ABSTRACT
Intracellular levels of the bacterial second messenger cyclic di-GMP (c-di-GMP) are controlled by antagonistic activities of diguanylate cyclases and phosphodiesterases. The phosphodiesterase PdeH was identified as a key regulator of motility in Escherichia coli, while deletions of any of the other 12 genes encoding potential phosphodiesterases did not interfere with motility. To analyze the roles of E. coli phosphodiesterases, we demonstrated that most of these proteins are expressed under laboratory conditions. We next isolated suppressor mutations in six phosphodiesterase genes, which reinstate motility in the absence of PdeH by reducing cellular levels of c-di-GMP. Expression of all mutant alleles also led to a reduction of biofilm formation. Thus, all of these proteins are bona fide phosphodiesterases that are capable of interfering with different c-di-GMP-responsive output systems by affecting the global c-di-GMP pool. This argues that E. coli possesses several phosphodiesterases that are inactive under laboratory conditions because they lack appropriate input signals. Finally, one of these phosphodiesterases, PdeL, was studied in more detail. We demonstrated that this protein acts as a transcription factor to control its own expression. Motile suppressor alleles led to a strong increase of PdeL activity and elevated pdeL transcription, suggesting that enzymatic activity and transcriptional control are coupled. In agreement with this, we showed that overall cellular levels of c-di-GMP control pdeL transcription and that this control depends on PdeL itself. We thus propose that PdeL acts both as an enzyme and as a c-di-GMP sensor to couple transcriptional activity to the c-di-GMP status of the cell.
IMPORTANCE Most bacteria possess multiple diguanylate cyclases and phosphodiesterases. Genetic studies have proposed that these enzymes show signaling specificity by contributing to distinct cellular processes without much cross talk. Thus, spatial separation of individual c-di-GMP signaling units was postulated. However, since most cyclases and phosphodiesterases harbor N-terminal signal input domains, it is equally possible that most of these enzymes lack their activating signals under laboratory conditions, thereby simulating signaling specificity on a genetic level. We demonstrate that a subset of E. coli phosphodiesterases can be activated genetically to affect the global c-di-GMP pool and thus influence different c-di-GMP-dependent processes. Although this does not exclude spatial confinement of individual phosphodiesterases, this study emphasizes the importance of environmental signals for activation of phosphodiesterases.
INTRODUCTION
The second messenger cyclic di-GMP (c-di-GMP) is a nearly ubiquitous small signaling molecule which greatly affects bacterial growth and behavior. In particular, c-di-GMP controls important cellular and behavioral processes in a wide range of bacteria, including motility and chemotaxis, surface colonization and the formation of communities, virulence and persistence, and cell cycle progression (for reviews, see references 1 to 3). The key enzymes involved in c-di-GMP metabolism are diguanylate cyclases (DGCs) (4) and phosphodiesterases (PDEs) (5). Together, DGCs and PDEs constitute one of the largest families of bacterial signaling proteins, with tens of thousands of members currently deposited in the protein databases. Contributing to an explanation for this enormous multiplicity and diversity is the observation that most bacteria contain multiple representatives, often a few tens, of these proteins (3). For example, the genomes of Escherichia coli K-12 strains contain genes encoding a total of 29 proteins harboring a GGDEF and/or EAL domain, the catalytic units of DGC and PDE enzyme activities, respectively (6). Moreover, throughout evolution, many of the formerly catalytic members of this family seem to have adopted novel functionalities as c-di-GMP effector proteins (7–11) or as protein interaction platforms that have lost the connection to their original effector altogether (12).
This caused some confusion in the field in the early years and raised the question of why bacteria evolved multiple DGCs and PDEs to control a small signaling molecule that likely shows rapid diffusion within bacterial cells, thereby providing limited options for signaling specificity. One possible explanation for this phenomenon is that individual representatives are expressed under specific environmental conditions or are specialized for specific cellular tasks which normally are kept separate from each other in either time or space (2). In the case of temporal sequestration, one would expect that only a subset of these enzymes is expressed at any given time or environmental situation. The other possibility is that cells express and display multiple members of this enzyme family to be able to rapidly respond to a diverse range of signaling inputs. In this case, one would expect that most or possibly all enzymes are expressed at any given time but that the majority of them are not active due to the absence of an input signal. In the past few years, the amount of information about biochemical and structural characteristics of DGCs and PDEs has increased rapidly (13–18). Despite such rapid progress, in vivo results often remain controversial. Considering that specific components of this signaling network might not be expressed or might not receive the appropriate stimuli to be active, genetic studies relying solely on mutant phenotypes will not give conclusive answers.
Here we address these questions by analyzing the expression and activities of multiple PDEs in E. coli K-12. This organism has a total of 16 EAL domain proteins, only 3 of which show obvious degeneration of consensus amino acid motifs required for catalytic activity (Fig. 1a and b). Among the other 13 proteins, only 7 have been characterized in detail and identified as PDEs (19–25). The functions of the other members of this family that potentially are able to catalyze c-di-GMP hydrolysis remain unclear. To identify additional candidate PDEs, we made use of a genetic approach by sequentially isolating activating gain-of-function mutations in specific members of the EAL domain proteins. Our analysis is based on some recent reports demonstrating that PdeH (YhjH), a highly active PDE that globally controls c-di-GMP levels in E. coli, is primarily responsible for motility control in this organism (19, 26, 27). The pdeH gene is coregulated with flagellar genes, and mutants lacking PdeH show increased c-di-GMP levels and poor motility. PdeH licenses flagellar motility in the exponential and early postexponential phases by keeping c-di-GMP levels low. Upon entry into stationary phase, c-di-GMP levels increase partially due to FlhDC-dependent downregulation of pdeH (28), leading to activation of the c-di-GMP effector protein YcgR, which interacts with the flagellar motor to curb its activity (27, 29). Thus, in growing E. coli cells, PdeH has a central role in maintaining cell motility by keeping the cellular concentration of c-di-GMP below a threshold level that is able to activate YcgR. The observation that pdeH mutants showed poor motility also suggested that under these conditions, no other PDE was expressed or active (enough) to functionally substitute for this PDE. We thus hypothesized that mutations activating any of the other PDEs would be able to restore the motility of the pdeH mutant. If so, this would then allow us to identify silent PDEs by genetically uncoupling their activities from the unknown signals that are normally required for their activation. We present genetic and biochemical evidence that a large fraction of the remaining potential PDEs can indeed be activated genetically to substitute for the function of PdeH. This argues in favor of the idea that these proteins are bona fide PDEs that are able to interfere with the general cellular pool of c-di-GMP and that, under laboratory conditions, these proteins lack the appropriate signal(s) to become active.
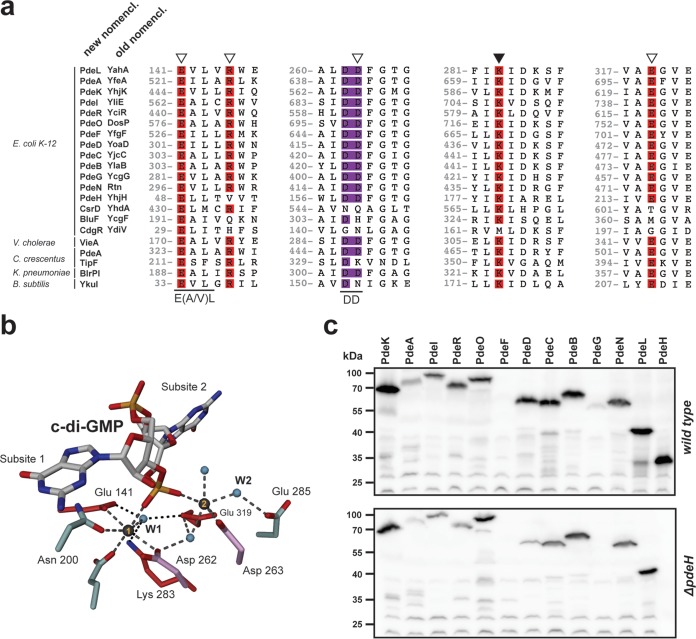
FIG 1.
Conservation of c-di-GMP-specific phosphodiesterases. (a) ClustalW alignment of PdeL with endogenous and exogenous phosphodiesterases and noncatalytic EAL domains. Regions containing residues involved in substrate binding (open triangles) and catalysis (closed triangle) are highlighted. Amino acid numbering refers to the numbering for PdeL. The double-aspartic-acid motif (DD) is displayed in purple, in analogy to panel b. (b) Atomic organization of the catalytic site of a PdeL monomer. The two essential metals are displayed as gray spheres. Conserved residues involved in metal coordination or catalysis are displayed in red. The double-aspartic-acid motif (Asp262 and Asp263; shown in purple) is involved in metal ion coordination as well as coordination of the catalytic water (small blue spheres). The same is true for the glutamic acid at position E141, which is part of the conserved E(A/V)L motif. (c) Immunoblot analysis of PDEs in E. coli wild-type and ΔpdeH mutant strains. PDEs were tagged at their C termini with a 3×Flag tag and were analyzed with an anti-Flag antibody. Cells were grown in tryptone broth (TB) at 37°C and harvested at an OD600 of 0.8. Note that in both strain backgrounds, all pde genes were expressed, with the exception of pdeF. PdeG levels were low in the wild type, and the protein seemed to be absent in the ΔpdeH mutant.
