ABSTRACT
The flagellar hook is a short tubular structure located between the external filament and the membrane-bound basal body. The average hook length is 55 nm and is determined by the soluble protein FliK and the integral membrane protein FlhB. Hook elongation is terminated by FliK-mediated cessation of hook protein secretion, followed by the secretion of filamentous proteins. This process is referred to as the substrate specificity switch. Switching of the secretion modes results from a direct interaction between the FliK C-terminal domain (FliKC) and the secretion gate in FlhB. FliKC consists of two α-helices and four β-strands. Loop 2 connects the first two β-sheets and contains a conserved sequence of 9 residues. Genetic and physiological analyses of various fliK partial deletion mutants pointed to loop 2 as essential for induction of a conformational change in the FlhB gate. We constructed single-amino-acid substitutions in the conserved region of loop 2 of FliK and discovered that the loop sequence LRL is essential for the timely switching of secretion modes.
IMPORTANCE Flagellar protein secretion is controlled by the soluble protein FliK. We discovered that the loop 2 sequence LRL in the FliK C terminus was essential for timely switching of secretion modes. This mechanism is applicable to type three secretions systems that secrete virulence factors in bacterial pathogens.
INTRODUCTION
One of the most important and mysterious regulatory processes is the control of size and length determination of biological structures. A few examples exist in which the molecular mechanism for length determination is known, including the tobacco mosaic virus, the bacteriophage lambda tail, and muscle thin filaments (1, 2). In recent years, the bacterial flagellum has been regarded as another example for which molecular elucidation of length control is possible (3–6).
The flagellum is a supramolecular structure for motility consisting of three substructures with distinctive functions. The basal body acts as a rotary motor, and the external filament works as a screw. The hook conveys torque generated at the basal body toward the filament (7). Hook length is controlled and averages approximately 55 nm (8, 9). The flagellar protein FliK controls hook length (3, 8, 10). FliK consists of 405 amino acids and is structurally divided into two distinct N- and C-terminal regions, FliKN and FliKC (11). The molecular length of FliKN is linearly correlated with hook length, while FliKC is necessary for switching the substrate specificity of protein secretion at FlhB (10, 12). Mutations in FliKC often result in hooks without filaments and uncontrolled lengths, called polyhooks (8, 11, 13). Although defects in FliKN lead to uncontrolled hook length, substrate switching eventually occurs, as long as FliKC is intact. This results in filament formation on polyhooks, referred to as polyhook-filaments (11, 14). Minamino et al. (13) performed site-directed mutagenesis and reported that 302Val, 304Ile, 335Leu, 401Val, and 405Ala of FliKC were critical for the switching process. Nuclear magnetic resonance (NMR) spectroscopy revealed that FliKC residues 268 to 352 consisted of two α-helices and four β-strands in the order of α1-β1-β2-β3-α2-β4 (15). Loop 2 between the β1 and β2 strands contains a conserved sequence of 9 residues (residues 292 to 300). From genetic analyses of fliK deletion mutants, we predicted that loop 2 might be essential for interaction with the secretion gate, FlhB (10).
Recently, we showed that induction of FliK from an inducible promoter resulted in the immediate switch in secretion mode for flagellar structures with hooks of greater than or equal to wild-type length in a chromosomally fliK-deleted mutant strain (16). That is, FliKC always interacted with the secretion gate when the hook was longer than 55 nm. We concluded that FliK uses the minimal length as a signal to switch secretion modes but does not measure hook length per se (17). In other words, FliK acts to prevent hook polymerization beyond an optimal functional length (55 nm). High-speed atomic force microscopy revealed that the molecular shape of intact FliK in solution was composed of two folded domains of differing sizes, linked by a flexible linker (17). The larger domain corresponded to FliKN, and the smaller domain corresponded to FliKC. Furthermore, the FliKN domain shape appeared to be more stable than that of the FliKC domain. These findings led us to our current model of the FliK molecular mechanism, developed from several preceding models (18–21). When FliKN extends out past the tip of a growing (but still short) hook, the FliKN domain folds quickly. The energy of folding facilitates secretion of the molecule through the central channel of the hook. Thus, the rapid secretion from the channel will minimize the probability of FliKC interaction with the FlhB gate, maintaining secretion of the hook. When FliKN remains within the central channel in the structures with longer hooks, FliKC interacts with FlhB and secretion switches to the filament (17).
