ABSTRACT
Sinorhizobium meliloti undergoes major lifestyle changes between planktonic states, biofilm formation, and symbiosis with leguminous plant hosts. In many bacteria, the second messenger 3′,5′-cyclic di-GMP (c-di-GMP, or cdG) promotes a sessile lifestyle by regulating a plethora of processes involved in biofilm formation, including motility and biosynthesis of exopolysaccharides (EPS). Here, we systematically investigated the role of cdG in S. meliloti Rm2011 encoding 22 proteins putatively associated with cdG synthesis, degradation, or binding. Single mutations in 21 of these genes did not cause evident changes in biofilm formation, motility, or EPS biosynthesis. In contrast, manipulation of cdG levels by overproducing endogenous or heterologous diguanylate cyclases (DGCs) or phosphodiesterases (PDEs) affected these processes and accumulation of N-Acyl-homoserine lactones in the culture supernatant. Specifically, individual overexpression of the S. meliloti genes pleD, SMb20523, SMb20447, SMc01464, and SMc03178 encoding putative DGCs and of SMb21517 encoding a single-domain PDE protein had an impact and resulted in increased levels of cdG. Compared to the wild type, an S. meliloti strain that did not produce detectable levels of cdG (cdG0) was more sensitive to acid stress. However, it was symbiotically potent, unaffected in motility, and only slightly reduced in biofilm formation. The SMc01790-SMc01796 locus, homologous to the Agrobacterium tumefaciens uppABCDEF cluster governing biosynthesis of a unipolarly localized polysaccharide, was found to be required for cdG-stimulated biofilm formation, while the single-domain PilZ protein McrA was identified as a cdG receptor protein involved in regulation of motility.
IMPORTANCE We present the first systematic genome-wide investigation of the role of 3′,5′-cyclic di-GMP (c-di-GMP, or cdG) in regulation of motility, biosynthesis of exopolysaccharides, biofilm formation, quorum sensing, and symbiosis in a symbiotic alpha-rhizobial species. Phenotypes of an S. meliloti strain unable to produce cdG (cdG0) demonstrated that this second messenger is not essential for root nodule symbiosis but may contribute to acid tolerance. Our data further suggest that enhanced levels of cdG promote sessility of S. meliloti and uncovered a single-domain PilZ protein as regulator of motility.
INTRODUCTION
Alpha-rhizobia are soil-dwelling alphaproteobacteria existing either in free-living states or in symbiosis with a leguminous plant host. Motile rhizobia undergo a major switch in lifestyle when establishing a biofilm or symbiosis with the host. In the symbiotic state, the bacteria inhabit root nodules and differentiate into polyploid bacteroids that fix nitrogen to the benefit of the host (1).
3′,5′-Cyclic di-GMP (c-di-GMP, or cdG) is a common second messenger in the bacterial kingdom that is known to promote the transition from a planktonic, motile to a sessile, frequently biofilm-associated, lifestyle (2, 3). cdG levels are controlled by diguanylate cyclases (DGCs) that synthesize cdG from two GTP molecules and by phosphodiesterases (PDEs) that degrade it (4). For the synthetic reaction, DGCs require an active site with a conserved GG(D/E)EF motif embedded in a GGDEF domain that can be inhibited by binding of cdG to a primary inhibition site (I-site) containing an RXXD motif. PDEs contain either an EAL or HD-GYP domain, which cleaves cdG to pGpG or two molecules of GMP, respectively. DGC and PDE domains are often coupled to diverse sensory input domains which regulate the enzymatic activities upon perception of environmental stimuli (3). cdG levels are sensed by effectors, such as PilZ- or GIL-domain-containing proteins, degenerate GGDEF domains, or riboswitches (5–9). Binding of cdG to PilZ domains involves RXXXR and DXSXXG motifs, whereas the I-site-like motif RXGD is required for its binding to GIL domains.
High intracellular cdG concentrations favor production of exopolysaccharides (EPS), fimbriae, pili, and adhesins that contribute to biofilm formation (3). Transition to a sessile lifestyle also includes inhibition of flagellar motility by cdG (10). In Escherichia coli, Salmonella enterica serovar Typhimurium, and Caulobacter crescentus, this regulation involves PilZ domain proteins (11, 12). A variety of other cellular functions, not directly involved in the switch between motile and sessile states, are also subject to regulation by cdG. Examples are virulence-associated processes in Vibrio, Yersinia, Xanthomonas, Salmonella, and Pseudomonas (13–17), quorum sensing (QS) in Vibrio and Xanthomonas (18, 19), and cell cycle control and cell differentiation in C. crescentus (20, 21).
While experimental manipulation of cdG levels indicated a role of cdG in regulation of the sessile-motile switch in alpha-rhizobia as well, to date only a few cdG-related genes have been studied in these symbiotic bacteria. In Rhizobium etli and Rhizobium leguminosarum, overproduction of a heterologous DGC enhanced EPS production, biofilm formation, and adhesion to plant roots and decreased symbiotic efficiency and swimming motility (22). In Sinorhizobium meliloti 102F34, overproduction of the E. coli cdG-binding protein BdcA resulted in biofilm dispersal and enhanced motility, presumably by lowering intracellular cdG levels (23). An initial incomplete screen of S. meliloti Rm1021 single-gene mutants in cdG-related genes identified 11 out of 14 tested genes as weakly affecting growth rate, motility, EPS production, and nodule occupancy (24). Recently, enhanced levels of cdG were shown to induce production of a novel S. meliloti Rm8530 mixed-linkage β-glucan, supposedly by binding of cdG to the C-terminal portion of the membrane-bound glycosyltransferase BgsA (25). So far, experimental hints for enzymatic activities of suggested DGCs and PDEs were reported only for two hybrid GGDEF-EAL proteins from R. etli (26).
Computational screening of alpha-rhizobial genomes identified a high number of cdG-related genes, with the highest number of 55 genes in Bradyrhizobium japonicum (27). The S. meliloti Rm1021 type strain (28) encodes 20 proteins containing either a GGDEF or EAL domain or both and two PilZ domain proteins (26, 27). In contrast to the presence of HD-GYP-domain-encoding genes in Rhizobium, Mesorhizobium, and Bradyrhizobium species, such genes were not found in sequenced Sinorhizobium genomes (27). In this study, we systematically explored the role of 22 cdG-related genes in swimming motility, EPS production, biofilm formation, and symbiotic efficiency of S. meliloti Rm2011. A mutant with deletions of 16 genes encoding GGDEF-domain-containing proteins did not produce detectable levels of cdG and was symbiotically potent. We report on genetic factors responsible for a cdG-mediated increase in biofilm formation and inhibition of motility in S. meliloti.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. This study was performed with S. meliloti Rm2011 (https://iant.toulouse.inra.fr/bacteria/annotation/cgi/rhime2011/rhime2011.cgi) which is very closely related to the Rm1021 type strain since both strains are streptomycin-resistant spontaneous derivatives of S. meliloti SU47 (29, 30). S. meliloti was grown at 30°C in tryptone-yeast extract (TY) medium (31), LB medium (32), modified yeast extract-mannitol (YM) medium (33), modified morpholinepropanesulfonic acid (MOPS)-buffered minimal medium (MM) (34), and nutrient-depleted 30% MM (nitrogen, carbon, and phosphate sources reduced to 30%). Phosphate-limiting MM contained 0.1 mM K2HPO4. Agrobacterium tumefaciens was grown in minimal glutamate mannitol (MGM) medium. Medium composition and antibiotic concentrations are provided in the supplemental material. Unless otherwise specified, isopropyl β-d-1-thiogalactopyranoside (IPTG) was added at 500 μM. Growth curves shown in Fig. S4 in the supplemental material were recorded using an Infinite F200 PRO fluorescence reader (Tecan) in 100-μl cultures in a flat-bottom 96-well plate incubated at 30°C with shaking.
Construction of strains and plasmids.
Constructs used in this work were generated using standard cloning techniques. The primers used are listed in Table S2 in the supplemental material. All constructs were verified by sequencing. Plasmids were transferred to S. meliloti by E. coli S17-1-mediated conjugation (35) as previously described (36).
For generation of single knockouts by plasmid integration, 280- to 385-bp internal gene fragments were cloned into suicide vector pK19mob2ΩHMB. Integration of the resulting constructs into the S. meliloti genome resulted in truncation of the target gene and inactivation of the downstream part of the transcription unit. Correct plasmid integrations were verified by PCR.
For construction of deletion mutants, gene-flanking regions of 650 to 770 bp were cloned into suicide plasmid pK18mobsacB. Resulting constructs were integrated into the S. meliloti genome, and transconjugants were subjected to sucrose selection as previously described (37). Gene deletions were verified by PCR. Multiple mutants were generated by sequential gene deletions. The genomes of Rm2011 ΔXVI and Rm2011 expR+ ΔXVI (strains with deletions of 16 of the 17 GGDEF-domain-encoding genes in Rm2011) as well as their parental strains Rm2011 and Rm2011 expR+ were resequenced. Total DNA for resequencing was purified using a DNeasy blood and tissue kit (Qiagen). DNA libraries for sequencing were generated by applying a Nextera XT DNA Library Preparation kit (Illumina), and sequencing was performed on a MiSeq Desktop Sequencer (Illumina) using a MiSeq reagent kit, version 2, for 2 × 250-bp paired-end reads (Illumina). The following numbers of reads were obtained: Rm2011, 4.5 × 106; Rm2011 ΔXVI, 4.8 × 106; Rm2011 expR+, 3.2 × 106; and Rm2011 expR+ ΔXVI, 4.4 × 106. After quality trimming, the reads were mapped with Bowtie2 (38) using the “very sensitive” preset and returning up to five matches per read (-k 5). Single-nucleotide polymorphism (SNP) detection was performed by applying the ReadXplorer software, version 2.1 (39), requiring 90% of variation, a minimum mismatch coverage of 15, and including only the single perfect match and single best match mapping classes.
To generate C-terminal translation fusions to enhanced green fluorescent protein (EGFP) at the native genomic location, about 700 bp of the 3′ portion of the coding region excluding the stop codon was cloned into suicide plasmid pK18mob2-EGFP, and resulting constructs were integrated into the S. meliloti genome.
The promoter-EGFP fusions were generated by insertion of approximately 300-bp fragments including the region upstream of the start codon and up to 10 first codons of the gene into the replicative low-copy-number plasmid pPHU231-EGFP or replicative medium-copy-number plasmid pSRKKm-EGFP. This generated an in-frame fusion of these first codons of the gene of interest to egfp.
Gene overexpression constructs were generated by insertion of the full-length coding sequence downstream of the IPTG-inducible T5 promoter and a Shine-Dalgarno sequence into the replicative medium-copy-number vector pWBT. For combined overexpression of two genes, the coding regions were inserted in tandem downstream of the T5 promoter in pWBT, with each coding region preceded by an identical Shine-Dalgarno sequence.
Constructs for production of His6-tagged McrA, McrA with R9A and R13A substitutions in the RXXXR motif (McrAAXXXA), and SMc00074 consisting of residues 390 to 970 (SMc00074390–970) were generated by insertion of the coding sequence into expression vector pWH844. For generation of the AXXXA substitutions encoded by mcrA (mcrAAXXXA) splicing by overlap extension PCR was applied.
Phenotype assays.
In phenotype assays of strains carrying pWBT-based constructs, gentamicin and IPTG (500 μM) were added to the medium to maintain the plasmid and to induce gene expression. Mucoidity was estimated on phosphate-limiting MM agar. Congo red staining was assayed on TY agar with 70 mg/liter Congo red. Calcofluor brightness was analyzed on LB agar with 200 mg/liter calcofluor. Fresh precultures grown on TY agar were resuspended in 0.9% NaCl to an optical density at 600 nm (OD600) of 1 (strains carrying pWBT-based overexpression constructs) or OD600 of 0.1 (all the other strains), and 50 μl was dropped onto the agar medium. Plates were incubated at 30°C and documented after 2 to 3 days.
Swimming motility was analyzed as follows: for each strain, 2 μl of stationary TY culture (similar growth confirmed by determination of OD600) was spotted onto three separate petri dishes containing 20% TY agar, with a final agar concentration of 0.3%, and dried for 5 min. Plates were incubated at 30°C and documented after 3 days. To quantify swimming motility, sizes of swimming halos of three replicates were measured using image software GIMP, version 2.
Biofilm formation was determined in flat-bottom polystyrene 96-well plates (Greiner) in 30% MM, in triplicates, as previously described (40). A preculture in the same medium was diluted to an OD600 of 0.2, with a final volume 100 μl, and grown without shaking for 48 h at 30°C. Growth was recorded by determining the OD600. The culture medium and nonattached cells were removed. Plate wells were washed with water, stained with 200 μl of 0.1% crystal violet solution for 30 min with gentle agitation, and washed twice with water. Stained attached cell material was dissolved in 200 μl of detaching solution (80% acetone–20% ethanol) for 20 min upon shaking. Biofilm formation was determined by quantification of crystal violet-bound attached cell material at the A570 and normalized to the OD600 of the culture.