Please note that throughout this report we use the systematic nomenclature for E. coli DGCs and PDEs that was recently proposed by Hengge et al. (30). To make it easier for the expert reader to adopt the new nomenclature, the corresponding traditional designations are listed in Fig. 1a and are highlighted in parentheses in the text.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. E. coli K-12 MG1655, obtained from Blattner et al. (31), and its derivatives were grown as indicated in the relevant sections. When needed, antibiotics were included at the following concentrations: 30 μg/ml chloramphenicol for plasmids, 20 μg/ml chloramphenicol for chromosomal chloramphenicol resistance cassettes, 12.5 μg/ml tetracycline, 50 μg/ml kanamycin, 100 μg/ml ampicillin for plasmids, and 30 μg/ml ampicillin for chromosomal ampicillin resistance cassettes.
DNA work. (i) PCR amplification.
Each PCR mixture contained the following: 1× polymerase buffer (NEB), a mix containing a 0.1 mM concentration of each deoxynucleoside triphosphate (dNTP), 0.3 μM forward primer, 0.3 μM reverse primer, 10.20 pg template DNA, and 0.7 μl Taq polymerase (NEB). For colony PCR, a single colony was picked up with a pipette tip and resuspended in the PCR mixture.
(ii) Gel electrophoresis.
Five microliters of PCR product was mixed with DNA loading dye, loaded into a 1% agarose gel supplemented with a 1:20,000 dilution of RedSafe DNA stain (iNtRON), and separated using 1× Tris-borate-EDTA (TBE) buffer. DNA was analyzed under UV light.
(iii) Sequencing.
Linear DNA was purified using NucleoSpin extract II (Macherey-Nagel). Sequencing reactions were carried out by Microsynth AG (Balgach, Switzerland). The sequences obtained were assembled and analyzed using 4Peaks.
(iv) Plasmid preparation.
Plasmid DNA was purified using a GenElute plasmid miniprep kit (Sigma) according to the commercial protocol.
(v) TSS transformation.
Transcription start site (TSS) transformation of plasmid DNA was carried out as previously described (32).
(vi) Electroporation.
For electroporation of purified linear DNA with a Bio-Rad GenPulser cuvette (1-mm diameter), the following electroporation settings were applied: 400 Ω, 1.75 kV, and 25 μF.
P1 phage lysate preparation and transduction.
P1 phage lysate preparations and transductions were carried out essentially as described by Miller (33).
λ-RED recombineering. (i) Chromosomal gene deletions and modifications.
Gene deletions were carried out essentially as described by Datsenko and Wanner (34), with the use of a comprehensive mutant library (Keio collection [35]) and P1-mediated transduction. Chromosomal 3×Flag tags were constructed according to the published method of Uzzau et al. (36). For unmodified strains, AB330 was used (see Table S1 in the supplemental material), whereas pKD46 was used for construction of 3×Flag-tagged versions of the motile suppressor mutants. Kanamycin resistance markers used for selection during strain construction were removed by site-specific recombination using pCP20, generating a short, “Frt” scar sequence which replaced the deleted gene or cotransduced kanamycin resistance marker (34).
(ii) Construction of lacZ promoter fusions.
The construction of chromosomal lacZ promoter fusions was constructed via λ-RED-mediated recombination essentially as described above. AB989 (see Table S1) was used as a template for construction of the reporter fusion. AB989 contains Prha-ccdB and a flanking kanamycin resistance cassette which is inserted upstream of the native lacZ locus. The donor PCR fragment harboring the promoter of interest was designed to site-specifically excise Prha-ccdB and integrate upstream of the lacZ open reading frame (ORF), generating a merodiploid translational fusion. Selection of successful integration events was achieved through growth at 30°C on minimal medium plates provided with 0.2% rhamnose supplemented with 0.5 μg/ml biotin. The fusion was transduced into strains of interest via P1 transduction.
Immunoblotting.
Cells were grown with shaking in tryptone broth (TB) at 37°C until an optical density at 600 nm (OD600) of 0.8. An equivalent of 1 ml of cells at an OD600 of 1.0 was pelleted and resuspended in 100 μl SDS Laemmli buffer. For detection of 3×Flag-tagged PdeA and its derivatives, the cells were resuspended in 30 μl SDS loading dye. Cells were lysed by boiling the sample at 98°C for 15 min. Eight microliters of the total cell extract was loaded onto a 12.5% SDS-polyacrylamide gel, and proteins were transferred by use of a Bio-Rad wet blot system. Proteins were detected with a 1:10,000 dilution of mouse anti-Flag monoclonal antibody (Sigma) and a 1:10,000 dilution of horseradish peroxidase (HRP)-conjugated rabbit anti-mouse secondary antibody (DakoCytomation, Denmark). Proteins were visualized by use of an enhanced chemiluminescence (ECL) detection reagent (PerkinElmer Life Sciences) on a photo film (Fuji) or gel imager (GE ImageQuant LAS 4000).
Suppressor screen.
The strains used for the genetic forward screen of individual PDEs are listed in Table S1 in the supplemental material. Each strain was transformed with pwspR. For each screen, 300 TB swarm plates supplemented with tetracycline were prepared, among which 100 were supplemented with 5 μM isopropyl-β-d-thiogalactopyranoside (IPTG) and 100 were supplemented with 20 μM IPTG. Single colonies of the screening strain harboring pwspR were applied to screening plates. The screening plates were incubated at 37°C. Over the course of a week, all plates displaying motile suppressor mutants showed visible flares spreading from the center of inoculation. The motile suppressor mutants were isolated and pooled in a liquid LB culture. A pool lysate was prepared and transduced into AB607 (ΔpdeH). Transductants were picked up and placed on TB swarm plates supplemented with kanamycin and 20 mM sodium citrate. After incubation at 37°C for 3 to 4 h, the motile suppressor mutants that appeared were restreaked, and the ORF of the PDE of interest was sequenced.
Video tracking.
Bacterial swimming speed measurements were carried out essentially as described by Boehm et al. (27). Briefly, bacteria were grown in TB at 37°C to an OD600 of 0.8. Cells were diluted 1:100 into fresh TB and applied to a coverslip that was attached to a glass slide with two-sided adhesive tape. Two videos of 30 s each were recorded at 15 frames per s with a video microscope and dark-field optics at a magnification of ×40. The acquired videos were imported into ImageJ 1.43 (NIH), and trajectories were calculated with the “2D particle tracker” plug-in. Velocities and statistical data were computed via a custom-made R script.
c-di-GMP measurements.
c-di-GMP measurements were performed according to the published procedure of Spangler et al. (37). Briefly, E. coli cells were grown in 5 ml TB at 37°C until an OD600 of 0.8. The culture was pelleted and washed in 300 μl ice-cold distilled H2O. After washing, the cell pellet was resuspended in 300 μl ice-cold extraction solvent (acetonitrile/methanol/distilled H2O, 40/20/20 [vol/vol/vol]). After incubating on ice for 15 min, the samples were boiled at 100°C for 15 min. After pelleting, the supernatant was transferred to a safe-lock tube, and the extraction procedure was repeated twice with 200 μl extraction solvent. Biological triplicates were performed for each tested bacterial strain. Measurements were performed in collaboration with the group of Volkhard Kaever (Institute of Pharmacology, Hannover, Germany) via high-pressure liquid chromatography–tandem mass spectrometry (HPLC-MS/MS). Measured values were mathematically converted into intracellular c-di-GMP concentrations (micromolar) per CFU.
Attachment assay.
Attachment assays were carried out as described by Boehm et al. (38). Briefly, 5 μl of a shaking overnight culture grown in TB at 37°C was used to inoculate 200 μl TB provided in a 96-well microtiter plate (Falcon, NJ). The plate was incubated statically at 30°C for 24 h. For quantification of cellulose-dependent attachment, cells were incubated statically in TB at room temperature for 24 h. After recording of the OD600 of the total biomass, the planktonic phase of the culture was discarded and the wells were washed with deionized water from a hose. The total attached biomass was stained with 300 μl 0.3% crystal violet (0.3% [wt/vol] in distilled H2O, 5% [vol/vol] 2-propanol, 5% [vol/vol] methanol) for 20 min. Subsequently, the plate was washed, and the remaining crystal violet-stained biomass was dissolved in 20% acetic acid for 20 min and quantified by measuring the OD600. Attachment was normalized to the initially measured total biomass.
Protein purification. (i) Strep II purification.
C-terminally Strep II-tagged wild-type and mutant variants of pdeL were cloned into a pET28a vector (Novagen) between the NcoI and NotI restriction sites. Proteins were overexpressed from plasmids in BL21-AI cells grown at 30°C in 2 liters of LB medium. At an OD600 of 0.6, the culture was induced with 0.1% l-arabinose. Cells were harvested at 4 h postinduction by centrifugation at 3,500 rpm for 30 min at 4°C. The cell pellet was resuspended in 8 ml buffer A (100 mM Tris-HCl, pH 8.0, 250 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM dithiothreitol [DTT]) including a tablet of Complete mini EDTA-free protease inhibitor (Roche) and a spatula tip of DNase I (Roche). Cells were lysed in a French press and the lysate cleared at 4°C in a table-top centrifuge for 40 min at full speed. The cleared supernatant was loaded onto 1 ml Strep-Tactin Superflow Plus resin (Qiagen). The supernatant was reloaded another two times before washing with a total of 60 ml buffer A. Protein was eluted as 500-μl aliquots with a total of 10 ml elution buffer A containing 2.5 mM d-desthiobiotin. Fractions with the highest protein concentrations were pooled.
(ii) Heparin purification.