The present study identified the structure-function determinants for conserved amino acids in FliKC that were predicted to play an important role in the folding of FliKC into a catalytically active domain. We made single-amino-acid substitutions in the conserved residues 296Pro and the neighboring 297Glu-298Glu. Following this, we constructed mutants that have substitutions in residues 292Leu-293Arg-294Leu. Substitutions of residues 296Pro-297Glu-298Glu were wild type, indicating that they were not involved in the switching of secretion modes. In contrast, substitutions of residues 292Leu-293Arg-294Leu gave rise to several polyhook mutants. We conclude that the sequence LXL (X being any amino acid) in loop 2 is essential for conformational switching of secretion modes.
MATERIALS AND METHODS
Strains and growth conditions.
The strains used in this study were derived from Salmonella enterica serovar Typhimurium. Cells were cultured in LB medium (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 1% [wt/vol] NaCl) or TY medium (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract) at either 30°C or 37°C.
Construction of amino acid substitutions in FliK.
The fliK coding region of the chromosome was replaced with a tetRA element using the λ-Red recombinant system (22). Briefly, the λ-Red expression plasmid pKD46, which is temperature sensitive for replication, was introduced into S. enterica serovar Typhimurium SJW1103 and selected on LB plates containing 100 μg/ml ampicillin at 30°C. The tetRA element was amplified by use of primers (Table 1) with KOD FX (Toyobo, Tokyo) and purified with a QIAquick gel extraction kit (Qiagen, Tokyo). To make electrocompetent cells, strains containing pKD46 were grown under inducing conditions in LB medium supplemented with ampicillin and 0.2% arabinose at 30°C until the optical density at 600 nm (OD600) reached 0.6 to 0.8. After treatment by electroporation, 1 ml LB medium was added into the cell-DNA mixture, and the mixture was shaken for 1 h at 30°C. Approximately 0.3 ml of cells was plated on LB containing 15 μg/ml tetracycline and incubated at 30°C. After confirming that the fliK coding region was replaced by the tetRA element and that the cells retained pKD46, the tetRA element was uniformly replaced with mutated fliK DNA fragments. Selection was carried out using tetracycline sensitivity at 37°C (23, 24). Mutated fliK DNA fragments were amplified using a four-primer PCR technique. Where necessary, the mutated region was PCR amplified for DNA sequencing to confirm that the sequence was correct.
TABLE 1.