Fluorescence measurements.
For promoter-EGFP assays, TY precultures were diluted 1:500 in 100 μl of TY medium or 30% MM supplemented with 100 μM IPTG and grown in 96-well plates at 30°C with shaking. EGFP fluorescence (excitation wavelength [λex] of 488 ± 9 nm; emission wavelength [λem] of 522 ± 20 nm; gain, 82) and growth (OD600) were recorded using an Infinite 200 Pro multimode reader (Tecan). Strains carrying the empty vector pPHU-EGFP were used for measuring background fluorescence. Relative fluorescence units (RFU), calculated as EGFP signals, were normalized to the OD600 (n = 2 to 3).
For determination of the emission ratio of yellow fluorescent protein for energy transfer/cyan fluorescent protein for energy transfer (YPet/CyPet), Rm2011 carrying the fluorescent resonance energy transfer (FRET) CyPet/YPet expression constructs and either the empty vector pSRKKm or pleD expression plasmid pSRKKm-PT5-pleD was used. Cells were grown in triplicates in 100 μl of MM with 500 μM IPTG in 96-well plates at 30°C with shaking. Fluorescence was determined using a Tecan Infinite 200 Pro multimode reader (λex of 425 ± 9 nm, λem1 of 476 ± 20 nm, λem2 of 526 ± 20 nm; gain, 82) and normalized to OD600.
Microscopy.
Microscopy of bacteria on 1% agarose pads was performed using a Nikon microscope Eclipse Ti-E with a differential interference contrast (DIC) CFI Apochromat TIRF oil objective (100×; numerical aperture of 1.49) and phase-contrast Plan Apo l oil objective (100×; numerical aperture, 1.45) with AHF HC filter sets F36-504 Texas Red (TxRed) (excitation band pass [ex bp] 562/40-nm, beam splitter [bs] 593-nm, and emission [em] bp 624/40-nm filters) and F36-525 EGFP (ex bp 472/30-nm, bs 495-nm, and em bp 520/35-nm filters). Images were acquired with an Andor iXon3 885 electron-multiplying charge-coupled-device (EMCCD) camera. For time-lapse analysis, MM agarose pads supplemented with 200 μM IPTG were used, and images were acquired every 20 min at 30°C.
Staining with Alexa Fluor 594-conjugated wheat germ agglutinin (WGA) (Life Technologies) was performed as described by Xu et al. (41) with modifications. One microliter of fluorescein (fl)-WGA stock solution (1 mg/ml) was added to 100-μl of stationary MM culture (500 μM IPTG), and the mixture was incubated for 20 min at room temperature (RT) and centrifuged at 6,000 × g. The pellets were washed twice with 1× phosphate-buffered saline (PBS) buffer. Bacteria were analyzed by microscopy using phase-contrast and TxRed settings.
Quantification of intracellular cdG.
Quantification of intracellular cdG of bacterial cells was performed as previously described (42). Briefly, nucleotides were extracted from cell pellets with 40% acetonitrile, 40% methanol, and 20% water; samples were dried and subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS). cdG was normalized to total protein, determined using Bradford reagent. Strains overproducing selected DGC/PDEs were grown in TY medium and induced with IPTG (500 μM) at an OD600 of 0.2 and harvested after 8 h of induction. Growth rates of the wild type were determined by measuring the OD600 in 20-min intervals for 80 min around the OD600 of harvest.
AHL detection assay.
Two replicate S. meliloti TY cultures were induced with 100 μM IPTG at an OD600 of 0.05 and grown for 24 h. Cultures were centrifuged for 10 min at 15,000 × g. Twenty microliters of supernatant was applied to an N-acyl-homoserine lactone (AHL) detection assay using A. tumefaciens NTL4(pZLR4) that was performed as previously described (36).
Protein purification.
Overnight cultures of E. coli M15/pREP4 strains carrying pWH844-SMc00507, pWH844-SMc00507-R9A/R13A, or pWH844-SMc00074390–970 were used to inoculate LB medium at an OD600 of 0.1 in flasks. At the OD600 of 0.6, 400 μM IPTG was added, and the cultures were further incubated with shaking at 140 rpm overnight at RT. Cultures were harvested by centrifugation (4,000 × g), resuspended in binding buffer (1.76 g/liter Na2HPO4 · 2H2O, 1.4 g/liter NaH2PO4 · H2O, 29.2 g/liter NaCl, 20 mM imidazole, 2 mM phenylmethylsulfonyl fluoride [PMSF], pH 7.4), and lysed using a French press (applied pressure, 1,000 lb/in2). After centrifugation for 50 min at 15,000 rpm at 4°C, supernatants were applied to His SpinTrap columns (GE Healthcare) and washed with binding buffer containing 100 mM imidazole. Proteins were eluted with 500 mM imidazole. Purity of isolated proteins was assessed by SDS-PAGE and Coomassie brilliant blue staining. Protein concentration was determined using Bradford reagent.
In vitro cdG binding assay.
cdG binding was assayed using a differential radial capillary action of ligand assay (DRaCALA) with purified protein and 32P-labeled c-di-GMP (6, 43). 32P-labeled c-di-GMP was prepared by incubating 10 μM His6-DgcA (C. crescentus) with 1 mM GTP/[α-32P]GTP (0.1 μCi/μl) in reaction buffer (50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 10 mM MgCl2) overnight at 30°C. The reaction mixture was then incubated with 5 units of calf intestine alkaline phosphatase (Fermentas) for 1 h at 22°C to hydrolyze unreacted GTP. The reaction was stopped by incubation for 10 min at 95°C followed by centrifugation (10 min, 20,000 × g, 20°C). The supernatant was used for the cdG binding assay. 32P-labeled cdG was mixed with a 20 μM concentration of His6-McrA or His6-McrAAXXXA and incubated for 10 min at RT in binding buffer (10 mM Tris, pH 8.0, 100 mM NaCl, 5 mM MgCl2). Ten microliters of this reaction mixture was transferred to a nitrocellulose membrane and allowed to dry prior to being exposed to a phosphorimaging screen (Molecular Dynamics). Data were collected and analyzed using a Storm 840 scanner (Amersham Biosciences) and Image Quant, version 5.2, software. For competition experiments, 0.4 mM unlabeled cdG (Biolog) or GTP (Sigma) was added.
In vitro DGC activity assay.
DGC activity was determined essentially as described previously (44). Briefly, assays were performed with 10 μM purified proteins (final concentration) in a final volume of 40 μl. Reaction mixtures were preincubated for 5 min at 30°C in reaction buffer (50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 10 mM MgCl2). DGC reactions were initiated by the addition of 1 mM GTP/[α-32P]GTP (0.1 μCi/μl) (Hartman Analytic) and incubated at 30°C for 0, 10, 20, 45, or 60 min. Reactions were then stopped by the addition of 1 volume of 0.5 M EDTA. Reaction products were analyzed by polyethyleneimine-cellulose thin-layer chromatography (TLC). Two-microliter aliquots were spotted onto TLC plates (Millipore), dried, and developed in 2:3 (vol/vol) 4 M (NH4)2SO4–1.5 M KH2PO4 (pH 3.65). Plates were dried prior to exposure to a phosphorimaging screen (Molecular Dynamics). Data were collected and analyzed using a Storm 840 scanner (Amersham Biosciences) and Image Quant, version 5.2, software.
Plant assays.
Medicago sativa cv. Eugenia seeds were surface sterilized by 95% sulfuric acid for 10 min, washed eight times with sterile water, and transferred to plant agar (45). The seeds were germinated in the dark overnight at 4°C and for 24 h at 30°C. Bacterial overnight TY cultures were diluted 1:10 with sterile 0.9% NaCl, and 100 μl of the suspensions was plated onto the lower half of plant agar plates (triplicates). For competition assays, test and control strains, of which only one carried a chromosomally integrated gentamicin resistance gene, were mixed 1:1. Four seedlings were placed onto each plate, and plants were incubated at 22°C and 90% humidity in an 18-h/6-h day/night rhythm for 4 weeks. To estimate nodulation kinetics, emerging nodules were counted every day. To analyze competitive nodulation, nodules were surface sterilized with 70% ethanol, washed three times with sterile water, and crushed. Different dilutions of these extracts were spread on TY agar containing either streptomycin or streptomycin and gentamicin. Nodule occupancy was determined by counting CFU of gentamicin-sensitive and -resistant strains. Nodules producing no gentamicin-resistant CFU were considered occupied by the gentamicin-sensitive strain, those with up to 60% gentamicin-resistant CFU were considered to have mixed occupancy, and those with more than 60% gentamicin-resistant CFU were considered to have predominant occupancy by the gentamicin-resistant strain.
RESULTS
Cellular cdG levels are higher in exponentially growing than in stationary-phase S. meliloti cells.
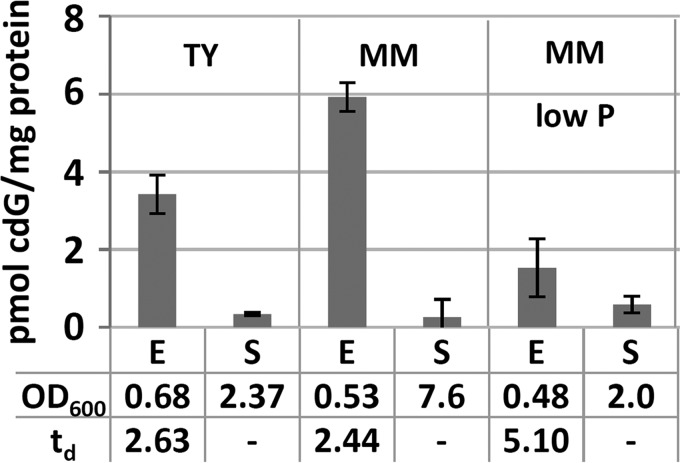
To determine conditions of cdG accumulation in S. meliloti Rm2011, intracellular cdG concentrations were determined in complex TY medium and MOPS-buffered phosphate-sufficient or phosphate-limiting minimal medium (MM) (Fig. 1). In all media, the cdG content of exponentially growing cells was 10- to 30-fold higher than that of cells in stationary phase. Moreover, intracellular cdG levels of exponential-phase cells approximately inversely correlated with growth rate. Cells growing in phosphate-sufficient MM or TY medium had about 2-fold shorter doubling times and 2- to 4-fold higher cdG levels than phosphate-limited cells.
FIG 1.
Quantification of cdG in Rm2011 grown in liquid TY and MM cultures. E, exponential growth phase; S, stationary growth phase; td, doubling time in hours. Error bars represent standard deviations of three biological replicates.
Individual or multiple knockout mutations in DCG- and PDE-encoding genes have no or only weak effects on motility and biofilm formation.
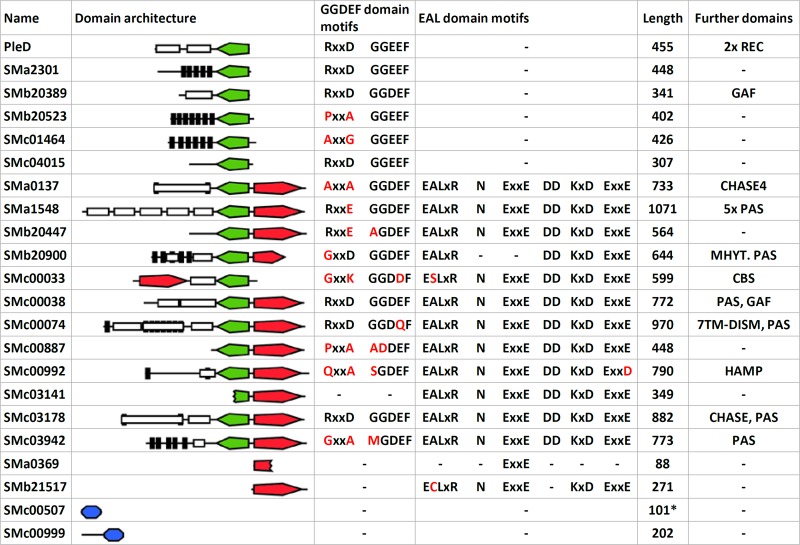
Fig. 2 shows an overview of the domain architecture of cdG-related proteins in S. meliloti Rm1021. Six proteins contain a GGDEF domain, 1 contains an EAL domain, and 11 contain both domains. SMa0369 codes for an internal fragment of an EAL domain, while the neighboring SMc03142 and SMc03141 encode the N-terminal and C-terminal parts of a putative bifunctional protein due to truncation of SMc03142 by a nonsense mutation at codon 196. Sequencing of the corresponding genome region confirmed this mutation in the Rm2011 genome. Eleven of the 17 GGDEF domain proteins contain a canonical GGDEF motif, whereas an RXXD I-site motif was found in seven proteins.