A 1-ml HiTrap heparin HP column (GE Healthcare) was washed with 10 ml distilled H2O followed by equilibration with 10 ml buffer A. The eluate from the Strep II purification was loaded three times. After loading, the column was washed with 10 ml buffer A followed by a washing step with 10 ml buffer B (100 mM Tris-HCl, pH 8.0, 300 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT). The protein was eluted in 500 μM fractions with a total of 10 ml buffer C (100 mM Tris-HCl, pH 8.0, 1 M NaCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT). The fractions containing the highest protein concentrations were pooled and dialyzed overnight against 1.5 liters of dialysis buffer (100 mM Tris-HCl, pH 8.0, 250 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT). The final protein concentration was recorded at 280 nm, and the content of copurified nucleotides was determined through the 260/280 nm ratio.
c-di-GMP hydrolysis assay and data fitting (phosphate sensor assay).
PdeL-catalyzed conversion of c-di-GMP to the linear pGpG dinucleotide was measured indirectly by a novel alkaline phosphatase (AP)/phosphate sensor online assay. In this assay, the terminal phosphate of the pGpG product is cleaved by the coupling enzyme AP (20 U/μl; 5 U in assay mixture; Roche), and the phosphate concentration is determined from the fluorescence increase through binding of phosphate to the phosphate sensor (0.5 μM in assay mixture; Life Technologies).
Dialysis buffer was used as the assay buffer. The assay was performed at a protein concentration of 100 nM and substrate concentrations ranging from 100 nM to 5 μM, in a final volume of 300 μl in a 5-mm by 5-mm cuvette (Hellma Analytics). Progress curves were recorded with a Jasco FP-6500 fluorescence spectrophotometer at 20°C. The instrument settings were as follows: bandwidth (excitation), 5 nm; bandwidth (emission), 5 nm; excitation wavelength, 430 nm; emission wavelength, 468 nm; response, 1 s; sensitivity, low; and data pitch, 2 s.
The measured progress curves of fluorescence increases were fitted to the following scheme:
| (1) |
| (2) |
| (3) |
with the measured relative fluorescence units (RFU) originating from the uncomplexed (RFU1) and complexed (RFU2) sensors, as follows: RFU = RFU1 + RFU2 = sc × PS + sc × gain × P-PS, where sc is the scaling factor.
By a sufficiently large concentration of AP, it was ensured that reaction 2 was not rate limiting. The equilibrium dissociation constant for PS (KD PS) was obtained by phosphate titration in the absence of enzymes. Fitting of the data with this kinetic model was done with a custom-built Python script using NumPy and SciPy libraries. The corresponding differential equations were integrated with the assumption that product formation is the rate-determining step. The kinetic parameters of the PDE (KD PDE and kcat) as well as the scaling parameters (sc and gain) were refined globally for each series of experiments measured with various substrate concentrations. An observed slight background increase with time was taken into account by addition of a linear term with locally refined parameters. Fitted progress curves as well as individual KM and kcat values are documented in Fig. S2 and Table S2 in the supplemental material.
Electrophoretic mobility shift assay (EMSA).
Cy3-labeled DNA probes were generated via either oligonucleotide annealing or PCR, using E. coli MG1655 as the template. Oligonucleotides used are indicated in the oligonucleotide list in Table S1 in the supplemental material. DNA (10 nM) was incubated with purified PdeL-Strep II (0, 200, 400, or 600 nM) for 10 min at room temperature in 10 μl buffer (50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 10 mM MgCl2, 10% glycerol, 1 mM DTT, 0.01% Triton X-100, 0.1 mg/ml bovine serum albumin [BSA], and 25 μg/ml λ-DNA). After electrophoresis on 8% polyacrylamide gels, DNA-protein complexes were analyzed using a Typhoon FLA 7000 imager (GE Healthcare).
β-Galactosidase assay.
Strains were grown in TB overnight at 37°C. The next day, cultures were diluted 1:1,000 in fresh medium and grown with shaking at 37°C to an OD600 of 0.8. An equivalent of 1 ml of culture at an OD600 of 1.0 was pelleted and resuspended in 1 ml Z buffer (75 mM Na2HPO4, 40 mM NaH2PO4, 1 mM KCl, 1 mM MgSO4; pH 7.0). One hundred microliters of 0.1% SDS was added together with 20 μl chloroform. The samples were vortexed for 20 s and then left on the bench to sediment until samples cleared up. Two hundred microliters of each sample was transferred to a 96-well plate. Twenty-five microliters of a 4-mg/ml o-nitrophenyl-β-d-galactopyranoside (ONPG) solution (dissolved in Z buffer) was added as the substrate. The β-galactosidase activity was measured in a plate reader at 405 nm (20 reads; fastest interval) and determined as the initial slope of the curve in the linear range. Experiments were carried out as biological triplicates.
RESULTS
Expression of PDEs in growing E. coli cells.
High levels of c-di-GMP generally obstruct flagellar motility in various microbes (27, 28, 39–41). As a consequence, PDEs play key roles in regulating cell motility (42, 43). In E. coli, the PDE PdeH appears to be the sole contributor to the maintenance of cell motility under laboratory conditions (27). This is surprising since the genomes of E. coli K-12 strains encode more than a dozen additional potential PDEs (6). One possibility is that most of these components are not expressed during growth under these conditions. Previous studies used microarrays and β-galactosidase reporter assays to demonstrate that, with the exception of pdeF (yfgF) and pdeG (ycgG), all genes encoding potential PDEs are actively transcribed (44, 45). To confirm this and to demonstrate that active transcription indeed results in the production of PDEs, chromosomal 3×Flag-tagged constructs were engineered for all potential pde genes in the E. coli strain MG1655. These were introduced into the wild type and a ΔpdeH mutant background, and protein levels were monitored in exponentially growing cells (OD600 of 0.5 to 0.8). As shown in Fig. 1c, most PDEs were readily detected. The only exceptions were PdeF and PdeG, the latter of which was present at low levels in the wild-type background but absent in the ΔpdeH strain. This confirmed previous results and indicated that these proteins failed to contribute to cell motility as a result of a lack of expression under these conditions. Rather, most of these PDEs may be present in the cell at high enough concentrations but may not interfere with motility control because of a lack of enzyme activity.
Motile suppressor mutants of a pdeH mutant identify activating mutations in alternative PDEs.
A ΔpdeH mutant is unable to swim effectively toward higher nutrient concentrations in motility plates. To isolate spontaneous motile suppressor mutants, the ΔpdeH mutant was inoculated onto the center of motility plates and incubated for an extended period, until visible “flares” were arising and spreading on the plates (Fig. 2a). It was shown previously that mutations in the gene encoding the motility regulator YcgR can restore motility under these conditions (27). Likewise, mutations in several genes encoding DGCs required for YcgR activation alleviate the motility block. We reasoned that activating mutations in “alternative” PDEs could also restore motility by countering high levels of c-di-GMP in the pdeH mutant. In order to enrich for such rare pde gain-of-function mutations, we first designed a tailored screening strain that reduced the likelihood of isolating mutations in known components of c-di-GMP-mediated motility control. To reduce the frequency of loss-of-function mutations in ycgR, a second chromosomal copy of ycgR was introduced into the ΔpdeH screening strain. In addition, the screening strain was equipped with a plasmid carrying a copy of wspR, the gene encoding the diguanylate cyclase WspR from Pseudomonas fluorescens. Expression of wspR from the Plac promoter maintains a threshold level of c-di-GMP that prevents motility even if one of the four active native DGCs is inactivated.
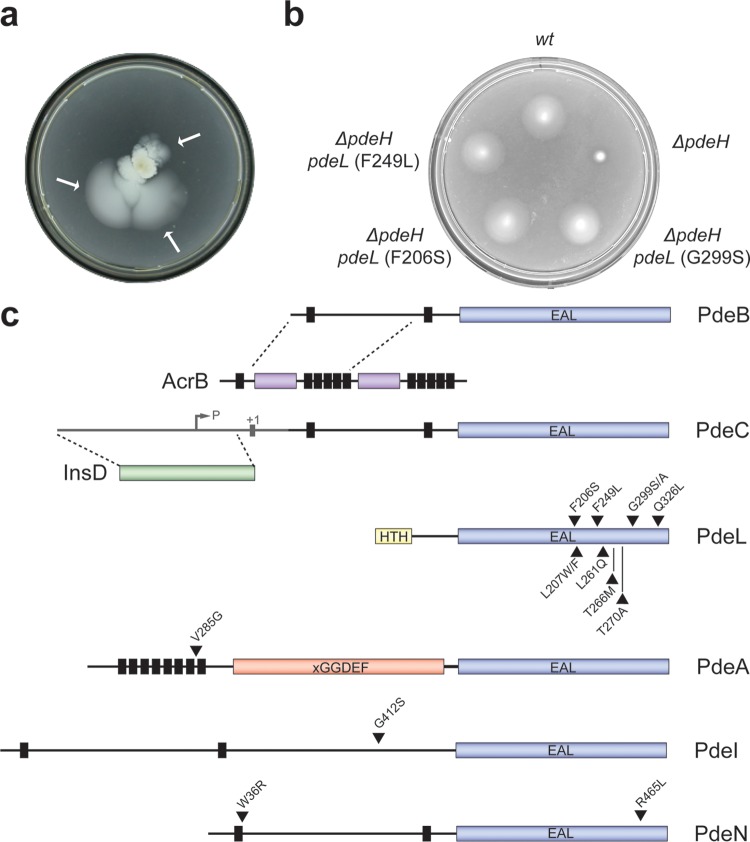
FIG 2.
