ABSTRACT
Bacterial evolution is accelerated by mobile genetic elements. To spread horizontally and to benefit the recipient bacteria, genes encoded on these elements must be properly regulated. Among the legionellae are multiple integrative conjugative elements (ICEs) that each encode a paralog of the broadly conserved regulator csrA. Using bioinformatic analyses, we deduced that specific csrA paralogs are coinherited with particular lineages of the type IV secretion system that mediates horizontal spread of its ICE, suggesting a conserved regulatory interaction. As a first step to investigate the contribution of csrA regulators to this class of mobile genetic elements, we analyzed here the activity of the csrA paralog encoded on Legionella pneumophila ICE-βox. Deletion of this gene, which we name csrT, had no observed effect under laboratory conditions. However, ectopic expression of csrT abrogated the protection to hydrogen peroxide and macrophage degradation that ICE-βox confers to L. pneumophila. When ectopically expressed, csrT also repressed L. pneumophila flagellin production and motility, a function similar to the core genome's canonical csrA. Moreover, csrT restored the repression of motility to csrA mutants of Bacillus subtilis, a finding consistent with the predicted function of CsrT as an mRNA binding protein. Since all known ICEs of legionellae encode coinherited csrA-type IV secretion system pairs, we postulate that CsrA superfamily proteins regulate ICE activity to increase their horizontal spread, thereby expanding L. pneumophila versatility.
IMPORTANCE ICEs are mobile DNA elements whose type IV secretion machineries mediate spread among bacterial populations. All surveyed ICEs within the Legionella genus also carry paralogs of the essential life cycle regulator csrA. It is striking that the csrA loci could be classified into distinct families based on either their sequence or the subtype of the adjacent type IV secretion system locus. To investigate whether ICE-encoded csrA paralogs are bona fide regulators, we analyzed ICE-βox as a model system. When expressed ectopically, its csrA paralog inhibited multiple ICE-βox phenotypes, as well as the motility of not only Legionella but also Bacillus subtilis. Accordingly, we predict that CsrA regulators equip legionellae ICEs to promote their spread via dedicated type IV secretion systems.
INTRODUCTION
Mobile DNA elements can spread advantageous traits among bacterial populations and contribute to the evolution of pathogens (1). Integrative conjugative elements (ICEs) are one type of mobile transposable element that encode their own type IV secretion system (T4SS) for transfer between strains and also the enzymes necessary to integrate the element into the host chromosome. ICEs have expanded the genetic repertoire of a number of bacterial pathogens by carrying a variety of cargo genes that confer advantageous traits for their hosts, enhancing fitness in specific environments. Some ICEs provide resistance to antibiotics, metal ions, and oxidative stress; others enhance nitrogen and chlorobenzoate metabolism, biofilm formation, and host cell infection (2–5). In addition to cargo genes are those encoding the elements' structural components. Multiple distinct classes of T4SSs mediate ICE transmission within and between species (6). This diversity may permit multiple ICEs to persist within one cell, since bacteria typically maintain only one of each T4SS type (7).
Bacteria coordinate expression of multiple extracellular structures, each with distinct purposes. L. pneumophila produces short pili for adherence or long T4SS conjugative pili for effector secretion or horizontal gene transfer but not both pili at the same time (8). Indeed, simultaneous expression of similar T4SS by L. pneumophila appears to be counterproductive, as activity of the core Dot/Icm T4SS is inhibited by the MobA component of a related RSF1010 conjugative plasmid (9). Accordingly, competition between ICEs likely provides selective pressure for regulatory circuits that ensure reciprocal expression of their T4SS machines.
The regulatory mechanisms governing the expression of ICE traits are as diverse as the ICEs themselves. For example, SXT from Vibrio cholerae and ICEBs1 from B. subtilis encode regulatory proteins that are cleaved or bound by host regulators induced by the bacterial SOS response (2, 10). Other ICEs are equipped with regulatory circuitries that act independently of host proteins. ICEclc from Pseudomonas aeruginosa and CTnDOT from Bacteroides thetaiotaomicron carry sensor proteins that respond to chlorobenzoate or tetracycline, respectively; these chemicals induce transcription of regulatory genes that drive expression of cargo loci that confer protection from these stressors (11, 12). The ICEs pLS20 from B. subtilis and pAD1 of Enterococcus faecalis rely on secreted peptides or pheromones to sense recipient cells in the environment and induce a signaling cascade that activates transcription of the conjugative pilus (13, 14). Finally, to regulate transfer, the Agrobacterium tumefaciens Ti conjugative plasmid responds to the secreted quorum sensing N-acyl-l-homoserine lactone molecule (15). In these ways, each regulatory system specifically activates ICE traits.
The environmental and opportunistic pathogen L. pneumophila carries multiple ICEs in its genome. For instance, the Philadelphia-1 strain of L. pneumophila harbors three distinct ICEs (16). One of these mobile elements, ICE-βox, enhances the pathogen's resistance to bleach and the β-lactam antibiotics oxacillin and penicillin (4). Strains that carry ICE-βox are also more infectious and tolerant of the macrophage phagocyte oxidase (4). Protecting the bacterium from these stresses presumably benefits ICE-βox by ensuring a healthy host competent for spread of the element. Thus, expression of the ICE-βox-encoded T4SS transmission machinery and cargo traits must be regulated in a manner that ensures ICE-βox maintenance in bacterial populations.
ICE-βox encodes a paralog of csrA, an essential regulator of L. pneumophila differentiation. The canonical CsrA is encoded in the “core” genome: the gene is ubiquitous and highly conserved in the Legionella genus and is not encoded within a horizontally acquired genetic element (17, 18). CsrA is an mRNA binding protein that is essential for L. pneumophila replication and represses numerous L. pneumophila transmissive-phase traits, including expression of substrates of the core Dot/Icm T4SS (19, 20). Similar to the dual CsrA system in P. aeruginosa (21), CsrA binds and represses expression of the mRNA encoding csrR, a newly identified csrA paralog in the L. pneumophila core genome, a design predicted to establish reciprocal expression profiles (22).
The legionellae also encode a large “accessory” genome: highly variable regions of the genome frequently composed of horizontally acquired elements, including ICEs, that are not conserved between strains and species (17, 18, 23–25). The ICE Trb-1 mobile element of L. pneumophila strain Corby also encodes a csrA paralog, lvrC (26). Mobility of ICE Trb-1 is repressed in laboratory conditions, as deletion of the lvrRABC regulatory region significantly increases the number of episomal ICE Trb-1 copies (26). No host phenotypic advantages of harboring ICE Trb-1 have been identified; thus, whether lvrRABC also regulates ICE Trb-1 cargo genes remains to be determined.
Because a paralog of the known pluripotent regulator csrA is found in every known L. pneumophila ICE, we sought to determine whether ICE csrA paralogs retain function as regulators. Given the observations that canonical CsrA represses multiple traits mediated by the core Dot/Icm T4SS (19, 20) and that ICE Trb-1 mobility is repressed by its lvrRABC locus (26), we investigated whether csrT, the csrA paralog on ICE-βox, is competent to regulate traits encoded by either its mobile element or the core L. pneumophila genome.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and reagents.
Legionella pneumophila strains were cultured in N-(2-acetamido)-2-aminoethanesulfonic acid (ACES; Sigma)-buffered yeast extract (AYE) broth supplemented with 100 μg/ml thymidine (AYET; Sigma) at 37°C. CFU were identified by plating on ACES-buffered charcoal yeast extract agar (CYE) supplemented with thymidine (CYET) and incubated at 37°C for 4 days (27). Bacteria cultured overnight in AYET were diluted and incubated overnight to obtain cells in exponential (E; optical density at 600 nm [OD600] of 1.2 to 1.8) or postexponential (PE; OD600 of 2 to 3.7) phases. When necessary for mutant or plasmid selection, the following antibiotics were added: gentamicin (Gibco) at 10 μg/ml, kanamycin (Kan; Sigma) at 25 μg/ml, or chloramphenicol (Cam; Fisher) at 5 μg/ml. IPTG (isopropyl-β-d-thiogalactopyranoside; Gold Biotechnology) was added to a final concentration of 200 μM unless otherwise indicated. Hydrogen peroxide (Sigma) was diluted in AYET to a final concentration of 2 mM. Strains, plasmids, and primers used in this study are summarized in Table S1 in the supplemental material.
Phylogeny and alignment.
The predicted amino acid sequences of the 34 ICE-associated csrA paralogs (see Table S2 in the supplemental material) were aligned using ClustalW2 (28). The alignment was used to generate a maximum-likelihood phylogenetic tree with PhyML using the LG substitution model, four substitution rate categories, and 100 bootstraps (29). The resulting tree was visualized using iTOL (30).
Using the order of csrA paralogs determined from the phylogeny, pairwise alignment of the T4SSs were generated using tBLASTx and EasyFig (31). Nucleotide sequences of the T4SS and annotation were taken directly from the NCBI Nucleotide database (see Table S3 in the supplemental material).
Induced expression constructs.