FIG 2.
Overview of S. meliloti Rm2011 cdG-related genes. Green, GGDEF domain; red, EAL domain; blue, PilZ domain. Differences from consensus sequence motifs are labeled in red. Domain architectures are drawn to scale. Black bars, predicted transmembrane helices; white boxes, other domains; length, protein length in amino acids; *, corrected based on experimental validation.
For phenotypic characterization, knockout mutants in 19 of the 20 putative DGC and/or PDE-encoding genes (see Fig. S1 in the supplemental material) were generated in S. meliloti Rm2011, either by plasmid integration (18 genes) or by deletion (SMa0369). We failed to knock out SMc00074, in accordance with Cowie et al. (46), who identified this gene as potentially essential. The mutants were characterized for production of EPS, swimming motility, and the ability to form a biofilm on an abiotic surface (see Fig. S1) and to establish symbiosis with the host plant Medicago sativa. S. meliloti produces two main EPS, succinoglycan (EPS I) and galactoglucan (EPS II) (47, 48). On rich medium, Rm2011 predominantly produces EPS I, while EPS II production is promoted by phosphate limitation (34, 49). To evaluate production of EPS by cells grown on rich medium, we applied staining with calcofluor, a fluorescent dye binding to EPS I and, to a lesser extent, to other EPS, and staining with Congo red binding to cellulose and amyloid compounds. Furthermore, mucoidy on phosphate-limiting MM was assessed.
Compared to the wild type, the mutants did not show apparent differences in EPS production or in their ability to swim on semisolid agar (see Fig. S1 in the supplemental material). Under the conditions tested, most of the mutants also appeared to be unaffected in biofilm formation. Only SMb20447, SMc00887, and SMc03942 mutants showed a 1.5- to 1.8-fold increase, while pleD, SMb20523, SMc01464, and SMc03178 mutants exhibited a 1.4- to 1.5-fold decrease in cell material attached to polystyrene. None of the single-gene mutants was impaired in symbiosis. Plants inoculated with the single mutants formed pink, nitrogen-fixing root nodules and appeared healthy, indistinguishable from plants inoculated with the wild type (data not shown).
Functional redundancy or compensatory effects may be responsible for the absence of strong phenotypes of the single-gene mutants. We therefore constructed mutants with multiple mutations in S. meliloti Rm2011 and its expR-sufficient derivative Sm2B3001, here referred to as Rm2011 expR+ (50). The latter was included in this study because Rm2011 carries a naturally occurring mutation in expR encoding the LuxR-type master transcription factor of QS regulation (51). Intracellular cdG levels of exponentially growing Rm2011 (4.25 ± 0.36 pmol/mg protein) and Rm2011 expR+ (4.50 ± 0.11 pmol/mg protein) in TY medium were similar, implying that under these conditions cdG content was not influenced by expR.
Starting with deletion of pleD, we sequentially deleted 16 of the 17 GGDEF-domain-encoding genes in Rm2011 and Rm2011 expR+, resulting in Rm2011 ΔXVI and Rm2011 expR+ ΔXVI, respectively (see Table S1 in the supplemental material). Since SMc00074 could not be deleted, the GGDEF-EAL-domain-containing portion of the encoded protein heterologously produced in E. coli was assayed for DGC activity (see Fig. S2). In agreement with the noncanonical GGDQF motif in the predicted GGDEF domain of SMc00074 (Fig. 2), DGC activity was not detected, whereas the positive-control protein DgcA from C. crescentus showed DGC activity. Exponentially growing Rm2011 ΔXVI and Rm2011 expR+ ΔXVI cells did not detectably produce cdG as assessed by our mass spectrometry-based assay; thus, these strains were considered cdG0. Genome resequencing of the S. meliloti cdG0 strains confirmed deletion of the 16 targeted genes and did not identify potential suppressor mutations that may circumvent a need for cdG. In the genome sequences we did not detect differences other than the targeted deletions when comparing Rm2011 to Rm2011 ΔXVI, and we identified only one SNP when comparing Rm2011 expR+ ΔXVI to Rm2011 expR+. This SNP causes a nonsense mutation in SMc02987 encoding the toxin of the putative SMc02987/SMc02988 toxin/antitoxin pair (52).
Assays for EPS production of the two cdG0 strains and the intermediates generated by the serial deletions did not reveal significant alterations in mucoidity or calcofluor brightness (see Fig. S3 in the supplemental material). In the Rm2011 background, serial deletions of GGDEF-domain-encoding genes did not alter motility on semisolid agar (see Fig. S3). Swimming motility of mutants in the background of the QS-sufficient strain was not tested because ExpR strongly induces production of EPS II, which hampers this assay. Biofilm formation was diminished about 2-fold in the Rm2011 ΔIV, ΔXII, and ΔXIII mutants (see Table S1 in the supplemental material) compared to the level in the wild type, whereas it was only slightly reduced in both cdG0 strains (see Fig. S3). Thus, contrary to expectations, the absence of cdG in these strains did not strongly affect any of the tested phenotypes that are typically associated with this second messenger. M. sativa plants inoculated with the cdG0 strains were indistinguishable from plants inoculated with the wild type with respect to the color of nodules and appearance of shoots (see Fig. S4A in the supplemental material). No significant differences in nodulation efficiency, nodule occupancy, or competitiveness between wild-type and cdG0 strains were observed (Fig. S4B and C). Thus, we conclude that preventing cdG synthesis does not significantly affect the symbiotic ability of S. meliloti.
Since we did not detect any strong phenotype of the cdG0 strains under standard growth conditions, their resistance to osmotic and acid stress was tested since EPS is an important factor for resistance to these stress factors (53, 54). Whereas growth of the mutants was not affected in the presence of 0.4 M NaCl, growth of Rm2011 cdG0 was significantly reduced at pH 5.7 (see Fig. S5A in the supplemental material). Complementation of Rm2011 cdG0 with pWBT-pleD restored growth at pH 5.7 (see Fig. S5B), implying that cdG or PleD may have a role in acid resistance of this QS-impaired strain.
Overexpression of cdG-related genes affects intracellular cdG levels, motility, biofilm formation, and production of EPS.
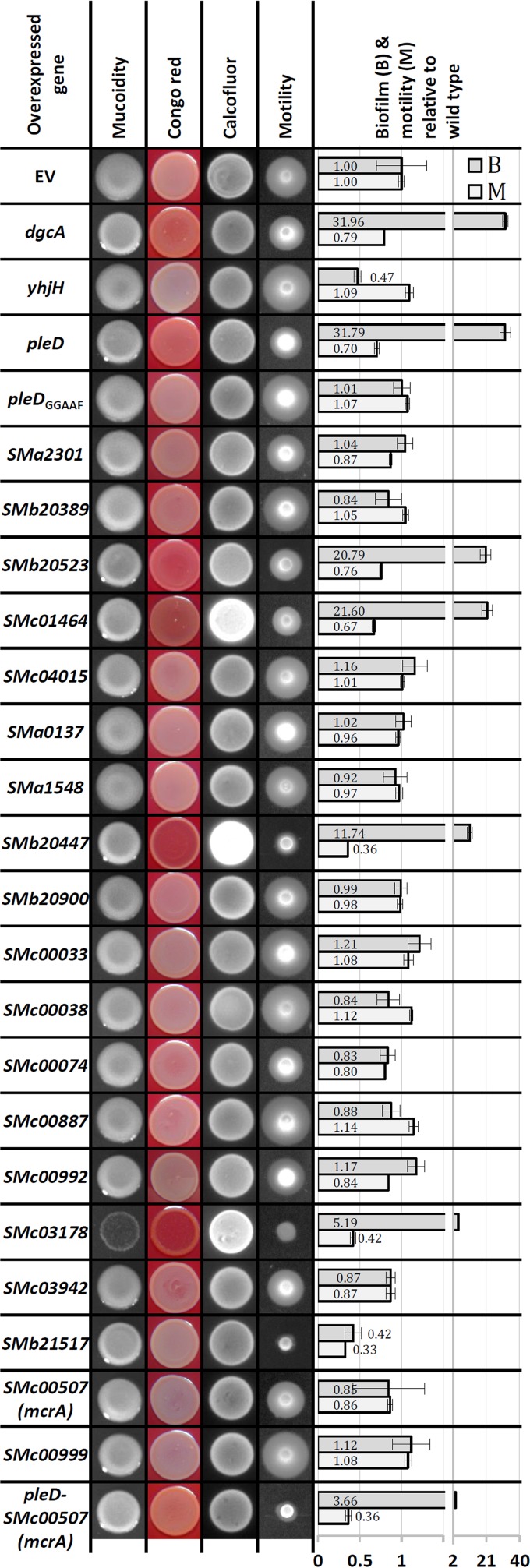
To functionally characterize S. meliloti cdG-related genes (Fig. 2) independent of their native expression conditions, IPTG-inducible overexpression constructs were generated in the medium-copy-number vector pWBT. Following the same strategy, we constructed overexpression plasmids of C. crescentus dgcA (55) and E. coli yhjH (56) encoding a well-characterized DGC and PDE, respectively. EPS production, motility, and biofilm formation of S. meliloti Rm2011 carrying these plasmids were characterized under conditions permitting overexpression of the plasmid-located cdG-related genes (Fig. 3).
FIG 3.
Phenotypic analysis of Rm2011 overexpressing the indicated cdG-related genes. EV, empty pWBT vector. The A570/OD600 mean value for Rm2011 pWBT was 0.259 ± 0.079 (set to 1). Error bars represent standard deviations of three biological replicates.
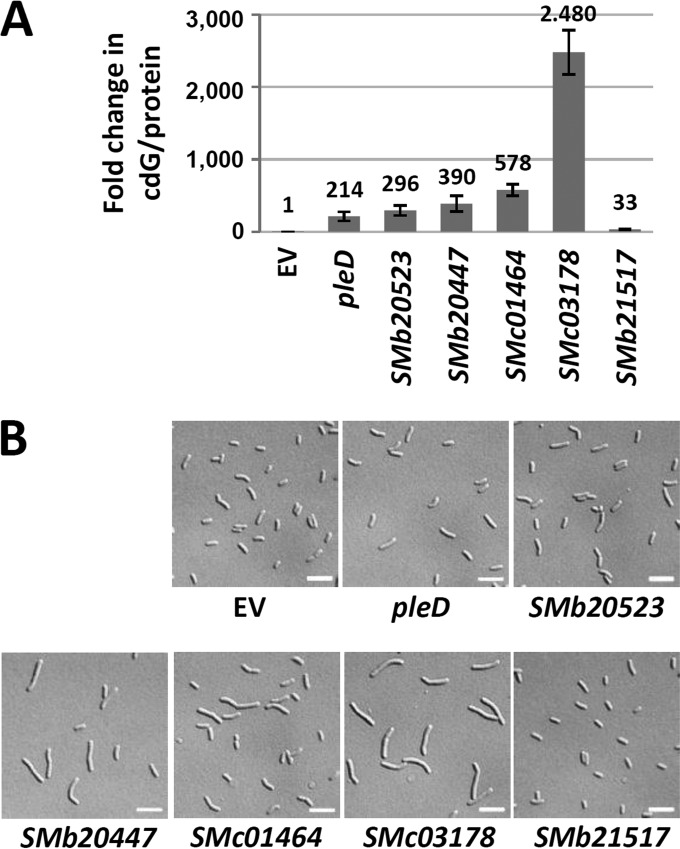
Overproduction of the heterologous DGC DgcA inhibited motility on semisolid agar, increased Congo red staining, and promoted biofilm formation. In contrast, overproduction of the heterologous PDE YhjH reduced biofilm formation 2-fold but did not affect motility. Inhibition of motility as well as increased biofilm formation and Congo red staining was observed upon overexpression of pleD, SMb20523, SMb20447, SMc01464, and SMc03178, all encoding putatively functional GGDEF domains (Fig. 3). This implies that these five S. meliloti genes may indeed encode functional DGCs. Mutation of the PleD GGDEF motif to GGAAF (pleDGGAAF) abolished the effects of pleD overexpression (Fig. 3), further supporting DGC activity of PleD and a cdG-dependent nature of the observed phenotypic changes. DGC activity of these five S. meliloti proteins is furthermore supported by a 214- to 2,480-fold increase in cdG content of cells overproducing these proteins compared to levels in the wild type (Fig. 4A). Interestingly, overexpression of the putative PDE gene SMb21517 inhibited both motility and biofilm formation, whereas the intracellular cdG concentration increased 33-fold (Fig. 3 and 4). This unexpected increase in cdG content is inconsistent with the observed reduction in biofilm formation. Taken together, these data strongly suggest that motility, production of Congo red-binding compounds, and biofilm formation are influenced by cdG in S. meliloti. Although we cannot exclude the possibility that these phenotypes were caused by effects of overexpression other than increased levels of cdG, the observation that overproduction of the heterologous DGC DgcA caused similar defects strongly supports this notion.