Isolation of alleles activating E. coli phosphodiesterases. (a) Selection for motile suppressor mutants of a nonmotile ΔpdeH mutant strain on a low-percentage agar plate. Independent suppressors were recovered from motile flares (arrows) after incubation on motility plates for several days at 37°C. (b) Mutations in pdeL restore the motility of a ΔpdeH mutant. Mutant alleles of pdeL are indicated. Motility was examined as described for panel a. wt, wild type. (c) Graphical representation of isolated pde suppressor variants. Vertical black bars represent transmembrane helices, c-di-GMP-specific phosphodiesterase domains (EAL) are depicted in blue, the LuxR-like DNA binding domain of PdeL is shown in yellow (HTH), and the degenerate cyclase domain (xGGDEF) of PdeA is shown in red. The positions of single amino acid substitutions are marked with black triangles.
With this strain, a continuous genetic forward screen was set up. First, activating mutations in one of the pde genes were isolated from a pool of spontaneous suppressor mutants. A kanamycin resistance cassette was introduced next to the corresponding pde gene on the chromosome. Suppressor mutations linked to this marker were then identified by cotransduction into a clean ΔpdeH background and by subsequent sequencing of the neighboring DNA regions. Second, the pde gene for which motility suppressor mutants were isolated was deleted from the chromosome. With the resulting mutant strain, a new round of selection for motile suppressor mutants was initiated to isolate mutations in one of the remaining pde genes. Successive rounds of selection resulted in the isolation of a total of 16 suppressor mutations in six individual PDEs (Fig. 2b and c). Closer examination revealed gene fusion events in both pdeB and pdeC. In the case of pdeB, a 5,846-bp deletion between two direct repeats (TTGATGTCATT) resulted in an in-frame fusion of pdeB with its upstream gene, acrB, encoding a subunit of the Acr multidrug efflux pump. The resulting protein was fused at amino acid 205 of AcrB and position 168 of PdeB, giving rise to a fusion protein of a size similar to that of PdeB. As shown in Fig. 3a, the overall level of the resulting fusion protein was strongly increased compared to that of the PdeB wild type. This increase likely resulted from the direct coupling of the truncated pdeB gene with the promoter of the acr operon. We reasoned that motility suppression results either from strong overexpression or from uncoupling of the respective catalytic domain of PdeB from its N-terminal regulatory region. Similarly, an IS element (ins mobile element) inserted into the promoter region of pdeC (28 bp upstream of the putative transcriptional start site of pdeC). As in the case of PdeB, this resulted in a strong upregulation of the overall level of PdeC (Fig. 3b), indicating a suppression mechanism similar to that described above.
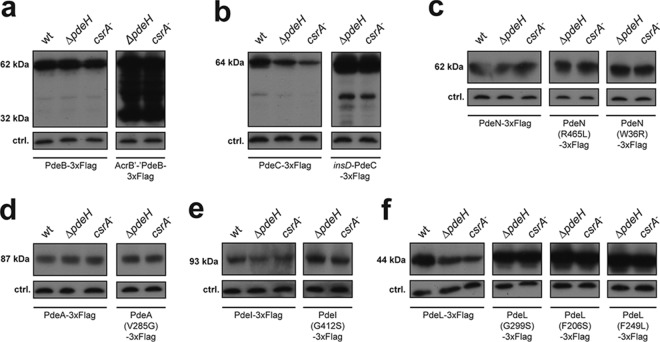
FIG 3.
Expression of mutant phosphodiesterases in E. coli. Immunoblot analysis was performed on the wild type and on strains with suppressor variants for detection of PdeB (a), PdeC (b), PdeN (c), PdeA (d), PdeI (e), and PdeL (f) carrying 3×Flag tags at their C termini. Proteins were analyzed in the following strains grown to exponential phase: wild type, ΔpdeH mutant (AB607), and csrA mutant (AB958). Suppressor variants of PdeB (a) and PdeC (b) showed strongly increased protein levels indicating derepression of their expression. In contrast, suppressor variants of PdeN, PdeA, and PdeI showed unaltered protein levels in all strain backgrounds tested. Of the 10 PdeL suppressor variants isolated, three were analyzed (G299S, F206S, and F249L). All variants showed increased protein levels in all genetic backgrounds tested. Note that protein levels of PdeC and PdeL differed in different genetic backgrounds.
Mutations resulting in single amino acid substitutions were identified in pdeL, pdeA, pdeI, and pdeN, arguing that the encoded proteins can be activated genetically (Fig. 2c). While substitutions in the pdeA-, pdeI-, and pdeN-encoded proteins localized to the EAL domain, to transmembrane regions, or to uncharacterized regions of the protein neighboring the EAL domain, mutations in the pdeL-encoded protein localized exclusively within the catalytic domain. This is in line with the observation that the soluble PdeL protein lacks a potential signal input domain and instead harbors a LuxR-type DNA binding domain. Levels of PdeA, PdeI, and PdeN proteins harboring suppressor mutations were unaltered compared to that of the wild type. Also, the cellular concentrations of these enzymes were similar in strains with different levels of c-di-GMP (Fig. 3c to e and 4). In contrast, levels of several PDEs were different in E. coli wild-type, ΔpdeH, and csrA mutant strains, indicating that their expression might be regulated by c-di-GMP itself (Fig. 3b and f). In line with this, a subset of the isolated PdeL suppressors revealed higher PdeL protein levels in all genetic backgrounds tested (Fig. 3f). This was not due to increased protein stability, as suppressor variants and wild-type PdeL showed very similar stabilities upon translation inhibition (see Fig. S1 in the supplemental material). Together with the finding that all mutations in PdeL mapped to the catalytic domain, this suggested that pdeL expression is autoregulated and possibly controlled by the overall cellular level of c-di-GMP.
FIG 4.
Swimming velocities of E. coli wild-type and phosphodiesterase mutant strains. Velocities of individual cells of the E. coli wild type (white), the ΔpdeH mutant (gray), and motile suppressor mutants of the ΔpdeH mutant (green) were scored. For statistical analysis, the Kruskal-Wallis rank sum test was applied. Swimming velocities of at least 76 single cells are shown as box plots. Boxes show the lower and upper quartiles. Black horizontal lines represent the median velocities. Dashed lines show extreme values, whereas small black squares represent individual outliers. Comparisons of motile suppressor mutants with the parental ΔpdeH strain all showed statistically significant differences (P < 0.05). Motile suppressor mutants showed swimming velocities restored to the levels observed for the wild type. Black bars represent intracellular c-di-GMP concentrations as measured by LC-MS/MS. A mutant lacking PdeH displayed a 10-fold-increased cellular c-di-GMP concentration (3.5 μM) compared to that of the wild type (0.31 μM). Motile suppressor mutants showed reduced intracellular c-di-GMP concentrations compared to their parental strain (ΔpdeH).
Together, these results indicate that E. coli possesses several PDEs that under normal conditions do not contribute to motility control but can be activated genetically to substitute for the role of the primary cellular PDE, PdeH.
Pde suppressor alleles restore motility by reducing intracellular c-di-GMP levels.
High levels of c-di-GMP interfere with flagellar motility via the YcgR effector protein. To demonstrate that the pde suppressor alleles do indeed reinstate the flagellar motor behavior of a ΔpdeH mutant by reducing levels of c-di-GMP, both single-cell trajectories and c-di-GMP concentrations were recorded for a selection of the isolated mutants. Dark-field microscopy tracking and subsequent computational analysis of the recorded trajectories determined the behavior of swimming bacteria. Measured trajectories of an exponentially growing ΔpdeH strain revealed swimming velocities of 3.4 to 6.1 μm/s (median, 4.1 μm/s), whereas a pdeH+ strain displayed velocities of 6.0 to 12.2 μm/s (median, 8.9 μm/s) (Fig. 4). Importantly, swimming velocities of all motile suppressor mutants were significantly higher than that of their isogenic ΔpdeH strain and were similar to velocities measured for the wild type. To complement these single-cell measurements, cellular c-di-GMP concentrations in cell populations of the same strains were quantified using LC-MS/MS technology (37). In accordance with earlier observations (27), levels of c-di-GMP were increased >10-fold in the ΔpdeH mutant (3.5 μM) compared to the wild type (0.31 μM). Importantly, all strains harboring mutations in PDEs showed a significant reduction of the intracellular c-di-GMP pool compared to their isogenic ΔpdeH mutant strain. While the reduction of c-di-GMP was moderate in some suppressor mutants, c-di-GMP levels were reduced to levels comparable to that of the wild type or, for one mutant, even below the detection limit (Fig. 4). Importantly, we observed a strong overall correlation between the reduction of the intracellular c-di-GMP levels and the measured swimming velocities (Fig. 4).
Together, these findings support the notion that the pde suppressor alleles increase the level and/or enzymatic activity of their respective PDE products, lowering the cellular concentration of c-di-GMP in the original ΔpdeH mutant and thereby restoring flagellar motor function.
Pde suppressor alleles reduce poly-GlcNAc levels and cellulose-dependent attachment.
