ABSTRACT
Glucose is known to inhibit the transport and metabolism of many sugars in Escherichia coli. This mechanism leads to its preferential consumption. Far less is known about the preferential utilization of nonglucose sugars in E. coli. Two exceptions are l-arabinose and d-xylose. Previous studies have shown that l-arabinose inhibits d-xylose metabolism in Escherichia coli. This repression results from l-arabinose-bound AraC binding to the promoter of the d-xylose metabolic genes and inhibiting their expression. This mechanism, however, has not been explored in single cells. Both the l-arabinose and d-xylose utilization systems are known to exhibit a bimodal induction response to their cognate sugar, where mixed populations of cells either expressing the metabolic genes or not are observed at intermediate sugar concentrations. This suggests that l-arabinose can only inhibit d-xylose metabolism in l-arabinose-induced cells. To understand how cross talk between these systems affects their response, we investigated E. coli during growth on mixtures of l-arabinose and d-xylose at single-cell resolution. Our results showed that mixed, multimodal populations of l-arabinose- and d-xylose-induced cells occurred at intermediate sugar concentrations. We also found that d-xylose inhibited the expression of the l-arabinose metabolic genes and that this repression was due to XylR. These results demonstrate that a strict hierarchy does not exist between l-arabinose and d-xylose as previously thought. The results may also aid in the design of E. coli strains capable of simultaneous sugar consumption.
IMPORTANCE Glucose, d-xylose, and l-arabinose are the most abundant sugars in plant biomass. Developing efficient fermentation processes that convert these sugars into chemicals and fuels will require strains capable of coutilizing these sugars. Glucose has long been known to repress the expression of the l-arabinose and d-xylose metabolic genes in Escherichia coli. Recent studies found that l-arabinose also represses the expression of the d-xylose metabolic genes. In the present study, we found that d-xylose also represses the expression of the l-arabinose metabolic genes, leading to mixed populations of cells capable of utilizing l-arabinose and d-xylose. These results further our understanding of mixed-sugar utilization and may aid in strain design.
INTRODUCTION
When bacteria are grown on a mixture of sugars, they will often consume the sugars one at a time in a defined hierarchy. The classic example is growth of Escherichia coli on glucose and lactose, where the cells will first consume the glucose and then the lactose. This process of ordered sugar consumption is known as carbon catabolite repression and has been studied in many species of bacteria (1–3). These studies have principally focused on the mechanisms governing the preferential consumption of glucose, which in the case of E. coli is known to involve the regulation of specific genes and metabolic fluxes (2, 4).
Far less is known about the mechanisms governing the preferential consumption of sugars other than glucose. One notable exception is the growth of E. coli on mixtures of l-arabinose and d-xylose (5–8), hereafter referred to simply as arabinose and xylose. Kang and coworkers first demonstrated that the xylose metabolic genes are repressed by arabinose (6). More recently, Desai and Rao investigated the mechanism for this hierarchy (5). Both the arabinose and xylose utilization pathways involve similar regulatory mechanisms (Fig. 1). The genes for these pathways are expressed only when their cognate sugar is present. AraC positively regulates the transcription of the arabinose metabolic and transporter genes in response to arabinose (9). Likewise, XylR positively regulates the transcription of the xylose metabolic and transporter genes in response to xylose (10). Desai and Rao found that arabinose-bound AraC binds to the promoter for the xylose metabolic genes and inhibits their expression, likely by a competitive mechanism (5). While this previous work established the mechanism governing the hierarchy between arabinose and xylose, it did not explain how individual cells behave.
FIG 1.
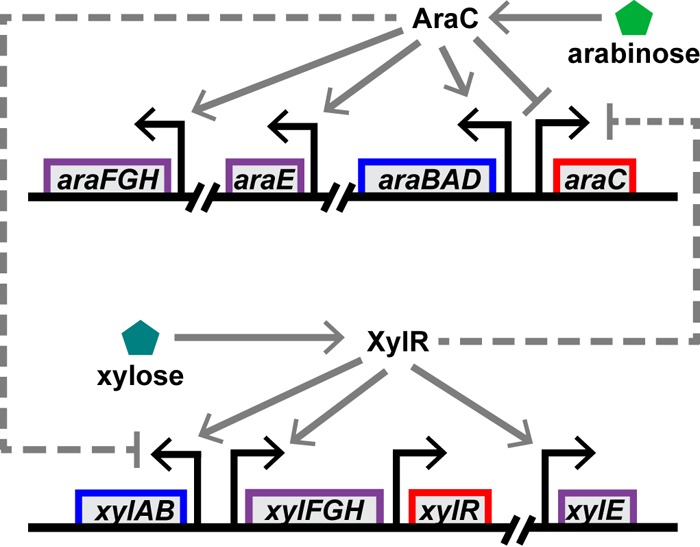
Regulation of the arabinose and xylose sugar utilization systems. Both systems are induced by their cognate sugar. Arabinose-bound AraC induces the expression of the arabinose metabolic genes (araBAD), high-affinity transporter genes (araFGH), and low-affinity transporter gene (araE) (9). In addition, arabinose-bound AraC represses its own expression. Xylose-bound XlyR induces the expression of the xylose metabolic genes (xylAB), high-affinity transporter genes (xylFGH) and, presumably, a low-affinity transport gene (xylE) (10). Xylose-bound XylR does not appear to regulate the PxylR promoter, though it may induce its expression due to transcription from the upstream PxylF promoter. In addition, the two systems are subject to transcriptional cross talk. Arabinose-bound AraC represses the expression of the xylose genes (5) and, as demonstrated in this study, xylose-bound XylR represses the expression of the arabinose genes. Furthermore, the arabinose and xylose systems are both repressed by glucose (mechanism not shown).
Both the arabinose and xylose genes are known to exhibit a bimodal/heterogeneous induction response to their cognate sugar (11–16). Of particular note, a previous study by Afroz and coworkers investigated the expression of the arabinose and xylose metabolic genes in E. coli in response to their cognate sugar (11). Using transcriptional fusions to green fluorescent protein, they measured expression from the ParaB and PxylA promoters at single-cell resolution by flow cytometry. They found that both pathways exhibited a bimodal induction response to their cognate sugar at intermediate concentrations, where the promoters were active in some cells but inactive in others. In the case of xylose, the response was all or nothing: increasing the concentrations of sugar simply increased the number of induced cells where the PxylA promoter was active. In the case of arabinose, the response was more complex. At low arabinose concentrations, the response was all or nothing. At higher concentrations, where the entire population was induced (ParaB promoter active), the response was graded: expression from the ParaB promoter increased with increasing arabinose concentrations.