FIG 4.
Effects of overexpression of DGC/PDE genes on cdG content and cell morphology. (A) Increase in cdG content of Rm2011 cells overexpressing the indicated genes relative to cells carrying the empty pWBT vector. Error bars represent standard deviations of three biological replicates. (B) Microscopy analysis of cells from the culture samples used for cdG quantification in panel A. Scale bar, 5 μm.
Effects of elevated cdG content on cell morphology were analyzed by microscopy (Fig. 4B). In these assays, we investigated cells harvested from exponentially growing cultures used for cdG quantification (Fig. 4A). Overexpression of pleD, SMb20523, and SMb21517 did not significantly affect cell morphology, whereas cells overexpressing SMb20447, SMc01464, and SMc03178 were elongated. The impact on cell morphology correlated well with the magnitude of increase in cdG concentration, with the highest cdG content resulting in the strongest cell elongation. On minimal and rich media, growth of cells overexpressing pleD, SMb20523, SMb20447, or SMc01464 was not significantly affected, whereas overexpression of SMc03178 notably inhibited growth (Fig. 3). This suggests that very high levels of cdG may interfere with cell proliferation.
The degree of motility impairment and biofilm formation upon overproduction of the putative DGCs did not correlate with cdG content (Fig. 3 and 4A). Strongest biofilm formation was observed upon pleD overexpression, which caused the smallest increase in cdG content. Thus, the regulatory pathways governing cdG-mediated control of motility and biofilm formation may respond to different concentration ranges of cdG. Additionally, cell elongation and impaired growth caused by very high cdG concentrations may also contribute to the observed phenotypes.
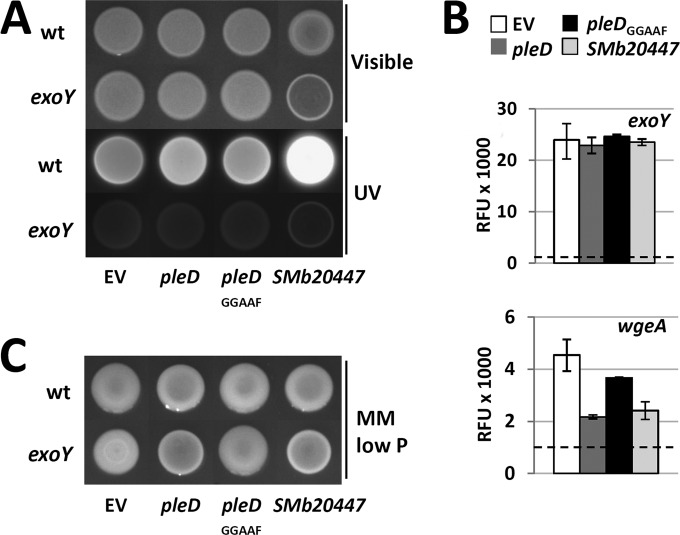
High cdG levels correlated with enhanced calcofluor brightness and Congo red staining of the agar cultures of SMb20447-, SMc01464-, and SMc03178-overexpressing strains (Fig. 3), implying that EPS production is positively regulated by cdG. Since calcofluor fluorescence is typically associated with enhanced EPS I production, we overexpressed SMb20447 in an exoY mutant, which is deficient in the galactosyltransferase initiating EPS I biosynthesis (57). The exoY background mutation abolished the calcofluor bright phenotype (Fig. 5A), confirming that it was caused by enhanced production of EPS I. Promoter activity of exoY was not affected by overexpression of SMb20447 (Fig. 5B). Thus, enhanced production of EPS I may be mediated by transcriptional regulation of other genes of this biosynthetic pathway or posttranscriptionally.
FIG 5.
Effects of overexpression of pleD, pleDGGAAF, or SMb20447 on EPS production. (A) Rm2011 wild-type (wt) and exoY mutant strains, both carrying the indicated overexpression constructs, grown on LB agar (Visible) and the associated calcofluor brightness (UV). (B) Activities of exoY and wgeA promoters upon overexpression of pleD, pleDGGAAF, or SMb20447 in Rm2011 measured using promoter-egfp fusions in stationary growth phase (45 h after inoculation). Dashed lines represent fluorescence of the cells carrying the empty vector with promoterless egfp. RFU, relative EGFP fluorescence units. Error bars indicate standard deviations of two to three biological replicates. (C) Rm2011 wild-type and exoY mutant strains, both carrying the indicated overexpression constructs, grown on phosphate-limiting MM agar. EV, empty pWBT vector.
We noted that mucoidity of agar cultures on phosphate-limiting MM was slightly reduced upon overexpression of dgcA, pleD, SMb20523, SMc01464, SMb20447, and SMc03178. This implies that the increase in cdG content may be inhibitory for EPS II biosynthesis. The mucoidity of an exoY mutant growing on phosphate-limiting MM is predominantly conferred by EPS II. In this mutant, the decrease in mucoidity upon overexpression of pleD or SMb20447 was even more apparent than in the wild-type background, whereas overexpression of pleDGGAAF had no effect (Fig. 5C). This further supports the assumption that EPS II production was negatively affected by overexpression of these DGC candidate genes. Promoter activity of wgeA, a gene involved in EPS II biosynthesis, was reduced to about one-third in pleD- and SMb20447-overexpressing stationary-phase cells (Fig. 5B). This suggests that either a direct or indirect effect of enhanced levels of cdG on transcription of EPS II biosynthesis genes contributes to reduced mucoidy.
PleD localizes to the old pole of the smaller daughter cell of asymmetrically dividing S. meliloti.
To learn about subcellular localization of the six DGC and PDE candidates that stood out in the overproduction assay (Fig. 3 and 4), we constructed C-terminal fusions of these proteins to EGFP. For this approach the encoding genes were replaced by the fusion constructs at their native genomic loci in Rm2011. Exponential- and stationary-phase cells of the resulting strains cultured in TY medium as well as in phosphate-sufficient and phosphate-limiting MM were analyzed by fluorescence microscopy (see Fig. S6 in the supplemental material). Out of these six fusion proteins only PleD-EGFP and SMc01464-EGFP produced a detectable fluorescence signal in cells cultured in the three media. SMb20447-EGFP fluorescence was detected only in cells growing in phosphate-limiting MM, and none of the strains with the remaining constructs produced a detectable fluorescence signal in any of the media tested.
Accumulation of PleD-EGFP in one or two predominantly polarly localized foci was observed in a considerable proportion of cells harvested from liquid cultures (Fig. 6A). We monitored the spatiotemporal localization of PleD-EGFP by time-lapse fluorescence microscopy (Fig. 6B). To unambiguously assign polar localization of PleD-EGFP to the mother or slightly smaller daughter cell of the asymmetrically dividing S. meliloti cells (58), the endogenous ParB was C-terminally tagged with mCherry in the pleD-egfp-expressing strain. ParB is known to bind close to the chromosomal origin of replication, and during chromosome segregation one copy moves from the old pole of the mother cell to the opposite pole (future old pole) of the emerging daughter cell (59, 60). Time-lapse microscopy revealed that during the S. meliloti cell cycle, a PleD-EGFP focus appeared in the daughter cell shortly before cells separated (Fig. 6B, arrow) and then disappeared and was no longer detectable at the time point when a second ParB-mCherry focus was visible at the new pole of the daughter cell.
FIG 6.
Spatiotemporal localization of PleD-EGFP during cell cycle. (A) Microscopy analysis of Rm2011 cells producing PleD-EGFP harvested from cultures in liquid TY medium in exponential (Exp) and stationary (Stat) growth phases. All images were taken using identical settings. Scale bar, 5 μm. (B) Time-lapse microscopy of Rm2011 carrying chromosomally integrated pleD-egfp and a parB-mCherry expression plasmid. Arrows point to PleD-EGFP foci overlapping ParB-mCherry foci. Green and red arrowheads point to PleD-EGFP and ParB-mCherry foci, respectively. Scale bar, 2 μm.
Elevated levels of cdG diminish biosynthesis of QS signal molecules.
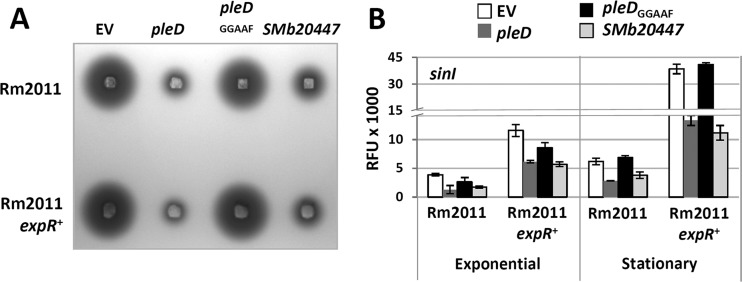
Links between cdG-dependent regulation and QS have previously been reported in several bacterial species (reviewed in references 61 and 62). We therefore asked if elevated concentrations of cdG affect N-acyl-homoserine lactone (AHL)-mediated QS signaling. The expression level of the AHL synthase gene sinI and overall AHL production were estimated in Rm2011 expR+ upon overexpression of pleD, pleDGGAAF, or SMb20447. Expression of sinI was assessed using a PsinI-egfp fusion, and AHL production was detected by the A. tumefaciens indicator strain NTL4 (pZLR4) (Fig. 7). PsinI activity and AHL levels decreased when pleD or SMb20447 was overexpressed, whereas overexpression of pleDGGAAF had no effect. These data suggest that cdG negatively regulates biosynthesis of QS signals. Repression of PsinI and reduction of AHL production were also observed in Rm2011 (Fig. 7). In this strain expR is disrupted by an insertion element (51). Therefore, this cdG-mediated regulation of sinI can be considered to be independent of expR. Moreover, in stationary-phase cultures of the Rm2011 cdG0 strain, sinI promoter activity and AHL abundance increased (see Fig. S7 in the supplemental material), further supporting a cdG-mediated negative regulation of AHL synthesis.
FIG 7.
Elevated cdG content negatively affects AHL production at the level of sinI transcription. (A) Semiquantitative detection of AHLs in S. meliloti stationary-phase culture supernatants by A. tumefaciens NTL4(pZLR4). (B) sinI promoter activity determined using a PsinI-egfp fusion. Exponential, 27 h after inoculation; stationary, 45 h after inoculation; EV, empty pWBT vector; RFU, relative EGFP fluorescence units. Error bars indicate standard deviations of three biological replicates.
cdG-stimulated biofilm formation is modulated by EPS.
The A. tumefaciens uppABCDEF cluster (see Fig. S8A in the supplemental material) controls biosynthesis of a unipolarly localized polysaccharide (UPP) adhesin strictly required for surface attachment (63). Surface contact-independent production of this polysaccharide is induced by elevated levels of cdG (41). This gene cluster is highly conserved among rhizobia, including S. meliloti and R. leguminosarum. In R. leguminosarum, it drives biosynthesis of a polar glucomannan which is not required for biofilm formation but which significantly promotes attachment to roots and root hair infection (64, 65).
Looking for target processes that may contribute to cdG-mediated regulation of biofilm formation in S. meliloti, we studied the role of the SMc01790-SMc01796 gene region showing homology to these gene clusters (see Fig. S8A in the supplemental material). A polar plasmid integration mutation in the S. meliloti uppE homolog SMc01792, also inactivating the downstream genes SMc01791 and SMc01790, failed to respond to pleD overexpression with strong biofilm formation (Fig. 8A). M. sativa plants inoculated with this mutant were indistinguishable from plants inoculated with the wild type with respect to the number and color of nodules and the appearance of shoots. Since SMc01794 promoter activity did not strongly alter upon pleD overexpression (see Fig. S8B), cdG-mediated regulation most likely occurs at the posttranscriptional level.
FIG 8.