Wild-type (WT) csrA (lpg0781), csrT (lpg2094), and csrA-22 (lpg1003) with C-terminal His6 tags were amplified from Lp02 genomic DNA using the primer sets ZA33+34, ZA11+12, and ZA15+16, respectively. The resultant PCR products were digested with SalI and HindIII and ligated into p206gent (32). These plasmids were then electroporated into Lp02 to generate strains MB1389, MB1383, and MB1435. We verified that adding 200 μM IPTG resulted in protein expression (data not shown) by Western analysis using a 1:5,000 dilution of anti-His(C-term)-horseradish peroxidase (HRP)-conjugated antibody and the Western blot protocol described below.
ΔcsrT mutant construction.
A csrT mutant was generated via recombineering as described previously (33). The csrT coding sequence plus its 500-bp 5′ and 3′ flanking region was amplified using primers (KF67 and KF70) (see Table S1 in the supplemental material). The resulting fragment was cloned into pGEM-T Easy (Promega) and electroporated into Escherichia coli DY330 where λ-red recombinase replaced the csrT region with a gentamicin (Gm) resistance cassette. The recombinant alleles were transformed into strain Lp02 (MB110) or Lp02 containing a marked ICE-βox (MB1353) to generate ΔcsrT mutant strains MB1409 and MB1507, respectively. Insertion of the mutant allele was confirmed by selection on CYET-Gm and screening by PCR.
Mice.
Six- to eight-week-old female A/J mice were purchased from Jackson Laboratories. Mice were housed in the University Laboratory Animal Medicine Facility at the University of Michigan Medical School under specific-pathogen-free conditions. The University Committee on Use and Care of Animals approved all experiments conducted in this study.
Hydrogen peroxide exposure assay.
Stress resistance was assessed by quantifying bacterial CFU after exposure to hydrogen peroxide. E-phase strains containing pcsrT (MB1383, MB1384, and MB1385) were cultured in AYET with or without IPTG induction overnight. After subculture to an OD600 of 1.0, cultures were treated with 2 mM H2O2 for 1 h at 37°C. The mean ± the standard error of the mean (SEM) percent survival was calculated from triplicate samples.
Intracellular growth and immunofluorescence microscopy.
Growth of bacteria in bone marrow-derived macrophages from A/J mice (Jackson Laboratories) was assessed as described previously (34). Strains were cultured to the PE phase with or without 200 μM IPTG overnight prior to infection. To achieve a similar multiplicity of infection (MOI), uninduced motile strains were added at bacteria/host ratio of 1, while induced nonmotile strains were added at ratio of 2. For induced samples, IPTG was included in the infection media at a concentration of 1 mM as described previously (19). Infection efficiency was calculated as follows: (macrophage-associated CFU at 90 min postinfection/input CFU) × 100.
Microscopy was performed by plating 3 × 105 macrophages on 12-mm glass coverslips overnight prior to infection with bacteria cultured as described above. At 90 min postinfection, macrophages were fixed and stained as described previously (35) using a 1:50 dilution of L. pneumophila primary antibody obtained from mouse monoclonal hybridoma cell line CRL-1765 (ATCC) and a 1:1,000 dilution of anti-mouse IgG antibody conjugated to Oregon Green (Molecular Probes). To quantify colocalization with the late endosomal and lysosomal marker LAMP-1, fixed macrophages were stained with a 1:500 dilution of rat anti-LAMP-1 (Santa Cruz) and a 1:1,000 dilution of anti-rat IgG antibody conjugated to Oregon Green (Molecular Probes). The DNA stain DAPI (4′,6′-diamidino-2-phenylindole) was applied in the ProLong Gold antifade reagent (Molecular Probes) used as a mounting medium. Scoring of degraded or LAMP-1-colocalized bacteria was performed by assessing 30 to 50 bacteria per coverslip in triplicate. NADPH oxidase experiments were performed using the pair of cell lines J774.16 (WT) and J774.D9 phox (deficient in gp91 subunit of NADPH oxidase) that were infected as described above (36).
Generation of marked LpgGI-1 strain.
The marked LpgGI-1 strain was generated by recombineering as described previously (4). The noncoding region between lpg1009 and lpg1010 was amplified by PCR using the primers 5′-CTGCGCCTACGTAAAACCC-3′ (forward, KF47) and 5′-GCAGACATGGGAGCGAG-3′ (reverse, KF50) and cloned into pGEM-T Easy (Promega). After electroporation into E. coli strain DY330, λ-red recombinase replaced the ICE-ice noncoding region with a kanamycin (Kan) resistance cassette as described previously (33). The recombinant alleles were transferred to strain Lp02 via natural transformation to generate strain MB1398. Insertion of the Kan cassette was confirmed by selection on CYET-Kan and by PCR.
Construction of donor strains for conjugation.
Strains containing the marked ICEs and the plasmids with IPTG-inducible csrT or csrA-22 genes were generated by mating as described previously (4, 37). Briefly, a culture of a strain with a marked ICE was mixed with a culture of the strain carrying a csr plasmid on CYET agar and incubated at 37°C for 2 h. The cell mixture was then transferred to CYET agar with appropriate antibiotics to select for isolates with the desired marked ICE and csr expression plasmid. Strains were confirmed by isolation streaking two times on CYET agar with appropriate antibiotics and by PCR.
Conjugation.
To analyze ICE-βox transfer by conjugation, the following thymidine auxotroph donor strains, each containing a chloramphenicol-resistance marked ICE-βox, were used: Lp02 (MB1353), csrT mutant (MB1507), Lp02/pcsrT (MB1465), and Lp02/pcsrA-22 (MB1499). Similarly, to analyze LpgGI-1 transfer by conjugation, the following thymidine auxotroph donor strains, each containing a Kan resistance marked LpgGI-1, were used: Lp02 (MB1398), Lp02/pcsrT (MB1500), and Lp02/pcsrA-22 (MB1501). Each donor strain was cultured in AYE with or without IPTG overnight to E phase. As a recipient, the thymidine prototroph JR32 (MB370) was also cultured in AYE overnight to E phase. 109 donor cells were mixed with 1010 recipient cells on 0.22-μm-pore-size filters (Millipore) placed on prewarmed CYET agar plates and incubated at 37°C for 2 h as described previously (4, 37). For strains in which ectopic csr expression was induced, 10 μl of 200 mM IPTG was placed under the filter. Serial dilutions of the mating mixture were plated on CYET-Cam (5 μg/ml) to select for donors and CYE-Cam to select for transconjugants. The transconjugation efficiency was calculated as (CFUtransconjugants/CFUdonor cells) × 100 in triplicate from three independent experiments.
Motility and flagellin analysis.
Wild-type (MB110), ΔflaA mutant (MB1390), and strains carrying pcsrA and pcsrT (MB1389 and MB1383) were cultured in AYET to E phase and then diluted to an OD600 of 0.2. Strains MB1389 and MB1383 were each divided between two tubes, and one was induced with 200 μM IPTG. Next, all strains were cultured at 37°C for ∼15 h to PE phase, and then motility was assessed qualitatively by observation of wet mounts by inverted phase microscopy. Aliquots of the cultures were subsequently normalized to an OD600 of 10 and then resuspended in 100 μl of Laemmli buffer (2% sodium dodecyl sulfate, 10% glycerol, 5% 2-mercaptoethanol, 0.005% bromophenol blue, 62.5 mM Tris-HCl [pH 6.8]). Samples were lysed by boiling for 5 min, and debris was pelleted at 13,000 rpm for 2 min. Proteins were separated on a 12% mini-Protean TGX precast gel (Bio-Rad), and a Precision Plus Kaleidoscope protein standard ladder (Bio-Rad) was used as a size marker and to verify transfer. To assess equal loading and transfer of each sample, Ponceau-S (Fisher) staining was performed on a duplicate membrane. Flagellin was detected using a 1:100 dilution of 2A5 rabbit monoclonal antibody (a gift from N. C. Engelberg, University of Michigan Medical School), a 1:3,000 dilution of goat anti-rabbit secondary IgG–HRP (Pierce), and the SuperSignal West Pico chemiluminescent substrate (Thermo Scientific).
Bacillus subtilis culture.
B. subtilis strains were cultured in Luria-Bertani (LB; 10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl per liter) broth or on LB plates fortified with 1.5% Bacto agar at 37°C. When appropriate, antibiotics were included at the following concentrations: 10 μg/ml tetracycline, 100 μg/ml spectinomycin, 5 μg/ml chloramphenicol, 5 μg/ml kanamycin, and 1 μg/ml erythromycin plus 25 μg/ml lincomycin. Finally, 1 mM IPTG (Sigma) was added to the medium when appropriate.
B. subtilis strain construction.
To generate the amyE::Physpank-csrALp spec complementation construct, PCR product containing the csrALp coding region was amplified from L. pneumophila chromosomal DNA using the primer pair 3520/3521. DNA fragments containing the IPTG-inducible Physpank promoter, a spectinomycin resistance cassette, and the amyE region was amplified from the strain DS4940 as the template using the primers 3177/3519 and 3170/3180. Next, the PCR products were ligated using Gibson assembly (38). The complementation constructs amyE::Physpank-csrX spec, amyE::Physpank-csrT spec, amyE::Physpank-csrA-22 spec, and amyE::Physpank-lvrC spec were generated in a similar way using PCR amplicons from L. pneumophila chromosomal DNA using the primer pairs 3522/3523, 3524/3525, 3526/3527, and 3528/3529, respectively.