The observation that the pde suppressor alleles restored motility in a ΔpdeH background by reducing the intracellular c-di-GMP concentration prompted us to test if this represents a general cellular response that can also interfere with other c-di-GMP-mediated processes. We have shown previously that poly-GlcNAc (PGA)-dependent biofilm formation is regulated posttranslationally by c-di-GMP (46). The pga operon encoding the poly-GlcNAc biosynthesis machinery is controlled by the carbon storage regulator CsrA. Inactivation of csrA leads to derepression of the pga genes and two genes encoding DGCs: dgcT (ycdT) and dgcZ (ydeH) (47, 48). As a consequence, a csrA mutant strain not only shows constitutive expression of PGA components but also displays a strong increase of the c-di-GMP level (5.35 μM) compared to that of the wild type (0.31 μM) (Fig. 5a). A mutant lacking both CsrA and DgcZ produces significantly less c-di-GMP and shows strongly reduced PGA-dependent attachment (46) (Fig. 5a). To assay the effect of the pde suppressor mutations on PGA-mediated attachment, mutant alleles were introduced into a csrA single mutant and a csrA ΔdgcZ double mutant. As shown in Fig. 5a, only pdeC and two of the pdeL alleles were able to effectively reduce attachment in the high-c-di-GMP background (csrA mutant). Apparently, in accordance with the capacity of restoring motility, only the most active PDE variants are able to reduce the level of c-di-GMP in this strain to a concentration range below the activation constant (Kact) of the PGA biosynthesis machinery (62 nM) (46). In contrast, when biofilm formation was assayed in the low-c-di-GMP background (csrA ΔdgcZ), all pde alleles showed a significant reduction of biofilm formation. The only suppressor allele that was not able to reduce PGA-dependent biofilm formation was PdeI(G412S) (Fig. 5b). However, because motile suppressor mutants were isolated at 37°C and biofilm assays were routinely carried out at 30°C, we tested if pdeI expression was temperature controlled. As shown in Fig. 5c, PdeI protein levels were indeed strongly temperature dependent, with the highest concentration reached at 42°C (Fig. 5c). In line with this observation, the pdeI(G412S) allele significantly reduced attachment of the csrA ΔdgcZ mutant at 37°C (Fig. 5c).
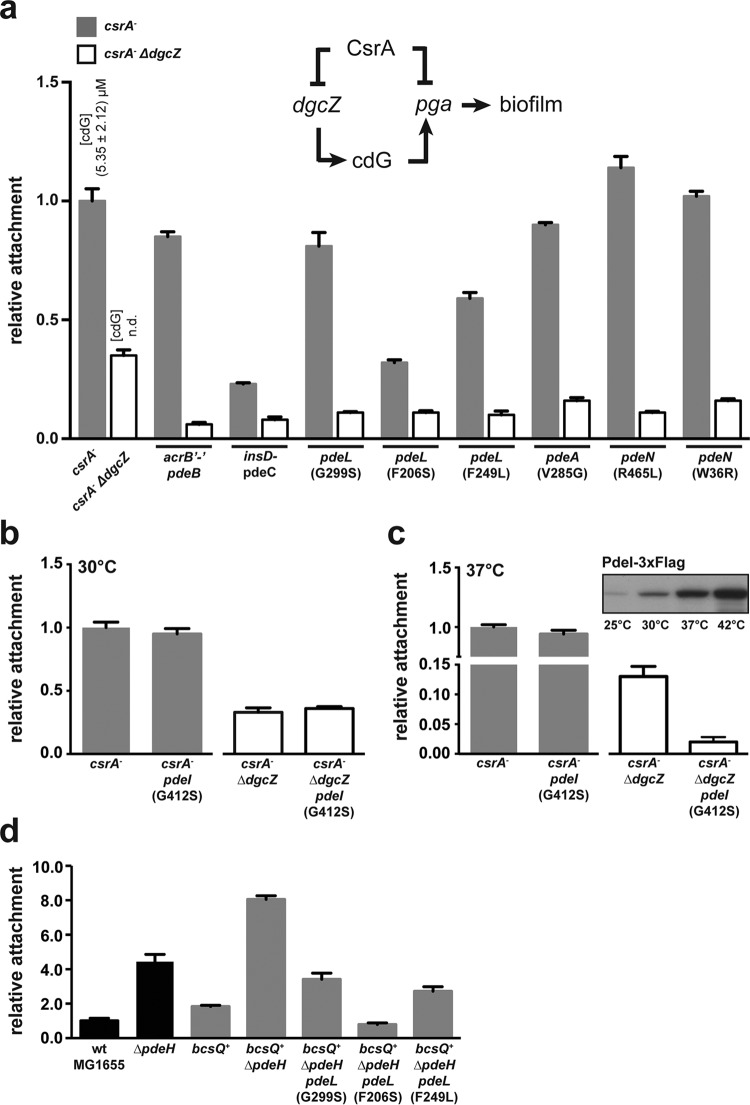
FIG 5.
Surface attachment of E. coli wild-type and phosphodiesterase mutant strains. (a) Relative surface attachment of E. coli csrA and csrA ΔdgcZ mutant strains harboring individual pde suppressor mutations, as indicated. Levels of c-di-GMP (cdG) in both mutant backgrounds are indicated (n.d., not detectable). A schematic of the regulatory network of PGA control is shown above the graph. Gray bars and white bars indicate relative levels of biofilm formation in the csrA and csrA ΔdgcZ strain backgrounds, respectively. Biofilm formation was examined at 30°C (b) and 37°C (c) for strains carrying wild-type pdeI and the pdeI(G412S) suppressor allele. Temperature-dependent expression of pdeI as measured by immunoblot analysis is shown in the inset of panel c. (d) Relative attachment of pdeL suppressor alleles (G299S, F206S, and F249L) in a cellulose-producing bcsQ+ ΔpdeH background. Black bars indicate strains harboring a single nucleotide polymorphism (SNP) in the bcsQ gene, and gray bars represent a “repaired” bcsQ gene (bcsQ+). Attachment is shown relative to that of the cellulose-deficient lab-adapted strain E. coli K-12 MG1655 of Blattner et al. (31). The assay was performed at room temperature.
While E. coli forms poly-GlcNAc biofilms in the host and at higher temperatures (49–51), it can form cellulose-based biofilms in the environment and at lower temperatures. Like that of poly-GlcNAc, production of cellulose is also stimulated by c-di-GMP (52). Many lab-adapted E. coli strains, including E. coli K-12 MG1655, are deficient in cellulose production. This is due to a single point mutation in the bcsQ gene, encoding cellulose synthase. Restoration of the bcsQ wild-type sequence results in proficient cellulose production (53). Introduction of a bcsQ wild-type allele into the cellulose-deficient strain MG1655 increased attachment about 2-fold. Deletion of pdeH in a bcsQ+ background increased attachment about 4-fold compared to that of the isogenic bcsQ+ strain (Fig. 5d). Deletion of pdeH in the cellulose-deficient MG1655 strain also led to a 4-fold increase in attachment compared to that of the wild type, arguing that other c-di-GMP-dependent systems contribute to biofilm formation in this strain. Importantly, when the three pdeL suppressor alleles (encoding G299S, F206S, and F249L mutations) were introduced into the bcsQ+ ΔpdeH background, cellulose-dependent attachment was strongly reduced, similar to the pattern observed for poly-GlcNAc-dependent biofilm formation (Fig. 5d).
These results strongly suggest that genetically activated variants of several PDEs have a profound effect on the cellular c-di-GMP concentration, which eventually becomes manifested in different c-di-GMP-responsive output systems.
PdeL suppressors show increased enzymatic activity.
To gain further insight into the suppression mechanisms that caused reduced levels of c-di-GMP, we investigated the specific in vitro activity of mutant phosphodiesterases. We chose three representative suppressor mutants of PdeL, since this is the only soluble cytoplasmic enzyme and because it was previously shown to be an active c-di-GMP-specific phosphodiesterase (18, 19). We overexpressed and purified PdeL wild-type and G299S, F206S, and F249L mutant proteins that carried a Strep II tag at the C terminus. To determine their activities, we developed a novel enzyme-coupled phosphate sensor-based assay that allows for sensitive real-time determination of c-di-GMP-specific phosphodiesterase activity (see the legend to Fig. 6 and Materials and Methods for details). PDE activity was determined at an enzyme concentration of 500 nM, with substrate concentrations ranging from 100 nM to 5 μM. While wild-type PdeL had a specific PDE activity (kcat/KM) of 0.14 μM−1 s−1, all three PdeL variants showed significantly increased turnover rates, ranging from 0.21 to 0.26 μM−1 s−1 (Fig. 6a).
FIG 6.
Enzyme activities and autoregulation of PdeL suppressor variants. (a) Specific phosphodiesterase activities of purified wild-type PdeL and mutant PdeL variants. The specific activities (kcat/KM [μM−1 s−1]) of PDEs were determined using an enzyme-coupled phosphate sensor assay (see Materials and Methods). In our assay, we applied an enzyme concentration of 100 nM and substrate concentrations ranging from 100 nM to 5 μM. All three PdeL mutants showed an increased turnover rate compared to that of the wild type (0.15 μM−1 s−1). (b) Relative β-galactosidase activities of ΔpdeH mutant strains carrying translational PpdeL-lacZ fusions at the native lacZ locus. A schematic of the reporter strain is shown at the top. The presence of pdeL suppressor alleles increased pdeL promoter activity about 5-fold. (c) The inset shows a partial alignment of the HTH domain sequence of E. coli PdeL and NarL. Lysine 192 of NarL (black arrow) is involved in DNA binding. EMSAs were performed with purified PdeL-Strep II (left panel) and PdeL(K60A)-Strep II (right panel) by using oligonucleotide 4991-7, containing the minimal PdeL binding region. (d) c-di-GMP regulates pdeL transcription in a PdeL-dependent manner. The promoter activity of pdeL was determined for the wild type, a strain exhibiting low levels of c-di-GMP [Δdgc(4)], and a strain with high levels of c-di-GMP (ΔpdeH). c-di-GMP levels of the respective strains are shown as green diamonds. The graph includes the pdeL promoter activity of a strain harboring the pdeL(K60A) allele. (e) Model of c-di-GMP-dependent pdeL transcription control. Enzymatic activity is depicted with a dashed arrow. c-di-GMP negatively regulates pdeL transcription through an unknown mechanism. The enhanced enzymatic activity of PdeL suppressor variants (PdeLmut) lowers the cellular levels of c-di-GMP and leads to pdeL transcription stimulation.
