denaturing conditions; we confirmed by circular dichroism (CD) analysis that these proteins acquired secondary structure elements after refolding. It has been reported that, in S. enterica, a histidine tag at the N terminus does not affect the protein function of rod components (17).
As observed in S. enterica (31), the rod components of R. sphaeroides showed a strong tendency to self-associate (Fig. 2). It was reported previously that FliE is especially prone to aggregation (31); here we showed that FliE forms very large homo-oligomers that can be only partially disrupted with NaCl, compared to the rest of the rod components. This is interesting, because it has been proposed that FliE is responsible for connecting the rod to the MS ring (17); therefore, this junction zone must be strong enough to transmit torque to the axial structure. The dissociating effect of NaCl was not expected, given that, in S. enterica, NaCl favored aggregation of the rod components (31). The dissociation of the homo-oligomers into smaller complexes with NaCl points to the fact that the interactions that support this homo-oligomerization are mainly electrostatic.
We observed that, even in the presence of NaCl, the rod proteins eluted in fractions that suggested the presence of oligomers. However, it was shown previously that anomalous elution profiles for these proteins indicated an elongated shape (25). This observation raises the possibility that the monomeric form could be present in some of our preparations.
The crystallographic structures of these proteins have not been resolved. Therefore, we modeled in silico the tertiary structures of the rod components of R. sphaeroides. The models we obtained using two different servers were very similar in all cases (data not shown). FliE is not structurally related to the rest of the rod proteins, as it does not show the typical architecture of the axial proteins. The modeled structure of FliE resembles the predicted structure of the inner rod component (PscI) of the injectisome type III secretion system (37).
The four rod proteins showed similar architectures, which we divided into two structural domains, i.e., the central region and the N and C termini, which are equivalent to the D0 and D1 domains, respectively, in the hook and filament proteins. The N- and C-terminal domains of the flagellin and the hook protein are structurally independent, since their elimination does not affect the folding of the rest of the protein (7, 12, 13, 33). To evaluate the role of the central domains of FlgF and FlgG in homo-oligomerization, we deleted the terminal domains of each protein, which yielded the fragments FlgF49–212 and FlgG47–227. We found that the mutant proteins were still able to form complexes with high molecular masses. Therefore, it is likely that the oligomerization process during rod assembly involves interactions between the central domains of different subunits of the same protein. Our findings are supported by the results of Chevance et al. (18), who reported mutations in the central domain of FlgG that affected the polymerization of this protein, yielding extremely long rods.
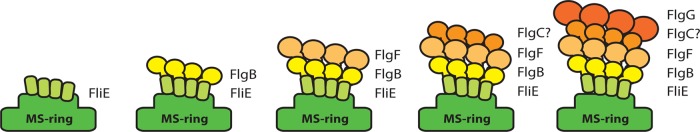
We also tested heteromeric interactions between the rod components. Based on the interactions detected by affinity blotting, we propose a model of the order in which these proteins are added into the rod in R. sphaeroides (Fig. 7). We confirmed a previously reported interaction between FliE and FlgB (17), and we also detected interactions between FlgB and FlgF and between FlgC and FlgG, which had not been reported previously. There is consensus regarding the localization of FlgG as the most distal rod component (18, 20). Therefore, we propose that FlgC is located in the boundary between FlgF and FlgG. Unfortunately, we were unable to detect the interaction between FlgF and FlgC. One possible explanation is that FlgC could be a checkpoint that senses FlgF polymerization. It has been suggested that the assembly of the rod is a cooperative process (15). Furthermore, a recent study showed, by cryo-electron microscopy, that the rod structure undergoes rearrangement and acquires the typical rod shape only after the addition of FlgF (38). It is known that the rod components belong to the same export specificity class (23). Our affinity blotting results suggest that the order in which the rod components are added into the overall rod structure is determined by their specific interactions. Finally, we found that the N- and C-terminal regions of FlgF and FlgG play decisive roles in heteromeric interactions. Since these regions project downward in our predicted three-dimensional models, as in flagellin and the hook protein, the N and C termini of one subunit may interact with the N and C termini of the underlying subunit (7).
FIG 7.
Model of flagellar rod assembly in R. sphaeroides. FliE subunits are assembled into the MS ring (16, 24). Direct evidence for an interaction between FliE and FlgB in S. enterica (17) and R. sphaeroides (this study) strongly suggests that subunits of FlgB are assembled into the distal end of the FliE zone. Based on the interactions we found in this work, we propose that FlgF subunits are added into the structure after FlgB. Subunits of FlgC may polymerize after FlgF and, finally, FlgG subunits are incorporated at the distal end of the rod (20). Question marks indicate that we were unable to detect the interaction between FlgF and FlgC.