All constructs were first introduced into the domesticated strain DS2569 by natural competence and then transferred to the 3610 and DS6530 background using SPP1-mediated generalized phage transduction (39).
B. subtilis swarm expansion assay.
Cells were cultured to mid-log phase at 37°C in LB and resuspended to an OD600 of 10 in pH 8.0 PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4) containing 0.5% India ink (Higgins). Freshly prepared LB medium containing 0.7% Bacto agar (25 ml/plate) was dried for 20 min in a laminar flow hood, centrally inoculated with 10 μl of the cell suspension, dried for another 10 min, and incubated at 37°C. The India ink demarks the origin of the colony, and the swarm radius was measured relative to the origin. For consistency, an axis was drawn on the back of the plate and swarm radii measurements were taken along this transect. For experiments including IPTG, cells were propagated in broth in the presence of IPTG, and IPTG was included in the swarm agar plates.
RESULTS
csrA paralogs and their associated T4SSs are genetically linked.
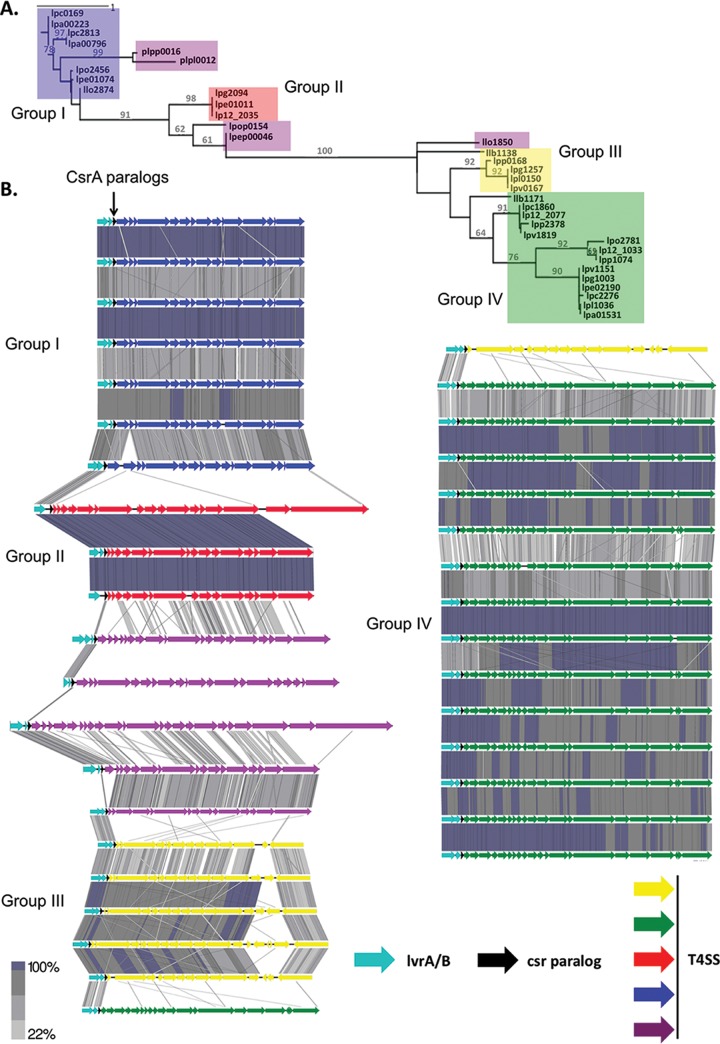
The Legionella pan-genome has been predicted or demonstrated to encode many ICEs (4, 40, 41). In each of the 34 legionellae ICEs identified (see Table S3 in the supplemental material), a csrA-like gene immediately precedes the locus encoding the T4SS. Previously, we identified distinct families of csrA-like genes based on their amino acid sequence similarity (22). Likewise, Legionella ICEs encode several different lineages of T4SSs (4, 26, 41–43). Therefore, we investigated whether particular lineages of ICE-encoded T4SSs were genetically linked to a distinct csrA paralog. To do so, we first determined the relationship of all 34 csrA paralogs (see Table S2 in the supplemental material) by generating a phylogenetic tree (Fig. 1A). Four distinct groups were apparent based on their amino acid sequence; only 5 of the 34 paralogs did not clearly fall into any group.
FIG 1.
Relationship of CsrA paralogs and T4SSs. (A) To determine the relationship between 34 ICE-associated csrA loci, a maximum-likelihood phylogenetic tree was generated from their predicted amino acid sequences. Four distinct clusters are boxed in blue (group 1, Trb), red (group II, Tra), yellow (group III, Lvh), and green (group IV, Lgi). The five protein sequences that did not clearly fall into any cluster are boxed in purple. Branches with at least 60% support from 100 bootstraps are labeled. (B) Pairwise translated nucleotide sequence identity (tBLASTx) of 34 T4SSs in the legionella pangenome was performed based on the relationship of their adjacent csrA paralogs as determined by the phylogeny in panel A. lvrA/B genes are teal arrows; csrA paralogs are black arrows and indicated by a pointer; T4SSs are colored and labeled by group to correspond with the boxes in panel A. Gray bars between T4SS operons indicate the percent identity as shown by the scale bar. For continuity, the bottom two T4SSs from the left column are reproduced as the top two in the right column.
To determine whether the T4SS lineages paired with a specific group of csrA paralogs, we next aligned the T4SSs associated with each of the csrA paralogs. Indeed, within each csrA paralog group, the associated T4SSs lineages were quite similar, whereas there was little similarity between T4SSs that are genetically linked to different csrA families (Fig. 1B). For instance, all the T4SSs associated with csrA paralogs in group IV aligned strongly and indeed included every T4SS in the LGI lineage identified by Wee et al. (41). On the other hand, the T4SSs in csrA group IV did not align with those of the Trb T4SS lineage (group I [42]), the Tra T4SS lineage (group II [4]), or the Lvh T4SS lineage (group III [43]). Whereas the csrA paralogs were genetically linked to their T4SSs, the same relationship did not exist between csrA paralogs and associated cargo regions (data not shown). The linkage of particular groups of csrA paralogs with specific T4SS lineages suggests that selective pressure(s) minimizes genetic drift between each csrA paralog and its associated T4SS locus. Accordingly, we investigated the functional relationship between ICE csrA regulators and their corresponding conjugation machinery.
Ectopic expression of csrA paralogs inhibits ICE transfer.
ICE conjugation is mediated by its T4SS (26, 44), and the ICE-encoded T4SS loci are genetically linked to an adjacent csrA allele (Fig. 1). Therefore, we tested whether CsrA proteins regulate ICE transfer. ICE-βox of L. pneumophila Philadelphia-1 carries the csrA paralog lpg2094, which we name here csrT (csrA paralog for ICE transfer) (4). To test the effect of csrT on ICE-βox conjugative transfer, we applied both loss- and gain-of-function approaches. First, we constructed a ΔcsrT::Gmr deletion mutant allele and a plasmid (pcsrT) on which csrT transcription can be induced by IPTG. To create donor strains, we transferred either the mutant ΔcsrT allele or pcsrT into MB1353, a derivative of the virulent Philadelphia-1 lab strain Lp02 that contains ICE-βox that has been marked with a Cam resistance cassette (4). Donor cells cultured with or without IPTG were mated with the recipient strain JR32, a lab derivative of Philadelphia-1 that does not contain ICE-βox (4).
In rich media at 37°C, deletion of csrT had no effect on conjugative transfer of ICE-βox (Fig. 2). On the other hand, ectopic expression of csrT reduced transconjugation efficiency ∼10-fold. IPTG treatment did not affect transfer of ICE-βox in a strain that did not contain a csrT expression vector, indicating that csrT expression itself inhibits ICE-βox transfer.
FIG 2.
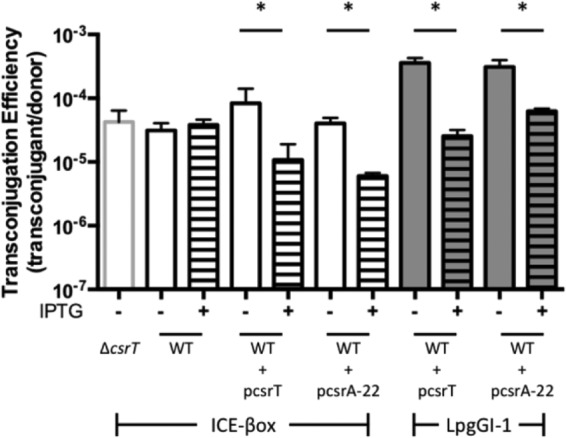
Ectopic csrA paralog expression reduces ICE transfer. The ICE-βox-null recipient strain (MB370) was mated with the following ICE-βox+ donor strains (white bars): ΔcsrT strain (MB1507), WT with no csr plasmid (MB1353), WT with pcsrT (MB1465), and WT with pcsrA-22 (MB1499). MB370 was also mated with the following LpgGI-1+ donor strains (gray bars): WT with pcsrT (MB1500) and WT with pcsrA-22 (MB1501). The strains were cultured prior to mating without (−) or with (+) IPTG to induce ectopic expression of CsrT. Mean transconjugation efficiency of the ICEs is expressed as (CFUtransconjugants/CFUdonor) ± the SEM of results from one representative of two independent experiments performed at least in duplicate. A Student t test was used to show whether the difference between uninduced and induced transconjugation efficiency was statistically significant (*, P < 0.05).