The response of these two sugar utilization systems at single-cell resolution has not yet been investigated in mixtures of arabinose and xylose. An open question is: if induction is bimodal, then is this also true for catabolite repression? In other words, if only a fraction of cells is induced by arabinose, then is the remaining fraction capable of being induced by xylose? And, can distinct populations of arabinose- and xylose-induced cells coexist at some intermediate concentrations of the two sugars? To answer these questions, we investigated the single-cell response of E. coli during growth on arabinose and xylose by using fluorescent protein reporters. Our results show that mixed populations of arabinose- and xylose-induced cells occur at some intermediate sugar concentrations. We also found that xylose inhibits the expression of the arabinose genes and that this repression occurs through XylR. Collectively, these results demonstrate that a strict hierarchy does not exist between arabinose and xylose, as previously thought, but rather that repression is reciprocal between these two sugar utilization systems.
MATERIALS AND METHODS
Media and growth conditions.
Luria-Bertani (LB) medium (10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter NaCl) was used for cell growth during strain and plasmid construction. Agar plates were prepared by adding 15 g/liter Bacto agar and antibiotics at the following concentrations: ampicillin at 100 μg/ml, chloramphenicol at 20 μg/ml, and kanamycin at 40 μg/ml. Cells were grown in M9 glycerol medium (6.8 g/liter Na2HPO4, 3 g/liter KH2PO4, 1 g/liter NH4Cl, 0.5 g/liter NaCl, 2 mM MgSO4, 100 μM CaCl2, 0.4% glycerol, and 0.001% thiamine hydrochloride) for the fluorescence measurement experiments. All growth experiments were performed at 37°C.
Bacterial strains and plasmid construction.
All strains and plasmids are listed in Tables 1 and 2, respectively. The ΔxylAB mutant strain (mutated in the genomic region from nucleotide [nt] 3725940 to 3728788) was constructed using the method of Datsenko and Wanner (17). The xylA′-mCherry reporter plasmid pSK376 was constructed by PCR amplification of the mCherry gene and ribosome binding site from the plasmid pKW667 (18) using the primers SK353F (5′-GAC CGA ATT CTA AAA GGA GGA GAA AAT G) and SK353R (5′-ATG GCT AGC CTT TGA GTG AGC TGA TAC CGC TC) and the PxylA promoter (genomic region from nt 3729038 to 3728713) from genomic DNA using the primers SK435F (5′-AGA GAG GTC GAC GAT TAC GAT TTT TGG TTT ATT TCT TGA TTT ATG ACC G) and SK435R (5′-AGA GAG GAA TTC ACG GAA TGC TAA CGG GTT TGA G). The two fragments were then cloned into the integrative plasmid pLC153 (18) by using the EcoRI/NheI and SalI/EcoRI restriction sites (restriction sites are underlined in the primer sequences). Similarly, the araB′-Venus reporter plasmid pSK459 was constructed by PCR amplification of the ParaB promoter (genomic region 70059 to 70386) using the primers SK380F (5′-AGA GAG GTC GAC ACT TTT CAT ACT CCC ACC ATT CAG AG) and SK380R (5′-AGA GAG GAA TTC CAT CCA AAA AAA CGG GTA TGG AGA AAC) and then cloning the fragment into the integrative plasmid pVenus (19) by using SalI and EcoRI restriction sites. The reporter plasmids pSK376 and pSK459 were integrated single copy into bacterial attachment sites for phages ϕ80 and λ, respectively, using the conditional-replication, integration, and modular (CRIM) method (20). The integrated plasmids were moved to a clean background via P1 transduction (21, 22).
TABLE 1.
E. coli strains used in this study
| Strain | Relevant characteristics | Source or reference |
|---|---|---|
| MG1655 | λ rph-1 (wild type) | CGSC |
| CR413 | ΔxylR::FRT | 5 |
| SK517 | ΔxylAB::FRT | |
| SK459 | attλ::[kan ParaB-Venus oriR6K] | |
| SK463 | attϕ80::[cm PxylA-mCherry oriR6K] | |
| SK504 | attλ::[kan ParaB-Venus oriR6K] attϕ80::[cm PxylA-mCherry oriR6K] | |
| SK518 | ΔxylR::FRT attλ::[kan ParaB-Venus oriR6K] | |
| SK519 | ΔxylAB::FRT attλ::[kan ParaB-Venus oriR6K] | |
| XW81 | attλ::[kan ParaC-Venus oriR6K] | |
| XW83 | attλ::[kan ParaE-Venus oriR6K] | |
| XW84 | attλ::[kan ParaF-Venus oriR6K] | |
| BL21(DE3) | F− ompT gal dcm hsdS(rB− mB−) λ(DE3) | GE HealthCare |
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| pVenus | kan attλ Venus oriR6K | 19 |
| pLC153 | cm attϕ80 oriR6K | 18 |
| pInt-ts | bla int oriR6K (helper plasmid for attλ integration) | 20 |
| pAH123 | bla int oriR6K (helper plasmid for attϕ80 integration) | 20 |
| pKW669 | cm PLtetO-1-mCherry colE1 | 18 |
| pmCherry | cm attϕ80 mCherry oriR6K | |
| pSK376 | cm attϕ80 PxylA-mCherry oriR6K | |
| pSK459 | kan attλ ParaB-Venus oriR6K | |
| pXW8 | kan attλ ParaC-Venus oriR6K | |
| pXW10 | kan attλ ParaE-Venus oriR6K | |
| pXW11 | kan attλ ParaF-Venus oriR6K | |
| pET-28(a) | kan lacI PT7-His6 T7 pBR322 | Novagen |
| pSK451 | kan lacI PT7-His6-xylR T7 pBR322 |
The araC′-Venus reporter plasmid pXW8 was constructed by PCR amplification of the ParaC promoter (genomic region from nt 70089 to 70386) by using the primers XW20F (5′-ACA CAC GTC GAC AGA GAG TTG CGA TAA AAA GCG) and XW20R (5′-ACA CAC GAA TTC ACT TTT CAT ACT CCC ACC ATT C) and then cloning the fragment into the integrative plasmid pVenus by using SalI and EcoRI restriction sites (underlined). The araE′-Venus reporter plasmid pXW10 was constructed by PCR amplification of the ParaE promoter (genomic region from nt 2980205 to 2980657) using the primers XW22F (5′-AGA GAG GTC GAC AAT TCT CGA CCG CAT TCC AG) and XW22R (5′-AGA GAG GAA TTC TTT TTC CTG CCA GCA GAG AG) and then cloning the fragment into the integrative plasmid pVenus using SalI and EcoRI restriction sites. The araF′-Venus reporter plasmid pXW11 was constructed by PCR amplification of the ParaF promoter (genomic region from nt 1984153 to 1984752) using the primers XW23F (5′-AGA GAG GTC GAC TTC ATT GAT GAA ATC AAT GTA ACT GC) and XW23R (5′-AGA GAG TCT AGA GGT TCT CTC CAG CTT TAG TGT C) and then cloning the fragment into the integrative plasmid pVenus using SalI and XbaI restriction sites. The reporter plasmids were integrated single copy into bacterial attachment sites for λ by using the CRIM method and then moved to a clean background via P1 transduction.