ACKNOWLEDGMENTS
We thank Héctor Miranda-Astudillo for help with size exclusion chromatography analysis. We also thank Diego González-Halphen and Bertha González-Pedrajo for helpful discussions, Teresa Ballado and Aurora Osorio for technical assistance, and the IFC Molecular Biology Unit for sequencing facilities.
M.O.-V. was a master's degree student from the Posgrado en Ciencias Bioquímicas, Universidad Nacional Autónoma de México, supported by a fellowship from CONACyT (grant 298834). This study was supported by grants from DGAPA/UNAM (grant IN204614) and CONACyT (grant 106081).
REFERENCES
- 1.Macnab RM. 2003. How bacteria assemble flagella. Annu Rev Microbiol 57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 2.Minamino T, Imada K, Namba K. 2008. Molecular motors of the bacterial flagella. Curr Opin Struct Biol 18:693–701. doi: 10.1016/j.sbi.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Yonekura K, Maki S, Morgan DG, DeRosier DJ, Vonderviszt F, Imada K, Namba K. 2000. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science 290:2148–2152. doi: 10.1126/science.290.5499.2148. [DOI] [PubMed] [Google Scholar]
- 4.Altegoer F, Schuhmacher J, Pausch P, Bange G. 2014. From molecular evolution to biobricks and synthetic modules: a lesson by the bacterial flagellum. Biotechnol Genet Eng Rev 30:49–64. doi: 10.1080/02648725.2014.921500. [DOI] [PubMed] [Google Scholar]
- 5.Minamino T, Namba K. 2004. Self-assembly and type III protein export of the bacterial flagellum. J Mol Microbiol Biotechnol 7:5–17. doi: 10.1159/000077865. [DOI] [PubMed] [Google Scholar]
- 6.Samatey FA, Imada K, Nagashima S, Vonderviszt F, Kumasaka T, Yamamoto M, Namba K. 2001. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410:331–337. doi: 10.1038/35066504. [DOI] [PubMed] [Google Scholar]
- 7.Yonekura K, Maki-Yonekura S, Namba K. 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 8.Aizawa SI, Vonderviszt F, Ishima R, Akasaka K. 1990. Termini of Salmonella flagellin are disordered and become organized upon polymerization into flagellar filament. J Mol Biol 211:673–677. doi: 10.1016/0022-2836(90)90064-S. [DOI] [PubMed] [Google Scholar]
- 9.Kuwajima G. 1988. Construction of a minimum-size functional flagellin of Escherichia coli. J Bacteriol 170:3305–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beatson SA, Minamino T, Pallen MJ. 2006. Variation in bacterial flagellins: from sequence to structure. Trends Microbiol 14:151–155. doi: 10.1016/j.tim.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Samatey FA, Matsunami H, Imada K, Nagashima S, Namba K. 2004. Crystallization of a core fragment of the flagellar hook protein FlgE. Acta Crystallogr D Biol Crystallogr 60:2078–2080. doi: 10.1107/S0907444904022735. [DOI] [PubMed] [Google Scholar]
- 12.Samatey FA, Matsunami H, Imada K, Nagashima S, Shaikh TR, Thomas DR, Chen JZ, Derosier DJ, Kitao A, Namba K. 2004. Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism. Nature 431:1062–1068. doi: 10.1038/nature02997. [DOI] [PubMed] [Google Scholar]
- 13.Fujii T, Kato T, Namba K. 2009. Specific arrangement of alpha-helical coiled coils in the core domain of the bacterial flagellar hook for the universal joint function. Structure 17:1485–1493. doi: 10.1016/j.str.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 14.Homma M, Kutsukake K, Hasebe M, Iino T, Macnab RM. 1990. FlgB, FlgC, FlgF and FlgG: a family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J Mol Biol 211:465–477. doi: 10.1016/0022-2836(90)90365-S. [DOI] [PubMed] [Google Scholar]
- 15.Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S. 1992. Morphological pathway of flagellar assembly in Salmonella typhimurium. J Mol Biol 226:433–446. doi: 10.1016/0022-2836(92)90958-M. [DOI] [PubMed] [Google Scholar]
- 16.Müller V, Jones CJ, Kawagishi I, Aizawa S, Macnab RM. 1992. Characterization of the fliE genes of Escherichia coli and Salmonella typhimurium and identification of the FliE protein as a component of the flagellar hook-basal body complex. J Bacteriol 174:2298–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minamino T, Yamaguchi S, Macnab RM. 2000. Interaction between FliE and FlgB, a proximal rod component of the flagellar basal body of Salmonella. J Bacteriol 182:3029–3036. doi: 10.1128/JB.182.11.3029-3036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chevance FF, Takahashi N, Karlinsey JE, Gnerer J, Hirano T, Samudrala R, Aizawa S, Hughes KT. 2007. The mechanism of outer membrane penetration by the eubacterial flagellum and implications for spirochete evolution. Genes Dev 21:2326–2335. doi: 10.1101/gad.1571607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones CJ, Macnab RM, Okino H, Aizawa S. 1990. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J Mol Biol 212:377–387. doi: 10.1016/0022-2836(90)90132-6. [DOI] [PubMed] [Google Scholar]