Next, we tested whether or not the observed repression of ICE-βox conjugation by ectopic expression of its cognate csr gene was allele specific. Ectopic expression of csrT also repressed conjugation of the ICE LpgGI-1 (41) whose T4SS is from the distinct LGI lineage (Fig. 1, group IV; Fig. 2). Likewise, ectopic expression of the csrA paralog associated with ICE LpgGI-1 (lpg1003; annotated in the L. pneumophila Philadelphia-1 genome as csrA-22), reduced conjugation not only of its cognate ICE LpgGI-1 but also of ICE-βox by approximately the same amount (∼10-fold reduction, Fig. 2). These data indicate that, when their expression is induced experimentally, the CsrA paralogs encoded on ICE-βox and ICE LpgGI-1 are promiscuous repressors of ICE conjugation.
ICE-βox excision is not sensitive to ectopic csrT expression.
Conjugative transfer requires that ICEs first direct their own excision from the host chromosome (45). Because ectopic csrT expression reduces ICE-βox conjugation (Fig. 2) and the lvrRABC locus inhibits excision of it ICE Trb-1 (26), we tested whether csrT expression also represses ICE-βox excision. To quantify excised and integrated copies of ICE-βox, quantitative PCR using specific primers was performed as described previously (4) using template DNA isolated from donor cells cultured at 37°C in rich medium to E and PE growth phases, with or without ITPG. ICE-βox excision was not affected by ectopic expression of csrT in either growth phase (data not shown). Therefore, in the laboratory conditions tested, ectopic expression of csrT represses conjugation at some step downstream of ICE-βox excision.
Ectopic csrT expression represses L. pneumophila resistance to hydrogen peroxide conferred by ICE-βox.
As another gauge of csrA paralog activity, we investigated whether either loss or ectopic expression of csrT modulates phenotypes conferred by ICE-βox. In L. pneumophila, ICE-βox enhances resistance to β-lactam antibiotics and oxidative stress, such as oxacillin and hydrogen peroxide (4). Therefore, we compared the capacity of ICE-βox to increase resistance to hydrogen peroxide when L. pneumophila either lacked or ectopically expressed csrT. Deletion of csrT did not affect bacterial survival; however, ectopic expression of WT csrT abrogated the capacity of ICE-βox to protect L. pneumophila from a 1 h exposure to 2 mM hydrogen peroxide (Fig. 3). In particular, ICE-βox+ donor and transconjugant strains induced to express csrT were ∼10-fold more susceptible to killing by hydrogen peroxide compared to the same strains cultured without IPTG induction of csrT. Ectopically expressed csrT reduced L. pneumophila survival by a mechanism that requires oxidative stress, since the yield of untreated strains was similar whether or not expression of pcsrT was induced with IPTG (Fig. 3). Thus, when expressed ectopically, csrT inhibits L. pneumophila resistance to hydrogen peroxide that is mediated by ICE-βox.
FIG 3.
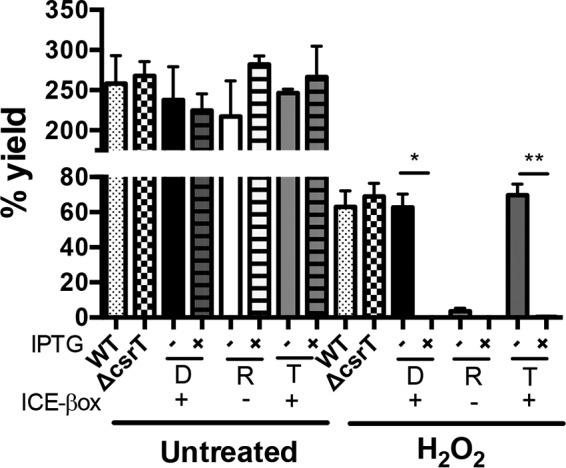
Ectopic csrT expression represses hydrogen peroxide resistance conferred by ICE-βox. CsrT abrogates the capacity of ICE-βox to protect L. pneumophila from hydrogen peroxide. E-phase cultures of WT Lp02 (WT, MB110), ΔcsrT mutant (MB1409), ICE-βox+ donor (D), ICE-βox− recipient (R), and ICE-βox+ transconjugant (T) strains that contained IPTG-inducible plasmid pcsrT (strains MB1383, MB1384, and MB1385, respectively) were cultured overnight without (−) or with (+) IPTG and then exposed to 2 mM H2O2 for 1 h. The mean percent survival ± the SEM was calculated from three independent experiments as (CFUposttreatment/CFUpretreatment) × 100. Student t tests were used to determine statistically significant differences between treated, pcsrT-induced ICE-βox+ strains and treated uninduced strains (*, P < 0.05; **, P < 0.01). Note that after induction of pcsrT expression, the yields of D, R, and T strains were <1%, values that are not visible on this scale.
Ectopic csrT expression represses L. pneumophila evasion of degradative lysosomes.
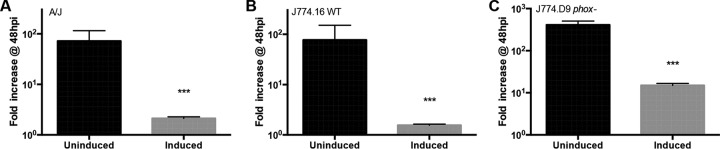
In addition to hydrogen peroxide, ICE-βox increases L. pneumophila protection from degradative lysosomes in restrictive C57BL/6 mouse macrophages (4). Therefore, as an independent measure of the capacity of csrT to regulate ICE-βox-mediated traits, we analyzed bacterial fate in macrophages, another source of oxidative stress (46). Because ectopic expression but not mutation of csrT affected hydrogen peroxide resistance (Fig. 3), the ΔcsrT mutant strain was not included in this assay. Strains that encode or lack ICE-βox and carry pcsrT were cocultured with permissive bone marrow-derived A/J mouse macrophages in the presence or absence of IPTG. After a 90-min infection, bacterial resistance to degradation was quantified by fluorescence microscopy.
When cultured in the absence of IPTG, >80% of the ICE-βox+ donor and transconjugant bacteria resisted degradation, similar to the ICE-βox+ PE-phase WT Lp02 control (Fig. 4A). However, when these same strains were induced to express csrT, the bacteria were degraded ca. 4 to 5 times more frequently (Fig. 4A), as evidenced by particulate L. pneumophila antigen scattered throughout the macrophage, similar to E-phase WT Lp02 (19) and ICE-βox− JR32 cells (4). The impact of csrT ectopic expression appeared to be mediated by ICE-βox, since IPTG treatment of bacteria that carried pcsrT but lacked ICE-βox did not measurably increase bacterial degradation.
FIG 4.