Primers used in this study
| fliK mutations | Sequenceb |
|
|---|---|---|
| Forward | Reverse | |
| Wild-type fliKa | TGTTAGCTGAAAACCGCATGG | ATCCAAATCGAGCGTTTGCTC |
| E297K,E298K | CGATTGCATCCGAAGAAGTTAGGTCAGGTG | CACCTGACCTAACTTCTTCGGATGCAATCG |
| E297R,E298R | CGATTGCATCCGCGTCGTTTAGGTCAGGTG | CACCTGACCTAAACGACGCGGATGCAATCG |
| E297D,E298D | CGATTGCATCCGGATGATTTAGGTCAGGTG | CACCTGACCTAAATCATCCGGATGCAATCG |
| E297P,E298P | CGATTGCATCCGCCACCATTAGGTCAGGTG | CACCTGACCTAATGGTGGCGGATGCAATCG |
| E297L,E298L | CGATTGCATCCGCTTCTTTTAGGTCAGGTG | CACCTGACCTAAAAGAAGCGGATGCAATCG |
| E297I,E298I | CGATTGCATCCGATTATTTTAGGTCAGGTG | CACCTGACCTAAAATAATCGGATGCAATCG |
| D309K,D310K | TCGCTCAAGCTGAAGAAGAATCAGGCGCAG | CTGCGCCTGATTCTTCTTCAGCTTGAGCGA |
| D309R,D310R | TCGCTCAAGCTGCGTCGTAATCAGGCGCAG | CTGCGCCTGATTACGACGCAGCTTGAGCGA |
| D309V,D310V | TCGCTCAAGCTGGTAGTAAATCAGGCGCAG | CTGCGCCTGATTTACTACCAGCTTGAGCGA |
| D309F,D310F | TCGCTCAAGCTGTTCTTCAATCAGGCGCAG | CTGCGCCTGATTGAAGAACAGCTTGAGCGA |
| D309I,D310I | TCGCTCAAGCTGATTATTAATCAGGCGCAG | CTGCGCCTGATTAATAATCAGCTTGAGCGA |
| L292L,R293L,L294L | AAAGCGCGCAGCTTCTACGTCATCCGGAA | TTCCGGATGACGTAGAAGCTGCGCGCTTT |
| L292L,R293L,L294R | AAAGCGCGCAGCTTCTACGTCATCCGGAA | TTCCGGATGACGTAGAAGCTGCGCGCTTT |
| L292R,R293L,L294L | AAAGCGCGCAGCGACTTCTACATCCGGAA | TTCCGGATGTAGAAGTCGCTGCGCGCTTT |
| L292R,R293L,L294R | AAAGCGCGCAGCTTCGACGTCATCCGGAA | TTCCGGATGACGTCGAAGCTGCGCGCTTT |
| L292L,R293R,L294R | AAAGCGCGCAGCGATTACGTCATCCGGAA | TTCCGGATGACGTAATCGCTGCGCGCTTT |
| L292R,R293L,L294R | AAAGCGCGCAGCGTCGATTACATCCGGAA | TTCCGGATGACGTCGACGCTGCGCGCTTT |
| L292L,R293R,L294R | AAAGCGCGCAGCGTCGACGTCATCCGGAA | TTCCGGATGTAGTAGAAGCTGCGCGCTTT |
| L292L,R293E,L294L | AAAGCGCGCAGCTTGAATTGCATCCGGAA | TTCCGGATGCAATTCAAGCTGCGCGCTTT |
| L292L,R293K,L294L | AAAGCGCGCAGCTTAAGTTGCATCCGGAA | TTCCGGATGCAACTTAAGCTGCGCGCTTT |
| L292L,R293H,L294L | AAAGCGCGCAGCTTCATTTGCATCCGGAA | TTCCGGATGCAAATGAAGCTGCGCGCTTT |
The promoters of fliK are located on fliJ (forward) and downstream fliK (reverse).
Boldface indicates DNA sequences changed for amino acid substitution.
Electron microscopy.
Samples were prepared as previously described (9, 15). Briefly, samples were negatively stained with 1 or 2% (wt/vol) phosphotungstic acid (pH 7.0) and observed with a JEM-1200EXII electron microscope (JEOL, Tokyo, Japan). Micrographs were taken at an accelerating voltage of 80 kV.
Hook length measurement.
To measure hook length, intact flagella were isolated and the hook basal bodies were prepared as previously described (16).
SDS-PAGE.
Proteins secreted into the medium were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described (9, 25, 26). The acrylamide concentration of the gel was 12.5%. Gels were stained with Coomassie brilliant blue (CBB).
Western blot analysis.
Samples were subjected to SDS-PAGE, and protein bands were transferred to polyvinylidene fluoride membranes. Polyclonal anti-FliC or anti-FliK antibodies were used for Western blot analysis (9).
PONDR methods.
Predictor of Naturally Disordered Regions (PONDR) is designed specifically for prediction of protein disorder (http://www.pondr.com). PONDR uses primary sequence data alone for calculation. Its accurate prediction of disordered regions of a protein naturally accompanies accurate prediction of ordered regions of the protein.