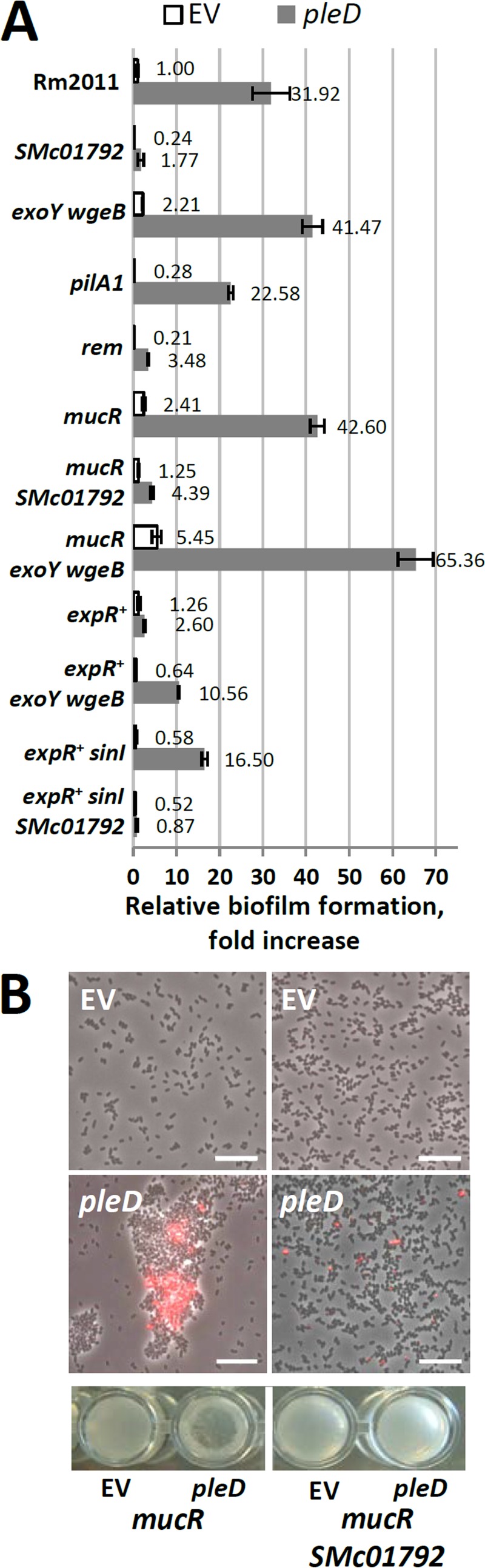
Biofilm formation and cell aggregation upon overexpression of pleD. (A) Biofilm formation in different genetic backgrounds relative to that of Rm2011 carrying the empty vector. EV, empty pWBT vector. The A570/OD600 mean value for Rm2011 pWBT was 0.162 ± 0.046 (set to 1). Error bars indicate standard deviations of three biological replicates. (B) Cell clumping and fl-WGA staining in stationary 30% MM cultures grown in shaken liquid culture. Microscopy images (top) and wells of a 96-well plate (bottom) are shown. Scale bar, 10 μm.
We further aimed at dissecting the contributions of EPS I, EPS II, and the biosynthetic product of the SMc01790-SMc01796 gene cluster to cdG-enhanced biofilm formation. Biofilm formation induced by pleD overexpression was enhanced in an exoY wgeB double mutant, deficient in production of EPS I and EPS II (57, 66) compared to that in the pleD-overexpressing Rm2011 wild type (Fig. 8A). The presence of an intact expR gene increases both EPS I and EPS II production (67, 68). Biofilm formation caused by pleD overexpression in Rm2011 expR+ was strongly reduced in comparison to the level in Rm2011 (Fig. 8A). Inactivation or strong reduction of EPS I and EPS II biosynthesis in Rm2011 expR+ by exoY wgeB double mutation or by knockout of the AHL synthase gene sinI (69), respectively, partially restored cdG-stimulated biofilm formation upon pleD overexpression (Fig. 8A). Biofilm formation of the pleD-overexpressing Rm2011 expR+ sinI strain was abolished by mutation of SMc01792, in the same way as in Rm2011 (Fig. 8A).
Mutation of mucR encoding a global transcriptional regulator leads to reduced production of EPS I and enhanced biosynthesis of EPS II (70). While EPS II produced by an expR+ strain is composed of low-molecular-weight and high-molecular-weight fractions, a mucR mutant primarily produces high-molecular-weight EPS II (71). In contrast to Rm2011 expR+, a mucR mutant responded to overexpression of pleD with strong biofilm formation, which was further enhanced upon mutation of exoY and wgeB (Fig. 8A). Mutation of SMc01792 strongly reduced biofilm formation by the pleD-overexpressing mucR mutant, further supporting an important role of the SMc01794-SMc01790 operon in cdG-enhanced surface attachment. Interestingly, pleD-overexpressing mucR cells clearly aggregated in agitated liquid cultures (Fig. 8B). Microscopy analysis confirmed the presence of large cell aggregates which were stainable with Alexa Fluor 594-conjugated wheat germ agglutinin (fl-WGA) binding to N-acetylglucosamine (GlcNAc), which is present in A. tumefaciens UPP (41) (Fig. 8B). Mutation of SMc01792 abolished cell clumping and strongly reduced fl-WGA staining (Fig. 8B).
We further asked if S. meliloti flagella and pili contribute to biofilm formation. Mutations in rem and flgH strongly reduced biofilm formation, compared to the wild-type level, of both the pleD-overexpressing strain and the strain carrying the empty vector (Fig. 8A; see also Fig. S9 in the supplemental material). The rem mutant is nonmotile due to deficiency in flagellar gene expression (72), while the polar miniTn5 insertion in flgH probably also disrupts transcription of the downstream genes fliL and fliP. The latter three genes encode membrane-associated components of the flagellar basal body. This implies that flagella or flagellar motility contributes to cdG-stimulated biofilm formation. In addition, a pilA1 mutant strain also showed reduced biofilm formation (Fig. 8A), suggesting a role of type IVb pili in this process.
PilZ domain protein SMc00507 negatively regulates motility.
The S. meliloti genome encodes two PilZ domain proteins (Fig. 2). The PilZ domains of both proteins contain conserved RXXXR and DXSXXG motifs separated by 21 amino acids in SMc00507 and by 19 amino acids in SMc00999. Whereas SMc00999 consists of an N-terminal domain of unknown function and a C-terminal PilZ domain, SMc00507 comprises only a single PilZ domain. SMc00507 was annotated to encode a protein of 112 amino acids (GenBank accession number CAC46339.2). However, we found the predicted start codon to be incorrect. The egfp reporter gene fused to this TTG codon was not expressed, whereas it was expressed when fused to the downstream ATG at position 34. This strongly suggests that this ATG is the genuine start codon of SMc00507 encoding a protein of 101 amino acids (see Fig. S10 in the supplemental material).
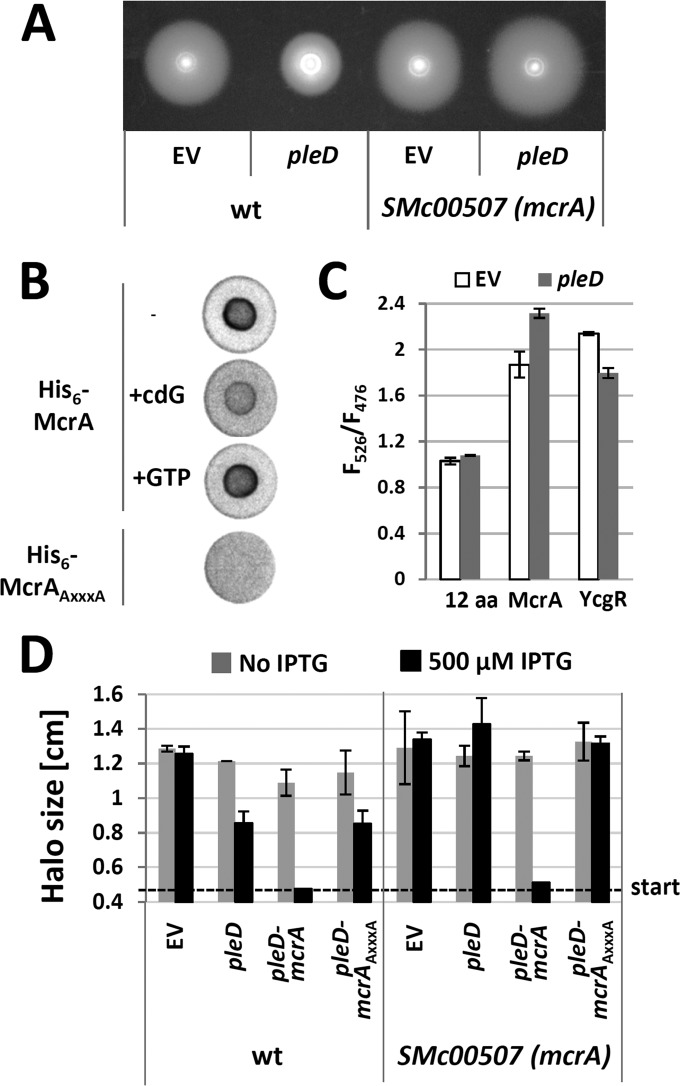
To learn about the role of S. meliloti PilZ domain proteins in cdG-dependent regulation, Rm2011 SMc00507 and SMc00999 single and double mutants were assayed for EPS production, motility, and biofilm formation (see Fig. S1 in the supplemental material). Under the conditions tested, no phenotypes could be assigned to these mutations. However, under conditions of elevated cdG levels caused by pleD overexpression, the SMc00507 SMc00999 double mutant was slightly hypermotile, in contrast to the Rm2011 wild type that showed reduced motility (see Fig. S11A). In contrast, this double mutant was not affected in pleD overexpression-enhanced Congo red staining and biofilm formation (Fig. S11A). Characterization of the SMc00507 and SMc00999 single mutants identified the mutation in SMc00507 to be responsible for enhanced motility upon pleD overexpression (Fig. 9A; see also Fig. S11B). Thus, SMc00507 encodes a putative cdG receptor protein involved in regulation of motility in S. meliloti, and this protein was therefore was named mcrA, for motility-associated cdG receptor A.
FIG 9.
Identification of McrA (SMc00507) as a cdG receptor mediating repression of swimming motility upon pleD overexpression. (A) Swimming motility of the Rm2011 wild type (wt) and mcrA mutant upon pleD overexpression. EV, empty pWBT vector. (B) DRaCALA with radiolabeled cdG and His6-McrA or His6-McrAAXXXA. (C) Detection of conformational change in McrA upon pleD overexpression in Rm2011 using the CyPet/YPet FRET biosensor. The negative control CyPet-12aa-YPet (12aa) and positive control (YcgR) were analyzed in parallel. EV, empty pSRKKm vector; F526, emission at 526 nm normalized to OD600 (excitation at 425 nm); F476, emission at 476 nm normalized to OD600 (excitation at 425 nm). Error bars indicate standard deviations of three biological replicates. (D) Swimming motility of wild-type Rm2011 and the mcrA mutant upon overexpression of pleD alone, in combination with mcrA, or in combination with mcrAAXXXA. EV, empty pWBT vector. Error bars indicate standard deviations of three biological replicates.
Binding of cdG by His6-McrA was tested by a differential radial capillary action of ligand assay (DRaCALA) (43) (Fig. 9B). Spotting of purified His6-McrA preincubated with radiolabeled cdG on the membrane produced an intense signal in the middle of the spotting area originating from protein-bound cdG, whereas a surrounding weak signal was produced by nonbound cdG. This characteristic pattern indicates binding of radiolabeled cdG by His6-McrA. Binding was outcompeted by nonlabeled cdG but not by nonlabeled GTP, supporting the idea that His6-McrA specifically binds cdG. Moreover, mutating the conserved N-terminal RXXXR motif by the amino acid exchanges R9A and R13A in His6-McrA (designated His6-McrAAXXXA) abolished cdG binding (Fig. 9B), supporting the idea that this motif is involved in cdG binding. We also attempted to mutate the conserved DXSXXG motif; however, the simultaneous amino acid exchanges D35A, S37A, and G40A rendered the protein insoluble (data not shown).
PilZ proteins were previously reported to undergo conformational changes upon cdG binding (4, 73). We tested for in vivo binding of cdG to McrA and a conformational change upon binding by employing a modified version of the fluorescence resonance energy transfer (FRET)-based cdG biosensor designed by Christen et al. (20). The original biosensor is composed of the cdG receptor protein YcgR flanked by CyPet and YPet fluorophores. A conformational change of YcgR upon cdG binding results in an altered YPet/CyPet emission ratio (526 nm/476 nm). In this biosensor, we replaced YcgR by McrA and produced this fusion protein in Rm2011 with and without pleD overexpression. The CyPet-YcgR-YPet and CyPet-12aa-YPet (CyPet and YPet with a 12-amino-acid linker) constructs were used as positive and negative controls, respectively. Compared to the Rm2011 wild-type background, in the pleD-overexpressing strain, the 526 nm/476 nm ratio of CyPet-12aa-YPet-mediated fluorescence did not change. In contrast, the 526 nm/476 nm ratio of CyPet-YcgR-YPet- and CyPet-McrA-YPet-mediated fluorescence decreased and increased, respectively (Fig. 9C). The former indicates that upon cdG binding to YcgR the expected conformational change moves the fluorophores farther apart (20), while the latter implies that cdG binding to McrA induces a conformational change moving the fluorophores closer together.