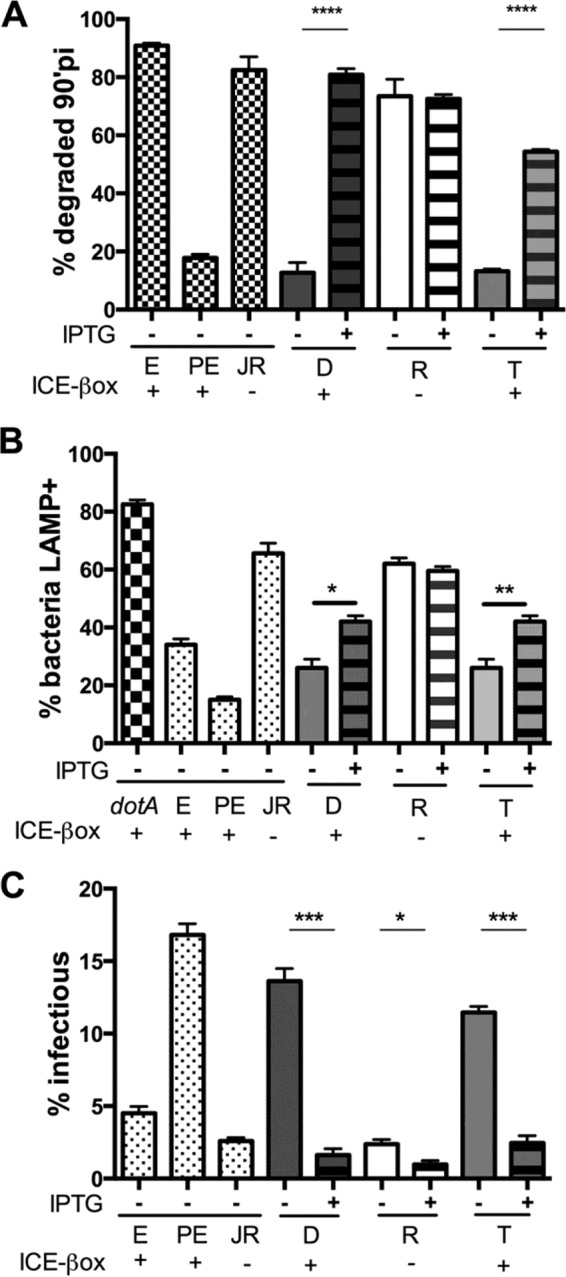
Ectopic csrT expression reduces L. pneumophila infection of macrophages. (A) CsrT induction increases L. pneumophila degradation in macrophages. E- and PE-phase strain ICE-βox+ Lp02 (MB110) and PE-phase ICE-βox null strain JR32 (JR; MB370) (hatched bars); ICE-βox donor (D), recipient (R), or transconjugant (T) strains containing the inducible pcsrT plasmid (MB1383, MB1384, and MB1385, respectively) were cultured overnight without (−) or with (+) IPTG and then incubated for 90 min with A/J macrophages at an MOI of 2 (induced) or 1 (uninduced). After fixation, bacterial integrity was quantified by immunofluorescence microscopy. Presented are the mean percents ± the SEM of total bacteria that were degraded from three independent experiments. Student t tests were used to determine that differences between induced and uninduced D and T strains are significant (P < 0.001). (B) Ectopic csrT expression increases L. pneumophila trafficking to LAMP-1+ compartments. Bacteria were cultured and macrophages were infected as for panel A, and then macrophages were stained with antibodies specific to Legionella and LAMP-1. Colocalization of L. pneumophila with LAMP-1 was quantified by immunofluorescence microscopy. Presented are the mean percents ± the SEM of L. pneumophila that colocalized with LAMP-1 determined from three coverslips in one experiment that is representative of three independent experiments. Student t tests were used to determine that differences between induced and uninduced ICE-βox+ D and T strains are significant (*, P < 0.05; **, P < 0.01). A ΔdotA mutant was included as a positive control for LAMP-1+ staining. (C) CsrT induction inhibits L. pneumophila infection of macrophages. A/J mouse macrophages were infected as described for panel A. After a 90-min infection, cells were lysed and CFU were enumerated. The percent infectious bacteria was calculated as (CFU at 90 min/CFU of input) × 100 and is expressed as the mean ± the SEM from three replicates in one experiment representative of three others. A Student t test was used to confirm whether differences between induced and uninduced strains are significant (*, P < 0.05; ***, P < 0.005).
As expected, when csrT was ectopically expressed by strains that carried ICE-βox, we also observed a modest but reproducible and statistically significant increase in colocalization of the late endosomal and lysosomal marker LAMP-1 with the intracellular bacteria (Fig. 4B) (35, 47). Again, ectopic expression had no appreciable effect on the isogenic strain that did not encode ICE-βox (Fig. 4B).
To verify our microscopic assays of bacterial fate in macrophages (Fig. 4A and B), we quantified the viability of cell-associated L. pneumophila. Permissive bone marrow-derived A/J mouse macrophages were infected with strains that carry pcsrT and encode or lack ICE-βox after culture with or without IPTG. After a 90-min infection, the fraction of input bacteria that were viable and cell associated was quantified. Consistent with our microscopy data, ectopic expression of csrT markedly reduced the cell-associated CFU for ICE-βox+ donor and transconjugant cultures (Fig. 4C). Therefore, ectopic expression of csrT is sufficient to inhibit two traits conferred by ICE-βox: resistance to hydrogen peroxide and to degradation in macrophages.
In addition, we noted that induction of pcsrT modestly decreased the already poor infectivity of the ICE-βox-negative recipient strain (Fig. 4C) (P < 0.05). This observation raised the possibility that, in addition to repressing ICE-βox-encoded traits (Fig. 3 and 4), ectopic csrT expression also affects components of the L. pneumophila core genome.
Ectopic csrT expression inhibits L. pneumophila growth in macrophages that encode or lack NADPH oxidase.
L. pneumophila containing the ICE-βox replicates more efficiently in macrophages than do bacteria that lack the element (4). Because ectopic expression of csrT represses ICE-βox-mediated oxidative stress resistance in broth (Fig. 3) and survival in macrophages (Fig. 4), we predicted csrT would also inhibit intracellular growth of L. pneumophila. To test the impact of csrT on intracellular replication, bone marrow-derived A/J mouse macrophages were infected with ICE-βox+ L. pneumophila carrying pcsrT and cultured with or without IPTG. After 48 h, bacterial yield was quantified and expressed as the fold increase relative to the number of intracellular bacteria present at 2 h, a method that accounts for differences in infectivity (Fig. 4C). Consistent with their poor survival of oxidative stress (Fig. 3) and macrophage infection (Fig. 4), after 48 h of coculture, bacteria that ectopically expressed csrT were recovered at a significantly reduced yield (∼100-fold, P < 0.005) (Fig. 5A).
FIG 5.
Ectopic csrT expression reduces L. pneumophila yield in macrophages that encode or lack NADPH oxidase. A/J mouse macrophages (A), J774.16 WT macrophage cells (B), and J774.D9 phox mutant macrophage cells (C) were infected with ICE-βox+ WT Lp02 carrying pcsrT (MB1383) that had been cultured overnight without or with IPTG induction at an MOI of 1 (uninduced) or 2 (induced). The results are expressed as the fold increase in CFU at 48 h postinfection (hpi) relative to the 2-h time point (CFU at 48 hpi/CFU at 2 hpi). Shown are means ± the SEM from one experiment that is representative of three others. In all macrophage backgrounds, Student t tests were used to confirm whether the differences in bacterial yield for uninduced and induced strains are significant (P < 0.005).
In macrophages, ICE-βox protects L. pneumophila from oxidative stress generated by the NADPH oxidase (4). Therefore, to test the contribution of the phagocyte oxidase to the poor yield of strains induced to express csrT, J774.16 WT and J774.D9 NADPH oxidase mutant macrophage cells were infected with ICE-βox+ strains carrying pcsrT and cultured with or without IPTG; after 48 h, the yield of intracellular bacteria was quantified. Much like the A/J mouse macrophage infection (Fig. 5A), the bacterial yield from J774.16 WT macrophages was significantly reduced by ectopic expression of csrT (∼80-fold less, P < 0.005, Fig. 5B). In addition, csrT expression reduced the yield of L. pneumophila in the J774.D9 NADPH oxidase mutants, although to a lesser extent (∼50-fold less, P < 0.005, Fig. 5C). Therefore, ectopic expression of csrT also inhibits L. pneumophila infection in macrophages by a mechanism(s) not solely dependent on either the macrophage NADPH oxidase (Fig. 5C) or ICE-βox (Fig. 4C).
Ectopic csrT expression inhibits motility and flagellar assembly in L. pneumophila.
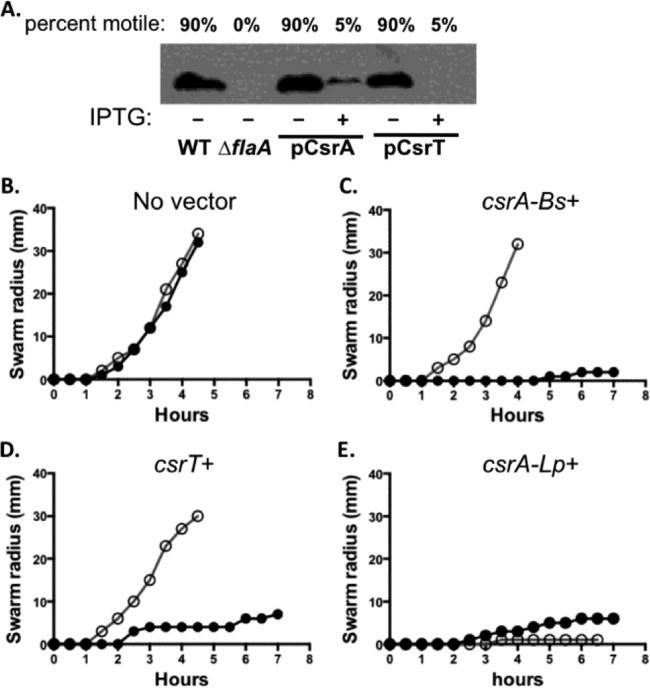
Maximal infection of macrophages by L. pneumophila requires evasion of the macrophage oxidative burst and endosomal pathway (35, 48), as well as bacterial motility, which increases contacts with host phagocytes (49). Canonical CsrA represses not only Dot/Icm T4SS activity but also motility (19, 20, 50), establishing a precedent that L. pneumophila motility and T4SSs can belong to the same regulon. Therefore, we investigated whether csrT expression represses motility, a broadly conserved trait encoded by the core genome. Indeed, when PE-phase L. pneumophila ectopically expressed csrT, only ∼ 5% of the bacteria were motile, compared to ca. 90% of uninduced control cells, as judged by phase microscopy (Fig. 6A). Repression of flagellin expression and motility by ectopically expressed csrT could not be accounted for by a general inhibition of L. pneumophila differentiation into the transmissive phase, since IPTG treatment of strains carrying pcsrT did not inhibit expression of other PE traits, including cell shortening, pigment production, and sodium sensitivity (data not shown).