PdeL suppressors enhance pdeL transcription.
Strikingly, strains expressing pdeL(G299S), pdeL(F206S), and pdeL(F249L) showed significantly higher PdeL protein levels than that of the isogenic pdeL wild-type strain (Fig. 3f). The observation that PdeL harbors an N-terminal LuxR-type DNA binding domain fused to its catalytic EAL domain led us to investigate whether pdeL expression is subject to autoregulation. To test this, we constructed a chromosomal reporter, fusing the entire intergenic region upstream of pdeL and downstream of betT to the lacZ gene. The fusion was engineered in the lacZ locus of the chromosome, leaving the original pdeL locus intact (Fig. 6b, inset). β-Galactosidase activity was then determined to compare pdeL promoter strengths in ΔpdeH strains harboring the pdeL alleles encoding the G299S, F206S, and F249L substitutions. All strains expressing activated mutant forms showed similar, about 5-fold increases of pdeL transcription compared to that in their isogenic strain (Fig. 6b). This suggested that pdeL transcription is autoregulated and that PdeL enzyme activity is coupled to the transcription of its own gene.
PdeL directly regulates its own expression in a c-di-GMP-dependent manner.
Promoter activity of pdeL could be linked directly to the enzymatic activity of PdeL, possibly through its DNA binding domain. Alternatively, the enzymatic activity of PdeL might influence pdeL transcription indirectly by modulating the cellular level of c-di-GMP. To distinguish between these two possibilities, we compared pdeL promoter activities in strains expressing a wild-type copy of PdeL but harboring distinct c-di-GMP concentrations. To this end, we used the MG1655 wild-type strain, the ΔpdeH mutant strain, and a strain [referred to as the Δdgc(4) strain] lacking four DGCs: DgcE (yegE), DgcN (yfiN), DgcO (yddV), and DgcQ (yedQ) (27). While wild-type MG1655 harbored intermediate cellular levels of c-di-GMP (0.31 μM), the ΔpdeH mutant showed high levels (>3 μM), and the Δdgc(4) mutant had very low levels of c-di-GMP as measured by LC-MS/MS (65 nM) (Fig. 6c).
Similar to that in strains harboring PdeL suppressors, pdeL promoter activity was increased in a strain lacking the four DGCs. In contrast, a strain lacking PdeH showed strongly reduced pdeL transcription. Together, these observations argued that pdeL transcription is controlled negatively by c-di-GMP and that the pdeL promoter is highly active when the cellular c-di-GMP concentration is very low. In principle, there are two possibilities to explain this regulatory behavior. Internal c-di-GMP levels could be sensed through an unknown transcription factor that modulates pdeL promoter strength accordingly. In this case, the role of PdeL and its enzyme activity in autoregulation would be entirely indirect, through the modulation of the cellular c-di-GMP pool. Alternatively, the role of PdeL could be more direct in that it not only is involved in c-di-GMP homeostasis but also acts as a sensor for the prevailing c-di-GMP concentration and, in response, directly regulates pdeL promoter strength, involving its DNA binding domain. To distinguish between these two possibilities, we performed EMSAs to test for binding of purified PdeL to its own promoter region. Due to the exceptional size of the region between pdeL and its upstream gene betT (874 bp), binding of PdeL to this region had to be tested by using a series of Cy3-labeled DNA probes of various lengths. This analysis yielded a minimal PdeL binding region of 24 bp, located 679 nucleotides upstream of pdeL, harboring an imperfect palindromic sequence (5′-TTC AAT AAG TTT AGT CTT ATT TAA) (Fig. 6c).
To corroborate these results, we aimed to construct a DNA binding-deficient mutant of PdeL which harbors an N-terminal LuxR-like helix-turn-helix (HTH) domain. Based on structural information of the LuxR-like domain of the response regulator NarL (54), we identified a conserved lysine at position 60 of PdeL, which in NarL interacts with DNA (Fig. 6c, inset). As shown in Fig. 6c, purified PdeL(K60A) failed to bind to the PdeL box as indicated above. Next, we determined pdeL promoter activity as a function of c-di-GMP levels in strains lacking PdeL. As shown in Fig. 6d, pdeL promoter activity was strongly reduced in the absence of PdeL, irrespective of the cellular concentration of c-di-GMP. Similarly, when the pdeL gene was replaced in the chromosome with a pdeL allele encoding the K60A mutation, pdeL promoter activity was abolished (Fig. 6d). Taken together, these experiments strongly argue that PdeL is an enzyme and a transcription factor stimulating its own expression in response to the prevailing c-di-GMP regimen in the cell.
DISCUSSION
A large variety of cellular processes in bacteria are dependent on c-di-GMP and are tuned during growth or behavioral processes by accurately regulated cellular levels of this second messenger. This requires tight and coordinated control of the enzymes producing or degrading c-di-GMP. Many of these enzymes contain N-terminal signal input domains to sense and integrate environmental cues. While some of these signals have been identified and include oxygen, NO, redox, light, and the availability of nutrients (15, 21, 55–58), the vast majority of input signals are unknown. It is thus not surprising that under controlled laboratory conditions, only a subset of these enzymes shows activity and contributes to known c-di-GMP-dependent cellular processes. We showed previously that from a total of 25 potential enzymes involved in c-di-GMP turnover, only four DGCs, DgcO (YddV), DgcQ (YedQ), DgcN (YfiN), and DgcE (YegE), and one PDE, PdeH (YhjH), contribute to the regulation of E. coli motility (27). A major player of this regulation is PdeH (YhjH), a soluble PDE that lacks a signal input domain and is coregulated with other flagellar genes to license cell motility and planktonic cell behavior (26). In contrast, deletions of any of the remaining 12 candidate PDEs encoded in the genomes of E. coli K-12 strains showed no effect on motility control (A. Boehm and U. Jenal, unpublished results). Several possibilities exist to explain this observation. Some of these proteins might not be expressed under laboratory conditions. If present, they might be sequestered to control specific cellular processes, or they might simply lack catalytic activity. Finally, they might require an appropriate stimulus to become operative.
Here we showed that most potential pde genes are expressed in E. coli, resulting in readily detectable protein levels. This indicated that these PDEs are present in an inactive state. This was corroborated by our findings that several of these components could be activated genetically to interfere with motility and biofilm control by lowering the overall levels of c-di-GMP in the cell. These experiments support the view that bacteria are equipped with an arsenal of sensors that allows bacteria to rapidly integrate a range of environmental signals to modulate the general c-di-GMP pool and thus to optimally adapt to their variable environments. This does not exclude the possibility that bacteria also tune the levels of these enzymes by altering transcriptional or translational control or as a result of differential protein stability. Also, bacteria likely express distinct sets of such sensory components for specific growth phases or environmental niches. This view is supported by the observation that in E. coli, several DGCs and PDEs are regulated by the stationary-phase sigma factor, σS (44). Similarly, we found that pdeI (yliE) expression is strongly temperature controlled and present at high concentrations only at temperatures well above 30°C. This argues that PdeI is part of an enzyme cocktail that is used primarily in the host environment. Finally, we were unable to isolate activating mutations in several of the remaining PDEs, including PdeK (YhjK), PdeR (YciR), PdeO (DosP), and PdeD (YoaD), despite applying strong selective pressure. It is possible that activating mutations in the relevant genes can be isolated in principle and that our genetic screen was not saturated. Likewise, the activities of some of these enzymes might simply be too weak, even in an activated state, to counter the relatively high cellular c-di-GMP levels of the ΔpdeH mutant strain. Alternatively, some of these components might be part of a specific spatial or structural organization that confines them to acting in a functionally restricted manner. This was recently proposed for PdeR (YciR). This enzyme was shown to form a signaling complex together with the diguanylate cyclase DgcM (YdaM) and the transcription factor MlrA. In this complex, PdeR seems to act both as an enzyme and as a local trigger of the transcriptional activity of MlrA, which drives the expression of CsgD, a central biofilm regulator activating the genes for cellulose matrix and curli fibers (20). While the mechanistic details of the PdeR transcription complex need to be worked out, this regulatory arrangement is consistent with a (locally) limited catalytic function of PdeR, thus offering a plausible explanation for why it was not picked up in our motility screen. Similarly, PdeO (DosP) was recently shown to be part of an RNA degradation complex, in which it seems to locally control RNA turnover in response to oxygen availability (59).
In this study, we used suppression analysis to identify PDE variants in E. coli that can substitute for the major PDE PdeH (YhjH). Using this genetic trick allowed us to bypass the requirement of individual input signals that are normally required to unleash the putative PDE activity. The fact that it is possible to isolate activating mutations in PDEs strongly argues that these enzymes exist in two distinct forms, an active and an inactive conformation, and that their activities are tightly controlled, possibly by switching between these two states. The nature of the mutations that lead to enhanced catalytic PDE activity thus reveals details about the specific mechanisms which these enzymes employ to control their own activity. In principle, several mechanisms to activate a PDE are conceivable. (i) Because PDEs are generally active as dimers (15, 18), an increase of the protein concentration by overexpression will shift the equilibrium toward the active dimeric state. Consistent with this, overexpression of PDEs (or DGCs) can indeed affect the global c-di-GMP pool of bacterial cells, regardless of their activation state (44, 55). (ii) Signal input domains might obstruct the substrate binding site of the catalytic domain or stabilize the enzyme in an inactive conformation. In this case, enzyme activation could result from a functional uncoupling of the two domains. (iii) Mutations within the enzymatic EAL domain may directly enhance specific catalytic PDE activity or change the equilibrium between putative inactive and active conformations toward the latter. In agreement with such a mechanism, we isolated several suppressor alleles encoding single amino acid changes within the EAL domain in PdeL and PdeN. These mutations likely represent true activating mutations.