Overexpression of mcrA in the Rm2011 wild type resulted in slightly reduced motility, whereas EPS and biofilm phenotypes were unaffected (Fig. 3). Furthermore, combined overexpression of pleD and mcrA but not mcrAAXXXA led to a nonmotile phenotype (Fig. 3 and 9D), strongly suggesting that cdG-bound McrA negatively controls motility. Combined pleD and mcrA overexpression in Rm2011 did not affect EPS production, whereas biofilm formation decreased about 10-fold compared to the level of Rm2011 overexpressing pleD alone (Fig. 3). This outcome is consistent with our observation that the nonmotile rem mutant is defective in pleD overexpression-mediated biofilm formation (Fig. 8A).
DISCUSSION
The role of cdG in S. meliloti.
Our systematic analysis of GGDEF-, EAL-, and PilZ-domain-encoding genes signified similarities but also differences of the role of cdG in S. meliloti compared to that in other bacteria. Single gene mutations in predicted cdG-related genes did not show striking changes in phenotypes typically associated with cdG, like EPS production, swimming motility, biofilm formation, and host infection. The only exception is the putatively essential gene SMc00074, predicted to encode an integral membrane protein with cytoplasmic EAL and inactive GGDEF domains. Single mutations in dgcA and dgcB encoding hybrid GGDEF-EAL domain proteins did not cause phenotypic changes in R. etli (26), a finding which is similar to our mutant screening data. However, our findings are in disagreement with those of Wang et al. (24), who reported weak changes in motility, symbiosis, and EPS production in 11 out of 14 single-gene mutants in GGDEF- and/or EAL-domain-encoding genes in S. meliloti Rm1021, which is closely related to the wild-type strain in our study. Slight differences between strains and assay conditions may account for this discrepancy.
While functional redundancies and compensatory effects may explain missing or weak effects of the single mutants in cdG-related genes, most unexpectedly the S. meliloti cdG0 strains also did not show salient phenotypes in the physiological processes tested, except for increased sensitivity to acid stress. The acid stress response of Rm2011 involves repression of motility and activation of EPS biosynthesis (74), processes that were affected by elevated levels of cdG. To our knowledge, S. meliloti is the only bacterium described so far which does not require cdG or GGDEF/EAL domain proteins for swimming motility. A C. crescentus cdG0 strain was nonmotile due to lack of flagella and impaired in surface attachment and cell differentiation, the latter impairment resulting in aberrant cell morphology (75). Deletion of all GGDEF-domain-encoding genes in Salmonella Typhimurium abolished virulence, resistance to desiccation, motility, biofilm formation, and synthesis of cellulose and fimbriae, while growth and resistance to acid, salt, or starvation stress were unaffected (16).
Since the cdG content was in the range of a few picomoles per milligram of total protein, similar to other bacteria studied (76–78), the phenotypic neutrality of the cdG0 strains is unlikely to be due to low abundance or absence of cdG in the wild type under the growth conditions applied in this study. However, we cannot exclude the possibility that the Rm2011 laboratory strain has lost sensory and regulatory pathways that stimulate enhanced cdG production. In S. meliloti, cdG content strongly dropped in the stationary phase. Growth-dependent alterations in cdG concentrations were also reported for E. coli showing higher levels of cdG at the transition to stationary phase than in the exponential and stationary phases (78). In contrast, levels of cdG in Myxococcus xanthus cells seem not to be growth phase regulated (76).
Nevertheless, in agreement with a classical cdG-mediated motile-sessile switch, elevated levels of cdG caused by overexpression of heterologous or native DGC-encoding genes resulted in enhanced biofilm formation coinciding with changes in EPS biosynthesis and reduced motility (summarized in Fig. 10). Our study strongly suggests that PleD, SMb20523, SMb20447, SMc01464, and SMc03178 have DGC activity in S. meliloti although we cannot exclude the possibility that overexpression of GGDEF- and/or EAL-encoding genes indirectly influence other DGCs or PDEs. Four of these proteins possess a canonical GG(D/E)EF motif, whereas SMb20447 has an AGDEF motif, as has been reported for the active DGC VCA0965 from Vibrio cholerae (79). Among the 20 proteins putatively associated with cdG synthesis, degradation, or binding, 11 have both complete GGDEF and EAL domains, including SMb20447 and SMc03178. Despite the presence of a well-conserved EAL domain, overproduction of either of these two proteins results in a strong increase in cdG levels. This suggests that under the conditions of overproduction, DGC activity of SMb20447 and SMc03178 is dominant. Control of the catalytic activities of such dual-function enzymes can be a key point of regulation. A regulatory control of both enzymatic functions has previously been reported for Rhodobacter sphaeroides BphG1, Vibrio parahaemolyticus ScrC, Legionella pneumophila Lpl0329, and A. tumefaciens DcpA (80–83). Unexpectedly, overproduction of the predicted PDE SMb21517, encoding a single EAL domain, resulted in a moderate increase in cdG content and inhibited motility. The same effects were observed upon overproduction of EAL-domain-containing or HD-GYP-domain-containing proteins in Legionella pneumophila or Pectobacterium atrosepticum (84, 85), indicating indirect effects on other DGCs or PDEs.
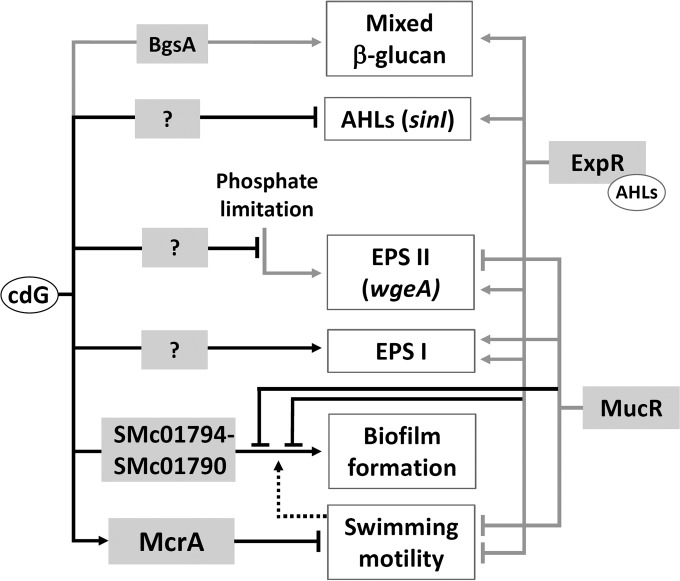
FIG 10.
Summary of cdG-mediated regulation in S. meliloti. Black lines show regulatory interactions described in this study, and gray lines denote previously identified regulatory links (25, 34, 49, 50, 68, 69). Biosynthesis of QS signals is negatively regulated at the level of sinI transcription. EPS I and EPS II biosynthesis is affected by cdG via unknown pathways. Phosphate limitation-induced activation of EPS II biosynthesis is reduced at elevated cdG levels. This includes a change in wgeA promoter activity. Biofilm formation is enhanced involving gene products of the SMc01794-SMc01790 operon and is negatively affected by the global regulators MucR and ExpR, which also regulate motility and EPS biosynthesis. Flagella contribute to biofilm formation. Repression of swimming motility involves binding of cdG to the PilZ-domain protein McrA.
Effect of cdG on the cell cycle.
Very high levels of cdG provoked S. meliloti cell elongation, which might have been directly caused by this second messenger or indirectly by exhausting the GTP pool. In C. crescentus, cdG and the single-domain response regulator DivK convergently regulate cell cycle progression (21). In this regulation, cdG-induced phosphatase activity of the cell cycle kinase CckA promotes replication initiation and cell cycle progression. The C-terminal part of the S. meliloti CckA, which contains the catalytic and REC domains, shares 47% identity with its C. crescentus ortholog, including residue Y514, shown to be important for binding of cdG (21). Thus, it is tempting to speculate that, at least at very high cdG concentrations, binding of cdG to CckA possibly also impacts the S. meliloti cell cycle.
In C. crescentus, PleD is involved in regulation of cell cycle progression and was shown to be responsible for uneven distribution of cdG in mother and daughter cells, with the mother cell containing more cdG (20). Localization of PleD to the stalked cell pole (the old pole of the mother cell) is believed to be associated with its DGC activity (86). Although the C. crescentus and S. meliloti PleD proteins share 51% identity, their modes of regulation and roles in the cell cycle may be different. This is substantiated by different localization patterns and mutant phenotypes. Localization of PleD in C. crescentus and S. meliloti differs in that it localizes to the old poles of the mother and daughter cells, respectively. Furthermore, the S. meliloti pleD mutant did not differ from the wild type in motility, while the C. crescentus pleD mutant was hypermotile due to defects in flagellum ejection and stalk morphogenesis (87). Similarly to the S. meliloti pleD mutant, an A. tumefaciens pleD deletion mutant was unaffected in motility and showed no obvious developmental defects (88). However, the pattern of flagellation differs between these organisms. Swimming motility of C. crescentus is mediated by a single polar flagellum localized at the new pole (89), while A. tumefaciens exhibits two flagella in a polar or subpolar tuft at one pole, plus one or two lateral flagella on each side (90), and peritrichous flagella drive motility of S. meliloti (91).
Modulation of QS by cdG.
Quorum sensing and cdG-dependent regulation are interconnected in several bacterial species. The most common mechanism is regulation of cdG biosynthesis or degradation by QS lowering cdG levels in the QS state (reviewed in references 61 and 62). Under the conditions tested, QS did not influence the total cdG content in S. meliloti, but both native and elevated levels of cdG negatively affected expression of the AHL synthase gene sinI and accumulation of AHLs in the growth medium. Similarly, the PDE activity of RpfR was required for normal levels of transcription of the AHL synthase gene cepI and AHL production in Burkholderia cenocepacia, whereas the factor mediating cdG-dependent negative regulation of the synthase gene remained unknown (92). Since the cdG content was higher in exponential- than in stationary-phase S. meliloti cells, negative regulation of AHL biosynthesis by cdG may serve as a fine-tuning mechanism attenuating AHL accumulation in rapidly growing cells.
The role of EPS in cdG-enhanced biofilm formation.
Our data strongly suggest that S. meliloti, just like A. tumefaciens (41, 63), employs a GlcNAc-containing UPP required for cdG-mediated cell aggregation and attachment to abiotic surfaces. The architectures of the S. meliloti SMc01796-SMc01790 and A. tumefaciens uppABCDEF biosynthetic gene clusters are similar, both encoding two polysaccharide export proteins, two proteins with unknown function, and two (A. tumefaciens) or three (S. meliloti) glycosyltransferases. GlcNAc polymers were also found to be important for surface attachment of C. crescentus as a component of the holdfast adhesin at the tip of the stalk (93, 94).
While this putative GlcNAc-containing UPP was essential for cdG-enhanced biofilm formation of S. meliloti, EPS I and/or EPS II seems to hamper this process, and the amount and composition of EPS may be relevant to this interference. This negative effect of EPS production on biofilm formation was clearly seen only under conditions of elevated levels of cdG. This is in agreement with EPS-deficient Rm1021 mutants which are unaffected in biofilm formation (95). In line with a negative role of EPS at least in initial steps of surface attachment and cell aggregation, overproduction of EPS I by an S. meliloti Rm1021 emrR mutant correlated with decreased biofilm formation (96). In contrast, extensive overproduction of EPS I may have a positive effect on biofilm formation, possibly by mechanisms not including the GlcNAc-containing UPP. An S. meliloti Rm1021 strain encoding a constitutively active variant of the receptor histidine kinase ExoS is nonmotile, extensively overproduces EPS I, and was reported to show stronger biofilm formation than the wild type (97).
Flagellar motility accelerates surface adhesion of various bacteria (reviewed in reference 98). Pili and flagella were suggested to mediate a first, reversible phase of surface attachment of C. crescentus (93). In S. meliloti, absence of flagella was reported to have a negative effect on biofilm formation (97). Our data suggest that cdG-enhanced biofilm formation of S. meliloti involves repression of swimming motility but also requires presence of pili and flagella.
A PilZ domain protein negatively regulates motility.
PilZ domain proteins were reported to negatively regulate motility in diverse bacteria like E. coli, C. crescentus, Azospirillum brasilense, and Pseudomonas fluorescens (11, 99, 100) or to stimulate it in Treponema denticola and Borrelia burgdorferi (101, 102). In this study, we identified McrA as a cdG receptor protein, able to repress swimming motility at elevated cdG levels. McrA is a small protein of 101 amino acids solely comprising a PilZ domain. Such architecture is not uncommon; however, not much is known about the functions of these proteins. DgrA and DgrB are single-domain PilZ proteins involved in cdG-dependent repression of motility in C. crescentus (11). DgrA binds cdG in vitro, and its overexpression results in decreased abundance of the flagellar rotation protein FliL via an unknown mechanism (11). Although no significant homology exists between McrA and DgrA, the congruent architecture comprising just a PilZ domain implies that their function likely relies on interactions with other proteins.