To construct the XylR expression vector with an N-terminal 6×His tag, the xylR gene was PCR amplified using the primers SK413F (5′-CAG CGC TAG CTT TAC TAA ACG TCA CCG CAT C) and SK411R (5′-GCT CGA ATT CTT ATT ACT ACA ACA TGA CCT CGC TAT TTA C) and then cloned into the pET-28(a) vector (Novagen) by using the NheI and EcoRI restriction sites (underlined).
Fluorescence assays.
Cells were grown overnight in M9 glycerol minimal medium supplemented with 0.2% glucose to an optical density at 600 nm (OD600) of ∼1. The overnight culture was diluted 1:106 and grown in M9 glycerol medium supplemented with various amounts of arabinose and xylose for 20 h to an OD600 of ∼0.002. This method has been shown to allow cells be in exponential phase for most of the incubation period (11). The cells were then analyzed using a BD LSR Fortessa flow cytometer. Fluorescence values for approximately 20,000 cells were collected using the fluorescein isothiocyanate channel (excitation at 488 nm, emission at 530/30 nm) for Venus and the phycoerythrin-Texas Red channel (excitation at 561 nm, emission at 610/20 nm) for mCherry-tagged fluorescent proteins. The population was gated using side scatter (SSC) and forward scatter (FSC) channels to distinguish cells from other debris. Data from the flow cytometry experiments were analyzed using the FCS Express program, version 4 (De Novo Software). The data were exported to Microsoft Excel 2010 and further processed in Origin Pro 9.0 to obtain density and histogram plots. The populations were segregated using the quadrant gate in the FCS Express version 4 software. Wild-type strain MG1655 was used as a negative control to set up the gate. All experiments were performed on three separate days; however, we have provided the histogram for only a single experiment, as little deviation was observed among the replicates.
Protein purification.
The 6×His-XylR expression vector was transformed into overexpression strain BL21(DE3) expressing T7 RNA polymerase and grown overnight in LB supplemented with 40 μg/ml kanamycin. The overnight culture was diluted 1:33 in fresh LB and grown to an OD600 of 0.6 before being induced with 1 mM isopropyl-β-d-1-thiogalactopyranoside. Cells were then grown for an additional 4 h at 37°C and harvested by centrifugation at 5,000 × g for 15 min. The cell pellet was resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole; pH 8) containing 1 mg/ml lysozyme and incubated on ice for 30 min. The cells were then sonicated (8 cycles of 10-s pulses at 2-min intervals) on ice. The cell lysate was separated from cells debris by centrifugation at 9,000 × g for 30 min. The lysate was then added to a half volume of Ni-nitrilotriacetic acid-agarose (Qiagen) and gently mixed on a rotary shaker at 4°C for 1 h. The mixture was then centrifuged at 1,000 × g for 5 min, and then the supernatant was discarded. The resin was washed twice with wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole; pH 8). Finally, the protein was eluted with 2 volumes (0.5 ml) of elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole; pH 8). The eluate was analyzed by SDS-PAGE.
Electrophoretic mobility shift assay.
DNA fragments for the PxylA (genomic region from nt 3729038 to 3728713), ParaC/ParaB (genomic region from nt 70059 to 70386), and PflgB (genomic region from nt 1129902 to 1130228) promoters were 32P-labeled in a phosphorylation reaction mixture containing 5 pmol DNA, 2 μl T4 polynucleotide kinase 10× buffer, 1 μl T4 polynucleotide kinase (New England BioLabs), and 2 μl of [γ-32P]ATP in a final volume of 20 μl. The mixture was incubated at 37°C for 20 min. The reaction was then quenched by adding 2 μl of 0.5 M EDTA. The binding reaction was performed by mixing 0.5 pmol 32P-labeled DNA, 0.5 μg salmon sperm DNA (Invitrogen), 10 mM Tris-HCl (pH 8.0), 50 mM KCl, 100 μg/ml bovine serum albumin (BSA), 10% glycerol, 1 mM dithiothreitol (DTT), 0.5 mM EDTA, 10 mM xylose (only when indicated), and 100 ng purified XylR protein (only when indicated) in a total volume of 50 μl. The mixture was incubated at room temperature for 30 min. The electrophoresis was carried out on a 5% native polyacrylamide gel in TBE buffer (10.8 g/liter Tris, 5.5 g/liter boric acid, 0.37 g/liter EDTA disodium salt) at room temperature. The gel was vacuum dried on a filter paper and placed on a phosphor screen overnight. The phosphor screen was scanned using a Storm 840 PhosphorImager.