Of the PDEs that were able to substitute for PdeH activity, PdeL is the best-characterized enzyme. In our study, we isolated 10 mutations affecting eight individual amino acid residues. Three of these were analyzed in detail and were shown to result in enhanced catalytic activity in vitro as well as enhanced pdeL expression. In principle, both properties could contribute to the observed suppression phenotype. At low substrate concentrations, i.e., below the observed KM of about 1 μM, the cellular turnover of c-di-GMP (catalyzed by PdeL) would be increased by a factor of about 10 for the suppressor mutants, as a consequence of a 2-fold increase in specific PDE activity and a 5-fold increase in expression. The increase in specific PDE activity of PdeL mutants is intriguing and warrants a closer analysis. Figure 7 shows the locations of all identified mutations that map to the two known wild-type PdeL EAL crystal structures (18). It is striking that none of the mutated sites are part of the active site that is located at the C-terminal end of the central β-barrel. This enforces the notion that it is not trivial to optimize the catalytic properties of an enzyme through a directed-evolution approach. Rather, the increase in activity may be due to subtle second- or higher-shell effects that are difficult to predict. Alternatively, the mutations may change the thermodynamic equilibrium between (at least) two global conformational states with distinct catalytic activities.
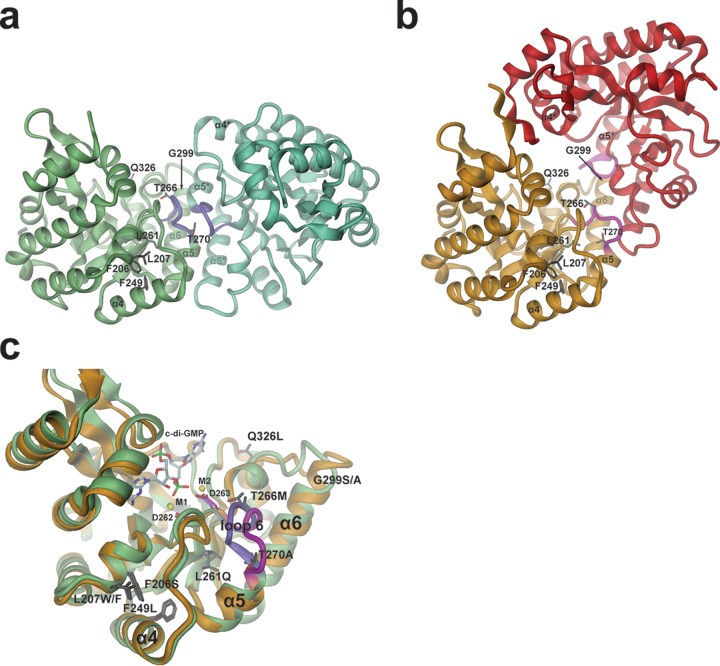
FIG 7.
Model for PdeL phosphodiesterase activation by suppressor mutations. (a and b) Two distinct EAL dimer structures are shown as obtained recently by X-ray crystallography (18). The sites of suppressor mutations are shown in full. The relative orientations of the dimers have been adjusted such that in each panel the left monomer is seen in the same orientation and position. In each case, the two monomers are distinguished by different colors. Purple and magenta coloring highlights the loop 6 region. (a) Structure of the canonical “open” PdeL EAL dimer as determined in the presence of magnesium (PDB code 4LYK). (b) Structure of the “closed” PdeL EAL dimer as determined in the presence of c-di-GMP/Ca2+ (PDB code 4LJ3). (c) Comparison of the PdeL EAL monomer structures of the “open” and “closed” dimers. Colors are the same as those shown in panels a and b (green with loop 6 in purple, “open” dimer; gold with magenta loop, “closed” dimer). In addition to the mutation sites (dark gray residues), the substrate c-di-GMP, the calcium ions M1 and M2, and metal-coordinating aspartates 262 and 263 at the end of β-strand 5 are highlighted. The latter precedes loop 6 and has been implicated in catalysis regulation (18).
We favor the second scenario, since it was recently shown that the EAL domain of PdeL exhibits exquisite inherent regulatory properties (18). The domain can adopt two states that differ drastically in their catalytic activity: a virtually inactive monomeric state and a catalytically competent dimeric state. Coupling of a quaternary state to the precise geometry of the active site, and thus to catalytic activity, appears to be mediated by the β5-α5 loop (loop 6) that constitutes a major part of the dimerization interface (Fig. 7), as also observed in other EAL structures (60). Intriguingly, it was also shown that the EAL domain of PdeL can adopt two distinct dimer conformations, both involving similar dimerization interfaces (formed mainly by loop 6 and helices α5 and α6) but showing drastically different relative monomer arrangements (“open” [Fig. 7a] and “closed” [Fig. 7b] dimers) (18). It has not yet been studied whether both kinds of dimers also exist in solution and, if so, what their relative catalytic activities and the equilibrium constant between them would be.
In light of this structural information, we propose that the fully characterized suppressor mutations (G299S, F206S, and F249L) shift the thermodynamic equilibrium of PdeL from an inactive or lowly active state (EAL domain monomer or dimer of low activity) toward the active state (highly active dimer). Structurally, the shift of the equilibrium would be due to different effects of the suppressor mutations on the two alternative dimerization interfaces (Fig. 7a and b). Indeed, amino acid 299 is part of dimerization helix α6, though the added side chain would not project directly toward the interface. The two phenylalanines, residues 206 and 249, are part of the hydrophobic core that is formed by the packing of helices α4 and α5 onto the central β-barrel. Upon mutation at these sites, the two helices may well shift relative to the β-barrel, causing a perturbation of that part of the interface, which is formed by the N terminus of α5 and the preceding β5-α5 loop (loop 6). Reassuringly, suppressor mutations F207W/F and L261Q map to the same hydrophobic core, and an increase of activity due to a similar mechanism is predicted. Residues T266 and T270 are part of loop 6 (Fig. 7c), the part of the structure that changes most between the two PdeL conformations. Thus, differential stabilization of the possibly more active conformation (or destabilization of the inactive conformation) is conceivable.
Our findings suggest that in addition to its enzymatic function, PdeL can also be a transcription factor and a sensor for c-di-GMP. We showed that increased activities of PdeL variants containing suppressor mutations result in increased levels of the respective proteins. The observation that the stability of these activated mutant variants was unaltered, together with the finding that the activity of the pdeL promoter was increased in the suppressor strains, strongly argued that PdeL exhibits autoregulation. Transcription of pdeL could respond directly to PdeL activity or conformation or could be controlled indirectly through cellular levels of c-di-GMP, which drop as a consequence of increased PdeL activity. The finding that the pdeL promoter is strongly upregulated in cells harboring low levels of c-di-GMP but inhibited at high c-di-GMP concentrations strongly argued for the latter. Finally, the observation that c-di-GMP-mediated regulation of pdeL promoter strength strictly depended on PdeL itself and on its intact DNA binding domain suggested that PdeL is able to sense cellular levels of c-di-GMP and, in response, tune its own expression. Considering the domain architecture of PdeL, it seems plausible that the EAL domain is involved in sensing c-di-GMP concentrations, while the LuxR-type DNA binding domain is likely required for transcriptional autoregulation. While the exact mechanism and physiological significance of this feedback control remain to be elucidated, it is notable that a similar mechanism was described recently, in which PdeR plays a role both as an enzyme and as a sensor for c-di-GMP (20). This example illustrates that an active phosphodiesterase can adopt additional functions to control gene expression in response to substrate availability. Thus, PdeL and PdeR are conceptually very similar in that both proteins “measure” c-di-GMP via an unknown mechanism and in turn regulate gene expression. But while PdeR engages in a signaling complex together with an independent transcription factor, PdeL apparently has evolved more independence by recruiting and directly coupling a DNA binding domain to its catalytic domain. Proteins coupling metabolite availability to gene expression control are common in bacteria and were termed trigger enzymes (61). It will be interesting to clarify the regulatory details and similarities of these systems and to analyze how widespread this phenomenon is among phosphodiesterases.
Supplementary Material
ACKNOWLEDGMENTS
We thank Volkhard Kaever from the Medizinische Hochschule Hannover for c-di-GMP measurements.
The Fellowship for Excellence International Ph.D. Program supported this work.
This project was initiated by A.B. (62). A.R., C.-S.H., S.O., A.B., U.J., and T.S. conceived and designed the experiments. A.R., C.-S.H., and S.O. performed the experiments. A.R., A.M., T.S., and U.J. analyzed the data. A.R., T.S., and U.J. wrote the paper.
This work is dedicated to our friend and scientist colleague Alexander Boehm.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00604-15.