Eliminating cdG-dependent McrA-mediated negative regulation of motility led to slight hypermotility at elevated cdG levels, which reveals an additional positive effect of cdG on motility. This observation is in line with a bipartite role of cdG in flagellar-based motility regulation proposed for S. Typhimurium and C. crescentus in that cdG may coordinate flagellar assembly and rotation (16, 73, 75). In a strikingly similar manner, overproduction of a heterologous DGC rendered an S. Typhimurium mutant in the PilZ domain-encoding ycgR gene hypermotile, and combined overproduction of this DGC and YcgR provoked a nonmotile phenotype in the same way as combined overexpression of pleD and mcrA in S. meliloti (73). An additional similarity is probably a conformational change triggered upon binding of cdG to YcgR (73) and McrA.
Consistent with the assumption of a common role of cdG in regulating the motile-sessile switch in bacteria, our findings indicate that high cdG levels favor a sessile lifestyle of S. meliloti. Yet no phenotypic differences were evident between mutants in 21 out of 22 cdG-related genes and the wild type, and only overexpression of seven of these genes resulted in phenotypic changes. Most unexpectedly, the S. meliloti mutants unable to produce cdG were not affected in processes typically controlled by this second messenger, including motility. Moreover, cdG was also not an important bacterial factor in root nodule symbiosis. This suggests that these proteins are not active under the conditions tested or that they have a role in processes not assayed in this study. It may also be indicative of overlapping regulatory controls, as has been reported for the regulatory pathway driving cell cycle progression in C. crescentus (21).
Supplementary Material
ACKNOWLEDGMENTS
We thank Vincent Franz for assistance with protein purification and plant assays, Pornsri Charoenpanich and Matthew McIntosh for sharing plasmids, Hemantha Kulasekara for providing the FRET-based cdG biosensor construct, and Annette Garbe for technical help with LC-MS/MS measurements.
Financial support from the German Research Foundation (Collaborative Research Centre 987 Microbial Diversity in Environmental Signal Response), the LOEWE program of the State of Hesse (Germany), and the Max Planck Society is acknowledged. We acknowledge technical assistance and access to resources supported by BMBF grant FKZ 031A533 within the de.NBI network.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00795-15.
REFERENCES
- 1.Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. 2007. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol 5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 3.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schirmer T, Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol 7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 5.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang X, Ahmad I, Blanka A, Schottkowski M, Cimdins A, Galperin MY, Römling U, Gomelsky M. 2014. GIL, a new c-di-GMP-binding protein domain involved in regulation of cellulose synthesis in enterobacteria. Mol Microbiol 93:439–452. doi: 10.1111/mmi.12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev 23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newell PD, Monds RD, O'Toole GA. 2009. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc Natl Acad Sci U S A 106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amikam D, Galperin MY. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 10.Wolfe AJ, Visick KL. 2008. Get the message out: cyclic-Di-GMP regulates multiple levels of flagellum-based motility. J Bacteriol 190:463–475. doi: 10.1128/JB.01418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christen M, Christen B, Allan MG, Folcher M, Jenö P, Grzesiek S, Jenal U. 2007. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc Natl Acad Sci U S A 104:4112–4117. doi: 10.1073/pnas.0607738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryjenkov DA, Simm R, Römling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem 281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 13.Tischler AD, Camilli A. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect Immun 73:5873–5882. doi: 10.1128/IAI.73.9.5873-5882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A 103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan RP, Fouhy Y, Lucey JF, Jiang BL, He YQ, Feng JX, Tang JL, Dow JM. 2007. Cyclic di-GMP signalling in the virulence and environmental adaptation of Xanthomonas campestris. Mol Microbiol 63:429–442. doi: 10.1111/j.1365-2958.2006.05531.x. [DOI] [PubMed] [Google Scholar]
- 16.Solano C, García B, Latasa C, Toledo-Arana A, Zorraquino V, Valle J, Casals J, Pedroso E, Lasa I. 2009. Genetic reductionist approach for dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Salmonella. Proc Natl Acad Sci U S A 106:7997–8002. doi: 10.1073/pnas.0812573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bobrov AG, Kirillina O, Ryjenkov DA, Waters CM, Price PA, Fetherston JD, Mack D, Goldman WE, Gomelsky M, Perry RD. 2011. Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol Microbiol 79:533–551. doi: 10.1111/j.1365-2958.2010.07470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waters CM, Lu W, Rabinowitz JD, Bassler BL. 2008. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J Bacteriol 190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrade MO, Alegria MC, Guzzo CR, Docena C, Rosa MC, Ramos CH, Farah CS. 2006. The HD-GYP domain of RpfG mediates a direct linkage between the Rpf quorum-sensing pathway and a subset of diguanylate cyclase proteins in the phytopathogen Xanthomonas axonopodis pv citri. Mol Microbiol 62:537–551. doi: 10.1111/j.1365-2958.2006.05386.x. [DOI] [PubMed] [Google Scholar]
- 20.Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, Miller SI. 2010. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328:1295–1297. doi: 10.1126/science.1188658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lori C, Ozaki S, Steiner S, Böhm R, Abel S, Dubey BN, Schirmer T, Hiller S, Jenal U. 2015. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature 523:236–239. doi: 10.1038/nature14473. [DOI] [PubMed] [Google Scholar]
- 22.Pérez-Mendoza D, Aragón IM, Prada-Ramírez HA, Romero-Jiménez L, Ramos C, Gallegos MT, Sanjuán J. 2014. Responses to elevated c-di-GMP levels in mutualistic and pathogenic plant-interacting bacteria. PLoS One 9:e91645. doi: 10.1371/journal.pone.0091645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Q, Zhang G, Wood TK. 2011. Escherichia coli BdcA controls biofilm dispersal in Pseudomonas aeruginosa and Rhizobium meliloti. BMC Res Notes 4:447. doi: 10.1186/1756-0500-4-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Xu J, Chen A, Zhu J, Yu G, Xu L, Luo L. 2010. GGDEF and EAL proteins play different roles in the control of Sinorhizobium meliloti growth, motility, exopolysaccharide production, and competitive nodulation on host alfalfa. Acta Biochim Biophys Sin (Shanghai) 42:410–417. doi: 10.1093/abbs/gmq034. [DOI] [PubMed] [Google Scholar]
- 25.Pérez-Mendoza D, Rodríguez-Carvajal M, Romero-Jiménez L, Farias GeA, Lloret J, Gallegos MT, Sanjuán J. 2015. Novel mixed-linkage β-glucan activated by c-di-GMP in Sinorhizobium meliloti. Proc Natl Acad Sci U S A 112:E757–E765. doi: 10.1073/pnas.1421748112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao S, Romdhane SB, Beullens S, Kaever V, Lambrichts I, Fauvart M, Michiels J. 2014. Genomic analysis of cyclic-di-GMP-related genes in rhizobial type strains and functional analysis in Rhizobium etli. Appl Microbiol Biotechnol 98:4589–4602. doi: 10.1007/s00253-014-5722-7. [DOI] [PubMed] [Google Scholar]
- 27.Chou SH, Galperin MY. 8 June 2015. Diversity of c-di-GMP-binding proteins and mechanisms. J Bacteriol doi: 10.1128/JB.00333-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galibert F, Finan TM, Long SR, Puhler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P, Bothe G, Boutry M, Bowser L, Buhrmester J, Cadieu E, Capela D, Chain P, Cowie A, Davis RW, Dreano S, Federspiel NA, Fisher RF, Gloux S, Godrie T, Goffeau A, Golding B, Gouzy J, Gurjal M, Hernandez-Lucas I, Hong A, Huizar L, Hyman RW, Jones T, Kahn D, Kahn ML, Kalman S, Keating DH, Kiss E, Komp C, Lelaure V, Masuy D, Palm C, Peck MC, Pohl TM, Portetelle D, Purnelle B, Ramsperger U, Surzycki R, Thebault P, Vandenbol M, et al. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 29.Casse F, Boucher C, Julliot J, Michel M, Dénarié J. 1979. Identification and characterization of large plasmids in Rhizobium meliloti using agarose gel electrophoresis. Microbiology 113:229–242. [Google Scholar]
- 30.Meade HM, Long SR, Ruvkun GB, Brown SE, Ausubel FM. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol 149:114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beringer JE. 1974. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol 84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 32.Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual, 4th ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 33.Tombolini R, Nuti MP. 1990. Poly(β-hydroxyalkanoate) biosynthesis and accumulation by different Rhizobium species. FEMS Microbiol Lett 60:299–304.2150658 [Google Scholar]
- 34.Zhan HJ, Lee CC, Leigh JA. 1991. Induction of the second exopolysaccharide (EPSb) in Rhizobium meliloti SU47 by low phosphate concentrations. J Bacteriol 173:7391–7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 36.Krol E, Becker A. 2014. Rhizobial homologs of the fatty acid transporter FadL facilitate perception of long-chain acyl-homoserine lactone signals. Proc Natl Acad Sci U S A 111:10702–10707. doi: 10.1073/pnas.1404929111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 38.Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hilker R, Stadermann KB, Doppmeier D, Kalinowski J, Stoye J, Straube J, Winnebald J, Goesmann A. 2014. ReadXplorer—visualization and analysis of mapped sequences. Bioinformatics 30:2247–2254. doi: 10.1093/bioinformatics/btu205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robledo M, Rivera L, Jiménez-Zurdo JI, Rivas R, Dazzo F, Velázquez E, Martínez-Molina E, Hirsch AM, Mateos PF. 2012. Role of Rhizobium endoglucanase CelC2 in cellulose biosynthesis and biofilm formation on plant roots and abiotic surfaces. Microb Cell Fact 11:125. doi: 10.1186/1475-2859-11-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu J, Kim J, Koestler BJ, Choi JH, Waters CM, Fuqua C. 2013. Genetic analysis of Agrobacterium tumefaciens unipolar polysaccharide production reveals complex integrated control of the motile-to-sessile switch. Mol Microbiol 89:929–948. doi: 10.1111/mmi.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burhenne H, Kaever V. 2013. Quantification of cyclic dinucleotides by reversed-phase LC-MS/MS. Methods Mol Biol 1016:27–37. doi: 10.1007/978-1-62703-441-8_3. [DOI] [PubMed] [Google Scholar]
- 43.Roelofs KG, Wang J, Sintim HO, Lee VT. 2011. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc Natl Acad Sci U S A 108:15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bordeleau E, Brouillette E, Robichaud N, Burrus V. 2010. Beyond antibiotic resistance: integrating conjugative elements of the SXT/R391 family that encode novel diguanylate cyclases participate to c-di-GMP signalling in Vibrio cholerae. Environ Microbiol 12:510–523. doi: 10.1111/j.1462-2920.2009.02094.x. [DOI] [PubMed] [Google Scholar]
- 45.Broughton WJ, Dilworth MJ. 1971. Control of leghaemoglobin synthesis in snake beans. Biochem J 125:1075–1080. doi: 10.1042/bj1251075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cowie A, Cheng J, Sibley CD, Fong Y, Zaheer R, Patten CL, Morton RM, Golding GB, Finan TM. 2006. An integrated approach to functional genomics: construction of a novel reporter gene fusion library for Sinorhizobium meliloti. Appl Environ Microbiol 72:7156–7167. doi: 10.1128/AEM.01397-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reinhold BB, Chan SY, Reuber TL, Marra A, Walker GC, Reinhold VN. 1994. Detailed structural characterization of succinoglycan, the major exopolysaccharide of Rhizobium meliloti Rm1021. J Bacteriol 176:1997–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Her GR, Glazebrook J, Walker GC, Reinhold VN. 1990. Structural studies of a novel exopolysaccharide produced by a mutant of Rhizobium meliloti strain Rm1021. Carbohydr Res 198:305–312. doi: 10.1016/0008-6215(90)84300-J. [DOI] [PubMed] [Google Scholar]
- 49.Bahlawane C, Baumgarth B, Serrania J, Rüberg S, Becker A. 2008. Fine-tuning of galactoglucan biosynthesis in Sinorhizobium meliloti by differential ExpG- (WggR-), PhoB-, and MucR-dependent regulation of two promoters. J Bacteriol 190:3456–3466. doi: 10.1128/JB.00062-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bahlawane C, McIntosh M, Krol E, Becker A. 2008. Sinorhizobium meliloti regulator MucR couples exopolysaccharide synthesis and motility. Mol Plant Microbe Interact 21:1498–1509. doi: 10.1094/MPMI-21-11-1498. [DOI] [PubMed] [Google Scholar]