RESULTS
As arabinose is known to inhibit the expression of the xylose metabolic genes, we sought to determine how this cross talk would affect the single-cell response of E. coli grown in a mixture of arabinose and xylose. In particular, would we observe mixed, multimodal populations of arabinose- and xylose-induced cells or would we observe some cells induced for arabinose but not xylose and other cells induced for xylose but not arabinose? To simultaneously measure the expression of the arabinose and xylose metabolic pathways in single cells, we constructed transcriptional fusions of the ParaB and PxylA promoters to the fluorescent proteins Venus (23) and mCherry (24). These constructs were then integrated single copy into the chromosome by using the phage λ and ϕ80 attachment sites (20). This design enabled us to simultaneously measure the expression from the ParaB and PxylA promoters in single cells by using flow cytometry. Following the protocol of Afroz and coworkers (11), the cells were grown for 20 h in M9 minimal medium supplemented with 0.4% glycerol and various amounts of arabinose and xylose, starting with a very low cell density to ensure that the cells were harvested during exponential phase.
We first investigated the response to a single sugar in order to validate our strains. As shown in Fig. 2, the cells exhibited a bimodal induction response to both sugars: coexisting populations of induced (promoter active) and uninduced (promoter inactive) cells were observed at intermediate sugar concentrations. As a control, we also measured fluorescence in a strain lacking both fluorescent proteins. The measured control level of fluorescence was similar to that observed for the uninduced population, indicating that both promoters were inactive in the absence of their cognate sugar (see Fig. S1 in the supplemental material). We also found that the xylose pathway exhibited an all-or-nothing response, whereas the arabinose pathway exhibited a more complex response: all or nothing at low concentrations and a graded response at higher ones (>1 μM). These results are entirely consistent with those of Afroz and coworkers, though we did see a somewhat graded response for xylose at high concentrations (>10 μM).
FIG 2.
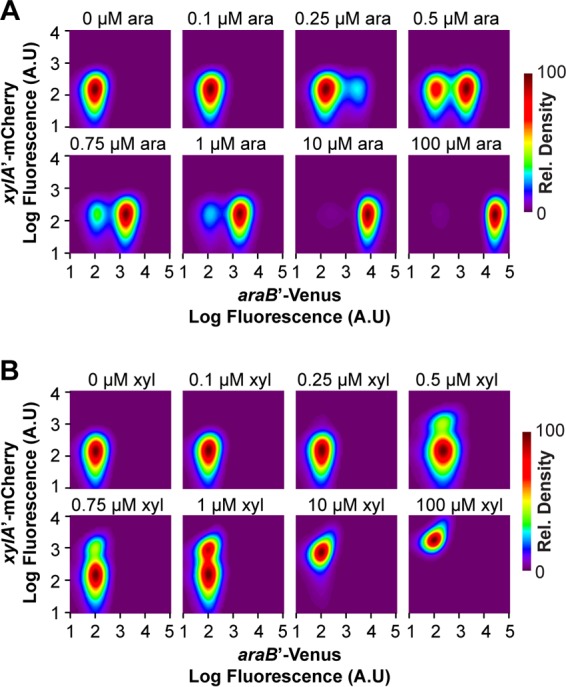
Both the arabinose and xylose systems exhibit a bimodal response to their cognate sugar. The response was measured in a single strain containing single-copy, chromosomal transcriptional fusions of the ParaB promoter to the Venus fluorescent protein and the PxylA promoter to the mCherry fluorescent protein. The density plots show the distributions of promoter activities in individual cells as determined based on Venus and mCherry fluorescence. Data were smoothed and normalized to a peak value of 100 to facilitate interpretation. The cells were grown in M9 minimal medium containing 0.4% glycerol and various concentrations of arabinose (A) or xylose (B) as indicated. Table S1 in the supplemental material lists the fractions of cells in the uninduced and induced states.
We next investigated the response to mixtures of sugars. Here we investigated two concentrations of arabinose, 0.5 μM in association with a mixed population (Fig. 3A) and 1 μM in association with a nearly induced population (Fig. 3B), and a range of xylose concentrations. At low concentrations (<0.5 μM), xylose did not have an effect, consistent with the single-sugar response (Fig. 2B). At intermediate xylose concentrations (0.5 to 1 μM) and 0.5 μM arabinose, we observed four distinct populations: one where both the ParaB and PxylA promoters were inactive, one where only the ParaB promoter was active, one where only the PxylA promoter was active, and one where both promoters were active (Fig. 3A). The same behavior was also observed with 1 μM arabinose (Fig. 3B), though the results were less pronounced because the population where only the ParaB promoter is active predominated, presumably due to repression of the PxylA promoter. These results indicated that the cells exhibit a mixed, multimodal response to arabinose and xylose, with some cells capable of consuming both sugars and others just one. At high xylose concentrations (>1 μM), the PxylA promoter was active in all cells. Interestingly, the fraction of cells where the ParaB promoter was active decreased. These results demonstrated that xylose can inhibit the expression of the arabinose genes. This inhibition became more apparent when expression from the ParaB promoter was averaged over the entire population (Fig. 4A). We note, however, that the inhibition by xylose was much weaker than inhibition by arabinose (Fig. 4B). In addition, high concentrations of xylose did not completely inhibit the utilization of arabinose, presumably due to the weak inhibition (data not shown).
FIG 3.
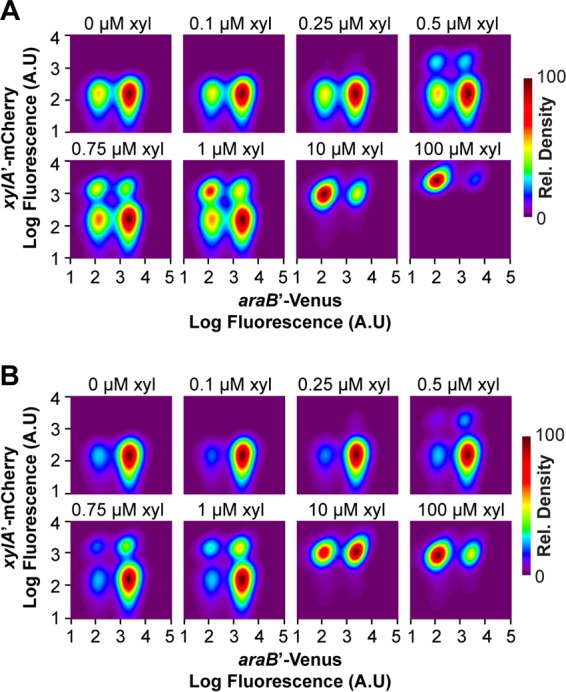
The arabinose and xylose systems exhibit a multimodal response to two sugars. The response was measured in a single strain containing single-copy, chromosomal transcriptional fusions of the ParaB promoter to the Venus fluorescent protein and the PxylA promoter to the mCherry fluorescent protein. The density plots show the distributions of promoter activities in individual cells as determined based on Venus and mCherry fluorescence. Data were smoothed and normalized to a peak value of 100 to facilitate interpretation. The cells were grown in M9 minimal medium containing 0.4% glycerol, 0.5 μM (A) or 1 μM (B) arabinose, and various concentrations of xylose, as indicated. Table S1 in the supplemental material lists the fractions of cells in the uninduced and induced states.