RESULTS
Loop 2 is important for FliK interaction with the secretion gate.
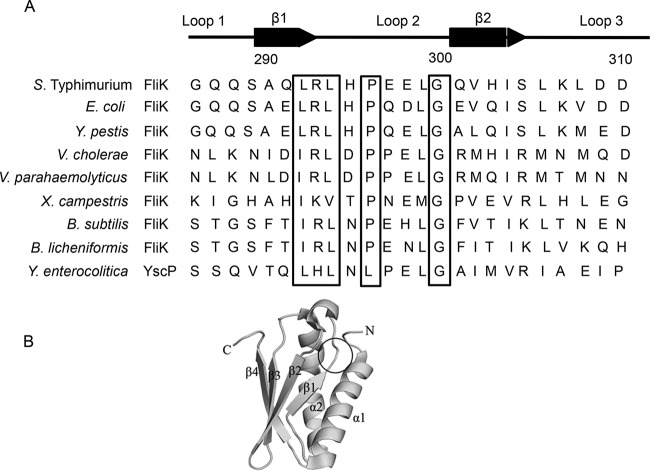
From previous studies, we suspected that the secretion specificity switch was between residues 289 and 298 of FliK. This amino acid sequence is located on loop 2 of the FliKC tertiary structure (10, 15). Alignment of the sequences in the region containing loop 2 in various species shows that several amino acids in loop 2 and its vicinity are well conserved (Fig. 1). Proline at position 296 (296Pro) and glycine at position 300 (300Gly) are commonly found in most species. The alternating arrangements of the hydrophobic and hydrophilic amino acids for residues 292 to 294 may be characteristic of loop 2. To identify the residues of functional importance in loop 2, we replaced the residue at the position of interest with all other amino acids. We then examined the behavior of the mutants on soft agar plates and by light microscopy. Their flagellar structures were also examined by electron microscopy.
FIG 1.
(A) Alignment of amino acid sequences of loop 2 and neighboring regions in FliKC from various species. YscP is an FliK homolog in the Yersinia enterocolitica type three secretion system (TTSS). The numbers marked correspond to the positions for the residues in FliK from S. Typhimurium. Secondary structures and loops are indicated above the amino acid sequence. Highly conserved amino acids in loop 2 are shown in boxes. Accession numbers of FliK homologs are as follows: S. Typhimurium, AAD15264; Escherichia coli, AAC75010; Yersinia pestis, AJK18643; Vibrio cholerae, CSA22037; Vibrio parahaemolyticus, AAD15917; Xanthomonas campestris, CEM58400; Bacillus subtilis, AGG60999; and Yersinia enterocolitica YscP, AAF70341. (B) Three-dimensional structure of FliK. The location of LRL is indicated by a circle. (Adapted from reference 15.)
(i) 296Pro.
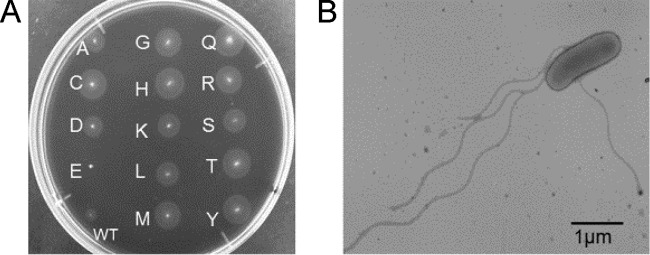
Because 296Pro was at the center of loop 2, we anticipated that it might be essential for loop formation. To test this, we replaced 296Pro with other amino acids (variants P296X) and examined swarming ability on a soft agar plate. All variants but one formed swarm rings as large as or even larger than those of the wild type (Fig. 2A). The one exception was the P296E mutant, which did not swarm (see Discussion). Electron microscopy showed several wild-type flagella on the cells of swarm-forming variants (Fig. 2B). These data indicated that FliK mutants functioned similarly to wild-type FliK. In short, 296Pro was not essential for FliK function.