- 51.Pellock BJ, Teplitski M, Boinay RP, Bauer WD, Walker GC. 2002. A LuxR homolog controls production of symbiotically active extracellular polysaccharide II by Sinorhizobium meliloti. J Bacteriol 184:5067–5076. doi: 10.1128/JB.184.18.5067-5076.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sevin EW, Barloy-Hubler F. 2007. RASTA-Bacteria: a web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol 8:R155. doi: 10.1186/gb-2007-8-8-r155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morris J, González JE. 2009. The novel genes emmABC are associated with exopolysaccharide production, motility, stress adaptation, and symbiosis in Sinorhizobium meliloti. J Bacteriol 191:5890–5900. doi: 10.1128/JB.00760-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geddes BA, González JE, Oresnik IJ. 2014. Exopolysaccharide production in response to medium acidification is correlated with an increase in competition for nodule occupancy. Mol Plant Microbe Interact 27:1307–1317. doi: 10.1094/MPMI-06-14-0168-R. [DOI] [PubMed] [Google Scholar]
- 55.Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, Meuwly M, Jenal U. 2006. Allosteric control of cyclic di-GMP signaling. J Biol Chem 281:32015–32024. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- 56.Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev 22:2434–2446. doi: 10.1101/gad.475808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reuber TL, Walker GC. 1993. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell 74:269–280. doi: 10.1016/0092-8674(93)90418-P. [DOI] [PubMed] [Google Scholar]
- 58.Hallez R, Bellefontaine AF, Letesson JJ, De Bolle X. 2004. Morphological and functional asymmetry in α-proteobacteria. Trends Microbiol 12:361–365. doi: 10.1016/j.tim.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 59.Mohl DA, Easter J, Gober JW. 2001. The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol Microbiol 42:741–755. [DOI] [PubMed] [Google Scholar]
- 60.Fogel MA, Waldor MK. 2006. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev 20:3269–3282. doi: 10.1101/gad.1496506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Srivastava D, Waters CM. 2012. A tangled web: regulatory connections between quorum sensing and cyclic di-GMP. J Bacteriol 194:4485–4493. doi: 10.1128/JB.00379-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kozlova EV, Khajanchi BK, Sha J, Chopra AK. 2011. Quorum sensing and c-di-GMP-dependent alterations in gene transcripts and virulence-associated phenotypes in a clinical isolate of Aeromonas hydrophila. Microb Pathog 50:213–223. doi: 10.1016/j.micpath.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu J, Kim J, Danhorn T, Merritt PM, Fuqua C. 2012. Phosphorus limitation increases attachment in Agrobacterium tumefaciens and reveals a conditional functional redundancy in adhesin biosynthesis. Res Microbiol 163:674–684. doi: 10.1016/j.resmic.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Laus MC, Logman TJ, Lamers GE, Van Brussel AA, Carlson RW, Kijne JW. 2006. A novel polar surface polysaccharide from Rhizobium leguminosarum binds host plant lectin. Mol Microbiol 59:1704–1713. doi: 10.1111/j.1365-2958.2006.05057.x. [DOI] [PubMed] [Google Scholar]
- 65.Williams A, Wilkinson A, Krehenbrink M, Russo DM, Zorreguieta A, Downie JA. 2008. Glucomannan-mediated attachment of Rhizobium leguminosarum to pea root hairs is required for competitive nodule infection. J Bacteriol 190:4706–4715. doi: 10.1128/JB.01694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Becker A, Rüberg S, Küster H, Roxlau AA, Keller M, Ivashina T, Cheng HP, Walker GC, Pühler A. 1997. The 32-kilobase exp gene cluster of Rhizobium meliloti directing the biosynthesis of galactoglucan: genetic organization and properties of the encoded gene products. J Bacteriol 179:1375–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Glazebrook J, Walker GC. 1989. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56:661–672. doi: 10.1016/0092-8674(89)90588-6. [DOI] [PubMed] [Google Scholar]
- 68.Glenn SA, Gurich N, Feeney MA, González JE. 2007. The ExpR/Sin quorum-sensing system controls succinoglycan production in Sinorhizobium meliloti. J Bacteriol 189:7077–7088. doi: 10.1128/JB.00906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Charoenpanich P, Meyer S, Becker A, McIntosh M. 2013. Temporal expression program of quorum sensing-based transcription regulation in Sinorhizobium meliloti. J Bacteriol 195:3224–3236. doi: 10.1128/JB.00234-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keller M, Roxlau A, Weng WM, Schmidt M, Quandt J, Niehaus K, Jording D, Arnold W, Pühler A. 1995. Molecular analysis of the Rhizobium meliloti mucR gene regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan. Mol Plant Microbe Interact 8:267–277. doi: 10.1094/MPMI-8-0267. [DOI] [PubMed] [Google Scholar]
- 71.González JE, Reuhs BL, Walker GC. 1996. Low molecular weight EPS II of Rhizobium meliloti allows nodule invasion in Medicago sativa. Proc Natl Acad Sci U S A 93:8636–8641. doi: 10.1073/pnas.93.16.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rotter C, Mühlbacher S, Salamon D, Schmitt R, Scharf B. 2006. Rem, a new transcriptional activator of motility and chemotaxis in Sinorhizobium meliloti. J Bacteriol 188:6932–6942. doi: 10.1128/JB.01902-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pultz IS, Christen M, Kulasekara HD, Kennard A, Kulasekara B, Miller SI. 2012. The response threshold of Salmonella PilZ domain proteins is determined by their binding affinities for c-di-GMP. Mol Microbiol 86:1424–1440. doi: 10.1111/mmi.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hellweg C, Pühler A, Weidner S. 2009. The time course of the transcriptomic response of Sinorhizobium meliloti 1021 following a shift to acidic pH. BMC Microbiol 9:37. doi: 10.1186/1471-2180-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abel S, Bucher T, Nicollier M, Hug I, Kaever V, Abel Zur Wiesch P, Jenal U. 2013. Bi-modal distribution of the second messenger c-di-GMP controls cell fate and asymmetry during the caulobacter cell cycle. PLoS Genet 9:e1003744. doi: 10.1371/journal.pgen.1003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Skotnicka D, Petters T, Heering J, Hoppert M, Kaever V, Søgaard-Andersen L. 29 June 2015. c-di-GMP regulates type IV pili-dependent-motility in Myxococcus xanthus. J Bacteriol doi: 10.1128/JB.00281-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blanka A, Düvel J, Dötsch A, Klinkert B, Abraham WR, Kaever V, Ritter C, Narberhaus F, Häussler S. 2015. Constitutive production of c-di-GMP is associated with mutations in a variant of Pseudomonas aeruginosa with altered membrane composition. Sci Signal 8:ra36. doi: 10.1126/scisignal.2005943. [DOI] [PubMed] [Google Scholar]
- 78.Spangler C, Böhm A, Jenal U, Seifert R, Kaever V. 2010. A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J Microbiol Methods 81:226–231. doi: 10.1016/j.mimet.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 79.Hunter JL, Severin GB, Koestler BJ, Waters CM. 2014. The Vibrio cholerae diguanylate cyclase VCA0965 has an AGDEF active site and synthesizes cyclic di-GMP. BMC Microbiol 14:22. doi: 10.1186/1471-2180-14-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tarutina M, Ryjenkov DA, Gomelsky M. 2006. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J Biol Chem 281:34751–34758. doi: 10.1074/jbc.M604819200. [DOI] [PubMed] [Google Scholar]
- 81.Ferreira RB, Antunes LC, Greenberg EP, McCarter LL. 2008. Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J Bacteriol 190:851–860. doi: 10.1128/JB.01462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Levet-Paulo M, Lazzaroni JC, Gilbert C, Atlan D, Doublet P, Vianney A. 2011. The atypical two-component sensor kinase Lpl0330 from Legionella pneumophila controls the bifunctional diguanylate cyclase-phosphodiesterase Lpl0329 to modulate bis-(3′-5′)-cyclic dimeric GMP synthesis. J Biol Chem 286:31136–31144. doi: 10.1074/jbc.M111.231340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Feirer N, Xu J, Allen KD, Koestler BJ, Bruger EL, Waters CM, White RH, Fuqua C. 2015. A pterin-dependent signaling pathway regulates a dual-function diguanylate cyclase-phosphodiesterase controlling surface attachment in Agrobacterium tumefaciens. mBio 6:e00156. doi: 10.1128/mBio.00156-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Levi A, Folcher M, Jenal U, Shuman HA. 2011. Cyclic diguanylate signaling proteins control intracellular growth of Legionella pneumophila. mBio 2:e00316–10. doi: 10.1128/mBio.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tan H, West JA, Ramsay JP, Monson RE, Griffin JL, Toth IK, Salmond GP. 2014. Comprehensive overexpression analysis of cyclic-di-GMP signalling proteins in the phytopathogen Pectobacterium atrosepticum reveals diverse effects on motility and virulence phenotypes. Microbiology 160:1427–1439. doi: 10.1099/mic.0.076828-0. [DOI] [PubMed] [Google Scholar]
- 86.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hecht GB, Newton A. 1995. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J Bacteriol 177:6223–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim J, Heindl JE, Fuqua C. 2013. Coordination of division and development influences complex multicellular behavior in Agrobacterium tumefaciens. PLoS One 8:e56682. doi: 10.1371/journal.pone.0056682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shapiro L, Agabian-Keshishian N, Bendis I. 1971. Bacterial differentiation. Science 173:884–892. doi: 10.1126/science.173.4000.884. [DOI] [PubMed] [Google Scholar]
- 90.Shaw CH, Loake GJ, Brown AP, Garrett CS, Deakin W, Alton G, Hall M, Jones SA, O'Leary M, Primavesi L. 1991. Isolation and characterization of behavioural mutants and genes of Agrobacterium tumefaciens. Microbiology 137:1939–1953. [Google Scholar]
- 91.Platzer J, Sterr W, Hausmann M, Schmitt R. 1997. Three genes of a motility operon and their role in flagellar rotary speed variation in Rhizobium meliloti. J Bacteriol 179:6391–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Deng Y, Lim A, Wang J, Zhou T, Chen S, Lee J, Dong YH, Zhang LH. 2013. cis-2-Dodecenoic acid quorum sensing system modulates N-acyl homoserine lactone production through RpfR and cyclic di-GMP turnover in Burkholderia cenocepacia. BMC Microbiol 13:148. doi: 10.1186/1471-2180-13-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li G, Brown PJ, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. 2012. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol Microbiol 83:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li G, Smith CS, Brun YV, Tang JX. 2005. The elastic properties of the Caulobacter crescentus adhesive holdfast are dependent on oligomers of N-acetylglucosamine. J Bacteriol 187:257–265. doi: 10.1128/JB.187.1.257-265.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rinaudi LV, Sorroche F, Zorreguieta A, Giordano W. 2010. Analysis of the mucR gene regulating biosynthesis of exopolysaccharides: implications for biofilm formation in Sinorhizobium meliloti Rm1021. FEMS Microbiol Lett 302:15–21. doi: 10.1111/j.1574-6968.2009.01826.x. [DOI] [PubMed] [Google Scholar]
- 96.Santos MR, Marques AT, Becker JD, Moreira LM. 2014. The Sinorhizobium meliloti EmrR regulator is required for efficient colonization of Medicago sativa root nodules. Mol Plant Microbe Interact 27:388–399. doi: 10.1094/MPMI-09-13-0284-R. [DOI] [PubMed] [Google Scholar]
- 97.Fujishige NA, Kapadia NN, De Hoff PL, Hirsch AM. 2006. Investigations of Rhizobium biofilm formation. FEMS Microbiol Ecol 56:195–206. doi: 10.1111/j.1574-6941.2005.00044.x. [DOI] [PubMed] [Google Scholar]
- 98.Karatan E, Watnick P. 2009. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev 73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Russell MH, Bible AN, Fang X, Gooding JR, Campagna SR, Gomelsky M, Alexandre G. 2013. Integration of the second messenger c-di-GMP into the chemotactic signaling pathway. mBio 4:e00001–13. doi: 10.1128/mBio.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Martínez-Granero F, Navazo A, Barahona E, Redondo-Nieto M, González de Heredia E, Baena I, Martín-Martín I, Rivilla R, Martín M. 2014. Identification of flgZ as a flagellar gene encoding a PilZ domain protein that regulates swimming motility and biofilm formation in Pseudomonas. PLoS One 9:e87608. doi: 10.1371/journal.pone.0087608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bian J, Liu X, Cheng YQ, Li C. 2013. Inactivation of cyclic di-GMP binding protein TDE0214 affects the motility, biofilm formation, and virulence of Treponema denticola. J Bacteriol 195:3897–3905. doi: 10.1128/JB.00610-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pitzer JE, Sultan SZ, Hayakawa Y, Hobbs G, Miller MR, Motaleb MA. 2011. Analysis of the Borrelia burgdorferi cyclic-di-GMP-binding protein PlzA reveals a role in motility and virulence. Infect Immun 79:1815–1825. doi: 10.1128/IAI.00075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.