FIG 4.
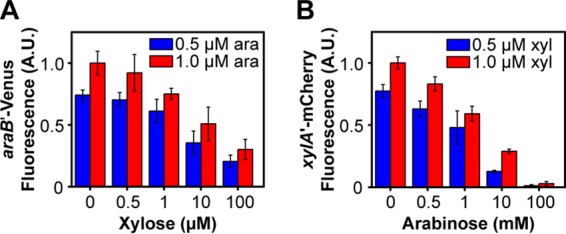
The arabinose and xylose systems are subject to transcriptional cross talk. (A) Xylose represses expression of the arabinose metabolic genes. (B) Arabinose represses expression of the xylose metabolic genes. The responses were measured in a single strain containing single-copy, chromosomal transcriptional fusions of the ParaB promoter to the Venus fluorescent protein and the PxylA promoter to the mCherry fluorescent protein. The cells were grown in M9 minimal medium containing 0.4% glycerol and the specified concentrations of arabinose and xylose. Note that repression by arabinose was stronger than repression by xylose. Fluorescence values were averaged from single-cell, flow cytometry data. Error bars denote the standard deviations from three experiments performed on separate days. Histograms for these data are provided in Fig. S2 in the supplemental material.
Desai and Rao (5) previously found that arabinose represses the xylose genes through AraC, where arabinose-bound AraC inhibits activation of the PxylA promoter by xylose-bound XylR (5). To determine if the reciprocal mechanism occurs with xylose, namely, that xylose-bound XylR binds and inhibits activation of the ParaB promoter by arabinose-bound AraC, we measured the response of the ParaB promoter in a ΔxylR mutant strain. No repression by xylose was observed (Fig. 5A). The results suggested that repression is XylR dependent, as opposed to xylose somehow inhibiting AraC. One caveat with this experiment is that the xylose utilization genes were not expressed in a ΔxylR mutant strain, suggesting that xylose may not be able to enter the cells due to the xylose transporters not being expressed. Numerous studies, however, have shown that the arabinose transporters are promiscuous and capable of taking up xylose (5, 25, 26). We also measured the response of the ParaB promoter in a ΔxylAB mutant strain to determine whether xylose was inhibiting the arabinose genes or some downstream metabolite. As shown in Fig. 5B, repression of the arabinose genes by xylose still occurred in the absences of xylose metabolism but the presence of xylose transport.
FIG 5.
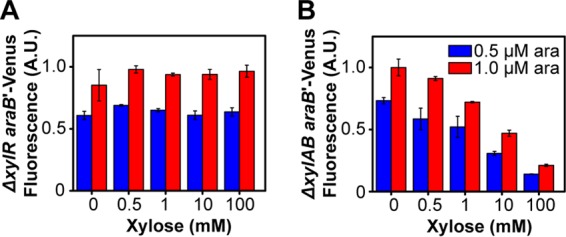
Xylose does not repress arabinose gene expression in the absence of XylR (ΔxylR mutant strain) (A) but does in the absence of xylose metabolism (ΔxylAB mutant strain) (B). The responses were measured in the specified strain containing a single-copy, chromosomal transcriptional fusion of the ParaB promoter to the Venus fluorescent protein. The cells were grown in M9 minimal medium containing 0.4% glycerol and the specified concentrations of arabinose and xylose. Fluorescence values were averaged from single-cell, flow cytometry data. Error bars denote the standard deviations from three experiments performed on separate days. Histograms for these data are provided in Fig. S3 in the supplemental material.
Based on the results above, XylR likely binds and competitively inhibits the arabinose promoters. To test this mechanism, we performed an electrophoretic mobility shift assay with purified XylR and a DNA fragment containing the divergently arranged ParaC and ParaB promoters. We also tested binding to the PxylA promoter as a positive control and binding to the PflgB promoter from the flagellar regulon as a negative control, because the latter was unlikely to be bound by XylR. As shown in Fig. 6A, XylR binds both the PxylA and ParaC/ParaB promoters in a xylose-dependent manner. In particular, a shift was observed only in the presence of XylR and xylose. Not surprisingly, no shift was observed with the PflgB promoter. These results demonstrate that xylose-bound XylR binds the divergent ParaC/ParaB promoter.
FIG 6.
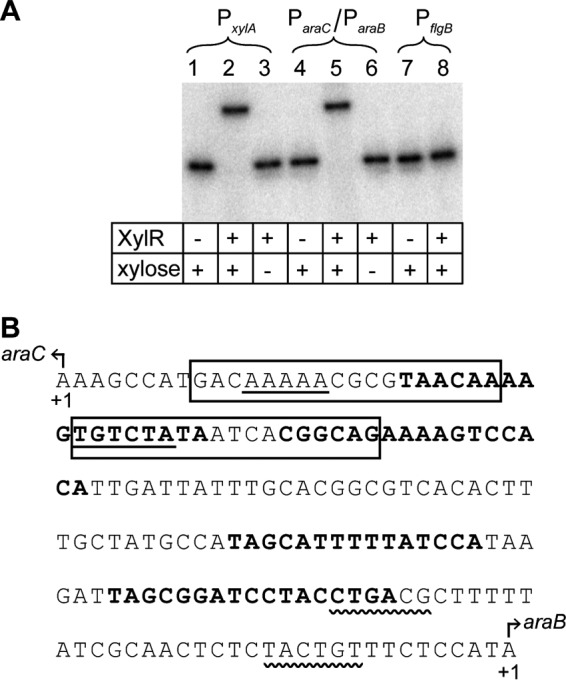
(A) XylR binds the ParaC/ParaB promoter in a xylose-dependent manner, as determined in an electrophoretic mobility shift assay. The PxylA promoter was included as a positive control, and the PflgB promoter was included as a negative control. (B) Nucleotide sequence of the ParaC/ParaB promoter region. The −10/−35 regions of ParaC and ParaB promoters are shown with straight and wavy underlines, respectively. The AraC binding sites are shown in boldface. These annotations were taken from RegulonDB (28). The putative XylR binding sites, as determined through sequence analysis, are boxed.