FIG 2.
Formation of flagella in P269X mutants. (A) Swarm rings formed by P296X variants on a soft agar plate. Letters at spots indicate the substituted amino acid (X) in P269X. (B) Electron microscopic image of P269A, which represents the other P269X variants. Bar, 1 μm.
(ii) 297Glu-298Glu.
Residue 296Pro of FliK is followed by two glutamic acids in tandem, 297Glu-298Glu. We suspected that these charged residues might play a crucial role in interaction with the secretion gate. Thus, we replaced the tandem residues EE with other amino acids that have different physicochemical properties. These were KK, RR, DD, LL, II, and PP. All mutants tested formed swarm rings similar to or larger than those of the wild type (data not shown). Their wild-type flagella were confirmed by electron microscopy. In short, tandem residues 297Glu-298Glu were not essential for FliK function.
(iii) 292Leu-293Arg-294Leu.
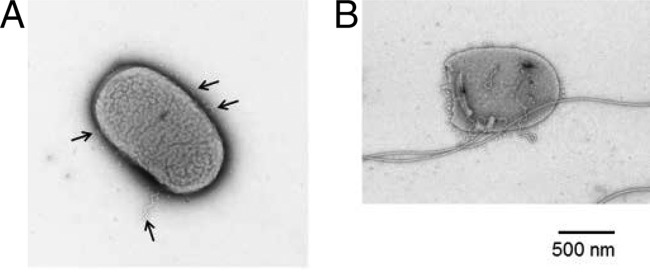
We then analyzed the three residues 292Leu-293Arg-294Leu preceding 296Pro. The sequence LRL is also well conserved among other species as a triplet: hydrophobic amino acid-positively charged amino acid-hydrophobic amino acid. In the construction of variants, we replaced L and R in all possible combinations: LLL, RLL, LRL (wild type), LLR, LRR, RLR, RRL, and RRR. As a result, we discovered two variants, LLL and the wild-type LRL, that exhibited the wild-type phenotype on soft agar plates, while the other variants did not swarm. We confirmed by electron microscopy that LLL and LRL produced wild-type flagella (data not shown).In contrast, the other variants produced either polyhooks or polyhook-filaments. For example, cells expressing LRR possessed several polyhooks per cell (Fig. 3A). We occasionally observed a small percentage of the overall cell population moving in a “jiggly” motion. This small percentage had only one polyhook-filament and several polyhooks per cell (Fig. 3B) (see Discussion). The other FliK variants, LLR, RLL, RLR, and RRR, exhibited numbers of polyhooks and polyhook-filaments similar to those of the LRR mutant. However, the RRL variant displayed as many polyhooks as LRR but produced more filaments (i.e., polyhook-filaments). Overall, 10% of cells had filaments, and we regularly observed a few filaments per cell (Table 2) (see Discussion). These data suggest that mutants with sequences other than LXL could not properly control hook length but were able to switch secretion modes. In short, these variant FliK molecules have lost function, indicating that the sequence LRL is important for FliK function (see Discussion).
FIG 3.
Electron microscopic images of the LRR mutant. (A) The cell possesses only polyhooks (arrows). (B) The cell possesses four polyhooks and one polyhook-filament. Bar, 500 nm.
TABLE 2.
Swimming ability and flagellum formation of LXL variants
| Variant | Flagella | Swimming cells in population (%) | Polyhook-filaments |
|---|---|---|---|
| LRL (wild type) | Wild-type flagella | ∼100 | − |
| LLL | Wild-type flagella | ∼100 | − |
| LEL | Wild-type flagella | ∼100 | − |
| LKL | Wild-type flagella | ∼100 | − |
| LHL | Wild-type flagella | ∼100 | − |
| RRR | Polyhooks | ∼2 | + |
| LRR | Polyhooks | ∼2 | + |
| RRL | Polyhooks | ∼10 | + |
| RLR | Polyhooks | ∼2 | + |
| RLL | Polyhooks | ∼2 | + |
| LLR | Polyhooks | ∼2 | + |
We then constructed mutants by replacing only the middle R with amino acids that have different physicochemical properties, e.g., LEL, LKL, and LHL. These variants were wild type (Table 2), confirming that LXL is essential for FliK function.