FIG 6.
Ectopic csrT expression inhibits motility via a conserved mechanism. (A) Western analysis of flagellin protein was performed on PE-phase cultures of L. pneumophila strain Lp02 carrying pcsrA (MB1389) or pcsrT (MB1383), induced (+) or not induced (−) with IPTG as indicated. Lp02 WT with no vector (MB110) was used as a positive control, and a flaA mutant (MB1390) was used as a negative control. The percentage of bacteria that were motile was judged by phase microscopy of wet mounts. Shown are results of one experiment that is representative of at least three others. (B to E) B. subtilis swarming motility was assessed on swarm agar plates every 30 min for WT (filled circles; strain 3610) and fliW sow3 mutants, which are insensitive to B. subtilis CsrA repression mutant (open circles; DS6530) with no vector (B), induced expression of B. subtilis csrA (DS4940 and DK1469) (C), induced expression of L. pneumophila csrT (DK677 and DK1471) (D), or induced expression of L. pneumophila csrA (DK675 and DK1470) (E). Plots represent the means of triplicate plates from one experiment.
To probe the mechanism of csrT inhibition of motility, we analyzed flagellin levels by Western analysis of L. pneumophila cultured with or without induced csrT expression. As a positive control, we induced ectopic expression of canonical csrA, a known repressor of the L. pneumophila flagellar operon (19, 20, 43). Similar to csrA, induction of csrT decreased flagellin protein levels (Fig. 6A). The observation that ectopic expression of either csrA or csrT inhibits flagellin production is consistent with the hypothesis that the family of ICE-associated csrA loci (Fig. 1) retain function as regulatory proteins.
csrT inhibits motility in B. subtilis via a conserved RNA-binding mechanism.
As in L. pneumophila, the B. subtilis CsrA protein inhibits bacterial motility (51). Because the CsrA-binding site on a specific mRNA target, hag, is defined in B. subtilis, we utilized this system to investigate whether CsrT of L. pneumophila ICE-βox can also regulate gene expression by binding mRNA. In particular, we exploited two B. subtilis mutants: the ΔfliW sow3 mutant, which is insensitive to CsrA repression of motility due to a point mutation in its CsrA binding site on the hag mRNA, which encodes flagellin, and the ΔfliW mutant, which lacks the protein that binds and antagonizes the CsrA repressor protein (51). Swarming motility by B. subtilis WT and ΔfliW sow3 mutant strains that ectopically expressed csrA paralogs of B. subtilis or L. pneumophila was quantified.
Induction of either B. subtilis csrA or L. pneumophila csrT inhibited motility in WT B. subtilis (Fig. 6C and D). Moreover, the mechanism of repression was specific, since neither B. subtilis csrA nor L. pneumophila csrT inhibited motility in the ΔfliW sow3 mutant, which lacks the mRNA binding site for the B. subtilis CsrA repressor (51) (Fig. 6C and D). Since a single point mutation in the hag mRNA was sufficient to abrogate CsrT repression of motility, this CsrA paralog encoded by ICE-βox likely binds to hag mRNA and inhibits its translation. Therefore, both bioinformatic (22) and genetic data (Fig. 2 to 6) are consistent with the model that CsrT functions as an RNA binding protein that regulates gene expression.
Like B. subtilis csrA and ICE-βox csrT, induction of canonical csrA of L. pneumophila also inhibited B. subtilis swarming (Fig. 6E). However, L. pneumophila csrA did so even when the hag mRNA binding site was mutated (Fig. 6E). Therefore, although the canonical CsrA repressor encoded by the core L. pneumophila genome does bind mRNA (22), its sequence specificity may be different from that of B. subtilis CsrA and L. pneumophila ICE-βox CsrT.
DISCUSSION
Legionellae must adapt to assault from diverse environmental and intracellular stresses, and horizontally acquired ICEs may increase the fitness of this opportunistic pathogen (4, 23–25, 52, 53). We demonstrate here that every L. pneumophila ICE identified to date encodes a csrA-like locus that is genetically linked to a particular T4SS lineage (Fig. 1; see Table S2 in the supplemental material). The csrA paralog encoded by L. pneumophila ICE-βox retains regulatory potential, as judged by the capacity of ectopically expressed csrT to repress not only conjugative transfer and oxidative stress resistance encoded by its cognate mobile element (Fig. 2 and 3) but also conjugation of a distinct lineage of ICE (group IV LpgGI-1) (Fig. 1 and 2), as well as L. pneumophila infection of macrophages (Fig. 4 and 5) and flagellin expression (Fig. 6A) and B. subtilis swarming motility (Fig. 6B to E).
By exploiting the well-characterized B. subtilis swarming motility pathway, we demonstrate that CsrT likely functions by binding mRNA (Fig. 6D). Since a point mutation of the CsrA binding site on the B. subtilis target mRNA was sufficient to relieve repression by ectopically expressed ICE-βox csrT, we deduce that CsrT protein binds mRNA by specifically recognizing the ANGGA consensus sequence for CsrA binding that several bacterial species use (51, 54–56). Indeed, the CsrT protein has retained many of the key residues that equip E. coli CsrA to bind mRNA (22, 57). Together, our bioinformatic, genetic and functional analyses indicate that CsrT is an RNA binding protein capable of regulating gene expression. Future biochemical experiments can verify that CsrT protein directly interacts with mRNA and identify its specific targets in L. pneumophila.
We favor the model developed for B. subtilis that CsrA proteins regulate nanomachineries (51) and speculate that the CsrA paralog encoded on each L. pneumophila ICE regulates its cognate T4SS machinery. B. subtilis controls flagella morphogenesis by a partner-switching mechanism comprised of the FliW regulator, a CsrA repressor, and flagellin (hag) subunits (51). It is striking that, just as fliW, csrA, and hag are encoded in tandem on the B. subtilis chromosome, every known L. pneumophila ICE carries a similar gene trio that encodes a putative regulatory protein (LvrA/B), a CsrA repressor, and a pilin structural protein (Fig. 1B). Accordingly, the hypothesis that, under physiological conditions, CsrT regulates assembly of the ICE-βox T4SS apparatus and CsrA-22 controls ICE LpgGI-1 warrants direct testing.
Although our conjugation experiments suggest that the csrA paralogs may retain some overlapping activity (Fig. 2), gene duplication events and subsequent genetic drift appear to have diversified the L. pneumophila family of core and ICE-associated csrA genes (22) (Fig. 1; see Table S2 in the supplemental material). For example, canonical CsrA represses expression of csrR by binding to its mRNA at an ANGGA motif that overlaps the start codon (22). Likewise, an ANGGA binding site on the hag RNA is required for CsrT of ICE-βox to repress B. subtilis motility (Fig. 6). However, canonical CsrA represses B. subtilis motility despite a point mutation in the ANGAA motif (Fig. 6), indicating divergence between these two L. pneumophila paralogs. Instead, canonical csrA is predicted to bind AGGA motifs in the mRNAs of multiple substrates of the dot/icm T4SS to regulate their expression; among this class of targets, ANGGA motifs are more rare (19, 20). Differences among the L. pneumophila csrA paralogs were also revealed by our phenotypic analysis. L. pneumophila motility is inhibited by ectopic expression of csrT (Fig. 6A) but not of csrA-22 or another ICE-associated csrA paralog, lpg1257 of ICE-Trb-1, both of whose protein sequences are less similar to canonical CsrA. Likewise, B. subtilis motility is inhibited by ectopic expression of three of the csrA paralogs encoded by L. pneumophila strain Lp02 (core csrA, ICE-βox csrT, and lpg1257) but not two others (core csrR and csrA-22 [22]) (see Table S2 in the supplemental material; also data not shown). Therefore, we speculate that genetic drift of individual ICE csrA paralogs may accommodate distinct regulatory demands of their corresponding T4SS class (Fig. 1), concomitantly relaxing their capacity to repress motility of their bacterial host.
Our bioinformatics analysis also predicts that some RNA targets of CsrT are encoded within its associated T4SS locus. Sequence alignment of 34 ICE-associated csrA paralogs revealed at least four distinct csrA gene families and CsrA-T4SS pairs (Fig. 1; see Table S2 in the supplemental material). The high degree of similarity within each of the four distinct csrA groups indicates that the ICE-encoded csrA loci are under selective pressure to maintain function. Moreover, the genetic linkage of each csrA paralog group to one of four previously identified T4SS lineages (Fig. 1B)—Tra (4), LGI (41), Trb (42), and Lvh (43)—predicts a functional relationship between ICE csrA paralogs and T4SSs. Whereas the specific amino acid sequence of each csrA paralog appears to be genetically constrained by the composition of the adjacent T4SS locus (Fig. 1), the same cannot be said for the associated cargo regions, which are diverse (data not shown). Therefore, we favor the model that CsrT of ICE-βox regulates its associated T4SS, rather than expression of ICE cargo genes.