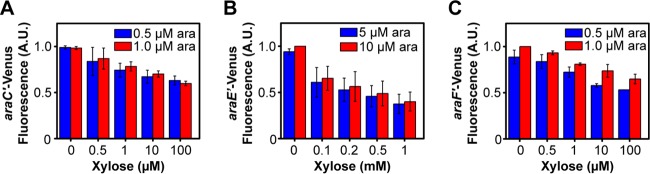
We next used the FIMO program (27) to search for likely XylR binding sites within the ParaC/ParaB promoter by using the consensus motif derived from known XylR binding sites within the PxylA and PxylF promoters (28). One putative XlyR binding site was identified (P < 0.005) that overlapped the ParaC promoter (Fig. 6B). These results suggested that XylR does not directly repress the ParaB promoter but rather indirectly represses it by inhibiting expression of AraC. To test this mechanism, we measured expression from the ParaC, ParaE, and ParaF promoters by using single-copy transcription fusions to the Venus fluorescent protein at different concentrations of xylose. With all three promoters, xylose was found to weakly inhibit their expression (Fig. 7). Because we also did not identify any clear XylR binding sites within the ParaE and ParaF promoters, these results suggest that xylose-bound XylR binds to the ParaC promoter and inhibits expression of AraC, which in turn reduces expression from the ParaB, ParaE, and ParaF promoters.
FIG 7.
Xylose represses the expression of arabinose transporters and the transcriptional activator AraC. The response was measured in a single strain containing a single-copy, chromosomal transcriptional fusion of the ParaC (A), ParaE (B), or ParaF (C) promoter to the Venus fluorescent protein. Fluorescence values were averaged from single-cell, flow cytometry data. Error bars denote the standard deviations from three experiments performed on separate days. Histograms for these data are provided in Fig. S4 in the supplemental material. Note that higher arabinose concentrations were required in order to induce expression from the ParaE promoter.
DISCUSSION
The original motivation for this study was to understand how individual E. coli cells behave when grown in mixtures of arabinose and xylose. Both sugar utilization systems exhibit bimodal responses (11–16), and our initial goal was to understand how catabolite repression affected the response of individual cells. Our data show that the response to the two sugars is multimodal and that the shape of this distribution is determined by the reciprocal regulation of these two sugar utilization systems. Multiple studies have shown that arabinose inhibits the utilization of xylose in E. coli by repressing the expression of the xylose metabolic genes (5, 6). The key finding in the present study is that xylose can also inhibit, albeit weakly, the expression of the arabinose metabolic genes. These results demonstrate that the hierarchy between arabinose and xylose is not fixed, as previously believed, but rather determined by the respective sugar concentrations. An immediate question is why.
The repression of xylose utilization by arabinose ostensibly makes sense, because E. coli grows at a faster rate on arabinose than it does on xylose due to differences in the route of transport employed. In particular, both the arabinose and xylose systems separately employ two similar pairs of transporters for sugar uptake, a low-affinity proton symporter and a high-affinity ATP-dependent transporter (Fig. 1). Arabinose is primarily transported through the more-energy-efficient AraE symporter, whereas xylose, for reasons unknown, is primarily transported through the more-energy-costly ATP-dependent XylFGH transporter, even at high sugar concentrations (25, 29). The resulting differences in growth rates fit the general pattern observed with hierarchical sugar consumption and presumably reflect the efficient allocation of metabolic resources by the cell (8). If xylose is in excess, however, then it presumably makes sense for the cells not to ignore it and to allocate their metabolic machinery in proportion to the availability of this alternate growth substrate. Indeed, both arabinose and xylose are principally derived from hemicellulose hydrolysates, where xylose is the more predominant sugar (30). Alternatively, mutual repression of these two sugar utilization systems could reflect a division-of-labor strategy, where some cells grow on arabinose and others grow on xylose. As our data show (Fig. 3), these mixed populations are observed during growth at intermediate sugar concentrations. Furthermore, we observed fewer cells induced for growth on both sugars than would be expected if the pathways operated independently of one another (see Table S1 in the supplemental material). Likely, individual cells can employ either strategy, because we observed that some cells adapted to growth on just one sugar and others adapted to growth on both. For either strategy, be it proportional allocation or division of labor, one would expect repression by arabinose to be stronger, because it is the better growth substrate. In other words, when the concentrations are equal, we would expect more cells to be adapted to growth on arabinose than on xylose. This in fact is what we observed.
While both the arabinose and xylose metabolic genes exhibit a bimodal response to their cognate sugar, the nature of this response is different. As first documented by Afroz and coworkers, the xylose response is all or nothing at all sugar concentrations, whereas the arabinose response is all or nothing at low sugar concentrations and graded at high sugar concentrations. While the physiological significance of these differences is not known (at least in response to a single sugar), they potentially explain why repression by arabinose is significantly stronger than repression by xylose: the arabinose genes exhibit a greater range of expression levels in individual cells than the xylose genes. Presumably, the degree of repression by arabinose-bound AraC also exhibits a greater range than xylose-bound XylR.