Structural stability of the FliKC domain.
Why is LXL critical for FliK function? To address this question, we used the PONDR structural prediction program to examine possible structural changes in FliKC resulting from substitutions of LXL. In the wild type, the segment containing β2 and the downstream loop sequence forms a stable structure (Fig. 4). In Fig. 4, portions above the vertical line of disorder score 0.5 indicate the location of domains and a reasonable stability of the connecting loop. In between α1-β1 and β2-β3-α2-β4 is a disordered gap consisting of loop 2 (Fig. 4, stars). Mutants composed of amino acids other than LXL (bottom row) had a deeper gap between the two domains (Fig. 4, arrows), suggesting a more destabilized FliKC structure. Higher stability at this location is essential for maintaining the β2 structure and consequently that of FliKC. Thus, we conclude that the sequence LXL is essential for stabilizing the entire FliKC structure.
FIG 4.
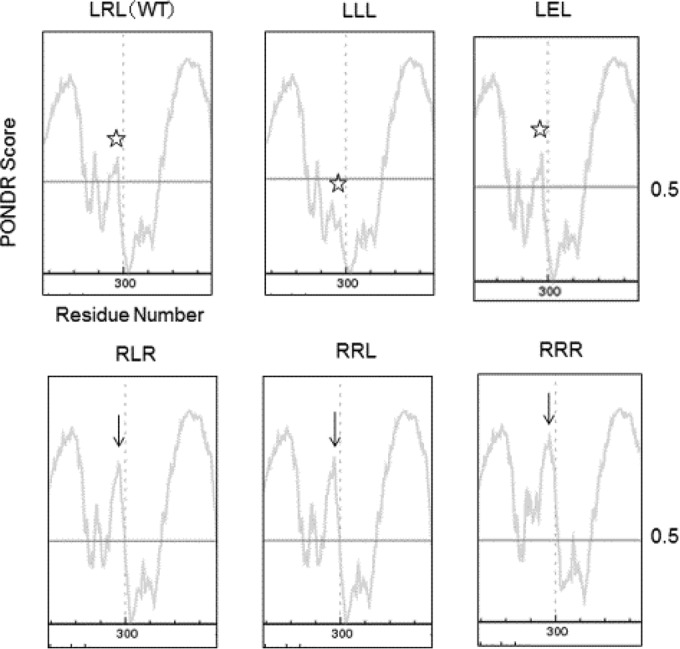
Graphic presentation of the disordered region in FliKC by PONDR prediction. Residues having PONDR scores exceeding a threshold of 0.5 are considered disordered. The disordered score of loop 2 (arrows) is higher in RLR, RRL, and RRR mutants than in LRL, LLL, and LEL (stars).
DISCUSSION
In order to identify the region essential for interaction with the FhlB secretion gate, we made single-amino-acid substitutions of several residues in loop 2. Most of the substitutions did not change FliK function. Only sequences that destroyed the LXL motif gave rise to polyhook mutants, emphasizing the essential role of the LXL sequence. It should be noted that our site-directed single-amino-acid substitutions were constructed in the chromosomal fliK locus to avoid plasmid overexpression artifacts (12).
Among variants P296X, only P296E mutant cells did not form swarm rings on soft agar plates. We further analyzed the P296E mutant and discovered that the mutant had a deletion rather than a single amino acid substitution. In the electron microscope, the deletion mutant produced polyhooks. Furthermore, the mutant was shorter than wild-type FliK as revealed by Western blotting (data not shown).