It is possible that all—or at least some—of the ICE-associated CsrA paralogs are capable of promiscuous interaction with mRNAs encoding multiple T4SSs or traits encoded in the core genome. When ectopically expressed, either csrT or csrA-22 (lpg1003) reduced transfer of ICE-βox or ICE-LpgGI-1 by ∼10-fold (Fig. 2), and csrT and csrA-22 (lpg1003) are members of distinct lineages (group IV and group II, respectively) (Fig. 1). In addition, when expressed ectopically, csrT slightly reduced the infectivity of L. pneumophila that lacked ICE-βox (Fig. 4C). Induced csrT expression also reduced the bacterial yield in the J774.D9 phagocyte oxidase mutant cell line (Fig. 5C), even though these cells are permissive for growth of L. pneumophila that do not encode ICE-βox (4). Furthermore, CsrT inhibits L. pneumophila flagellin expression and motility (Fig. 6A), a broadly conserved trait that contributes to virulence (19). The reduced motility of cultures that ectopically express csrT likely contributes to their poor infectivity (Fig. 4C), since amotile L. pneumophila are less infectious in cell culture due to decreased contacts with macrophages (49). Inhibition of flagellin production by ectopically expressed CsrT may simply reflect its ancestry as a homologue of canonical CsrA, a repressor of L. pneumophila motility (19). It is also formally possible that the capacity of CsrT to repress motility increases fitness of ICE-βox, since bacteria conjugate more efficiently when stationary.
However, a limitation of our phenotypic analyses is the reliance on ectopic expression of csrA paralogs (Fig. 2 to 6), rather than loss of function. In rich medium at 37°C, excision of ICE-βox, expression of its T4SS genes, and the conjugation efficiency are quite low (data not shown and Fig. 2). If csrT is not naturally expressed under the conditions tested, deletion of csrT would have no observable phenotype. Ectopic expression bypasses the need to know a gene's natural inducers or regulatory circuit; nevertheless, it is a blunt genetic tool. For example, ectopic expression is likely to confound the nuances of local concentrations of protein and RNA in the bacterial cell that are known to be important for CsrA protein interaction with its RNA targets. CsrA proteins have different affinities for mRNA targets based on sequence of the RNA binding site and side chains of the CsrA protein (54, 56). The local concentration of protein and RNA therefore dictate whether high- or low-affinity interactions occur. Indeed, local concentrations of mRNAs are not constant in the intracellular space of bacteria (58). Moreover, CsrA proteins function as dimers and can also form complexes with other regulatory proteins (51, 54). Therefore, induced expression of csrA paralogs from a multicopy vector may enable protein-RNA or protein-protein interactions that do not occur in nature. Accordingly, at present we can definitively conclude only that csrT has the capacity to inhibit multiple traits encoded by ICE-βox and the core genome. Identifying laboratory conditions that are optimal for csrT expression and the bona fide targets of CsrT regulation is a goal of future work.
Using bioinformatic analysis (Fig. 1) (22), we describe a new large class of csrA-like genes associated with T4SSs in ICEs never before characterized in Legionella or any other bacterial genus. The genetic linkage of these paralogs with T4SSs strongly predicts a functional relationship between the two. Furthermore, the observation that ectopic expression of csrT inhibits phenotypes as diverse as ICE conjugation and L. pneumophila and B. subtilis motility not only demonstrates that CsrT is a functional paralog of CsrA but also raises intriguing questions as to why csrA regulators are ubiquitous among L. pneumophila ICEs. By analogy to B. subtilis' partner-switching mechanism that regulates flagellum morphogenesis (51), we postulate that the assembly of ICE T4SS pili are governed by interactions between LvrA/B, CsrA, and pilin proteins. It is also noteworthy that, in L. pneumophila, CsrA is an essential pluripotent posttranscriptional repressor of transmission traits that the intracellular pathogen requires to replicate in vitro and in macrophages and amoebae (19, 20). Moreover, CsrA directly binds and represses translation of csrR mRNA, which encodes a second broadly conserved core csrA paralog that enhances L. pneumophila survival in water (22). Accordingly, it is feasible that some ICE csrA paralogs increase the fitness of their mobile elements by retaining the capacity to manipulate core life cycle switches of its host bacterium. In either case, a posttranscriptional regulator would equip the ICE to load its host cell with a pool of transcripts whose translation can be quickly derepressed when environmental conditions favor T4SS assembly and ICE spread. Thus, we postulate that conserved csrA-like regulators equip Legionella ICEs to coordinate expression of traits that ensure their maintenance within bacterial populations.
Supplementary Material
ACKNOWLEDGMENTS
We thank Paul Carlson for technical guidance, Phil Hanna's lab for the use of equipment, and Renee Tsolis for the J774 cell lines.
This study was supported by the Endowment for the Basic Sciences at the University of Michigan Medical School (M.S.S.), two University of Michigan Rackham Merit Fellowships (K.J.F. and Z.D.A.), the Molecular Mechanisms of Microbial Pathogenesis training program (NIH T32 AI007528; K.J.F.), the Cellular Biotechnology training program (NIH T32 GM008353; Z.D.A.), and the NIH R01 (grant GM093030 to D.B.K.).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00732-15.
REFERENCES
- 1.Wozniak RA, Waldor MK. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 2.Beaber JW, Hochhut B, Waldor MK. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez-Blanco A, Lemos ML, Osorio CR. 2012. Integrating conjugative elements as vectors of antibiotic, mercury, and quaternary ammonium compound resistance in marine aquaculture environments. Antimicrob Agents Chemother 56:2619–2626. doi: 10.1128/AAC.05997-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flynn KJ, Swanson MS. 2014. Integrative conjugative element ICE-betaox confers oxidative stress resistance to Legionella pneumophila in vitro and in macrophages. mBio 5:e01091–14. doi: 10.1128/mBio.01091-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koraimann G, Wagner MA. 2014. Social behavior and decision making in bacterial conjugation. Front Cell Infect Microbiol 4:54. doi: 10.3389/fcimb.2014.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juhas M, Crook DW, Hood DW. 2008. Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell Microbiol 10:2377–2386. doi: 10.1111/j.1462-5822.2008.01187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frost LS, Koraimann G. 2010. Regulation of bacterial conjugation: balancing opportunity with adversity. Future Microbiol 5:1057–1071. doi: 10.2217/fmb.10.70. [DOI] [PubMed] [Google Scholar]
- 8.Stone BJ, Abu Kwaik Y. 1998. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect Immun 66:1768–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segal G, Shuman HA. 1998. Intracellular multiplication and human macrophage killing by Legionella pneumophila are inhibited by conjugal components of IncQ plasmid RSF1010. Mol Microbiol 30:197–208. doi: 10.1046/j.1365-2958.1998.01054.x. [DOI] [PubMed] [Google Scholar]
- 10.Bose B, Auchtung JM, Lee CA, Grossman AD. 2008. A conserved anti-repressor controls horizontal gene transfer by proteolysis. Mol Microbiol 70:570–582. doi: 10.1111/j.1365-2958.2008.06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sentchilo V, Ravatn R, Werlen C, Zehnder AJ, van der Meer JR. 2003. Unusual integrase gene expression on the clc genomic island in Pseudomonas sp. strain B13. J Bacteriol 185:4530–4538. doi: 10.1128/JB.185.15.4530-4538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Shoemaker NB, Salyers AA. 2004. Regulation of a Bacteroides operon that controls excision and transfer of the conjugative transposon CTnDOT. J Bacteriol 186:2548–2557. doi: 10.1128/JB.186.9.2548-2557.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bose B, Grossman AD. 2011. Regulation of horizontal gene transfer in Bacillus subtilis by activation of a conserved site-specific protease. J Bacteriol 193:22–29. doi: 10.1128/JB.01143-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clewell DB. 2011. Tales of conjugation and sex pheromones: A plasmid and enterococcal odyssey. Mob Genet Elements 1:38–54. doi: 10.4161/mge.1.1.15409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haudecoeur E, Faure D. 2010. A fine control of quorum-sensing communication in Agrobacterium tumefaciens. Commun Integr Biol 3:84–88. doi: 10.4161/cib.3.2.10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao C, Benhabib H, Ensminger AW. 2013. Phylogenetic reconstruction of the Legionella pneumophila Philadelphia-1 laboratory strains through comparative genomics. PLoS One 8:e64129. doi: 10.1371/journal.pone.0064129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chien M, Morozova I, Shi S, Sheng H, Chen J, Gomez SM, Asamani G, Hill K, Nuara J, Feder M, Rineer J, Greenberg JJ, Steshenko V, Park SH, Zhao B, Teplitskaya E, Edwards JR, Pampou S, Georghiou A, Chou IC, Iannuccilli W, Ulz ME, Kim DH, Geringer-Sameth A, Goldsberry C, Morozov P, Fischer SG, Segal G, Qu X, Rzhetsky A, Zhang P, Cayanis E, De Jong PJ, Ju J, Kalachikov S, Shuman HA, Russo JJ. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966–1968. doi: 10.1126/science.1099776. [DOI] [PubMed] [Google Scholar]