While transcriptional cross talk provides one mechanism to explain catabolite repression among nonglucose sugars, other mechanisms have also been proposed. Aidelberg and coworkers recently investigated the selective consumption of six non-phosphotransferase system (PTS) sugars (8). They observed a hierarchy in the expression of the genes associated with α-lactose, l-arabinose, d-xylose, d-sorbitol, l-rhamnose, and d-ribose metabolism in E. coli. The hierarchy, with lactose at the top and ribose at the bottom, matches the ordering of the growth rates supported by these sugars. In other words, a sugar is preferred to another if it supports faster growth. These results demonstrate that catabolite repression is widespread among non-PTS sugars. In addition to discovering this hierarchy, Aidelberg et al. also found that the cyclic AMP (cAMP) receptor protein-cAMP complex (CRP-cAMP) differentially activates the promoters for these metabolic genes, where the relative degree of activation follows the same hierarchy. This observation is significant, because previous studies have demonstrated that cAMP synthesis is inversely proportional to the growth rate of the cell (31–33). Thus, a cell growing on lactose will produce less cAMP than one growing on any one of the other five sugars. Based on these findings, those authors proposed an alternate model for catabolite repression among non-PTS sugars that was based on sequential activation as opposed to one based on competitive inhibition. According to this model, the metabolic genes for the less-preferred sugar are not expressed, because cAMP concentrations are too low to induce their expression due to the faster growth supported by the preferred sugar. While this model is appealing, it does not explain the selective utilization of arabinose and xylose. For one, the present study demonstrates that the hierarchy between these two sugars is not fixed but rather is determined by their relative concentrations. Furthermore, Desai and Rao demonstrated that arabinose still inhibits xylose gene expression in a mutant (ΔaraBAD strain) unable to metabolize arabinose (5). Moreover, we found in the present study that xylose inhibited arabinose gene expression in a mutant (ΔxylAB strain) unable to metabolize xylose (Fig. 5B). These results clearly demonstrate that catabolite repression is not due to differential activation by CRP-cAMP but instead transcriptional cross talk; how else can xylose (or arabinose) repress arabinose (or xylose) gene expression when xylose (or arabinose) is not being metabolized? Our results, however, do not invalidate their general model, as only two sugars were considered in the present study. In addition, the relative degree of activation of the arabinose and xylose genes by CRP-cAMP, as reported by Aidelberg and coworkers, is small and substantially less than many other sugars investigated in their work. It is unlikely to result in the hierarchical expression of these two sets of metabolic genes. Whether the differences are sufficiently great to govern the hierarchical expression of other sugar genes is unknown.
In addition to identifying a new facet to the coregulation of arabinose and xylose metabolism, the present work may also aid efforts to produce chemical and fuels from plant biomass. Plant biomass is a renewable and low-carbon feedstock for many value-added compounds. Fermentation of the constituent sugars using engineered microorganisms provides one promising route for the conversion of plant biomass to chemicals and fuels (34, 35). After glucose, xylose and arabinose are the next-most-abundant sugars in plant-derived hydrolysates. For the fermentation process to be economic and efficient, the microorganisms need to be able to use all of these sugars and, ideally, at the same time. Not surprisingly, numerous design strategies have been proposed to enable simultaneous sugar utilization in E. coli as well as other bacteria and yeast. Most of these efforts have focused on the coutilization of glucose and another sugar (36). Less effort has been directed toward the coutilization of nonglucose sugar (7, 37–39). In the context of this work, the work of Groff and coworkers (37) is notable. They engineered an E. coli strain capable of simultaneously consuming arabinose and xylose by overexpressing XylR. Presumably, the increased concentration of XylR counterbalances inhibition by arabinose-bound AraC, thus enabling the expression of both sets of metabolic genes. In the course of designing this strain, they found that arabinose utilization was inhibited when they overexpressed XylR from too strong of a promoter. They hypothesized that high levels of XylR repress expression of the arabinose metabolic and transport genes. The present work validates their hypothesis and furthers shows that xylose can repress arabinose gene expression even under native conditions. It also shows that regulation of these two sugar utilization systems is not as simple as previously believed, and further engineering will be required to design optimal coutilizing strains. An open question concerns how the individual cells of the strain engineered by Groff and coworkers actually behave. Are they in fact simultaneously consuming the two sugars or are they separating into two balanced populations selectively consuming the two sugars? The present work demonstrates that both scenarios are possible. An additional question is whether one scenario would be preferred over the other in the context of industrial fermentations; in particular, are generalists preferred to specialists? Nature seems to prefer both. More work is required to resolve these issues.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ahmet Badur and Hanna Walukiewicz from the University of Illinois for assistance with protein purification and the gel shift assays. All flow cytometry experiments were performed using the Roy J. Carver Biotechnology Center (CBC) facility at the University of Illinois.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00709-15.
REFERENCES
- 1.Magasanik B. 1961. Catabolite repression. Cold Spring Harbor Symp Quant Biol 26:249–256. doi: 10.1101/SQB.1961.026.01.031. [DOI] [PubMed] [Google Scholar]
- 2.Gorke B, Stulke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 3.Postma PW, Lengeler JW, Jacobson GR. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev 57:543–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kremling A, Geiselmann J, Ropers D, de Jong H. 2015. Understanding carbon catabolite repression in Escherichia coli using quantitative models. Trends Microbiol 23:99–109. doi: 10.1016/j.tim.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Desai TA, Rao CV. 2010. Regulation of arabinose and xylose metabolism in Escherichia coli. Appl Environ Microbiol 76:1524–1532. doi: 10.1128/AEM.01970-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang HY, Song S, Park C. 1998. Priority of pentose utilization at the level of transcription: arabinose, xylose, and ribose operons. Mol Cells 8:318–323. [PubMed] [Google Scholar]