- 20.Okino H, Isomura M, Yamaguchi S, Magariyama Y, Kudo S, Aizawa SI. 1989. Release of flagellar filament-hook-rod complex by a Salmonella typhimurium mutant defective in the M ring of the basal body. J Bacteriol 171:2075–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonifield HR, Yamaguchi S, Hughes KT. 2000. The flagellar hook protein, FlgE, of Salmonella enterica serovar Typhimurium is posttranscriptionally regulated in response to the stage of flagellar assembly. J Bacteriol 182:4044–4050. doi: 10.1128/JB.182.14.4044-4050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee HJ, Hughes KT. 2006. Posttranscriptional control of the Salmonella enterica flagellar hook protein FlgE. J Bacteriol 188:3308–3316. doi: 10.1128/JB.188.9.3308-3316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirano T, Minamino T, Namba K, Macnab RM. 2003. Substrate specificity classes and the recognition signal for Salmonella type III flagellar export. J Bacteriol 185:2485–2492. doi: 10.1128/JB.185.8.2485-2492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minamino T, Macnab RM. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J Bacteriol 181:1388–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saijo-Hamano Y, Namba K, Oosawa K. 2000. A new purification method for overproduced proteins sensitive to endogenous proteases. J Struct Biol 132:142–146. doi: 10.1006/jsbi.2000.4311. [DOI] [PubMed] [Google Scholar]
- 26.Harlow E, Lane D. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 27.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Raman S, Vernon R, Thompson J, Tyka M, Sadreyev R, Pei J, Kim D, Kellogg E, DiMaio F, Lange O, Kinch L, Sheffler W, Kim BH, Das R, Grishin NV, Baker D. 2009. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins 77(Suppl 9):89–99. doi: 10.1002/prot.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall RA. 2004. Studying protein-protein interactions via blot overlay or Far Western blot. Methods Mol Biol 261:167–174. [DOI] [PubMed] [Google Scholar]
- 31.Saijo-Hamano Y, Uchida N, Namba K, Oosawa K. 2004. In vitro characterization of FlgB, FlgC, FlgF, FlgG, and FliE, flagellar basal body proteins of Salmonella. J Mol Biol 339:423–435. doi: 10.1016/j.jmb.2004.03.070. [DOI] [PubMed] [Google Scholar]
- 32.Saijo-Hamano Y, Matsunami H, Namba K, Imada K. 2013. Expression, purification, crystallization and preliminary X-ray diffraction analysis of a core fragment of FlgG, a bacterial flagellar rod protein. Acta Crystallogr Sect F Struct Biol Cryst Commun 69:547–550. doi: 10.1107/S1744309113008075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samatey FA, Imada K, Vonderviszt F, Shirakihara Y, Namba K. 2000. Crystallization of the F41 fragment of flagellin and data collection from extremely thin crystals. J Struct Biol 132:106–111. doi: 10.1006/jsbi.2000.4312. [DOI] [PubMed] [Google Scholar]
- 34.Toker AS, Macnab RM. 1997. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J Mol Biol 273:623–634. doi: 10.1006/jmbi.1997.1335. [DOI] [PubMed] [Google Scholar]
- 35.Hirano T, Minamino T, Macnab RM. 2001. The role in flagellar rod assembly of the N-terminal domain of Salmonella FlgJ, a flagellum-specific muramidase. J Mol Biol 312:359–369. doi: 10.1006/jmbi.2001.4963. [DOI] [PubMed] [Google Scholar]
- 36.Minamino T, MacNab RM. 2000. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol Microbiol 35:1052–1064. doi: 10.1046/j.1365-2958.2000.01771.x. [DOI] [PubMed] [Google Scholar]
- 37.Monlezun L, Liebl D, Fenel D, Grandjean T, Berry A, Schoehn G, Dessein R, Faudry E, Attree I. 2015. PscI is a type III secretion needle anchoring protein with in vitro polymerization capacities. Mol Microbiol 96:419–436. doi: 10.1111/mmi.12947. [DOI] [PubMed] [Google Scholar]
- 38.Zhao X, Zhang K, Boquoi T, Hu B, Motaleb MA, Miller KA, James ME, Charon NW, Manson MD, Norris SJ, Li C, Liu J. 2013. Cryoelectron tomography reveals the sequential assembly of bacterial flagella in Borrelia burgdorferi. Proc Natl Acad Sci U S A 110:14390–14395. doi: 10.1073/pnas.1308306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sockett RE, Foster JCA, Armitage JP. 1990. Molecular biology of the Rhodobacter sphaeroides flagellum. FEMS Symp 53:473–479. [Google Scholar]