- 18.Cazalet C, Jarraud S, Ghavi-Helm Y, Kunst F, Glaser P, Etienne J, Buchrieser C. 2008. Multigenome analysis identifies a worldwide distributed epidemic Legionella pneumophila clone that emerged within a highly diverse species. Genome Res 18:431–441. doi: 10.1101/gr.7229808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molofsky AB, Swanson MS. 2003. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol Microbiol 50:445–461. doi: 10.1046/j.1365-2958.2003.03706.x. [DOI] [PubMed] [Google Scholar]
- 20.Nevo O, Zusman T, Rasis M, Lifshitz Z, Segal G. 2014. Identification of Legionella pneumophila effectors regulated by the LetAS-RsmYZ-CsrA regulatory cascade, many of which modulate vesicular trafficking. J Bacteriol 196:681–692. doi: 10.1128/JB.01175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marden JN, Diaz MR, Walton WG, Gode CJ, Betts L, Urbanowski ML, Redinbo MR, Yahr TL, Wolfgang MC. 2013. An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 110:15055–15060. doi: 10.1073/pnas.1307217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbott ZD, Yakhnin H, Babitzke P, Swanson MS. 2015. csrR, a paralog and direct target of CsrA, promotes Legionella pneumophila resilience in water. mBio 6:e00595. doi: 10.1128/mBio.00595-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Connor TJ, Adepoju Y, Boyd D, Isberg RR. 2011. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc Natl Acad Sci U S A 108:14733–14740. doi: 10.1073/pnas.1111678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cazalet C, Rusniok C, Bruggemann H, Zidane N, Magnier A, Ma L, Tichit M, Jarraud S, Bouchier C, Vandenesch F, Kunst F, Etienne J, Glaser P, Buchrieser C. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet 36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- 25.Gomez-Valero L, Rusniok C, Jarraud S, Vacherie B, Rouy Z, Barbe V, Medigue C, Etienne J, Buchrieser C. 2011. Extensive recombination events and horizontal gene transfer shaped the Legionella pneumophila genomes. BMC Genomics 12:536. doi: 10.1186/1471-2164-12-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lautner M, Schunder E, Herrmann V, Heuner K. 2013. Regulation, integrase-dependent excision, and horizontal transfer of genomic islands in Legionella pneumophila. J Bacteriol 195:1583–1597. doi: 10.1128/JB.01739-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feeley JC, Gibson RJ, Gorman GW, Langford NC, Rasheed JK, Mackel DC, Baine WB. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J Clin Microbiol 10:437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 29.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 30.Letunic I, Bork P. 2011. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalebroux ZD, Yagi BF, Sahr T, Buchrieser C, Swanson MS. 2010. Distinct roles of ppGpp and DksA in Legionella pneumophila differentiation. Mol Microbiol 76:200–219. doi: 10.1111/j.1365-2958.2010.07094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bryan A, Harada K, Swanson MS. 2011. Efficient generation of unmarked deletions in Legionella pneumophila. Appl Environ Microbiol 77:2545–2548. doi: 10.1128/AEM.02904-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swanson MS, Isberg RR. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun 63:3609–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swanson MS, Isberg RR. 1996. Identification of Legionella pneumophila mutants that have aberrant intracellular fates. Infect Immun 64:2585–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldberg M, Belkowski LS, Bloom BR. 1990. Regulation of macrophage function by interferon-gamma. Somatic cell genetic approaches in murine macrophage cell lines to mechanisms of growth inhibition, the oxidative burst, and expression of the chronic granulomatous disease gene. J Clin Invest 85:563–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogel JP, Andrews HL, Wong SK, Isberg RR. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 38.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 39.Yasbin RE, Young FE. 1974. Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol 14:1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doleans-Jordheim A, Akermi M, Ginevra C, Cazalet C, Kay E, Schneider D, Buchrieser C, Atlan D, Vandenesch F, Etienne J, Jarraud S. 2006. Growth-phase-dependent mobility of the lvh-encoding region in Legionella pneumophila strain Paris. Microbiology 152:3561–3568. doi: 10.1099/mic.0.29227-0. [DOI] [PubMed] [Google Scholar]
- 41.Wee BA, Woolfit M, Beatson SA, Petty NK. 2013. A distinct and divergent lineage of genomic island-associated Type IV Secretion Systems in Legionella. PLoS One 8(12):e82221. doi: 10.1371/journal.pone.0082221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glockner G, Albert-Weissenberger C, Weinmann E, Jacobi S, Schunder E, Steinert M, Hacker J, Heuner K. 2008. Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int J Med Microbiol 298:411–428. doi: 10.1016/j.ijmm.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 43.Segal G, Russo JJ, Shuman HA. 1999. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol Microbiol 34:799–809. doi: 10.1046/j.1365-2958.1999.01642.x. [DOI] [PubMed] [Google Scholar]
- 44.Ilangovan A, Connery S, Waksman G. 2015. Structural biology of the Gram-negative bacterial conjugation systems. Trends Microbiol 23:301–310. doi: 10.1016/j.tim.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 45.Waldor MK. 2010. Mobilizable genomic islands: going mobile with oriT mimicry. Mol Microbiol 78:537–540. doi: 10.1111/j.1365-2958.2010.07365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klebanoff SJ. 2005. Myeloperoxidase: friend and foe. J Leukoc Biol 77:598–625. doi: 10.1189/jlb.1204697. [DOI] [PubMed] [Google Scholar]
- 47.Andrews HL, Vogel JP, Isberg RR. 1998. Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway. Infect Immun 66:950–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berger KH, Merriam JJ, Isberg RR. 1994. Altered intracellular targeting properties associated with mutations in the Legionella pneumophila dotA gene. Mol Microbiol 14:809–822. doi: 10.1111/j.1365-2958.1994.tb01317.x. [DOI] [PubMed] [Google Scholar]
- 49.Molofsky AB, Shetron-Rama LM, Swanson MS. 2005. Components of the Legionella pneumophila flagellar regulon contribute to multiple virulence traits, including lysosome avoidance and macrophage death. Infect Immun 73:5720–5734. doi: 10.1128/IAI.73.9.5720-5734.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rasis M, Segal G. 2009. The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, posttranscriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol Microbiol 72:995–1010. doi: 10.1111/j.1365-2958.2009.06705.x. [DOI] [PubMed] [Google Scholar]
- 51.Mukherjee S, Yakhnin H, Kysela D, Sokoloski J, Babitzke P, Kearns DB. 2011. CsrA-FliW interaction governs flagellin homeostasis and a checkpoint on flagellar morphogenesis in Bacillus subtilis. Mol Microbiol 82:447–461. doi: 10.1111/j.1365-2958.2011.07822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bandyopadhyay P, Liu S, Gabbai CB, Venitelli Z, Steinman HM. 2007. Environmental mimics and the Lvh type IVA secretion system contribute to virulence-related phenotypes of Legionella pneumophila. Infect Immun 75:723–735. doi: 10.1128/IAI.00956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miyamoto H, Yoshida S, Taniguchi H, Shuman HA. 2003. Virulence conversion of Legionella pneumophila by conjugal transfer of chromosomal DNA. J Bacteriol 185:6712–6718. doi: 10.1128/JB.185.22.6712-6718.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vakulskas CA, Potts AH, Babitzke P, Ahmer BM, Romeo T. 2015. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol Mol Biol Rev 79:193–224. doi: 10.1128/MMBR.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dubey AK, Baker CS, Romeo T, Babitzke P. 2005. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 11:1579–1587. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duss O, Michel E, Diarra dit Konte N, Schubert M, Allain FH. 2014. Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition. Nucleic Acids Res 42:5332–5346. doi: 10.1093/nar/gku141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mercante J, Edwards AN, Dubey AK, Babitzke P, Romeo T. 2009. Molecular geometry of CsrA (RsmA) binding to RNA and its implications for regulated expression. J Mol Biol 392:511–528. doi: 10.1016/j.jmb.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montero Llopis P, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, Jacobs-Wagner C. 2010. Spatial organization of the flow of genetic information in bacteria. Nature 466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berger KH, Isberg RR. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol 7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 60.Wiater LA, Sadosky AB, Shuman HA. 1994. Mutagenesis of Legionella pneumophila using Tn903 dlllacZ: identification of a growth-phase-regulated pigmentation gene. Mol Microbiol 11:641–653. doi: 10.1111/j.1365-2958.1994.tb00343.x. [DOI] [PubMed] [Google Scholar]
- 61.Mukherjee S, Bree AC, Liu J, Patrick JE, Chien P, Kearns DB. 2015. Adaptor-mediated Lon proteolysis restricts Bacillus subtilis hyperflagellation. Proc Natl Acad Sci U S A 112:250–255. doi: 10.1073/pnas.1417419112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.