- 7.Hernandez-Montalvo V, Valle F, Bolivar F, Gosset G. 2001. Characterization of sugar mixtures utilization by an Escherichia coli mutant devoid of the phosphotransferase system. Appl Microbiol Biotechnol 57:186–191. doi: 10.1007/s002530100752. [DOI] [PubMed] [Google Scholar]
- 8.Aidelberg G, Towbin BD, Rothschild D, Dekel E, Bren A, Alon U. 2014. Hierarchy of non-glucose sugars in Escherichia coli. BMC Syst Biol 8:133. doi: 10.1186/s12918-014-0133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schleif R. 2000. Regulation of the L-arabinose operon of Escherichia coli. Trends Genet 16:559–565. doi: 10.1016/S0168-9525(00)02153-3. [DOI] [PubMed] [Google Scholar]
- 10.Song S, Park C. 1997. Organization and regulation of the D-xylose operons in Escherichia coli K-12: XylR acts as a transcriptional activator. J Bacteriol 179:7025–7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Afroz T, Biliouris K, Kaznessis Y, Beisel CL. 2014. Bacterial sugar utilization gives rise to distinct single-cell behaviours. Mol Microbiol 93:1093–1103. doi: 10.1111/mmi.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fritz G, Megerle JA, Westermayer SA, Brick D, Heermann R, Jung K, Radler JO, Gerland U. 2014. Single cell kinetics of phenotypic switching in the arabinose utilization system of E. coli. PLoS One 9:e89532. doi: 10.1371/journal.pone.0089532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khlebnikov A, Datsenko KA, Skaug T, Wanner BL, Keasling JD. 2001. Homogeneous expression of the PBAD promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology 147:3241–3247. doi: 10.1099/00221287-147-12-3241. [DOI] [PubMed] [Google Scholar]
- 14.Morgan-Kiss RM, Wadler C, Cronan JE Jr. 2002. Long-term and homogeneous regulation of the Escherichia coli araBAD promoter by use of a lactose transporter of relaxed specificity. Proc Natl Acad Sci U S A 99:7373–7377. doi: 10.1073/pnas.122227599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegele DA, Hu JC. 1997. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc Natl Acad Sci U S A 94:8168–8172. doi: 10.1073/pnas.94.15.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Megerle JA, Fritz G, Gerland U, Jung K, Radler JO. 2008. Timing and dynamics of single cell gene expression in the arabinose utilization system. Biophys J 95:2103–2115. doi: 10.1529/biophysj.107.127191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu K, Rao CV. 2010. The role of configuration and coupling in autoregulatory gene circuits. Mol Microbiol 75:513–527. doi: 10.1111/j.1365-2958.2009.07011.x. [DOI] [PubMed] [Google Scholar]
- 19.Saini S, Pearl JA, Rao CV. 2009. Role of FimW, FimY, and FimZ in regulating the expression of type i fimbriae in Salmonella enterica serovar Typhimurium. J Bacteriol 191:3003–3010. doi: 10.1128/JB.01694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haldimann A, Wanner BL. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol 183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomason LC, Costantino N, Court DL. 2007. E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol Chapter 1:Unit 1.17. doi: 10.1002/0471142727.mbo117s79. [DOI] [PubMed] [Google Scholar]
- 22.Lennox ES. 1955. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 23.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 24.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 25.Khankal R, Chin JW, Cirino PC. 2008. Role of xylose transporters in xylitol production from engineered Escherichia coli. J Biotechnol 134:246–252. doi: 10.1016/j.jbiotec.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Novotny CP, Englesberg E. 1966. The L-arabinose permease system in Escherichia coli B/r. Biochim Biophys Acta 117:217–230. doi: 10.1016/0304-4165(66)90169-3. [DOI] [PubMed] [Google Scholar]
- 27.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salgado H, Peralta-Gil M, Gama-Castro S, Santos-Zavaleta A, Muniz-Rascado L, Garcia-Sotelo JS, Weiss V, Solano-Lira H, Martinez-Flores I, Medina-Rivera A, Salgado-Osorio G, Alquicira-Hernandez S, Alquicira-Hernandez K, Lopez-Fuentes A, Porron-Sotelo L, Huerta AM, Bonavides-Martinez C, Balderas-Martinez YI, Pannier L, Olvera M, Labastida A, Jimenez-Jacinto V, Vega-Alvarado L, Del Moral-Chavez V, Hernandez-Alvarez A, Morett E, Collado-Vides J. 2013. RegulonDB v8.0: omics data sets, evolutionary conservation, regulatory phrases, cross-validated gold standards and more. Nucleic Acids Res 41:D203–D213. doi: 10.1093/nar/gks1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hasona A, Kim Y, Healy FG, Ingram LO, Shanmugam KT. 2004. Pyruvate formate lyase and acetate kinase are essential for anaerobic growth of Escherichia coli on xylose. J Bacteriol 186:7593–7600. doi: 10.1128/JB.186.22.7593-7600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carroll A, Somerville C. 2009. Cellulosic biofuels. Annu Rev Plant Biol 60:165–182. doi: 10.1146/annurev.arplant.043008.092125. [DOI] [PubMed] [Google Scholar]
- 31.You C, Okano H, Hui S, Zhang Z, Kim M, Gunderson CW, Wang YP, Lenz P, Yan D, Hwa T. 2013. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature 500:301–306. doi: 10.1038/nature12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peebo K, Valgepea K, Maser A, Nahku R, Adamberg K, Vilu R. 2015. Proteome reallocation in Escherichia coli with increasing specific growth rate. Mol Biosyst 11:1184–1193. doi: 10.1039/c4mb00721b. [DOI] [PubMed] [Google Scholar]
- 33.Hermsen R, Okano H, You C, Werner N, Hwa T. 2015. A growth-rate composition formula for the growth of E. coli on co-utilized carbon substrates. Mol Syst Biol 11:801. doi: 10.15252/msb.20145537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JW, Na D, Park JM, Lee J, Choi S, Lee SY. 2012. Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat Chem Biol 8:536–546. doi: 10.1038/nchembio.970. [DOI] [PubMed] [Google Scholar]
- 35.Lee SK, Chou H, Ham TS, Lee TS, Keasling JD. 2008. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr Opin Biotechnol 19:556–563. doi: 10.1016/j.copbio.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 36.Vinuselvi P, Kim MK, Lee SK, Ghim CM. 2012. Rewiring carbon catabolite repression for microbial cell factory. BMB Rep 45:59–70. doi: 10.5483/BMBRep.2012.45.2.59. [DOI] [PubMed] [Google Scholar]
- 37.Groff D, Benke PI, Batth TS, Bokinsky G, Petzold CJ, Adams PD, Keasling JD. 2012. Supplementation of intracellular XylR leads to coutilization of hemicellulose sugars. Appl Environ Microbiol 78:2221–2229. doi: 10.1128/AEM.06761-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nichols NN, Dien BS, Bothast RJ. 2001. Use of catabolite repression mutants for fermentation of sugar mixtures to ethanol. Appl Microbiol Biotechnol 56:120–125. doi: 10.1007/s002530100628. [DOI] [PubMed] [Google Scholar]
- 39.Jarmander J, Hallstrom BM, Larsson G. 2014. Simultaneous uptake of lignocellulose-based monosaccharides by Escherichia coli. Biotechnol Bioeng 111:1108–1115. doi: 10.1002/bit.25182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.