In the multiply substituted FliK (except LXL), there were swimming cells present. By searching these swimming cells by microscopy and surveying the number of filaments by electron microscopy at a low magnification, we estimated rates of swimming cells among the population. The variants LRR, LLR, RLL, and RLR showed that less than 2% of the cell population had just one filament. Polyhooks without filaments were observed more often (4×) than those with filaments present. It is known that the fliK knockout mutant exhibits a leaky motility phenotype, suggesting that the FlhB secretion gate can spontaneously undergo a conformational change in the absence of FliK. Thus, the fliK knockout mutant cells produce not only polyhooks but also polyhook-filaments, albeit very few (0.4%) (27). The rate of appearance of polyhook-filaments in the mutants mentioned above was similar to the rate of appearance of polyhook-filaments in the fliK knockout mutant. We conclude that these FliK mutants are not functional. In contrast, the RRL mutant produced swimming cells that were more than 10% of the cell population and accordingly formed more polyhook-filaments than the former two. We do not know why only the variant RRL has more filaments than RRR, LRR, RLR, RLL, and LLR. These data indicate that the variants other than LXL possess FliKs with either deteriorated or no function. In the future, we will attempt to directly observe the molecular shape of variant FliK molecules.
Polyhook mutants in previous studies were mostly deletion mutants (8, 10–13). There are only two examples of single-amino-acid substitutions that resulted in polyhooks: S319Y and R336G (11). Residue 319Ser is located at the end of β3 followed by loop 4 and is structurally close to the LRL region. Minamino et al. (13) isolated suppressor mutants of S319Y: four were intragenic suppressors in FliKC, and one intergenic suppressor was in FlhB. Among the four intragenic suppressors, three (M317I, P320S, and P320Q) were neighbors of the original mutation (S319Y) and one (P296L) was on loop 2 (13), further supporting our view that the sequence LRL is the site of interaction with the FhlB secretion gate. The residue 336Arg is located in the middle ofα2. How this residue affects FliK interaction with the secretion gate is presently not known.
ACKNOWLEDGMENTS
We thank Shino Mizuno for technical assistance in the early stages of the study, Shogo Nakano for drawing the FliK three-dimensional structure, and Tomoko Kubori for encouragement. We also thank Linda Kenney for correcting the English.
REFERENCES
- 1.Katsura I. 1987. Determination of bacteriophage lambda tail length by a protein ruler. Nature 327:73–75. doi: 10.1038/327073a0. [DOI] [PubMed] [Google Scholar]
- 2.Labeit S, Gibson T, Lakey A, Leonard K, Zeviani M, Knight P, Wardale J, Trinick J. 1991. Evidence that nebulin is a protein-ruler in muscle thin filaments. FEBS Lett 282:313–316. doi: 10.1016/0014-5793(91)80503-U. [DOI] [PubMed] [Google Scholar]
- 3.Hughes KT. 2012. Flagellar hook length is controlled by a secreted molecular ruler. J Bacteriol 194:4793–4796. doi: 10.1128/JB.00343-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes KT. 2012. Rebuttal: mystery of FliK in length control of the flagellar Hook. J Bacteriol 194:4801. doi: 10.1128/JB.06454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aizawa SI. 2012. Mystery of FliK in length control of the flagellar hook. J Bacteriol 194:4798–4800. doi: 10.1128/JB.06239-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aizawa SI. 2012. Rebuttal: flagellar hook length is controlled by a secreted molecular ruler. J Bacteriol 194:4797. doi: 10.1128/JB.06333-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aizawa S-I. 2009. Flagella, p 393–403. In Schaechter M. (ed), Encyclopedia of microbiology. Elsevier, Oxford, United Kingdom. [Google Scholar]