REFERENCES
- 1.Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet 40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 2.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem 280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 5.Hengge R, Gründling A, Jenal U, Ryan R, Yildiz F. 8 June 2015. Bacterial signal transduction by c-di-GMP and other nucleotide second messengers. J Bacteriol doi: 10.1128/JB.00331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol 65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev 23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Navarro MVAS, De N, Bae N, Wang Q, Sondermann H. 2009. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure 17:1104–1116. doi: 10.1016/j.str.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis NJ, Cohen Y, Sanselicio S, Fumeaux C, Ozaki S, Luciano J, Guerrero-Ferreira RC, Wright ER, Jenal U, Viollier PH. 2013. De- and repolarization mechanism of flagellar morphogenesis during a bacterial cell cycle. Genes Dev 27:2049–2062. doi: 10.1101/gad.222679.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newell PD, Boyd CD, Sondermann H, O'Toole GA. 2011. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol 9:e1000587. doi: 10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tschowri N, Busse S, Hengge R. 2009. The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev 23:522–534. doi: 10.1101/gad.499409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 13.Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci U S A 101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De N, Pirruccello M, Krasteva PV, Bae N, Raghavan RV, Sondermann H. 2008. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol 6:e67. doi: 10.1371/journal.pbio.0060067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barends TRM, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I. 2009. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459:1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- 16.Minasov G, Padavattan S, Shuvalova L, Brunzelle JS, Miller DJ, Baslé A, Massa C, Collart FR, Schirmer T, Anderson WF. 2009. Crystal structures of YkuI and its complex with second messenger cyclic di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains. J Biol Chem 284:13174–13184. doi: 10.1074/jbc.M808221200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tchigvintsev A, Xu X, Singer A, Chang C, Brown G, Proudfoot M, Cui H, Flick R, Anderson WF, Joachimiak A, Galperin MY, Savchenko A, Yakunin AF. 2010. Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases. J Mol Biol 402:524–538. doi: 10.1016/j.jmb.2010.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sundriyal A, Massa C, Samoray D, Zehender F, Sharpe T, Jenal U, Schirmer T. 2014. Inherent regulation of EAL domain-catalyzed hydrolysis of second messenger cyclic di-GMP. J Biol Chem 289:6978–6990. doi: 10.1074/jbc.M113.516195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. 2013. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control. EMBO J 32:2001–2014. doi: 10.1038/emboj.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tuckerman JR, Gonzalez G, Sousa EHS, Wan X, Saito JA, Alam M, Gilles-Gonzalez M-A. 2009. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry 48:9764–9774. doi: 10.1021/bi901409g. [DOI] [PubMed] [Google Scholar]
- 22.Gu H, Furukawa K, Breaker RR. 2012. Engineered allosteric ribozymes that sense the bacterial second messenger cyclic diguanosyl 5′-monophosphate. Anal Chem 84:4935–4941. doi: 10.1021/ac300415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacey MM, Partridge JD, Green J. 2010. Escherichia coli K-12 YfgF is an anaerobic cyclic di-GMP phosphodiesterase with roles in cell surface remodelling and the oxidative stress response. Microbiology 156:2873–2886. doi: 10.1099/mic.0.037887-0. [DOI] [PubMed] [Google Scholar]
- 24.Zheng Y, Sambou T, Bogomolnaya LM, Cirillo JD, McClelland M, Andrews-Polymenis H. 2013. The EAL domain containing protein STM2215 (rtn) is needed during Salmonella infection and has cyclic di-GMP phosphodiesterase activity. Mol Microbiol 89:403–419. doi: 10.1111/mmi.12284. [DOI] [PubMed] [Google Scholar]
- 25.Brombacher E, Baratto A, Dorel C, Landini P. 2006. Gene expression regulation by the Curli activator CsgD protein: modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J Bacteriol 188:2027–2037. doi: 10.1128/JB.188.6.2027-2037.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ko M, Park C. 2000. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J Mol Biol 303:371–382. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- 27.Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 28.Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev 22:2434–2446. doi: 10.1101/gad.475808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell 38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hengge R, Galperin MY, Ghigo J-M, Gomelsky M, Green J, Hughes KT, Jenal U, Landini P. 6 July 2015. Systematic nomenclature for GGDEF and EAL domain-containing c-di-GMP turnover proteins of Escherichia coli. J Bacteriol doi: 10.1128/JB.00424-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Heine S, Entian M, Sauer K, Frankenberg-Dinkel N. 2013. NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by an MHYT domain-coupled phosphodiesterase. J Bacteriol 195:3531–3542. doi: 10.1128/JB.01156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi Y, Rao F, Luo Z, Liang Z-X. 2009. A flavin cofactor-binding PAS domain regulates c-di-GMP synthesis in AxDGC2 from Acetobacter xylinum. Biochemistry 48:10275–10285. doi: 10.1021/bi901121w. [DOI] [PubMed] [Google Scholar]
- 33.Cao Z, Livoti E, Losi A, Gärtner W. 2010. A blue light-inducible phosphodiesterase activity in the cyanobacterium Synechococcus elongatus. Photochem Photobiol 86:606–611. doi: 10.1111/j.1751-1097.2010.00724.x. [DOI] [PubMed] [Google Scholar]
- 34.Zähringer F, Lacanna E, Jenal U, Schirmer T, Boehm A. 2013. Structure and signaling mechanism of a zinc-sensory diguanylate cyclase. Structure 21:1149–1157. doi: 10.1016/j.str.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 35.Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA. 2011. Cyclic di-GMP activation of polynucleotide phosphorylase signal-dependent RNA processing. J Mol Biol 407:633–639. doi: 10.1016/j.jmb.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 36.Rao F, Qi Y, Chong HS, Kotaka M, Li B, Li J, Lescar J, Tang K, Liang Z-X. 2009. The functional role of a conserved loop in EAL domain-based cyclic di-GMP-specific phosphodiesterase. J Bacteriol 191:4722–4731. doi: 10.1128/JB.00327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boehm A, Steiner S, Zaehringer F, Casanova A, Hamburger F, Ritz D, Keck W, Ackermann M, Schirmer T, Jenal U. 2009. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol Microbiol 72:1500–1516. doi: 10.1111/j.1365-2958.2009.06739.x. [DOI] [PubMed] [Google Scholar]
- 38.Commichau FM, Stülke J. 2008. Trigger enzymes: bifunctional proteins active in metabolism and in controlling gene expression. Mol Microbiol 67:692–702. [DOI] [PubMed] [Google Scholar]
- 39.Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 40.Chung CT, Niemela SL, Miller RH. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci U S A 86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller JH. 1992. A short course in bacterial genetics—a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 42.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc Natl Acad Sci U S A 98:15264–15269. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spangler C, Böhm A, Jenal U, Seifert R, Kaever V. 2010. A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J Microbiol Methods 81:226–231. doi: 10.1016/j.mimet.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 46.Simm R, Morr M, Kader A, Nimtz M, Römling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 47.Wolfe AJ, Visick KL. 2008. Get the message out: cyclic-di-GMP regulates multiple levels of flagellum-based motility. J Bacteriol 190:463–475. doi: 10.1128/JB.01418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krasteva PV, Fong JCN, Shikuma NJ, Beyhan S, Navarro MVAS, Yildiz FH, Sondermann H. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kulasekara BR, Kamischke C, Kulasekara HD, Christen M, Wiggins PA, Miller SI. 2013. c-di-GMP heterogeneity is generated by the chemotaxis machinery to regulate flagellar motility. eLife 2:e01402. doi: 10.7554/eLife.01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, Laub MT, Baker TA, Jenal U. 2011. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol Cell 43:550–560. doi: 10.1016/j.molcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber H, Pesavento C, Possling A, Tischendorf G, Hengge R. 2006. Cyclic-di-GMP-mediated signalling within the sigma network of Escherichia coli. Mol Microbiol 62:1014–1034. doi: 10.1111/j.1365-2958.2006.05440.x. [DOI] [PubMed] [Google Scholar]
- 52.Sommerfeldt N, Possling A, Becker G, Pesavento C, Tschowri N, Hengge R. 2009. Gene expression patterns and differential input into curli fimbriae regulation of all GGDEF/EAL domain proteins in Escherichia coli. Microbiology 155:1318–1331. doi: 10.1099/mic.0.024257-0. [DOI] [PubMed] [Google Scholar]
- 53.Steiner S, Lori C, Boehm A, Jenal U. 2013. Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J 32:354–368. doi: 10.1038/emboj.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. 2005. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol 56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- 55.Jonas K, Edwards AN, Simm R, Romeo T, Römling U, Melefors Ö. 2008. The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol 70:236–257. doi: 10.1111/j.1365-2958.2008.06411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Palaniyandi S, Mitra A, Herren CD, Lockatell CV, Johnson DE, Zhu X, Mukhopadhyay S. 2012. BarA-UvrY two-component system regulates virulence of uropathogenic E. coli CFT073. PLoS One 7:e31348. doi: 10.1371/journal.pone.0031348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoong P, Cywes-Bentley C, Pier GB. 2012. Poly-N-acetylglucosamine expression by wild-type Yersinia pestis is maximal at mammalian, not flea, temperatures. mBio 3:e00217-12. doi: 10.1128/mBio.00217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen K-M, Chiang M-K, Wang M, Ho H-C, Lu M-C, Lai Y-C. 2014. The role of pgaC in Klebsiella pneumoniae virulence and biofilm formation. Microb Pathog 77:89–99. doi: 10.1016/j.micpath.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 59.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 60.Serra DO, Richter AM, Hengge R. 2013. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J Bacteriol 195:5540–5554. doi: 10.1128/JB.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maris AE, Sawaya MR, Kaczor-Grzeskowiak M, Jarvis MR, Bearson SMD, Kopka ML, Schröder I, Gunsalus RP, Dickerson RE. 2002. Dimerization allows DNA target site recognition by the NarL response regulator. Nat Struct Biol 9:771–778. doi: 10.1038/nsb845. [DOI] [PubMed] [Google Scholar]
- 62.Boos W, Parkinson JS, Jenal U, Vogel J, Søgaard-Andersen L. 2013. Alexander Böhm (1971–2012). Mol Microbiol 88:219–221. doi: 10.1111/mmi.12198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.