- 8.Hirano T, Yamaguchi S, Oosawa K, Aizawa SI. 1994. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J Bacteriol 176:5439–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uchida K, Dono K, Aizawa SI. 2013. Length control of the flagellar hook in a temperature-sensitive flgE mutant of Salmonella enterica serovar Typhimurium. J Bacteriol 195:3590–3595. doi: 10.1128/JB.00302-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shibata S, Takahashi N, Chevance FF, Karlinsey JE, Hughes KT, Aizawa SI. 2007. FliK regulates flagellar hook length as an internal ruler. Mol Microbiol 64:1404–1415. doi: 10.1111/j.1365-2958.2007.05750.x. [DOI] [PubMed] [Google Scholar]
- 11.Williams AW, Yamaguchi S, Togashi F, Aizawa SI, Kawagishi I, Macnab RM. 1996. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J Bacteriol 178:2960–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minamino T, Saijo-Hamano Y, Furukawa Y, González-Pedrajo B, Macnab RM, Namba K. 2004. Domain organization and function of Salmonella FliK, a flagellar hook-length control protein. J Mol Biol 341:491–502. doi: 10.1016/j.jmb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 13.Minamino T, Ferris HU, Morioya N, Kihara M, Namba K. 2006. Two parts of the T3S4 domain of the hook-length control protein FliK are essential for the substrate specificity switching of the flagellar type III export apparatus. J Mol Biol 362:1148–1158. doi: 10.1016/j.jmb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Hirano T, Shibata S, Ohnishi K, Tani T, Aizawa SI. 2005. N-terminal signal region of FliK is dispensable for length control of the flagellar hook. Mol Microbiol 56:346–360. doi: 10.1111/j.1365-2958.2005.04615.x. [DOI] [PubMed] [Google Scholar]
- 15.Mizuno S, Amida H, Kobayashi N, Aizawa SI, Tate S. 2011. The NMR structure of FliK, the trigger for the switch of substrate specificity in the flagellar type III secretion apparatus. J Mol Biol 409:558–573. doi: 10.1016/j.jmb.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 16.Uchida K, Aizawa SI. 2014. The flagellar soluble protein FliK determines the minimal length of the hook in Salmonella enterica serovar Typhimurium. J Bacteriol 196:1753–1758. doi: 10.1128/JB.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kodera N, Uchida K, Ando T, Aizawa SI. 2015. Two-ball structure of the flagellar hook-length control protein FliK as revealed by high-speed atomic force microscopy. J Mol Biol 427:406–414. doi: 10.1016/j.jmb.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Keener JP. 2005. A model for length control of flagellar hooks of Salmonella typhimurium. J Theor Biol 234:263–275. doi: 10.1016/j.jtbi.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 19.Keener JP. 2010. A molecular ruler mechanism for length control of extended protein structures in bacteria. J Theor Biol 263:481–489. doi: 10.1016/j.jtbi.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 20.Evans LD, Poulter S, Terentjev EM, Hughes C, Fraser GM. 2013. A chain mechanism for flagellum growth. Nature 504:287–290. doi: 10.1038/nature12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erhardt M, Singer HM, Wee DH, Keener JP, Hughes KT. 2011. An infrequent molecular ruler controls flagellar hook length in Salmonella enterica. EMBO J 30:2948–2961. doi: 10.1038/emboj.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bochner BR, Huang HC, Schieven GL, Ames BN. 1980. Positive selection for loss of tetracycline resistance. J Bacteriol 143:926–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maloy SR, Nunn WD. 1981. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol 145:1110–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komoriya K, Shibano N, Higano T, Azuma N, Yamaguchi S, Aizawa SI. 1999. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol Microbiol 34:767–779. doi: 10.1046/j.1365-2958.1999.01639.x. [DOI] [PubMed] [Google Scholar]
- 26.Mizusaki H, Takaya A, Yamamoto T, Aizawa SI. 2008. Signal pathway in the salt-activated expression of the SPI1/ type III secretion system in Salmonella enterica serovar Typhimurium. J Bacteriol 190:4624–4631. doi: 10.1128/JB.01957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aizawa SI. 2013. Flagella. In Schaechter M. (ed), Brennan's online encyclopedia of genetics, 2nd ed. Elsevier, Academic Press, Oxford, United Kingdom. [Google Scholar]